Abstract
We designed ROS-activated cytotoxic agents (RACs) that are active against AML cancer cells. In this study, the mechanism of action and synergistic effects against cells coexpressing the AML oncogenes MLL-AF9 fusion and FLT3-ITD were investigated. One RAC (RAC1) had an IC50 value of 1.8 ± 0.3 µm, with ninefold greater selectivity for transformed cells compared to untransformed cells. Treatment induced DNA strand breaks, apoptosis, and cell cycle arrest. Proteomics and transcriptomics revealed enhanced expression of the pentose phosphate pathway, DNA repair, and pathways common to cell stress. Western blotting confirmed repair by homologous recombination. Importantly, RAC1 treatment was synergistic in combination with multiple pathway-targeting therapies in AML cells but less so in untransformed cells. Together, these results demonstrate that RAC1 can selectively target poor prognosis AML and that it does so by creating DNA double-strand breaks that require homologous recombination.
Keywords: acute myeloid leukemia, cytotoxicity, homologous recombination, mechanism of action, ROS
Introduction
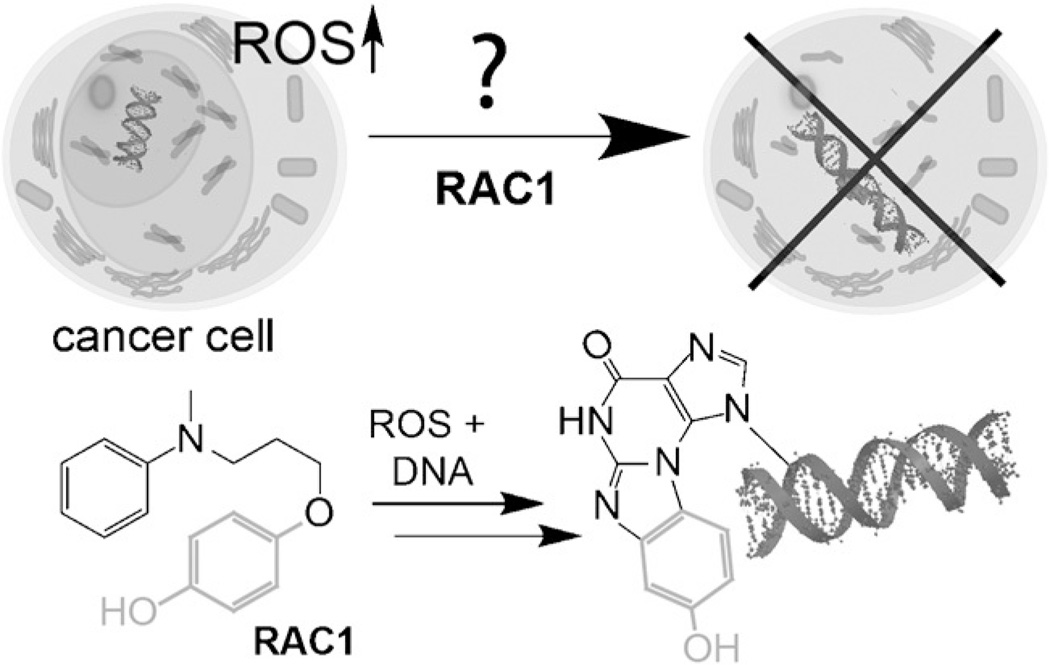
Agents that alter or modify DNA play a central role as anticancer agents.[1] The development of these agents has taken a back seat to the development of kinase inhibitors and antibody therapies due to their exquisite selectivity. For example, the success of targeting chronic myelogenous leukemia harboring the unique fusion protein made by a t(9;22)(q34;q11) translocation with imatinib (Gleevec) is well known.[2] Interestingly, despite decades of effort, agents that modify, bind, or limit DNA metabolism still hold a coveted position in the treatment of cancers. In acute myeloid leukemia (AML), the standard of care remains daunorubicin, a DNA intercalator, and cytosine arabinoside, a DNA metabolism inhibitor. The reason for the success of DNA alteration is simple. In principle, the formation of a single or few alterations or inhibitory events within the three billion base pair genome can be sufficient to cause cancer cell apoptosis due to lack of repair, which represents a reaction yield of 10−8.[3, 4] Recently, the development of DNA-altering agents has been extended to conjugation with cell-targeting antibodies, as in the case of the CD33-targeted antibody in gemtuzumab ozogamicin.[5] We used an alternative and complementary approach based on selective activation by reactive oxygen species (ROS) to design next-generation smart DNA modifiers, termed ROS-activated cytotoxic agents (RACs).[6, 7] In this manuscript, we investigate the role and mechanism of DNA repair in treated cells to better understand the DNA modifying action of these novel RAC agents (Figure 1).
Figure 1.
Mechanism of ROS-activated cytotoxic agents (RACs). AML is thought to be addicted to elevated levels of ROS; thus, we designed new agents that are activated by ROS and selective to AML. In this manuscript we examine the cellular mechanism of one such agent. Bottom shows known chemical reaction that RAC1 undergoes.
Selective activation of a DNA-modifying agent is a means to access the benefits of DNA damage while enhancing agent selectivity. This is a common, yet underappreciated, strategy in many natural products. Classically, the natural product mitomycin C is selectively activated by reduction into a potent DNA crosslinking agent within cancer cells.[8] More recently, the diazo-containing class of natural products, including lomaiviticins and kinamycins, were found to damage DNA in a free-radical- mediated activation process.[9] Our designs take advantage of elevated ROS in select cancers. Not every cancer has elevated ROS, but for those that do, ROS activation represents a powerful means to activate a DNA modifying agent. Current data shows that AML is a cancer with elevated ROS.[10, 11] For example, AML cells have high levels of NADPH oxidases that generate superoxide ROS to enhance growth.[12, 13] Excessive ROS production is associated with transformation by FLT3 upon internal tandem duplication of the receptor.[14, 15] Furthermore, the activity of several AML-associated tyrosine kinases and transcription factors, including Stat5, PI3-kinase, and Src, is altered by ROS.[16–18]
The compound under investigation, RAC1, has interesting chemical reactivity that led us to further investigate its mechanism of action (Figure 1, bottom). First, oxidation leads to a potent electrophile that a DNA arylamine (guanine, cytosine, adenine) can attack by 1,2-addition, followed by Michael addition and elimination to yield an unusual hydroxy-benzethenoguanine adduct. Based on this chemical mechanism, it is unlikely to be a DNA crosslinker, but nevertheless elicits a 50% loss in cell viability at 700 nm in AML cells. DNA repair is a complex process mediated by multiple different mechanisms. For DNA modifying agents, induction of double-strand breaks is a sought-after mechanism, as formation of such lesions are highly cytotoxic. In this manuscript, we examine the mechanism of cellular cytotoxicity of RACs against AML cells and explore the likely mechanism of DNA repair.
