Summary
Defects in DNA repair have been linked to cognitive decline with age and neurodegenerative disease. Yet the mechanisms that protect neurons from genotoxic stress remain largely obscure. In this report, we characterize the roles of the NAD+-dependent deacetylase, SIRT1, in the neuronal response to DNA double-strand breaks (DSBs). We show that SIRT1 is rapidly recruited to DSBs in postmitotic neurons, where it exhibits a synergistic relationship with ATM. SIRT1 recruitment to breaks is ATM-dependent; however, SIRT1 also stimulates ATM auto-phosphorylation and activity and stabilizes ATM at DSB sites. Upon DSB induction, SIRT1 also binds the neuroprotective class I histone deacetylase, HDAC1. We show that SIRT1 deacetylates HDAC1 and stimulates its enzymatic activity, which is necessary for DSB repair through the nonhomologous end-joining (NHEJ) pathway. HDAC1 mutants that mimic a constitutively acetylated state render neurons more susceptible to DNA damage, whereas pharmacological SIRT1 activators that promote HDAC1 deacetylation also reduce DNA damage in two mouse models of neurodegeneration. We propose that SIRT1 is an apical transducer of the DSB response and that SIRT1 activation offers an important therapeutic avenue in neurodegeneration.
Once formed during early development, neurons are retained for life and are therefore faced with the challenge of maintaining a stable genome for long periods of time. DNA damage, which perturbs genomic stability, has been linked to cognitive decline in the aging human brain1 and mutations in DNA repair genes manifest profoundly with neurological implications2. Recent studies suggest that DNA damage is also elevated in disorders such as Alzheimer’s disease (AD) and amyotrophic lateral sclerosis (ALS)3–5. However, the precise mechanisms connecting DNA damage with neurodegeneration remain poorly understood.
Sirtuins are NAD+-dependent lysine deacetylases that modulate a number of biological processes that are highly relevant to aging and neurodegeneration6. Previously, we reported that overexpression of SIRT1, the archetypal mammalian sirtuin, confers significant protection against neuronal loss in the transgenic CK-p25 mouse model of neurodegeneration7; however, the mechanisms underlying this protection were unclear. CK-p25 mice express a truncated fragment of the CDK5-activating partner, p35, in an inducible and forebrain-specific manner8 and p25 induction systematically recapitulates various neurodegenerative pathologies, including the accumulation of amyloid-β peptides, neurofibrillary tau tangles, reduced synaptic density, and neuronal atrophy in the forebrain8, 9. Interestingly, further characterization of CK-p25 mice revealed that the appearance of DNA double-strand breaks (DSBs) precedes all other pathological symptoms in these mice10. To understand how SIRT1 is able to suppress neuronal loss, we therefore directly characterized the functions of SIRT1 in the neuronal DNA DSB response.
SIRT1 is essential for DSB signaling and DNA repair in neurons
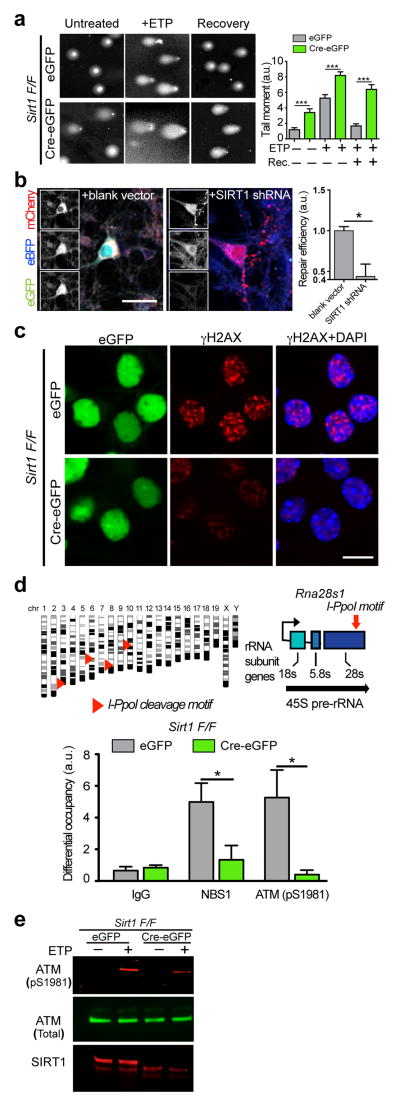
To determine whether SIRT1 is essential for genomic stability in neurons, we transduced neurons cultured from Sirt1 floxed (F/F) mouse embryos with a lentiviral vector carrying the Cre recombinase (Cre-eGFP) to delete Sirt1 (Supplementary Fig. 1a), and assessed DNA damage levels using the single cell electrophoresis assay (comet assay)11. A significant fraction of Sirt1 F/F neurons transduced with Cre-eGFP (hereafter referred to as Sirt1 KO neurons) displayed comet tails even without treatment with an exogenous DNA damaging agent (Fig. 1a). In the presence of the DSB-inducing drug, etoposide, Sirt1 KO neurons displayed longer “tail moments” compared to controls (Fig. 1a). These results suggest that neurons lacking SIRT1 are more susceptible to DNA damage. In addition, whereas “tail moments” in etoposide-treated control neurons were significantly reduced after recovery for 16 h, Sirt1 KO neurons continued to display long comet tails, suggesting that Sirt1 KO neurons are also deficient in DSB repair (Fig. 1a). To verify this, we utilized a reporter assay system (Supplementary Figs. 1b and 1c)12 in which reconstitution of a functional GFP gene indicates successful DSB repair through the nonhomologous end-joining (NHEJ) pathway. In this assay, the number of GFP+ neurons was significantly reduced upon SIRT1 knockdown (Fig. 1b), confirming that SIRT1 is necessary for NHEJ-mediated DSB repair in neurons.
Figure 1. SIRT1 is necessary for initial DSB signaling events and DNA repair in neurons.
a, Sirt1 F/F neurons were infected with lentiviral vectors carrying either a functional Cre recombinase (Cre-eGFP) or a non-functional Cre (eGFP) were treated with 5μM etoposide for 1h, and were either allowed to recover for 16h in the absence of etoposide or lysed immediately. DNA damage was then assessed using the comet assay. Graph indicates “comet tail moments” (***p< 0.001, n = at least 50 per condition, one-way ANOVA). b, Cultured primary neurons were transfected with a pre-digested NHEJ reporter construct (see also Supplementary Figs. 1b and 1c) together with either scrambled shRNA or SIRT1 shRNA and the number of GFP+ cells were assessed to indicate NHEJ-mediated repair (* p < 0.05, unpaired t-test). c, Sirt1 F/F neurons infected as in a were treated with either vehicle or 2μM etoposide, following which the cells were fixed and stained with antibodies to γH2AX. d, A synthetic, inducible system encoding the rare-cutting homing endonuclease, I-PpoI, was used to generate DSBs at defined regions within the genomes of primary Sirt1 F/F neurons transduced as in a. I-PpoI cleavage sites in the Rna28s1 locus are depicted (top). Results from ChIP experiments measuring the recruitment of ATM (pS1981) and NBS1 to cleavage sites within the Rna28s1 locus (bottom) (*p < 0.05, student’s t-test). e, Sirt1 F/F neurons infected as in a were treated with either vehicle or 5μM etoposide, and levels of phosphorylated ATM were compared by western blotting.
Interestingly, despite the elevated levels of DNA damage (Fig. 1a), phosphorylation of H2AX (γH2AX) was reduced (by at least 30%) in Sirt1 KO neurons that were challenged with etoposide (Fig. 1c). Similar results were obtained with neurons pre-treated with the SIRT1 inhibitor, sirtinol (Supplementary Fig. 1d), and together these findings suggested that initial events in DSB signaling could be disrupted in the absence of SIRT1 activity. To address this possibility, we first employed the rare-cutting homing endonuclease, I-PpoI, to generate DSBs at defined loci within the genome of cultured primary neurons13, 14. Chromatin immunoprecipitation (ChIP) following I-PpoI induction revealed that the occupancy of phosphorylated ATM and NBS1, key DSB sensors/transducers, are attenuated at chromatin proximal to I-PpoI-generated DSBs in Sirt1 KO neurons (Fig. 1d). ATM is rapidly activated through auto-phosphorylation upon DSB induction and in turn, coordinates signaling events at DSBs through phosphorylation of its numerous targets, including H2AX15. In addition to being deficient in ATM recruitment, levels of phosphorylated ATM were also significantly reduced in etoposide-treated Sirt1 KO neurons compared to controls (Fig. 1e), indicating that SIRT1 is also essential for ATM activity. Taken together, these results suggest that SIRT1 is an apical component of the neuronal DSB response whose activity is critical for the initial sensing and signaling from DSBs and for their eventual repair through NHEJ.
