Abstract
The SCN5A gene encodes cardiac sodium channel Nav1.5 and causes lethal ventricular arrhythmias/sudden death and atrial fibrillation (AF) when mutated. MicroRNAs (miRNAs) are important post-transcriptional regulators of gene expression, and involved in the pathogenesis of many diseases. However, little is known about the regulation of SCN5A by miRNAs. Here we reveal a novel post-transcriptional regulatory mechanism for expression and function of SCN5A/Nav1.5 via miR-192-5p. Bioinformatic analysis revealed that the 3′-UTR of human and rhesus SCN5A, but not elephant, pig, rabbit, mouse, and rat SCN5A, contained a target binding site for miR-192-5p and dual luciferase reporter assays showed that the site was critical for down-regulation of human SCN5A. With Western blot assays and electrophysiological studies, we demonstrated that miR-192-5p significantly reduced expression of SCN5A and Nav1.5 as well as peak sodium current density INa generated by Nav1.5. Notably, in situ hybridization, immunohistochemistry and real-time qPCR analyses showed that miR-192-5p was up-regulated in tissue samples from AF patients, which was associated with down-regulation of SCN5A/Nav1.5. These results demonstrate an important post-transcriptional role of miR-192-5p in post-transcriptional regulation of Nav1.5, reveal a novel role of miR-192-5p in cardiac physiology and disease, and provide a new target for novel miRNA-based antiarrhythmic therapy for diseases with reduced INa.
Keywords: Atrial fibrillation, Cardiac sodium channel Nav1.5, SCN5A, MicroRNA, MiR-192-5p
1. Introduction
Nav1.5 is the primary voltage-gated sodium channel in the heart and responsible for the fast depolarization and generation of the cardiac action potential (AP) and conduction of the cardiac AP [1]. It is encoded by the SCN5A gene on chromosome 3p21 [1]. SCN5A is one of the first two genes identified for long QT syndrome (LQTS), an inherited cardiac arrhythmia [2,3,4,5]. LQTS mutations in SCN5A act by a gain-of-function mechanism, mostly by generating late persistent sodium currents [6]. On the other hand, loss-of-function mutations were later identified by us and others in SCN5A in patients with Brugada syndrome (BrS), cardiac conduction defects, sudden infant death, sick sinus syndrome, dilated cardiomyopathy and atrial fibrillation (AF) [7,8,9,10]. Moreover, INa density was reduced in AF patients [11] and a dog model for AF [12], but the underlying molecular mechanism is unknown. Reduced Nav1.5 expression and INa were also detected in the atria in a mouse model for catecholamine-induced VT with the RyR2-P2328S gain-of-function mutation [13]. Similarly, reduced INa density has also been associated with more common cardiovascular diseases such as heart failure [14, 15,16] and myocardial ischemia [17], but the underlying molecular mechanism is unknown either.
Post-transcriptional regulation by microRNAs (miRNAs) may be an attractive, putative mechanism for the observed reduction of INa in AF, heart failure, and myocardial ischemia [18]. MiRNAs are small, non-coding RNAs (about 22 nucleotide long) that regulate gene expression through translational repression or mRNA degradation, typically by binding to their targeted sites located in the 3′ untranslated regions (3′-UTRs) of mRNAs [19,20]. For example, miR-1, miR-133, miR590, miR-208a and miR-328 have been found to be significantly associated with AF and heart failure [21,22]. Many miRNAs that regulate the expression of cardiac ion channels have been reported. For example, miR-1 regulates KCNJ2, CACNA1C, and GJA1; miR-133 regulates ERG; miR-328 regulates CACNA1C and CACNB1; miR-212 regulates KCNJ2; miR-208a regulates GJA5 [23]. In this study, we found that cardiac sodium channel gene SCN5A was regulated by miR-192-5p. MiR-192-5p was first identified by Lagos Quintana et al. [24] and later by Lim et al. [25]. MiR-192-5p was shown to be highly expressed in kidney and hepatocellular carcinoma tissue samples [26,27], but its expression in cardiac tissues was not reported. In this study, we reveal that miR-192-5p binds to the 3′-UTR of human SCN5A to negatively regulate the expression of Nav1.5 and reduce INa density. We further show that the expression of miR-192-5p is significantly reduced in human AF tissues compared with normal tissue samples and associated with down-regulation of SCN5A/Nav1.5.
2. Methods
2.1. Study subjects
This study was approved by the appropriate local institutional review boards on human subject research at the First Affiliated Hospital of Xiamen University and Huazhong University of Science and Technology and conformed to the guidelines set forth by the Declaration of Helsinki. Written informed consent was obtained from the participants. We studied left atrial appendage tissue samples from AF patients with rheumatic valve disease and control subjects with rheumatic valve disease alone for the expression levels of miR-192-5p and SCN5A/Nav1.5. The demographic and clinical characteristics of study subjects are shown in Table 1.
Table 1.
Clinical and demographic characteristics of study subjects.
| ID | Gender | Age | Height (cm) | Weight (kg) | BMI (kg/m2) | Systolic (mm Hg) | Diastolic (mm Hg) | AF | Other cardiovascular disease | Test |
|---|---|---|---|---|---|---|---|---|---|---|
| O2 | F | 49 | 158 | 47 | 0.30 | 110 | 72 | No | Mitral stenosis and incompetence | qPCR |
| 7 | F | 27 | 163 | 50 | 0.31 | 122 | 71 | No | Mitral stenosis and incompetence | qPCR |
| 5 | F | 40 | 160 | 56 | 0.35 | 130 | 60 | No | Aortic valve stenosis and incompetence | qPCR |
| Y2 | F | 60 | – | – | – | 105 | 70 | Yes | Heart valve disease | qPCR |
| 3 | F | 43 | 163 | 57 | 0.35 | 112 | 73 | Yes | Heart valve disease | qPCR |
| 8 | F | 61 | 161 | 53 | 0.33 | 130 | 82 | Yes | Heart valve disease | qPCR |
| A | F | 51 | 156 | 53 | 0.34 | 104 | 88 | Yes | Heart valve disease | ISH Immunohis |
| B | F | 43 | – | – | – | – | – | Yes | Heart valve disease | ISH Immunohis |
| M | M | 62 | – | – | – | – | – | Yes | Congenital heart disease, ventricular septal defect | ISH Immunohis |
| L | F | 48 | 145 | 40 | 0.28 | 100 | 65 | No | Heart valve disease | ISH Immunohis |
| G | F | 35 | 160 | 54 | 0.34 | 102 | 74 | No | Heart valve disease | ISH Immunohis |
| F | M | 73 | – | – | – | 123 | 60 | No | Heart valve disease | ISH Immunohis |
qPCR, real-time quantitative RT-PCR analysis; ISH, in situ hybridization; Immunohis, immunohistochemical analysis.
2.2. Mutational analysis by sequencing analysis
Extraction of human genomic DNA from blood samples was as described previously by us [28]. All exons, intron–exon boundaries and 5′ and 3′ UTRs of SCN5A were amplified by polymerase chain reactions (PCR) using primers published before [29]. The PCR products were purified by agarose gel electrophoresis, followed by direct sequencing using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Life Technologies).
2.3. Bioinformatic analysis
The sequences at SNP rs41310757 and in its flanking regions were analyzed for potential binding sites for miRNAs by bioinformatic analyses. We used several online miRNA binding site prediction software programs with different algorithms, including TargetScan (http://www.targetscan.org/), RNA Hybrid (http://bibiserv.tenchfak.uni-bielefeld.de/rnahybird), miRWalk (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/), DIANA-microT v3.0 (http://diana.cslab.ece.ntua.gr/microT/), and microRNA.org (http://www.microrna.org/microrna/getGeneForm.do).
