Abstract
The aim of this study was to unravel the molecular pathogenesis of an unusual retinitis pigmentosa (RP) phenotype observed in a Turkish consanguineous family. Homozygosity mapping revealed two candidate genes, SAMD7 and RHO. A homozygous RHO mutation c.448G > A, p.E150K was found in two affected siblings, while no coding SAMD7 mutations were identified. Interestingly, four non-coding homozygous variants were found in two SAMD7 genomic regions relevant for binding of the retinal transcription factor CRX (CRX-bound regions, CBRs) in these affected siblings. Three variants are located in a promoter CBR termed CBR1, while the fourth is located more downstream in CBR2. Transcriptional activity of these variants was assessed by luciferase assays and electroporation of mouse retinal explants with reporter constructs of wild-type and variant SAMD7 CBRs. The combined CBR2/CBR1 variant construct showed significantly decreased SAMD7 reporter activity compared to the wild-type sequence, suggesting a cis-regulatory effect on SAMD7 expression. As Samd7 is a recently identified Crx-regulated transcriptional repressor in retina, we hypothesize that these SAMD7 variants might contribute to the retinal phenotype observed here, characterized by unusual, recognizable pigment deposits, differing from the classic spicular intraretinal pigmentation observed in other individuals homozygous for p.E150K, and typically associated with RP in general.
Inherited retinal dystrophies (RDs) affect millions of individuals worldwide. Retinitis pigmentosa (RP, MIM[268000]) is the most prevalent form of RD, with a prevalence of 1 in 4000 and accounting for approximately half of the RD cases. It is a progressive disease that typically presents with degeneration of the rod photoreceptors, followed by loss of cone photoreceptor function. Most patients experience night blindness as an initial symptom. Subsequently, a gradual constriction of the peripheral visual field occurs, often followed by loss of central vision. However, the clinical presentation of RP is highly variable, which is also reflected in an impressive genetic heterogeneity. Inheritance can be autosomal dominant (30–40% of cases), autosomal recessive (50–60%) or X-linked (5–15%)1,2. More than 70 disease causing genes have been associated with the molecular pathogenesis of RP today, most of which account for only a small percentage of cases3. One exception is the rhodopsin (RHO) gene, accounting for 30–40% of autosomal dominant RP (adRP) in the North American population3,4. In contrast to the numerous mutations with an autosomal dominant (AD) inheritance described in this gene, only a few recessive mutations have been reported so far5,6,7,8,9,10. Rhodopsin, which is a photosensitive molecule located in the membranes of the outer segments of rod photoreceptors, consists of a rod-specific opsin bound to the chromophore 11-cis-retinal. After absorption of a photon, this chromophore undergoes a conformational change, triggering the phototransduction cascade, converting light into electrical signals11.
Despite the large number of RP disease genes that already have been identified, still 40–50% of patients remain without molecular genetic diagnosis1. Some of these patients are assumed to have a mutation in a novel, yet to be identified disease gene. The current emergence of whole exome/genome sequencing (WES/WGS) studies in RDs greatly facilitates in identifying these genes12,13,14,15,16. However, analysis of WES and WGS data is still challenging and additional filtering strategies are needed. One strategy that has been shown to be effective is the use of a Crx-based cis-regulatory dataset13,17,18. This dataset consists of a genome-wide map of binding regions of the key retinal transcription factor Crx thereby identifying genes that are specifically regulated by this transcription factor17. One of these genes is Samd7 (sterile alpha motif domain containing 7), which is highly expressed in the adult murine retina and localizes to the outer nuclear layer of the retina, containing the photoreceptor nuclei17,19. It has been suggested that this gene encodes a transcriptional repressor, involved in fine-tuning of Crx-regulated gene expression19. Although this is an excellent candidate RD disease gene, no SAMD7 mutations have been reported in humans so far.
Besides patients with coding mutations in novel disease genes, other patients remaining without molecular genetic diagnosis could have a yet unidentified mutation in already known RD disease genes, albeit located in functional elements outside the protein coding part of the gene. Examples are mutations in the 5′ untranslated region (UTR), the core promoter of the gene, the 3′ UTR containing microRNA binding sites, deep intronic regions influencing splicing or in more distant regulatory elements such as enhancers20,21,22,23,24. Several examples of non-coding mutations in such elements have been reported in RD, including a deletion of the first non-coding exon of EYS25, LCA526 and PCDH1527 containing the promoter of the gene; a non-coding mutation in the first non-coding exon of EYS25; mutations in the 5′ untranslated region of NMNAT128, deep intronic variants in the ABCA429,30,31, OFD132, CEP29033, USH2A34 and PROM135 genes leading to alternate splicing; and a mutation in the seed region of microRNA-20436, and very recently variations in a DNase I hypersensitivity site upstream of PRDM1337.
Here, we report on the molecular pathogenesis in a Turkish consanguineous autosomal recessive RP (arRP) family with unusual nummular retinal pigmentation. The RHO mutation E150K was found in a homozygous state in the affected individuals as well as homozygous non-coding SNPs of SAMD7. Specifically, these variants were found in non-coding CRX-bound regions (CBRs) associated with the SAMD7 gene. Luciferase experiments and electroporation assays in mouse retinal explants revealed an effect of these variants on SAMD7 expression, possibly contributing to the phenotype observed in this family.
Results
Clinical evaluation
Six family members of a Turkish consanguineous family participated in this study: two affected siblings displaying severe RP (IV:5 and IV:8), two healthy siblings (IV:3 and IV:9) and their mother (III:2), and a fifth sibling (IV:6) with a minor subclinical expression of disease (Fig. 1a).