SIRT1 physically interacts with HDAC1
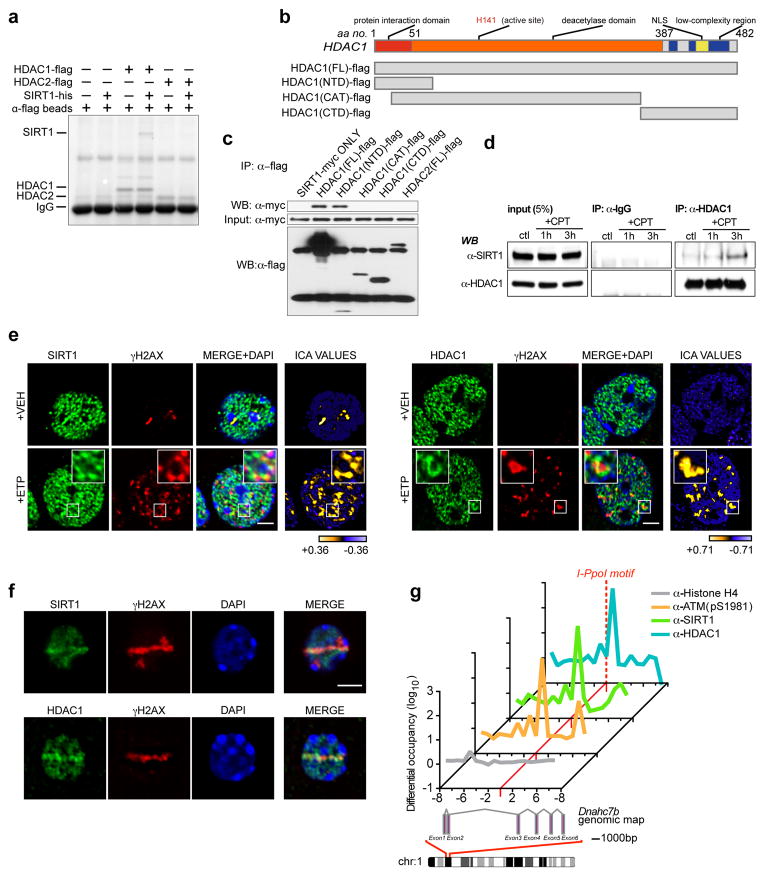
Previously, we showed that overexpression of the class I histone deacetylase, HDAC1, could also suppress neuronal loss in CK-p25 mice10. Because both SIRT1 and HDAC1 were able to suppress neuronal loss in the same mouse model, we reasoned that the two proteins might work collaboratively to promote genomic stability in neurons. Incubation of purified recombinant HDAC1-flag with recombinant SIRT1-his, followed by the precipitation of HDAC1 with anti-flag conjugated agarose beads, co-precipitated SIRT1, suggesting a direct physical interaction between SIRT1 and HDAC1 (Fig. 2a). Notably, HDAC2, which shares ~ 85% similarity with HDAC1, was unable to bind SIRT1 under the same conditions (Fig. 2a). Next, we generated and expressed multiple flag-tagged fragments of HDAC1 (Fig. 2b) and mapped the interaction to the N-terminal domain (NTD) of HDAC1 (Fig. 2c). Interestingly, immunoprecipitation of endogenous HDAC1 indicated only a weak binding with endogenous SIRT1 (Fig. 2d), but this interaction was considerably strengthened upon induction of DNA damage (Fig. 2d).
Figure 2. SIRT1 and HDAC1 physically interact and localize to DSB sites in neurons.
a, Recombinant SIRT1-his was incubated with either HDAC1-flag or HDAC2-flag, following which HDAC1 and HDAC2 were precipitated with anti-flag conjugated agarose beads, and their ability to retain SIRT1 was assessed. b, Diagram illustrating HDAC1 fragment constructs for interaction mapping. FL – full-length; CTD – carboxy-terminal domain; CAT – catalytic domain; NTD – amino-terminal domain. c, The indicated flag-tagged fragments were expressed together with SIRT1-myc, and the flag-tagged proteins were immunoprecipitated and blotted with antibodies against myc. d, HT22 cells were treated with camptothecin (CPT; 1μM), precipitated with antibodies against HDAC1, and blotted with antibodies against SIRT1. e, Etoposide-treated primary neurons treated were fixed and stained with antibodies against either SIRT1 or HDAC1, and γH2AX. Right-most panel illustrates Intensity Correlation Analysis (ICA). Pixels from input channel co-varying positively with corresponding signal from γH2AX channel are indicated in yellow, while negatively co-varying pixels are indicated in blue. Scale bar = 3μm. f, Primary neurons were subjected to sub-nuclear, laser-generated DNA lesioning and stained with antibodies against either SIRT1 or HDAC1, and γH2AX. Scale bar = 3μm. g, The rare-cutting homing endonuclease, I-PpoI, was used to generate DNA DSBs at defined genetic loci in mouse cortical neurons. Recruitment of the indicated proteins at a unique cleavage site located between exons 2 and 3 in the Dnahc7b gene was then assessed by ChIP. Primers were designed at regular 1kb intervals, spanning 10kb both 3’ and 5’ to the I-PpoI consensus site (red dotted line).
Because an interaction between SIRT1 and HDAC1 is stimulated upon DNA damage, we inquired whether SIRT1 and HDAC1 localize to sites of DNA DSBs in neurons. Colocalization analysis of cultured primary neurons revealed that SIRT1 and HDAC1 exhibit a punctate distribution pattern devoid of chromocenters (sub-nuclear compartments consisting of densely packed chromatin) and nucleoli in mouse primary neurons. Upon treatment with etoposide, a significant fraction of SIRT1 and HDAC1 co-localized with γH2AX foci (Fig. 2e), suggesting that both SIRT1 and HDAC1 are present at sites of DSBs in neurons. Next, we used microirradiation to generate sub-nuclear DSBs within the nuclei of individual Hoechst-stained primary neurons (Supplementary Figs. 2a and 2b). Following microirradiation, both SIRT1 and HDAC1 exhibited strong enrichment within lesioned regions that were marked with γH2AX (Fig. 2f). In addition, we again utilized the I-PpoI system and targeted a unique I-PpoI cleavage site within the Dnahc7b locus on chromosome I for ChIP analysis. As before (Fig. 1f), a strong enrichment in phosphorylated ATM was detected at chromatin proximal to the I-PpoI cleavage site following I-PpoI induction (Fig. 2g). Importantly, SIRT1 and HDAC1 levels were also enriched at damage proximal chromatin following DSB generation, with ATM, SIRT1, and HDAC1 displaying the strongest accumulation immediately 3’ to the cleavage site (Fig. 2g). Together, these results suggest that SIRT1 and HDAC1 are recruited to sites of DNA DSBs in neurons.
