Purpose and Appropriate Sample Types
This panel was developed to quantify natural killer (NK) cell subsets in Rhesus macaques (Macaca mulatta) during SIVmac239 infection induced pathogenesis. It includes markers to monitor changes in the activation/proliferation phenotype of up to 12 NK cell populations. The performance of the staining was tested on cryopreserved lymph node samples, and on fresh and cryopreserved peripheral blood mononuclear cells (PBMC) isolated from EDTA anti-coagulated blood. The panel can be used to characterize NK cells in a range of normal and pathologic conditions of this species and can be easily adapted to stain samples from various tissues.
Background
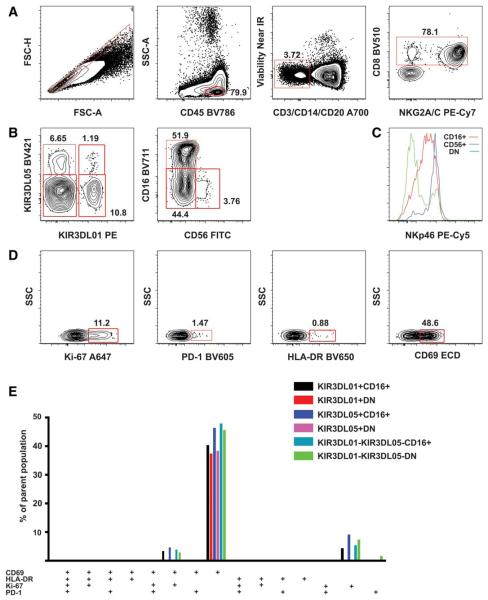
NK cells are cytotoxic effector cells of the innate immune system. They are triggered by combined signals of activating and inhibitory receptors. In humans, NK cells are typically defined by the expression of CD56 (neural cell adhesion molecule—NCAM) and CD16 (Fcγ receptor IIIa) antigens with cells positive for both markers representing the overwhelming majority of the population. A subset of human NK cells express CD8 antigen. In the blood of healthy Rhesus macaques, CD16 and CD56 are co-expressed primarily by monocytes. The majority (>85%) of circulating NK cells are CD16 single positive. CD56 single positive, CD16/CD56 double positive, or double negative cells only represent a small fraction of the cells. In macaques, the total population of NK cells is defined by the expression of CD8αα chain and NKG2A/C antigens on CD3 negative lymphocytes (Fig. 1a) (1,2). The population can be further divided according to the expression of killer cell Ig-like (KIR) receptors. Currently, up to four NK cell populations can be delineated by two well-characterized KIRs—KIR3DL01 and KIR3DL05—in an individual animal (Fig. 1b) (3,4). Each subpopulation can be further divided into CD16+, CD56+, and CD16-CD56− subsets, altogether up to 12 subsets. Chronic simian immunodeficiency virus (SIV) infection induces a significant increase of the absolute number of NK cells and a major shift in the NK cell subset distribution (5). The change in the subset composition can be characterized by a pronounced increase of the CD16/CD56 double negative population. The CD56 single positive population always remains below 5%. The three subpopulations possess different NKp46 (an NK cell stimulatory protein) expression profile (Fig. 1c), which shows animal-to-animal variation and may change depending on the stage of SIV-infection (5,6).
Figure 1.
Gating strategy for OMIP-028. Ficoll-purified PBMC of a healthy Rhesus macaque was stained with the NK cell proliferation/activation panel as detailed in the Supporting Information. A: With the initial sequential gating, we excluded doublets and larger cell aggregates (singlets defined by forward scatter area and height), dead cells (Amine reactive dye−), T cells (CD3−), B cells (CD20−), and monocytes (CD14−) within the lymphocyte (CD45+) gate. CD8+ and NKG2A/C+ were used to delineate the total NK cell population. B: Four subsets were identified according to the expression of KIR3DL01 and KIR3DL05 receptors within the NK cell population. All four subsets were further divided into CD16+, CD56+, and CD16−CD56− (DN) populations. C: The expression pattern of stimulatory receptor/differentiation marker NKp46 was compared between the three NK cell populations using histograms. D: We defined the activation/proliferation marker positive cell subsets within all CD16+, CD56+, and CD16−CD56− NK cell populations. E: Further subsets as defined by the combined expression pattern of the four activation markers were quantified by Boolean gating and displayed in a bar graph. Only subsets above 0.6% of the parent population, containing at least 10 events are displayed. Subsets within the CD56+ populations did not reach this cutoff level. In healthy animals, the differentiation state of circulating NK cells was characterized by three phenotypes regardless of KIR3DL or CD16 expression. To assure that all four activation markers were informative we stained samples from a limited number of SIV-mac239-infected animals. We found a more diversified NK cell population in these samples; therefore, we decided to monitor the emergence of the novel populations using all four markers (Supporting Information Fig. 8).
