Abstract
Bathyal cold seeps are isolated extreme deep-sea environments characterized by low species diversity while biomass can be high. The Håkon Mosby mud volcano (Barents Sea, 1,280 m) is a rather stable chemosynthetic driven habitat characterized by prominent surface bacterial mats with high sulfide concentrations and low oxygen levels. Here, the nematode Halomonhystera hermesi thrives in high abundances (11,000 individuals 10 cm−2). Halomonhystera hermesi is a member of the intertidal Halomonhystera disjuncta species complex that includes five cryptic species (GD1-5). GD1-5’s common habitat is characterized by strong environmental fluctuations. Here, we compared the transcriptomes of H. hermesi and GD1, H. hermesi’s closest relative. Genes encoding proteins involved in oxidative phosphorylation are more strongly expressed in H. hermesi than in GD1, and many genes were only observed in H. hermesi while being completely absent in GD1. Both observations could in part be attributed to high sulfide concentrations and low oxygen levels. Additionally, fatty acid elongation was also prominent in H. hermesi confirming the importance of highly unsaturated fatty acids in this species. Significant higher amounts of transcription factors and genes involved in signaling receptor activity were observed in GD1 (many of which were completely absent in H. hermesi), allowing fast signaling and transcriptional reprogramming which can mediate survival in dynamic intertidal environments. GC content was approximately 8% higher in H. hermesi coding unigenes resulting in differential codon usage between both species and a higher proportion of amino acids with GC-rich codons in H. hermesi. In general our results showed that most pathways were active in both environments and that only three genes are under natural selection. This indicates that also plasticity should be taken in consideration in the evolutionary history of Halomonhystera species. Such plasticity, as well as possible preadaptation to low oxygen and high sulfide levels might have played an important role in the establishment of a cold-seep Halomonhystera population.
Keywords: transcriptomics, adaptive radiation, phenotypic plasticity, evolutionary biology, cryptic species, nematodes
Introduction
One of the most important goals in evolutionary biology is to comprehend how and why living organisms diversify (Pfennig et al. 2010). Dispersal allows organisms to colonize new suitable habitats and found new populations. Speciation in the new habitat has its origin in the establishment of reproductive barriers between source and founder population (Coyne and Orr 2004). While gene flow between populations is often considered as a homogenizing evolutionary force that can prevent conspecific populations from diverging (Walsh and Mena 2013), ecological selection can also initiate speciation in the presence of gene flow (Seehausen et al. 2014). Therefore, restriction of gene flow and ecological differentiation between habitats are two drivers which can mediate speciation, thereby, paving the way for adaptive evolution of newly founded populations (Mayr 1963).
Adaptive evolution is the process in which natural selection will select for advantageous alleles that provide a higher fitness to the organism thereby causing changes in allele frequencies in a population. Adaptive evolution is influenced by mutation rate, recombination, genetic drift, selection, etc. (reviewed in Olson-Manning et al. 2012) each of which cause genetic patterns that are traceable in sequence data. In addition, individuals will respond to changes in their environment by remodeling their transcriptome, allowing individuals to quickly respond to environmental conditions. This is referred to as phenotypic plasticity where one genotype can produce multiple phenotypes. While some studies define phenotypic plasticity as nonheritable (Ellner et al. 2011), gene expression can also be regulated by epigenetic modifications which are heritable (Surani et al. 1993; Cubas et al. 1999; Chong and Whitelaw 2004; Bird 2007; Goldberg et al. 2007). In the case of cryptic species, that is, morphologically identical but genetically distinct species, the degree of plasticity in morphological traits is limited, despite the fact that they can be found in quite distinct habitats (Vrijenhoek 2009). Understanding the biology of cryptic species in different environments requires information on genetic adaptive traits and the degree of plasticity in other traits than morphology (Gittenberger and Gittenberger 2011).
Cryptic diversity is common in marine environments, which may be attributed to the use of chemical cues for mate recognition (Stanhope et al. 1992; Palumbi 1994; Lonsdale et al. 1998). The existence of cryptic species has been described in different orders of marine nematodes (Derycke et al. 2005, 2007, 2010), usually the most abundant and diverse Metazoa in marine benthic communities (Heip et al. 1985; Lambshead and Boucher 2003). In this work, we focus on the genus Halomonhystera Andrássy (2006) belonging to the order of the Monhysterida which is currently situated outside the five clades (I–V) proposed by Blaxter (2011) and placed in clade 5c based on the numerical system of van Megen et al. (2009). The genus has a widespread geographical distribution and has been identified from deep-sea environments (Van Gaever et al. 2006; Portnova et al. 2010) and intertidal regions (Trotter and Webster 1983; Vranken, Herman, et al. 1988; Mokievsky et al. 2005; Derycke et al. 2007). Nuclear and mitochondrial sequence data led to the discovery of five cryptic Halomonhystera disjuncta species (GD1-5) with only limited morphometric differences in the Western Scheldt estuary (Derycke et al. 2007; Fonseca et al. 2008). Halomonhystera was also reported as the dominant nematode genus in the sulfide-rich bacterial mats of the Nyegga pockmark (Nordic Norwegian margin, 730 m) (Portnova et al. 2010) and the Håkon Mosby mud volcano (HMMV, 1,280 m, Barent Sea slope) (Van Gaever et al. 2006). However, a recent study revealed subtle morphological and clear genetic differences between the HMMV and intertidal Halomonhystera species (Van Campenhout et al. 2013): uncorrected p-distances between HMMV and GD1 ranged between 19.1% and 25.2% for the cytochrome oxidase c subunit I gene part, whereas uncorrected p-distances of the nuclear 18S gene ranged between 2.4% and 2.9% and the nuclear ITS-D2D3 concatenated genes showed a divergence of 9.5% (Van Campenhout et al. 2013). The HMMV Halomonhystera has consequently been described as a new species, Halomonhystera hermesi (Tchesunov et al. 2015). Phylogenetic analysis revealed a close relationship between H. hermesi and both GD1 and GD4 (Van Campenhout et al. 2013). For Halomonhystera, a deep-sea invasion from intertidal regions has been hypothesized followed by adaptive evolution as a result of absence of gene flow between both environments (Van Campenhout et al. 2013) and strong selective forces in the deep-sea habitat. GD1 is hypothesized to share the most recent ancestor with H. hermesi as it appears to be more resistant to bathyal seep conditions compared with GD2-3 (Van Campenhout et al. 2014). In addition, previous studies have revealed a high tolerance of H. disjuncta to low food- and high heavy metal concentrations (Vranken et al. 1985, 1989; Vranken, Tire, et al. 1988). These studies clearly highlight that in addition to the genetic differences between GD1 and H. hermesi, the intertidal species also have the plasticity to survive deep-sea conditions. Halomonhystera hermesi thrives at the HMMV, a cold, stable chemosynthetically based deep-sea environment (Van Gaever et al. 2006). The microbial mats, the habitat of H. hermesi, are characterized by limited oxygen penetration (∼1 mm) and high sulfide concentrations (up to 4 mM) (de Beer et al. 2006; Van Gaever et al. 2006). Even though episodic events such as extensive methane venting and mud flows occur, the upflow velocity in the bacterial mats is limited to 0.3–1.0 m yr-1 (de Beer et al. 2006). In contrast, GD1 thrives on decaying algae in the Western Scheldt, an ephemeral habitat with daily fluctuations in temperature, light, salinity, and frequent inundation due to tidal activity. We, therefore, consider the HMMV habitat to be more stable compared with the intertidal habitat of GD1.
The detailed knowledge on the biology and phylogenetic relationships of these two species, together with their occurrence in well characterized and distinct habitats render them an excellent system to study the molecular mechanisms allowing species to thrive in different habitats. To this end, we compared the transcriptome between the intertidal (GD1) and deep-sea nematode (H. hermesi) related species. Evaluation of gene expression will contribute to our understanding of which pathways are important in both species with respect to their environment. Additionally, pinpointing genes that are under natural selection will contribute to our understanding of adaptive evolution of both species. Moreover, its intertidal occurrence subjects GD1 to strong physical, chemical, and biological gradients indicating that it can rapidly adjust to environmental changes. Organisms in fluctuating environments must constantly sense and adapt to environmental changes and must be able to quickly respond to these changes. This may be achieved by transcriptional regulation (Kussell and Leibler 2005). In contrast, stable environments generally favor specialist species adapted to that specific niche (Miner 2005). In view of the clear differences in (a)biotic conditions in both habitats, we expected to find substantial differences in the molecular pathways activated in both habitats. More specifically, we expected to find one/more genes expressed and more active pathways in the intertidal than in the deep-sea habitat, and two/an overrepresentation of pathways involved in transcription and environmental information processing in the intertidal GD1 species.
Materials and Methods
Sample Collection
Deep-sea sediment samples were collected, using a TV-guided multicorer, from the active methane seeping Håkon Mosby mud volcano (HMMV; situated west of the Barents Sea (72.1°N 14.73°E) at an average water depth of 1,280 m) during the Maria S. Merian cruise 2010 (MSM 16/2). Core samples were immediately sliced from 0 to 10 cm (with a 1-cm interval), instantly frozen in liquid nitrogen on board and stored at −80 °C.
Intertidal monospecific GD1 nematodes reared in the lab were transferred to rehydrated (salinity of 25 practical salinity unit [psu]) defaunated macroalgae (Fucus vesiculosis) and allowed to grow for approximately 4 weeks in three litter bags (mesh size 200 µm) at 8 °C and a salinity of 25 psu. Following this period, litter bags were positioned in the Paulina salt marsh (Western Scheldt, The Netherlands, at 51° 20.9′N, 3° 43.5′E) allowing nematodes to acclimatize to the environment for 72 h before retrieval. Macroalgae were washed over two stacked sieves (top sieve: 1 mm, bottom sieve: 32 µm) with sieved (32 µm) seawater and fauna was retained on the 32 µm sieve. Meiofauna from the sieve was then collected with sterile artificial seawater (25 psu) and replicate samples were immediately frozen in liquid nitrogen and stored at −80 °C.