Results and Discussion
A RAC agent shows selective toxicity against a common type of AML
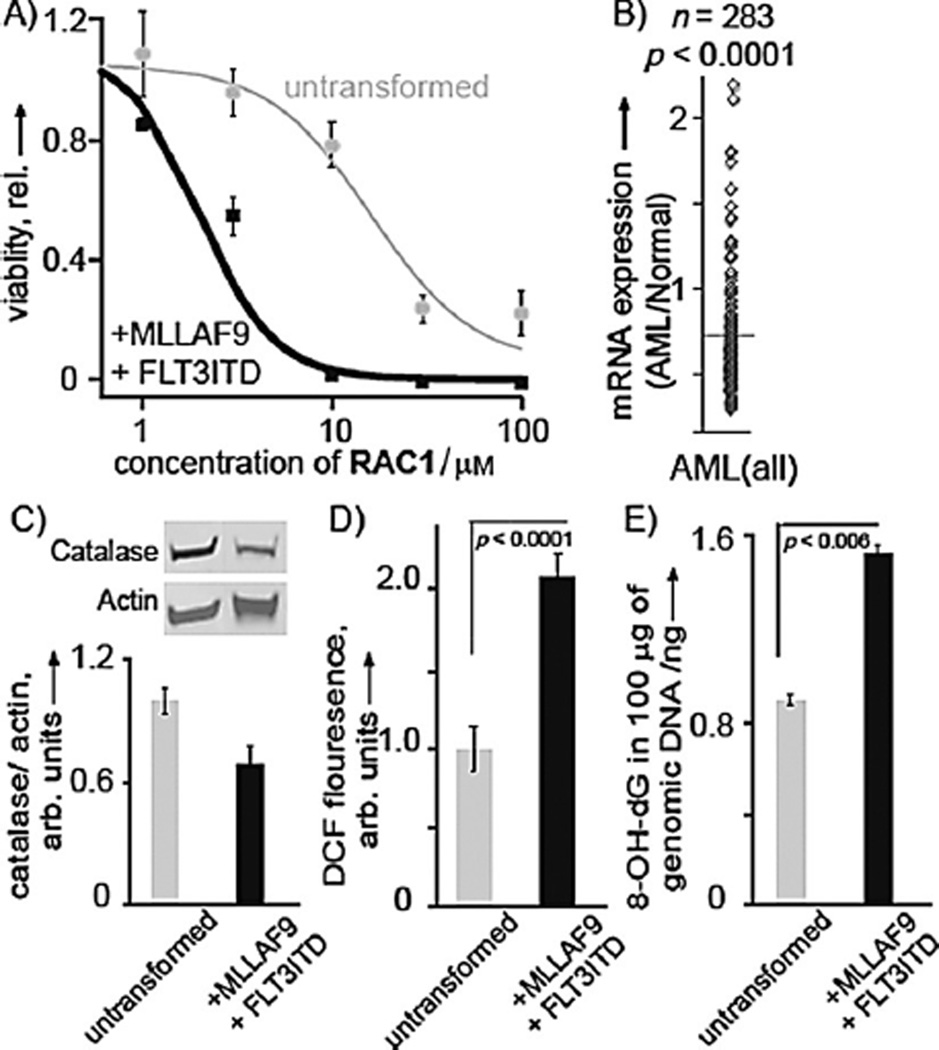
To test the efficacy of the RAC agent, we used primary human CD34+ blood stem cells that were transformed with the MLLAF9 fusion protein and internal tandem duplication of FLT3 (MLL-AF9 ITD). This transformation represents a good model for poor prognosis AML.[19] RAC1 showed highly selective cytotoxicity against this transformed cell line with an IC50 value of 1.8 ± 0.3 µm, whereas normal CD34+ blood cells showed an IC50 of 16.1 ± 0.5 µm (Figure 2A). We next investigated key markers of oxidative stress between the AML cells and untransformed cells. We first investigated the levels of the key antioxidant protein catalase in these two cell lines. The protein concentration of catalase was chosen for interrogation as data mining in HemaExplorer, a new curated database of mRNA expression in AML and normal cells,[20] showed a strong reduction in catalase mRNA expression in AML cells from patients (p < 0.0001; Figure 2B), with a median reduction of 29 %. Cells were grown to modest density, protein was extracted, and relative expression of catalase to β-actin was quantified (Figure 2C). It was found that untransformed cells had an expression of 1.0 ± 0.07, whereas the AML cells had a statistically significant decrease to 0.68 ± 0.09 (p < 0.04). A key question was to determine if poor prognosis AML displayed excessive ROS. Basal ROS level measurement by DCF assay revealed a 2.1-fold higher (p < 0.0001) levels of ROS in AML cells compared to untransformed cells (Figure 2D). Next, we investigated if ROS led to mutagenic DNA lesions by quantifying 8-oxo-7,8-dihydro-2’-deoxyguanosine by ELISA (Figure 2E). In agreement with our hypothesis, the data showed that untransformed cells had a relative 8-oxo-7,8-dihydro-2’-deoxyguanosine concentration of 1.0 ± 0.03, whereas the AML cells had a concentration of 1.71 ± 0.04 (p < 0.006). Not only did transformation lower the level of antioxidants, but it also enhanced DNA oxidation. Thus, poor prognosis AML is a viable target for RAC1 due to its elevated ROS status.
Figure 2.
Selective activity of RAC1. A) RAC1 displayed highly selective activity against an AML cell line expressing both MLL-AF9 and FLT3-ITD. Viability of AML cells (black) and untransformed blood stem cells (gray) is shown. B) Extracted data from HemeExplorer[20] indicating that patient AML samples have reduced catalase antioxidant mRNA. C) Transformation with MLL-AF9 and FLT3-ITD leads to more oxidative stress, as measured by a reduction in the antioxidant enzyme catalase by western blot analysis. D) Basal ROS levels in AML and untransformed cells as measured by DCF assay. E) An increase in 8-oxo-7,8-dihydro-2’-deoxyguanosine, as measured by ELISA. Results are presented as means ± SEM from biological triplicates.
Treatment causes DNA strand breaks and apoptosis
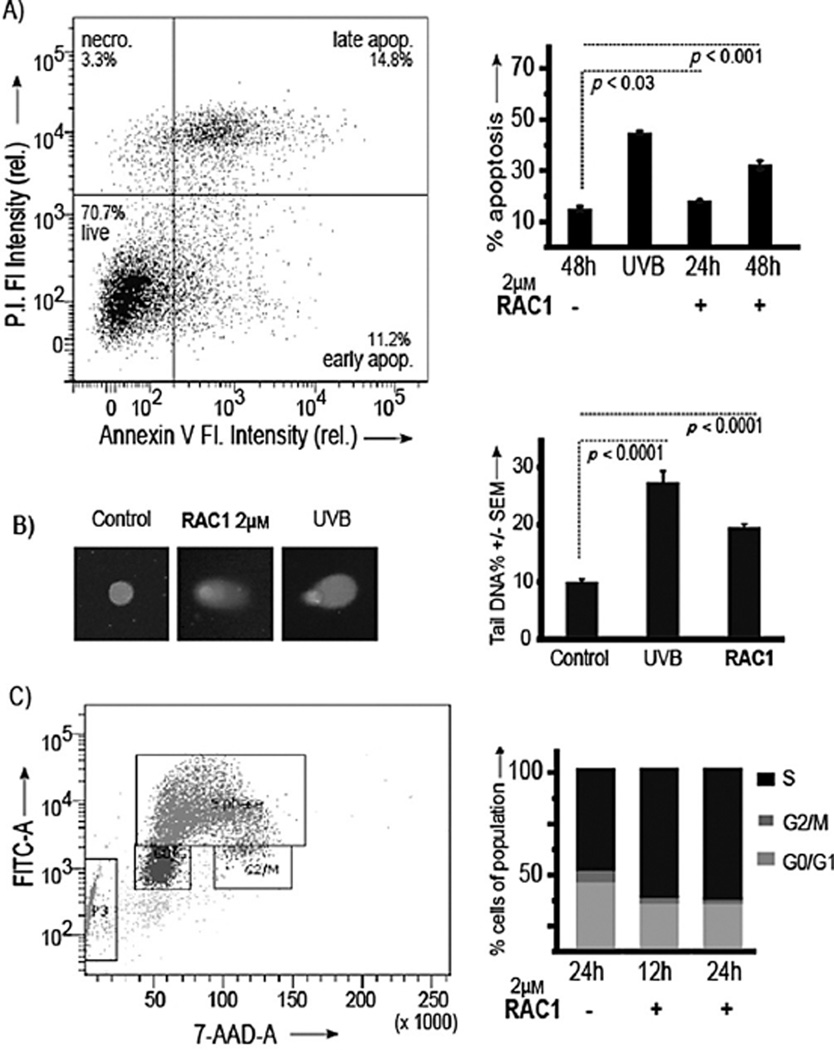
Apoptosis is the expected outcome for a reactive cytotoxic agent like RAC1. To determine whether cell death by RAC1 was due to the induction of apoptosis, annexin V/PI staining was assessed. Unstressed cells, treated with vehicle for 48 h, showed 13.9 ± 0.9 percent of the cells in apoptosis. As a positive control, AML cells were irradiated in the UVB region (280–315 nm). Irradiation increased the percent apoptosis to 43.2 ± 0.5 (p < 0.001). Treatment with RAC1 led to an 1.2-fold increase in the percent of apoptosis (17.0 ± 0.3 %; p < 0.03) at 24 h and 2.2-fold (30.8 ± 1.5%; p < 0.001) at 48 h (Figure 3A).