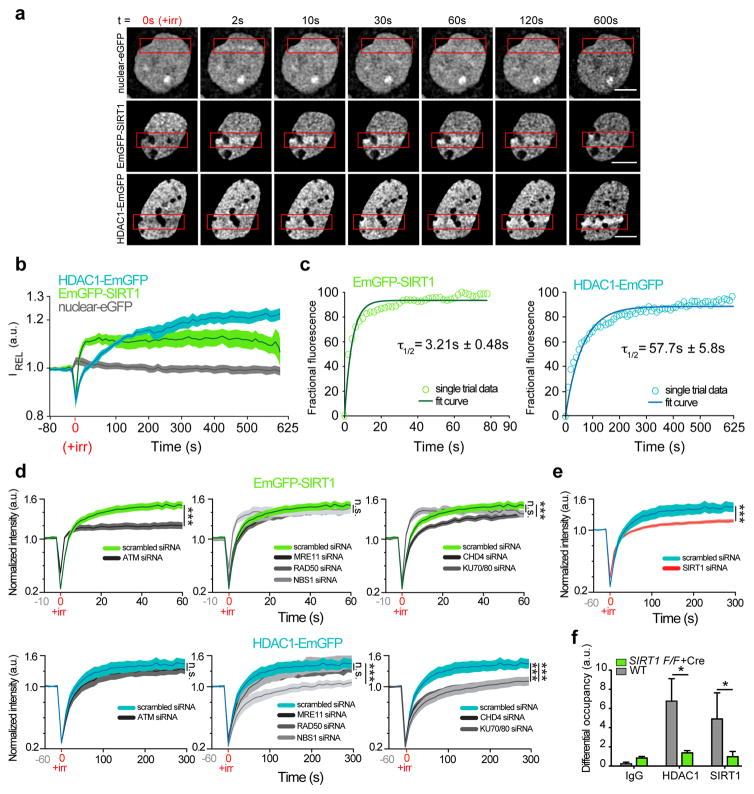
To further appreciate the dynamics of SIRT1 and HDAC1 recruitment to DSB sites, we micro-irradiated Hoechst-stained primary neurons expressing either EmGFP-SIRT1 or HDAC1-EmGFP and monitored their localization to sites of laser-induced DSBs as a function of time. An increase in EmGFP-SIRT1 in laser-lesioned regions was detected almost immediately after DSB induction (τ1/2 = 3.21 ± 0.48 s) (Figs. 3a–3c), whereas HDAC1-EmGFP accumulation became apparent by ~ 60 s (τ1/2 = 57.7 ± 5.8 s), indicating that SIRT1 localizes to DSB sites with faster kinetics than HDAC1. Next, we individually knocked down various known DSB sensors and assessed their effects on SIRT1 and HDAC1 dynamics at laser-induced DSBs in live postmitotic neurons. The recruitment of SIRT1 to DSBs was strictly ATM dependent and knockdown of ATM caused a marked reduction in both the maximal intensity and the kinetics of SIRT1 accrual at DSBs (Fig. 3d and Supplementary Fig. 2c). Thus, SIRT1 and ATM exhibit a mutually dependent relationship, with SIRT1 being essential for ATM stability at DSBs and ATM activity following DSB induction (Figs. 1d–1f), and ATM being necessary to recruit SIRT1 to DSBs. On the other hand, the accumulation of HDAC1 was most severely affected by the knockdown of NBS1 and KU70/80 (Fig. 3d), with the latter also causing HDAC1 to accrue with slower kinetics (Supplementary Fig. 2c). Moreover, HDAC1 is likely recruited as part of the NuRD (nucleosome remodeling and deacetylase) complex, since knockdown of CHD4, an integral component of the NuRD complex, also severely disrupted HDAC1 localization to DSBs (Fig. 3d)16. Knockdown of individual DSB sensors had no effect on the expression of either EmGFP-SIRT1 or HDAC1-EmGFP (Supplementary Fig. 2d).
Figure 3. SIRT1 stabilizes HDAC1 at sites of DNA DSBs in neurons.
a, Time-lapse images of primary cortical neurons that were transfected with a vector carrying nuclear-eGFP, EmGFP-SIRT1, or HDAC1-EmGFP, and subjected to sub-nuclear, laser-generated DNA lesioning using a confocal microscope equipped with a 405 nm laser. Red boxes indicate damage ROI. Scale bar = 5 μm. b, Quantification of relative fluorescence intensity (IREL) as a function of time at lesioned ROIs for neurons expressing nuclear-eGFP (grey; n=13, **r=0.37 Pearson corr.), HDAC1-EmGFP (blue; n=15, ***r=0.98 Pearson corr.), or EmGFP-SIRT1 (green; n=11, ***r=0.77 Pearson corr.). c, Modeling and regression analysis of time-lapse data. Empty circles (green-SIRT1; blue-HDAC1) indicate data from a single trial plotted against the fitted curve (solid lines). d, Relative fluorescence intensity (IREL) as a function of time at lesioned ROIs for neurons expressing either EmGFP-SIRT1 (Top) or HDAC1-EmGFP (Bottom) together with the indicated siRNAs (* denotes p < 0.05; one-way ANOVA; the scrambled siRNA traces are reproduced in each graph for SIRT1 and HDAC1 respectively for convenience of visualization). e, Quantification of relative fluorescence intensity (IREL) as a function of time at lesioned ROIs for neurons expressing either HDAC1-EmGFP together with either a scrambled siRNA or a SIRT1 siRNA (*** p < 0.001, unpaired t-test). f, DSBs were generated using the I-PpoI-ER system in WT and Sirt1 KO primary neurons, and the recruitment of the indicated proteins to cleavage sites within the Rna28s1 locus was assessed using ChIP as in Fig. 1d (* p < 0.05, unpaired t-test).
Overall, distinct proteins seemed to govern SIRT1 and HDAC1 recruitment to DSBs. However, because an interaction between SIRT1 and HDAC1 is enhanced upon DNA damage and SIRT1 localizes to DSBs with faster kinetics than HDAC1, we tested the effect of SIRT1 knockdown on HDAC1 localization to laser-generated DSBs. In neurons transfected with SIRT1 siRNAs, HDAC1 localization to laser-induced DSBs was markedly reduced (Fig. 3e), suggesting that in addition to the above factors, SIRT1 is also essential for HDAC1 recruitment. In corroboration with this finding, HDAC1 enrichment at I-PpoI-generated DSBs was also significantly diminished in Sirt1 KO neurons (Fig. 3f). Together, these experiments describe the dynamics of SIRT1 and HDAC1 in response to DSB formation in living neurons, unveil a novel synergistic relationship between SIRT1 and ATM that is critical for signaling at DSBs, and identify a new interaction between SIRT1 and HDAC1 that helps recruit HDAC1 to DSBs.
SIRT1 deacetylates HDAC1 and stimulates its enzymatic activity
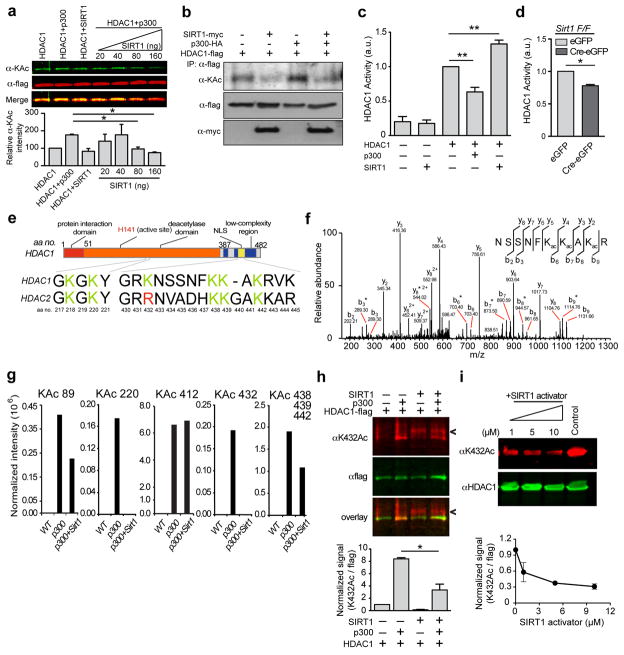
While an important function of the physical interaction between SIRT1 and HDAC1 could be to facilitate the recruitment of HDAC1 to DSB sites (Figs. 3e and 3f), HDAC1 is itself acetylated by the p300/CBP acetyltransferase, and acetylation of HDAC1 inhibits its deacetylase activity17. We therefore posited that the interaction of SIRT1 with HDAC1 might lead to the deacetylation and activation of HDAC1. Incubation of recombinant HDAC1 with increasing amounts of p300 recapitulated the previous observation that p300 acetylates HDAC1 (Supplementary Fig. 3a)17, whereas titration of SIRT1 in the HDAC1-p300 reactions decreased the acetylation of HDAC1 in a dose-dependent manner (Fig. 4a). In addition, whereas overexpression of HDAC1 together with p300 resulted in increased HDAC1 acetylation, co-expression of SIRT1 caused marked HDAC1 deacetylation (Fig. 4b), suggesting that SIRT1 can deacetylate HDAC1.
Figure 4. SIRT1 deacetylates HDAC1 at residue K432 and stimulates its enzymatic activity.