Nematode Retrieval and RNA Isolation
The frozen first deep-sea sediment slice (0–1 cm) of three locations (Station MSM16-2_802-1, 72° 0.17′N 14° 43.88′E; Station MSM16-2_829-1, 72° 0.16′ N, 14° 43.94′ E; Station MSM16-2_831-1, 72° 0.14′ N 14° 43.94′ E) and all intertidal samples were washed separately over a 32 µm-mesh sieve. Halomonhystera nematodes were extracted by density gradient centrifugation at 4 °C and 3,000 × g, using Ludox (a colloidal silica polymer; specific gravity 1.18) as a flotation medium (Heip et al. 1985). β-mercaptoethanol, in a final concentration of 0.143 M, was added to avoid RNA breakdown. Nematodes were captured on a 32 µm sieve after centrifugation and washed again with a sterile formamide-β-mercaptoethanol (0.143 M) solution and finally retained in the same solution. Replicate samples of both deep-sea and intertidal samples contained approximately 1,500–2,000 nematodes. To minimize the effect of different life stages, only adult H. hermesi and GD1 nematodes of each replicate were morphologically identified and randomly and manually picked out using a binocular microscope in a 4 °C climate room. Samples from GD1 or H. hermesi were pooled, prior to RNA extraction, into 50 µl RLT buffer (RNeasy Mini kit, Qiagen Inc., Düsseldorf, Germany) to which we added β-mercaptoethanol in a final concentration of 0.143 M. Each sample (deep sea and intertidal) contained approximately 5,000 nematodes and such pooling of replicates was a necessary practical consideration to obtain sufficient RNA.
The cuticle and cell membranes of nematodes and cells were disrupted by vortexing with beads (1 min at 5,000 rpm). Total RNA was extracted using the RNeasy Mini kit (Qiagen Inc., Düsseldorf, Germany) according to the manufacturer’s instructions. Genomic DNA was removed by on-column digest with DNase I. RNA was quantified by measuring the optical density (OD) at 260 nm using a NanoDrop spectrophotometer (2000 UV-Vis, Thermo Fisher Scientific, Inc.). RNA purity was assessed at an absorbance ratio of OD260/280 and OD260/230. RNA integrity was analyzed on a 1% agarose gel stained with ethidium bromide.
cDNA Library Construction, Illumina Sequencing, and Quality Filtering
Samples were sent to Bio S&T Inc. (Canada) to perform cDNA synthesis and normalization allowing us to sequence low expressed genes. Briefly, the cDNA library was constructed with about 8 µg of total RNA using a modified Clonetech SMART cDNA Library Construction kit using oligo(dT) primers. Double stranded cDNAs were obtained by primer extension and purified. To increase the ability to sequence low expressed transcripts, cDNAs were normalized, reamplified, and purified (Bio S&T, Canada). Further library construction and sequencing were performed by Genomics Core, UZ Leuven, Belgium. Briefly, the cDNA was sheared using the Covaris M220 machine with a maximum fragment length of 800 bp and a fragment size peak around 450–500 bp. cDNA libraries were end repaired, and NEXTflex adaptors from BIOO Scientific were ligated using the SPRIworks robot (Beckman) followed by an AMPure bead purification to remove adaptor dimers. Libraries were amplified using a 12 cycle polymerase chain reaction step with NEXTflex primer mix. An additional AMPure bead purification was performed. The resulting cDNA libraries were pooled and paired-end sequenced on a single Illumina MiSeq 2*250 bp lane. Because of a low read diversity within deep-sea reads (troubling for the nucleotide incorporation imaging software), this cDNA library was mixed in with other samples and sequenced on the same Illumina MiSeq 2*250 bp machine. Data from both runs were pooled.
Low quality read ends (<Q20) were trimmed with FastX 0.0.13 (http://hannonlab.cshl.edu/fastx_toolkit, last accessed November 1, 2015). MiSeq adapters were trimmed only at the end (at least 10 bp overlap and 90% match) with cutadapt 1.0 (Martin 2011). Using FastX 0.0.13 and ShortRead 1.14.4 (Morgan et al. 2009), short reads (length <50 bp), poly A-reads (more than 90% of the bases equal A), ambiguous reads (containing N), low quality reads (more than 50% of the bases <Q25), and artifact reads (all but three bases in the read equal one base type) were removed in succession. Using bowtie 2.0.0-beta5 (Langmead et al. 2009), reads that align to PhiX (an adapter-ligated library used as a control for Illumina sequencing runs) or human hg19 were identified and removed. Because of quality drop for all reverse reads after 150 bp, these reads were all cut after 150 bp. Finally, cDNA primers were removed using the CLC Genomics Workbench 6.0.3 trim sequences tool. The raw sequencing reads were deposited at the Short Read Archive of the National Center for Biotechnology Information (NCBI) under bioproject SRP050898 containing biosamples SRS748702 (Run SRR1657929) and SRS748703 (Run SRR1657930).
De Novo Assembly and BLAST
De novo assembly was conducted using the CLC Genomics Workbench 6.0.3 de novo assembly tool according to the manufacturer’s preset features. Contigs shorter than 200 bp were removed. To remove redundant transcripts and retain a set of contigs each representing a putatively unique isoform (unigenes), the open-source program CD-HIT-EST (Li and Godzik 2006) was used to cluster contigs at a 95% sequence identity. The largest contig of each cluster was retained in the data set. The coding sequences (CDS) were predicted using the open reading frame (ORF)-predictor server (Min et al. 2005) with a 200 bp CDS cutoff.
BLAST + (Camacho et al. 2009) was used to subject the remaining unigenes to a similarity search against NCBI’s nonredundant (nr) database (standard genetic code) using the BLASTx algorithm (Altschul et al. 1990), with a cutoff e-value of ≤ 10−3 and a maximum of 20 BLAST hits. The top BLASTx hit was used to assign taxonomy to each unigene using a custom perl script (Get_classification.pl, Supplementary Material online). Unigenes classified as Nematoda were selected for further analysis.
Functional Annotation (KEGG) and Pathway Reconstruction
Prediction of pathways expressed in both data sets was done by using known metabolic and signaling pathways previously found in Eukaryotes and nematodes using the Kyoto Encyclopedia of Genes and Genomes (KEGG) automated Annotation Server (KAAS) (Moriya et al. 2007). Unigenes were submitted to KAAS with the preset Eukaryotes. Caenorhabditis briggsae, Brugia malayi, Loa loa, and Trichinella spiralis were added as additional sequence templates; the option “basis of single-best hit” was selected. KAAS annotates transcripts with KEGG orthology (KO) identifiers representing an orthologous group of genes linked to an object in the KEGG pathways and BRITE hierarchy (Mao et al. 2005; Moriya et al. 2007). Unigenes from both transcriptomes with the same KO identifier, that is, functional annotation, were considered to be shared between both species.
The KO identifiers were mapped against KEGG pathways to reconstruct pathways (http://www.genome.jp/kegg/mapper.html, last accessed April 30, 2015). KEGG pathways involved in human diseases were not taken into further consideration. KO identifiers were additionally mapped against BRITE hierarchical classifications of protein families.
To extract pathways that are over- and underrepresented in the deep-sea and intertidal habitats, respectively, KEGG pathways and protein families of unique unigenes from each transcriptome were determined separately using the Cytoscape (Shannon et al. 2003) plugin BINGO (Maere et al. 2005) with both transcriptomes combined as a reference set. P values were false-discovery rate (FDR)-corrected.
Functional Gene Ontology Annotation
To retrieve Gene Ontology (GO) terms describing biological processes, molecular functions, and cellular components (Ashburner et al. 2000), the publicly available BLAST2GO-platform (Conesa et al. 2005) was used. Annotations were conducted for unigenes with default parameters (e-value ≥ 10−6, Annotation cutoff ≥55, and a GO weight of 5). BLAST2GO was also used to retrieve InterPro (conserved patterns in sequences) annotations and merged with GO terms for a wide functional range cover in annotation.
Because some unigenes without KO annotation displayed a GO annotation, the following step was taken to increase the amount of previously defined shared unigenes (based on KO identifier). Unigenes without KO identifier but with a GO annotation and a CDS of at least 200 bp from one transcriptome were aligned against both the shared unigenes and unigenes without KEGG annotation of the other transcriptome using the PROmer pipeline of the MUMmer 3.0 software (Kurtz et al. 2004) with default parameters. Unigenes with an in-frame PROmer hit, minimum similarity of 75% and an alignment length of minimum 300 bp were additionally considered as shared. Determining overrepresented GO-terms in unique unigenes was performed as described before. Similarly, we used BINGO to determine whether each complete GO data set was enriched in level 2 GO terms compared with both data sets combined.
Positive Selection, Amino Acid Composition, and Codon Usage
Orthologs present in both transcriptomes were determined by selecting unigenes with a CDS of minimal 200 bp which were then aligned and compared using the PROmer pipeline with default parameters. We only retained in-frame PROmer hits (similarity ≥ 75% and an alignment length of 300 bp) that matched the same KO identifier. The full length CDS of filtered ortologous gene pairs were then aligned using ClustalW (Thompson et al. 1994), with default parameters, provided in the Molecular Evolutionary Genetics Analysis version 6.0 (MEGA6) software package (Tamura et al. 2013) and uncorrected p-distances were calculated.
Positive selection was tested using the ratios of the rates of nonsynonymous substitutions per nonsynonymous sites (Ka) to the number of synonymous substitutions per synonymous site (Ks) using KaKs_Calculator 2.0 (Wang et al. 2010) with the γ-MYN model. This model incorporates previously unaddressed dynamic features of evolving DNA sequences in transition/transversion rate, nucleotide frequency, and unequal transitional substitution. The model further accounts for DNA sequences unequal substitution rates among different sites (Wang et al. 2009).