2.4. Cell lines, plasmids and miRNAs
HCT116 cells (human colorectal carcinoma cell), SW620 cells (human colorectal adenocarcinoma cell) and HEK293 cells (human embryonic kidney cell)were purchased from ATCC (American Type Culture Collection, Rockville, MD, USA). Cells were cultured in Dulbecco’s Modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS, Gibco Life Technologies, Gaithersburg, MD, USA) in a humidified incubator with 5% CO2 at 37 °C.
We amplified a region of the 3′-UTR of SCN5A that contains the predicted miR-192-5p target binding site using human genomic DNA. The PCR product included two restriction sites, Sac I at the left end and Hind III at the right end. The PCR product was cut with the two enzymes and sub-cloned into the multiple cloning site of the pMIR-REPORT luciferase vector (Applied Biosystems, Foster City, CA, USA) (Fig. 1A). Two reporter genes were created. Reporter gene pMIR-REPORT-SCN5A-3′ UTR-T contains the wild type binding sequence for miR-192-5p, whereas pMIR-REPORT-SCN5A-3′UTR-C contains a variant, a T to C substitution (single nucleotide polymorphism or SNP rs41310757), at the miR-192-5p binding sequence. The sequences of primers are 5′ACTCTGGCCCCCTATTGTCT3′ for the forward primer and 5′CACCCTTGTGTCCTTTGGTT3′ for the reverse primer.
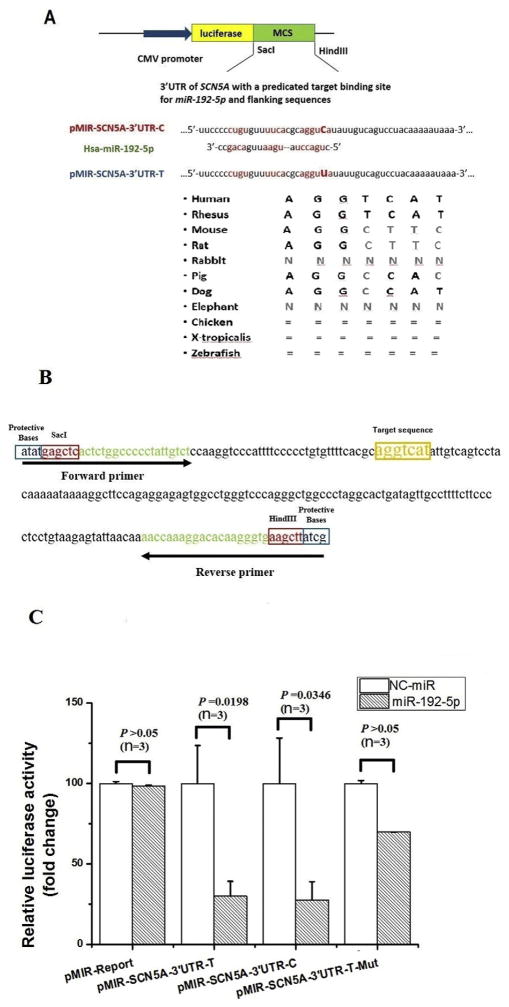
Fig. 1.
MiR-192-5p down-regulates expression of the SCN5A gene by targeting the 3′-UTR. A. Bioinformatic analysis identified a potential binding site for miR-192-5p at the 3′-UTR of SCN5A. A diagram showing luciferase reporter constructs containing the miR-192-5p targeting sequences with the C allele or T allele of rs41310757 is shown. The miR-192-5p “seed region” and the predicted miR-192-5p target sequences are indicated. The evolutionary conservation of the miR-192-5p binding sequences at the 3′-UTR of SCN5A is shown for several different species at the bottom. B. The sequence of the 3′-UTR fragment of human SCN5A, which contains the target binding sequence of miR-192-5p and was subcloned into the pMIR-REPORT luciferase vector. Forward and reverse primers used for PCR-amplification of the 3′-UTR fragment of SCN5A are shown. C. Effects of miR-192-5p mimics on the pMIR-SCN5A-3′UTR luciferase reporters compared with negative control mimics (NC-control) transfected into HCT116 cells. Three luciferase reporters were analyzed, including the pMIR-SCN5A-3′UTR luciferase reporter containing the C allele of rs41310757, the reporter with the T allele, and the reporter with the miR-192-5p target sequence (aggtcat) deleted. Luciferase activities were calculated as the ratio of firefly/Renilla activities and normalized to the negative control mimic group. Results were obtained from three independent experiments. The data was normalized to NC-miR. Data is shown as means ± SD.
The putative target binding site of miR-192-5p at the 3′-UTR of human SCN5A, AGGTCAT, was deleted from pMIR-REPORT-SCN5A-3′ UTR-T by overlapping PCR-based mutagenesis as previously described [30]. This generated a mutant reporter, pMIR-REPORT-SCN5A-3′UTRT-Mut.
The pHL3 expression plasmid for SCN5A was described previously [31,32,33]. The whole 3′-UTR of SCN5A with the miR-192-5p binding site was then sub-cloned into pHL3 by GeneWiz (Suzhou, Jiangsu, China) and used for Western blot and electrophysiological studies.
MiR-192-5p mimics and negative control miRNA mimics were purchased from Guangzhou RioboBio (Guangzhou, Guangdong, China). The miR-192-5p mimics were chemically synthesized double-stranded RNA molecules with the sequence identical to the mature miR-192-5p sequence. When the mimics were transfected into cells, it would be recognized and processed by Argonaute (AGO) and RNA-induced silencing complex (RISC) into single-stranded, functional miR-192-5p. The negative control miRNA mimics were miR-mimics with sequences that do not bind any mRNA in humans.
2.5. Dual luciferase reporter assays
HCT116 cells were cultured in 24-well plates for 24 h and transfected with 200 ng of either pMIR-REPORT-SCN5A-3′UTR-T or pMIR-REPORT-SCN5A-3′UTR-C and 100 nM of miR-192-5p mimics or negative control NC-miRNA mimics, along with 20 ng of the pRL-TK vector containing the Renilla luciferase gene (Promega, Madison, WI, USA). The transfection was carried out using 2 μl of lipofectamine 2000 and 500 μl of Opti-MEM reduced serum medium according to the manufacturer’s the protocol (Gibco Life Technologies, Gaithersburg, MD, USA). Cells were harvested 48 h after transfection and lysed using 1× passive lysis buffer (Promega, Madison, WI, USA). Luciferase assays were performed as described by us previously [34] and using the Dual-Glo luciferase assay kit (Gibco Life Technologies, Gaithersburg, MD, USA). The ratio of firefly over Renilla luciferase activities was calculated and considered as the final luciferase activity value. Each assay was performed in triplicate and repeated at least three times.
2.6. Quantitative real time RT-PCR analysis
To examine the effect of miR-192-5p on the expression of SCN5A mRNA, we searched literature for a cell line with expression of SCN5A and found that SW620 cells expressed a high level of endogenous Nav1.5 protein [35]. Therefore, SW620 cells were cultured in 24-well plates for 24 h and transfected with 100 nM of miR-192-5p mimics or NC-miRNA mimics using lipofectamine 2000 and Opti-MEM reduced serum medium according to the manufacturer’s the protocol (Gibco Life Technologies, Gaithersburg, MD, USA). Cells were collected 48 h after transfection and lysed using RNAiso plus (TaKaRa, Dalian, China). Total RNA was isolated from cells using the Trizol reagent (TaKaRa, Dalian, China) and converted into cDNA by reverse transcription using oligo(dT)15 with the First-Strand cDNA Synthesis kit (Toyobo, Japan).