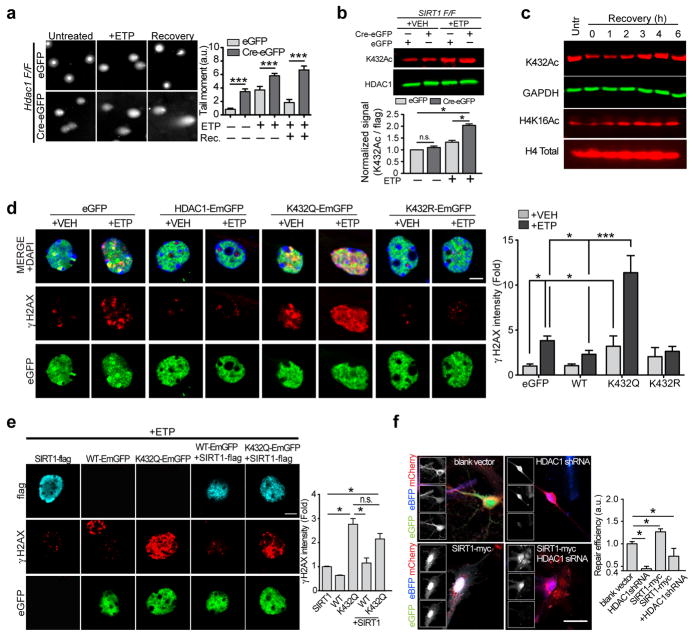
a, Recombinant SIRT1 was titrated over a fixed amount of p300 and HDAC1 and the acetylation of HDAC1 was assessed using western blotting (* p < 0.05, one-way ANOVA). b, HDAC1-flag, p300-HA, and SIRT1-myc were expressed in HT22 cells. HDAC1 was immunoprecipitated, and its acetylation status was probed using an anti-acetyl lysine antibody. c, HDAC1 was pre-incubated with either p300 or with SIRT1 as in (A) and (B), and its enzymatic activity was examined using a fluorescence based HDAC enzymatic activity assay (Supplementary Fig. 3b) (** p < 0.01, one-way ANOVA). d, Sirt1 F/F neurons were infected with Cre-eGFP and eGFP lentiviral vectors, following which HDAC1 was immunoprecipitated, and its activity was measured as in c (*p < 0.05, unpaired t-test). e, Diagram depicting the acetylated lysine residues in HDAC1 and HDAC2. f, Annotated MS/MS spectrum of Lys acetylated peptide “NSSNFKacKacAKacR” that identified lysine acetylation sites on K438, K439 and K441 of HDAC1 upon reaction with p300. b and y ions represents collision-induced peptide fragment ions containing N- or C-terminal respectively. “*” indicates fragment ions with neutral loss of amine. g, Label-free quantification for each lysine acetylation site using protein-abundance normalized peptide precursor ion intensity shows that lysine acetylation abundance on p300-treated HDAC1 decreased upon the addition of SIRT1 to the reaction. h, Recombinant SIRT1 was incubated together with p300 and HDAC1 as in a, and acetylation of HDAC1 was assessed using an antibody specific to acetylated K432. (Arrowhead shows a non-specific cross-reacting band in the lanes containing recombinant SIRT1; * p < 0.05, unpaired t-test). i, HEK293T cells were treated with the indicated concentrations of SIRT1 activator (Compound#10) for 12 h and the HDAC1 acetylation at K432 was assessed.
We next determined whether SIRT1-mediated deacetylation of HDAC1 affects its enzymatic activity. In a fluorescence-based reporter assay (Supplementary Fig. 3b), incubation of recombinant HDAC1 with p300 caused about a 40% reduction in HDAC1 activity, whereas its activity was stimulated by about 30% in the presence of SIRT1 (Fig. 4c). In addition, immunoprecipitated HDAC1 from Sirt1 KO neurons displayed a significant deficit in enzymatic activity compared to HDAC1 precipitated from control neurons (Fig. 4d). Conversely, SIRT1 overexpression resulted in a stimulation of HDAC1 activity (Supplementary Fig. 3c). Together, these results suggest that SIRT1-mediated deacetylation of HDAC1 stimulates its activity.
To specifically determine the residues within HDAC1 that are deacetylated by SIRT1, we incubated recombinant HDAC1 with either p300 alone or with p300 and SIRT1 as before and subjected the reaction mixtures to analysis by mass spectrometry. In the presence of p300 alone, HDAC1 was readily acetylated at residues K89, 220, 412, 432, 438, 439, and 441 (Figs. 4e–4g). Label-free quantification indicated that the addition of SIRT1 resulted in decreased acetylation on all sites except K412 (Fig. 4g). Moreover, acetylation in two of these residues, K220 and K432, was not detected in the mass spectrometry assay, suggesting significant deacetylation activity of SIRT1 on the two acetyl-lysine residues (Fig. 4g). An antibody that specifically recognizes acetylated K432 of HDAC118 and quantitative western further confirmed the ability of SIRT1 to deacetylate HDAC1 at this residue (Fig. 4h). Similarly, treatment of 293T cells with a pharmacological SIRT1 activator (compound #10)19 decreased HDAC1 acetylation at K432 in a dose dependent manner (Fig. 4i). Furthermore, in a computationally predicted tertiary structure of HDAC1 (Supplementary Fig. 3d), the proximity of the region containing K432 to the NTD indicates that the binding of SIRT1 at the NTD could allow for deacetylation of residues at the C-terminus.
Intriguingly, of all the acetylatable residues in HDAC1, K432 was shown to be particularly important for its enzymatic activity and mutation of this residue to an acetyl-mimetic glutamine (K432Q) nearly completely abolished HDAC1 activity17. Moreover, whereas the remaining lysines are conserved between HDAC1 and the closely related HDAC2, K432 in HDAC1 is occupied instead by arginine in HDAC2 (Fig. 4e), and only HDAC1, but not HDAC2 can be acetylated by p30017, 20. Taken together, these results suggest that in addition to facilitating the recruitment of HDAC1 to DSB sites, SIRT1 also exists in an enzyme-substrate relationship with HDAC1, wherein SIRT1 deacetylates HDAC1 at a critical lysine residue, K432, thereby stimulating its enzymatic activity.
SIRT1-mediated deacetylation of HDAC1 is essential for DSB repair in neurons
To understand whether HDAC1 deacetylation has a role in DSB signaling and repair in neurons, we characterized the effects of HDAC1 loss in these processes. In comet assays, Hdac1 KO neurons displayed longer “tail moments” than controls and like Sirt1 KO neurons (Fig. 1a), were unable to recover from etoposide-induced DSBs (Fig. 5a). This suggests that neurons become more susceptible to DSBs in the absence of HDAC1 and that HDAC1 is essential for DSB repair in neurons. In contrast to SIRT1 however, HDAC1 had no effect on ATM auto-phosphorylation (Supplementary Fig. 4a) and γH2AX intensity was increased in Hdac1 KO neurons compared to controls following etoposide treatment (Supplementary Fig. 4b). Similar results were obtained in cultured primary neurons expressing a catalytically inactive HDAC1, HDAC1H141Y (Supplementary Fig. 4c). Thus, although HDAC1 is essential for DNA repair in neurons, initial events in DSB signaling such as ATM and H2AX phosphorylation, do not require HDAC1 activity. These results are consistent with the notion that HDAC1 functions downstream of SIRT1 in the DSB response.
Figure 5. Deacetylation of HDAC1 is essential for DSB repair in neurons.
a, Hdac1 F/F neurons were infected and treated with etoposide and DNA damage was assessed using the comet assay as in Fig. 1a (***p< 0.001, one-way ANOVA). b, Primary neurons cultured from SIRT1F/F embryos were infected as in Fig. 1a. The neurons were then treated with 5μM etoposide for 2 h and the acetylation of HDAC1 at K432 was assessed using western blots (*p < 0.05, one-way ANOVA). c, Cultured primary neurons (DIV7) were treated with 5μM etoposide for 30 min. Cells were then lysed either immediately or following recovery after etoposide washout for the indicated times. The lysates were electrophoresed and the indicated acetylation marks were probed by western blotting. d, Primary neurons were transfected with the indicated vectors and treated with etoposide (2μM) for 1 h, after which the cells were fixed and stained with antibodies against γH2AX. Scale bar = 10μm (* p < 0.05, *** p < 0.001, one-way ANOVA). e, Cultured primary neurons expressing flag-SIRT1 together with either HDAC1-EmGFP or HDAC1K432Q-EmGFP were treated with etoposide (2μM) for 1 h, following which the cells were fixed and stained as in d. Scale bar = 15μm; Quantification shown below (* p < 0.05, one-way ANOVA). f, Cultured primary neurons expressing either SIRT1 together with HDAC1 shRNA were transfected with the pre-digested NHEJ reporter construct and the number of GFP+ cells were assessed as a measure of DNA repair using NHEJ (*p < 0.05, one-way ANOVA).