The alignment of orthologous gene pairs was also used to analyze GC content, amino acid (AA) composition, and codon usage. Amino acid frequencies were determined by summing the occurrence of each AA per translated CDS (tCDS) and dividing it through the total amount of AA present for each tCDS. Codon frequencies per AA were obtained by summing the occurrence of each single codon per tCDS and dividing it through the respective AA count of the tCDS. GC content, AA composition, and codon frequencies per gene were determined using a custom perl script (AA_counter.pl and Codon_counter.pl, Supplementary Material online) and averaged over all unigenes. To investigate whether significant differences were present between deep-sea and intertidal transcriptomes, mean values of both H. hermesi and GD1 corresponding orthologous unigenes were subtracted. The H. hermesi and GD1 data sets were then pooled and randomly permuted into two equal groups after which GC content, AA- and codon frequencies were calculated and subtracted from each other. This was repeated 100,000 times with custom R-scripts (GC_permutation.R, AA_permutation.R and Codon_permutation.R, Supplementary Material online) in R version 3.0.2 (R Core Team 2013) and the obtained values were seen as a null distribution for the GC content, codon, and AA frequencies. The P value is then calculated as the fraction of how many times the permuted difference is equal or more extreme than the observed difference and was FDR-corrected. If the observed difference fell outside the range of the null distribution, no P values could be calculated and P values were considered to be <E−6.
Differential Gene Expression between Shared Unigenes
Deep-sea and intertidal contigs with a nematode top-BLAST hit were selected and used to generate a single new assembly using the de novo assembly tool in CLC Genomics Workbench 6.0.3. The assembly was treated in the same manner as described above (CD-HIT-EST clustering, BLASTx and functional annotation). Cleaned deep-sea and intertidal reads were mapped against the full assembly using the RNA-seq tool in CLC Genomics Workbench 6.0.3. (minimum length fraction = 0.9 and minimum similarity fraction = 0.8). Values for relative expression were based on mean RPKM (Reads Per Kilo-base of exon model per Million mapped reads) values. A statistical analysis on proportions (Kal’s test) was performed and P values were FDR-corrected. The RNA-seq analysis resulted in five data sets: 1) uniquely expressed genes in H. hermesi, 2) uniquely expressed genes in GD1, 3) shared unigenes similarly expressed in both species, 4) differentially overexpressed unigenes in H. hermesi, and 5) differentially overexpressed unigenes in GD1. Data set 1 and 2 were not analyzed further as this was already performed for the complete transcriptome. To increase the validity of the three other RNA-seq data sets, only unigenes with a KO identifier which was present in both original nematode transcriptomes and present in only one RNA-seq data set were retained. Over- and underrepresented KEGG pathways, protein families, and GO annotations of differentially expressed genes were determined as described above.
Results
Sequencing Results
Illumina MiSeq 2*250 bp cDNA sequencing generated a total of 3.47 and 2.47 Gb for the deep-sea and intertidal samples respectively (supplementary table S1, Supplementary Material online). After quality assessment and data filtering, 4,605,081 and 3,105,224 pair-end reads remained for deep-sea and intertidal samples, respectively, (supplementary table S1, Supplementary Material online). De novo assembly yielded a lower amount of deep-sea contigs (57,636) than intertidal contigs (72,678). In terms of unigenes, 54,767 deep-sea and 68,537 intertidal unigenes with a respective N50 of 933 and 720 bp were found. The size of the deep-sea unigenes ranged from 200 to 149,303 bp with a mean length of 750 whereas the intertidal unigenes ranged from 200 to 8,783 bp with a mean length of 618 bp (supplementary table S1, Supplementary Material online). It must be noted that the large unigenes within the deep-sea assembly are most likely partially assembled prokaryotic chromosomes. ORF prediction (>200 bp CDS in length) was very similar for deep-sea (81.45%) and intertidal (83.47%) unigenes.
BLAST Results and Classification
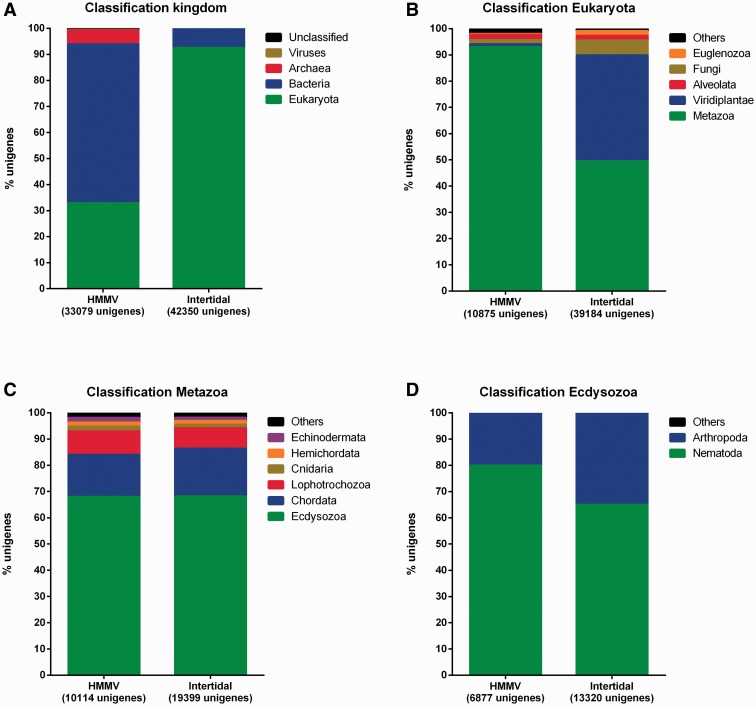
BLASTx results showed that 33,079 (60.40%) deep-sea and 42,409 (61.89%) intertidal unigenes had at least one BLAST result with an e-value ≤ 10−3. Surprisingly, a substantial portion of the unigenes did not give a nematode top-blast hit. Instead, most deep-sea unigenes classified as Bacteria (61.01%) whereas intertidal unigenes were mostly classified as Eukaryotes (92.52%) (fig. 1A). Of the intertidal unigenes that were classified as Eukaryota, only 49.51% could be assigned to the Metazoa (fig. 1B). The remaining part (40.31%) mainly belonged to Viridiplantae. In contrast, the deep-sea eukaryote unigenes were nearly all assigned to the Metazoa (93%; fig. 1B). Within the Metazoa, the composition of both data sets was highly similar: 67.99% deep-sea and 68.15% intertidal unigenes could be assigned to the Ecdysozoa, 16.02% deep-sea and 18.19% intertidal unigenes were assigned to the Chordata, and 8.95% deep-sea and 7.80% intertidal unigenes were designated as Lophotrochozoa (fig. 1C). The majority of the Ecdysozoa unigenes were classified as nematodes: 5,498 (79.95%) unigenes for the deep-sea and 8,588 (64.96%) for the intertidal data set (fig. 1D). Overall, 16.62% and 20.28% of the deep-sea and intertidal unigenes, respectively, were assigned to nematodes.
Fig. 1.—
Top BLAST classification of HMMV and intertidal unigenes resulting from the de novo transcriptome assembly. Classification was based on the top-BLAST hit (BLASTx) after removal of redundant and non-ORF contigs. (A) Kingdom classification, (B) Eukaryota classification, (C) Metazoa classification, and (D) Ecdysozoa classification. Taxonomic groups containing less than1% of the total amount of unigenes were combined with unigenes without a classification and/or an environmental classification, and plotted under unclassified (A) or others (B–D).
Nematode Assembly
Based on their nematode top-BLAST hit 5,498 deep-sea (H. hermesi) and 8,588 intertidal (GD1) unigenes were retained for further analysis. Halomonhysterahermesi unigenes had a maximum length of 7,235 bp and an average length of 943.07 bp. This was comparable to GD1 unigenes which had a maximum length of 8,783 bp and an average length of 837.39 bp (table 1). The amount of unigenes with a CDS longer than 200 bp was lower for H. hermesi (5,227) than for GD1 (8,102). GC content of the H. hermesi unigenes was higher than that of the GD1 unigenes (56.11% vs. 48.71%, respectively).
Table 1.
Summary of BLAST Results, Assembly Statistics, ORF Prediction, and Annotation of Unigenes with a Nematode Top BLAST Hit of Two Targeted Locations and Species: HMMV (deep sea, Halomonhystera hermesi) and Western Scheldt (intertidal, GD1)
| Stage | Deep Sea | Intertidal | Total | |
|---|---|---|---|---|
| BLAST result | Unigenes with at least one BLAST result | 33,079 | 42,409 | 75,429 |
| Nematode assembly | Contigs with nematode top BLAST hit (unigenes) | 5,498 | 8,588 | 14,086 |
| N50 length (bp) | 1,378 | 1,169 | ||
| Max length (bp) | 7,253 | 8,783 | ||
| Average length (bp) | 943.07 | 837.39 | ||
| Average coverage per base | 14.07 | 20.43 | ||
| GC content unigenes (%) | 56.11% | 48.71% | ||
| ORF prediction | Unigenes with predicted ORF (CDS ≥ 200bp) | 5,227 | 8,102 | 13,329 |
| KEGG annotation | Unigenes assigned to KO identifier | 2,985 | 4,548 | 7,533 |
| Unique KO identifiers | 1,648 | 1,246 | 2,894 | |
| Go annotation | Mapped to GO terms | 5,014 | 7,822 | 12,836 |
| Unigenes annotated with GO terms | 3,657 | 5,636 | 9,293 |
Functional Annotation of the Full Transcriptomes
Using the KAAS, 2,985 H. hermesi unigenes (54.29%) were assigned to 1,648 KO identifiers, whereas 4,548 GD1 unigenes (52.96%) were assigned to 2,166 KO identifiers. Both transcriptomes shared 1,246 KO identifiers corresponding to 2,499 H. hermesi- and 3,244 GD1 unigenes. In addition, 402 KO identifiers were uniquely found in H. hermesi (corresponding to 487 unigenes), and 920 KO identifiers were uniquely found in GD1 (corresponding to 1,304 unigenes). These genes are further addressed as uniquely expressed unigenes. The KO identifiers were mapped to 245 shared pathways, and to three and ten unique pathways for H. hermesi and GD1, respectively (full data set in supplementary table S2, Supplementary Material online). However, only 1–3 unigenes were mapped to these unique pathways. The same three pathways were most represented in deep-sea and intertidal nematodes: “protein processing in endoplasmic reticulum,” “endocytosis,” and “spliceosome” (table 2). Of all shared pathways, 40 had a higher amount of unigenes mapped for H. hermesi than for GD1 (supplementary table S2, Supplementary Material online). “Oxidative phosphorylation” was the pathway with the highest differences in mapped proteins, that is, eight additional H. hermesi unigenes were mapped against this pathway compared with GD1. A higher amount of GD1 unigenes were mapped to 156 pathways compared with H. hermesi. The highest amount of additional GD1 unigenes (i.e., 23) mapped against the Neuroactive ligand-receptor interaction pathway. Equal amounts of GD1 and H. hermesi unigenes were mapped against 49 pathways (supplementary table S2, Supplementary Material online). At the level of protein families (full data in supplementary table S3, Supplementary Material online), “enzymes” was by far the most abundant category for both species (table 2).