Total RNA samples were also isolated from left atrial appendage samples from three AF patients (Y2, 3, 8) and three non-AF control patients (O2, 7, 5) (Table 1) using the Trizol reagent (TaKaRa, Dalian, China). Total RNA was converted into cDNA as described above.
Real time qPCR analysis was carried out using the FastStart Universal SYBR Green Master kit (Roche Applied Science, Mannheim, Germany) in a 10 μl reaction volume on an ABI 7900 Genome Analyzer System. The reaction system contained 2 μl of cDNA template, 5 μl of SYBR green mix (including ROX), 200 nM of forward primer and 200 nM of reverse primer (a total volume of 10 μl). Human GAPDH was used as an internal standard. The primers for RT-PCR analyses were listed in Table 2. The PCR products were verified by melting curve analysis and the results were analyzed using the 2−ΔΔCt method as described [36]. Each assay was performed in triplicate and repeated at least three times.
Table 2.
The primer sequences for RT-PCR analysis of SCN5A and control GAPDH.
| Gene | Forward | Reverse |
|---|---|---|
| SCN5A | CCAGATCTCTATGGCAATCCA | GAATCTTCACAGCCGCTCTC |
| GAPDH | AGTCAGCCGCATCTTCTT | GCCCAATACGACCAAATCC |
Expression of miR-192-5p was carried out for left atrial appendage samples from three AF patients and three non-AF controls (Table 1) using the stem-loop real-time PCR as described previously [37]. Total RNA was isolated as described above. Bulge-loop™ miRNA qRT-PCR Primer Sets (one RT primer and a pair of qPCR primers for each set) specific for miR-192-5p were purchased from RiboBio (Guangzhou, Guangdong, China). MiRNA bulge-loop was reverse transcribed with the First-Strand cDNA Synthesis kit (Invitrogen, Carlsbad, CA, USA) using the specific miR-192-5p RT primer. Real time qPCR analysis was carried out as described for SCN5A above except that the reaction volume was 20 μl and the internal control was U6 small RNA.
2.7. Electrophysiological studies
HEK293 cells are the most commonly used cells for electrophysiological studies of cardiac sodium channels. HEK293 cells were cultured at 37 °C under 5% CO2 in high glucose DMEM media supplemented with 10% FBS (Invitrogen). When cell culture was 70–80% confluent in 3.5-cm plates, transfection was carried out with 2 μg of pcDNA3-SCN5A-whole 3′UTR and 200 ng of pEGFP-N1 with miR-192-5p mimics or negative control NC-miRNA mimics using lipofectamine 2000 and Opti-MEM reduced serum medium. Cells were cultured for 48 h and EGFP-positive cells were selected for patch-clamp recordings. We recorded sodium current at room temperature using a Multiclamp 700B amplifier (Axon Instruments, Sunnyvale, CA). The patch pipette (tip resistance was 2–3MΩ) was filled with following solutions as described previously [3,38,39]: 20 mM NaCl, 130 mM CsCl, 10 mM HEPES, and 10mMEGTA, pH 7.2 (adjusted with CsOH). The components of bath solution was 70 mM NaCl, 80 mM CsCl, 5.4 mM KCl, 2 mM CaCl2, 1mM MgCl2, 10 mM HEPES, and 10 mM glucose, pH 7.3 (adjusted with CsOH). All chemicals were purchased from Sigma (Madison, WI, USA). The junction potential, capacitance and series resistance were automatically compensated in the whole cell configuration. The holding potential was maintained at −120 mV, and the voltage clamp protocols were as described previously. The sodium currents were filtered at 5 kHz, sampled at 50 kHz, and stored on a desktop computer equipped with an AD converter (Digidata 1440A, Molecular Devices). All current measurements were normalized using the cell capacitance. The Clamp fit 10.2 (Axon Instruments), Excel (Microsoft), and Origin 85 (Microcal Software) were used for data acquisition and analysis.
2.8. Western blot analysis
Cells were cultured and transfected as described above. Transfected cells were collected and incubated in ice-cold TNEN lysis buffer (in mmol/l: 50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 2.0 mM EDTA, 1.0% Nonidet P-40) with 1 mini tab of EDTA-free protease inhibitors (Roche) and 1 mmol/l PMSF (phenylmethylsulfonyl fluoride) for 30 min at 4 °C. The insoluble fraction was pelleted by centrifugation at 12,000 ×g for 15 min at 4 °C. 100 μl of supernatant was mixed with 20 μl of 6× Laemmli buffer (0.3 mol/l Tris–HCl, 6% SDS, 60% glycerol, 120 mmol/l dithiothreitol (DDT) and proprietary pink tracking dye), and heated at 37 °C for 10 min. Then, 20 μl of samples was subjected to SDS-PAGE. After electrophoresis, proteins were transferred onto a 0.45 μm polyvinylidene fluoride (PVDF) membrane (Millipore). The membrane was probed with an anti-Nav1.5 rabbit polyclonal antibody (Alomone, ASC-005), followed by incubation with a HRP-conjugated secondary goat anti-rabbit antibody (Millipore). The protein signal was visualized by a Super Signal West Pico Chemiluminescent substrate according to the manufacturer’s instructions (Pierce Chemical Co., Rockford, Illinois, USA). Human GAPDH was used as loading control. Each assay was performed in triplicate and repeated at least three times.
2.9. In situ hybridization analysis (ISH)
ISH for miR-192-5p was carried out using a Dig-labeled miR-192-5p antisense probe. The sequence of the probe was 5′-GGCTGTCAATTCATAGGTCAG-3′. Human left atrial appendage samples from three AF patients (A, B, M) and three non-AF controls (L, G, F) (Table 1) were fixed in 4% paraformaldehyde (PFA) and paraffin-embedded. Serial sections of 5 μm were cut and hybridized in 100 ml hybridization buffer with 50 pM of denatured Dig-labeled probe at 42 °C overnight. The sections were then washed with 1× SSPE buffer (8.765 g NaCl, 1.38 g NaH2PO4 and 0.37 g EDTA–Na2·2H2O dissolved with 500 ml DEPC H2O, pH 7.4) and incubated with an anti-DIG-AP antibody in 1×blocking buffer (2% sheep serum and 0.1% Triton X-100 dissolved with 100 mM Tris–HCl and 150 mM NaCl, pH 7.5) for 2 h. The sections were then stained with 45 μg Nitroblue tetrazolium (NBT) in 10 ml TNM-50 (50 ml Tris, pH 7.5, 10 ml 5 M NaCl and 25 ml 1 M MgCl2 dissolved with 500ml DEPC H2O, pH 9.0) for 12 h and imaged under a microscope (LEICA). The florescent optical density for miR-192-5p was quantified using a computer image analysis program (Image J) and analyzed for differences between the AF group and the control group by Student’s t-test.
2.10. Immunohistochemical analysis
Immunohistochemistry was performed for Nav1.5 with human left atrial appendage samples from three AF patients (A, B, M) and three non-AF controls (L, G, F) (Table 1) as described [37]. A polyclonal anti-Nav1.5 antibody from Sigma-Aldrich (St. Louis, MO, USA) was used for immunohistochemistry staining.
2.11. Statistical analysis
Experimental data from at least three independent experiments was presented as means ± standard deviation (S.D.). Statistical analysis was performed with Student’s t-test using SPSS version 17.0 software (SPSS, Chicago, IL, USA). A P value of 0.05 or less was considered to be significant.
