Abstract
Background
The beak deformity (crossed beaks) was found in some indigenous chickens of China, such as Beijing-You (BJY), Qingyuan Partridge, and Huxu Chickens. Birds with deformed beaks have reduced feed intake and drinking, impeded growth rate, and poor production performance. Beak deformity reduces the economy of poultry industry and affects animal welfare as well. The genetic basis of this malformation remains incompletely understood. LOC426217, also named claw keratin-like, was the most up-regulated gene in the deformed beaks from a previous digital gene expression (DGE) analysis and was selected as an important candidate gene for further analysis.
Results
In the present study, quantitative real-time PCR (qRT-PCR) was firstly performed to determine the expression pattern of LOC426217 gene in deformed and normal beaks to verify the DGE results. Tissue-specific expression profile of this gene in 14 tissues was also determined using qRT-PCR. The LOC426217 was amplified from the genomic DNA of 171 deformed and 164 normal beaks, and sequenced to detect the single nucleotide polymorphisms (SNPs). The results showed that LOC426217 was significantly high-expressed in the deformed beaks, which was in good agreement with the DGE results. This gene was specifically high-expressed in beaks than other tissues. Eight SNPs were detected in LOC426217: -62G > T, 24 T > C, 36G > C, 192A > T, 204C > T, 222 T > C, 285G > T, and 363 T > C. Genotype frequency of G-62 T, T24C, G36C, T222C, and T363C loci was significant different between deformed and normal beaks. Haplotype analysis revealed one block with SNPs T24C and G36C, and one block with SNPs A192T, C204T, T222C, and G285T in normal birds, while the block with SNPs G36C and A192T in deformed ones.
Conclusions
It was concluded from these results that the over-expression of LOC426217 in the beak maybe related to the malformation. The polymorphisms of LOC426217 gene were associated with the beak deformity trait where the SNPs of G-62 T, T24C, G36C, T222C, and T363C loci maybe used as markers. The specific haplotype block in deformed birds may be a potential linkage marker for this trait.
Electronic supplementary material
The online version of this article (doi:10.1186/s12863-016-0353-x) contains supplementary material, which is available to authorized users.
Keywords: Beijing-you chickens, Beak deformity, LOC426217, SNPs, Haplotype
Background
The beak is an external structure of birds, consisting of the upper and lower mandibles covered with a thin keratinized layer of epidermis [1]. It is used for many important activities such as feeding, drinking, fighting, and preening. In addition to striking morphological differences between species, beak deformities of different forms (noticeably elongated, crossed, bent at right angles) have been documented in many wild birds [2–7]. Frequencies of 1 % to 3 % of beak deformity (normally a crossed beak) were found in various indigenous chickens of China, such as Beijing-You (BJY) (studied here), Silkies, Qingyuan Partridge, and Huxu Chickens. Chickens with deformed beaks have reduced feed intake and growth rate. Therefore, beak deformity represents an economic as well as an animal welfare problem in poultry industry. According to our observations in a BJY population, in the absence of known environmental factors contributing to the malformation, birds with deformed beaks present consistently in each generation and cannot be eliminated from a population simply on the basis of the phenotype. This indicated the genetic effects underlying this trait. Studies have been performed to identify the teratogenic genes or molecular genetic background of beak deformity. Previously recognized genetic factors associated with beak deformity include some knwon genes such as fibroblast growth factor 8 (FGF8) [8], bone morphogenetic protein 4 (BMP4) [9–11], calmodulin (CaM) [12], and ALX homeobox 1 (ALX1) [13]. The over-expression of homeobox A1 (HOXA1) and homeobox D3 (HOXD3) may result in beak deformity in chicks [14].
Sets of differently expressed genes in the deformed and normal beaks have been detected using digital gene expression (DGE) profile analysis based on high-sequencing technology. Of these genes, LOC426217, also known as claw keratin-like gene, was the most up-regulated in the deformed beak (log2-Ratio (deformed/normal) = 10.91) [15]. Located on GGA 25, LOC426217 is a member of the keratin family, containing 417 base pairs with only one exon (Fig. 1). Keratin is crucial for maintaining normal cell morphology involved in the cytoskeleton remodeling keratin filaments and cytoskeletal signaling pathways. Change of its structure results in dysmorphic cells [16]. The cytoskeleton is a complex of intracellular proteins that contribute to shape, support, and movement of cells [17]. Up to now, less study was reported about this gene in chickens. Although highlighted in the DGE analysis, further study of this gene is still needed to verify its roles in beak malformation.
Fig. 1.

The molecular structure map of LOC426217 gene. Note: White box: promoter region; black box: exon encoding amino acids; Number: number of base pairs
In the present study, qRT-PCR was used to detect the relative expression of LOC426217 gene in the deformed and normal beaks, to verify the results of DGE profiling. Tissue expression profile of this gene was also determined in 14 tissues of the birds. Eventually, LOC426217 was amplified and sequenced to seeking the SNPs and haplotypes related with the beak malformation.
Methods
Animals and samples collection
The Institutional Animal Care and Use Committee at Institute of Animal Science, Chinese Academy of Agricultural Sciences (IAS, CAAS) approved all procedures involving the use of animals. All efforts were made to minimize the suffering of animals following the animal care guidelines [18]. The animals used in this study came from a pure-line stock of a local breed (Beijing-You) kept by IAS, CAAS (Beijing, China). They were incubated contemporaneously and housed under the same conditions.