Figure 3.
Treatment leads to apoptosis and DNA damage. A) Left: Representative flow cytometer plot. Right: Assessment of percent apoptosis upon treatment with RAC1 in AML. B) Left: Representative images of AML cells after treatment with RAC1 and single cell electrophoresis. Right: Quantification of strand breaks upon treatment with RAC1. C) Left: Representative cell cycle analysis plot. Right: Assessment of percent of cells in each cell cycle phase upon treatment with RAC1 in AML. Results are presented as means ± SEM from biological triplicates.
Importantly, we showed direct evidence of cellular DNA damage. DNA damage and repair strand breaks induced by RAC1 were assessed by single cell gel electrophoresis (comet assay). AML cells were treated with RAC1 (2 µm) for 4 h and, as a positive control, cells were irradiated in the UVB region (280–315 nm; Figure 3B). Untreated cells had 10.0 ± 0.5% DNA in the tail. Irradiation resulted in an increase to 27.5 ± 1.9% DNA in the tail (p < 0.0001). Treatment with RAC1 led to an increase to 19.5 ± 0.6% DNA in the tail, indicating a twofold increase over untreated cells (p < 0.0001). Together, we conclude that upon failure to repair the DNA damage induced by RAC1, cells undergo death by apoptosis.
To investigate cell cycle perturbations by RAC1, flow cytometry analysis with a BrdU cell cycle assay was performed. The data are shown in Figure 3C. Unstressed cells, treated with vehicle for 24 h, showed 38.7 ± 0.3, 54.7 ± 1.6, and 6.5 ± 0.2% cells in the G0/G1, S, and G2M phases, respectively. Addition of RAC1, followed by 12h of incubation, led to 27.2 ± 3.1, 70.0 ± 4.6, and 2.8 ± 0.9% of cells in the G0/G1, S, and G2M phases, respectively, with the change in S phase increasing more than 15% (p < 0.001) and a large loss in the AML cells in the G2M phase. Similar analysis after 24 h showed 27.0 ± 2.2, 71.0 ± 3.3, and 1.9 ± 1.2% of cells in the G0/G1, S, and G2M phases, respectively, over 24 h. This further confirmed that cells were arrested in the S phase (p < 0.001) and could not reach the G2M phase. Thus, AML cells tried to synthesize damaged DNA (Figure 2C) but failed, leading to strand breaks and apoptosis (Figure 3A and B). These data left important questions regarding the DNA repair mechanism.
High-throughput methods to elucidate cellular responses
In order to assess the mechanism of action of RAC1 in an unbiased manner, a quantitative proteomics approach was used. As the data in Figure 3 showed elevated damage to genomic DNA as a consequence of RAC1, the proteomics analysis was targeted to isolated nuclei. This has the added advantage of enriching DNA damage response proteins that are generally low in concentration and/or are activated by post-translational modification, thereby providing the best opportunity to detect differences in these proteins between controls and treatments. The Supporting Information (Figure S1, Table S1) shows that nuclear isolation enriched the nuclear marker, lamin A, by ninefold. We chose the 24 h time point for analysis as expression changes, rather than post-translational modification, occurred on this time-scale. Interestingly, because of these criteria, we were able to elucidate key protein pathway changes.
Nuclear proteins extracted from AML cells after treatment with RAC1 or vehicle control were separated by 2D gel electrophoresis from pI 3–10 and in the 10–300 kD range. Image analysis revealed that 12 proteins (p < 0.05) changed in concentration in response to treatment, with ten proteins increasing and two decreasing. Figure S2 shows the example of the 2D gel image and an example of a protein spot that increased in concentration after RAC1 treatment (circled). Of the twelve differentially expressed proteins, eleven were successfully identified by MALDI-TOF/TOF and MS/MS analysis. The characteristics of all identified proteins, including protein name, NCBI accession number, fold change and ANOVA p value, are all summarized in Table 1.
Table 1.
Nuclear proteins with altered expression.
| Protein | Accession no | Fold change | ANOVA P | Role |
|---|---|---|---|---|
| VCP/p97 | gil6005942 | 3.4 | 0.002 | essential DSBR factor |
| LGALS3 | gil34234 | 3 | 0.007 | adhesion and growth |
| CPSF2 | gil34101288 | 2.8 | 0.003 | pre m-RNA processing |
| RPLP0 | gil4506667 | 2.4 | 0.001 | structural constituent of ribosome |
| VIM | gil62414289 | 2.2 | 0.013 | nucleus stability |
| HSP90B1 | gil15010550 | 2.1 | 0.006 | chaperone/protein folding |
| HSP90 | gil306891 | 1.9 | 0.001 | chaperone/stabilizes PolB |
| HSP90 α2 | gil61656603 | 1.9 | 0.001 | as above |
| TALDO1 | gil5803187 | 1.7 | 0.009 | PPP metabolism: ribose-phosphate |
| HMG-1 | gil478813 | −7.4 | 0.002 | unknown |
| IRF8 | gil4504567 | −1.7 | 0.0001 | apoptosis through Bax/Fas |
Significantly regulated proteins can be categorized into two groups: proteins involved in DNA damage repair, and proteins involved in cancer cell survival mechanisms/ROS homeostasis. Among the proteins that increased upon treatment, transitional endoplasmic reticulum ATPase (VCP/p97) showed the highest fold change of 3.4 (p < 0.003). The protein VCP/p97 activates the nuclear factor-kB signaling pathway and has recently been under intense study.[21–25] Functionally, VCP/p97 remodels multimeric protein complexes that have multiple ubiquitins, like nucleosome core particles, to initiate DNA repair. VCP/p97 is essential for double-strand break repair in an RNF8- and ubiquitin-dependent manner, where it facilitates the recruitment of BRCA1 and the tumor suppressor 53BP1 to DNA lesions. VCP/97 has also been associated with repair by both nonhomologous end joining and homologous recombination.[26, 27] This is interesting, given that RAC1 is not a crosslinking agent.
Next, expression of HSP90B1, HSP90, and HSP90 α2 increased upon treatment by 2.1-fold (p < 0.007), 1.9-fold (p < 0.002), and 1.9-fold (p < 0.002), respectively. A recent SILAC experiment revealed their essential role in DNA damage response.[28] Laminin-binding proteins appear to act as adaptors for the chromatin organization, gene expression, epigenetic regulation and modulation of signaling pathways; therefore, it was not surprising that LGALS3 expression increased threefold, due to the large amount of stress cells undergo upon treatment with RAC1.[29] Additionally, VIM increased by 2.2-fold (p < 0.02). This protein functions to fortify nuclei structure to maintain survival. Within this set of proteins, we noted the key connection between double-strand break repair, especially BRCA1-mediated repair, that we will further evaluate directly, as many DNA repair proteins are modulated by post-transcriptional modification.