For analysis of the data from real-time RT-PCR analysis, we calculated the means for RQ values from the 3 wells and then compare the means from three independent experiments between two different groups with Student’s t-test. For Western blot analysis, the images from three independent experiments were scanned with Quantity One 4.6.8 (Basic) (Bio-Rad, Hercules, California, USA) and quantified. The density of the Nav1.5 protein band for the miR-192-5p treatment group was calibrated to that for the control sample (NC-miR). The means from three independent experiments were compared between two different groups with Student’s t-test.
3. Results
3.1. Identification of miR-192-5p as a potential posttranscriptional regulator of cardiac sodium channel gene SCN5A
During mutational screening of the SCN5A gene in a panel of 45 BrS patients, we identified a T to C variant in the 3′-UTR of SCN5A (754 bps from the stop codon), which appeared to be a common polymorphism (SNP rs41310757). The minor allele frequency (MAF) of the rs41310757 variant was 37.30% in the BrS group, which was higher than that in a group of 100 Chinese Han controls (31.2%) (P = 0.04). The MAF of the rs41310757 variant was similar to that in the NCBI database for the East Asian population (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=41310757) (32.50%). Bioinformatic analysis of the sequences flanking SNP rs41310757 revealed a potential binding site for miR-192-5p (Fig. 1A). The miR-192-5p binding site on SCN5A shows Watson–Crick matches to the seed region of miR-192-5p (Fig. 1A). The mature sequence of miR-192-5p and potential binding regions are shown in Fig. 1A. Interestingly, the putative miR-192-5p target binding sequences at the 3′UTR of human SCN5A are not evolutionarily conserved. High conservation was observed only with rhesus and dogs, but not with the pig, elephant, rabbit, mouse, rat, chicken, zebrafish and Xenopus tropicalis (Fig. 1A). The data supports the hypothesis that human SCN5A may be regulated by miR-192-5p.
3.2. Analysis of regulation of miR-192-5p on the expression of SCN5A using pMIR-REPORT luciferase reporters
Based on the data from the bioinformatic analysis described above, we hypothesized that miR-192-5p could bind to the 3′-UTR of SCN5A and regulate the expression of SCN5A. To test the hypothesis, we cloned a 194 bp region that contains the predicted miR-192-5p binding site and flanking sequences at the 3′-UTR of SCN5A into the pMIR-REPORT luciferase vector, resulting in a reporter gene pMIR-SCN5A-3′UTR-T with the T allele of SNP rs41310757 and pMIR-SCN5A-3′UTR-C with the C allele (Fig. 1A, B).
Each reporter was co-transfected with miR-192-5p mimics and negative NC-control mimics into cells and luciferase assays were carried out. Compared to NC-control mimics, over-expression of miR-192-5p (i.e. miR-192-5p mimics) reduced luciferase activities by 3.3-fold (P = 0.0198) for the pMIR-SCN5A-3′UTR-T reporter gene (Fig. 1C). The miR-192-5p mimics reduced the activity from pMIR-SCN5A-3′UTR-C by 3.8-fold (P = 0.0346) (Fig. 1C). The data suggests that miR-192-5p negatively regulates the activity of pMIR-SCN5A-3′UTR luciferase reporters containing the miR-192-5p target binding site. We also analyzed the luciferase activity from a mutant pMIR-SCN5A-3′UTR-T reporter gene with the putative miR-192-5p target sequence (AGGTCAT) deleted (pMIR-SCN5A-3′UTR-T-Mut). No significance difference was found between miR-192-5p mimics and NC-control mimics. The data suggests that miR-192-5p negatively regulates the activity of pMIR-SCN5A-3′ UTR luciferase reporters by targeting the miR-192-5p target binding sequence (AGGTCAT).
There was no statistical significant difference between pMIR-SCN5A-3′UTR-T and pMIR-SCN5A-3′UTR-C with or without miR-192-5p mimics (P = 0.444 and 0.351, respectively) (Fig. 1B). The data suggests that the T to C substitution (SNP rs41310757) at the miR-192-5p binding site does not affect the expression SCN5A, consistent with the finding that it is a benign polymorphism.
3.3. MiR-192-5p negatively regulates expression of SCN5A at both mRNA and protein levels
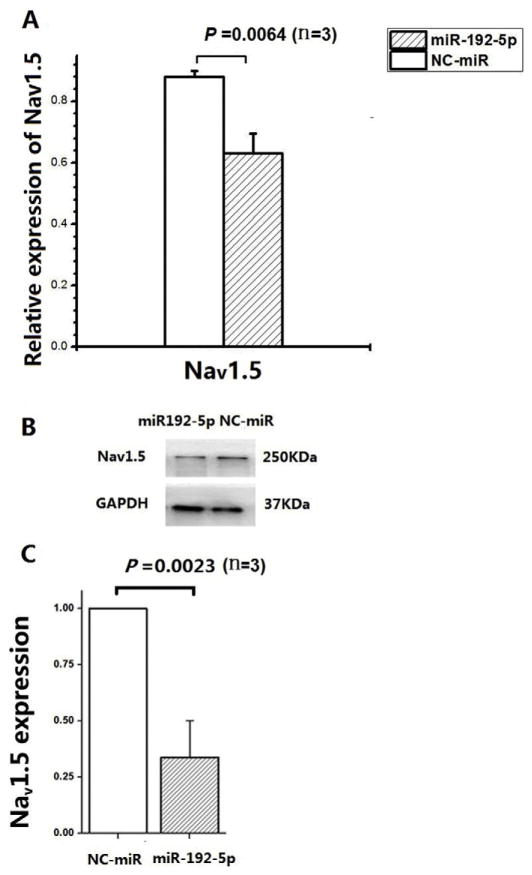
To further validate the finding that miR-192-5p regulates the expression of SCN5A, we assessed the expression level of SCN5A mRNA using real-time qPCR analysis in SW620 cells with a relatively high expression level of endogenous SCN5A. SW620 cells were transfected with miR-192-5p mimics or NC-control miRNA mimics. Real-time qPCR analysis showed that compared to control mimics, miR-192-5p mimics significantly reduced the level of SCN5A mRNA, although the magnitude of reduction was only 17% (P = 0.0064) (Fig. 2A).
Fig. 2.
MiR-192-5p negatively regulates expression of SCN5A mRNA or Nav1.5 protein. A. MiR-192-5p overexpression with mimics significantly reduced the expression level of SCN5A mRNA in SW620 cells compared with negative control mimics (NC-control) by real time qPCR analysis. GAPDH was used as internal control and the fold change was calculated by RQ values (2−ΔΔCt). B, C. MiR-192-5p overexpression with mimics significantly reduced the expression level of Nav1.5 protein in SW620 cells compared with negative control mimics (NC-control) by Western blot analysis. GAPGH was used as loading control. The images of Western blot analysis shown in (B) were scanned, quantified and plotted in (C). The data was normalized to NC-miR. Data is shown as means ± SD.
We then further examined the regulation of Nav1.5 encoded by SCN5A at the protein level using Western blot analysis. Protein extracts were isolated from SW620 cells transfected with miR-192-5p mimics or NC-control miRNA mimics. Western blotting analysis showed that miR192-5p mimics significantly reduced the expression level of Nav1.5 protein by 66% compared with the negative control mimics (P= 0.0023) (Fig. 2B and C).
Taken together, these results indicated that miR-192-5p significantly down-regulated the expression of cardiac sodium channel mRNA (SCN5A) and protein (Nav1.5). The regulation is more on the protein level than on them RNA level, 66% and 17%, respectively. These features are consistent with the general regulatory characteristics by miRNAs.