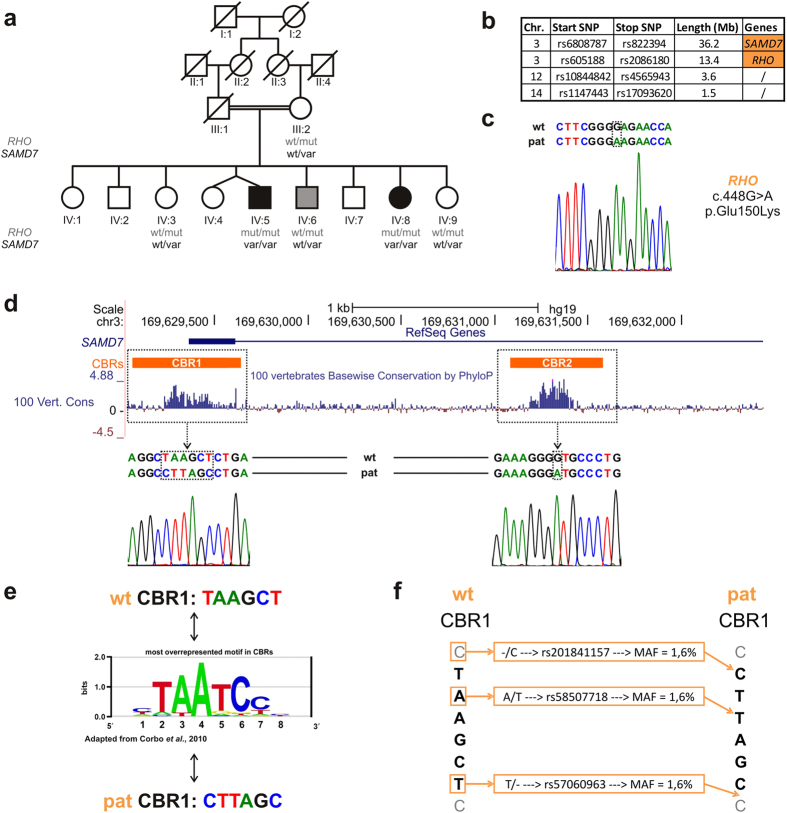
Figure 1. Identification of a homozygous RHO mutation and non-coding SAMD7 variants.
(a) Pedigree of the family. Six family members participated in this study and were clinically and genetically investigated. The genotype of each individual for the RHO mutation is indicated in grey, while the genotype for the SAMD7 variants is shown in black. The affected siblings IV:5 and IV:8 are homozygous for both variants, while all other family members are heterozygous carriers. One of these siblings IV:6 displays minor subclinical manifestations, more specifically pigmentary anomalies on fundus photography and subjective complaints of night blindness. (b) Homozygosity mapping. SNP chip analysis in the affected siblings revealed four shared homozygous regions, harboring the SAMD7 and the RHO genes. Both genes are located on chr. 3, but are not genetically linked as they are 40 Mb apart. (c) RHO mutation. Electropherogram showing the homozygous RHO c.448G > A, p.(Glu150Lys) identified in the affected siblings. Segregation of this mutation is depicted in Fig. 1a. (d) SAMD7-associated regulatory variants. UCSC genome browser view of the SAMD7 locus (http://genome.ucsc.edu/). The horizontal blue line in the upper part of the figure represents the structure of the SAMD7 gene, here only showing the first non-coding exon and a part of the adjacent intron. The orange blocks indicate the SAMD7-associated CBRs, with CBR1 located in the promoter region and CBR2 in the first intron. The next track represents the PhyloP conservation scores across 100 vertebrates, showing high conservation of both CBRs. The electropherograms show the homozygous variants located in the CBRs identified in both affected siblings. Segregation analysis of these variants is represented in Fig. 1a. (e) Disruption of CRX-binding motif. The middle part depicts the most overrepresented motif in CBRs, as reported by Corbo et al.17 While the wt CBR1 sequence is very similar to this CRX-binding motif, the variants identified in this study seem to disrupt it completely. (f) CBR1 variation. Comparison of the wt and the variant sequences shows that the variation in CBR1 consists of three SNPs with MAF 1.6%: rs201841157, rs58507718 and rs57060963 leading to an insertion, transversion and deletion event, respectively.
The first affected family member (IV:5), a 49 year-old male, was diagnosed at the age of 20, after previously suffering from night blindness. His best corrected visual acuity (BCVA) is 20/50 in the right eye (RE) and 20/100 in the left eye (LE), with a concentric constriction of Goldmann visual fields to the central 5°. A color vision defect in the blue-yellow axis as well as a subcapsular cataract were observed. On fundus photography, major retinal atrophy was visible, with vascular attenuation and a relatively preserved macula. In the retinal periphery a remarkable aspect of pigmentation was seen. Apart from classic spicular intraretinal pigmentations, conglomerates of grouped nummular pigment deposits were present (Fig. 2c).
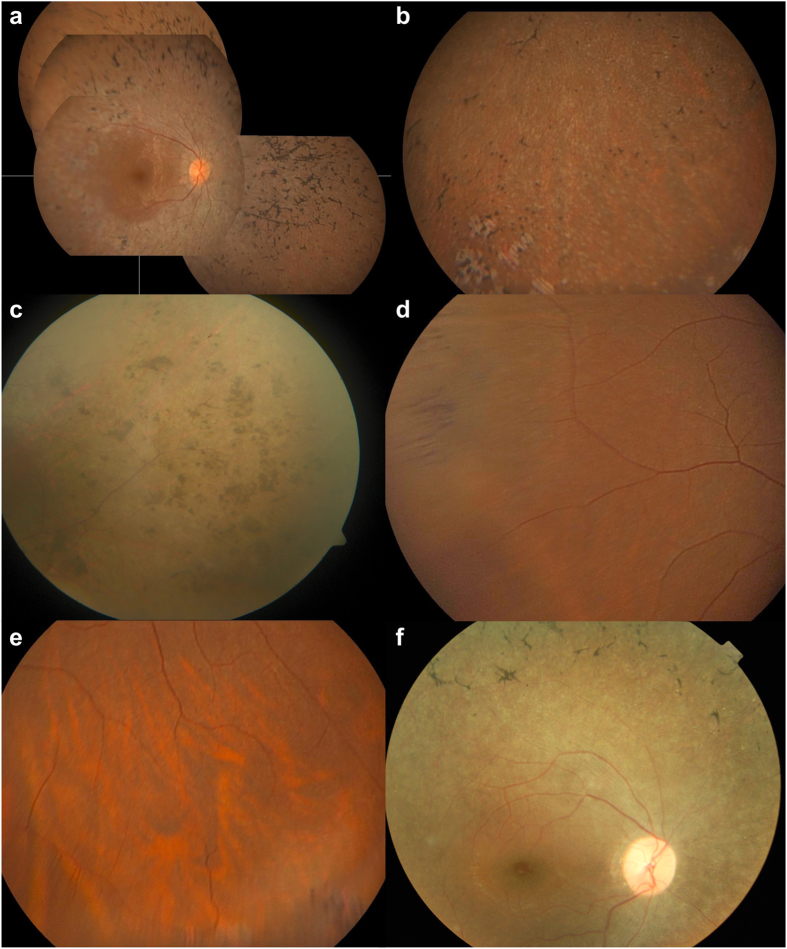
Figure 2. Fundus appearances.