The lower mandibles of the beaks were collected from 18 BJY chickens of 56 days of age: 9 with crossed beaks and 9 with normal beaks. Total RNA of the lower mandibles of the crossed and normal beaks above was collected for the verification of DGE profiling results using quantitative real-time PCR (qRT-PCR).
Three normal birds of 56 days of age were killed by stunning and exsanguination. Tissues samples including bursa of fabricius, beak, brain, breast, feather, heart, kidney, thigh, liver, lung, skin, small intestine, stomach, and testicle (50–100 mg) were rapidly collected and snap-froze in liquid nitrogen and storage at -80 °C. The RNA of these samples was used to determine the tissue expression profile of LOC426217.
Blood samples were collected from the brachial vein by venipuncture from 171 beak- deformed birds (deformed) and 164 normal ones (control). Based on the case-control study design, we selected these birds according to the phenotype of the birds without family structure. The beak-deformity birds were collected from two generations. The normal birds were selected randomly from the same generation. DNA was isolated from the blood samples and stored at -20 °C for the detection of SNPs located in LOC426217 gene.
DNA and RNA extraction and reverse transcription (RT)
Genomic DNA (gDNA) was extracted from blood samples using phenol-chloroform. Total RNA was isolated at 4 °C using the Trizol reagent (Invitrogen, USA). Any residual gDNA and protein were removed with Dnase I (TaKaRa, Japan) and RNA clean kit (TIANGEN, China). The purified RNA was dissolved (200-400 ng/mL, OD260/OD280 = 1.8–2.0), and stored at -80 °C. Total RNA was used for RT (in 20 μL final volume) following the manufacturer's instruction (Promega, USA). The cDNA was stored at -80 °C for subsequent qRT-PCR.
PCR amplification and qRT-PCR
PCR amplification was performed using PCR Gene Amplifier (Bio-Rad, USA) in a total volume of 25 μL which contains 12 μL of 2 × Taq PCR StarMix (GenStar, China), 1 μL (10 pmol) of each primer (Table 1), 1 μL of gDNA (50 ng) and 10 μL of ddH2O. After an initial denaturing for 2 min at 95 °C, there were 35 cycles of amplification (94 °C for 30 s, 60 °C for 30 s, and 72 °C for 90 s) and extension for 5 min at last. PCR products were detected by 1 % agarose gel electrophoresis for 15 min 120 V, stained with ethidium bromide, examined under UV light, and photographed. The PCR products were then sequenced by BGI (Beijing, China).
Table 1.
Gene-specific primers used in PCR
| Gene | Primer sequence | Product length (bp) | Tm (°C) | GenBank No. |
|---|---|---|---|---|
| LOC426217 | F: AGTCCTCTATCCAGCTTCCT | 806 | 60 | NC_006112.2 |
| R: GAGTAGGCAGTCAGAGCTTG |
To validate the DGE results, qRT-PCR was performed to determine the expression of LOC426217 in the 9 deformed and 9 normal beaks using the ABI 7500 Real-time Detection System (Applied Biosystems, USA) and TaKaRa DRR018A reagents. Each 20 μL PCR mixture contained10 μL of SYBR Premix Ex Taq™ II, 0.8 μL (10 pM) of each primer (Table 2), 0.4 μL of ROX Reference Dye II (50×), 2 μL of cDNA (100 ng) and 6 μL of ddH2O. After an initial denaturing for 30 s at 95 °C, there were 40 cycles of amplification (95 °C for 5 s and 60 °C for 32 s), followed by thermal denaturing to generate melting curves to verify amplification specificity. β-actin was amplified in the same plates as endogenous control. Samples were assayed in triplicate for standard curves. PCR efficiency of the LOC426217 gene and β-actin was consistent. cDNA from normal beaks served as a standard control for tissue-specific expression profile study. The amplification efficiency of transcripts of interest and the internal standard (β-actin) were consistent. Dissociation curves verified that amplification was specific.
Table 2.
Gene-specific primers used in qRT-PCR
SNPs filter and genotyping
PCR amplification product of LOC426217 gene was then directly sequenced by BGI company (Beijing, China) using Sanger sequencing methods [19]. The SNPs and amino acids were determined and filtered by the software DNAStar (Version 5.01).
Statistical analysis
The relative abundance of transcripts was calculated from 2−ΔΔCT [20]. All data presented graphically are means ± SEM. The significance level was P < 0.05 or P < 0.01. Student’s t-tests were used to evaluate the relative expression differences of LOC426217 between the RNA samples of deformed and normal beaks. The ANOVA procedure of SAS 8.0 was used to assess the differences expression of LOC426217 in all the tissues. All SNPs were checked for Hardy-Weinberg Equilibrium (HWE) in both groups (P > 0.05 means equilibrium). Allele frequency, genotype frequency, and polymorphism information content (PIC, PIC < 0.25: low polymorphism; 0.25 < PIC < 0.5: moderate polymorphism; PIC > 0.5: high polymorphism) were calculated by PopGene (Version 1.31). Chi-square tests were used to evaluate the genotype frequency differences between deformed and normal beaks. Benjamini & Hochberg method was used for the Bonferroni correction [21]. Linkage Disequilibrium (LD) pattern for the SNPs genotyped was plotted using Haploview (Version 4.2). The sliding window method was used to generate different haplotypes between two groups [22, 23].