The next two proteins described are involved in stress response, and potentially ROS reduction, as a means for survival. IRF8 is a key tumor suppressor protein and regulates the expression of BAX and FAS.[30] An increase of 1.7-fold (p > 0.009) in TALDO1 (transaldolase) expression could be beneficial for cells in two ways. Transaldolase has a well-known role in the pentose phosphate pathway.[31] Upregulation of this pathway seems to be a critical response, as the pentose phosphate pathway synthesizes ribose precursors for DNA synthesis, which would be of obvious benefit in combating the DNA damage function of RAC1. The pentose phosphate pathway is known to be activated upstream by phosphorylation of ATM, a critical DNA repair protein. It is possible that TALDO1 could also lead to increased glycolysis and NADPH production instead of ROS-generating oxidative phosphorylation.[32] NADPH is a coenzyme for glutathione reductase needed to convert oxidized glutathione (GSSG) to its reduced form (GSH). We have potentially identified an important cellular adaption mechanism that can act as a survival response for AML cancer cells by lowering the activation of RAC1 and synthesizing more precursors for DNA repair/synthesis.[31] From the proteomics data, we obtained two critical responses: increased double-strand break repair and movement towards pentose phosphate metabolism to make ribose.
Transcriptomics of differentially expressed mRNAs upon treatment
RNA-Seq was carried out to characterize the expression of mRNAs upon treatment with RAC1. Three treated and three control mRNAs were validated for integrity by using an Agilent Bioanalyzer 2100. Sequencing results showed that more than 1000 genes were differentially expressed with p < 0.01 (Figure S3). These genes were analyzed by using the ToppGene Suite[33] in order to identify the pathways implicated by the differentially expressed genes. The biological pathways associated with treatment-induced mRNA production changes are summarized in Table 2.
Table 2.
Biological pathways (fold change > 2; p < 0.01) from ToppGene analysis.
| ID | Name | p value | Genes from input/ annotation |
|---|---|---|---|
| Upregulated | |||
| GO:0006952 | response | 6.517 × 10−30 | 133/1515 |
| GO:0006955 | immune response | 8.569 × 10−29 | 126/1416 |
| GO:0009611 | response to wounding | 1.142 × 10−19 | 101/1255 |
| GO:0002682 | regulation of immune system process | 3.197 × 10−18 | 96/1212 |
| GO:0045087 | innate immune response | 4.817 × 10−18 | 79/883 |
| Downregulated | |||
| GO:0006334 | nucleosome assembly | 2.078 × 10−14 | 16/143 |
| GO:0031497 | chromatin assembly | 9.157 × 10−14 | 16/157 |
| GO:0034728 | nucleosome organization | 2.201 × 10−13 | 16/166 |
| GO:0065004 | protein–DNA complex assembly | 7.786 × 10−13 | 16/180 |
The pathways involved in upregulated mRNAs are described. Out of 1300 genes, 133 (p ≤ 10–29) were involved in the defense response pathway. Also, there were significantly increased transcript levels for genes participating in immune response (126/1416, p ≤ 10–28), wound response (101/1255, p ≤ 10–18), regulation of immune system processes (96/1212, p ≤ 10–17), and innate immune response (79/883, p ≤ 10–17). For example, CREB3L3, CYGB, and AQP1 were only expressed upon treatment. These genes are part of stress responses, and CYGB is well-known to protect cells exposed to oxidative damage.[34] This implies the involvement of classic ROS/cell damage stress responses upon treatment, as immune responses and wound healing involve ROS and cell growth, respectively. Interestingly, pathways involving nucleosome assembly (16/143, p ≤ 10–13), chromatin assembly or disassembly (16/182, p ≤ 10–12), nucleosome organization (16/166, p ≤ 10–12), and protein–DNA complex assembly (16/180, p ≤ 10–12) were enriched among downregulated genes, implicating cell cycle arrest as a result of agent treatment as well as the activation of cellular apoptosis as a result of failure of damage repair. Further investigation of expression changes of individual genes involved in the DNA damage repair pathway was carried out. Interestingly, several poly(ADP-ribose) polymerase (PARP) genes were upregulated. These enzymes are responsible PARP polymerization at DNA damage sites. The identified genes were PARP3, PARP9, PARP10, PARP12, and PARP14 and were upregulated by 2.1-, 2.4-, 2.0-, 3.7-, and 1.6-fold, respectively, following RAC1 treatment. Though the biological functions of most PARP enzymes are still unknown, a recent study showed that PARP3 has a functional role in DNA repair.[35] It was also reported that PARP12 has a functional role in stress response.[36] Additionally expression of damage-specific DNA binding protein 2 (DDB2) was upregulated twofold. mRNA levels of a major non-homologous end joining (NHEJ) pathway protein, XRCC4, were downregulated 0.5-fold, which aligned with our proteomics analysis that showed BRCA1-mediated responses, which are part of a homologous recombination pathway. CDKN1A (p21) expression was upregulated sevenfold, again implicating cell cycle arrest in response to RAC1. Further, our transcriptomics data showed the upregulation of LGALS3 and VIM and the downregulation of IFR-8, supporting our proteomics data and showing a robust connection between the two techniques. Transcriptomics also showed an increased expression of pentose phosphate pathway enzyme ATP:d-ribose 5-phosphotransferase. Together, this data correlates with the proteomics data and supports further investigation into DNA damage response.
Western blotting to identify double-strand break repair mechanism
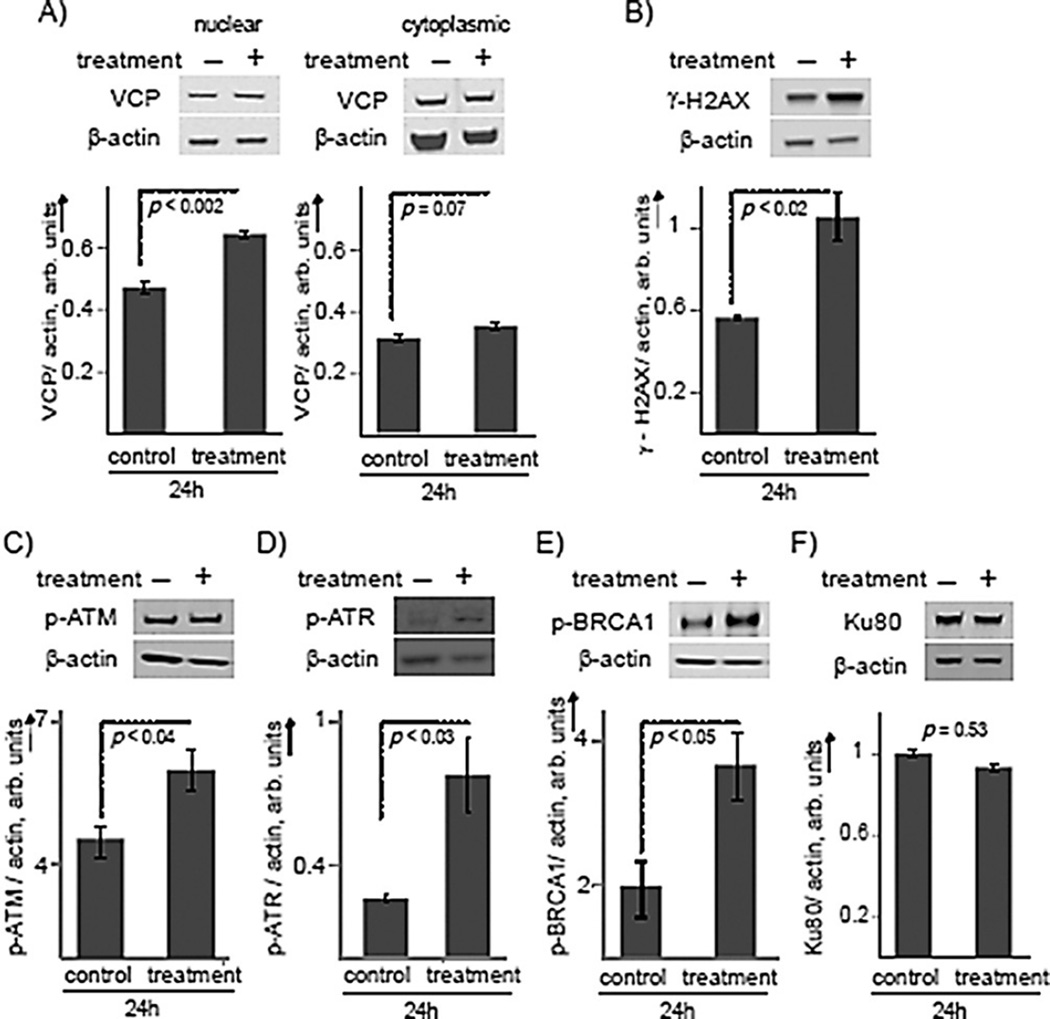
Proteomic analysis revealed that VCP increased upon treatment. However, this ATPase has many roles and can eject any polyubiquitylated protein substrate. Thus, we wanted to ensure its increase could potentially be involved in DNA repair and chromatin remodeling functions. Western blotting after treatment of AML cells showed an elevation in VCP concentration of 1.37 ± 0.03-fold (p < 0.002) within the nucleus, but increased expression was not observed in the cytoplasm (p < 0.07; Figure 4A). Thus, increased VCP activity might be localized to the nucleus in response to DNA damage, necessitating DNA repair and chromatin remodeling.
