Abstract
Chloroplast (cp) DNA is thought to originate from the ancestral endosymbiont genome and is compacted to form nucleoprotein complexes, cp nucleoids. The structure of cp nucleoids is ubiquitously observed in diverse plants from unicellular algae to flowering plants and is believed to be a multifunctional platform for various processes, including cpDNA replication, repair/recombination, transcription, and inheritance. Despite its fundamental functions, the protein composition for cp nucleoids in flowering plants was suggested to be divergent from those of bacteria and algae, but the evolutionary process remains elusive. In this research, we aimed to reveal the evolutionary history of cp nucleoid organization by analyzing the key organisms representing the three evolutionary stages of eukaryotic phototrophs: the chlorophyte alga Chlamydomonas reinhardtii, the charophyte alga Klebsormidium flaccidum, and the most basal land plant Marchantia polymorpha. To clarify the core cp nucleoid proteins in C. reinhardtii, we performed an LC-MS/MS analysis using highly purified cp nucleoid fractions and identified a novel SAP domain-containing protein with a eukaryotic origin as a constitutive core component. Then, homologous genes for cp nucleoid proteins were searched for in C. reinhardtii, K. flaccidum, and M. polymorpha using the genome databases, and their intracellular localizations and DNA binding activities were investigated by cell biological/biochemical analyses. Based on these results, we propose a model that recurrent modification of cp nucleoid organization by eukaryotic factors originally related to chromatin organization might have been the driving force for the diversification of cp nucleoids since the early stage of green plant evolution.
Keywords: chloroplast, nucleoid, evolution, endosymbiosis
Introduction
Plants are thought to have descended from a eukaryotic ancestor that acquired a once free-living photosynthetic prokaryote, closely related to the present-day cyanobacteria, which led to the emergence of chloroplasts (cps; plastids) (Gray 1992; Timmis et al. 2004; Bowman et al. 2007; Bogorad 2008; Keeling 2010). Over time, a drastic transfer of genetic materials from the endosymbiont cyanobacteria (ancestral cps) to the eukaryotic host genome occurred, which is thought to have been the driving force that reduced cp autonomy and increased nuclear dominance, thereby establishing a robust endosymbiotic relationship (Timmis et al. 2004; Bogorad 2008). The extant cp genomes are small, encoding only ∼120–200 genes (<10% of full-fledged cyanobacteria) (Timmis et al. 2004), but cp-encoded genes remain critical for photosynthesis, gene expression, and cp biogenesis (Allen 2003; Timmis et al. 2004; Stern et al. 2010).
cpDNA is not naked but is packaged into cpDNA–protein complexes, cp nucleoids (Kuroiwa 1991; Sakai et al. 2004; Pfalz and Pfannschmidt 2013, 2015; Powikrowska et al. 2014). Cp nucleoids can be visualized as dot-like structures in cps by staining with DNA-specific fluorochromes such as 4′, 6-diamidino-2-phenylindole (DAPI) or SYBR Green I, and are ubiquitously observed in diverse taxa of plants and algae (supplementary fig. S1). Cp nucleoids are thought to be the functional unit for cpDNA replication, inheritance, and transcription (Kuroiwa 1991; Sakai et al. 2004; Pfalz and Pfannschmidt 2013, 2015; Powikrowska et al. 2014).
To understand the composition of core cp nucleoid proteins, biochemical and proteomic analyses have been conducted. The core cp nucleoid proteins are regarded as proteins that are sufficiently abundant to coat cpDNA and co-localize to cp nucleoids via DNA-binding activity, which corresponds to the DNA-inheritance subdomain according to a previous report (Pfalz and Pfannschmidt 2013). In the primitive red alga Cyanidioschyzon merolae, histone-like protein (hlp), which is the ortholog of the bacterial nucleoid protein, heat unstable (hu), was found in the cp genome and histone-like protein (HLP) is a major component of cp nucleoids, validating an evolutionary continuity between cp nucleoids and bacterial nucleoids (Kobayashi et al. 2002). HLP was also found in cp nucleoids in the green alga Chlamydomonas reinhardtii, in which the HLP gene was encoded in the nuclear genome (Karcher et al. 2009).
However, in land plants, HLP genes have not been found in any of the sequenced cp genomes nor in any of the sequenced nuclear genomes. Instead, various core cp nucleoid proteins have been reported, including sulfite reductase (SiR) (Sato et al. 2001), Whirly (pTAC1) (Krupinska et al. 2014), pTAC3 (SAP domain protein) (Pfalz et al. 2006; Majeran et al. 2012; Yagi et al. 2012), and Switch/sucrose non-fermentable complex B (SWIB)-4 (Melonek et al. 2012), indicating a discontinuity or fundamental divergence of the cp nucleoid organization between algae and land plants (Sato 2001; Yagi and Shiina 2014). This divergence of the protein composition for DNA compaction in cps is in marked contrast to the situation in the nucleus and mitochondria, where structural proteins are well conserved among eukaryotic organisms (Chen and Butow 2005; Stros et al. 2007; Annunziato 2008).
Elucidating the precise evolutionary process forming the cp nucleoid structures from the endosymbiont bacterium into those of flowering plants, is hindered by the limited knowledge of the cp proteins’ composition in algae and basal land plants. In this research, we started with a proteomic analysis of cp nucleoids in the chlorophyte alga C. reinhardtii. Among identified proteins, a protein with a SAP domain exclusively co-localized with cp nucleoids. The SAP domain-containing protein was distantly related to pTAC3, suggesting an evolutionary continuity in the cp nucleoid structures of green plants. In contrast, SWIB domain-containing proteins and Whirly, which form the core of cp nucleoids in flowering plants, were not detected constitutively in the cp nucleoids of C. reinhardtii. We searched for homologous genes of cp nucleoid proteins in two other model plants representing key evolutionary stages: the charophyte alga Klebsormidium flaccidum, and the most basal land plant Marchantia polymorpha. These protein localizations and DNA-binding activities were investigated using cell biological/biochemical analyses, including indirect immune-fluorescence microscopy and electrophoretic mobility shift assays. Based on these results, we propose a model that recurrent modification of cp nucleoid organization by eukaryotic factors originally related to chromatin organization might have been the driving force for the diversification of cp nucleoids since the early stage of green plant evolution.
Materials and Methods
Plant Culture Conditions
Chlamydomonas reinhardtii was cultured in Tris-acetate-phosphate (TAP) medium on a shaker at 120 rpm at 23 °C under an illumination of 30 μmol/m2 s1 with a photoperiod of 12 h light and 12 h dark cycle.
Klebsormidium flaccidum (NIES-2285) was cultured in BCDATG liquid medium at 23 °C under continuous light (10 µmol/m2 s1).
Marchantia polymorpha (Takaragaike-1) was maintained asexually. Plants were cultured using half-strength Gamborg’s B5 medium supplemented with 0.5 g/l MES and 1.3% (w/v) agar. The pH was adjusted to 5.7 with KOH before autoclaving.
Vector Constructions
Primers used in this study are listed in supplementary table S2. YFP (Venus) was a generous gift from Dr Ralph Bock (Max-Planck-Institute). PCR was performed using the proof-reading enzyme KOD-Plus (Toyobo Life Science, Osaka, Japan). The PCR products were separated using 1.2% agarose gel electrophoresis, and were gel-purified. To generate the PSADp::YFP vector (pNYAN), the pGenD vector was digested with NdeI and EcoRI, and then the YFP gene was amplified with the primer pair pNYANF and pNYANR and was cloned. The PCR products were cloned into the linearized pNYAN vector using the Infusion technique (Takara Bio Inc., Shiga, Japan).
Nuclear Transformation of C. reinhardtii
Nuclear transformation was performed using the Gene Pulser X cell electroporation system (Bio-Rad Laboratories, CA, USA). For transformations, 1 µg of plasmid was introduced into the cells. Colonies were obtained on TAP agar plates supplemented with 10 µg/ml paromomycin (Nacalai Tesque, Kyoto, Japan) and then transferred individually to fresh TAP agar plates containing paromomycin, and maintained separately.
Isolation of cps in C. reinhardtii
Cps were isolated using an airbrush as previously described (Nishimura et al. 2002) with minor modifications. The cell wall-less mutant CW15 (a generous gift from Dr Fukuzawa, Kyoto University, Japan) was used to facilitate the isolation of cps. Cells were harvested and resuspended in 30 mM sucrose solution at room temperature. Then, cells were broken by an airbrush (HP-62B, OLYMPOS, Japan) connected to an air regulator and air compressor (TC-65-21, OLYMPOS) at a pressure of 3.0 kg/cm2. Cell lysates were collected in a pre-cooled 1-l beaker. After centrifugation at 800 × g at 4 °C for 3 min, the precipitated cps were washed four times with suspension buffer (0.3 M sucrose, 5% polyethylene glycol 6,000, 1.2 mM HEPES-KOH at pH 6.8, 1 mM MgSO4, and 1.5 mM spermidine).
Isolation of cp Nucleoids in C. reinhardtii
Isolated cps was suspended in suspension buffer containing 30% Percoll and overlaid on top of a discontinuous Percoll gradient (45–80% Percoll in suspension buffer) in 15-ml tubes. After centrifugation by a swinging-bucket rotor (2,000 × g, 4 °C, 40 min), the green band at the interface between the 45% and 80% layers of Percoll was collected. The cp fraction was diluted 4-fold with suspension buffer and then layered onto a discontinuous sucrose density gradient (3 ml each of 80%, 40%, and 20% sucrose in suspension buffer) in 15-ml tubes, and centrifuged by a swinging-bucket rotor (2,000 × g, 4 °C, 40 min). Green bands of plastids at the 80–40% sucrose interface (ca. 2 ml) were recovered, diluted to 6 ml with TAN buffer (20 mM Tris–HCl [pH 7.6], 0.5 mM EDTA, 7 mM 2-mercaptoethanol, 1.2 mM spermidine, 0.4 mM Phenylmethylsulfonyl fluoride [PMSF]). Four hundred microliters of 20% Nonidet P-40 were added into the cps suspension while stirring to a final concentration to 2% on ice and left stirring for 30 min. The solution was centrifuged at 100 × g for 5 min to remove starch grains and debris. The supernatant was filtered through the nylon mesh with 5-μm pores (Millipore Corporation, Billerica, Massachusetts, USA), and the filtrate was centrifuged (20,000 × g, 4 °C, 45 min) to sediment the plastid nucleoids. The sedimented plastid nucleoids were resuspended in 200 μl of TAN buffer, divided into aliquots, quickly frozen, and stored at −80 °C.