Results
Verification for LOC426217 gene of DGE results
To verify the previous DGE analysis, where LOC426217 was the most up-regulated in the deformed beaks, qRT-PCR was used to estimate the expression of this gene in 9 deformed and 9 normal beaks. As shown in Fig. 2, the relative expression of LOC426217 in deformed beaks was significantly higher than that in normal ones (P < 0.05). This was in good agreement with the DGE analysis.
Fig. 2.
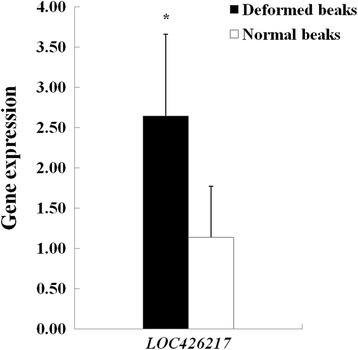
Relative expression of LOC426217 in 9 deformed and 9 normal beaks. Note: * means significant difference between two groups (P < 0.05)
Tissue expression profile of LOC426217
As shown in Fig. 3, LOC426217 gene was hardly expressed in brain, heart, bursa, or small intestine. The relative expression in beak was significantly higher than that in other tissues (P < 0.01). The results revealed that this gene maybe specifically expressed in beak tissue. In order to seek the structural changes of this gene, DNA sequencing was carried out subsequently.
Fig. 3.
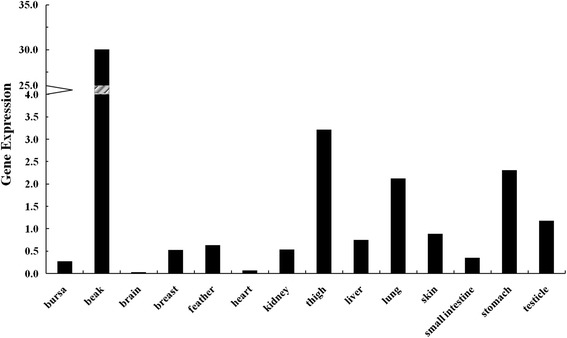
Relative expression of LOC426217 gene in 14 tissues of the chicken
DNA sequencing and SNPs detection
LOC426217 gene of 171 beak-deformed and 164 normal birds was amplified and sequenced. Eight SNPs were detected, including one locus in promoter region: -62G > T, and seven loci in coding region: 24 T > C, 36G > C, 192A > T, 204C > T, 222 T > C, 285G > T, and 363 T > C (Fig. 4). The loci in coding region were synonymous mutations resulting with no amino acid changing. These SNPs were presented in both beak-deformed and normal birds. The 18 birds used in the qRT-PCR analysis for DGE verification were also sequenced and their genotypes were shown in Table 3 and Additional file 1: Table S1.
Fig. 4.
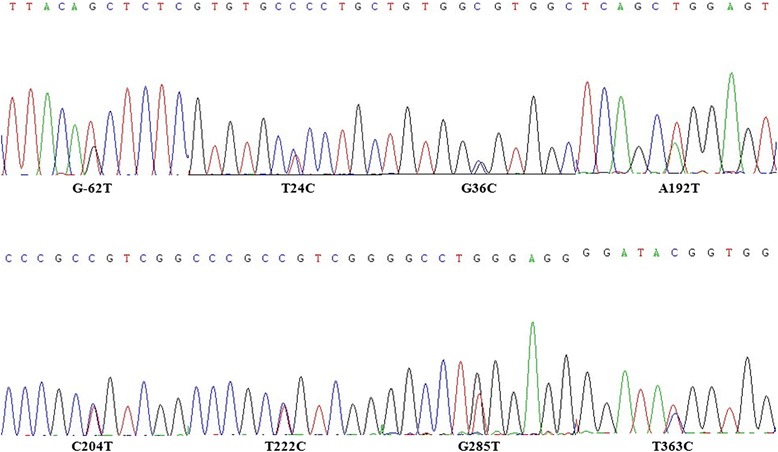
Sequencing map of the mutation loci in LOC426217 in chicken
Table 3.
Genotypes of the birds used for qRT-PCR analysis for validation of Digital Gene Expression results
| Trait | No. | SNP Loci | |||||||
|---|---|---|---|---|---|---|---|---|---|
| G-62 T | T24C | G36C | A192T | C204T | T222C | G285T | T363C | ||
| Deformed | 1 | TT | TC | GC | AT | CT | TT | GT | CC |
| 2 | TT | TC | GC | AT | CT | TT | GT | TT | |
| 3 | TT | CC | CC | AA | CT | TC | GG | TT | |
| 4 | GT | TC | GC | AA | CC | TC | GG | TT | |
| 5 | GT | TC | GC | AA | CC | TC | GG | TT | |
| 6 | TT | TT | GG | AA | TT | TT | GG | TT | |
| 7 | TT | TT | GG | AT | CC | TT | GT | CC | |
| 8 | TT | TT | GG | AT | CC | TT | GT | TT | |
| 9 | GT | TC | GC | AT | CC | TC | GT | TT | |
| Normal | 10 | TT | TC | GC | AT | CC | TC | GT | TT |
| 11 | TT | CC | CC | AA | TT | TT | GG | TT | |
| 12 | TT | TC | GC | AA | CT | TC | GG | TT | |
| 13 | TT | TC | GC | AA | TT | TT | GG | TT | |
| 14 | TT | TC | GC | AT | CT | TT | GT | CC | |
| 15 | TT | TC | GC | AT | CT | TT | GT | CC | |
| 16 | TT | TC | GC | AA | CT | TC | GG | TT | |
| 17 | TT | CC | CC | AA | CT | TC | GG | TT | |
| 18 | TT | TC | GC | AT | CC | TC | GT | TT | |
Genetic diversity analysis
Genetic diversity was analyzed by PopGene (Version 1.31). All SNPs were firstly checked for HWE. In the normal birds (n = 164), T24C, G36C, A192T, T222C, G285T, and T363C were not in agreement with the HWE (P < 0.05), while in the birds with a deformed beak (n = 171), T24C, G285T, and T363C were not in agreement with the HWE. Two loci (G-62 T and C204T) of normal beaks and two loci (G-62 T and T363C) of deformed ones were low polymorphism (PIC < 0.25) (Tables 4 and 5).