Table 2.
Top 20 Selected KEGG Metabolic Pathways of Putative Proteins Mapped Using KEGG Database
| Full transcriptome |
Unique unigenes |
RNAseq unigenes |
||||||
|---|---|---|---|---|---|---|---|---|
| H. hermesi | GD1 | H. hermesi | GD1 | OE in H. hermesi | OE in GD1 | EE | ||
| KEGG pathways | Protein processing in endoplasmic reticulum | 52 | 60 | 11 | 19 | 9 | 10 | 0 |
| Endocytosis | 46 | 60 | 10 | 24 | 4 | 2 | 1 | |
| Spliceosome | 44 | 54 | 9 | 19 | 5 | 2 | 0 | |
| Oxidative phosphorylation | 43 | 35 | 17 | 9 | 6 | 3 | 1 | |
| Rap1 signaling pathway | 41 | 52 | 7 | 18 | 6 | 1 | 0 | |
| Purine metabolism | 38 | 53 | 8 | 23 | 2 | 0 | 1 | |
| Focal adhesion | 38 | 41 | 7 | 10 | 3 | 1 | 0 | |
| MAPK signaling pathway | 37 | 45 | 11 | 19 | 4 | 3 | 0 | |
| Carbon metabolism | 37 | 39 | 8 | 10 | 9 | 2 | 3 | |
| Lysosome | 37 | 37 | 5 | 5 | 1 | 1 | 1 | |
| Regulation of actin cytoskeleton | 36 | 46 | 5 | 15 | 5 | 2 | 1 | |
| PI3K-Akt signaling pathway | 35 | 50 | 5 | 20 | 3 | 4 | 1 | |
| cAMP signaling pathway | 35 | 43 | 3 | 11 | 4 | 2 | 0 | |
| Oxytocin signaling pathway | 34 | 38 | 4 | 8 | 3 | 2 | 0 | |
| RNA transport | 33 | 42 | 10 | 19 | 0 | 2 | 1 | |
| Ras signaling pathway | 32 | 42 | 9 | 19 | 0 | 3 | 1 | |
| AMPK signaling pathway | 32 | 33 | 7 | 8 | 3 | 4 | 0 | |
| cGMP—PKG signaling pathway | 31 | 35 | 3 | 7 | 2 | 4 | 0 | |
| Insulin signaling pathway | 29 | 41 | 5 | 17 | 8 | 1 | 0 | |
| Adrenergic signaling in cardiomyocytes | 28 | 26 | 3 | 1 | 3 | 3 | 0 | |
| Protein families | Enzymes | 708 | 901 | 161 | 354 | 59 | 44 | 14 |
| Exosome | 160 | 188 | 22 | 50 | 12 | 21 | 9 | |
| Transporters | 123 | 144 | 22 | 43 | 7 | 6 | 0 | |
| Protein kinases | 111 | 139 | 19 | 47 | 10 | 12 | 3 | |
| Chromosome | 105 | 157 | 27 | 79 | 3 | 10 | 3 | |
| Spliceosome | 93 | 118 | 21 | 46 | 12 | 6 | 0 | |
| Peptidases | 91 | 111 | 20 | 40 | 8 | 6 | 1 | |
| Protein phosphatases and associated proteins | 75 | 95 | 17 | 37 | 4 | 4 | 0 | |
| Mitochondrial biogenesis | 65 | 77 | 21 | 33 | 8 | 4 | 1 | |
| Ubiquitin system | 65 | 102 | 22 | 59 | 3 | 2 | 0 | |
| Ribosome biogenesis | 59 | 78 | 16 | 35 | 11 | 10 | 0 | |
| Transcription machinery | 57 | 74 | 10 | 27 | 2 | 5 | 2 | |
| Chaperones and folding catalysts | 51 | 54 | 12 | 15 | 7 | 10 | 0 | |
| Ion channels | 49 | 61 | 14 | 26 | 2 | 0 | 1 | |
| Transcription factors | 43 | 71 | 17 | 45 | 1 | 1 | 0 | |
| Cytoskeleton proteins | 43 | 52 | 8 | 17 | 3 | 3 | 2 | |
| DNA repair and recombination proteins | 36 | 52 | 12 | 28 | 2 | 3 | 1 | |
| Transfer RNA biogenesis | 34 | 46 | 11 | 23 | 1 | 3 | 1 | |
| Proteasome | 33 | 30 | 10 | 7 | 3 | 7 | 0 | |
| Cell adhesion molecules and their ligands | 32 | 48 | 7 | 23 | 1 | 2 | 0 | |
Note.—Data of the full nematode transcriptome, the unique unigenes, equally(EE)-, and overexpressed (OE) RNAseq unigenes for Halomonhystera hermesi and GD1 is represented.
Over/Underrepresentation of KEGG Pathways and Protein Families in Uniquely Annotated H. hermesi- and GD1 Unigenes
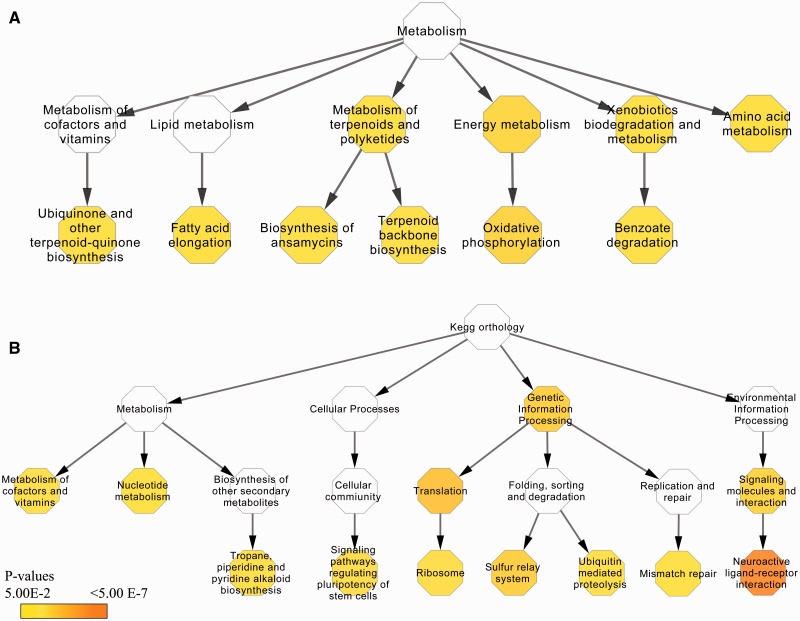
Unigenes that were uniquely found in H. hermesi or GD1 were considered to be characteristic of the deep-sea and intertidal environment, respectively, and were used to investigate pathways that were over/underrepresented in a particular habitat. Six KEGG pathways: “oxidative phosphorylation,” “ubiquinone” and “other terpenoid-quinone biosynthesis,” “benzoate degradation,” “terpenoid backbone biosynthesis,” “biosynthesis of ansamycins” and “fatty acid elongation” were significantly overrepresented in H. hermesi unique unigenes (P values < 0.05), all belonging to the metabolism category (fig. 2A). The unique unigenes were most enriched in the oxidative phosphorylation (P = 2.92E−3) belonging to the overrepresented energy metabolism class (P = 4.01E−3; fig. 2A). A closer inspection of our data revealed that the observed overrepresentation was due to NADH dehydrogenase (ubiquinone) 1 alpha/beta subcomplex subunits, several F- andV- type ATPase subunits, and succinate dehydrogenase, involved in the electron transport chain at the inner mitochondrial membrane. Three KEGG pathways were significantly underrepresented: “tight junction,” “circadian entrainment,” and “salivary secretion.” For GD1, over- and underrepresented pathways in which unique unigenes were involved belonged to a wider spectrum of molecular pathways compared with the deep-sea transcriptome (fig. 2B). The most significant overrepresented pathway in GD1 uniquely annotated unigenes belonged to the neuroactive ligand-receptor interaction pathway (p = 2.24E−7; fig. 2B) and was mostly the result of G-protein coupled receptors in the GD1 transcriptome which were not expressed in H. hermesi. Six other overrepresented pathways were “tropane,” “piperidine and pyridine alkaloid biosynthesis,” “signaling pathways regulating pluripotency of stem cells, ribosome,” “sulfur relay system,” “ubiquitin mediated proteolysis,” and “mismatch repair.” Many more pathways were underrepresented in GD1 compared with underrepresented H. hermesi pathways (supplementary table S4, Supplementary Material online). The top 3 were “fatty acid metabolism” (P = 1.07E−4), “tight junctions” (P = 3.54E−4), and “fatty acid biosynthesis” (P = 7.01E−4).
Fig. 2.—
Visualization of overrepresented KEGG pathways in unique H. hermesi (A) and GD1 (B) unigenes. Each hexagon represents either a KEGG pathway or higher order category according to BRITE hierarchy. White hexagons are not significant categories while colored ones represent the FDR P values.