3.4. MiR-192-5p reduces the density of cardiac sodium current
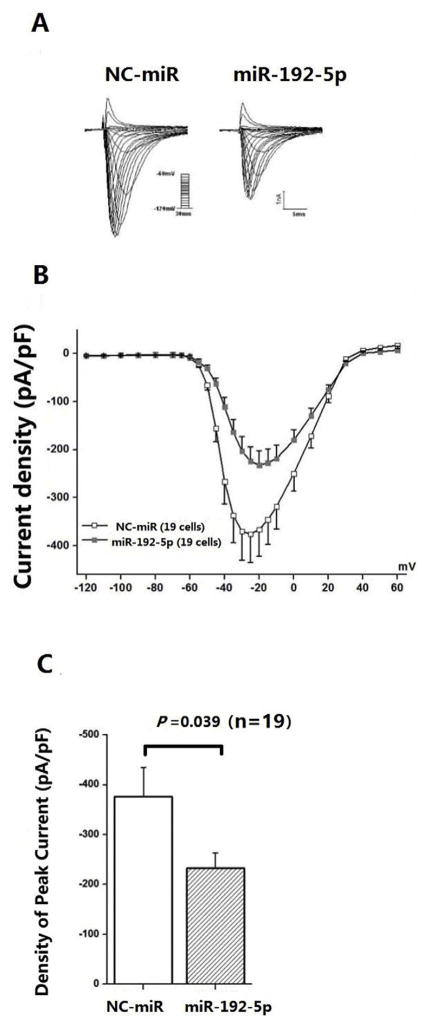
Nav1.5 is the sodium channel subunit that generates sodium current in the heart. The sodium current is commonly analyzed by patch-clamping of HEK293 cells transfected with the expression plasmid for SCN5A. We co-transfected HEK293 cells with 2 μg of pcDNA3-SCN5A-whole 3′UTR and 200 ng of pEGFP-N1 together with miR-192-5p mimics or negative control miRNA mimics and selected cells with a green signal for recordings of sodium current. Fig. 3A shows representative whole-cell sodium currents recorded from HEK293 cells transfected with a plasmid expressing human SCN5A with its whole 3′UTR and pEGFP-N1 (marker for successful transfection) together with miRNA mimics. Compared with NC-control miRNA mimics, cells expressing the miR-192-5p mimics had a markedly decreased sodium current density (peak current normalized to cell capacitance, pA/pF, n = 19 cells/group) under test potentials. As shown in Fig. 3C, compared with NC-control miRNA mimics, miR-192-5p mimics significantly reduced the sodium current density recorded at −25 mV by 40% (−375.63 ± 59.14 pA/pF vs. −224.21 ± 30.83 pA/pF).We also carried out Western blot analysis to measure the expression level of Nav1.5. Consistent with electrophysiological results, miR-192-5p mimics significantly reduced the expression level of Nav1.5 in transfected HEK293 cells (data not shown).
Fig. 3.
MiR-192-5p significantly reduces the sodium current density. A. Representative traces for sodium currents from HEK293 cells co-transfected with pcDNA3-SCN5A-whole-3′UTR and negative control mimics (NC-control) (left) or miR-192-5p mimics (right). The current protocol is depicted in the inset. B. Current–voltage (I–V) relation for sodium currents. The average sodium current density is significantly smaller in cells transfected with miR-192-5p mimics than those transfected with NC-control mimics. C. Histogram of sodium current densities at −20 mV. Data is in means ± SEM.
3.5. Correlation of increased miR-192-5p expression and down-regulation of SCN5A/Nav1.5 in human AF tissue
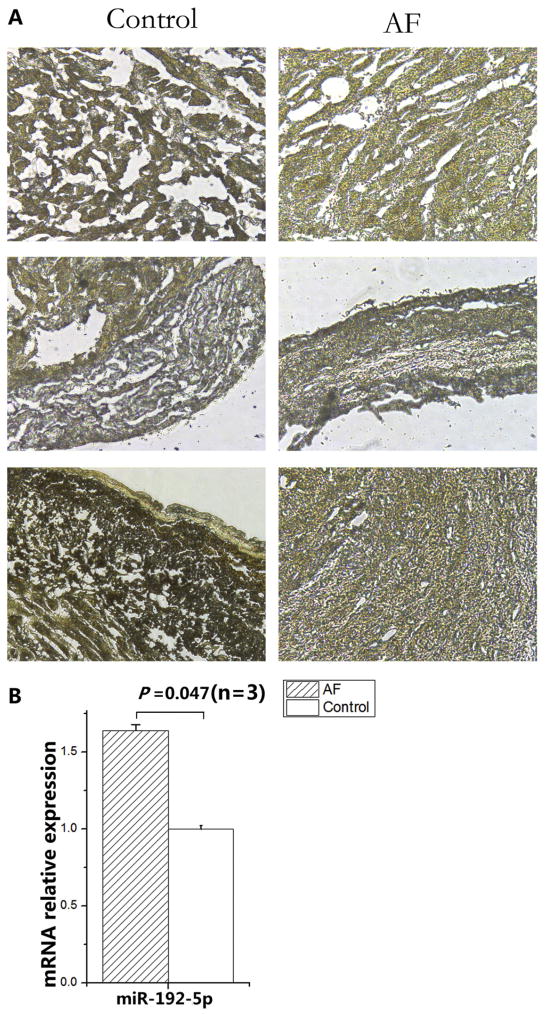
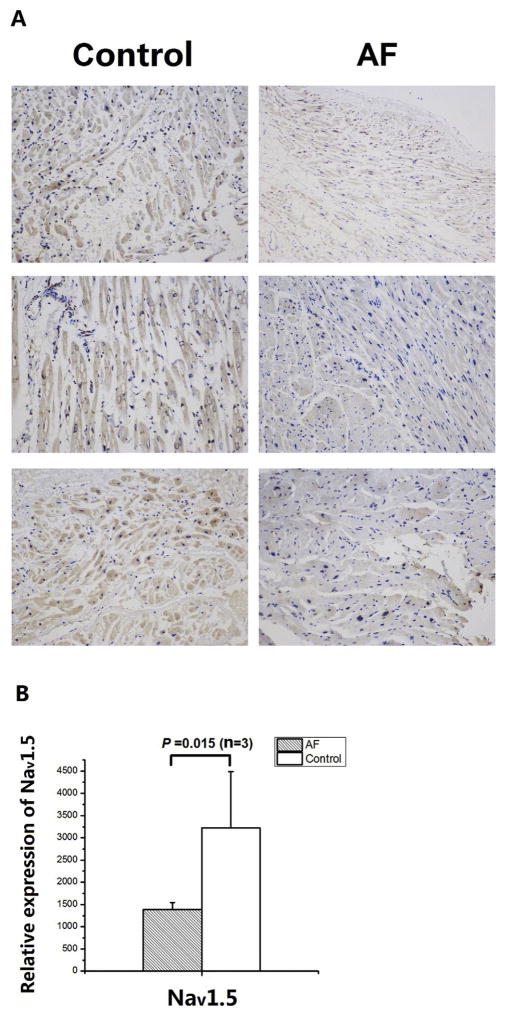
A previous study with right atrial appendages from AF patients showed that the expression level of the Nav1.5 protein and the peak sodium current density were reduced [11]. Therefore, we hypothesized that the expression level of miR-192-5p, the negative regulator of SCN5A, may be altered in AF. To test the hypothesis, we studied six left appendage tissue samples, three of which were from AF patients and three were from normal non-AF controls (Table 1).We analyzed the expression of miR-192-5p using in situ hybridization with a miR-192-5p anti-sense oligonucleotide probe. As shown in Fig. 4, miR-192-5p expression was significantly up-regulated in AF samples compared with non-AF control samples by 1.6-fold (P = 0.047). Consistent with up-regulation of miR-192-5p, the expression level of Nav1.5 was significantly down-regulated (P = 0.015) (Fig. 5). Therefore, there was a correlation between up-regulation of miR-192-5p and down-regulation of Nav1.5 in AF.