Table 4.
Genetic diversity analysis of LOC426217 in normal birds
| Loci | Genotype | Genotype frequency (n) | Allele | Allele frequency | χ 2 value (P value) | PIC |
|---|---|---|---|---|---|---|
| G-62 T | GG | 0.00(0) | G T |
0.13 0.87 |
3.837 (0.050) | 0.205 |
| GT | 0.27 (44) | |||||
| TT | 0.73 (120) | |||||
| T24C | TT | 0.36 (59) | T C |
0.65 0.35 |
10.32 (0.001)* | 0.353 |
| TC | 0.57 (94) | |||||
| CC | 0.07 (11) | |||||
| G36C | GG | 0.48 (78) | G C |
0.72 0.28 |
5.48(0.019)* | 0.324 |
| GC | 0.48 (79) | |||||
| CC | 0.04 (7) | |||||
| A192T | AA | 0.53 (86) | A T |
0.76 0.24 |
10.58 (0.001)* | 0.301 |
| AT | 0.46 (76) | |||||
| TT | 0.01 (2) | |||||
| C204T | CC | 0.70 (115) | C T |
0.83 0.17 |
0.13 (0.721) | 0.237 |
| CT | 0.27 (44) | |||||
| TT | 0.03 (5) | |||||
| T222C | TT | 0.55 (90) | T C |
0.76 0.24 |
6.71(0.0096)* | 0.295 |
| TC | 0.43(71) | |||||
| CC | 0.02 (3) | |||||
| G285T | GG | 0.54(89) | G T |
0.76 0.24 |
9.17 (0.0025)* | 0.295 |
| GT | 0.45 (73) | |||||
| TT | 0.01(2) | |||||
| T363C | TT | 0.70(115) | T C |
0.79 0.21 |
36.58 (<0.0001)* | 0.277 |
| TC | 0.18 (29) | |||||
| CC | 0.12(20) |
Note: χ 2 value means the test values of different genotypes to Hardy-Weinberg equilibrium. * (P < 0.05) means the loci were not in agreement with the Hardy-Weinberg equilibrium
Table 5.
Genetic diversity analysis of LOC426217 in beak-deformed birds
| Loci | Genotype | Genotype frequency (n) | Allele | Allele frequency | χ 2 value (P value) | PIC |
|---|---|---|---|---|---|---|
| G-62 T | GG | 0.02 (3) | G T |
0.10 0.90 |
1.37 (0.242) | 0.163 |
| GT | 0.16(28) | |||||
| TT | 0.82 (140) | |||||
| T24C | TT | 0.24(41) | T C |
0.535 0.465 |
5.80 (0.016)* | 0.374 |
| TC | 0.59(101) | |||||
| CC | 0.17 (29) | |||||
| G36C | GG | 0.32 (54) | G C |
0.594 0.406 |
3.77 (0.050) | 0.366 |
| GC | 0.55(95) | |||||
| CC | 0.13 (22) | |||||
| A192T | AA | 0.56(96) | A T |
0.760 0.240 |
1.33 (0.249) | 0.298 |
| AT | 0.40 (68) | |||||
| TT | 0.04(7) | |||||
| C204T | CC | 0.59(101) | C T |
0.781 0.219 |
1.98 (0.160) | 0.284 |
| CT | 0.38(65) | |||||
| TT | 0.03 (5) | |||||
| T222C | TT | 0.50 (85) | T C |
0.699 0.301 |
0.34 (1.562) | 0.332 |
| TC | 0.40(69) | |||||
| CC | 0.10 (17) | |||||
| G285T | GG | 0.57 (97) | G T |
0.772 0.228 |
4.38 (0.036)* | 0.290 |
| GT | 0.41(70) | |||||
| TT | 0.02(4) | |||||
| T363C | TT | 0.82 (140) | T C |
0.848 0.152 |
104.165 (<0.0001)* | 0.225 |
| TC | 0.06 (10) | |||||
| CC | 0.12(21) |
Note: χ 2 value means the test values of different genotypes to Hardy-Weinberg equilibrium.* (P < 0.05) means the loci were not in agreement with the Hardy-Weinberg equilibrium
Genotype frequency differences in the beak-deformed and normal birds
Chi-square tests were used to evaluate the genotype frequency differences of SNP loci in LOC426217 gene between two groups with a Bonferroni correction for the P-values. As shown in Table 6, the genotype frequency of G-62 T locus showed significant difference between two groups (P < 0.05), while the genotype frequencies of T24C, G36C, T222C and T363C loci showed highly significant differences (P < 0.01). There was no significant difference for the rest SNPs (P > 0.05).