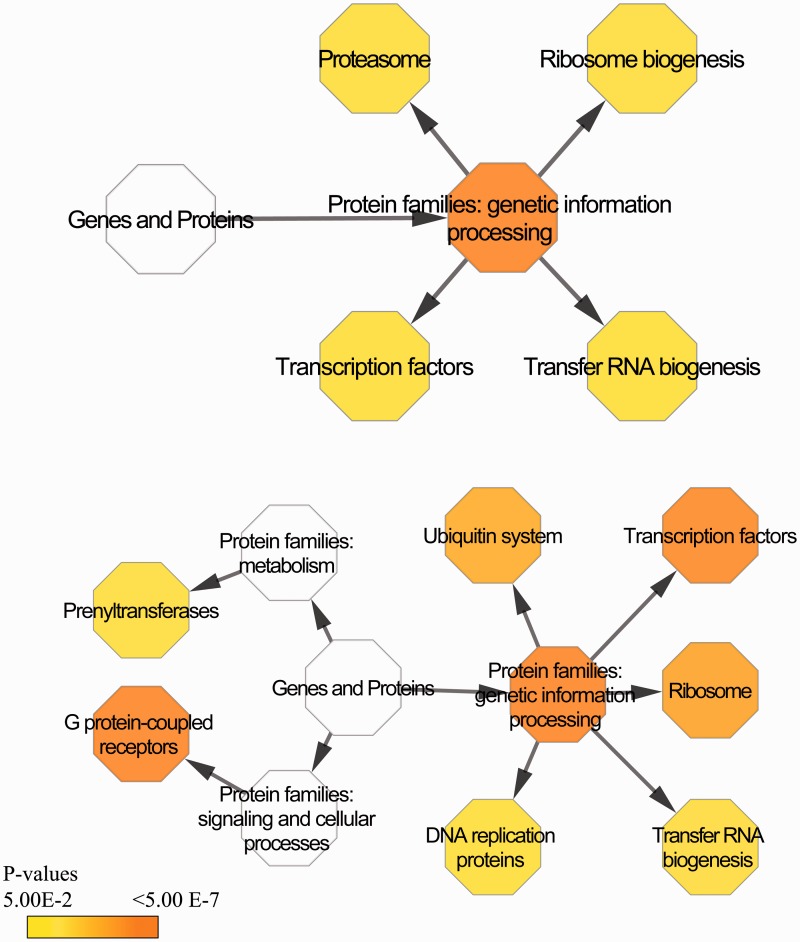
Only protein families involved in genetic information processing were significantly (P = 4.42E−5) overrepresented in H. hermesi unique genes as a result of overrepresented unique unigenes involved in “transcription factors,” “proteasome,” “t-RNA biogenesis,” and “ribosome biogenesis” (fig. 3A), whereas exosome involved proteins were underrepresented. The two most overrepresented protein families in GD1 unique unigenes were G-protein-coupled receptors (P = 5.26E−16) and transcription factors (P = 3.32E−9; fig. 3B). Except for the overrepresentation of prenyltransferases, all other overrepresented protein families are involved in genetic information processing (P = 3.43E−11).
Fig. 3.—
Overrepresented protein families (BRITE mapping) in unique H. hermesi (A) and GD1 (B) unigenes. Parent-child relations are represented by arrows between hexagons. Hexagons’ colors represent FDR P values from white, being not significant, to red, being highly significant.
GO Classification of the Full Transcriptomes
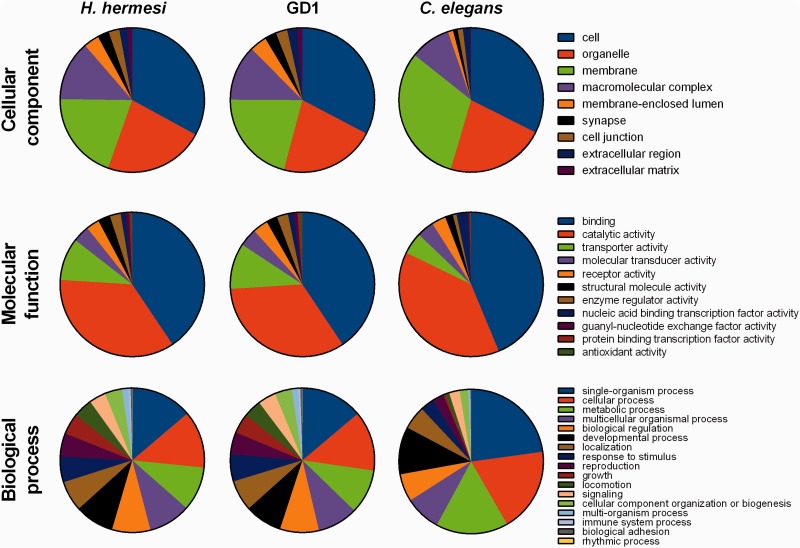
GO terms were assigned to 3,657 (72.94%) H. hermesi and 5,636 (72.05%) GD1 unigenes which could be classified into three categories: cellular component, molecular function, and biological process (fig. 4). The BINGO analysis of each GO data sets separate compared with both data sets combined did not reveal any overrepresented level 2 GO-terms. As such, GO annotation were highly similar between intertidal and deep-sea samples and reflected very well annotation patterns observed in the nematode Caenorhabditis elegans (depicted from geneontology.org). GO-annotation yielded 6,108 H. hermesi- and 9,591 GD1 GO cellular component annotations, 5,086 H. hermesi and 7,417 GD1 molecular function annotations, and 15,981 H. hermesi and 23,692 GD1 GO annotations for biological process. Within the cellular component category 32.95% and 32.69% of GD1 and H. hermesi unigenes were assigned to “Cell,” followed by “Organelle” (22.48% and 21.45%), “Membrane” (19.78% and 20.95%) and “Macromolecular Complex” (13.43% and 12.45%) (fig. 4). GO term assignment of H. hermesi- and GD1 unigenes, respectively, within the molecular function category, was dominated by “Binding” (40.59% and 40.61%) and “Catalytic activity” (35.40% and 33.39%) (fig. 4). “Receptor activity” and “nucleic acid binding transcription factor activity” were significantly overrepresented in GD1 unique unigenes (P values = 3.18E−5 and 1.80E−6, respectively). Finally, within the biological process category 12 GO-terms captured more than 90% of the assignments of which “Single-Organism Process,” “Cellular Process,” and “Metabolic Process” were the three largest classes for both transcriptomes (fig. 4).
Fig. 4.—
GO annotation (Level 2) based on transcriptomic data of H. hermesi, GD1, and C. elegans. Unigenes were classified into three categories: cellular components, molecular functions, and biological processes. Caenorhabditis elegans data were inferred from geneontology.org.
Overrepresentation of GO Terms in Unique Unigenes
The addition of selected PROmer hits resulted into 2,492 H. hermesi- and 4,752 GD1 unique unigenes. Halomonhysterahermesi unique unigenes only showed overrepresented GO terms from the molecular function category (P values in table 3) related to “enzyme inhibitor- and peptidase regulator activity” (supplementary fig. S1, Supplementary Material online). Much more overrepresented GO terms in GD1 unique unigenes were found (supplementary table S5, Supplementary Material online). Several GO terms involved in transcription, for example, transcription factor complex (cellular component, supplementary fig. S2, Supplementary Material online), sequence-specific DNA binding (molecular function, supplementary fig. S3, Supplementary Material online), and regulation of transcription (biological process, supplementary fig. S4, Supplementary Material online), were overrepresented in GD1 (P values in table 3). Moreover, overrepresented GO terms dealing with signaling pathways, for example, receptor- and kinase activity were prominent in both the molecular function and biological process category for GD1.
Table 3.