Figure 4.
RAC1 induced activation of DNA double-strand repair, likely through homologous recombination. A) Western blot of VCP in the nucleus (left) and in the cytoplasm (right) of AML cells treated with RAC1 (2 µm) for 24 h. B) Western blot analysis for γ-H2AX in AML cells. C) Phosphorylated ATM, D) phosphorylated ATR, E) phosphorylated BRCA1, and F) Ku80 in AML cells treated with RAC1 for 24 h. Results are presented as means ± SEM from biological triplicates.
Double-strand break repair is a complex pathway involving mainly homologous recombination (HR) and NHEJ.[37] Following the detection of a double-strand break, histone protein H2AX is phosphorylated to form γ-H2AX. This phosphorylation is one of the most common signals for double-strand break repair and signals to initiate the repair of double-strand breaks. The fate of the double-strand break repair pathway will be decided by the enrichment of key proteins, ku70/ku80 and phospho-BRCA1, at the damage sites. Ku80 repair is characteristic of NHEJ, whereas BRCA1 is characteristic of HR.[38]
In this study, western blot analysis was carried out to quantify the key proteins in the double-strand break repair pathway in response to treatment with RAC1. The nuclear proteins were extracted after 24 h of treatment with RAC1 in AML cells and from mock-treated cells. Figure 4B shows that the phosphorylated H2AX variant, γ-H2AX, increased 1.9 ± 0.2-fold (p < 0.02) upon RAC1 treatment compared to vehicle treatment (Figure 4B). We have also found that treatment of untransformed cells did not show a significant increase (Figure S4). Next, we evaluated the phosphorylation status of ATM. This protein not only regulates DNA repair through phosphorylation but is also an obvious link between stress and shunting glycolysis to produce ribose precursors. Western blotting showed an increase of 1.3 ± 0.1-fold (p < 0.04) in phosphorylation of ATM compared to mock-treated cells (Figure 4C). Upon treatment, phosphorylation of ATR significantly increased by 3.1 ± 1.1-fold (p < 0.03; Figure 4D) Thus, it is clear that DNA double-strand break repair was initiated.
To differentiate HR from NHEJ, we chose to investigate the key proteins BRCA1 and ku80. A clear differentiation can be observed by investigating phosphorylation of BRCA1 and the concentration of Ku80 for HR and NHEJ, respectively. BRCA1 is a promiscuous protein with well-known interactions with RAD51 and other proteins within HR. Furthermore, the activation/ phosphorylation of BRCA1 occurs through the auto-phosphorylated ATM after sensing the damaged DNA.[39] The role of Ku80 in NHEJ is well-characterized.[40] The Ku complex can bind to the broken end of DNA with high affinity in a sequence-independent manner, and at the break site, it can interact with several factors and enzymes to allow end joining.[41] BRCA1 phosphorylation was enhanced 1.9 ± 0.2-fold (p < 0.05) upon treatment with RAC1 (Figure 4E). In contrast, treatment with RAC1 did not change Ku80 levels within the nucleus, as we observed a concentration change of 0.93 ± 0.08 and a p value of 0.53 (Figure 4F). We conclude that HR, and not NHEJ, is the preferred DNA double-strand repair mechanism following treatment with RAC1.
Synergy with on-pathway compounds
Proteomics, transcriptomics, and western blotting indicated phospho-ATM-dependent HR DNA repair as a key response/ survival mechanism of AML cells upon treatment with RAC1. Importantly, phospho-ATM regulates the identified pentose phosphate pathway and DNA double-strand break repair.[42] The standard of care agents also target the DNA repair pathway, as daunorubicin will cause DNA double-strand breaks and should require phospho-ATM, whereas cytarabine (AraC) is a multi-modal agent but is thought to principally act by limiting the synthesis of nucleotides needed for DNA synthesis. We therefore expect to see synergy with RAC1 in combination with specific inhibitors affecting these pathways in AML cells.
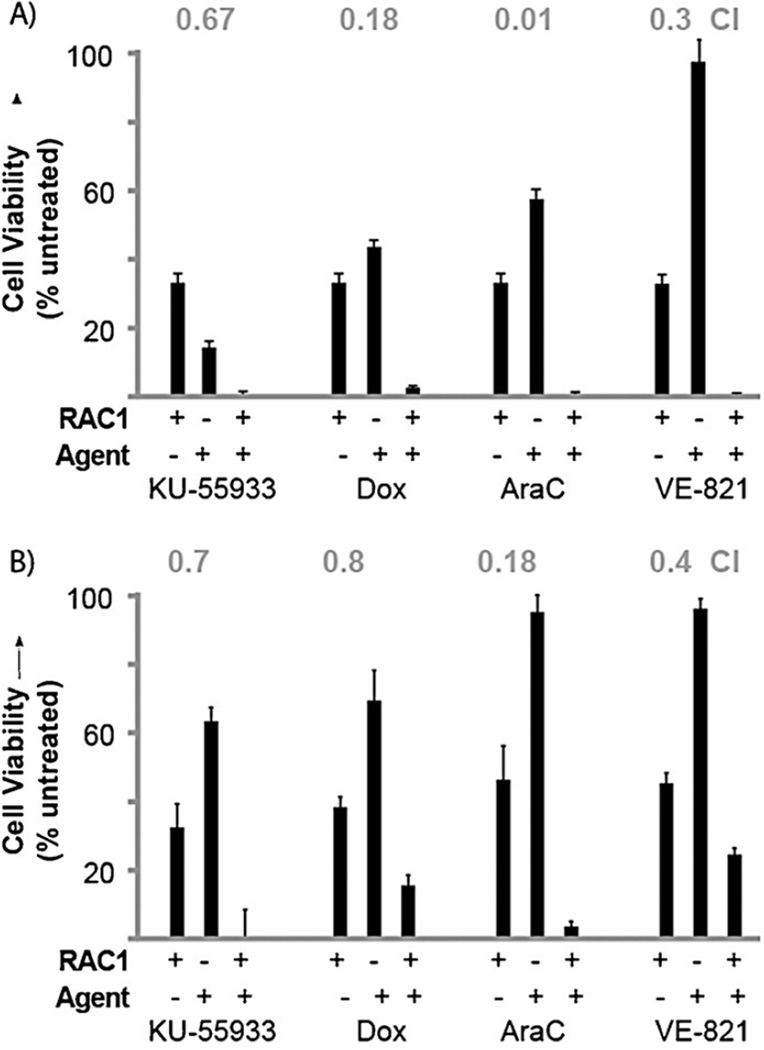
Several inhibitors were examined for synergy. KU-55933 was selected, as it is a known inhibitor for ATM that limits phosphorylation.[43] VE-821, a potent and selective ATP-competitive inhibitor of ATR, was also examined.[44] In addition, we analyzed the DNA-damaging agents doxorubicin and AraC. The first step in synergy measurements is to elucidate the IC50 values so that dose effects can be quantified. As such, the IC50 values for KU-55933, VE-821, doxorubicin, and AraC were 40 µm, 10 µm, 500 ± 5 nm, and 1.4 ± 0.2 µm, respectively, in AML cells. Cells were then treated at doses to elicit ~50% viability with each of these agents, including RAC1 (Figure 5A). The combinational indexes (CIs) were calculated by using CompuSyn software.[45] As shown in Figure 5A, the combination of RAC1 and KU-55933 resulted in significant growth inhibition and showed a synergistic effect (CI < 1) with a CI value of 0.67. Doxorubicin and AraC showed dramatic synergy, with CI values of 0.18 and 0.01, respectively. VE-821 showed a strong synergistic effect, with a CI value of 0.3. Synergistic effects were also investigated in normal CD34+ cells with the same inhibitors. In normal cells, KU-55933 and doxorubicin showed synergy, with CI values of 0.7 and 0.8, respectively. AraC showed a strong synergy, with a CI value of 0.18, and VE-821 showed synergy as well, with a CI value of 0.4. Please note that viability in the combination is almost completely lost, indicating very apparent synergy. These data strongly show that the synergistic effects of RAC1 with KU-55933, VE-821, and the standard of care compounds doxorubicin and AraC are specific to transformed AML cells and not their CD34+ counterparts. Thus, these experiments open possibilities for combinatorial therapy with RAC1.