Table 6.
Genotypes frequency comparison of all the loci between beak-deformed and normal birds
| Loci | Trait | Genotype | χ 2 value | P-value | Corrected P-value | ||
|---|---|---|---|---|---|---|---|
| AA | AB | BB | |||||
| G-62 T | normal deformity | 0 | 44 | 120 | 7.951 | 0.019 | 0.030* |
| 3 | 28 | 140 | |||||
| T24C | normal deformity | 59 | 94 | 11 | 11.450 | 0.003 | 0.008** |
| 41 | 101 | 29 | |||||
| G36C | normal deformity | 78 | 79 | 7 | 13.453 | 0.001 | 0.008** |
| 54 | 95 | 22 | |||||
| A192T | normal deformity | 86 | 76 | 2 | 3.627 | 0.163 | 0.186 |
| 96 | 68 | 7 | |||||
| C204T | normal deformity | 115 | 44 | 5 | 4.809 | 0.090 | 0.120 |
| 101 | 65 | 5 | |||||
| T222C | normal deformity | 90 | 71 | 3 | 9.829 | 0.007 | 0.014* |
| 85 | 69 | 17 | |||||
| G285T | normal deformity | 89 | 73 | 2 | 0.928 | 0.629 | 0.629 |
| 97 | 70 | 4 | |||||
| T363C | normal deformity | 115 | 29 | 20 | 11.591 | 0.003 | 0.008** |
| 140 | 10 | 21 | |||||
Note: χ 2 value means the tested values of different genotypes between two groups. * means significant difference between two groups (P < 0.05). ** means highly significant difference between two groups (P < 0.01). Corrected P-value using Benjamini & Hochberg method [21] was used for the Bonferroni correction
Haplotype analysis
In the normal birds, two haplotype blocks of LOC426217 were identified: one block with SNPs T24C and G36C, and one block with SNPs A192T, C204T, T222C, and G285T. In the birds with a deformed beak, one block with SNP G36C and A192T was identified (Table 7 and Fig. 5). The haplotype blocks of deformed birds were different from those of the normal ones.
Table 7.
Haplotype frequencies in the normal- and deformed-beaks birds
| Normal | Deformed | ||||
|---|---|---|---|---|---|
| Block1 | Block2 | Block | |||
| Haplotype | Frequency | Haplotype | Frequency | Haplotype | Frequency |
| TG | 0.646 | ACTG | 0.357 | CA | 0.406 |
| CC | 0.284 | ACCG | 0.235 | GA | 0.354 |
| CG | 0.070 | TCTT | 0.235 | GT | 0.240 |
| ATTG | 0.165 | ||||
Fig. 5.
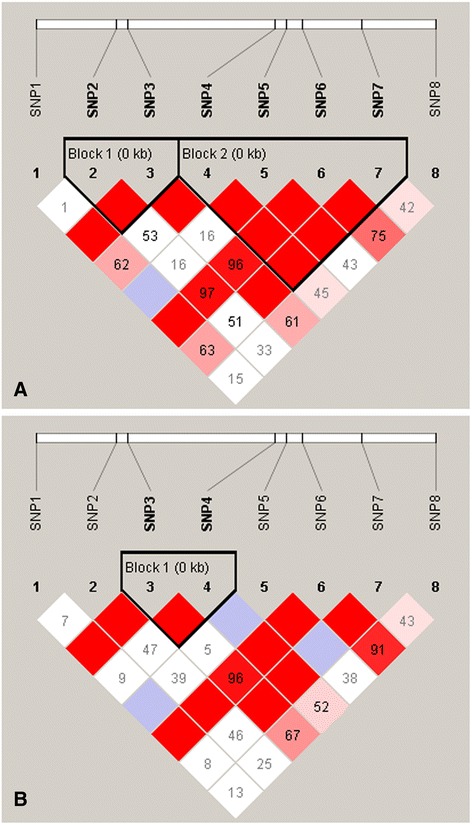
Haplotype blocks of LOC426217 in the normal (a) and deformed (b) beaks birds. SNP1: G-62 T; SNP2: T24C; SNP3: G36C; SNP4: A192T; SNP5: C204T; SNP6: T222C; SNP7: G285T; SNP8: T363C. In the normal birds, two haplotype blocks were identified: one block with SNPs T24C and G36C, and one block with SNPs A192T, C204T, T222C, and G285T. In the birds with a deformed beak, one block with SNP G36C and A192T was identified
Discussion
The molecular genetic mechanism underlying beak deformity is likely to be very complex. Beak deformities of different forms (noticeably elongated, crossed, bent at right angles) have been documented in many wild birds. The molecular mechanism of beak deformity trait is not clear yet. For the wild birds, it is difficult to obtain the individuals for genetic study. Beak deformity was also found in various indigenous chickens and it is easy to collect individuals. This made the chicken a perfect model for the genetic study of this defect. Based on the previous DGE profiling and bioinformatics analyses, we identified a cluster of differentially expressed genes (DEGs) in the deformed and normal beaks. Some of the DEGs were quite extreme, especially the LOC426217 gene in the present study.