Significantly Overrepresented GO Terms of Unique H. hermesi and GD1 Unigenes in Comparison to the Whole Transcriptome of Both Species
| GO Category | Species | GO-ID | FDR Corrected P Value | Description |
|---|---|---|---|---|
| Cellular component | GD1 | GO:0005667 | 1.2165E-2 | Transcription factor complex |
| Molecular function | H. hermesi | GO:0030414 | 5.2388E-5 | Peptidase inhibitor activity |
| GO:0004857 | 8.0869E-5 | Enzyme inhibitor activity | ||
| GO:0004866 | 1.1168E-4 | Endopeptidase inhibitor activity | ||
| GO:0061134 | 1.3406E-4 | Peptidase regulator activity | ||
| GO:0061135 | 1.4138E-4 | Endopeptidase regulator activity | ||
| GO:0004867 | 2.5001E-4 | Serine-type endopeptidase inhibitor activity | ||
| GD1 | GO:0004930 | 1.2256E-15 | G-protein coupled receptor activity | |
| GO:0043565 | 2.0797E-7 | Sequence-specific DNA binding | ||
| GO:0038023 | 1.4593E-6 | Signaling receptor activity | ||
| GO:0004872 | 3.9906E-6 | Receptor activity | ||
| GO:0004888 | 5.1755E-6 | Transmembrane signaling receptor activity | ||
| GO:0001071 | 2.2160E-5 | Nucleic acid binding transcription factor activity | ||
| GO:0003700 | 2.2160E-5 | Sequence-specific DNA binding transcription factor activity | ||
| GO:0005272 | 6.6765E-5 | Sodium channel activity | ||
| GO:0009022 | 2.8690E-4 | tRNA nucleotidyltransferase activity | ||
| GO:0042302 | 3.0103E-4 | Structural constituent of cuticle | ||
| GO:0004871 | 4.5351E-4 | Signal transducer activity | ||
| GO:0060089 | 4.5351E-4 | Molecular transducer activity | ||
| GO:0030971 | 4.5661E-4 | Receptor tyrosine kinase binding | ||
| Biological process | GD1 | GO:0051171 | 4.4070E-3 | Regulation of nitrogen compound metabolic process |
| GO:0019219 | 4.4070E-3 | Regulation of nucleobase-containing compound metabolic process | ||
| GO:0008033 | 1.0951E-2 | tRNA processing | ||
| GO:1903506 | 1.0951E-2 | Regulation of nucleic acid-templated transcription | ||
| GO:0006355 | 1.0951E-2 | Regulation of transcription, DNA-templated | ||
| GO:2001141 | 1.0951E-2 | Regulation of RNA biosynthetic process | ||
| GO:0043628 | 1.0951E-2 | ncRNA 3′-end processing | ||
| GO:0007186 | 1.4586E-2 | G-protein coupled receptor signaling pathway |
Positive Selection, Amino Acid Composition, and Codon Usage
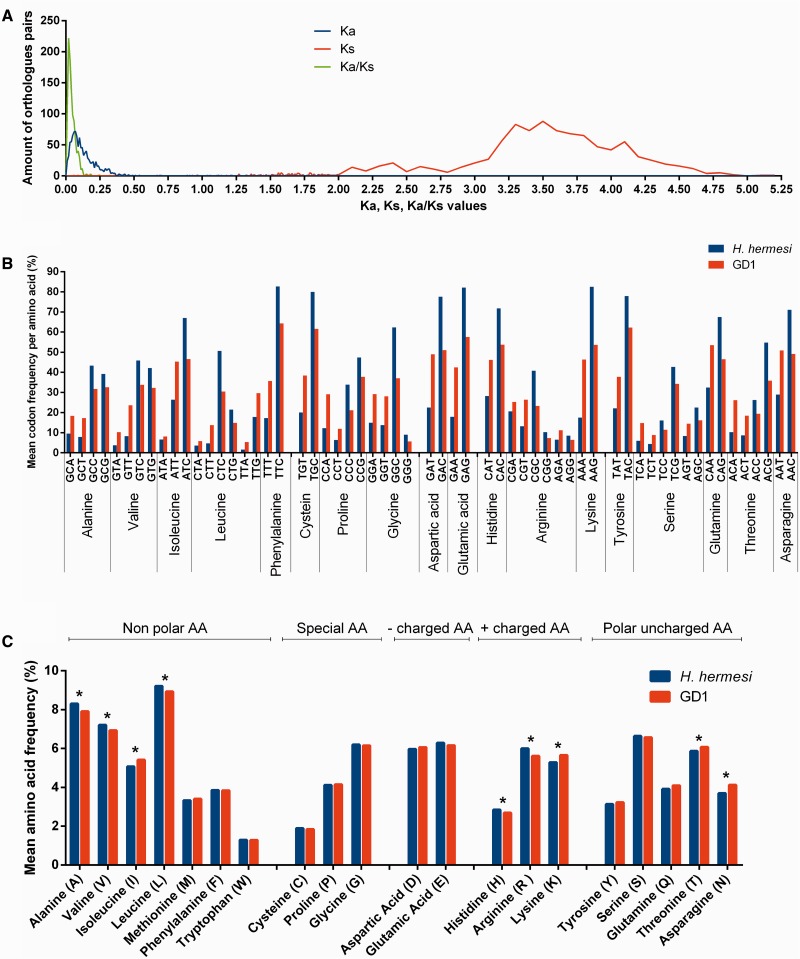
Filtering PROmer hits based on similarity (≥75%) and alignment length (≥300 bp) resulted into 1,053 hits with the same functional annotation in both data sets. GC content of H. hermesi unigenes (58.97%) was significantly higher (P < E−6) compared with corresponding GD1 sequences (50.96%). Genetic divergence (uncorrected p-distances) between orthologous pairs ranged from 8.3% to 40.1%. Mean values of Ka, Ks and Ka/Ks ratio (fig. 5A) were 0.125, 3.266 and 0.0453, respectively. Of the orthologous pairs only three had a Ka/Ks ≥ 1 which were mapped to three different KEGG pathways: “proteasome (20S proteasome subunit beta 3),” “Spliceosome (splicing factor U2AF 65 kDa subunit),” and “tight junctions (Myosin heavy chain).”
Fig. 5.—
Alignment of H. hermesi and GD1 orthologous pairs were used to determine (A) Ka, Ks and Ka/Ks ratio, (B) amino acid frequencies, and (C) codon frequencies. Significant difference between amino acid frequencies are indicated with an asterisk. All codon frequency differences were always significantly different between both species.
In both transcriptomes, GC rich codons were more frequently used, but this was especially true for the deep-sea transcriptome (fig. 5B). All codon differences were significant (supplementary table S6, Supplementary Material online). The relative frequency of amino acids (AAs) with GC-rich codons (Alanine, and Arginine) was significantly higher (both P values = 0.00105) in H. hermesi (fig. 5B). In contrast, GD1 had significantly higher relative frequency of AAs with AU-rich codons (fig. 5C) such as Isoleucine (P = 0.00105), Lysine (P = 0.00672), and Asparagine (P < E-6). In addition, GD1 had higher frequencies of Threonine (P = 0.01467), while relative frequencies of Valine (P = 0.00544), Histidine (P = 0.0117), and Leucine (P = 0.0129) were higher in H. hermesi. Summary statistics of the null distribution and P values can be found in supplementary table S7, Supplementary Material online.
Differential Expression of Shared Genes between Intertidal and Deep-Sea Nematodes
The de novo assembly, based on H. hermesi- and GD1 unigenes collectively, resulted in 3,345 unigenes with a CDS larger 200 bp. We identified 740 and 1,693 unigenes exclusive to H. hermesi and GD1, respectively, and 1,014 unigenes expressed in both species of which 845 were significantly differentially expressed (Kal’s Z-test). After applying the KO filtering on the 1,014 unigenes, we retained 142 and 118 unigenes that were differentially overexpressed in H. hermesi and GD1, respectively, whereas 90 were equally expressed.
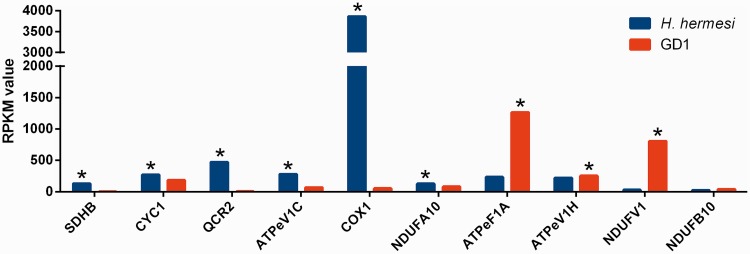
We further focus on those genes that are involved in KEGG pathways and protein families that were shown to be over/underrepresented, allowing us to now compare gene expression levels of shared genes in these categories. Seven shared unigenes that are involved in transcription had a significantly higher gene expression for GD1 compared with four H. hermesi overexpressed unigenes. Out of nine shared unigenes involved in oxidative phosphorylation, six revealed to have higher gene expression levels for H. hermesi (fig. 6). We observed very high gene expression differences for cytochrome c oxidase subunit I (COX1) between both species. Moreover, two subunits of the electron transport chain complex III, ubiquinol-cytochrome c reductase cytochrome c1 subunit (CYC1), and ubiquinol-cytochrome c reductase core subunit 2 (QCR2) had significantly higher gene expression values in H. hermesi.
Fig. 6.—
Expressed unigenes involved in oxidative phosphorylation for H. hermesi and GD1. Significant differences (P < 0.05) are indicated with an asterisk. SDHB, succinate dehydrogenase (ubiquinone) iron-sulfur subunit; CYC1, ubiquinol-cytochrome c reductase cytochrome c1 subunit; QCR2, ubiquinol-cytochrome c reductase core subunit 2; ATPeV1C, V-type H+-transporting ATPase subunit C; COX1, cytochrome c oxidase subunit 1; NDUFA10, NADH dehydrogenase (ubiquinone) 1 alpha subcomplex subunit 10; ATPeF1A, F-type H+-transporting ATPase subunit alpha; ATPeV1H, V-type H+-transporting ATPase subunit H; NDUFV1, NADH dehydrogenase (ubiquinone) flavoprotein 1; NDUFB10, NADH dehydrogenase (ubiquinone) 1 beta subcomplex subunit 10.
Discussion
Halomonhystera hermesi and GD1 have a very similar morphology but were considered different species based on their genetic differentiation at mitochondrial and nuclear loci (Van Campenhout et al. 2013). The phylogenetic position of H. hermesi amidst the intertidal species suggests that H. hermesi has originated from shallow water relatives followed by adaptive evolution in the deep sea. However, no typical cold-seep adaptations such as ecto- and/or endosymbionts or sulfur crystal were observed (Van Gaever et al. 2006, 2009), which suggests that adaptations are situated at the molecular level. Our results revealed that both species use the same molecular pathways to cope and survive in their distinct environment. Nevertheless, we found a higher amount of unigenes and active pathways in GD1, and most shared unigenes were differentially expressed between both species. Gene expression can be influenced by environmental factors (Lobo 2008) and our results revealed that different pathways were overrepresented in each environment suggesting that these pathways are of high importance to survive in the respective environment. The experimental procedure to collect specimens from either habitat may have added additional differences in gene expression between both habitats, and we may also have missed some very low expressed genes due to the normalization step. In addition, sex ratio differences between both populations might have influenced our results. Nevertheless, we believe that our approach is the most solid and feasible way for comparing the transcriptomes of both species in such different habitats.
Transcription and Signaling Receptor Activity Are More Strongly Expressed in Intertidal Environments
Our data disclosed a higher amount of unigenes, more overrepresented GO terms and a wider spectrum of molecular overrepresented pathways in GD1. Transcription factors were clearly more overrepresented in GD1: 71 transcription factors, based on BRITE protein families, and 20 basal transcription factors, based on KEGG pathways, were found in GD1 whereas 43 transcription factors (protein families) and 11 basal transcription factors were detected in H. hermesi. Moreover, of the shared genes most unigenes involved in transcription had higher gene expression values for GD1 compared with H. hermesi. This suggests that the activation and expression of transcription factors is much more prominent in species that inhabit fluctuating environments. Intertidal mudflats, the preferred habitat for GD1, are highly dynamic systems that are characterized by rapid changes in environmental variables (de Brouwer et al. 2006). Organisms in fluctuating environments must constantly adapt their behavior to survive (Kussell and Leibler 2005) and can perform faster transcriptional reprogramming (New et al. 2014). The ability to rapidly respond to changing environmental features further relies on sensing these changes (Lopez-Maury et al. 2008). Other prominent overrepresented GO terms in GD1 nematodes were the G protein-coupled receptors (GPCRs). Moreover, more than three times more GPCRs were identified in GD1 compared with H. hermesi. The high amount of GPCRs further resulted in an enrichment of the neuroactive ligand-receptor interaction class (environmental information processing). GPCRs consist of chemosensory receptors which mediate most cellular responses to hormones and neurotransmitters and are able to detect extracellular chemical stimuli converting them to intracellular responses (Mombaerts 1999). The enrichment in GPCRs is a clear indication of a heightened sensorial and neural activity of GD1 compared with H. hermesi. This, and the higher amount of transcription factors and higher gene expression values of shared genes in GD1 illustrate the importance of species to swiftly detect and respond environmental changes to be able to live in variable environments.