We next assessed the status of HDAC1 acetylation in Sirt1 KO neurons following etoposide treatment. Whereas HDAC1 acetylation at K432 was increased in etoposide-treated control neurons, this increase was far more pronounced in Sirt1 KO neurons (Fig. 5b), suggesting that the acetylation of HDAC1 is modulated in response to DSB formation and that SIRT1 maintains HDAC1 in a deacetylated and active state in neurons. However, given that HDAC1 is essential for DSB repair, we found it peculiar that the acetylation of HDAC1 is elevated following DSB formation. To further clarify this matter, we briefly treated cultured primary neurons with etoposide (30 min) followed by washout and recovery in etoposide-free media. Lysates were then prepared at hourly intervals and HDAC1 acetylation at K432 was monitored as a function of time post-etoposide treatment. Interestingly, compared to untreated controls, neurons treated with etoposide for 30 min showed a reduction in K432 acetylation (Fig. 5c). This trend continued until 1 h after etoposide washout, following which time acetylated K432 levels began to rise again, surpassing acetylation levels in untreated controls by about 4 h post-washout (Fig. 5c). Furthermore, this pattern of HDAC1 acetylation mirrored changes in the acetylation of H4K16 (Fig. 5c), a previously identified HDAC1 target21 and levels of H4K16 acetylation were increased in Hdac1 KO neurons (Supplementary Fig. 4d).
To further understand the significance of HDAC1 deacetylation in the DSB response, we overexpressed eGFP-tagged variants of HDAC1 carrying either a K432Q (acetylation mimetic) or a K432R (acetylation resistant) mutation in cultured primary neurons. In control neurons expressing eGFP alone, etoposide treatment readily triggered the formation of γH2AX foci, and consistent with previous observations, overexpression of HDAC1-EmGFP caused a reduction in γH2AX foci (Fig. 5d). In contrast, neurons expressing the K432Q mutant displayed γH2AX foci even in the absence of etoposide treatment, and a dramatic increase in the presence of etoposide (Fig. 5d). On the other hand, neurons expressing the K432R mutant displayed a modest reduction in γH2AX intensity compared to controls, indicating that constitutive acetylation of HDAC1 renders neurons more susceptible to genotoxic insults, especially DSBs.
Because SIRT1 stimulates HDAC1 through deacetylation, we predicted that the acetyl-mimetic HDAC1K432Q mutant would also be refractory to the effects of SIRT1 overexpression. In the presence of etoposide, neurons overexpressing SIRT1 showed a significant reduction in γH2AX foci compared to controls; however, SIRT1 overexpression had little effect of γH2AX levels in neurons also expressing the HDAC1K432Q mutant (Fig. 5e). Similar to SIRT1 overexpression, treatment of neurons with the pharmacological SIRT1 activator also caused a reduction in γH2AX intensity (Supplementary Figs. 4e and 4f) and neurons expressing the K432Q and K432R mutants were also refractory to SIRT1 activator-mediated reduction in γH2AX intensity (Supplementary Fig. 4f). Mutation of the lysine residue to glutamine had no effect on the ability of HDAC1 to bind SIRT1 (Supplementary Fig. 4g). However, expression of the NTD fragment of HDAC1, which would compete with endogenous HDAC1 for SIRT1 binding, resulted in increased γH2AX intensity compared to controls (Supplementary Fig. 4f). Also, SIRT1 activator treatment could not directly stimulate the activity of purified recombinant HDAC1 in the absence of SIRT1 (Supplementary Fig. 4g). Additionally, we again utilized the fluorescence-based NHEJ reporter system, wherein we transfected the pre-digested reporter construct in neurons expressing SIRT1 together with either a control shRNA or HDAC1 shRNA. SIRT1 overexpression in neurons stimulated NHEJ-mediated DSB repair, as indicated by the increase in GFP+ cells compared controls (Fig. 5f). However, this increase was attenuated in neurons expressing HDAC1shRNA (Fig. 5f). Taken together, these results suggest that the ability of SIRT1 to protect against DNA damage and stimulate DNA repair requires it to interact with and deacetylate HDAC1.
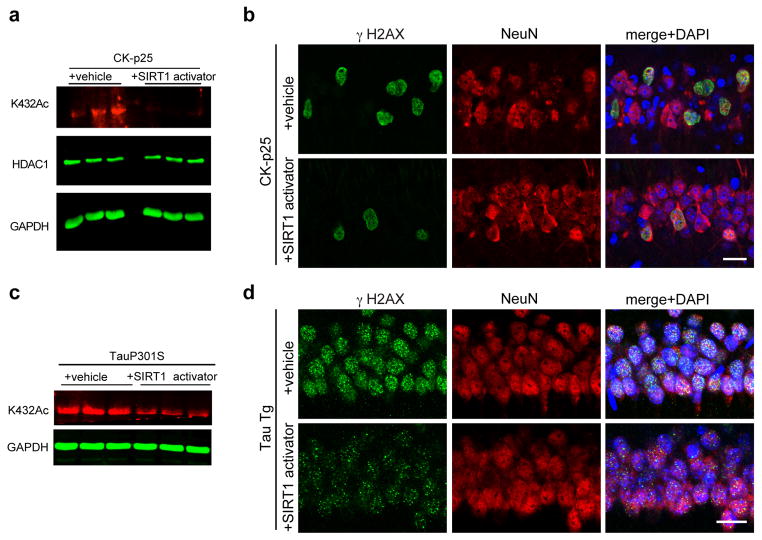
Based on these observations, we assessed whether SIRT1 activation also affects DSB levels and HDAC1 acetylation in neurodegenerating CK-p25 mice by orally administering the pharmacological SIRT1 activator to CK-p25 mice. Western blot analysis of hippocampal lysates after six weeks of p25 induction revealed a sharp reduction in HDAC1K432 acetylation in CK-p25 mice that were treated with the SIRT1 activator compared to CK-p25 mice treated with a vehicle control (Fig. 6a). At the same time, while vehicle-treated CK-p25 mice displayed a marked increase in γH2AX-positive cells in the hippocampus, γH2AX-positive cells were reduced by about 40% in CK-p25 mice administered with the SIRT1 activator (Fig. 6b). These results suggest a strong correlation between SIRT1 activation, HDAC1 deacetylation, and a reduction in DNA DSBs. Additionally, administration of the SIRT1 activator was also able to reduce HDAC1 acetylation and γH2AX intensity in two-month old human tau transgenic (P301S Tg) mice22 (Figs. 6c and 6d). Together, these results highlight the therapeutic potential of SIRT1 activators against neurodegeneration.
Figure 6. Pharmacological SIRT1 activation can protect neurons against DNA damage in vivo.
a, 6-weeks induced CKIIα-p25 mice were administered either vehicle or 30mg/kg compound#10. Brain lysates were then prepared and the acetylation of HDAC1 at K432 was assessed using quantitative western blotting. b, Representative immunohistochemical images depicting γH2AX and NeuN staining in 6-weeks induced CKIIα-p25 mice that were either administered compound#10 or vehicle as described in Methods. c, Two-month old Tau P301S Tg mice were treated with SIRT1 activator and hippocampal lysates were prepared. The acetylation of HDAC1 at K432 was then assessed using western blotting. d, Two-month old P301S Tg mice were administered either vehicle or 30mg/kg compound#10 through oral gavage (5 mice per group). Injections were performed once daily for two weeks. Following this, the mice were sacrificed, and their brains were sectioned and stained with antibodies against γH2AX and NeuN.
Discussion
Overall, our data suggests that SIRT1 primes the cellular response to DNA DSBs by stimulating the activities of ATM and HDAC1. Upon being recruited to DSBs in an ATM-dependent manner, SIRT1 in turn stimulates ATM auto-phosphorylation and ATM recruitment to DSB sites, and thereby primes the cellular DSB signaling cascade (Supplementary Fig. 5). Following DSB induction, the MRE11-RAD50-NBS1 complex is known to activate ATM via an interaction between NBS1 and ATM23. Interestingly, SIRT1 also interacts with and deacetylates NBS124, and NBS1 recruitment to DSBs is also compromised in Sirt1 KO neurons (Figure 1d). It is therefore intriguing to consider the SIRT1-NBS1 interaction as a potential mechanism of ATM activation. However, ATM is also known to be an acetylated protein25 and an equally interesting possibility involves SIRT1-mediated deacetylation of ATM being important for ATM autophosphorylation and activity.