Fig. 4.
Up-regulation of miR-192-5p expression in tissue samples from AF patients compared with non-AF controls by in situ hybridization. Representative images of in situ hybridization are shown on the left. The images were scanned, quantified and plotted on the right. The data was normalized to non-AF control. Data is shown as means ± SD.
Fig. 5.
Down-regulation of Nav1.5 expression in tissue samples from AF patients compared with non-AF controls by immunohistochemistry with an anti-Nav1.5 antibody. A. The immunohistochemical images from three AF patients and three non-AF patients. B. The immunohistochemical images in (A) were scanned, quantified and graphed. Data is shown as means ± SD.
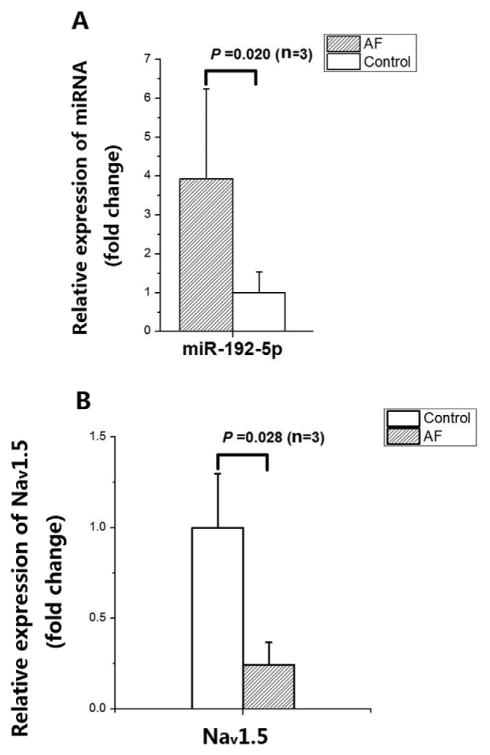
To confirm the correlation between up-regulation of miR-192-5p and down-regulation of Nav1.5 in AF, we studied another independent set of AF samples and non-AF controls (left appendages) (Table 1) by real-time qPCR analysis. As shown in Fig. 6A, miR-192-5p expression was significantly up-regulated in AF samples compared with non-AF control samples by 3.9-fold (P = 0.020). The expression of SCN5A was significantly down-regulated in AF samples compared with control samples by 3.3-fold (P = 0.028) (Fig. 6B). Together, the data suggests that up-regulation of miR-192-5p is associated with down-regulation of Nav1.5 in AF.
Fig. 6.
Correlation between up-regulation of miR-192-5p expression and down-regulation of SCN5A mRNA expression in tissue samples from AF patients compared with non-AF controls by qRT-PCR analysis. A. Up-regulation of miR-192-5p expression in tissue samples from three AF patients compared with three non-AF controls. B. Down-regulation of SCN5A expression in tissue samples from three AF patients compared with three non-AF controls. The data was normalized to non-AF control. Data is shown as means ± SD.
4. Discussion
Cardiac sodium channel gene SCN5A is critical to the generation of cardiac action potentials and, when mutated, causes many cardiac arrhythmias and sudden death. Therefore, the regulation of SCN5A expression and function must be in a delicate balance in cells and human bodies. In this study, we report a novel post-transcriptional regulatory mechanism for SCN5A expression and function. Our view of how target genes are regulated has been dramatically altered by the important role of miRNAs in the regulation of post-transcriptional protein expression. In this study, we report that miR-192-5p is involved in the negative regulation of expression of both SCN5A mRNA and Nav1.5 protein and in reducing the density of peak sodium current generated by Nav1.5. Several pieces of evidence strongly support the new finding. First, luciferase assays from Luc-SCN5A-3′-UTR reporter genes with the miR-192-5p binding site showed that miR-192-5p mimics significantly decreased the expression of SCN5A by 3.3-fold compared with negative control miRNA mimics (Fig. 1C).When the miR-192-5p binding site was deleted from the Luc-SCN5A-3′-UTR reporter, the effect of miR-192-5p mimics was lost (Fig. 1C). Second, real time qPCR analysis and Western blot analysis showed that miR-192-5p mimics significantly reduced the levels of both SCN5A mRNA and Nav1.5 protein compared with negative control miRNA mimics (Fig. 2A). Third, miR-192-5p mimics significantly reduced the sodium current density by about 40% compared with NC control mimics (Fig. 3). Together, our study demonstrates an important role of miRNAs in the posttranscriptional regulation of SCN5A. Moreover, because SCN5A is a target for anti-arrhythmic therapies, miR-192-5p becomes a target for anti-arrhythmic therapies.
Very little is known about the function of miR-192-5p. It was reported that miR-192-5p was usually down-regulated in tumors [27, 40] and in patients with wet age-related macular degeneration (plasma miRNA levels) [41]. In contrast, in this study we found that miR-192-5p was significantly up-regulated in tissue samples from AF patients by both in situ hybridization and real-time qRT-PCR analysis (Figs. 4 and 6). Therefore, miR-192-5p may play a role in several different human diseases.
AF is the most common arrhythmia encountered at the clinical setting and causes more than 15% of stroke cases. Reduced expression of SCN5A/Nav1.5 and reduction of INa density have been reported in AF [11]. In this study, we found that Nav1.5 expression was significantly down-regulated in AF left atrial appendage samples compared with non-AF control samples by immunohistochemistry (Fig. 5). Similarly, real-time qPCR analysis showed that the expression of SCN5A was also significantly down-regulated (Fig. 6B). Interestingly, the expression level of miR-192-5p was significantly up-regulated in AF samples compared with non-AF controls (Figs. 4 and 6A). Considering our demonstration that miR-192-5p directly down-regulates the expression of SCN5A/Nav1.5, our data suggests that up-regulation of miR-192-5p may be one of the causes for down-regulation of SCN5A/Nav1.5 in AF tissue samples.
Reduction of INa density was reported not only in AF, but also in heart failure and myocardial ischemia. The cardiac diseases with reduced INa density may be potentially treated by modalities that increase INa density or SCN5A expression. Our finding that SCN5A is the first downstream target of miR-192-5p in the heart makes it an attractive target to modulate the expression of SCN5A and INa density in diseased states. For example, inhibitors of miR-192-5p may be used to increase SCN5A expression or enhance density of INa, and treat patients with down-regulation of SCN5A or reduced INa density as in AF, heart failure, myocardial ischemia, BrS and other arrhythmias.
Houria et al. recently showed that miR-219 also regulated the expression of SCN5A, however, it increased the expression of both SCN5A mRNA and Nav1.5 protein as well as INa in HL1 cells derived from mouse atria [42]. MiRNAs normally down-regulates the expression of its downstream target genes either by inducing mRNA degradation or by inhibiting translation. Thus, it is surprising that miR-219 increases the expression of SCN5A/Nav1.5, thus, the underlying mechanism needs further clarification. On the contrary, here we show that miR-192-5p down-regulates the expression of SCN5A/Nav1.5. Therefore, the finding of regulation of SCN5A/Nav1.5 by miR-192-5p in this study distinguishes the regulation by miR-219 into two aspects. First, as with almost all miRNAs, miR-192-5p down-regulates the expression of SCN5A/Nav1.5 and INa. Second, the miR-192-5p binding sequences at the 3′-UTR of SCN5A are not evolutionarily conserved in the elephant, rabbit, pig, mouse, rat and other species (Fig. 1A). Therefore, regulation of SCN5A/Nav1.5/INa by miR-192-5p may be specific for some species including humans.