Activated/proliferating NK cells can express CD69, human leukocyte antigen-DR (HLA-DR), PD-1, and Ki-67 antigens to various extents (Fig. 1d). The combination of these markers defines 16 phenotypically distinct subsets probably representing different stages of maturation (Fig. 1e). With the present staining panel, we wanted to provide a tool to follow SIV infection-induced changes of NK cell subpopulations at a more refined level than previously described.
We had to work around two major limitations: scarcity of cross-reactive antibodies recognizing certain Rhesus macaque antigens, and limited number of reagents binding macaque KIR receptors. Mamu-KIR3DL01 is recognized only by the PE and APC conjugated version of monoclonal antibody NKVFS1. KIR3DL05 binds to a select number of Mamu-A1*00201 MHC-I allele restricted SIVmac239 epitope tetramers. Amongst these reagents the gag GY9 tetramer provided the best separation of the positive subsets (4). The GY9 tetramer is produced in BV421, PE, or APC conjugated forms by the NIH Tetramer Core Facility, all of which yield excellent staining. In Rhesus samples, the single available cross-reacting antibody—clone Z199—binds to both the activating NKG2C and the inhibitory NKG2A receptor (7). It can only be obtained in PE, PE-Cy7, and APC conjugated forms. Both APC and PE-Cy7 labeled versions provided excellent results. Of the two monoclonal antibody clones against the proliferation marker Ki-67 (B56 and Ki-67), only the Alexa Fluor 647 form of clone B56 yielded interpretable data. Finally, our only option of a cross-specific antibody for NKp46—clone BAB281—is available in PE, PECy5, and PE-Cy7 conjugates. Our anchor reagents therefore became KIR3DL05 (GY9) BV421, KIR3DL01 PE, NKp46 PECy5, NKG2A/C PECy7, and Ki-67 Alexa Fluor 647.
There are multiple choices for cross-reacting antibody clones against the CD8α chain. Since this protein is expressed abundantly on the cell surface, we selected a conjugate—the BV510 labeled SK1 clone—with good separation and least concern for broadening into other channels. In Rhesus macaques, the most frequently used clone against CD16 is the 3G8. It is available in 20 different fluorochrome-labeled versions from which we tested the V500, Pacific Blue, PE, PerCP-Cy5.5, and the BV711 varieties. All of the tested antibodies presented a clear separation between positive and negative populations. Since the Pacific Blue channel was reserved for KIR3DL05 BV421, the PE for KIR3DL01 PE and the PerCPCY5.5 channel for NKp46 PECy5, we chose the BV711 conjugate. Tissue resident NK cell populations contain larger proportions of CD56 positive subsets than the circulating population (5). Whether this distribution shifts during acute or chronic infection remains to be determined. We tested three clones of CD56-specific antibodies conjugated either to FITC (clone B159), Vio-Bright FITC (clone AF12-7H3), PE-Cy7 (clone NCAM16.2), or Alexa Fluor 700 (clone B159). Although the PE-Cy7-labeled NCAM16.2 clone possessed the best stain index we chose the FITC labeled clone B159 for the final panel based on prioritization of detector blue A for NKG2A/C PE-Cy7 (Supporting Information Fig. 3).
Recently increased PD-1 expression on NK cells was implicated in better HIV-1 control (8). Therefore, we were interested to see whether similar correlation could be observed in SIV-infected Rhesus macaques. The sole cross-reactive PD-1-specific clone EH12.2H7 is available in more than ten fluorochromeconjugated versions including six Brilliant Violet dye-conjugated forms. Since this antigen is not abundant on the cell surface it was an ideal candidate for the BV605 conjugate. This fluorochrome has a second emission peak at 650 so there is a possibility of significant spillover into the Violet C channel. The low intensity staining of the PD-1 BV605 conjugate provided us with an acceptable staining pattern with no fluorescence spillover. There are four cross-reactive monoclonal antibodies against the very early activation antigen CD69. We tested the PE-CF594 labeled FN50 and the ECD labeled TP1.55.3 clones. Both of them yielded similar results regarding the stain index and frequency of positive events. Finally, HLA-DR was shown to be present on a subset of NK cells characterized by enhanced degranulation (9). This antigen is expressed approximately one log lower on NK cells compared to B cells or monocytes. There is an abundance of cross-reactive HLA-DR-specific monoclonal antibodies in more than 50 fluorochrome-conjugated forms. We evaluated clone L243 in APC-Cy7 and BV650 labeled form. Both versions showed clear separation of HLA-DR positive NK cell populations. Since we reserved detector Red A for the live/dead discrimination we opted for the BV650 conjugate.
Supplementary Material
Table 1.
Summary table for OMIP-028
| Purpose | Characterize the activation and proliferation phenotype of various NK cell subsets |
| Species | Macaca mulatta |
| Cell types | Fresh PBMC and lymph node samples |
| Cross references | OMIP-007 (10) |
Table 2.