(a,b) Right eye of affected sibling IV:8, fundus montage images (a), periphery (b). Typical aspect of RP: retinal atrophy with vascular attenuation and relatively preserved macula. Note variation in morphology of pigment deposits: aspect of classic spicular pigmentary changes alternate with unusual nummular types and several well-delineated areas of punched-out retinal atrophy, as seen here in inferior periphery. (c) Fundus of left eye of affected sibling IV:5, periphery. Note conglomerates of grouped nummular intraretinal pigmentations in superotemporal periphery in IV:5. (d,e). Fundus of right eye of sibling IV:6 with mild subclinical expression of disease, periphery. Multiple zones of peripheral intraretinal pigment migration as mild expression of RP; note difference in aspect of lesions with both spicular and nummular pigmentations combined. (f) Fundus of the index patient of Pakistani family RP218. Classic aspect of outer retinal atrophy with spicular intraretinal pigmentation without the unusual and distinct grouped nummular pigmentation seen in the family studied here.
The other affected sibling (IV:8), a 44 year-old female, suffers from night blindness and visual field constriction since she was 15 years old. Her BCVA is 20/40 in both eyes, with a concentric constriction of the visual fields to less than 10°. Bilateral posterior subcapsular cataracts were also observed. Fundus imaging showed retinal atrophy, albeit less severe than in the other affected sibling (IV:5). As in her affected brother (IV:5), particular small, nummular intraretinal pigmentations were seen, although not grouped in her. In the periphery, several well-delineated areas of punched-out retinal atrophy were present (Fig. 2a,b).
Extensive ophthalmological examination of two additional siblings (IV:3 and IV:9) and the mother (III:2), was entirely normal.
The phenotype of individual IV:6, a 47-year old male, is particularly interesting. BCVA was measured at 20/30 in the RE and 20/25 in the LE. Visual fields were normal. He claimed to suffer from night blindness, which unfortunately could not be confirmed as the patient did not allow to perform a full field electroretinogram (ERG). Anterior segments were normal. Fundoscopy showed multiple zones of peripheral intraretinal pigmentation with either a spicular or nummular aspect, as well as multiple white dots, representing minor subclinical manifestations (Fig. 2d,e).
Identification of homozygous RHO mutation E150K and non-coding SAMD7 variants
To identify the underlying genetic cause of RD in this Turkish consanguineous family, homozygosity mapping was performed in the affected siblings IV:5 and IV:8. This revealed several shared homozygous regions, containing the SAMD7 and RHO genes located on two different non-contiguous regions on chromosome 3 (Fig. 1b). Sanger sequencing of the coding regions of RHO in one of the affected siblings led to the identification of a rare, yet previously described missense mutation in a homozygous state: c.448G > A, p.(Glu150Lys), also known as E150K (Fig. 1c)6,8,10. Segregation analysis showed that this mutation is homozygously present in the affected siblings IV:5 and IV:8 and that all other family members are heterozygous carriers (Fig. 1a).
In parallel, sequencing of all coding and non-coding exons of SAMD7 did not reveal any pathogenic changes. However, subsequent analysis of regulatory regions of SAMD7 in one affected sibling revealed four different variations located in CBRs. Segregation analysis demonstrated that the affected siblings IV:5 and IV:8 are homozygous for these variants, while all other investigated family members are heterozygous carriers (Fig. 1a). SAMD7 has two previously functionally validated CBRs, one located in the promoter region, CBR1, and the other, CBR2, located in the intronic region between the first two non-coding exons of the gene17,19. The homozygous variants are known single nucleotide polymorphisms (SNPs), having a minor allele frequency of 1.6% according to dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/). Three variants (rs201841157, rs58507718 and rs57060963) are located in CBR1 and consist of an insertion, transversion and deletion event, possibly disrupting a CRX-binding sequence, while the last variant (rs73040228) was found in CBR2 (Fig. 1d–f).
For comparison, family members of two previously described Pakistani arRP families segregating the same E150K RHO mutation8 were investigated for the presence of the non-coding SAMD7 variants identified in this study. However, not surprisingly, none of these SAMD7 variants were detected in both families. In addition, we performed haplotype analysis of the RHO region using the flanking markers described by Azam et al. This revealed no differences between the disease haplotype here and the previously reported disease-associated haplotype of these two Pakistani families (Supplementary Fig. 1)6,8.
SAMD7 localization in human retina
Previously, Samd7 expression has been demonstrated in cross sections of the adult murine retina, showing predominant expression in the photoreceptor nuclei19. To investigate the retinal localization of the human SAMD7 protein, immunohistochemical analyses of human post mortem retinas were performed. This showed a similar expression pattern as in murine retina, with a distinctive staining pattern in the photoreceptor nuclei (Fig. 3a).
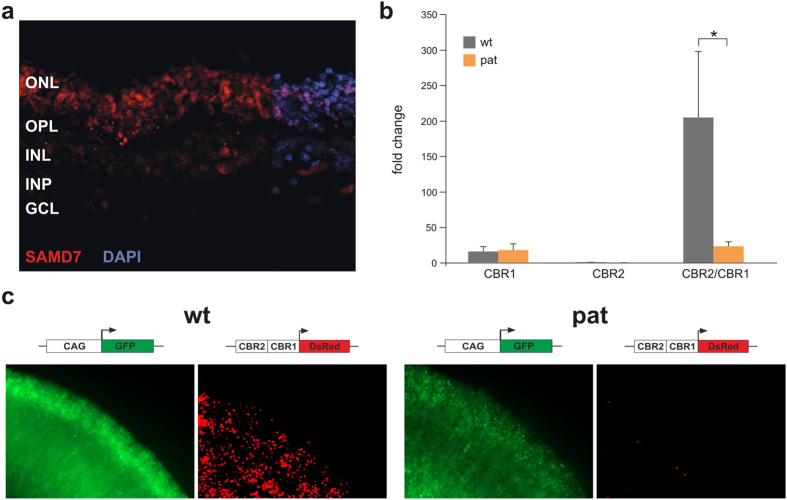
Figure 3. Retinal immunostaining of SAMD7 and transcriptional activity of SAMD7 variants.