There is a limitation with the present study. The sample size for the human AF study was small due to difficulties in collecting human tissue samples. Future studies with larger sample sizes are needed to further validate the finding on the up-regulation of miR-192-5p in AF. Moreover, the up-regulation of miR-192-5p may be only one mechanism for the reduction of SCN5A expression and INa in AF. There may be other plausible mechanisms for the reduction of INa. For example, Eleonora and Herren showed that Ca2+/calmodulin-dependent kinase II (CaMKII) can also regulate INa [43].
In conclusion, our results in this study demonstrate that the expression of cardiac sodium channel gene SCN5A and its function can be regulated at the posttranscriptional level by miR-192-5p. Importantly, we showed that the expression of miR-192-5p was up-regulated in AF patients, which may promote the arrhythmia by down-regulating the expression of SCN5A. This study provides important insights into the posttranscriptional regulation of cardiac ion channels by miRNAs and suggests that miR-192-5p may be an attractive target for novel antiarrhythmia therapies for treating cardiac diseases with reduced SCN5A expression and decreased INa density as in AF, heart failure, myocardial ischemia, BrS and other arrhythmias.
Acknowledgments
This study was supported by the Chinese National Basic Research Programs (973 Programs 2013CB531101), National Natural Science Foundation of China grants 81170090, 31430047 and NSFC-J1103514, Hubei Province’s Outstanding Medical Academic Leader Program, National Institutes of Health/National Heart, Lung, and Blood Institute grants R01 HL121358 and R01 HL126729, Natural Science Foundation of Hubei Province (2014CFB216) and Specialized Research Fund for the Doctoral Program of Higher Education from the Ministry of Education of the People’s Republic of China.
Footnotes
Competing interests
None.
Author contributions
Y-Y. Z., Y. H., Z-J.W.,M-C. Z., Y-F. H., Z-P. Z. and Y-X. H. carried out the experimental work. W-H. L. and Z-R. H. provided and assisted in the clinical sample analysis. Q. K. W., Z-R. H., K.-T.L., Y-Y. Z., Y. H., Z-J. W., M-C. Z., Y-F. H., Z-P. Z., Y-X.H., Q.C., and X. T. analyzed data and provided advice. Y-Y. Z., Y. H., Q. K.W. and Z-R. H. designed experiments. Q. K. W., Y-Y. Z. and Y.H. wrote the manuscript. Z-R. H., Q.C. and Q. K.W. obtained funding for this project, and directed and supervised the research. Z-R. H. and Q. K.W. are co-corresponding authors.
References
- 1.Gellens ME, George AL, Chen L, Chahine M, Horn R, Barchi RL, Kallen RG. Primary structure and functional expression of the human cardiac tetrodotoxin-insensitive voltage-dependent sodium channel. Proc Natl Acad Sci. 1992;89:554–558. doi: 10.1073/pnas.89.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Curran ME, Atkinson DL, Ewart AK, Morris CA, Leppert MF, Keating MT. The elastin gene is disrupted by a translocation associated with supravalvular aortic stenosis. Cell. 1993;73:159–168. doi: 10.1016/0092-8674(93)90168-p. [DOI] [PubMed] [Google Scholar]
- 3.Wang Q, Shen J, Li Z, Timothy K, Vincent GM, Priori SG, Schwartz PJ, Keating MT. Cardiac sodium channel mutations in patients with long QT syndrome, an inherited cardiac arrhythmia. Hum Mol Genet. 1995;4:1603–1607. doi: 10.1093/hmg/4.9.1603. [DOI] [PubMed] [Google Scholar]
- 4.Wang Q, Shen J, Splawski I, Atkinson D, Li Z, Robinson JL, Moss AJ, Towbin JA, Keating MT. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell. 1995;80:805–811. doi: 10.1016/0092-8674(95)90359-3. [DOI] [PubMed] [Google Scholar]
- 5.Watanabe H, Ohkubo K, Watanabe I, Matsuyama TA, Ishibashi-Ueda H, Yagihara N, Shimizu W, Horie M, Minamino T, Makita N. SCN5A mutation associated with ventricular fibrillation, early repolarization, and concealed myocardial abnormalities. Int J Cardiol. 2013;165:e21–e23. doi: 10.1016/j.ijcard.2012.10.074. [DOI] [PubMed] [Google Scholar]
- 6.Dumaine R, Wang Q, Keating MT, Hartmann HA, Schwartz PJ, Brown AM, Kirsch GE. Multiple mechanisms of Na+ channel-linked long-QT syndrome. Circ Res. 1996;78:916–924. doi: 10.1161/01.res.78.5.916. [DOI] [PubMed] [Google Scholar]
- 7.Herren AW, Bers DM, Grandi E. Post-translational modifications of the cardiac Na channel: contribution of CaMKII-dependent phosphorylation to acquired arrhythmias. Am J Physiol Heart Circ Physiol. 2013;305:H431–H445. doi: 10.1152/ajpheart.00306.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Modell SM, Lehmann MH. The long QT syndrome family of cardiac ion channelopathies: a HuGE review. Genet Med. 2006;8:143–155. doi: 10.1097/01.gim.0000204468.85308.86. [DOI] [PubMed] [Google Scholar]
- 9.Hedley PL, Jørgensen P, Schlamowitz S, Moolman-Smook J, Kanters JK, Corfield VA, Christiansen M. The genetic basis of Brugada syndrome: a mutation update. Hum Mutat. 2009;30:1256–1266. doi: 10.1002/humu.21066. [DOI] [PubMed] [Google Scholar]
- 10.van Hoeijen DA, Blom MT, Tan HL. Cardiac sodium channels and inherited electrophysiological disorders: an update on the pharmacotherapy. Expert Opin Pharmacother. 2014;15:1875–1887. doi: 10.1517/14656566.2014.936380. [DOI] [PubMed] [Google Scholar]
- 11.Sossalla S, Kallmeyer B, Wagner S, Mazur M, Maurer U, Toischer K, Schmitto JD, Seipelt R, Schondube FA, Hasenfuss G, Belardinelli L, Maier LS. Altered Na(+) currents in atrial fibrillation effects of ranolazine on arrhythmias and contractility in human atrial myocardium. J Am Coll Cardiol. 2010;55:2330–2342. doi: 10.1016/j.jacc.2009.12.055. [DOI] [PubMed] [Google Scholar]
- 12.Gaspo R, Bosch RF, Bou-Abboud E, Nattel S. Tachycardia-induced changes in Na+ current in a chronic dog model of atrial fibrillation. Circ Res. 1997;81:1045–1052. doi: 10.1161/01.res.81.6.1045. [DOI] [PubMed] [Google Scholar]
- 13.King JH, Wickramarachchi C, Kua K, Du Y, Jeevaratnam K, Matthews HR, Grace AA, Huang CL, Fraser JA. Loss of Nav1.5 expression and function in murine atria containing the RyR2-P2328S gain-of-function mutation. Cardiovasc Res. 2013;99:751–759. doi: 10.1093/cvr/cvt141. [DOI] [PubMed] [Google Scholar]
- 14.Valdivia CR, Chu WW, Pu J, Foell JD, Haworth RA, Wolff MR, Kamp TJ, Makielski JC. Increased late sodium current in myocytes from a canine heart failure model and from failing human heart. J Mol Cell Cardiol. 2005;38:475–483. doi: 10.1016/j.yjmcc.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 15.Shang LL, Pfahnl AE, Sanyal S, Jiao Z, Allen J, Banach K, Fahrenbach J, Weiss D, Taylor WR, Zafari AM. Human heart failure is associated with abnormal Cterminal splicing variants in the cardiac sodium channel. Circ Res. 2007;101:1146–1154. doi: 10.1161/CIRCRESAHA.107.152918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Remme CA, Bezzina CR. Review: sodium channel (dys)function and cardiac arrhythmias. Cardiovasc Ther. 2010;28:287–294. doi: 10.1111/j.1755-5922.2010.00210.x. [DOI] [PubMed] [Google Scholar]