Microscopic Observation
Cells were fixed in 3.7% formaldehyde and stained with 0.5 μg/μl DAPI (Nacalai Tesque). To stain the DNA in living cells, SYBR green I was added to produce a final dilution of 1:2,000. The stained cells were observed under UV excitation for DAPI or blue excitation for SYBR Green I using a fluorescence/differential interference (DIC) microscope (BX51; Olympus) connected to a charge-coupled device camera (DP71; Olympus). Confocal laser scanning microscopy of M. polymorpha was performed using a Zeiss LSM780 (Carl Zeiss AG, Oberkochen, Germany).
Antibody Preparation
pQE80l (Qiagen, Venlo, Netherlands) vectors harboring the cDNA sequences encoding CreCNS, CreHLP, CreWhirly, CreSWIB2, KfHLP, KfpTAC3, KfWhirly and KfSWIB were prepared and transformed into the Escherichia coli BL21 strain and selected on Luria–Bertani (LB) agar medium containing 50 µg/ml carbenicillin (Nacalai Tesque). The culture was grown in LB medium at 37 °C, and isopropyl β-D-1-thiogalactopyranoside (IPTG) was added at OD600 0.7–1 to a final concentration of 1 mM. Proteins were purified using Ni-NTA agarose (Qiagen) following the manufacturer’s instructions. To raise antibodies, the purified recombinant proteins were injected to mice five times every 2 weeks.
Indirect Immunofluorescence Microscopy in C. reinhardtii
Proteins were detected by immunofluorescence staining according to a previously described method (Nishimura et al. 2002). Cells were harvested and fixed in 3.8% formaldehyde on ice for 10 min. After washing with TBS, the cells were gently permeabilized with 0.1% Triton X-100 on ice for 20 min. They were then resuspended in 5% bovine serum albumin buffer containing 0.25% Tween 20 in Tris Buffered Saline (TBS), and kept on ice for 30 min. The cells were then incubated on ice for 1 h with the primary antibody at a 1:1,000 dilution. After washing, the cells were resuspended in TBS buffer on ice for 30 min. The secondary antibody (Alexa Fluor 488 Goat Anti-Mouse IgG) was added at a 1:1,000 dilution and incubated on ice for 1 h. The cells were washed, stained with 1% DAPI, and observed by fluorescence microscopy under UV (for DAPI) or blue (for Alexa) light.
Indirect Immunofluorescence Microscopy in K. flaccidum
Cells were collected by centrifugation (1,800 × g, 2 min, 4 °C) from 6 ml of liquid culture and frozen in liquid nitrogen. Pellets were thawed in a water bath (42 °C, 1 min) and resuspended in −20 °C acetone containing 1% formaldehyde (6 ml). After 24 h of incubation at −30 °C, cells were collected by centrifugation (1,800 × g, 2 min, 4 °C), and 3 ml supernatant was replaced by 3 ml TBS. Then, cells were centrifuged again and 5 ml supernatant was replaced by 5 ml TBS. After another round of centrifugation, all of the supernatant was replaced by 6 ml TBS containing 0.1% Triton X-100 and incubated for 20 min on ice. The cells were then incubated on ice for 1 h with the primary antibody at a 1:1,000 dilution. After washing, the cells were resuspended in TBS buffer on ice for 30 min. The secondary antibody (goat anti-mouse IgG conjugated with Alexa Fluor 488) was added at a 1:1,000 dilution and incubated on ice for 1 h. Then, the cells were rinsed with TBS buffer and observed by fluorescence microscopy.
Co-immunoprecipitation of Nucleoids
Isolated cps or cp nucleoids were suspended with Lysis buffer (150 mM NaCl, 1% Triton X-100, 50 mM Tris–HCl [pH 8.0]) and incubated at 4 °C for 2 min on ice. Co-immunoprecipitations (Co-IPs) were performed using μMACS epitope tag protein isolation kits (Miltenyi Biotec, Bergish Gladbach, Germany). For negative control, co-IPs were also performed in the lysates of cps or cp nucleoids isolated from cells that do not express HLP-YFP. Twenty microliters of YFP beads were added to each sample tube and incubated for 15 min on ice. Samples were applied to μ columns and rinsed with 1.2 ml of Wash Buffer I (150 mM NaCl, 1% Igepal CA-630, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris–HCl [pH 8.0]) and 100 μl of Wash Buffer II (20 mM Tris–HCl [pH 7.5]). IPs were collected by adding 70 μl of Elusion Buffer (50 mM Tris–HCl [pH 6.8], 50 mM DTT, 1% SDS, 1 mM EDTA, 0.005% bromophenol blue, 10% glycerol). IPs were separated using the precast gradient gel E-T520L (ATTO Corp., Tokyo, Japan) and stained using the silver stain kit Sil-Best Stain One (Nacalai Tesque).
Peptide Preparation for MS/MS Analysis
The IP products were separated using ready-made 7.5% (w/v) SDS-polyacrylamide gels (ATTO). The separated proteins were sliced into five pieces of equal length. Each gel slice was washed twice with HPLC-grade water containing 60% (v/v) acetonitrile/50 mM ammonium bicarbonate. Next, the gel slice was incubated in 10 mM DTT/50 mM ammonium bicarbonate for 45 min at 56 °C, followed by 55 mM iodoacetamide/50 mM ammonium bicarbonate for 30 min at room temperature. The incubated gel slices were washed twice with HPLC-grade water containing 60% (v/v) acetonitrile/50 mM ammonium bicarbonate and dried in a vacuum concentrator. The dried gel slices were treated with 2 µl of 10 ng/µl trypsin (MS grade gold; Promega, Japan) dissolved in 50 mM ammonium bicarbonate and incubated at 37 °C for 16 h. The digested peptides were transferred to clean tubes. The gel slices were treated twice with 20 µl of a solution containing 0.2% (v/v) formic acid (Wako, Osaka, Japan) and 50% (v/v) acetonitrile, and the extracts were collected, dried in a vacuum concentrator, and dissolved in a solution containing 0.1% (v/v) formic acid/5% (v/v) acetonitrile. The solution was filtered using Ultrafree-MC Centrifugal Filters (PVDF 0.45 μm; Merck Millipore, Billerica, MA, USA) to eliminate gel contamination.
LC-MS/MS Analysis and Database Searching
LC-MS/MS analysis was performed using a HTC-PAL/Paradigm MS4 system coupled to a LTQ Orbitrap XL (Thermo Fisher Scientific Inc., Waltham, MA USA) mass spectrometer. Trypsin-digested peptides were loaded on the L-column (100 µm internal diameter, 15 cm; CERI) using a Paradigm MS4 HPLC pump (Michrome BioResources/Bruker, Billerica, MA USA) and an HTC-PAL autosampler (CTC analytics AG, Zwingen, Switzerland). Buffers were 0.1% (v/v) acetic acid and 2% (v/v) acetonitrile in water (Solvent A), and 0.1% (v/v) acetic acid and 90% (v/v) acetonitrile in water (Solvent B). A linear gradient from 5% to 45% Solvent B was applied for 26 min, and peptides eluted from the L-column were introduced directly into a LTQ Orbitrap XL mass spectrometer with a flow rate of 500 nl/min and a spray voltage of 2.0 kV. All of the events for the MS scan were controlled and acquired using Xcalibur software version 2.0.7 (Thermo Fisher Scientific Inc.). The range of the MS scan was m/z 400–1,500, and the top three peaks were subjected to MS/MS analysis. Obtained spectra were compared with a protein database (Chlamydomonas genomic information v5.3.1) using the MASCOT server (version 2.4). The mascot search parameters were as follows: set the threshold at 0.05 for the ion-score cutoff, peptide tolerance at 10 ppm, MS/MS tolerance at ± 0.5 Da, peptide charge of 2+ or 3+, trypsin as enzyme allowing up to one missed cleavage, carbamidomethylation on cysteine as a fixed modification, and oxidation on methionine as a variable modification.
Electrophoretic Mobility Shift Assay
Recombinant proteins were expressed as fusion proteins with a 6×His tag using pQE80l. E. coli cells were lysed using lysozyme (Nacalai Tesque) and fusion proteins were purified with Ni-NTA Agarose (Qiagen). Fusion proteins were mixed with 5 µl of 100 bp DNA ladder marker (WATSON Co., Ltd., Kobe, Japan). After a 2-h incubation at room temperature, the mixtures were electrophoresed in a 1.2% agarose gel and stained by ethidium bromide.
DNA Condensation Assays in E. coli Cells
The full coding sequence of the CreHLP and partial coding sequence of CreCNS and KfpTAC3 genes were cloned into the pQE80 vector (Qiagen) for the transformation in E. coli DH5α cells. Cells were grown in LB medium containing 50 µg/ml carbenicillin at 37 °C, and IPTG was added at OD600 0.4–0.6 to a final concentration of 1 mM. Four hours after induction, cells were stained with DAPI for fluorescence microscopy.
Phylogenetic Analysis
HLP homologs were collected with searches using the BLAST algorithm against public databases. The sequences were aligned using ClustalW in MEGA 5.0 (Tamura et al. 2011). The full lengths of HLP homologs were used for the phylogenetic analyses. A Bayesian inference was performed using MrBayes version 3.2 (Ronquist et al. 2012). One million generations were completed, and trees were collected every 5,000 generations, after discarding trees corresponding to the first 25% (burn-in), to generate a consensus phylogenetic tree. Bayesian posterior probabilities were estimated as the proportion of trees sampled after burn-in. A Maximum Likelihood phylogenetic tree was constructed using morePhyML with the default conditions (Criscuolo 2011).