Validation and tissue expression profile
In order to validate the reliability and accuracy of DGE results, qRT-PCR was performed. The results showed that the expression of LOC426217 in the deformed beaks was significantly higher than that in the normal beaks, which was in great agreement with DGE analysis. The result of tissue expression profile also revealed that this gene was specifically expressed in beak tissue. LOC426217 is a member of the keratin family [24]. Keratin is a key gene family for maintaining normal cell morphology [16]. Variation of keratin structure can lead to beak deformity [25]. It is also an intermediate filament protein that has essential functions in maintaining the structural integrity of epidermis and its appendages [26], presumably including the beak. In addition, keratin is the main composition of the chicken beak. This may be the main reason for its high expression in beak tissue. According to our early observation of beak anatomy, the lower mandibles of the beaks were abnormal/asymmetry. It was reported that avian keratin disorder could result in gross over growth of the rhamphotheca [7]. The beak deformity caused by the excessive growth of one side of the lower mandibles maybe a result of abnormal high expression of LOC426217.
SNPs and haplotypes associated with beak deformity
Based on the case-control study design, the investigated birds used for sequencing were selected from the pure line BJY population according to the beak phenotype of the birds with no family structures. Since the percentage of beak-deformity birds in the chicken population was 1 %-3 %, it was not easy to collect enough birds for gene sequencing. The beak-deformity birds were collected from two generations (about 5000 birds in each generation) and the normal birds were selected randomly from the same generation.
Using direct sequencing, eight SNPs located in the LOC426217 gene were detected, including one locus in promoter region and seven loci in coding region. These loci were found in both birds with deformed and normal beaks. Four loci (T24C, G36C, T222C, and T363C) were previously released on the database of NCBI (http://www.ncbi.nlm.nih.gov) while the rest four SNPs found in this study were new. Furthermore, five SNPs, individuals with different genotypes showed significant differences between beak-deformed and normal birds (P < 0.05). Previous research demonstrated that promoter mutation is a kind of the mutations which can enhance or reduce the expression level of a gene and lead to different phenotypes [27, 28]. In this current study, one SNP (-G62T) was detected in the promoter region and the genotype frequency was significant different between two groups. This mutation could possibly affect the start, expression time and level of this gene. As shown in Table 3, the genotype of 9 normal birds were all TT, while 3 out of the 9 beak-deformed were TG. Further validation study with larger sample size is needed to validate the mRNA expression profile of different genotypes. Similarly, the mutations in coding region are also very important. As we all know, exon is a nucleotide sequence in DNA that carries the code for the final mRNA molecule and thus defines the amino acid sequence during protein synthesis. If the base pairs changed in coding region, the structure and function of protein could possibly change, such as missense mutation or silent mutation [29–32]. In this study, all the loci in coding region were synonymous mutations with no amino acid and protein changing. Synonymous mutations, which do not alter the protein sequence, have also been shown to affect protein function [33, 34] and play key roles in human diseases [35]. Therefore, the mutations detected here may also affect this gene function. Sometimes, the phenotype altering or diseases occurring was not associated with one single SNP changing but related to several SNPs. Global patterns of DNA sequence variation (haplotypes) defined by common SNPs have important implications for identifying disease and traits [36, 37]. A previous study showed that the ALX1 haplotype has contributed to diversification of beak shapes among the Darwin’s finches and, thereby, to an expanded utilization of food resources [13]. In this present study, different haplotypes were analyzed between two groups. It indicated that the beak deformity trait might be related to the specific haplotype block which was only existed in beak-deformed chickens. To sum up, SNPs and haplotypes described here were interesting and worthy of further study.
Conclusions
To the best of our knowledge, this is the first time that LOC426217 was studied as an important candidate gene for beak deformity in birds. The over-expression of LOC426217 may be the cause of beaks malformation. The genotype frequency of SNPs at G-62 T, T24C, G36C, T222C, and T363C loci showed significant differences between deformed and normal birds, and might be used as candidate SNP markers for this trait. The specific haplotype block in the deformed group could be served as a potential linkage marker for this trait. Further functional verification studies like over-expression or RNA interfere of LOC426217 during the embryonic development are required to reveal its roles in the beak malformation.
Availability of supporting data
All the supporting data are included in the manuscript as well as additional files in the supplementary section.
Acknowledgements
This work was supported by 1. National Natural Science Foundation of China (31501949) (http://www.nsfc.gov.cn/); 2. Special fund for basic scientific research business of central public research institutes (2014ywf-yb-3). 3. National High Technology Research and Development Program (863 Program) (2011AA100305); and 4. Agricultural Science and Technology Innovation Program (ASTIP-IAS04), Chinese Academy of Agricultural Sciences, China (http://www.caas.cn/en). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- ALX1
ALX homeobox 1
- BJY
Beijing-You
- BMP4
Bone morphogenetic protein 4
- CaM
Calmodulin
- DEGs
Differentially expressed genes
- DGE
Digital gene expression
- FGF8
Fibroblast growth factor 8
- gDNA
Genomic DNA
- HOXA1
Homeobox A1
- HOXD3
Homeobox D3
- HWE
Hardy-Weinberg Equilibrium
- LD
Linkage Disequilibrium
- PIC
Polymorphism information content
- qRT-PCR
Quantitative real-time PCR
- RT
Reverse transcription
- SNPs
Single nucleotide polymorphisms
Additional files
Genotypes of all the 171 beak-deformed and 164 normal birds in this study (XLSX 26 kb)
The ARRIVE Guidelines Checklist Animal Research: Reporting In Vivo Experiments. (DOCX 661 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JLC, HB and JZ conceived and designed the experiments. HB, JZ and NL performed the experiments. DLL, FGX and YLL participated in data collection and sequence alignment. HB and YS analyzed the data. HB and YS drafted the manuscript. JLC, YS and HB revised the manuscript critically. All authors read and approved the final manuscript.