- 17.Lau DH, Clausen C, Sosunov EA, Shlapakova IN, Anyukhovsky EP, Danilo P, Rosen TS, Kelly C, Duffy HS, Szabolcs MJ. Epicardial border zone overexpression of skeletal muscle sodium channel SkM1 normalizes activation, preserves conduction, and suppresses ventricular arrhythmia an in silico, in vivo, in vitro study. Circulation. 2009;119:19–27. doi: 10.1161/CIRCULATIONAHA.108.809301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo X, Yang B, Nattel S. MicroRNAs and atrial fibrillation:mechanisms and translational potential. Nat Rev Cardiol. 2015;12:80–90. doi: 10.1038/nrcardio.2014.178. [DOI] [PubMed] [Google Scholar]
- 19.Divakaran V, Mann DL. The emerging role of microRNAs in cardiac remodeling and heart failure. Circ Res. 2008;103:1072–1083. doi: 10.1161/CIRCRESAHA.108.183087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruan K, Fang X, Ouyang G. MicroRNAs: novel regulators in the hallmarks of human cancer. Cancer Lett. 2009;285:116–126. doi: 10.1016/j.canlet.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 21.Pan ZW, Lu YJ, Yang BF. MicroRNAs: a novel class of potential therapeutic targets for cardiovascular diseases. Acta Pharmacol Sin. 2010;31:1–9. doi: 10.1038/aps.2009.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, Zhang Y, Xu C, Bai Y, Wang H. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13:486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- 23.Kim GH. MicroRNA regulation of cardiac conduction and arrhythmias. Transl Res. 2013;161:381–392. doi: 10.1016/j.trsl.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA (New York NY) 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 26.Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, Natarajan R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-β-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci. 2007;104:3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng S, Cong S, Zhang X, Bao X, Wang W, Li H, Wang Z, Wang G, Xu J, Du B. MicroRNA-192 targeting retinoblastoma 1 inhibits cell proliferation and induces cell apoptosis in lung cancer cells. Nucleic Acids Res. 2011;39:6669–6678. doi: 10.1093/nar/gkr232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang F, Xu C-Q, He Q, Cai J-P, Li X-C, Wang D, Xiong X, Liao Y-H, Zeng Q-T, Yang Y-Z. Genome-wide association identifies a susceptibility locus for coronary artery disease in the Chinese Han population. Nat Genet. 2011;43:345–349. doi: 10.1038/ng.783. [DOI] [PubMed] [Google Scholar]
- 29.Wang Q, Li Z, Shen J, Keating MT. Genomic organization of the human SCN5A gene encoding the cardiac sodium channel. Genomics. 1996;34:9–16. doi: 10.1006/geno.1996.0236. [DOI] [PubMed] [Google Scholar]
- 30.Wang P, Yang Q, Wu X, Yang Y, Shi L, Wang C, Wu G, Xia Y, Yang B, Zhang R, Xu C, Cheng X, Li S, Zhao Y, Fu F, Liao Y, Fang F, Chen Q, Tu X, Wang QK. Functional dominant-negative mutation of sodium channel subunit gene SCN3B associated with atrial fibrillation in a Chinese GeneID population. Biochem Biophys Res Commun. 2010;398:98–104. doi: 10.1016/j.bbrc.2010.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wan X, Wang Q, Kirsch GE. Functional suppression of sodium channels by β-subunits as a molecular mechanism of idiopathic ventricular fibrillation. J Mol Cell Cardiol. 2000;32:1873–1884. doi: 10.1006/jmcc.2000.1223. [DOI] [PubMed] [Google Scholar]
- 32.Wan X, Chen S, Sadeghpour A, Wang Q, Kirsch GE. Accelerated inactivation in a mutant Na+ channel associated with idiopathic ventricular fibrillation. Am J Physiol Heart Circ Physiol. 2001;280:H354–H360. doi: 10.1152/ajpheart.2001.280.1.H354. [DOI] [PubMed] [Google Scholar]
- 33.Wang Q, Chen S, Chen Q, Wan X, Shen J, Hoeltge G, Timur A, Keating M, Kirsch G. The common SCN5A mutation R1193Q causes LQTS-type electrophysiological alterations of the cardiac sodium channel. J Med Genet. 2004;41:e66. doi: 10.1136/jmg.2003.013300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan C, Ouyang P, Timur AA, He P, You SA, Hu Y, Ke T, Driscoll DJ, Chen Q, Wang QK. Novel roles of GATA1 in regulation of angiogenic factor AGGF1 and endothelial cell function. J Biol Chem. 2009;284:23331–23343. doi: 10.1074/jbc.M109.036079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baptista-Hon DT, Robertson F, Robertson G, Owen S, Rogers G, Lydon E, Lee N, Hales T. Potent inhibition by ropivacaine of metastatic colon cancer SW620 cell invasion and NaV1.5 channel function. Br J Anaesth. 2014;113:i39–i48. doi: 10.1093/bja/aeu104. [DOI] [PubMed] [Google Scholar]
- 36.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 37.Zhou B, Ma R, Si W, Li S, Xu Y, Tu X, Wang Q. MicroRNA-503 targets FGF2 and VEGFA and inhibits tumor angiogenesis and growth. Cancer Lett. 2013;333:159–169. doi: 10.1016/j.canlet.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 38.Zeng Z, Zhou J, Hou Y, Liang X, Zhang Z, Xu X, Xie Q, Li W, Huang Z. Electrophysiological characteristics of a SCN5A voltage sensors mutation R1629Q associated with Brugada syndrome. PLoS One. 2013;8:e78382. doi: 10.1371/journal.pone.0078382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang P, Yang Q, Wu X, Yang Y, Shi L, Wang C, Wu G, Xia Y, Yang B, Zhang R. Functional dominant-negative mutation of sodium channel subunit gene SCN3B associated with atrial fibrillation in a Chinese GeneID population. Biochem Biophys Res Commun. 2010;398:98–104. doi: 10.1016/j.bbrc.2010.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiang Y, Song Y, Wang Z, Liu Z, Gao P, Liang J, Zhu J, Xing C, Xu H. microRNA- 192, -194 and-215 are frequently downregulated in colorectal cancer. Exp Ther Med. 2012;3:560–566. doi: 10.3892/etm.2011.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ertekin S, Yıldırım Ö, Dinç E, Ayaz L, ŞFidancı B, Tamer L. Evaluation of circulating miRNAs in wet age-related macular degeneration. Mol Vis. 2014;20:1057. [PMC free article] [PubMed] [Google Scholar]
- 42.Daimi H, Lozano-Velasco E, Haj Khelil A, Chibani JB, Barana A, Amoros I, Gonzalez de la Fuente M, Caballero R, Aranega A, Franco D. Regulation of SCN5A by microRNAs: miR-219 modulates SCN5A transcript expression and the effects of flecainide intoxication in mice. Heart Rhythm. 2015;12:1333–1342. doi: 10.1016/j.hrthm.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 43.Grandi E, Herren AW. CaMKII-dependent regulation of cardiac Na(+) homeostasis. Front Pharmacol. 2014;5:41. doi: 10.3389/fphar.2014.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]