(a) Human retinal SAMD7 localization. Representative fluorescent images of horizontal cross-sections of human retina stained with anti-SAMD7 antibody (red, 1:250). Retinal counterstaining was performed with 4′,6-diamidino-2-phenylindole (DAPI) (blue). SAMD7 immunoreactivity is predominantly detected in the photoreceptor nuclei, located in the ONL. ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer. (b) Luciferase experiments. Luciferase assays were performed in HEK cells, using different SAMD7 reporter constructs. The first construct consisted of CBR1 cloned in the pGL4.10 reporter vector containing a luciferase expressing gene without promoter. For the second construct CBR2 was cloned in the pGL3 promoter vector, upstream of a luciferase reporter gene and a minimal basal promoter. Finally, the third construct was obtained by cloning CBR2 upstream of CBR1 in the pGL4.10 vector. Two types of each construct were created, one containing the wt sequence of a healthy control, the other one consisting of the patient (pat) sequences containing the SAMD7 CBR variants. Cis-regulatory activity could be demonstrated for the CBR1 constructs, while for CBR2 only very little luciferase expression could be measured. In both cases, no significant difference in luciferase expression has been observed between patient and control. However, when combining CBR2 and CBR1, luciferase expression increases for the control construct, while for the patient construct there is a significant decrease in expression. *corresponds with p < 0.05 after a two-sample t-test. (c) Electroporation reporter assays in mouse retinal explants. As a confirmation for the luciferase experiments, electroporation assays were carried out for a CBR2/CBR1 construct in mouse retinal explants. Control and variant CBR2/CBR1 were cloned in a dsRed expressing vector without basal promoter and electroporated into isolated retinas of P0 mice. After eight days of in vitro culture, retinas were harvested, fixed and imaged, confirming that the combined cis-regulatory activity of CBR2/CBR1 is lost for the patient constructs.
Assessment of the transcriptional activity of the SAMD7 variants
Cis-regulatory activity of the two murine wild-type (wt) CBRs associated with Samd7 has been demonstrated previously by electroporation assays in mouse retinal explants17,19. Here, in order to gain insights into a regulatory effect on SAMD7 transcription of the SNPs found in CBR1 and CBR2, we wanted to study the cis-regulatory effect of the human SAMD7 CBRs, both for wt CBRs and for variant CBRs of the patients.
First, both wt and variant human CBR1 and CBR2 were cloned in a luciferase expression vector to assess the activity of the CBRs by luciferase assays. CBR1 is located in the proximal promoter region of SAMD7 and was therefore cloned immediately upstream of the luciferase reporter gene. As CBR2 is located in a more distant enhancer region outside the promoter, it was cloned upstream of a luciferase reporter gene carrying a minimal basal promoter. A third construct consisted of CBR2 cloned upstream of CBR1, in order to assay the combined regulatory activity of CBR2 upstream of CBR1. As depicted in Fig. 3b, no significant difference in luciferase expression could be demonstrated between patient and wt constructs for CBR1 and CBR2 separately. However, variant constructs for which CBR2 was cloned upstream of CBR1 showed a significant 8.8 times decrease in luciferase activity compared to the corresponding wt constructs [wt mean = 204.77, variant mean = 23.31, p = 0.0281] (Fig. 3b).
To confirm these results, electroporation assays in mouse retinal explants have been performed with vectors containing the combined CBRs. CBR1 was cloned upstream of a dsRed reporter gene, followed by upstream cloning of CBR2. Electroporation of this construct in mouse retinal explants substantiated the luciferase data. Indeed, the combined CBR2/CBR1 variant construct again showed a markedly decreased SAMD7 reporter activity compared with the wild type CBR2/CBR1 construct (Fig. 3c).
Discussion
The aim of this study was to unravel the molecular pathogenesis of an atypical retinitis pigmentosa phenotype (RP) observed in a Turkish consanguineous family. Homozygosity mapping followed by candidate gene analysis revealed a rare missense mutation in the RHO gene, c.448G > A p.(Glu150Lys), also known as E150K, and four non-coding variants in retina-specific regulatory regions of the SAMD7 gene.
The E150K RHO mutation was described for the first time by Kumaramanickavel et al. in a large Indian consanguineous family. The four living affected siblings were homozygous for this mutation and displayed typical RP. Two other siblings were heterozygous carriers and displayed no symptoms6. Azam et al. reported two additional Pakistani arRP families segregating the same RHO mutation in homozygous state in affected family members and in heterozygous state in asymptomatic individuals8. Recently, this mutation was also reported in a homozygous state in affected individuals of another Pakistani family with classic RP, with no information about heterozygous carriers10. Additionally, this mutation has been extensively studied by Zhang et al. in E150K knock-in mice. Homozygous KK mice displayed early-onset retinal degeneration, while heterozygous EK mice showed a delayed-onset milder retinal degeneration. Hence, the authors state that the heterozygous RHO E150K-associated retinopathy should rather be classified as slowly progressing adRP instead of pure arRP and encourage all human patients carrying this mutation heterozygously to have a follow-up monitoring of their retinal function38. So far, no human symptomatic heterozygous carriers of E150K have been reported in literature however.