Figure 5.
Synergy studies of on-pathway inhibitors with RAC1. A) AML cells B) Normal CD34+ cells. Cell viability was measured by MTT assay after 48 h of treatment and compared to IC50 values of single agent treatment for quantification. CI values (grey, above) were calculated by using CompuSyn software. Results are presented as means ± SEM from biological triplicates.
Conclusion
Targeting increased ROS in cancer by using ROS-activated agents is a promising new approach that is gaining attention.[46, 47] In this study, we report induction of HR as a predominate mechanism of DNA repair upon treatment with RAC1. Proteomics and transcriptomics were used to generate leads regarding the mechanism of action. Interestingly, proteomics of the nuclear fraction led to direct evidence not only of HR but also of activation of the pentose phosphate pathway, which synthesizes ribose for DNA synthesis after repair, consistent with the proposed mechanism. These pathways were further validated by synergy studies. Cells were almost completely eliminated when inhibitors, added at doses that individually had suboptimal effects alone, were combined with RAC1 (Figure 5). Importantly, these effects were specific to cells transformed with MLL-AF9 and FLT3-ITD, as untransformed counterparts showed reduced synergy in all cases.
In this manuscript, several important liabilities for poor prognosis AML were observed that we will highlight. Importantly, expression of the oncogenes MLL-AF9 and FLT3-ITD in these cells resulted in enhanced DNA oxidation and reduced antioxidant enzyme levels, which likely leads to a higher mutation rate and potentially more ROS signaling. We show that RACs exploit this key liability for selective anti-AML activity. Collectively, these data highlight that RAC1 is able to reach the nucleus, activate through ROS, and cause lethal DNA double-strand breaks. These cells entered arrest in the S phase of the replication cycle, indicating that DNA synthesis was attempted but could not be completed. Comparing this interesting feature of RAC1 to literature examples shows that this type of genotoxic arrest does occur in some cases, with ATR, ATM, and BRCA1 being implicated in the biological mechanism.[48] This is interesting, as most of a cell’s ROS will be in the mitochondria, endoplasmic reticulum, and cytosol. Despite this, modification of DNA occurred as shown through induction of strand breaks and double-strand repair activation. Taken together with our last publication,[7] we infer that the formation of adducts at dG, dC, and dA, along with a bulky hydroxy-benzethenoguanine adduct (Figure 1), is enough of a challenge to these cells that the lesion is difficult to repair and requires double-strand break repair during synthesis. This was validated by quantitative proteomics, which gave a strong indication of the mechanism, along with RNA-Seq results. In addition, the proteomics data showed that TALDO1 was increased in response to RAC1. This change shows how quickly these AML cells can shift metabolism to generate the ribose-phosphate precursors needed for DNA synthesis after repair (within 24 h). The synergy studies, predicated on the findings from our proteomic and transcriptomic analyses, demonstrated strong cooperativity of RAC1 with standard of care compounds for AML. This study paves the way for in vivo efficacy experiments, while at the same time showing that the design of other ROS-activated agents to limit metabolic shifts in cancer cell metabolism could be a promising approach.
Experimental Section
Cell culture and cytotoxicity
Human umbilical cord blood (UCB) and transformed AML cells were cultured in IMDM supplemented with 20% FBS.[49] UCB required SCF, IL-3, IL-6, Flt-3L, and TPO growth factors at 10 ngmL−1. Cytotoxicity analysis was carried out as previously described.[6] All R values were greater than 0.98, and standard errors of the three biological replicates were less than 20%. Unless otherwise stated, all RAC1 incubations occurred at 2 µm for 24 h.
Western blot analysis
AML cells were grown to a density of 1 × 106 cells mL−1. Samples were treated with RAC1 for 24 h. Nuclear protein was extracted by using NE-PER (Thermo Scientific), according to the manufacturer’s protocol. Protein (10 µg per well) was separated by denaturing PAGE and transferred onto nitrocellulose membrane by using an Invitrogen iBlot instrument, according to the manufacturer’s instructions. All western blot data are expressed as relative to actin. Images were analyzed by using a LI-COR imager according to standard methods. All fluorescently labeled antibodies were from LI-COR. Assays for catalase were performed with whole cell lysates.
Diacetate fluorescence (DCF) assay
Cells in active growth phase were pelleted and dissolved in 1 × HBSS to a final concentration of 5 × 105 cells mL−1. Cells (100 µL) were added to each well in a 96-well plate and incubated at 37 °C and 5% CO2 for 30 min. HBSS (1 ×, 100 µL) was used as a blank. Then 2’,7’-dichlorofluorescein diacetate (DCFH-DA; 100 µL of 20µm solution) in 1 × HBSS was added to each well. The fluorescence increase was measured (λex= 485 nm; λem=535 nm) with a microplate reader after 30 min incubation at 37°C and 5% CO2. Care was taken to control extended light exposure, as DCF can undergo light-induced oxidation. Data are reported as the mean ± standard deviation of eight replicates. An unpaired t-test was performed to determine significance.
8-Oxo-7,8-dihydro-2’-deoxyguanosine ELISA assay
Genomic DNA from UCB and AML cells was isolated by using a genomic DNA purification kit (Promega), according to the manufacturer’s instructions. The concentration of DNA in each DNA extract was quantified by using agarose gel electrophoresis. DNA was digested as described previously.[50] After digestion, enzymes were removed by centrifugation, and the supernatant was used for 8-oxo-7,8-dihydro-2’-deoxyguanosine quantification by using an Oxiselect oxidative DNA damage ELISA kit (Cell Biolabs), according to the manufacturer’s instructions.
Apoptosis assay
Cells were plated and treated with RAC1 (2 µm) or irradiated with 200 mJ cm−2 UVB. After the desired post-treatment time, cells were harvested and stained with APC-Annexin (BD Pharmingen) and propidium iodide (Sigma Chemical). Samples were analyzed by flow cytometry immediately after staining on BD LSR, and the data were analyzed by CELLQuest software.