Nuclear Transformation in M. polymorpha
Nuclear transformation was performed essentially as previously described (Kubota et al. 2013), and Agrobacterium tumefaciens harboring the pMpGWB108 vector was used (Ishizaki et al. 2015). Apical portions, about 2–3 mm from the tip, including meristem, were removed from 14-day-old thalli. The thalli were further cut into four pieces and grown on half-strength Gamborg’s B5 medium containing 1% (w/v) sucrose and 1.3% agar under 50–60 mmol/m2 s1 continuous white fluorescent light at 22 °C for 3 days to promote regeneration. In all, 15–20 regenerating plantlets and 1 ml of Agrobacterium culture, prepared as described above, were co-cultivated in 50 ml of M51C medium containing 2% sucrose and 100 mM acetosyringone in a 200-ml flask under continuous white light (50–60 mmol/m2 s1) with agitation at 130 rpm at 22 °C for 3 days. The plantlets were washed five to six times with sterilized water, then soaked in sterilized water containing 1 g/l of cefotaxime (Claforan; Sanofi-Adventis, Tokyo) for 30 min. The plantlets were transferred directly to half-strength B5 agar medium containing 1.3% agar, 10 mg/l of hygromycin, and 100 mg/l of cefotaxime, and incubated under continuous white light (40 mmol/m2 s1) at 22 °C.
Results
The Novel Eukaryotic SAP Domain Protein CreCNS is the Constitutive cp Nucleoid Component in C. reinhardtii
To clarify the core components of algal cp nucleoids, we conducted a proteomic analysis of cp nucleoids in C. reinhardtii. To purify cp nucleoid fractions, we used a Co-IP technique. Prokaryotic HLP, which is an abundant core component of the cp nucleoid in algae, was used as a bait protein (supplementary figs. S2–S4). Consistent with a previous report (Karcher et al. 2009), we confirmed the localization of HLP in C. reinhardtii (CreHLP) using a construct that consisted of PsaD promoter and the CreHLP genomic sequence (CreHLPg) fused with YFP (PsaDp::CreHLPg-YFP) (supplementary fig. S3). The signal was not lost during the biochemical procedure used to purify cp nucleoids, reflecting the tight interaction between CreHLP and cpDNA (supplementary fig. S3). These observations confirmed that HLP is a core component of cp nucleoids in C. reinhardtii. Intact cps and isolated cp nucleoids were suspended in lysis buffer containing the non-ionic detergent Triton X-100, and Co-IPs were performed using αYFP magnetic beads (supplementary figs. S4B and C). The Co-IP products purified from isolated cps and cp nucleoids were independently analyzed using LC-MS/MS. HLP was detected as a protein with high values in the IP products purified from isolated cps and cp nucleoids. In this proteomic analysis, 197 proteins were identified and 131 proteins were predicted to localize to cps or mitochondria according to the TargetP program (supplementary fig. S4D; supplementary tables S1 and S2). Of the identified proteins containing putative transit peptides, we focused on 15 proteins that were detected in the IP products from isolated cps and isolated cp nucleoids (table 1).
Table 1.
The proteins identified in the IP products against isolated cps and isolated cp nucleoids. CPs (IP products against isolated cps) and CPNs (IP products against isolated cp nucleoids) represent the respective scores in the proteomic analysis
| JGI accession no. | Protein name | Score |
||
|---|---|---|---|---|
| Mass | CPs | CPNs | ||
| Cre06.g259100 | Unknown | 13,8477 | 2,959 | 1310 |
| Cre03.g158900 | Dihydrolipoamide acetyltransferase | 49,879 | 1,294 | 36 |
| Cre15.g635650 | SAP domain protein (CNS) | 281,717 | 1,282 | 280 |
| Cre06.g285400 | Histone-Like Protein (HLP) | 17,846 | 1,263 | 764 |
| Cre12.g497350 | Unknown | 190,123 | 742 | 129 |
| Cre08.g378850 | Ribose-phosphate pyrophosphokinase | 44,707 | 561 | 65 |
| Cre09.g416850 | Unknown | 165,149 | 392 | 285 |
| Cre06.g250100 | Heat shock protein 70B | 72,081 | 136 | 62 |
| Cre12.g553250 | Phosphofructokinase | 55,365 | 95 | 298 |
| Cre03.g199000 | Phototropin | 81,857 | 84 | 98 |
| Cre17.g734450 | Plastid ribosomal protein L19 | 16,643 | 79 | 55 |
| Cre07.g335400 | ABC TRANSPORTER ATP-BINDING PROTEIN YDIF | 70,195 | 57 | 227 |
| Cre10.g436550 | Low-CO2-inducible protein | 32,295 | 41 | 348 |
| Cre06.g283950 | Chlorophyll a/b binding protein of LHCII | 27,202 | 30 | 25 |
| Cre13.g606800 | DUF218 domain | 34,300 | 23 | 20 |
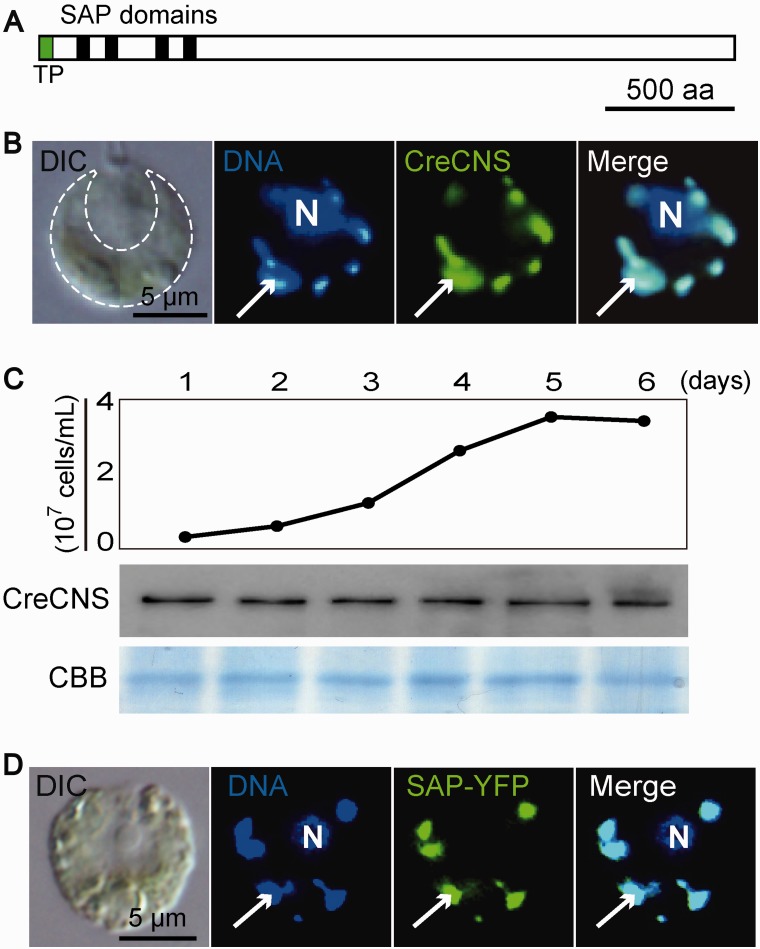
To investigate the actual localization of the candidates for cp nucleoid proteins identified in our proteomic analysis, we performed immunofluorescence staining or expressed chimeric YFP-fusion proteins. Most of the candidates were detected in stroma region in cps and did not show dot-like distribution overlapping cp nucleoids (supplementary fig. S5). However, one protein was exclusively co-localized to cp nucleoids (fig. 1B). Database searches using PROSIT (Sigrist et al. 2013) and SMART (Schultz et al. 1998) predicted that this protein contains four SAP domains in the N-terminal region (fig. 1A). A SAP domain may form the DNA-binding domain found in proteins involved in nuclear architecture, transcription, RNA processing, and DNA repair (Aravind and Koonin 2000), indicating its eukaryotic origin. Therefore, we designated this cp nucleoid protein as Cp Nucleoid SAP (CNS) domain-containing protein. An immunoblotting analysis showed that the CreCNS accumulation level was constant throughout the exponential to stationary growth phases (fig. 1C).
Fig. 1.—
CreCNS is the constitutive cp nucleoid protein. (A) Schematic model of CreCNS. Black and green boxes represent SAP domain and transit peptides, respectively. (B) Differential interference contrast microscopy (DIC) shows a C. reinhardtii cell containing a single green cup-shaped cp (indicated by the dashed line). Indirect immunofluorescence microscopic imaging of an individual cell showing the localization of CreCNS to the cp nucleoids. Nuclear (N) and cpDNA were stained with DAPI. The CreCNS protein was detected as green signals using an anti-CreCNS primary antibody and Alexa488-conjugated secondary antibody. Arrows indicate a cp nucleoid. (C) Time-course study of cellular CreCNS accumulation analyzed by immunoblotting. Cells were cultured under continuous light. The Coomassie Brilliant Blue (CBB) stained gel is a loading control. (D) The chimeric CreCNS protein (SAP-YFP) is localized to cp nucleoids in an individual cell of C. reinhardtii. SAP-YFP was detected as green signals in indirect immunofluorescence microscopy using an anti-YFP primary antibody and Alexa488-conjugated secondary antibody.
The genomic sequence encoding the N-terminal region, including four SAP domains, was cloned and fused to the YFP sequence. The chimeric construct was expressed in C. reinhardtii cells under the control of the PsaD promoter. RT-qPCR showed that the expression level of the chimeric construct (PsaDp::SAP-YFP) is modestly (∼2–3 times) higher than the intrinsic CreCNS gene (supplementary fig. S6A). The expression of SAP-YFP did not disturb the gene expression levels in cps (psbD, atpA, psaB, and petA), cpDNA copy number, or photo-autotrophic growth (supplementary fig. S6B–D). Under the microscope, the YFP signal was clearly detected at cp nucleoids, indicating that the cp-targeting peptide and the N-terminal region containing four SAP domains are sufficient for the cp nucleoid localization of CreCNS (Fig. 1D).