Oxidative Phosphorylation and Fatty Acid Metabolism Are More Strongly Expressed in Deep-Sea Environments
Oxidative phosphorylation is the mitochondrial process in which the energy released by the transfer of electrons over electron transport carriers is used to establish a proton gradient which in turn is employed to produce energy under the form of ATP (Voet et al. 2006). Our results show that this process is more strongly expressed in H. hermesi and 43 H. hermesi unigenes were mapped against the oxidative phosphorylation compared with 35 mapped GD1 unigenes. Interestingly, we have identified a number of overexpressed genes, part of complex III (electron transport chain) in H. hermesi. The HMMV, the habitat of H. hermesi, is a chemosynthetic environment characterized by high hydrogen sulfide concentrations (Van Gaever et al. 2006). Hydrogen sulfide is a toxin that is capable of impairing biological processes in Metazoa. The toxicity of sulfide is mainly a result of inhibition of aerobic respiration by inhibition of cytochrome c oxidase (NRC 1979) and thus interfering with cellular respiration (Somero et al. 1989; Vismann 1991). Interestingly, the most overexpressed gene in H. hermesi involved in oxidative phosphorylation was cytochrome c oxidase c subunit I which might be a countermeasure to overcome sulfide poisoning. Sulfide can also be oxidized in the mitochondria and electrons can be incorporated into the electron transport chain by ubiquinone or ubiquinol-cytochrome-c oxidoreductase (complex III) (Powell and Somero 1986; Russell et al. 1989; Völkel and Grieshaber 1996). A higher expression of specific complex III genes might, therefore, be an important adaptation to sulfidic environments. However, oxidative phosphorylation is also influenced by a variety of other abiotic factors such as temperature (Toffaletti et al. 2003), pressure (Theron et al. 2000), salinity (Li et al. 2014), and oxygen levels (Mustroph et al. 2010). Oxygen levels rapidly deplete beneath 1 mm sediment depth at the HMMV (de Beer et al. 2006) while H. hermesi was identified up to 10 cm sediment depth. The effect of low oxygen levels include the induction of fermentation and glycolysis (Mustroph et al. 2010), influence mitochondria fusion (Tondera et al. 2009), mitochondrial biogenesis (Qin et al. 2014), and mitochondrial organelle number and volume (Gupta 2002), which implies that mitochondria are key players in the adaptation to low oxygen concentrations. Furthermore, the AMP-activated protein kinase signaling pathway, which is rapidly induced by low oxygen levels (Laderoute et al. 2006) was underrepresented in GD1 suggesting that oxygen levels are important. In summary, our results indicate that sulfide- and oxygen concentrations are presumably the most important environmental features explaining overrepresentation of genes involved in oxidative phosphorylation in H. hermesi unigenes.
The deep-sea species further showed a clear response in fatty acid elongation compared with GD1. We observed an overrepresentation of the fatty acid elongation pathway whereas the fatty acid metabolism was underrepresented in GD1 both in KEGG and GO annotations. A recent study revealed that H. hermesi has higher proportions of highly unsaturated fatty acids (HUFAs, chain length ≥ 20 carbons and ≥ 3 double bond) due to increased proportions of eicosapentaenoic acid (EPA–20:5ω3) and the presence of docosahexanoic acid (DHA–22:6ω3) which was absent in GD1 (Van Campenhout J and Vanreusel A submitted for publication). These HUFAs are believed to be essential in maintaining membrane fluidity under pressure (Yano et al. 1997) and docosahexaenoic acid (DHA–C22:6ω3) was hypothesized to be the key to homeoviscous adaptations to depth and temperature changes in vertically migrating planktonic copepods (Pond et al. 2014). Given the absence of these HUFAs in the food source of H. hermesi (Van Gaever et al. 2009), the nematode has most likely the ability to synthesize these HUFAs. EPA, and DHA can be assimilated into triglyceride forms and stored until they are required for incorporation in the membrane, beta-oxidation, signalizing activity, etc. (Williams and Burdge 2006; Reisner 2012). Our results at the molecular level now confirm that these HUFAs are indeed important to deep-sea adaptation.
GC Content, Amino Acid Composition, and Codon Usage Differs between Deep-Sea and Intertidal Species
Our results revealed that the genetic divergence (uncorrected p-distances) between orthologous pairs ranged from 8.3% to 40.1%. This observation further confirms the identity of both species and is influenced by the significant higher GC content in H. hermesi than in GD1 orthologous pairs. To date, many hypothesis such as directional mutation pressure (Sueoka 1988), metabolism (Martin 1995), CDS length (Oliver and Marin 1996), nitrogen fixation, genome size (Musto et al. 2006), DNA polymerase III subunit alpha (Zhao et al. 2007), and environmental pressure (Foerstner et al. 2005) have been postulated to explain varying GC contents. The GC content of both Halomonhystera species is correlated with codon usage bias, especially at the third codon position and with amino acid content, a pattern that has been frequently observed (Bernardi 1985; Wilquet and Van de Casteele 1999; Basak et al. 2004; Foerstner et al. 2005). Furthermore, nucleotide bias can have a dramatic effect on the amino acid composition of encoded proteins (Lobry 1997; Singer and Hickey 2000) and it is currently believed that amino acid composition is more correlated to GC-content of the genome than to environment (Moura et al. 2013). In addition, arginine and alanine have been postulated as the “most piezophilic amino acids,” playing a role in adaptation to the piezophilic lifestyle in organisms (Di Giulio 2005; Jun et al. 2011; Pradel et al. 2013). One possible reason is the cost of ATP synthesis imposed on H. hermesi by low oxygen levels, low temperatures, and high sulfide concentrations. Whatever the driving force behind the GC content differences might be, it is clear that the different environments indirectly result in different codon usage with relatively small differences in amino acid abundances.
Few Gene Were Under Selection whereas Many Genes Were Uniquely Expressed
Adaptation depends on the natural selection of individuals with higher fitness thereby increasing their chance to produce more viable offspring (Darwin 1859). We could detect three genes that are subjected to positive selection. The 20S proteasome subunit beta 3 is part of the large 20S proteasome (28 subunits) the barrel shaped cylindrical core of the 26S proteasome, which degrades ubiquitinated proteins in an ATP-dependent process (Voet et al. 2006). This subunit has no catalytic activity (Jung et al. 2009) and its exact role in deep-sea adaptation remains unknown. The Splicing factor U2AF 65 kDA subunit is part of the U2 auxiliary factor important in mRNA splicing by recognizing the polypyrimidine tract at the 3′-end of introns (Wahl et al. 2009). U2AF65 preferentially binds on uridine-rich sites with frequent interruptions by cytidine residues but variation in the tract sequences and length affects the efficiency of splice site recognition (Singh et al. 1995). Halomonhysterahermesi has a higher GC content which might constrain the presence of uridine rich tracts in mRNA impairing the U2AF65 binding efficiency. Positive selection on U2AF65 might, therefore, be an indirect result of environmental factors acting on GC content and can be essential to better recognize splice sites. In addition, mechanisms such as alternative splicing are known to be a response to external stimuli and play an important role in gene function diversification and regulation (Stamm 2002; Ramani et al. 2011) but knowledge on its true role in adaptation remains scant.
The “myosin heavy chain” is involved in tight junctions and its amino acid composition can adaptively alter its pressure sensitivity (Morita 2008, 2010). Tight junctions, underrepresented in GD1, establish physiological barriers that regulate the movement of ions, small solutes, and fluid between tissue compartments (Gonzalez-Mariscal et al. 2003). Both osmotic pressure and hydrostatic pressure are known to affect tight junctions (Tokuda et al. 2009; Gunzel and Yu 2013), suggesting that pressure and salinity may be important drivers for the adaptive changes in both species. In contrast to the low number of genes detected to be under positive selection, we observed a high rate of synonymous substitutions for those genes that were identified in both species. This highlights that new mutations which give rise to amino acid changes are removed from the populations by purifying selection, and that these genes are under high functional constraint. However, the molecular basis of adaptation is not only restricted to CDS but also includes noncoding regulatory sequences, for example, promoters (Lopez-Maury et al. 2008) and enhancers (Kamachi et al. 2009). Functionally important nontranslated sequences have been hypothesized to also be subject to positive and purifying selection (Andolfatto 2005). We currently lack information on these regulatory sequences and further research is required.
The low amount of genes under selection can be the result of several possibilities. First, increase in synonymous substitution rates may obscure detection of genes under positive selection (Erixon and Oxelman 2008). Second, it has been suggested that also synonymous sites are subject of nonneutral evolution (Chamary et al. 2006), but our analysis did not accounted for this. Third, we lack any information on adaptive evolution of uniquely expressed genes which might be under selection. Fourth, under the hypothesis that H. hermesi has an intertidal ancestor, this ancestor might have been preadapted to certain deep-sea conditions. Intertidal sediments, as well as decaying macroalgae, show similar environmental characteristics as a cold-seep environment such as hypoxic/anoxic conditions and high sulfide concentration as well as low temperatures in the winter and temporary high salinity levels are not out of the ordinary. As such, several genes might have been preadapted allowing synonymous mutations to accumulate.