In addition to activating ATM, SIRT1 participates in conjunction with KU70/KU80, NBS1, and the NuRD complex to stabilize HDAC1 at DSB sites. Furthermore, SIRT1 deacetylates and stimulates HDAC1, thus facilitating the dynamic regulation of HDAC1 activity that is essential for DSB repair through NHEJ. Our data are consistent with the notion that HDAC1 functions in DNA repair by affecting chromatin configuration through epigenetic modification. For instance, acetylation of H4K16 and H3K56 decrease in an HDAC1/HDAC2-dependent manner following DSB induction and the inability to deacetylate these residues results in a more relaxed chromatin and decreased DNA repair21, 26. However, an “open” chromatin configuration is also important for repair because it allows repair proteins to easily access damaged sites26. We propose a dynamic model in which “closed” chromatin configuration (likely mediated by HDAC1 and other proteins) in the initial stages after DSB formation would allow for the broken DNA ends to be retained in close proximity and for transcriptional silencing in their vicinity, following which controlled “opening” of the chromatin would grant access to repair and signaling factors. The dynamic modulation of HDAC1 activity through its acetylation and the biphasic pattern of H4K16 acetylation (Fig. 5c)21 are consistent with such a model. We demonstrate here that one arm of such modulation is conferred by SIRT1, which deacetylates and activates HDAC1. The other arm, likely involving an acetyltransferase that inactivates HDAC1, awaits discovery. Several HATs, including p300, hMOF, and TIP60 are known to function in the DNA DSB response27–29. Moreover, these HATs are also deacetylated by SIRT1, and interestingly, SIRT1-mediated deacetylation was shown to inhibit their HAT activity30, 31. The significance of these interactions in chromatin organization and signaling at DSBs is only beginning to be unraveled and while our work emphasizes the significance of the SIRT1-HDAC1 connection in DSB repair, the relative significance of SIRT1-mediated deacetylation of the above substrates warrants further investigation.
In neurodegenerative disorders such as AD, Parkinson’s disease, and ALS, the major risk factor is age itself32. Microarray analysis of post-mortem human brain samples revealed that genes encoding for synaptic transmission, learning, and memory are downregulated after age 45, and that this is associated with elevated levels of oxidative damage in the promoters of the downregulated genes1. In addition to this, DNA DSBs and an upregulation of DNA damage response genes precede the appearance of all other AD-like neuropathological hallmarks in CK-p25 mice10, and elevated levels of DNA strand breaks were observed in the AD brain itself3. Together, these results raise the intriguing possibility that the accrual of DNA damage with age could underlie the pathological changes associated with neurological disease. Interestingly, SIRT1 is known to directly modulate synaptic plasticity and memory formation33, and SIRT1 redistribution in response to chronic DNA damage is thought to underlie some of the transcriptional changes in the aging brain34. Considering these observations therefore, the benefits conferred by pharmacological activation of SIRT1 could be significant.
Online Methods
Mouse strains, expression constructs, shRNA constructs, and virus generation
Hdac1 F/F and Sirt1 F/F mice were the kind gifts of Dr. Eric N. Olson and Dr. Frederick W. Alt, respectively and were as described35, 36. Murine HDAC1 and SIRT1 were sub-cloned into pcDNA6.2/C-EmGFP Gateway Vector (Invitrogen cat. no. V355-20) and pcDNA6.2/N-EmGFP Gateway Vector (Invitrogen cat. no. V356-20), resulting in C-terminal and N-terminal fusion proteins respectively. HDAC1 fragments were constructed according to functional protein domains as determined by bioinformatic analysis with PFAM and as described previously37. The Stratagene QuikChange Site-Directed Mutagenesis Kit (Stratagene cat. no. 200518) was used to generate HDAC1 mutants mimicking either a constitutively acetylated or non-acetylatable state at amino acid position 432 (K432Q and K432R, respectively). Because fusion of several different affinity tags to the N-terminus of HDAC1 was shown to interfere with its catalytic activity, all HDAC1 fusion constructs were generated as C-terminal fusions. HDAC1 and SIRT1 shRNA constructs and catalytic residue mutants were as previously described7, 10. The HA-ER-I-PpoI overexpression construct was obtained from the lab of Dr. Michael Kastan (St. Jude’s Children’s Research Hospital)13, 14, and was modified for production of lentivirus by subcloning into a lentiviral backbone containing T2A-RFP under the control of the PGK promoter. Lentiviral constructs, lenti-Cre and lenti-ΔCre, were the kind gift of Dr. Richard Huganir (Johns Hopkins University) and were as reported previously38. Pooled siRNA oligonucleotides targeting mouse Sirt1 (sc-40987), Hdac1 (sc-29344), Atm (sc-29762), Mre11 (sc-37396), Nbs1 (sc-36062), Rad50 (sc-37398), and control scrambled siRNA (sc-37007) were obtained from Santa Cruz Biotechnologies. Pooled siRNA oligonucleotides targeting mouse Ku70 (EMU067171), and Ku80 (EMU053211) were obtained from Sigma-Aldrich. Pooled siRNA oligonucleotides targeting mouse Chd4 (L-052142-00-0005) were obtained from Dharmacon.
Antibodies
HDAC1 antibodies against acetylated HDAC1 (K432Ac) were generated from rabbits injected with acetylated c-terminal peptide (peptide sequence: GEGGRK(Ac)NSSNF). Antibodies used were for staining were as follows: anti-HDAC1 1.T9 (Abcam; ab31263) 1:1000, anti-SIRT1 (Abcam; ab7343) 1:1000, anti- γH2AX (Millipore; 05-636) 1:1000, anti-flag M2 (Sigma; F1804) 1:1000. Antibodies used for western blots were as follows: anti-flag M2 (Sigma; F1804) 1:1000, anti-c-myc 9E10 (Thermo; MA1-980) 1:1000, anti-HDAC1 (Thermo; PA1-860) 1:1000, anti-Sir2α (Millipore; 07-131) 1:1000, anti-acetyl-lysine (Millipore; 05-515) 1:500, anti-HA (Millipore; ab3254) 1:1000, anti-HDAC1 (K432ac) 1:1000, Anti-ATM (pS1981)1:1000 (Abcam; ab36810), Anti-ATM 1:1000 (Abcam; ab2618) . Antibodies used for ChIP and IP experiments were all used at an amount of 2μg per IP reaction, and are as follows: anti-Histone H4, pan (Millipore; 04-858), anti-ATM (pS1981) (Abcam; ab36810), anti-HDAC1 1.T9 (Abcam; ab31263), anti-NBS1 (Novus; NB100-60654), anti-Sir2a (Millipore; 07-131), anti-flag M2 (Sigma; F1804).
Cell culture and transfection/infection
Cell lines used for experimentation were either Human Embryonic Kidney 293T cells (HEK293T) (ATCC; CRL-11268) or an immortalized mouse hippocampal cell line; HT2239. All cell lines were maintained in Dulbecco’s Modified Eagle Media (DMEM; Gibco, 10566) supplemented with L-glutamine (5mM), penicillin/streptomycin, and FBS. For live-imaging experiments, phenol red-free DMEM (Gibco; 31053) was used as a media substitute. Cell lines were maintained at standard environmental conditions (97% humidity, 5% CO2, 37°C).
For primary neurons, dissociated cortical neurons dissected from E16-E18 Swiss-Webster mice were plated at a density of 500,000 cells/well for 24-well plates, 2 million cells/plate for 35 mm glass bottom plates, and 15 million cells/plate for 10 cm plates. The plates were coated beforehand by incubation with poly D-lysine (0.05 mg/ml) and laminin (0.005 mg/ml) for 1 hour at 37°C, followed by washing twice with dH2O. Neurons were maintained in neurobasal media (NB) (Gibco; 21103) supplemented with L-glutamine (5mM), penicillin/streptomycin, and B27 neuronal additive. Phenol red-free NB (Gibco; 12348) was used as a substitute for neuron cultures used in live-imaging experiments.
Cell lines and primary neuron cultures were transiently transfected using Lipofectamine 2000 reagent (Invitrogen; 11668) for at least 1 hour in media lacking antibiotics, after which cells were washed in warmed media and given at least 24 hours to allow for construct expression prior to usage. For live-imaging experiments using siRNA, 0.75μg siRNA was cotransfected with either HDAC1-EmGFP, or EmGFP-SIRT1 given 48 hours to allow for sufficient knockdown prior to imaging. For viral gene transduction, virus was added directly to the culture media. I-PpoI-ER was induced by adding 4-OHT (Sigma; H7904) to a final concentration of 1μM and incubated for 6 hours prior to fixation.
Western blotting and immunoprecipitation
For both primary neurons and cell lines, 1.5–2 × 106 cells were washed once with PBS and lysed for 10 min on ice in RIPA buffer (150 mM NaCl, 1% IGEPAL, 0.5% NaDOC, 0.1% SDS, 50 mM Tris pH 8.0, supplemented with protease inhibitors) on the plates in which they were originally grown. Cells were then collected by scraping and rotated for 30 min at 4°C. Supernatant was collected following centrifugation (13,000 rpm, 10 min, 4°C). For each sample, SDS protein loading buffer was added to 1X and boiled (95°C, 10 min) before loading onto a 10% SDS-PAGE gel. Gels were transferred to PVDF membranes (200 mA constant current) amd blocked with 3% BSA in PBS-T for one hour prior to application of primary antibodies. Membranes were visualized either with electro-chemiluminescence and autoradiographic film detection, or with the LiCor Odyssey quantitative western imaging system.