Most RHO mutations have a dominant mode of inheritance. Only a few recessive mutations have been reported: apart from the E150K mutation, only three other recessive mutations have been described, two of them associated with subtle retinal involvement in heterozygous carriers. Rosenfeld et al. identified the first RHO mutation with autosomal recessive inheritance, c.745G > T, introducing a premature stop codon, p.(Glu249*), in a RP patient of French-Canadian descent originating from a consanguineous marriage. Interestingly, ERGs performed in both parents and heterozygous siblings showed decreased light sensitivity of rod photoreceptors in heterozygous carriers, although no visual field loss could be detected5. The second mutation is located in the consensus sequence of the splice donor site of intron 4 (c.936 + 1G > T) and presumably leads to defective splicing of the gene. This mutation was first reported by Rosenfeld et al. heterozygously in a healthy control individual, only presenting a subtle abnormality in rod function based on ERG, suggesting an autosomal recessive (AR) inheritance of the mutation5. Subsequently, Macke et al. and Jacobson et al. reported about an adRP family where all four affected family members are heterozygous for this splice site mutation. Disease manifestation varied greatly with age in this family, as the oldest patient had severely decreased visual acuity and undetectable ERG, while the youngest one had a normal fundus, visual acuity and ERG, only showing rod sensitivity loss. This led to the hypothesis of AD inheritance, with a delayed effect of the mutation39,40. Later, Rosenfeld et al. reported two additional families with a total of 25 heterozygous carriers of whom only one 45-year old has been diagnosed with RP during adolescence. Ophthalmological examinations in 11 of the 24 asymptomatic carriers revealed only subtle rod abnormalities, even in four individuals older than 65 years of age. According to the authors, these findings seem to exclude an AD inheritance pattern (penetrance would only be 4%), leaving the possibility of AR/digenic inheritance due to yet unidentified mutations or the involvement of mutations in a different RD gene41. Finally, the mutation was identified for the first time in a homozygous state in a South African family, substantiating a recessive effect of this mutation7. Finally, Kartasasmita et al. identified a homozygous nonsense mutation, c.482G > A [p.(Trp161*)] in two Indonesian families with arRP. Asymptomatic heterozygous family members were found to have a normal fundus, although a slight delay and decrease of b-wave in scotopic ERG response was seen9.
The family we describe here is the fifth family reported to segregate the E150K RHO mutation, where the affected individuals IV:5 and IV:8 are homozygous, while all other siblings are heterozygous carriers. Of note, one of these siblings (IV:6) displays minor subclinical manifestations, more specifically pigmentary anomalies on fundus photography and subjective complaints of night blindness. However, no remarkable decrease in visual acuity could be objectivized. These findings are of interest in light of the delayed-onset and milder phenotype observed in heterozygous EK mice and the difficult genotype-phenotype correlations for the other recessively inherited RHO mutations described above. Interestingly, this individual is the first heterozygous carrier of the E150K mutation in whom subtle pigment deposits could be detected on fundoscopy, although no fundus anomalies were seen in his two heterozygous, asymptomatic sisters. Unfortunately, all family members were reluctant to undergo an ERG, which can be seen as a drawback of this study. Performing an ERG in the heterozygous mildly symptomatic and asymptomatic individuals would allow us to detect subtle decreases in light sensitivity, as seen for the other recessively inherited RHO mutations.
The non-coding variants identified in the SAMD7 region add another level of complexity to the genotype-phenotype correlation in this family. As for the RHO mutation, the affected family members IV:5 and IV:8 are homozygous for the four variants, while all other siblings and their mother are heterozygous carriers. This is the first report of human SAMD7-associated non-coding variants, for which an effect on transcriptional regulation has been demonstrated by luciferase experiments and electroporation assays in retinal explants. In addition, we demonstrated, for the first time, immunostaining of SAMD7 in human retina. As a potential transcriptional repressor, involved in fine-tuning of CRX-regulated gene expression19, reduced or abolished SAMD7 expression could alter the expression of other RD genes. Even though the primary genetic defect has been identified in this family, specifically a well studied missense mutation in RHO, a modifying effect of the regulatory SAMD7 variants on the observed phenotype cannot be excluded. Interestingly, in addition to the spicular pattern of pigment typically seen in RP, distinct conglomerates of grouped nummular pigmentations and several well-delineated areas of punched-out atrophy could be observed in the two affected siblings here. Comparison of the fundus photographs of these Turkish patients with those of the index patient of Pakistani family RP21 segregating the E150K RHO mutation8, but not the SAMD7 variants, revealed a classic RP pigmentation and absence of the distinct nummular retinal pigmentation in the Pakistani patient. These phenotypic differences between patients with the same primary mutation might be attributed to modifying genetic factors, like the SAMD7 variants we identified in this study. While the same atypical, recognizable retinal pigmentation was also seen in the extreme periphery in the heterozygous brother, both healthy heterozygous sisters and their mother appear to have completely normal fundus photographs. This means that the subtle fundus abnormalities seen in the heterozygous brother cannot only be explained by the RHO and SAMD7 genotypes, assuming the presence of other modifying factors.
At present, little is known about modifying genes in human RDs. While some examples have been described for mice and dogs42,43,44,45,46, the first report in humans dealt with a known AIPL1 mutation p.(Arg302Leu) identified as a potential modifier allele in a patient with Leber congenital amaurosis (LCA) and prominent maculopathy carrying two CRB1 mutations47. Later, a variant in RPGRIP1L has been associated with the development of RD in individuals with ciliopathies caused by mutations in other genes48; AHI1 mutations were presented as potential neurological modifiers of CEP290-related disease49; in addition to previous studies showing that penetrance of PRPF31 mutations in adRP correlates with the expression level of the remaining wt PRPF31 allele50,51, CNOT3 was proposed as a modifier of PRPF31 mutations in RP with incomplete penetrance52,53; and more recently it has been suggested that a specific NMNAT1 variant could act as a modifier in other genetic subtypes of LCA54. In this study we showed a potential cis-regulatory role of upstream non-coding SNPs on SAMD7 expression, which might contribute to the retinal phenotype observed here. Further studies are needed however to provide more evidence for a primary or modifying role of respectively SAMD7 coding and non-coding variation in RD pathogenesis.
In conclusion, we identified the rare missense mutation E150K in the RHO gene in a Turkish consanguineous RD family, together with non-coding variants impairing cis-regulatory activity of SAMD7-associated CBRs, which might contribute to the phenotype observed in this family, characterized by a specific unusual pigmentation, in addition to classic RP characteristics.
Methods
Clinical evaluation
Six family members of a Turkish consanguineous arRP family participated in this study: five siblings (IV:3, IV:5, IV:6, IV:8 and IV:9) and their mother (III:2). All family members underwent an ophthalmological examination, consisting of visual acuity measurement, slit lamp examination and fundoscopy. We completed the examination with Goldmann visual field and optical coherence tomography for the two affected siblings (IV:5 and IV:8) and sibling IV:6. Informed consent was obtained, and research protocols adhered to the tenets of the Declaration of Helsinki and were approved by the ethical committee of Ghent University (PA2011/022).