Reagents used in OMIP-028
| specificity | clone | fluorochrome | purpose |
|---|---|---|---|
| Live/Dead | N/A | Near infrared | Viability |
| CD45 | D058-1283 | BV786a | hematopoietic cell lineage |
| CD3 | SP34-2 | Alexa Fluor 700 | Exclusion |
| CD20 | 2H7 | Alexa Fluor 700 | |
| CD14 | M5E2 | Alexa Fluor 700 | |
| CD8 | SK1 | BV510 | NK subsets |
| CD16 | 3G8 | BV711 | |
| CD56 | B159 | FITC | |
| NKG2A/C | Z199 | PE-Cy7 | |
| KIR3DL01 | NKVFS1 | PE | |
| KIR3DL05 | GY9 tetramer | BV421 | |
| NKp46 | BAB281 | PE-Cy5 | NK cell differentiation |
| CD279 (PD-1) | EH12.2H7 | BV605 | |
| CD69 | TP1.55.3 | ECDb (PE-Texas Red) | Activation |
| HLA-DR | L243 | BV650 | |
| Ki-67 | B56 | Alexa Fluor 647 | Proliferation |
BV=Brilliant Violet.
ECD=energy coupled dye (PE-Texas Red).
ACKNOWLEDGMENTS
Animals were handled in accordance with the standards of the American Association for the Accreditation of Laboratory Animal Care (AAALAC).
Grant sponsor: NIH, Grant numbers: 5P51OD011106-53; R01 AI095098
Footnotes
Additional Supporting Information may be found in the online version of this article.
Literature Cited
- 1.Hermes M, Albrecht C, Schrod A, Brameier M, Walter L. Expression patterns of killer cell immunoglobulin-like receptors (KIR) of NK-cell and T-cell subsets in Old World monkeys. PLoS One. 2013;8:e64936. doi: 10.1371/journal.pone.0064936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biassoni R, Fogli M, Cantoni C, Costa P, Conte R, Koopman G, Cafaro A, Ensoli B, Moretta A, Moretta L, et al. Molecular and functional characterization of NKG2D, NKp80, and NKG2C triggering NK cell receptors in rhesus and cynomolgus macaques: Monitoring of NK cell function during simian HIV infection. J Immunol. 2005;174:5695–5705. doi: 10.4049/jimmunol.174.9.5695. [DOI] [PubMed] [Google Scholar]
- 3.Colantonio AD, Bimber BN, Neidermyer WJ, Jr, Reeves RK, Alter G, Altfeld M, Johnson RP, Carrington M, O’Connor DH, Evans DT. KIR polymorphisms modulate peptide-dependent binding to an MHC class I ligand with a Bw6 motif. PLoS Pathog. 2011;7:e1001316. doi: 10.1371/journal.ppat.1001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schafer JL, Colantonio AD, Neidermyer WJ, Dudley DM, Connole M, O’Connor DH, Evans DT. KIR3DL01 recognition of Bw4 ligands in the rhesus macaque: Maintenance of Bw4 specificity since the divergence of apes and Old World monkeys. J Immunol. 2014;192:1907–1917. doi: 10.4049/jimmunol.1302883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reeves RK, Gillis J, Wong FE, Yu Y, Connole M, Johnson RP. CD16- natural killer cells: Enrichment in mucosal and secondary lymphoid tissues and altered function during chronic SIV infection. Blood. 2010;115:4439–4446. doi: 10.1182/blood-2010-01-265595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mandelboim O, Lieberman N, Lev M, Paul L, Arnon TI, Bushkin Y, Davis DM, Strominger JL, Yewdell JW, Porgador A. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature. 2001;409:1055–1060. doi: 10.1038/35059110. [DOI] [PubMed] [Google Scholar]
- 7.LaBonte ML, Choi EI, Letvin NL. Molecular determinants regulating the pairing of NKG2 molecules with CD94 for cell surface heterodimer expression. J Immunol. 2004;172:6902–6912. doi: 10.4049/jimmunol.172.11.6902. [DOI] [PubMed] [Google Scholar]
- 8.Taborda N, Hernandez JC, Laiole J, Juno JA, Kimani J, Rugeles MT, Fowke KR. Low expression of activation and inhibitory molecules on NK cells and CD4+ T cells is associated with viral control. AIDS Res Hum Retroviruses. 2015;31:636–640. doi: 10.1089/AID.2014.0325. [DOI] [PubMed] [Google Scholar]
- 9.Evans JH, Horowitz A, Mehrabi M, Wise EL, Pease JE, Riley EM, Davis DM. A distinct subset of human NK cells expressing HLA-DR expand in response to IL-2 and can aid immune responses to BCG. Eur J Immunol. 2011;41:1924–1933. doi: 10.1002/eji.201041180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Currier JR, Eller MA. OMIP-007: Phenotypic analysis of human natural killer cells. Cytometry A. 2012;81A:447–449. doi: 10.1002/cyto.a.22033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.