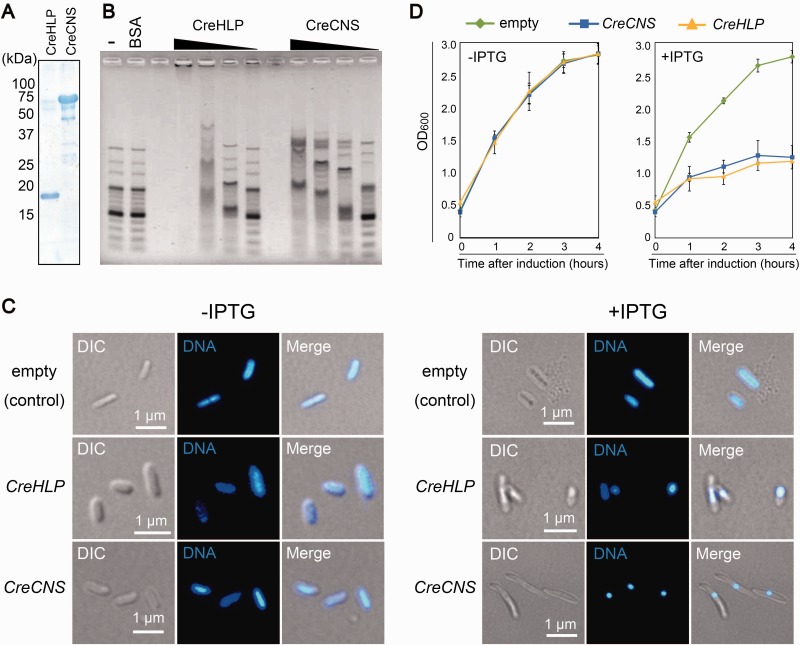
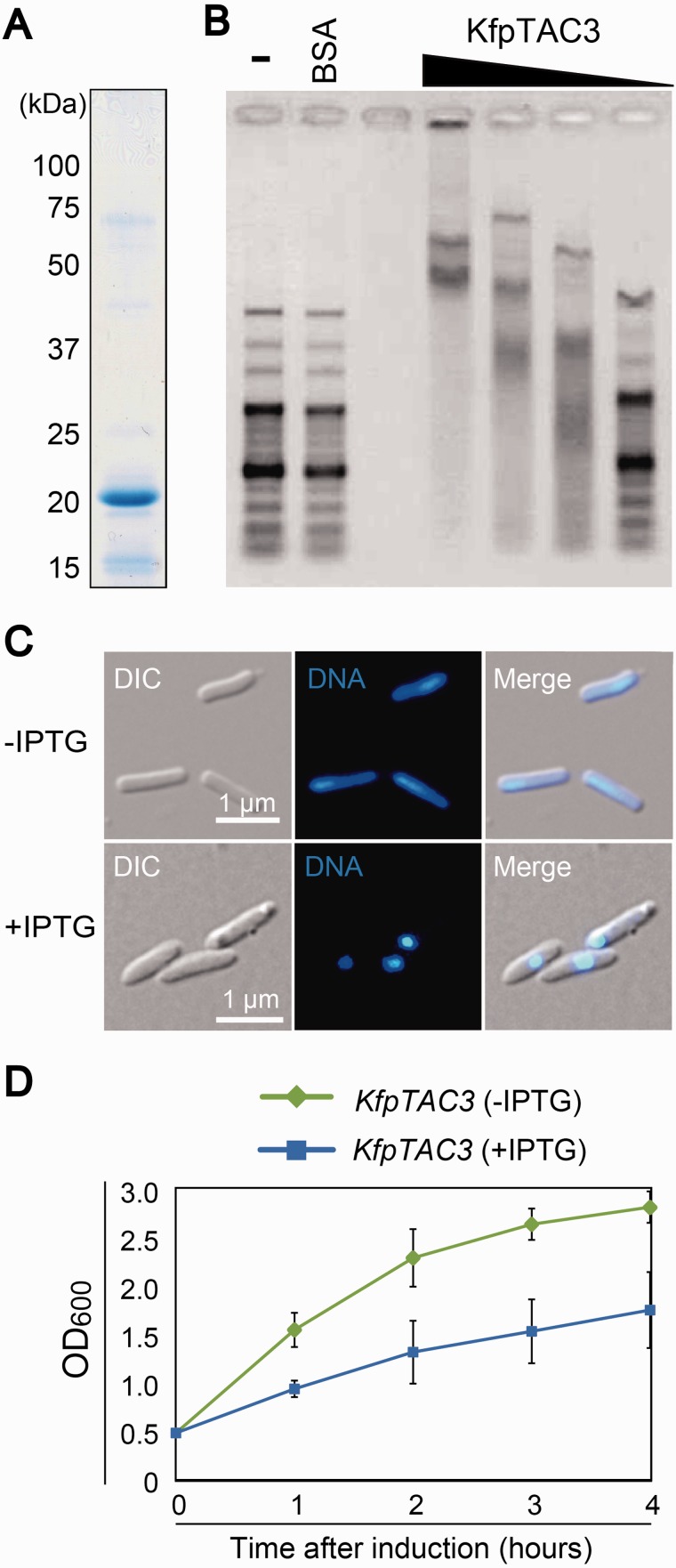
To assess the DNA-binding activity of the SAP domains, an electrophoretic mobility shift assay was performed. A recombinant protein containing the SAP domains (560 aa) was synthesized in E. coli (supplementary fig. S7) and purified in its native condition (Fig. 2A). As a control, a recombinant CreHLP protein (full length) was also prepared (Fig. 2A). The purified recombinant proteins (80, 40, 20, and 10 pmol) were incubated with a DNA ladder marker that contained linear DNA fragments of various sizes (100 bp to 3 kb) and analyzed by agarose gel electrophoresis (Fig. 2B). As with CreHLP, increasing concentrations of the recombinant protein containing the SAP domain yielded slower-migrating DNA–protein complexes, indicating the non-sequence-specific binding of the SAP domains with the DNA. Furthermore, to investigate whether CreCNS has an effect on bacterial nucleoid structure, E. coli cells that over-accumulated the fusion protein were stained with DAPI and observed under fluorescence microscopy. In the cells over-accumulating CreCNS and CreHLP, a drastic shrinkage of the bacterial nucleoids was observed, whereas no such change was detected in control cells (Fig. 2C). In parallel, E. coli cells showed a reduced proliferation after the induction of CreCNS and CreHLP overexpression (Fig. 2D). The results indicate that prokaryotic CreHLP and eukaryotic CreCNS co-localize to cp nucleoids via DNA-binding activity and coat the entire cpDNA.
Fig. 2.—
CreCNS is a DNA binding protein that compacts nucleoids in E. coli. (A) Coomassie Brilliant Blue-stained gel after SDS-PAGE of purified partial CreCNS and CreHLP proteins (80 pmol each). (B) Electrophoretic mobility shift assay of CreCNS and CreHLP proteins. The 100-bp DNA ladder was incubated with various amounts (80, 40, 20, and 10 pmol) of purified recombinant protein. (C) DH5α E. coli cells were transformed with the pQE80l vector containing the sequence of the CreCNS or CreHLP gene, or with the empty vector. Cells were grown in LB medium containing 50 µg/ml carbenicillin at 37 °C. At an OD600 of 0.4–0.6, CreCNS and CreHLP overexpression was induced with 1 mM IPTG for 4 h. The bacterial nucleoids were stained with DAPI. (D) Effect of nucleoid condensation mediated by CreCNS and CreHLP on DH5α E. coli cells. Escherichia coli cells were transformed with the pQE80l vector containing the sequence of the CreCNS or CreHLP gene, or with the empty vector. Cells were grown at 37 °C, and OD600 was measured 0–4 h after induction with 1 mM IPTG. Error bars represent standard deviation. There are three independent data points.
SWIB and Whirly are not Constitutive Core Components of cp Nucleoids in C. reinhardtii
We looked for algal homologs of the land plants’ core cp nucleoid proteins in the unicellular alga C. reinhardtii. Here, we focused on SWIB (Melonek et al. 2012), Whirly (Krupinska et al. 2014), and SiR (Sato et al. 2001; Chi-Ham et al. 2002) as representative core cp nucleoid proteins.
Our database analyses identified two candidate genes encoding SWIB domain-containing proteins in C. reinhardtii, and they were tentatively designated as CreSWIB1 (Cre02.g118250) and CreSWIB2 (Cre10.g448700). CreSWIB1 and CreSWIB2 were predicted to encode a 31.4 kDa protein with two SWIB domains and a 66.9 kDa protein with a single SWIB domain, respectively (supplementary fig. S8A). A phylogenetic analysis of two CreSWIB genes with SWIB genes in Arabidopsis thaliana showed that neither of them belonged to Group 1 (supplementary fig. S8B). The subcellular localization of CreSWIB1 was further studied by introducing the constructs encoding the protein fused with yellow fluorescent protein (CreSWIB1-YFP). The YFP signal was specifically detected in the nucleus (supplementary fig. S8C). Because the YFP signal was not detected for CreSWIB2, probably because of the low expression level, CreSWIB2 was stained with immunofluorescence using a specific antibody raised against the CreSWIB2 recombinant protein. CreSWIB2 was detected in the cytosol (supplementary fig. S8C). In C. reinhardtii, neither CreSWIB1 nor CreSWIB2 was a component of cp nucleoids.