Molecular genetic evaluation
Identity-by-descent and homozygosity mapping
This was performed in the two affected siblings by genome-wide single-nucleotide polymorphism chip analysis using the HumanCytoSNP-12 BeadChip platform (Illumina, San Diego, CA). Identity-by-descent regions (>1 Mb) were identified using PLINK software55 integrated in ViVar56. Resulting homozygous regions were ranked according to their length and number of consecutive homozygous single-nucleotide polymorphisms, as described by Coppieters et al57.
Sanger sequencing of candidate genes
Primers for the coding exons of RHO (NM_000539.3), both coding and non-coding exons of SAMD7 (NM_182610.2) and the SAMD7-associated CBRs were designed using Primer3Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi). All primer sequences can be found in Supplementary Table 1. Sanger sequencing was performed on an ABI3730XL genetic analyzer using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). Sequences were analyzed with Seqscape software (Applied Biosystems). All genomic positions are based on genome build GRCh37/hg19.
Haplotype analysis
Primers were designed for the three SNPs used in the haplotype analysis described by Azam et al. (rs789231, rs2855557 and rs2625961), followed by Sanger sequencing and determination of the genotype for each SNP in all family members of the Turkish family and the described Pakistani family RP218. Primer sequences and SNP details are listed in Supplementary Table 1 and 2.
SAMD7 immunohistochemistry in human retina
Retinal samples of human donors
Retinal samples of human donors were obtained from the Eye Bank of the Center of Ophthalmology, University of Cologne, Germany. The research followed the tenets of the Declaration of Helsinki. After dissection of the anterior segment, the remaining tissue included the posterior pole. Remaining vitreous humor was removed to obtain retinal tissue before further processing.
Immunohistochemistry
For immunofluorescence analysis, horizontal human retinal cryo-sections were fixed with 4% paraformaldehyde and rinsed with PBS. Sections were then rehydrated in PBS and preincubated with 1% dried milk in PBS and 0.01% Tween 20 to reduce nonspecific immunoreactivity. Overnight incubation with the primary anti-SAMD7 Q-12 antibody (sc100141, Santa Cruz Biotechnology, Dallas, TX) was performed at 4 °C in PBS containing 2% BSA, 0.02% NaN3 and 0.1% Triton X-100. To estimate the specificity of the primary antibody, control stainings without primary antibody were performed in parallel. After washing in PBS, samples were labeled for 1 h at room temperature with the secondary anti-rabbit antibody conjugated to Alexa594 (red) (Dianova, Hamburg, Germany). Nuclei counter-staining was performed with 0.1 mg/ml DAPI (4′,6-diamidino-2-phenylindole) in PBS (Molecular Probes, Life Technologies, Frankfurt, Germany) for 10 min at room temperature. The cryo-sections were mounted with fluorescent mounting medium (Dako Cytomation, Hamburg, Germany) and viewed with a Zeiss Axio Imager.M2 fluorescence microscope equipped with ApoTome.2 (Carl Zeiss, Jena, Germany). Microscopic pictures were analyzed with ZEN software (Carl Zeiss, Jena, Germany).
Functional evaluation of SAMD7 variants
Luciferase assays
Human CBR1 and CBR2 were PCR amplified in a healthy control individual and one of the affected siblings, leading to a 594 bp and 694 bp amplicon, respectively. The primer sequences are listed in Supplementary Table 1. Using NheI and BglII restriction enzyme (RE) sites CBR1 was cloned in the pGL4.10 reporter vector (Promega, Madison, WI) containing the luciferase expressing gene luc2 without promoter. CBR2 was cloned in the pGL3 promoter vector (Promega), upstream of a luciferase reporter gene luc + and a SV40 promoter, using the KpnI and NheI REs. Finally, using the same set of RE sites CBR2 was cloned upstream of CBR1 in the pGL4.10 vector. All clones were validated using vector-specific and internal primers. The SAMD7 reporter vectors were co-transfected with either a CRX-expressing vector (pcDNA4 CRX) or a control vector (pcDNA4HisMaxA) in HEK cells. HEK cells do not normally drive photoreactive gene expression and will need Crx co-transfection to drive luciferase expression. Transfection was carried out in 12 well plates using 100 μL of serum-free medium, 0,2 μg of vector DNA and 3 μL of TransIT®-LT1 Transfection Reagent (Mirus, Madison, WI) per vector per well. After 48 h, luciferase activity was measured on an Infinite F200 Pro plate reader (Tecan, Crailsheim, Germany), using 20 μL of cell lysate and 100 μL of luciferase assay reagent (Promega) per well. Luciferase activity was measured as fold change compared to empty plasmid. All assays have been performed in three independent experiments, using three replicates in each experiment.
Electroporation assays
As a confirmation for the luciferase assays, electroporation assays were carried out with CBR2/CBR1 vectors. CBR1 was cloned in a dsRed expressing vector without basal promoter using XbaI and KpnI RE sites, followed by upstream cloning of CBR2 with SalI and XbaI REs. DNA cocktails containing the SAMD7 reporter vectors and a pCAG-GFP vector as an electroporation control were electroporated in isolated retinas of P0 mice. After eight days of in vitro culture, retinas were harvested, fixed and imaged. The detailed protocol used for the electroporation assays has been described previously58.
Additional Information
How to cite this article: Van Schil, K. et al. Autosomal recessive retinitis pigmentosa with homozygous rhodopsin mutation E150K and non-coding cis -regulatory variants in CRX-binding regions of SAMD7. Sci. Rep. 6, 21307; doi: 10.1038/srep21307 (2016).
Supplementary Material
Acknowledgments
We are most grateful to the family members who participated in this study. This work was supported by grants from the Research Foundation Flanders (FWO) (FWO 3G079711 to E.D.B.), by Ghent University Special Research Fund (BOF15/GOA/011), by Belspo IAP project P7/43 (Belgian Medical Genomics Initiative: BeMGI) to E.D.B., by a grant from Funds for Research in Ophthalmology (FRO) to K.V.S., and by grants from the DFG (La1203/8-1) and the Hans und Marlies Stock Foundation to T.L. Supported by a faculty mobility grant (Faculty of Medicine and Health Sciences, Ghent University), K.V.S. performed part of the research in the lab of T.L. (Cologne, Germany). E.D.B. and B.P.L. are Senior Clinical Investigators of the FWO. K.V.S. is doctoral fellow from the institute for Innovation by Science and Technology (IWT).