Single cell electrophoresis assay (comet assay)
Single-cell electrophoresis was performed as previously described by Song and colleagues.[51] Briefly, cells were grown to a density of 1 × 106 cells mL−1 and were treated with RAC1 (2 µm) for 4 h. As a positive control, cells were treated with 105 mJ cm−2 UVB and incubated for 4 h. Cells were embedded in 1 % low-melting-point agarose gel and electrophoresed. The extension of each tail moment, defined as the product of DNA in the tail and the mean distance of its migration, was analyzed with a computerized image analysis system (TriTek CometScore Freeware). The tail moment values obtained from a minimum of 50 randomly selected cells from each slide were expressed as the mean value ± SEM, and data were analyzed by ANOVA (p ≤ 0.05).
Cell cycle assay
Cells were plated and treated with RAC1 (2 µm). After the desired post-treatment time, cells were harvested and fixed according to the manufacturer’s protocol. BrdU uptake was measured by FACS (FITC BrdU Flow kit, BD Biosciences).
Proteomics
Nuclear extracts (150 µg) from three control samples (control) and three experimental samples (treatment) in NE-PER buffer were sonicated with a probe to shear any residual DNA. Buffer exchange was then performed on the samples with four cycles of GE destreak rehydration buffer (100 µL, GE Healthcare Life Sciences), followed by centrifugation using an Amicon Ultra 3 kDa centrifugal filter (EMD Millipore). The samples were then loaded in the first dimension on GE 18 cm 3–10NL IPG strips (GE Healthcare Life Sciences) for a total of 57.5 kVh. Reduction and alkylation were performed with DTT and iodoacetamide as previously described.[52] The strips were run in the second dimension (12.5% SDS PAGE) at 5 W per gel for 30 min, then at 17 W per gel until the dye front reached the end of the gel. After fixing and silver staining, 16 bit tiff images were obtained. The images were analyzed by Nonlinear Dynamics Progenesis Same Spots software (Durham, NC). The gels were grouped into control versus treatment, and automatic spot outlines were manually edited. The software calculates fold change, ANOVA p value, q value, and power for each spot, based on all gels in the experiment. Spots were picked on the basis of having a fold change greater than 1.3, an ANOVA p < 0.05, and consistency within a group (power > 0.8).
The spots determined to be significantly different between groups were excised from the gel and digested with trypsin (Promega), followed by extraction, desalting, and identification by MALDI TOF-TOF on an ABSCIEX 4800 operated in reflector positive mode as previously described. Twenty-two proteins were identified by searching the resultant peptide MS/MS sequencing data of the top 20 peptides against the NCBInr Homo sapiens database by using Mascot software (Matrix Science). Variable modifications of carbamidomethyl cysteine, deamidated asparagine, or glutamine, and oxidation of methionine with a maximum of two missed cleavages were used. The peptide mass tolerance was set to 125 ppm and the fragment mass tolerance to 0.8 Da. A minimum of two confident peptides was required for each protein identification reported.
RNA-Seq analysis
After 24 h of treatment, RNA was collected by using a total RNA isolation kit (Qiagen). After normalization to 50 ng µL−1, samples were submitted to the University of Cincinnati (UC) Transcriptomics core. At the core facility, samples were analyzed for degradation by using a Bioanalyzer 1200. After validating the integrity, samples were converted to cDNA by using a PrepX mRNA Library kit (WaferGen), an Apollo 324 NGS automatic library prep system, and Superscript III reverse transcriptase (Lifetech, Grand Island, NY). Amplified cDNA was quantified by using a Kapa Library Quantification kit (Kapabiosystem, Woburn, MA) following standard methods, and sequence reads were aligned to the genome by using the standard Illumina sequence analysis pipeline, which was analyzed by the UC Statistical Genomics and Systems Biology Core to yield fold change and ANOVA scores. Next, mRNAs with significant changes (> twofold and p < 0.01) were clustered into pathways by using ToppGene.[33]
Combination studies
Response curves, combinational indices, and dose reduction indices were generated for all treatments and time points by using CompuSyn software (Paramus, NJ), according to the manufacturer’s instructions; s = dosed as listed. Conditions were the same as for cytotoxicity assays.
Supplementary Material
Acknowledgments
This work was supported by an National Institutes of Health (NIH)/National Cancer Institute (NCI) grant (R21A185370 to E.J.M.). The Genomics, Epigenomics, and Sequencing Core is supported in part by a Center for Environmental Genetics grant (NIEHS P30-ES006096). We would also like to acknowledge an Institutional Clinical and Translational Science Award from the NIH/National Center for Research Resources (NCRR; 1L1RR026314-01). This work was supported by a Translational Trials Development and Support Laboratory Award from the United States Public Health Service (USPHS;(MO1 RR 08084) and a Center of Excellence in Molecular Hematology P30 award (DK090971) to J.C.M. J.C.M. is a Leukemia and Lymphoma Society Scholar. This work was further supported by through the University of Cincinnati Millennium Scholar Fund and the Cincinnati Children’s Research Foundation to K.D.G.
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/cbic.201500304.
References
- 1.Shrivastav N, Li DY, Essigmann JM. Carcinogenesis. 2010;31:59–70. doi: 10.1093/carcin/bgp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shah NP, Tran C, Lee FY, Chen P, Norris D, Sawyers CL. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 3.Greenberg MM. Curr. Opin. Chem. Biol. 2014;21:48–55. doi: 10.1016/j.cbpa.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKibbin PL, Fleming AM, Towheed MA, Van Houten B, Burrows CJ, David SS. J Am. Chem. Soc. 2013;135:13851–13861. doi: 10.1021/ja4059469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu AM, Senter PD. Nat. Biotechnol. 2005;23:1137–1146. doi: 10.1038/nbt1141. [DOI] [PubMed] [Google Scholar]
- 6.Bell-Horwath TR, Vadukoot AK, Thowfeik FS, Li G, Wunderlich M, Mulloy JC, Merino EJ. Bioorg. Med. Chem. Lett. 2013;23:2951–2954. doi: 10.1016/j.bmcl.2013.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones AR, Bell-Horwath TR, Li G, Rollmann SM, Merino EJ. Chem. Res. Toxicol. 2012;25:2542–2552. doi: 10.1021/tx300337j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siddique YH, Ara G, Beg T, Gupta J, Afzal M. Exp. Toxicol. Pathol. 2010;62:503–508. doi: 10.1016/j.etp.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 9.Colis LC, Woo CM, Hegan DC, Li ZW, Glazer PM, Herzon SB. Nat. Chem. 2014;6:504–510. doi: 10.1038/nchem.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pei SS, Minhajuddin M, Callahan KP, Balys M, Ashton JM, Neering SJ, Lagadinou ED, Corbett C, Ye HB, Liesveld JL, O’Dwyer KM, Li Z, Shi L, Greninger P, Settleman J, Benes C, Hagen FK, Munger J, Crooks PA, Becker MW, Jordan CT. J Biol. Chem. 2013;288:33542–33558. doi: 10.1074/jbc.M113.511170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Godfrey R, Arora D, Bauer R, Stopp S, Mueller JP, Heinrich T, Boehmer S-A, Dagnell M, Schnetzke U, Scholl S, Ostman A, Boehmer F-D. Blood. 2012;119:4499–4511. doi: 10.1182/blood-2011-02-336446. [DOI] [PubMed] [Google Scholar]