Contributor Information
Hao Bai, Email: bhowen1027@163.com.
Yanyan Sun, Email: yanyansun2014@163.com.
Jing Zhu, Email: jingjing19870220@163.com.
Nian Liu, Email: liunian.14@163.com.
Dongli Li, Email: lidongli20002004@163.com.
Fuguang Xue, Email: xueguang123@126.com.
Yunlei Li, Email: mailyunlei@163.com.
Jilan Chen, Email: chen.jilan@163.com.
References
- 1.Seki Y, Bodde SG, Meyers MA. Toucan and hornbill beaks: a comparative study. Acta Biomater. 2010;6(2):331–343. doi: 10.1016/j.actbio.2009.08.026. [DOI] [PubMed] [Google Scholar]
- 2.Rintoul DA. Beak deformity in a brown-headed cowbird, with notes on causes of beak deformities in birds. Kansas Ornithol Soc Bull. 2005;56:29–32. [Google Scholar]
- 3.Clabaut C, Herrel A, Sanger TJ, Smith TB, Abzhanov A. Development of beak polymorphism in the African seedcracker, Pyrenestes ostrinus. Evol Dev. 2009;11(6):636–646. doi: 10.1111/j.1525-142X.2009.00371.x. [DOI] [PubMed] [Google Scholar]
- 4.Handel CM, Pajot LM, Matsuoka SM, Hemert CV, Terenzi J, Talbot SL, Mulcahy DM, Meteyer CU, Trust KA. Epizootic of beak deformities among wild birds in Alaska: an emerging disease in North America? Auk. 2010;127(4):882–898. doi: 10.1525/auk.2010.10111. [DOI] [Google Scholar]
- 5.Hemert CV, Handel CM. Beak deformities in Northwestern Crows: evidence of a multispecies epizootic. Auk. 2010;127(4):746–751. doi: 10.1525/auk.2010.10132. [DOI] [Google Scholar]
- 6.Demery ZP, Chappell J, Martin GR. Vision, touch and object manipulation in Senegal parrots Poicephalus senegalus. Proc Biol Sci. 2011;278:3687–3693. doi: 10.1098/rspb.2011.0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Hemert C, Handel CM, O'Hara TM. Evidence of accelerated beak growth associated with avian keratin disorder in black-capped chickadees (Poecile atricapillus) J Wildlife Dis. 2012;48(3):686–694. doi: 10.7589/0090-3558-48.3.686. [DOI] [PubMed] [Google Scholar]
- 8.MacDonald ME, Abbott UK, Richman JM. Upper beak truncation in chicken embryos with the cleft primary palate mutation is due to an epithelial defect in the frontonasal mass. Dev Dynam. 2004;230(2):335–349. doi: 10.1002/dvdy.20041. [DOI] [PubMed] [Google Scholar]
- 9.Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Bmp4 and morphological variation of beaks in Darwin’s finches. Science. 2004;305(5689):1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- 10.Wu P, Jiang T, Suksaweang S, Widelitz RB, Chuong C. Molecular shaping of the beak. Science. 2004;305(5689):1465–1466. doi: 10.1126/science.1098109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu P, Jiang TX, Shen JY, Widelitz RB, Chuong CM. Morphoregulation of avian beaks: comparative mapping of growth zone activities and morphological evolution. Dev Dynam. 2006;235(5):1400–1412. doi: 10.1002/dvdy.20825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abzhanov A, Kuo WP, Hartmann C, Grant BR, Grant PR, Tabin CJ. The calmodulin pathway and evolution of elongated beak morphology in Darwin’s finches. Nature. 2006;442(7102):563–567. doi: 10.1038/nature04843. [DOI] [PubMed] [Google Scholar]
- 13.Lamichhaney S, Berglund J, Almén MS, Maqbool K, Grabherr M, Martinez-Barrio A, Promerová M, Rubin C, Wang C, Zamani N, Grant BR, Grant PR, Webster MT, Andersson L. Evolution of Darwin/’s finches and their beaks revealed by genome sequencing. Nature. 2015;518(7539):371–375. doi: 10.1038/nature14181. [DOI] [PubMed] [Google Scholar]
- 14.Jaszczak K, Malewski T, Parada R, Malec H. Expression of Hoxal and Hoxd3 genes in chicken embryos with exencephaly. J Anim Feed Sci. 2006;15(3):463. [Google Scholar]
- 15.Bai H, Zhu J, Sun Y, Liu RR, Liu N, Li DL, Wen J, Chen JL. Identification of genes related to beak deformity of chickens using digital gene expression profiling. PLoS One. 2014;9:e107050. doi: 10.1371/journal.pone.0107050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinert PM. Structure, function, and dynamics of keratin intermediate filaments. J Invest Dermatol. 1993;100(6):729–734. doi: 10.1111/1523-1747.ep12475665. [DOI] [PubMed] [Google Scholar]
- 17.Frixione E. Recurring views on the structure and function of the cytoskeleton: a 300-year epic. Cell Motil Cytoskel. 2000;46(2):73–94. doi: 10.1002/1097-0169(200006)46:2<73::AID-CM1>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 18.Institute for Laboratory Animal Research (US), Division on Earth and Life Studies (US), National Research Council (US). Wang JF, Zhou Y, Liu JH, Lu JQ, Ge LJ (Translation). Guide for the Care and Use of Laboratory Animals: Eighth Edition. Shanghai Scientific & Technical Publishers (SSTP), 2012.