Footnotes
Author Contributions K.V.S. performed the genetic analyses. E.D.B. supervised the genetic analyses. K.V.S., A.A. and K.D. performed the functional analyses. M.K. and T.L. supervised the functional analyses. F.D. and B.P.L. performed the clinical analyses. M.A. performed the genetic and clinical analyses in the Pakistany family. R.Q. supervised the genetic and clinical analyses in the Pakistany family. K.V.S., E.D.B. and F.D. wrote the main manuscript text, all authors reviewed the manuscript.
References
- Hartong D. T., Berson E. L. & Dryja T. P. Retinitis pigmentosa. Lancet 368, 1795–1809 (2006). [DOI] [PubMed] [Google Scholar]
- Daiger S. P., Sullivan L. S. & Bowne S. J. Genes and mutations causing retinitis pigmentosa. Clin Genet. 84, 132–141 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daiger S. P., Sullivan L. S., Bowne S. J. & Rossiter B. J. F. RetNet. Retinal Information Network. Date of access: 28/09/2015 at https://sph.uth.edu/retnet/ (1996).
- Dryja T. P., Hahn L. B., Cowley G. S., Mcgee T. L. & Berson E. L. Mutation spectrum of the rhodopsin gene among patients with autosomal dominant retinitis pigmentosa. Proc Natl Acad Sci USA. 88, 9370–9374 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld P. J. et al. A null mutation in the rhodopsin gene causes rod photoreceptor dysfunction and autosomal recessive retinitis pigmentosa. Nat Genet. 1, 209–213 (1992). [DOI] [PubMed] [Google Scholar]
- Kumaramanickavel G. et al. Missense rhodopsin mutation in a family with recessive RP. Nat. Genet. 8, 10–11 (1994). [DOI] [PubMed] [Google Scholar]
- Greenberg J., Roberts L. & Ramesar R. A rare homozygous rhodopsin splice-site mutation: the issue of when and whether to offer presymptomatic testing. Ophthalmic Genet. 24, 225–232 (2003). [DOI] [PubMed] [Google Scholar]
- Azam M. et al. A homozygous p.Glu150Lys mutation in the opsin gene of two Pakistani families with autosomal recessive retinitis pigmentosa. Mol. Vis. 15, 2526–2534 (2009). [PMC free article] [PubMed] [Google Scholar]
- Kartasasmita A. et al. A novel nonsense mutation in rhodopsin gene in two Indonesian families with autosomal recessive retinitis pigmentosa. Ophthalmic Genet. 32, 57–63 (2011). [DOI] [PubMed] [Google Scholar]
- Saqib M. A. N. et al. Homozygosity mapping reveals novel and known mutations in Pakistani families with inherited retinal dystrophies. Sci. Rep. 5, 9965, doi: 10.1038/srep09965 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Soest S., Westerveld A., de Jong P. T., Bleeker-Wagemakers E. M. & Bergen A. A. Retinitis pigmentosa: defined from a molecular point of view. Surv Ophthalmol. 43, 321–334 (1999). [DOI] [PubMed] [Google Scholar]
- Züchner S. et al. Whole-exome sequencing links a variant in DHDDS to retinitis pigmentosa. Am. J. Hum. Genet. 88, 201–206 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozgül R. K. et al. Exome sequencing and cis-regulatory mapping identify mutations in MAK, a gene encoding a regulator of ciliary length, as a cause of retinitis pigmentosa. Am. J. Hum. Genet. 89, 253–264 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiguchi K. M. et al. Whole genome sequencing in patients with retinitis pigmentosa reveals pathogenic DNA structural changes and NEK2 as a new disease gene. Proc. Natl. Acad. Sci. USA 110, 16139–16144 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila-Fernandez A. et al. Whole-exome sequencing reveals ZNF408 as a new gene associated with autosomal recessive retinitis pigmentosa with vitreal alterations. Hum. Mol. Genet. 24, 4037–4048 (2015). [DOI] [PubMed] [Google Scholar]
- Haer-Wigman L. et al. Non-syndromic retinitis pigmentosa due to mutations in the mucopolysaccharidosis type IIIC gene, heparan-alpha-glucosaminide N-acetyltransferase (HGSNAT). Hum. Mol. Genet. 24, 3742–3751 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbo J. C. et al. CRX ChIP-seq reveals the cis-regulatory architecture of mouse photoreceptors. Genome Res. 20, 1512–1525 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmann T. et al. Nonsense mutations in FAM161A cause RP28-associated recessive retinitis pigmentosa. Am. J. Hum. Genet. 87, 376–381 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlawatsch J. et al. Sterile alpha motif containing 7 (samd7) is a novel crx-regulated transcriptional repressor in the retina. PLoS One 8, e60633 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B. E. et al. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest A. R. R. et al. A promoter-level mammalian expression atlas. Nature 507, 462–470 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R. et al. An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanoski C., Glass C., Stunnenberg H., Wilson L. & Almouzni G. Epigenomics: Roadmap for regulation. Nature 518, 314–316 (2015). [DOI] [PubMed] [Google Scholar]
- Weinhold N. et al. Gene regulation. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science 347, 1010–1014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberger T. et al. Increasing the yield in targeted next-generation sequencing by implicating CNV analysis, non-coding exons and the overall variant load: the example of retinal dystrophies. PLoS One 8, e78496 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander A. I. Den et al. Mutations in LCA5 , encoding the ciliary protein lebercilin , cause Leber congenital amaurosis. Nat. Genet. 39, 889–895 (2007). [DOI] [PubMed] [Google Scholar]
- Guédard S. Le, Faugère V., Malcolm S., Claustres M. & Roux A. Large genomic rearrangements within the PCDH15 gene are a significant cause of USH1F syndrome. Mol Vis. 26, 102–107 (2007). [PMC free article] [PubMed] [Google Scholar]
- Coppieters F. et al. Hidden genetic variation in LCA9 - associated congenital blindness explained by 5′UTR mutations and copy number variations of NMNAT1. Hum Mutat. 36, 1188–1196 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T. A. et al. Non-exomic and synonymous variants in ABCA4 are an important cause of Stargardt disease. Hum Mol Genet. 22, 5136–5145 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauwens M. et al. An augmented ABCA4 screen targeting noncoding regions reveals a deep intronic founder variant in Belgian Stargardt patients. Hum Mutat. 36, 39–42 (2015). [DOI] [PubMed] [Google Scholar]