- 12.Hole PS, Zabkiewicz J, Munje C, Newton Z, Pearn L, White P, Marquez N, Hills RK, Burnett AK, Tonks A, Darley RL. Blood. 2013;122:3322–3330. doi: 10.1182/blood-2013-04-491944. [DOI] [PubMed] [Google Scholar]
- 13.Irwin ME, Rivera-Del Valle N, Chandra J. Antioxid. Redox Signaling. 2013;18:1349–1383. doi: 10.1089/ars.2011.4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Y, Lu W, Chen G, Wang P, Chen Z, Zhou Y, Ogasawara M, Trachootham D, Feng L, Pelicano H, Chiao PJ, Keating MJ, Garcia-Manero G, Huang P. Cell Res. 2012;22:399–412. doi: 10.1038/cr.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reddy MM, Fernandes MS, Salgia R, Levine RL, Griffin JD, Sattler M. Leukemia. 2011;25:281–289. doi: 10.1038/leu.2010.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sallmyr A, Fan J, Datta K, Kim K-T, Grosu D, Shapiro P, Small D, Rassool F. Blood. 2008;111:3173–3182. doi: 10.1182/blood-2007-05-092510. [DOI] [PubMed] [Google Scholar]
- 17.Ke K, Sul OJ, Choi EK, Safdar AM, Kim ES, Choi HS. Am. J. Physiol. Endocrinol. Metab. 2014;307:E61–E70. doi: 10.1152/ajpendo.00044.2014. [DOI] [PubMed] [Google Scholar]
- 18.Zhu QS, Xia L, Mills GB, Lowell CA, Touw IP, Corey SJ. Blood. 2006;107:1847–1856. doi: 10.1182/blood-2005-04-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wunderlich M, Mizukawa B, Chou FS, Sexton C, Shrestha M, Saunthararajah Y, Mulloy JC. Blood. 2013;121:e90–e97. doi: 10.1182/blood-2012-10-464677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bagger FO, Rapin N, Theilgaard-Monch K, Kaczkowski B, Thoren LA, Jendholm J, Winther O, Porse BT. Nucleic Acids Res. 2013;41:D1034–D1039. doi: 10.1093/nar/gks1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamamoto S, Tomita Y, Nakamori S, Hoshida Y, Nagano H, Dono K, Umeshita K, Sakon M, Monden M, Aozasa K. J Clin. Oncol. 2003;21:447–452. doi: 10.1200/JCO.2003.06.068. [DOI] [PubMed] [Google Scholar]
- 22.Tsujimoto Y, Tomita Y, Hoshida Y, Kono T, Oka T, Yamamoto S, Nonomura N, Okuyama A, Aozasa K. Clin. Cancer Res. 2004;10:3007–3012. doi: 10.1158/1078-0432.ccr-03-0191. [DOI] [PubMed] [Google Scholar]
- 23.Dai RM, Chen E, Longo DL, Gorbea CM, Li CC. J Biol. Chem. 1998;273:3562–3573. doi: 10.1074/jbc.273.6.3562. [DOI] [PubMed] [Google Scholar]
- 24.Asai T, Tomita Y, Nakatsuka S-i, Hoshida Y, Myoui A, Yoshikawa H, Aozasa K. Cancer Sci. 2002;93:296–304. doi: 10.1111/j.1349-7006.2002.tb02172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magnaghi P, D’Alessio R, Valsasina B, Avanzi N, Rizzi S, Asa D, Gasparri F, Cozzi L, Cucchi U, Orrenius C, Polucci P, Ballinari D, Perrera C, Leone A, Cervi G, Casale E, Xiao Y, Wong C, Anderson DJ, Galvani A, Donati D, O’Brien T, Jackson PK, Isacchi A. Nat. Chem. Biol. 2013;9:548–556. doi: 10.1038/nchembio.1313. [DOI] [PubMed] [Google Scholar]
- 26.Dantuma NP, Acs K, Luijsterburg MS. Exp. Cell Res. 2014;329:9–17. doi: 10.1016/j.yexcr.2014.08.025. [DOI] [PubMed] [Google Scholar]
- 27.Luijsterburg MS, van Attikum H. Curr. Opin. Cell Biol. 2012;24:439–447. doi: 10.1016/j.ceb.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Sharma K, Vabulas RM, Macek B, Pinkert S, Cox J, Mann M, Hartl FU. Mol. Cell. Proteomics. 2012;11:M111.014654. doi: 10.1074/mcp.M111.014654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson KL, Foisner R. Cold Spring Harbor Perspect. Biol. 2010;2:a000554. doi: 10.1101/cshperspect.a000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang J, Hu X, Zimmerman M, Torres CM, Yang D, Smith SB, Liu K. J Immunol. 2011;187:4426–4430. doi: 10.4049/jimmunol.1101034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perl A, Hanczko R, Telarico T, Oaks Z, Landas S. Trends Mol. Med. 2011;17:395–403. doi: 10.1016/j.molmed.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sosa V, Moline T, Somoza R, Paciucci R, Kondoh H, Me LL. Ageing Res. Rev. 2013;12:376–390. doi: 10.1016/j.arr.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 33.Chen J, Bardes EE, Aronow BJ, Jegga AG. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thuy LTT, Matsumoto Y, Thuy TTV, Hai H, Suoh M, Urahara Y, Motoyama H, Fujii H, Tamori A, Kubo S, Takemura S, Morita T, Yoshizato K, Kawada N. Am. J. Pathol. 2015;185:1045–1060. doi: 10.1016/j.ajpath.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 35.Boehler C, Gauthier LR, Mortusewicz O, Biard DS, Saliou J-M, Bresson A, Sanglier-Cianferani S, Smith S, Schreiber V, Boussin F, Dantzer F. Proc. Natl. Acad. Sci. USA. 2011;108:2783–2788. doi: 10.1073/pnas.1016574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Welsby I, Hutin D, Gueydan C, Kruys V, Rongvaux A, Leo O. J Biol. Chem. 2014;289:26642–26657. doi: 10.1074/jbc.M114.589515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chapman JR, Taylor MR, Boulton SJ. Mol. Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 38.Srivastava M, Raghavan SC. Chem. Biol. 2015;22:17–29. doi: 10.1016/j.chembiol.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 39.Gullotta F, De Marinis E, Ascenzi P, di Masi A. Curr. Med. Chem. 2010;17:2017–2048. doi: 10.2174/092986710791233698. [DOI] [PubMed] [Google Scholar]
- 40.Fell VL, Schild-Poulter C. Mutation Res. Rev. Mutat. Res. 2015;763:15–29. doi: 10.1016/j.mrrev.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 41.Moon C, Moon HY, Kim CS. Korean J. Urol. 2007;48:976–983. [Google Scholar]
- 42.Tsouko E, Khan AS, White MA, Han JJ, Shi Y, Merchant FA, Sharpe MA, Xin L, Frigo DE. Oncogenesis. 2014;3:e103. doi: 10.1038/oncsis.2014.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 44.Reaper PM, Griffiths MR, Long JM, Charrier JD, Maccormick S, Charlton PA, Golec JM, Pollard JR. Nat.Chem. Biol. 2011;7:428–430. doi: 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- 45.Chou T-C. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 46.Peng X, Gandhi V. Ther. Deliv. 2012;3:823–833. doi: 10.4155/tde.12.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen W, Balakrishnan K, Kuang Y, Han Y, Fu M, Gandhi V, Peng X. J Med. Chem. 2014;57:4498–4510. doi: 10.1021/jm401349g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bartek J, Lukas C, Lukas J. Nat. Rev. Mol. Cell Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- 49.Mulloy JC, Cammenga J, MacKenzie KL, Berguido FJ, Moore MA, Nimer SD. Blood. 2002;99:15–23. doi: 10.1182/blood.v99.1.15. [DOI] [PubMed] [Google Scholar]
- 50.Boysen G, Collins LB, Liao S, Luke AM, Pachkowski BF, Watters JL, Swenberg JA. J Chromatogr. B. 2010;878:375–380. doi: 10.1016/j.jchromb.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song X, Mosby N, Yang J, Xu A, Abdel-Malek Z, Kadekaro AL. Pigment Cell Melanoma Res. 2009;22:809–818. doi: 10.1111/j.1755-148X.2009.00615.x. [DOI] [PubMed] [Google Scholar]
- 52.Eismann T, Huber N, Shin T, Kuboki S, Galloway E, Wyder M, Edwards MJ, Greis KD, Shertzer HG, Fisher AB, Lentsch AB. Am. J. Physiol. Gastrointest Liver Physiol. 2009;296:G266–G274. doi: 10.1152/ajpgi.90583.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.