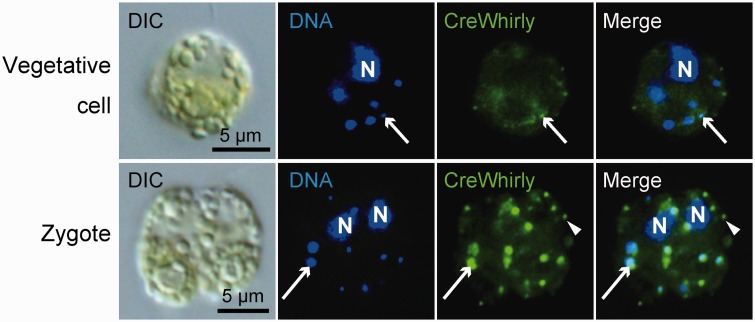
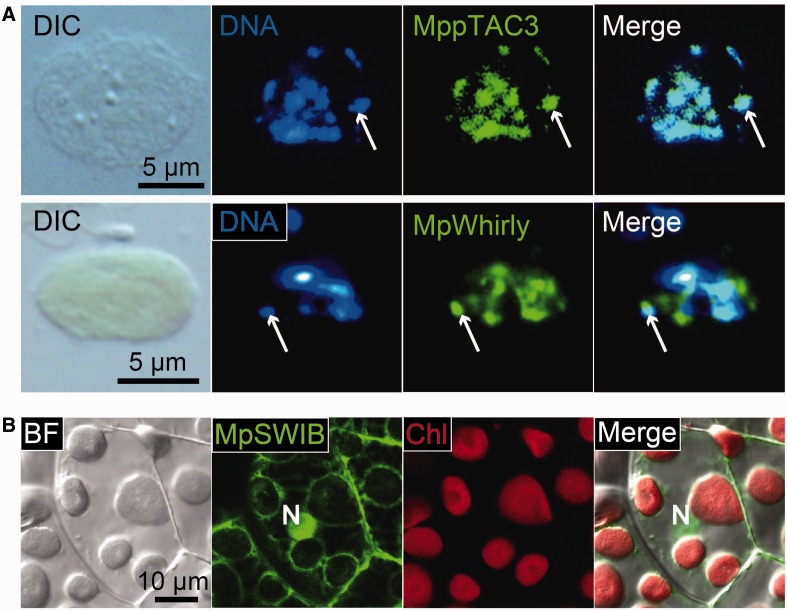
We studied the possibility that Whirly is a constitutive core component of cp nucleoids in C. reinhardtii. In C. reinhardtii, our database search identified a single gene homologous to Whirly (supplementary fig. S9). This gene has previously been annotated as an early zygote-specific expressed gene 18 (EZY18) (Kubo et al. 2008). To confirm the localization and expression profile, we performed immunofluorescence staining using an antibody raised against the CreWhirly protein (Fig. 3). In vegetative cells, while faint dot-like fluorescent signals were observed at some of the cp nucleoids, most of the signals were detected in the stroma of cps. The extra-nucleoid Whirly may function in RNA metabolism (Melonek et al. 2010). In zygotes, clear fluorescent signals were emitted from cp nucleoids, but not from all of the cp nucleoids. Furthermore, Whirly in stroma regions was frequently observed as puncta that did not overlap cp nucleoids (Fig. 3).
Fig. 3.—
CreWhirly is the zygote-specific cp nucleoid protein. Indirect immunofluorescence microscopy using a specific antibody in a vegetative or zygotic cell. Nuclear (N) and cpDNA were stained with DAPI. The CreCNS protein was detected as green signals using an anti-CreWhirly primary antibody and Alexa488-conjugated secondary antibody. Arrows indicate a cp nucleoid. Arrowheads indicate the puncta of extra-nucleoid Whirly.
Previous immuno-transmission electron microscopic analyses in C. reinhardtii, demonstrated that SiR proteins localize near the pyrenoid, a proteinaceous structure found in algal cps (Patron et al. 2008), indicating that it may not associate with cp nucleoids in C. reinhardtii.
Cp Nucleoid Proteins in the Chlorophyte Alga K. flaccidum
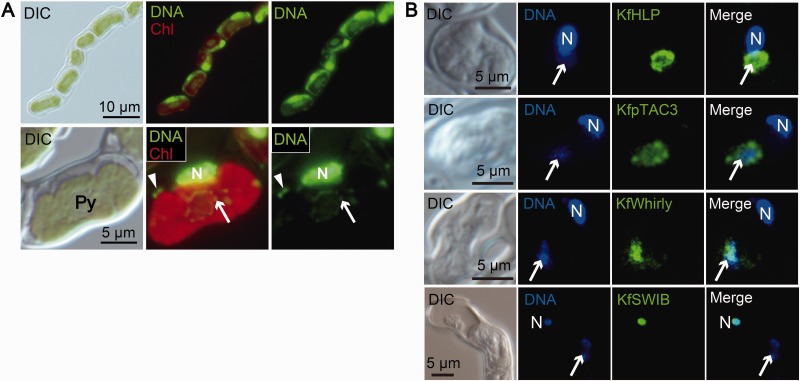
We determined that core cp nucleoids in C. reinhardtii contain prokaryotic CreHLP (Karcher et al. 2009; supplementary fig. S3) and eukaryotic CreCNS proteins (figs. 1–3). To clarify the evolution of cp nucleoids from green algae to land plants, we next focused on the filamentous terrestrial alga K. flaccidum, which is a charophyte alga, the closest relatives of land plants (Lewis and McCourt 2004). Recently, the whole genome sequence of K. flaccidum was reported and it is now regarded as one of the best model organisms for elucidating the early transitional events from aquatic algae to land plants (Hori et al. 2014). Our investigation of the K. flaccidum genome database in pursuit of homologs for possible cp nucleoid proteins identified one HLP, one Whirly, and one Group1 SWIB domain-containing protein, as well as a protein with a SAP domain (pTAC3) (supplementary figs. S2, S9–S11). pTAC3 was originally identified in the proteome analysis of transcriptionally active plastid chromosome (TAC) fraction (Pfalz et al. 2006) and its association with cp nucleoids has been shown in A. thaliana (Yagi et al. 2012) and Zea mays (Majeran et al. 2012). These homologous genes were designated as KfHLP, KfWHY1, KfpTAC3, and KfSWIB, respectively. To confirm the localization of these candidate proteins experimentally, specific antibodies were prepared for indirect immunofluorescence staining. In K. flaccidum, cp nucleoids were observed as dot-like structures surrounding the pyrenoids (Fig. 4A). The immunofluorescence signals for KfHLP proteins were detected as punctate structures that precisely co-localized with the cp nucleoids. While KfpTAC3 and Whirly proteins co-localized with cp nucleoids, immunofluorescence signals were also detected in the area surrounding the nucleoids. On the other hand, KfSWIB protein signal was detected in the cell nucleus (Fig. 4B).
Fig. 4.—
HLP, pTAC3, and Whirly are localized to cp nucleoids in K. flaccidum. (A) Nuclear, cp and mt DNA were stained with SYBR Green I in living cells. Red is the autofluorescence emitted from chlorophyll. N represents a nucleus. Arrows and arrowheads represent cp and mt nucleoids, respectively. Cp nucleoids are localized surrounding a pyrenoid. (B) Indirect immunofluorescence microscopy. Nuclear and cp DNA were stained with DAPI. N represents a nucleus.
To examine the DNA-binding activity of KfpTAC3, a recombinant protein containing the SAP domain of KfpTAC3 was prepared and analyzed using an electrophoretic mobility shift assay (Fig. 5A and supplementary fig. S12). The result indicated that the KfpTAC3 recombinant protein bound to DNA in vitro without apparent sequence specificity (Fig. 5B). Furthermore, overexpression of full-length KfpTAC3 caused the drastic shrinkage of E. coli nucleoids in vivo (Fig. 5C), inhibiting proliferation (Fig. 5D). These results support the possibility that KfpTAC3 could be a DNA-packaging protein in the cp nucleoids of K. flaccidum.
Fig. 5.—
KfpTAC3 binds dsDNA nonspecifically and condenses nucleoids in E. coli. (A) Coomassie Brilliant Blue-stained gel after SDS-PAGE of purified partial KfpTAC3 recombinant proteins (80 pmol). (B) Electrophoretic mobility shift assay of partial KfpTAC3 recombinant proteins. The 100-bp DNA ladder was incubated with various amounts of purified fusion protein (80, 40, 20, and 10 pmol). (C) DH5α E. coli cells with the pQE80l vector containing the sequence of the full-length KfpTAC3 gene or with the empty vector for control. KfpTAC3 overexpression was induced with 1 mM IPTG for 4 h. The bacterial nucleoids were stained with DAPI. (D) Effect of nucleoid condensation mediated by KfpTAC3 on DH5α E. coli cells. Escherichia coli cells expressing the pQE80l vector containing the KfpTAC3 gene were grown at 37 °C, and after the addition of IPTG, the OD600 was measured at different time points. Error bars represent standard deviation. There are three independent data points.
These results suggest that prokaryotic KfHLP and eukaryotic KfWhirly and KfpTAC3, but not KfSWIB, are core components of cp nucleoids in the charophyte alga K. flaccidum.
Cp Nucleoids Consist of Eukaryotic Components in M. polymorpha
We focused on liverwort, which is one of the earliest diverging lineages of land plants, thus constituting a sister group to all other land plants. M. polymorpha is a common liverwort with a worldwide distribution and is an extensively investigated model system. Because they belong to a clade of the most basal plant lineage, the liverwort M. polymorpha was analyzed to elucidate the evolution of land plants (Bowman et al. 2007).
A search of the M. polymorpha genome database identified one copy of Whirly, pTAC3, and Group1 SWIB homologs, which are designated as MpWhirly, MppTAC3, and MpSWIB, respectively (supplementary figs. S9–S11). However, no genes homologous to prokaryotic HLP were detected. To investigate the subcellular localization of cp nucleoid protein candidates, chimeric YFP proteins were expressed in a construct consisting of the Elongation Factor promoter and the full-length cDNA sequence fused to the YFP sequence (EFp::MppTAC3-YFP, EFp::MpWhirly-YFP, and EF::MpSWIB-YFP) in M. polymorpha thalli. Isolated cps were stained with DAPI and observed using fluorescence microscopy. These observations revealed that MppTAC3 proteins were strictly co-localized with cp nucleoids in M. polymorpha (Fig. 6A). Although MpWhirly co-localized with cp nucleoids, a fluorescence signal was also detected in the extra-nucleoid region (Fig. 6A). In contrast to MppTAC3 and MpWhirly, the MpSWIB protein was mainly localized to the nucleus and the cytosol (Fig. 6B). These results suggest that the prokaryotic HLP might have been lost and replaced by eukaryotic components, such as MppTAC3 or MpWhirly. Meanwhile, MpSWIB (Group1) may not be a component of cp nucleoids but may function as a nuclear factor in M. polymorpha.