- Bax N. M. et al. Heterozygous deep-intronic variants and deletions in ABCA4 in persons with retinal dystrophies and one exonic ABCA4 variant. Hum Mutat. 36, 43–47 (2015). [DOI] [PubMed] [Google Scholar]
- Webb T. R. et al. Deep intronic mutation in OFD1, identified by targeted genomic next-generation sequencing, causes a severe form of X-linked retinitis pigmentosa (RP23). Hum. Mol. Genet. 21, 3647–3654 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Hollander A. I. et al. Mutations in the CEP290 (NPHP6) Gene Are a Frequent Cause of Leber Congenital Amaurosis. Am J Hum Genet. 79, 556–561 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaché C. et al. Usher syndrome type 2 caused by activation of an USH2A pseudoexon: implications for diagnosis and therapy. Hum Mutat. 33, 104–108 (2012). [DOI] [PubMed] [Google Scholar]
- Mayer A. K. et al. Homozygosity mapping and whole-genome sequencing reveals a deep intronic PROM1 mutation causing cone-rod dystrophy by pseudoexon activation. Eur J Hum Genet, doi: 10.1038/ejhg.2015.144. (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte I. et al. MiR-204 is responsible for inherited retinal dystrophy associated with ocular coloboma. Proc Natl Acad Sci USA. 112, E3236–3245 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small K. W. et al. North Carolina macular dystrophy is caused by dysregulation of the retinal transcription factor PRDM13. Ophthalmology, doi: 10.1016/j.ophtha.2015.10.006 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N. et al. Autosomal recessive retinitis pigmentosa E150K opsin mice exhibit photoreceptor disorganization. 123, 121–137 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macke J. P. et al. Identification of Novel Rhodopsin Mutations Responsible for Retinitis Pigmentosa: Implications for the Structure and Function of Rhodopsin. Am J Hum Genet. 53, 80–89 (1993). [PMC free article] [PubMed] [Google Scholar]
- Jacobson S. G. et al. Phenotypes of Stop Codon and Splice Site Rhodopsin Mutations Causing Retinitis Pigmentosa. Invest Ophthalmol Vis Sci. 35, 2521–2534 (1994). [PubMed] [Google Scholar]
- Rosenfeld P. J., Hahn L. B., Sandberg M. A., Dryja T. P. & Berson E. L. Low Incidence of Retinitis Pigmentosa Among Heterozygous Carriers of a Specific Rhodopsin Splice Site Mutation. Invest Ophthalmol Vis Sci. 36, 2186–2192 (1995). [PubMed] [Google Scholar]
- Samardzija M., Wenzel A., Naash M., Remé C. E. & Grimm C. Rpe65 as a modifier gene for inherited retinal degeneration. Eur J Neurosci. 23, 1028–1034 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyadera K., Kato K., Boursnell M., Mellersh C. S. & Sargan D. R. Genome-wide association study in RPGRIP1(−/−) dogs identifies a modifier locus that determines the onset of retinal degeneration. Mamm Genome. 23, 212–223 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox D. M. et al. An allele of microtubule-associated protein 1A (Mtap1a) reduces photoreceptor degeneration in Tulp1 and Tub Mutant Mice. Invest Ophthalmol Vis Sci. 53, 1663–1669 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz N. M. et al. Modifier genes as therapeutics: the nuclear hormone receptor Rev Erb alpha (Nr1d1) rescues Nr2e3 associated retinal disease. PLoS One. 9, e87942 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellissier L. P. et al. CRB2 acts as a modifying factor of CRB1-related retinal dystrophies in mice. Hum Mol Genet. 23, 3759–3571 (2014). [DOI] [PubMed] [Google Scholar]
- Yzer S. et al. CRB1 heterozygotes with regional retinal dysfunction: implications for genetic testing of leber congenital amaurosis. Invest Ophthalmol Vis Sci. 47, 3736–3744 (2006). [DOI] [PubMed] [Google Scholar]
- Khanna H. et al. A common allele in RPGRIP1L is a modifier of retinal degeneration in ciliopathies. Nat Genet. 41, 739–745 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppieters F. et al. Genetic screening of LCA in Belgium: predominance of CEP290 and identification of potential modifier alleles in AHI1 of CEP290-related phenotypes. Hum Mutat. 31, E1709–1766 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vithana E. N. et al. Expression of PRPF31 mRNA in patients with autosomal dominant retinitis pigmentosa: a molecular clue for incomplete penetrance? Invest Ophthalmol Vis Sci. 44, 4204–4209 (2003). [DOI] [PubMed] [Google Scholar]
- Rivolta C. et al. Variation in retinitis pigmentosa-11 (PRPF31 or RP11) gene expression between symptomatic and asymptomatic patients with dominant RP11 mutations. Hum Mutat. 27, 644–653 (2006). [DOI] [PubMed] [Google Scholar]
- Venturini G., Rose A. M., Shah A. Z., Bhattacharya S. S. & Rivolta C. CNOT3 is a modifier of PRPF31 mutations in retinitis pigmentosa with incomplete penetrance. PLoS Genet. 8, e1003040 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A. M. et al. Dominant PRPF31 mutations are hypostatic to a recessive CNOT3 polymorphism in retinitis pigmentosa: a novel phenomenon of “linked trans-acting epistasis”. Ann Hum Genet. 78, 62–71 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemiatkowska A. M. et al. Novel compound heterozygous NMNAT1 variants associated with Leber congenital amaurosis. Mol Vis. 20, 753–759 (2014). [PMC free article] [PubMed] [Google Scholar]
- Purcell S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sante T. et al. ViVar: a comprehensive platform for the analysis and visualization of structural genomic variation. PLoS One. 9, e113800 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppieters F. et al. Identity-by-descent-guided mutation analysis and exome sequencing in consanguineous families reveals unusual clinical and molecular findings in retinal dystrophy. Genet Med. 16, 671–680 (2014). [DOI] [PubMed] [Google Scholar]
- Hsiau T. H.-C. et al. The cis-regulatory logic of the mammalian photoreceptor transcriptional network. PLoS One. 2, e643 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.