- 19.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 − ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B. 1995;57:289–300. [Google Scholar]
- 22.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 23.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 24.Eckhart L, Dalla VL, Jaeger K, Ballaun C, Szabo S, Nardi A, Buchberger M, Hermann M, Alibardi L, Tschachler E. Identification of reptilian genes encoding hair keratin-like proteins suggests a new scenario for the evolutionary origin of hair. Proc Natl Acad Sci U S A. 2008;105(47):18419–18423. doi: 10.1073/pnas.0805154105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D'Alba L, Saranathan V, Clarke JA, Vinther JA, Prum RO, Shawkey MD. Colour-producing β-keratin nanofibres in blue penguin (Eudyptula minor) feathers. Biol Lett-UK. 2011;7(4):543–546. doi: 10.1098/rsbl.2010.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Törmä H. Regulation of keratin expression by retinoids. Dermato-endocrinol. 2011;3(3):136–140. doi: 10.4161/derm.15026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang GB, Liu XP, Ma T, Xu L, Wang HZ, Li ZT, Guo XM, Xu Q, Chen GH. A mutation in the NLRC5 promoter limits NF-κB signaling after Salmonella Enteritidis infection in the spleen of young chickens. Gene. 2015;568:117–123. doi: 10.1016/j.gene.2015.05.023. [DOI] [PubMed] [Google Scholar]
- 28.Mirhafez SR, Avan A, Pasdar A, Kazemi E, Ghasemi F, Tajbakhsh A, Tabaee S, Ferns GA, Ghayour-Mobarhan M. Association of tumor necrosis factor-α promoter G-308A gene polymorphism with increased triglyceride level of subjects with metabolic syndrome. Gene. 2015;568:81–84. doi: 10.1016/j.gene.2015.05.019. [DOI] [PubMed] [Google Scholar]
- 29.Stanford PM, Halliday GM, Brooks WS, Kwok JB, Storey CE, Creasey H, Morris JG, Fulham MJ, Schofield PR. Progressive supranuclear palsy pathology caused by a novel silent mutation in exon 10 of the tau gene. Brain. 2000;123(5):880–893. doi: 10.1093/brain/123.5.880. [DOI] [PubMed] [Google Scholar]
- 30.Houston RD, Cameron ND, Rance KA. A melanocortin-4 receptor (MC4R) polymorphism is associated with performance traits in divergently selected large white pig populations. Anim Genet. 2004;35(5):386–390. doi: 10.1111/j.1365-2052.2004.01182.x. [DOI] [PubMed] [Google Scholar]
- 31.Hou JX, An XP, Han P, Peng JY, Cao BY. Two missense mutations in exon 9 of caprine PRLR gene were associated with litter size. Anim Genet. 2015;46(1):87–90. doi: 10.1111/age.12223. [DOI] [PubMed] [Google Scholar]
- 32.Salomons GS, Bok LA, Struys EA, Pope LL, Darmin PS, Mills PB, Clayton PT, Willemsen MA, Jakobs C. An intriguing “silent” mutation and a founder effect in antiquitin (ALDH7A1) Ann Neurol. 2007;62(4):414–418. doi: 10.1002/ana.21206. [DOI] [PubMed] [Google Scholar]
- 33.Ramamurthi KS, Schneewind O. A synonymous mutation in Yersinia enterocolitica yopE affects the function of the YopE type III secretion signal. J Bacteriol. 2005;187(2):707–715. doi: 10.1128/JB.187.2.707-715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet. 2011;12(10):683–691. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- 35.Gartner JJ, Parker SC, Prickett TD, Dutton-Regester K, Stitzel ML, Lin JC, Davis S, Simhadri VL, Jha S, Katagiri N. Whole-genome sequencing identifies a recurrent functional synonymous mutation in melanoma. Proc Natl Acad Sci U S A. 2013;110(33):13481–13486. doi: 10.1073/pnas.1304227110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patil N, Berno AJ, Hinds DA, Barrett WA, Doshi JM, Hacker CR, Kautzer CR, Lee DH, Marjoribanks C, McDonough DP. Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science. 2001;294(5547):1719–1723. doi: 10.1126/science.1065573. [DOI] [PubMed] [Google Scholar]
- 37.Salman M, Dasgupta S, Cholendra A, Venugopal PN, Lakshmi GL, Xaviour D, Rao J, D'Souza CJ. MTNR1B gene polymorphisms and susceptibility to Type 2 Diabetes: a pilot study in South Indians. Gene. 2015;566(2):189–193. doi: 10.1016/j.gene.2015.04.064. [DOI] [PubMed] [Google Scholar]