Fig. 6.—
pTAC3 and Whirly were localized to cp nucleoids in M. polymorpha. (A) Cps isolated from the cells expressing MppTAC3-YFP or MpWhirly-YFP chimeric proteins. CpDNA was stained with DAPI. (B) Confocal laser microscopy image of the cells expressing MpSWIB-YFP. Red is the fluorescence emitted from chlorophyll. The YFP fluorescence signal was detected mainly in the nucleus and cytosol, not in the cps.
Discussion
The cp nucleoid consists of cpDNA and various, mostly uncharacterized, proteins. cpDNA undoubtedly originated from an ancestral cyanobacterial genome, but the origin of the cp nucleoid proteins is unclear. Our cell biological/biochemical characterization, which focused on a green alga (C. reinhardtii), a charophyte alga (K. flaccidum), and a liverwort (M. polymorpha), as well as comparative genome informatics, showed that the recurrent modification of cp nucleoid organization by eukaryotic factors originally related to chromatin organization might have been the driving force for the diversification of cp nucleoids since the early stage of green plant evolution.
In bacteria, genomic DNA is compacted into a limited space with a variety of proteins. In E. coli, major nucleoid-associated proteins, including H-NS, HU, Fis, IHF, and StpA, can each bind up to hundreds of specific sites per chromosome. HU, Fis, IHF, and StpA were uniformly distributed throughout the nucleoids. In contrast, H-NS, a global transcriptional silencer that formed two compact clusters per chromosome and was driven by the oligomerization of DNA-bound H-NS, played a key role in the global chromosomal organization (Wang et al. 2011). Interestingly, bacterial nucleoid proteins are not highly conserved among bacterial species, except for HU (Dillon and Dorman 2010; Grove 2011).
In a eukaryotic cell, nuclear DNA, cpDNA, and mitochondrial (mt) DNA have their own DNA packing systems that consist of numerous proteins. In the nucleus, many structural proteins, called histones, organize nuclear DNA into chromosomes (Annunziato 2008). This structure is highly conserved among eukaryotic cells. In mitochondria, the core component of mt nucleoids, are mainly nonhistone, high-mobility group (HMG) box proteins that show homology to the DNA-binding HMG proteins of nuclear chromatin (Diffley and Stillman 1991; Sasaki et al. 2003; Ekstrand, et al. 2004). These positively charged mtDNA-binding proteins contain two HMG boxes and are evolutionarily conserved from yeast to humans (Chen and Butow 2005; Stros et al. 2007), suggesting that putative prokaryotic mt nucleoid proteins from ancestral bacterium were completely replaced by eukaryote-type HMG box proteins at the early in the establishment of endosymbiosis. Meanwhile, mt HMG proteins have other functions in addition to their proposed roles in mtDNA packaging. They were initially identified as transcription factors that facilitate the assembly and promoter recognition of the mt transcriptional machinery (Fisher and Clayton 1988; Parisi and Clayton 1991; McCulloch and Shadel 2003). The mt HMG proteins, therefore, show dual functions, with some DNA-binding specificity that is related to the transcriptional function, as well as nonspecific DNA-binding properties related to packing mtDNA molecules.
HLP as a Prokaryotic Component of Cp Nucleoids
HLP is an important prokaryotic component used to elucidate the evolution of cp nucleoids. In the primitive red alga Cyanidioschyzon merolae, the hlp gene is encoded in the cp genome. Interestingly, in the cryptomonad Guillardia theta, which evolved via the secondary symbiosis of a red alga, the hlp gene is also found in the cp genome (Wang and Liu 1991; Gray 1992; Douglas and Penny 1999). In the glaucophyte Cyanophora paradoxa, the HLP gene was identified in the nuclear genome, suggesting that the translocation of the hlp gene from the cp genome to the nuclear genome occurred after the three groups (Glaucophyta, Rhodophyta, and Viridiplantae) branched (Price et al. 2012). Our phylogenetic analysis of algal HLPs indicates that KfHLP is more similar to glaucophytan or rhodophytan HLPs than to chlorophytan HLPs (supplementary fig. S13). Also, the analysis indicates that KfHLP evolved from the endosymbiont cyanobacteria, and it forms a monophyletic group with other chlorophytan HLPs (supplementary fig. S13). However, a previous phylogenetic analysis indicated that CreHLP is not related to the cyanobacterial or rhodophytan HLP proteins (Sato et al. 2003). There are two possible explanations for this phylogenetic relationship of algal HLPs. One is that only chlorophytan HLPs evolved at relatively rapid rates. Alternatively, chlorophytes might have acquired the gene for HLP from an unknown prokaryote during the early stage of chlorophytan evolution, as Sato et al. (2003) proposed.
SAP Domain-containing Protein
pTAC3 was originally identified as a component of the TAC fraction (Pfalz et al. 2006) and has been proposed to be an essential transcription factor that interacts with the bacterial-type plastid RNA polymerase (Yagi et al. 2012). Additionally, Majeran et al. (2012) suggested that pTAC3 was a good candidate for a core component of cp nucleoids because in their proteomic analysis, pTAC3 was highly abundant and nearly exclusively observed in nucleoids from the base, tip, and young leaves of maize. In this study, we identified pTAC3 homologs in K. flaccidum and M. polymorpha, and confirmed that they are localized to the cp nucleoid (Figs. 4 and 6). We also showed that KfpTAC3 has nonspecific DNA-binding activity in vitro and DNA-packing activity in E. coli (Fig. 5), supporting the idea that pTAC3 is a core component of cp nucleoids. On the other hand, in K. flaccidum, we also detected immunofluorescence signal of pTAC3 in the extra-cp nucleoid region (Fig. 4B). pTAC3 might be a bifunctional protein that is involved not only in the core of cp nucleoids but also in the regulation of gene expression (Yagi et al. 2012), in a manner similar to the mt HMG box protein, Abf2/TFAM (Kukat et al. 2011).
CreCNS was identified by our proteomic analysis as one of the most abundant proteins in the cp nucleoids of C. reinhardtii (table 1, supplementary tables S1 and S2). CreCNS contains a cp transit peptide and four SAP domains at the N-terminal region, which were sufficient for its localization to cp nucleoids (Fig. 1). The SAP domain region showed nonspecific DNA-binding activity in vitro (Fig. 2B) and DNA-packing activity in E. coli (Fig. 2C), indicating that CreCNS could be one of the core components of cp nucleoids in C. reinhardtii. CNS and pTAC3 might have evolved from a common eukaryotic ancestral gene because both proteins have similar functions in DNA packing with the same SAP DNA-binding domain. Our analysis using the BLAST algorithm identified homologous CNS genes in two green algae, Volvox carteri (Prochnik et al. 2010) and Coccomyxa subellipsoidea (Blanc et al. 2012), but not in other chlorophytes. Chlorophytan SAP domain-containing proteins might have evolved uniquely and at relatively rapid rates.
Whirly
Whirly proteins form a small protein family, mainly in the plant kingdom (Desveaux et al. 2005), and can localize to the nucleus, plastids, or mitochondria (Desveaux et al. 2005; Krupinska et al. 2013; Pfalz and Pfannschmidt 2015).
Whirly proteins preferentially bind to single-stranded DNA/RNA. Previous reports have indicated diverse functions for Whirly proteins. Whirly in eudicots has been reported to function as a nuclear transcription factor in salicylic acid signaling (Desveaux et al. 2000, 2004, 2005) or as a negative regulator of telomere length (Yoo et al. 2007). As for the functions in cps, Whirly 1 and 3 were found in the proteome study of TAC isolated from Arabidopsis cps (Pfalz et al. 2006). Whirly proteins are likely to be important for cp biogenesis, and the disruption of the genes leads to defective plastid development (Prikryl et al. 2008; Marechal et al. 2009). T-DNA insertion knock-out lines in A. thaliana revealed that Whirly could be responsible for maintaining the stability of the cp genome by inhibiting aberrant recombination events depending on micro homology (Marechal et al. 2009; Cappadocia et al. 2010). In maize, Whirly proteins are probably tethered to the thylakoid membrane by interacting with cpDNA, and they promote the splicing of cp atpF group II introns (Prikryl et al. 2008). Using a green fluorescent protein fusion, Krause et al. (2005) revealed that Whirly 1 proteins were co-localized with cp nucleoids in A. thaliana. Melonek et al. (2010) performed high-resolution imaging and reported that only a minor part of Whirly colocalizes with cp nucleoids. The localization pattern of Whirly was consistent with the biochemical assay, which showed that Whirly is not included in the core TAC fraction, called TAC-II (Melonek et al. 2010). A recent reverse genetic analysis using RNAi knocked-down lines in the barley Hordeum vulgare revealed that the suppression of Whirly caused an aberrant morphology in a subset of nucleoids in cps. Furthermore, overexpression of Whirly in E. coli cells exhibited a higher compaction of bacterial nucleoids, suggesting that Whirly has a function in compacting a subpopulation of cp nucleoids (Krupinska et al. 2014). Interestingly, Isemer et al. (2012) showed that recombinant Whirly1 could translocate from cps to the nucleus. Foyer et al. (2014) proposed that Whirly1 functions as a messenger that perceives and reports the redox status and other information from cps to the nucleus.