For immunoprecipitations, 1mg total protein lysate was used for each condition and brought to a total volume of 500μl with RIPA buffer. The appropriate antibody was then added and the mixture was incubated on a rotator overnight at 4°C. As required, protein A/G-conjugated agarose beads (GE Health Science; 17-5280/17-0618) or flag M2 affinity gel (Sigma; A2220) were equilibrated with RIPA buffer, blocked overnight with 3% BSA, and washed prior to adding to samples. For all immunoprecipitations, a total volume of 30μl bead slurry was used per reaction mix. The reaction mixtures were incubated for 1 hour at 4°C, following which they were washed 4 times, and denatured by boiling (95°C, 10 min) in RIPA buffer containing 1X SDS sample buffer.
Immunostaining, image acquisition, and analysis
For immunostaining, cells were fixed by incubating with 4% PFA for 10 min at room temperature, and incubated with blocking buffer (5% normal donkey serum, 0.3% TritonX-100 in PBS) for 1 h. Primary antibodies were diluted in blocking buffer and incubated with cells overnight at 4°C. Primary antibodies were visualized using appropriate secondary antibodies conjugated to Cy2, Cy3, and Cy5 fluorescent dyes. Images for all fixed cells were acquired on a Zeiss LSM510 laser-scanning confocal microscope, and subsequently deconvolved using theoretical PSFs generated in ImageJ, along with the Tikhonov-Miller iterative image restoration algorithm, which has been implemented in the DeconvolutionLab plugin written for ImageJ40. To assess and analyze images in a quantitative and unbiased manner, CellProfiler automated image analysis software41 was trained to measure per-cell γH2AX signal from a minimum of one hundred cells per condition for all imaging experiments. For experiments utilizing tagged proteins, CellProfiler was trained to consider transfected cells exclusively for quantification. Colocalization was assessed using Intensity Correlation Analysis computation, which highlights pixels either negatively, or positively co-varying between two input channels. A plugin implementing this technique has been written as a plugin for ImageJ, and is freely available for download (http://www.macbiophotonics.ca/imagej/colour_analysis).
Image processing and analysis for microirradiation live-imaging
All data acquired from microirradiation time-lapses was processed according to the above mentioned iterative deconvolution strategy prior to analysis. Normalized relative fluorescence (IREL) within the 2μm2 rectangular strip ROI was quantified for each time point using a computation that compensates for both background signal and fluorescence loss due to observational bleaching42. τ1/2 was extracted from IREL values by first converting them into fractional fluorescence values as described previously43, and Prism5 was used to plot and fit curves to this data.
HDAC1 structural modeling and tertiary structure prediction
The I-TASSER (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) computational protein structure prediction algorithm was used to generate a 3-dimensional predicted model for HDAC144, 45. The mouse HDAC1 (NP_032254) amino acid sequence, and crystal structure from the ancient HDAC1 ancestor, HDLP (RCSB PDB ID: 1C3P)46, were used as inputs for processing by the I-TASSER algorithm. Output structure coordinates were processed and formatted in PyMol, and amino acids were color-coded to correspond to the HDAC1 functional domain schematic.
In vivo drug administration
Mice were administered oral doses of 30 mg/kg SIRT1 activator (compound#10) for 4 weeks. Dosage determination was based on pharmacokinetic studies that optimized for brain penetrance and minimal side effects. Oral gavage was performed using 1.5-inch, curved, 20-gauge, stainless steel feeding needles with a 2.25-mm ball (Braintree Scientific). Twice-daily gavage treatments were performed between the hours of 0800 to 1000 and 1600 to 1900.
Microirradiation and live imaging
Microirradiation was performed as described previously47 on DIV7 mouse primary cortical neurons using a Zeiss LSM710 inverted laser scanning confocal microscope. Cells were maintained in a controlled and stable environment for the duration of the imaging sessions. At least 24 hours were allowed in transfected cells for construct expression.
Chromatin immunoprecipitation
For ChIP, 1.5 × 106 cells were subjected to a two-step dual cross-linking procedure as described previously48, 49. Purified DNA was analyzed by q-PCR using a BioRad CFX-96 quantitative thermocycler, and SsoFast EvaGreen Low-ROX qPCR SuperMix (BioRad). Data was analyzed using the ΔΔCT method (Applied Biosystems).
Single cell electrophoresis (comet) assays
Cultured primary neurons of were treated with 5μM etoposide for 1h and either allowed to recover for 16 h or dissociated immediately using 0.025% Trypsin-EDTA. Dissociated neurons were embedded in a thin layer of low-melting agarose (0.5%), lysed, and subjected to single-cell gel electrophoresis under alkaline conditions11.
In vitro binding and acetylation/deacetylation reactions
For binding reactions, anti-flag-conjugated agarose beads (Sigma) were resuspended in 100μl binding buffer (50mM Tris-HCl [pH 7.5 at 25°C], 5mM MgCl2, 1mM EDTA, 100mM NaCl, 0.05% NP-40, 10% glycerol). HDAC1-flag and SIRT1-his (1μg each) were combined in reaction buffer (50 mM Tris-HCl [pH 7.5 at 25°C], 1mM DTT, 137mM NaCl, 2.7mM KCl, 4mM MgCl2, 0.1mM EDTA, 1mM NAD+, 10% glycerol) and incubated at 37°C for 1h. Each reaction was then supplemented with 30μl bead slurry and 90μl of binding buffer, and incubated in a rotator overnight at 4°C. The beads were then pelleted, washed, boiled in Laemmli SDS-PAGE loading dye, and analyzed by SDS-PAGE. In acetylation reactions (30μl), 100ng recombinant HDAC1-flag was incubated together with recombinant p300-HA in a buffer containing 50mM Tris [pH 8.0 at 25°C], 1mM DTT, 1mM PMSF, 0.1mM EDTA, 50nM acetyl-CoA, and 10% glycerol were incubated at 30°C for 1h. For deacetylation reactions, acetylation reactions were first performed as described above, following which the reactions were supplemented with NaCl (137mM final), KCl (2.7mM final), MgCl2 (4mM final), NAD+ (1mM final), and recombinant SIRT1-his, and incubated for 1h at 37°C. HDAC1 enzymatic activity was measured using a fluorimetric assay as described in Haumaitre et. al. (2008). Prior to measuring HDAC1 activity, acetyl CoA and NAD+ in reactions were removed through dialysis and 5mM NAM was added to ensure SIRT1 inactivation.
LC-MS analysis
Gel bands containing HDAC1 were excised and in-gel digested using a protocol previously described50. The tryptic peptides were solubilized in HPLC buffer A (0.1% formic acid in water) and loaded onto a capillary HPLC column (10 cm, 75 um ID) self-packed with Jupiter C12 resin (Phenomenex). Samples were separated using 90 min linear gradient of 5%–30% HPLC buffer B (0.1% formic acid in acetonitrile) and analyzed by LTQ Orbitrap Velos mass spectrometer (Thermofisher Scientific Inc.). The MS/MS data were analyzed with Mascot search engine (Matrix Science, v2.1) with a Mascot cutoff score of 10 and p<0.05 and with subsequent manual verification. Label-free quantification was performed for each Lys acetylation site based on peptide precursor ion intensity normalized by protein abundance ratios.
Statistical Methods
Standard statistical methods consistent with previous reports employing similar assays, techniques, and methods were utilized to analyze all data11, 14, 21, 47. No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications10, 13, 21. Analysis of γH2AX intensity in CK-p25 and TauP301S following SIRT1 activator treatment were performed blind to the conditions of the experiment. For most other analysis, data collection and analysis were not performed blind to experimental conditions. Data were collected and processed randomly and appropriately blocked. Data distribution was assumed to be normal but this was not formally tested.
Supplementary Material
Acknowledgments
We would like to thank the following: Dr. Frederick W. Alt (Harvard Medical School) and Dr. Eric N. Olson (UT Southwestern Medical Center) for providing the Sirt1 F/F and Hdac1 F/F mice, respectively, Dr. Richard Huganir (Johns Hopkins University) for providing the lentiviral Cre constructs, Vipin Suri, James Ellis and George Vlasuk (Sirtris) for providing compound#10, and together with Johannes Gräff, for critical comments on the manuscript, Dr. Michael Kastan (St. Jude’s Medical Research Hospital) for gifting the I-PpoI-ER construct, and Dr. Vera Gorbunova (University of Rochester) for providing the NHEJ constructs. This work was supported by NIH PO1 grant AG27916 and the Glenn award for research in biological mechanisms of aging to L.-H.T., NIH grant R01 HL095674 to Y.Q., and NIH grant U54 RR020389 to Y.Z. M.M.D. was supported by NIH Training Grants T32 GM007484 and T32 MH081728.
Footnotes
Author Contribution
This study was designed by MMD, RM and L-HT, and directed and coordinated by L-HT. MMD and RM planned and performed most of the experiments. LP maintained the Sirt1 F/F and Hdac1 F/F mice, and together with BA, helped with the microirradiation experiments. YC and YZ conducted the mass spectrometry analysis of HDAC1 acetylation. DK and JG performed some preliminary experiments with CK-p25 mice, and JG performed the compound#10 treatment and subsequent analysis of CK-p25 mice. P-CP contributed to statistical analysis and quantification of several experiments, and YQ developed and provided the HDAC1K432Ac antibody. RM, MMD, and L-HT wrote the manuscript, with critical input from all the authors.
References
- 1.Lu T, et al. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 2.Rass U, Ahel I, West SC. Defective DNA repair and neurodegenerative disease. Cell. 2007;130:991–1004. doi: 10.1016/j.cell.2007.08.043. [DOI] [PubMed] [Google Scholar]
- 3.Adamec E, Vonsattel JP, Nixon RA. DNA strand breaks in Alzheimer's disease. Brain Res. 1999;849:67–77. doi: 10.1016/s0006-8993(99)02004-1. [DOI] [PubMed] [Google Scholar]
- 4.Ferrante RJ, et al. Evidence of increased oxidative damage in both sporadic and familial amyotrophic lateral sclerosis. J Neurochem. 1997;69:2064–2074. doi: 10.1046/j.1471-4159.1997.69052064.x. [DOI] [PubMed] [Google Scholar]
- 5.Nouspikel T, Hanawalt PC. When parsimony backfires: neglecting DNA repair may doom neurons in Alzheimer's disease. Bioessays. 2003;25:168–173. doi: 10.1002/bies.10227. [DOI] [PubMed] [Google Scholar]
- 6.Gan L, Mucke L. Paths of convergence: sirtuins in aging and neurodegeneration. Neuron. 2008;58:10–14. doi: 10.1016/j.neuron.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim D, et al. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer's disease and amyotrophic lateral sclerosis. Embo J. 2007;26:3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cruz JC, Tseng HC, Goldman JA, Shih H, Tsai LH. Aberrant Cdk5 activation by p25 triggers pathological events leading to neurodegeneration and neurofibrillary tangles. Neuron. 2003;40:471–483. doi: 10.1016/s0896-6273(03)00627-5. [DOI] [PubMed] [Google Scholar]
- 9.Cruz JC, et al. p25/cyclin-dependent kinase 5 induces production and intraneuronal accumulation of amyloid beta in vivo. J Neurosci. 2006;26:10536–10541. doi: 10.1523/JNEUROSCI.3133-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim D, et al. Deregulation of HDAC1 by p25/Cdk5 in neurotoxicity. Neuron. 2008;60:803–817. doi: 10.1016/j.neuron.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 12.Seluanov A, Mittelman D, Pereira-Smith OM, Wilson JH, Gorbunova V. DNA end joining becomes less efficient and more error-prone during cellular senescence. Proc Natl Acad Sci U S A. 2004;101:7624–7629. doi: 10.1073/pnas.0400726101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 14.Berkovich E, Monnat RJ, Jr, Kastan MB. Assessment of protein dynamics and DNA repair following generation of DNA double-strand breaks at defined genomic sites. Nat Protoc. 2008;3:915–922. doi: 10.1038/nprot.2008.54. [DOI] [PubMed] [Google Scholar]
- 15.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 16.Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. Embo J. 2010;29:3130–3139. doi: 10.1038/emboj.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu Y, et al. HDAC1 acetylation is linked to progressive modulation of steroid receptor-induced gene transcription. Mol Cell. 2006;22:669–679. doi: 10.1016/j.molcel.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 18.Yang T, et al. Acetylation of histone deacetylase 1 regulates NuRD corepressor complex activity. J Biol Chem. 2012;287:40279–40291. doi: 10.1074/jbc.M112.349704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai H, et al. SIRT1 activation by small molecules: kinetic and biophysical evidence for direct interaction of enzyme and activator. J Biol Chem. 2010;285:32695–32703. doi: 10.1074/jbc.M110.133892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo Y, et al. Trans-regulation of histone deacetylase activities through acetylation. J Biol Chem. 2009;284:34901–34910. doi: 10.1074/jbc.M109.038356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller KM, et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17:1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshiyama Y, et al. Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron. 2007;53:337–351. doi: 10.1016/j.neuron.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 23.Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–7748. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- 24.Yuan Z, Zhang X, Sengupta N, Lane WS, Seto E. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol Cell. 2007;27:149–162. doi: 10.1016/j.molcel.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ball AR, Jr, Yokomori K. Damage site chromatin: open or closed? Curr Opin Cell Biol. 2011;23:277–283. doi: 10.1016/j.ceb.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan ZM, et al. Role for p300 in stabilization of p53 in the response to DNA damage. J Biol Chem. 1999;274:1883–1886. doi: 10.1074/jbc.274.4.1883. [DOI] [PubMed] [Google Scholar]
- 28.Ikura T, et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 29.Sykes SM, et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bouras T, et al. SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. J Biol Chem. 2005;280:10264–10276. doi: 10.1074/jbc.M408748200. [DOI] [PubMed] [Google Scholar]
- 31.Peng L, et al. SIRT1 negatively regulates the activities, functions, and protein levels of hMOF and TIP60. Mol Cell Biol. 2012;32:2823–2836. doi: 10.1128/MCB.00496-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yankner BA, Lu T, Loerch P. The aging brain. Annu Rev Pathol. 2008;3:41–66. doi: 10.1146/annurev.pathmechdis.2.010506.092044. [DOI] [PubMed] [Google Scholar]
- 33.Gao J, et al. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oberdoerffer P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montgomery RL, et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev. 2007;21:1790–1802. doi: 10.1101/gad.1563807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng HL, et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takamiya K, Mao L, Huganir RL, Linden DJ. The glutamate receptor-interacting protein family of GluR2-binding proteins is required for long-term synaptic depression expression in cerebellar Purkinje cells. J Neurosci. 2008;28:5752–5755. doi: 10.1523/JNEUROSCI.0654-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis JB, Maher P. Protein kinase C activation inhibits glutamate-induced cytotoxicity in a neuronal cell line. Brain Res. 1994;652:169–173. doi: 10.1016/0006-8993(94)90334-4. [DOI] [PubMed] [Google Scholar]
- 40.Vonesch C, Unser M. A fast thresholded landweber algorithm for wavelet-regularized multidimensional deconvolution. IEEE Trans Image Process. 2008;17:539–549. doi: 10.1109/TIP.2008.917103. [DOI] [PubMed] [Google Scholar]
- 41.Carpenter AE, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dundr M, Misteli T. Measuring dynamics of nuclear proteins by photobleaching. Curr Protoc Cell Biol. 2003;Chapter 13(Unit 13):15. doi: 10.1002/0471143030.cb1305s18. [DOI] [PubMed] [Google Scholar]
- 43.Snapp EL, Altan N, Lippincott-Schwartz J. Measuring protein mobility by photobleaching GFP chimeras in living cells. Curr Protoc Cell Biol. 2003;Chapter 21(Unit 21):21. doi: 10.1002/0471143030.cb2101s19. [DOI] [PubMed] [Google Scholar]
- 44.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Finnin MS, et al. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 1999;401:188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- 47.Kruhlak MJ, Celeste A, Nussenzweig A. Monitoring DNA breaks in optically highlighted chromatin in living cells by laser scanning confocal microscopy. Methods Mol Biol. 2009;523:125–140. doi: 10.1007/978-1-59745-190-1_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nowak DE, Tian B, Brasier AR. Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. Biotechniques. 2005;39:715–725. doi: 10.2144/000112014. [DOI] [PubMed] [Google Scholar]
- 49.Zeng PY, Vakoc CR, Chen ZC, Blobel GA, Berger SL. In vivo dual cross-linking for identification of indirect DNA-associated proteins by chromatin immunoprecipitation. Biotechniques. 2006;41:694, 696–698. doi: 10.2144/000112297. [DOI] [PubMed] [Google Scholar]
- 50.Zhao Y, Zhang W, White MA. Capillary high-performance liquid chromatography/mass spectrometric analysis of proteins from affinity-purified plasma membrane. Anal Chem. 2003;75:3751–3757. doi: 10.1021/ac034184m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.