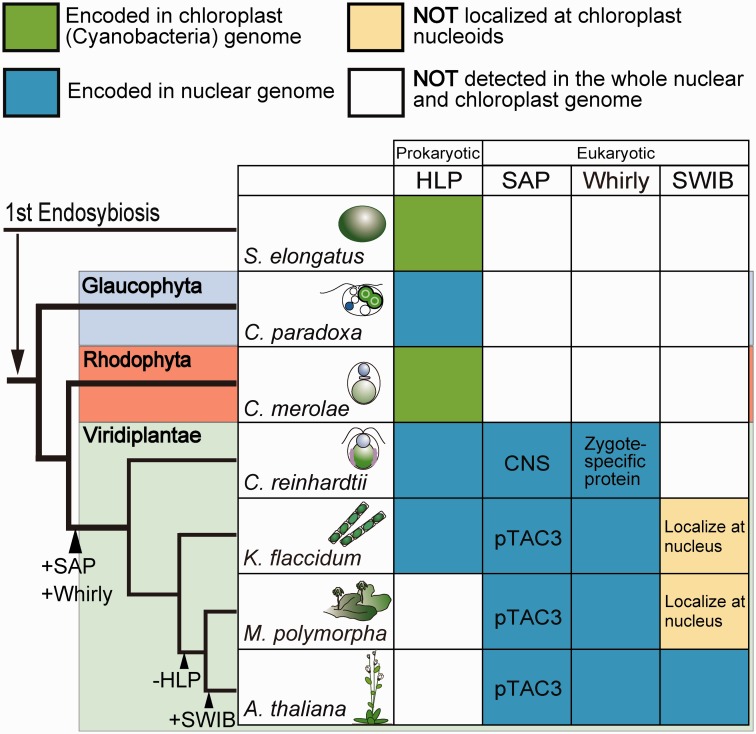
We identified genes homologous to Whirly in C. reinhardtii, K. flaccidum, and M. polymorpha, and determined whether the proteins are localized to cp nucleoids (Figs. 3, 4, and 6). Meanwhile, no homologous genes were found in Synechococcus elongates, C. paradoxa, or C. merolae (Fig. 7). These results suggest that the ancestral Whirly might have evolved as a eukaryotic component of cp nucleoids in green algae. In C. reinhardtii vegetative cells, however, the Whirly protein signals at the cp nucleoids were faint but became intense in the early zygotes (Fig. 3). This observation is consistent with a previous report showing that the expression of Whirly (EZY18) was drastically up-regulated in the young zygotes (Kubo et al. 2008). We note that the Whirly protein was not detected in all cp nucleoids and punctate immunofluorescence signals were also observed in extra-nucleoid (stroma) regions in the zygote. Although the actual function of Whirly in C. reinhardtii zygotes remains elusive, this expression and localization patterns imply that the roles of Whirly proteins in zygotes may be distinct from those in vegetative cells. The extra-nucleoid Whirly protein was also observed in K. flaccidum and M. polymorpha (Figs. 4 and 6). The elusive and complex localization of Whirly may represent its multifunctionality. Whirly in a subpopulation of cp nucleoids may function as the core (Krupinska et al. 2014), whereas Whirly in the extra-nucleoid region may regulate cp gene expression levels (Melonek et al. 2010) or function as a messenger in retrograde signaling (Isemer et al. 2012).
Fig. 7.—
Schematic view of the alternation of cp nucleoid components during plant evolution. The tree topology is based on Archibald (2009) and Becker and Marin (2009). Branch lengths do not represent phylogenetic distances.
SWIB
The SWIB domain is conserved in a subunit of the ATP-dependent chromatin-remodeling complex in the nucleus (SWI/SNF). An in silico analysis revealed homology between the SWIB and MDM2 domains (Bennett-Lovsey et al. 2002), which is an oncoprotein that tightly binds p53 tumor suppressor. MDM2 binds p53 at its transactivation domain to block its ability to activate transcription, promotes the nuclear export of p53, and may enhance p53 degradation by serving as a ubiquitin ligase (Michael and Oren 2003). Although no direct structural or biochemical information is available for the SWIB domain, it may also be involved in protein–protein interactions. The SWIB domain can be found almost exclusively in eukaryotes, with the exception of Chlamydia trachomatis, a parasitic bacterium. In C. trachomatis, the SWIB domain is fused to the C-terminus of DNA topoisomerase. Being a parasitic bacteria, C. trachomatis probably acquired the SWIB domain gene from a mammalian host by horizontal gene transfer (Stephens et al. 1998). In A. thaliana, SWIB-4 localizes to cp nucleoids, has a histone H1 domain adjacent to the SWIB domain, and has DNA-binding activity (Melonek et al. 2012), indicating that it would be one of the core architectural proteins of cp nucleoids.
In our research, we identified Group 1 SWIB genes in K. flaccidum and M. polymorpha (supplementary fig. S11). We determined their cellular localizations in the nucleus and in the cytosol, and none were detected in cp nucleoids, indicating that their ancestral functions might have been involved in regulating the nuclear chromatin structure or some the other nuclear genes (Figs. 4 and 6). These results imply that cp nucleoid-type SWIB proteins evolved through gene duplication and were conserved only among seed plants.
Evolutionary Scenario of Cp Nucleoid Proteins
Existing planta consists of three major groups: Glaucophyta, Rhodophyta (red algae), and Viridiplantae (green plants) (Archibald 2009; Becker and Marin 2009). Under a microscope, cp nucleoids may appear as similar dot-like structures in algae, liverworts, mosses, and seed plants (supplementary fig. S1); however, our research suggested the diversity of the cp nucleoid’s protein composition among plant lineages, with an overall trend in which eukaryotic factors originally related to nuclear chromatin remodeling have modified cp nucleoid organization since early stage of green plants. (Fig. 7). This process can be divided into three discrete steps: first, immediately after the branching of the three major groups, the ancestor of green plants acquired the eukaryotic SAP domain-containing and Whirly proteins for cp nucleoid organization. Secondly, the HLP gene, which has a bacterial origin (Haselkorn and Rouviere-Yaniv 1976) and may have been the original core component of cp nucleoids in algae, was lost at the initial stage of land plant evolution. Finally, seed plants gained other eukaryotic components for cp nucleoids, such as SWIB domain-containing proteins, for the further remodeling of cp nucleoids (Fig. 7). The cp nucleoids could be unique hybrids of prokaryotic and eukaryotic proteins that cannot be understood by simple analogies with bacterial nucleoids.
A similar situation was found in the gene expression machinery in cps. Recent genomic information reveals no homologs of bacterial transcription factors or bacterial nucleoid proteins are encoded in seed land plants, indicating the possibility that bacterial transcription factors might have been replaced by eukaryotic machinery during the land plant evolution (Yagi and Shiina 2014).
Cp nucleoids may have had the same fate as mt nucleoids because mt HMG box proteins, which are probably of eukaryotic origin, function as the major factors that govern mtDNA copy number, mt nucleoid structures, and mt gene expression levels in most eukaryotes, whereas no bacterial factors have been found in mt nucleoids (Sato 2001; Kukat and Larsson 2013).
Our understanding of the cp nucleoid is still at a preliminary stage, and further extensive analyses are required in the future, to elucidate the evolutionary history of cp nucleoid structures in more detail. To do so, the experimental conditions for the purification of cp nucleoids should be optimized, mutants defective in regulating cp nucleoid organization should be isolated, and crystal structures of cp nucleoid proteins should be resolved. These analyses would illuminate the further details and biological significance of cp nucleoid evolution that underlies the diversity of cp nucleoid compositions of green plants.
Supplementary Material
Supplementary figures S1–S13 and tables S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Dr Osami Misumi, Dr Tsuneyoshi Kuroiwa, Dr Masaki Odahara, Dr Atsushi Sakai, Dr Ryuji Tsugeki, Dr Haruko Kuroiwa, and Dr Kentaro Tamura (Kyoto University) for insightful and thoughtful advice. We are also grateful to Yumiko Sakai (Kyoto University) for her technical assistance in microscopy. This work was supported, in part, by the Ministry of Education, Culture, Sports, Science, and Technology of Japan grants to T.S. and Y.N., the Funding Program for Next Generation World-Leading Researchers (NEXT Program: GS015), Grant-in-Aid for Challenging Exploratory Research (26650111), Grant for Basic Research Projects from the Sumitomo Foundation, Core stage back-up program from Kyoto University to Y.N, Strategic Research Foundation Grant-aided Project for Private Universities to Y.N., and Grant-in-Aid for JSPS Fellows (grant 26 · 786 to Y.K.).
Literature Cited
- Allen JF. (2003). The function of genomes in bioenergetic organelles. Philos Trans R Soc Lond B Biol Sci. 358:19–37; discussion 37–18. doi:10.1098/rstb.2002.1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziato A. (2008). DNA packaging: nucleosomes and chromatin. Nat Educ. 1:26. [Google Scholar]
- Aravind L, Koonin EV. (2000). SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 25:112–114. [DOI] [PubMed] [Google Scholar]
- Archibald JM. 2009. The puzzle of plastid evolution. Curr Biol. 19:R81–R88. [DOI] [PubMed] [Google Scholar]
- Becker B, Marin B. 2009. Streptophyte algae and the origin of embryophytes. Ann Bot. 103:999–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett-Lovsey R, Hart SE, Shirai H, Mizuguchi K. 2002. The SWIB and the MDM2 domains are homologous and share a common fold. Bioinformatics 18:626–630. [DOI] [PubMed] [Google Scholar]
- Blanc G, et al. 2012. The genome of the polar eukaryotic microalga Coccomyxa subellipsoidea reveals traits of cold adaptation. Genome Biol. 13:R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogorad L. 2008. Evolution of early eukaryotic cells: genomes, proteomes, and compartments. Photosynth Res. 95:11–21 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Floyd SK, Sakakibara K. 2007. Green genes-comparative genomics of the green branch of life. Cell 129:229–234 [DOI] [PubMed] [Google Scholar]
- Cappadocia L, et al. 2010. Crystal structures of DNA-Whirly complexes and their role in Arabidopsis organelle genome repair. Plant Cell 22:1849–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XJ, Butow RA. 2005. The organization and inheritance of the mitochondrial genome. Nat Rev Genet. 6:815–825 [DOI] [PubMed] [Google Scholar]
- Chi-Ham CL, Keaton MA, Cannon GC, Heinhorst S. 2002. The DNA-compacting protein DCP68 from soybean chloroplasts is ferredoxin:sulfite reductase and co-localizes with the organellar nucleoid. Plant Mol Biol. 49:621–631. [DOI] [PubMed] [Google Scholar]
- Criscuolo A. 2011. morePhyML: improving the phylogenetic tree space exploration with PhyML 3. Mol Phylogenet Evol. 61:944–948. [DOI] [PubMed] [Google Scholar]
- Desveaux D, Despres C, Joyeux A, Subramaniam R, Brisson N. 2000. PBF-2 is a novel single-stranded DNA binding factor implicated in PR-10a gene activation in potato. Plant Cell 12:1477–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desveaux D, Marechal A, Brisson N. 2005. Whirly transcription factors: defense gene regulation and beyond. Trends Plant Sci. 10:95–102 [DOI] [PubMed] [Google Scholar]
- Desveaux D, et al. 2004. A “Whirly” transcription factor is required for salicylic acid-dependent disease resistance in Arabidopsis. Dev Cell 6:229–240. [DOI] [PubMed] [Google Scholar]
- Diffley JF, Stillman B. 1991. A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc Natl Acad Sci U S A. 88:7864–7868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon SC, Dorman CJ. 2010. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol. 8:185–195 [DOI] [PubMed] [Google Scholar]
- Douglas SE, Penny SL. 1999. The plastid genome of the cryptophyte alga, Guillardia theta: complete sequence and conserved synteny groups confirm its common ancestry with red algae. J Mol Evol. 48:236–244. [DOI] [PubMed] [Google Scholar]
- Ekstrand MI, et al. 2004. Mitochondrial transcription factor a regulates mtDNA copy number in mammals. Hum Mol Genet. 13:935–944. [DOI] [PubMed] [Google Scholar]
- Fisher RP, Clayton DA. 1988. Purification and characterization of human mitochondrial transcription factor 1. Mol Cell Biol. 8:3496–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Karpinska B, Krupinska K. 2014. The functions of WHIRLY1 and REDOX-RESPONSIVE TRANSCRIPTION FACTOR 1 in cross tolerance responses in plants: a hypothesis. Philos Trans R Soc Lond B Biol Sci. 369:20130226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW. 1992. The endosymbiont hypothesis revisited. Int Rev Cytol. 141:233–357. [DOI] [PubMed] [Google Scholar]
- Grove A. 2011. Functional evolution of bacterial histone-like HU proteins. Curr Issues Mol Biol. 13:1–12. [PubMed] [Google Scholar]
- Haselkorn R, Rouviere-Yaniv J. 1976. Cyanobacterial DNA-binding protein related to Escherichia coli HU. Proc Natl Acad Sci U S A. 73:1917–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, et al. 2014. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat Commun. 5:3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isemer R, et al. 2012. Recombinant Whirly1 translocates from transplastomic chloroplasts to the nucleus. FEBS Lett. 586:85–88 [DOI] [PubMed] [Google Scholar]
- Ishizaki K, et al. 2015. Development of Gateway Binary Vector Series with Four Different Selection Markers for the Liverwort Marchantia polymorpha. PLoS One 10(9):e0138876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcher D, Koster D, Schadach A, Klevesath A, Bock R. 2009. The Chlamydomonas chloroplast HLP protein is required for nucleoid organization and genome maintenance. Mol Plant 2:1223–1232. [DOI] [PubMed] [Google Scholar]
- Keeling PJ. 2010. The endosymbiotic origin, diversification and fate of plastids. Philos Trans R Soc Lond B Biol Sci. 365:729–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, et al. 2002. Detection and localization of a chloroplast-encoded HU-like protein that organizes chloroplast nucleoids. Plant Cell 14:1579–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause K, et al. 2005. DNA-binding proteins of the Whirly family in Arabidopsis thaliana are targeted to the organelles. FEBS Lett. 579:3707–3712. [DOI] [PubMed] [Google Scholar]
- Krupinska K, Melonek J, Krause K. 2013. New insights into plastid nucleoid structure and functionality. Planta 237:653–664. [DOI] [PubMed] [Google Scholar]
- Krupinska K, et al. 2014. WHIRLY1 is a major organizer of chloroplast nucleoids. Front Plant Sci. 5:432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T, et al. 2008. Characterization of novel genes induced by sexual adhesion and gamete fusion and of their transcriptional regulation in Chlamydomonas reinhardtii. Plant Cell Physiol. 49:981–993. [DOI] [PubMed] [Google Scholar]
- Kubota A, Ishizaki K, Hosaka M, Kohchi T. 2013. Efficient Agrobacterium-mediated transformation of the liverwort Marchantia polymorpha using regenerating thalli. Biosci Biotechnol Biochem. 77:167–172. [DOI] [PubMed] [Google Scholar]
- Kukat C, Larsson NG. 2013. mtDNA makes a U-turn for the mitochondrial nucleoid. Trends Cell Biol. 23:457–463. [DOI] [PubMed] [Google Scholar]
- Kukat C, et al. 2011. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc Natl Acad Sci U S A. 108:13534–13539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa T. 1991. The replication, differentiation, and inheritance of plastids with emphasis on the concept of organelle nuclei. Int Rev Cytol. 128:1–62. [Google Scholar]
- Lewis LA, McCourt RM. 2004. Green algae and the origin of land plants. Am J Bot. 91:1535–1556. [DOI] [PubMed] [Google Scholar]
- Majeran W, et al. 2012. Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: a new conceptual framework for nucleoid functions. Plant Physiol. 158:156–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal A, et al. 2009. Whirly proteins maintain plastid genome stability in Arabidopsis. Proc Natl Acad Sci U S A. 106:14693–14698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch V, Shadel GS. 2003. Human mitochondrial transcription factor B1 interacts with the C-terminal activation region of h-mtTFA and stimulates transcription independently of its RNA methyltransferase activity. Mol Cell Biol. 23:5816–5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melonek J, Matros A, Trosch M, Mock HP, Krupinska K. 2012. The core of chloroplast nucleoids contains architectural SWIB domain proteins. Plant Cell 24:3060–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melonek J, et al. 2010. Whirly1 in chloroplasts associates with intron containing RNAs and rarely co-localizes with nucleoids. Planta 232:471–481. [DOI] [PubMed] [Google Scholar]
- Michael D, Oren M. 2003. The p53-Mdm2 module and the ubiquitin system. Semin Cancer Biol. 13:49–58. [DOI] [PubMed] [Google Scholar]
- Nishimura Y, et al. 2002. An mt(+) gamete-specific nuclease that targets mt(-) chloroplasts during sexual reproduction in C. reinhardtii. Genes Dev. 16:1116–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi MA, Clayton DA. 1991. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science 252:965–969. [DOI] [PubMed] [Google Scholar]
- Patron NJ, Durnford DG, Kopriva S. 2008. Sulfate assimilation in eukaryotes: fusions, relocations and lateral transfers. BMC Evol Biol. 8:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmuller R. 2006. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18:176–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Pfannschmidt T. 2013. Essential nucleoid proteins in early chloroplast development. Trends Plant Sci. 18:186–194. [DOI] [PubMed] [Google Scholar]
- Pfalz J, Pfannschmidt T. 2015. Plastid nucleoids: evolutionary reconstruction of a DNA/protein structure with prokaryotic ancestry. Front Plant Sci. 6:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powikrowska M, Oetke S, Jensen PE, Krupinska K. 2014. Dynamic composition, shaping and organization of plastid nucleoids. Front Plant Sci. 5:424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DC, et al. 2012. Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science 335:843–847. [DOI] [PubMed] [Google Scholar]
- Prikryl J, Watkins KP, Friso G, van Wijk KJ, Barkan A. 2008. A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res. 36:5152–5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochnik SE, et al. 2010. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 329:223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A, Takano H, Kuroiwa T. 2004. Organelle nuclei in higher plants: structure, composition, function, and evolution. Int Rev Cytol. 238:59–118. [DOI] [PubMed] [Google Scholar]
- Sasaki N, et al. 2003. Glom is a novel mitochondrial DNA packaging protein in Physarum polycephalum and causes intense chromatin condensation without suppressing DNA functions. Mol Biol Cell. 14:4758–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N. 2001. Was the evolution of plastid genetic machinery discontinuous? Trends Plant Sci. 6:151–155. [DOI] [PubMed] [Google Scholar]
- Sato N, Nakayama M, Hase T. 2001. The 70-kDa major DNA-compacting protein of the chloroplast nucleoid is sulfite reductase. FEBS Lett. 487:347–350. [DOI] [PubMed] [Google Scholar]
- Sato N, Terasawa K, Miyajima K, Kabeya Y. 2003. Organization, developmental dynamics, and evolution of plastid nucleoids. Int Rev Cytol. 232:217–262. [DOI] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 95:5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist CJ, et al. 2013. New and continuing developments at PROSITE. Nucleic Acids Res. 41:D344–D347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens RS, et al. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754–759. [DOI] [PubMed] [Google Scholar]
- Stern DB, Goldschmidt-Clermont M, Hanson MR. 2010. Chloroplast RNA metabolism. Annu Rev Plant Biol. 61:125–155. [DOI] [PubMed] [Google Scholar]
- Stros M, Launholt D, Grasser KD. 2007. The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell Mol Life Sci. 64:2590–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W. 2004. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 5:123–135. [DOI] [PubMed] [Google Scholar]
- Wang SL, Liu XQ. 1991. The plastid genome of Cryptomonas phi encodes an hsp70-like protein, a histone-like protein, and an acyl carrier protein. Proc Natl Acad Sci U S A. 88:10783–10787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Li GW, Chen C, Xie XS, Zhuang X. 2011. Chromosome organization by a nucleoid-associated protein in live bacteria. Science 333:1445–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y, Ishizaki Y, Nakahira Y, Tozawa Y, Shiina T. 2012. Eukaryotic-type plastid nucleoid protein pTAC3 is essential for transcription by the bacterial-type plastid RNA polymerase. Proc Natl Acad Sci U S A. 109:7541–7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y, Shiina T. 2014. Recent advances in the study of chloroplast gene expression and its evolution. Front Plant Sci. 5:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HH, Kwon C, Lee MM, Chung IK. 2007. Single-stranded DNA binding factor AtWHY1 modulates telomere length homeostasis in Arabidopsis. Plant J. 49:442–451. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.