Finally, many genes were uniquely expressed in both species. However, caution is required for the correct interpretation of our results as we here compared two species (without genomic data available) from two different environments and sampled only a single point in time. As such, it remains to be investigated whether the uniquely expressed genes are the result of adaptive evolution of, for example, regulatory sequences, or are in fact the result of environmental signals acting at the level of gene expression regulation at the time of sampling. Our results, combined with the tolerance of GD1 for bathyal cold-seep conditions (Van Campenhout et al. 2014), do suggest that plasticity should be taken into account. It remains to be determined to what extent plasticity or preadaptation of intertidal nematodes played a role in adaptive evolution of cold-seep nematodes.
Conclusion
In our study, the transcriptomes of two phylogenetically closely related marine nematode species from contrasting environments (deep sea and intertidal) have been compared. We revealed that most pathways are shared between both habitats, but several pathways and genes were differentially expressed in different habitats. Oxidative phosphorylation and fatty acid metabolism/elongation appeared to be of higher importance in the deep sea. In addition, we observed a higher GC content in H. hermesi resulting in higher amino acid frequencies with GC-rich codons affecting codon usage between H. hermesi and GD1 orthologous. Meanwhile the dynamic feature of GD1’s habitat was reflected in higher amounts of transcription factors, more strongly expressed signaling receptor activity, and a higher amount of active pathways and genes. Many genes were uniquely expressed in one species and only few genes show a signal that natural selection has been involved in their evolution. This indicates that Halomonhystera species might be preadapted to specific environmental conditions such as low oxygen levels and high sulfide concentrations.
Supplementary Material
Supplementary material is available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank chief scientist A. Boetius and project LOOME coordinator D. De Beer for the sampling opportunity during the RV Maria S. Merian campaign MSM 16/2. The authors acknowledge the captain and the crew of RV “Maria S. Merian” for their support with work at sea. The authors thank IFREMER (France), Woods hole oceanographic institution (MA, USA, AUV “Sentry”) and the Renard Centre of Marine Geology (Ghent University, Belgium, ROV “genesis”). Financial support was provided by the FP7 EU projects Hotspot Ecosystem Research and Man’s Impact On European Seas (Grant number 226354), Managing Impacts of Deep-sea resource exploitation (Grant number 603418) and European Sea Observatory NETwork (Grant number. 036851), and by the DFG for the Maria S. Merian cruise MSM 16/2.
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Andolfatto P. 2005. Adaptive evolution of non-coding DNA in Drosophila. Nature 437:1149–1152. [DOI] [PubMed] [Google Scholar]
- Andrássy I. 2006. Halomonhystera, a new genus distinct from Geomonhystera Andrássy, 1981 (Nematoda: Monhysteridae). Meiofauna Marina 15:11–24. [Google Scholar]
- Ashburner M, et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak S, Banerjee T, Gupta SK, Ghosh TC. 2004. Investigation on the causes of codon and amino acid usages variation between thermophilic Aquifex aeolicus and mesophilic Bacillus subtilis. J Biomol Struct Dyn. 22:205–214. [DOI] [PubMed] [Google Scholar]
- Bernardi G. 1985. Codon usage and genome composition. J Mol Evol. 22:363–365. [DOI] [PubMed] [Google Scholar]
- Bird A. 2007. Perceptions of epigenetics. Nature 447:396–398. [DOI] [PubMed] [Google Scholar]
- Blaxter M. 2011. Nematodes: the worm and its relatives. PLoS Biol. 9:e1001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, et al. 2009. BLAST plus: architecture and applications. BMC Bioinformatics 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamary JV, Parmley JL, Hurst LD. 2006. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat Rev Genet. 7:98–108. [DOI] [PubMed] [Google Scholar]
- Chong SY, Whitelaw E. 2004. Epigenetic germline inheritance. Curr Opin Genet Dev. 14:692–696. [DOI] [PubMed] [Google Scholar]
- Conesa A, et al. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. 2004. Speciation. Sunderland (MA): Sinauer Associates. [Google Scholar]
- Cubas P, Vincent C, Coen E. 1999. An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401:157–161. [DOI] [PubMed] [Google Scholar]
- Darwin C. 1859. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. London: John Murray. [PMC free article] [PubMed] [Google Scholar]
- de Beer D, et al. 2006. In situ fluxes and zonation of microbial activity in surface sediments of the Hakon Mosby Mud Volcano. Limnol Oceanogr. 51:1315–1331. [Google Scholar]
- de Brouwer J, Neu TR, Stal LJ. 2006. On the function of secretion of extracellular polymeric substances by benthic diatoms and their role in intertidal mudflats: a review of recent insights and views In: Jea Kromkamp, editor. Functioning of microphytobenthos in estuaries: Proceedings of the Colloquium. Amsterdam: Koninklijke Nederlandse Akademie van Wetenschappen Verhandelingen, Afd. Natuurkunde (Tweede Reeks) p. 45–61. [Google Scholar]
- Derycke S, et al. 2005. Mitochondrial DNA variation and cryptic speciation within the free-living marine nematode Pellioditis marina. Mar Ecol Prog Ser. 300:91–103. [Google Scholar]
- Derycke S, et al. 2007. Spatiotemporal analysis of population genetic structure in Geomonhystera disjuncta (Nematoda, Monhysteridae) reveals high levels of molecular diversity. Mar Biol. 151:1799–1812. [Google Scholar]
- Derycke S, et al. 2010. Linking DNA sequences to morphology: cryptic diversity and population genetic structure in the marine nematode Thoracostoma trachygaster (Nematoda, Leptosomatidae). Zool Script. 39:276–289. [Google Scholar]
- Di Giulio M. 2005. A comparison of proteins from Pyrococcus furiosus and Pyrococcus abyssi: barophily in the physicochemical properties of amino acids and in the genetic code. Gene 346:1–6. [DOI] [PubMed] [Google Scholar]
- Ellner SP, Geber MA, Hairston NG. 2011. Does rapid evolution matter? Measuring the rate of contemporary evolution and its impacts on ecological dynamics. Ecol Lett. 14:603–614. [DOI] [PubMed] [Google Scholar]
- Erixon P, Oxelman B. 2008. Whole-Gene Positive Selection, Elevated Synonymous Substitution Rates, Duplication, and Indel Evolution of the Chloroplast clpP1 Gene. Plos One 3:e1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foerstner KU, von Mering C, Hooper SD, Bork P. 2005. Environments shape the nucleotide composition of genomes. EMBO Rep. 6:1208–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca G, Derycke S, Moens T. 2008. Integrative taxonomy in two free-living nematode species complexes. Biol J Linn Soc. 94:737–753. [Google Scholar]
- Gittenberger A, Gittenberger E. 2011. Cryptic, adaptive radiation of endoparasitic snails: sibling species of Leptoconchus (Gastropoda: Coralliophilidae) in corals. Organ Divers Evol. 11:21–41. [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. 2007. Epigenetics: a landscape takes shape. Cell 128:635–638. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Mariscal L, Betanzos A, Nava P, Jaramillo BE. 2003. Tight junction proteins. Prog Biophys Mol Biol. 81:1–44. [DOI] [PubMed] [Google Scholar]
- Gunzel D, Yu ASL. 2013. Claudins and the modulation of tight junction permeability. Physiol Rev. 93:525–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta BKS. 2002. Modern Foraminifera. Dordrecht (Germany): Kluwer Academic Publishers. [Google Scholar]
- Heip C, Vincx M, Vranken G. 1985. The ecology of marine nematodes. Oceanogr Mar Biol. 23:399–489. [Google Scholar]
- Jun X, et al. 2011. Complete genome sequence of the obligate piezophilic hyperthermophilic archaeon Pyrococcus yayanosii CH1. J Bacteriol. 193:4297–4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung T, Catalgol B, Grune T. 2009. The proteasomal system. Mol Aspects Med. 30:191–296. [DOI] [PubMed] [Google Scholar]
- Kamachi Y, et al. 2009. Evolution of non-coding regulatory sequences involved in the developmental process: reflection of differential employment of paralogous genes as highlighted by Sox2 and group B1 Sox genes. Proc Jpn Acad Ser B Phys Biol Sci. 85:55–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, et al. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussell E, Leibler S. 2005. Phenotypic diversity, population growth, and information in fluctuating environments. Science 309:2075–2078. [DOI] [PubMed] [Google Scholar]
- Laderoute KR, et al. 2006. 5'-AMP-activated protein kinase (AMPK) is induced by low-oxygen and glucose deprivation conditions found in solid-tumor microenvironments. Mol Cell Biol. 26:5336–5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambshead PJD, Boucher G. 2003. Marine nematode deep-sea biodiversity - hyperdiverse or hype? J Biogeogr. 30:475–485. [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg S. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li EC, et al. 2014. Transcriptome sequencing revealed the genes and pathways involved in salinity stress of Chinese mitten crab, Eriocheir sinensis. Physiol Genomics. 46:177–190. [DOI] [PubMed] [Google Scholar]
- Li WZ, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. [DOI] [PubMed] [Google Scholar]
- Lobo I. 2008. Environmental influences on gene expression. Nat Edu. 1:39 [Google Scholar]
- Lobry JR. 1997. Influence of genomic G+C content on average amino-acid composition of proteins from 59 bacterial species. Gene 205:309–316. [DOI] [PubMed] [Google Scholar]
- Lonsdale DJ, Frey MA, Snell TW. 1998. The role of chemical signals is copepod reproduction. J Mar Syst. 15:1–12. [Google Scholar]
- Lopez-Maury L, Marguerat S, Bahler J. 2008. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat Rev Genet. 9:583–593. [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. 2005. BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in biological networks. Bioinformatics 21:3448–3449. [DOI] [PubMed] [Google Scholar]
- Mao XZ, Cai T, Olyarchuk JG, Wei LP. 2005. Automated genome annotation and pathway identification using the KEGG orthology (KO) as a controlled vocabulary. Bioinformatics 21:3787–3793. [DOI] [PubMed] [Google Scholar]
- Martin AP. 1995. Metabolic-rate and directional nucleotide substitution in animal mitochondrial-DNA. Mol Biol Evol. 12:1124–1131. [DOI] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17:10–12. [Google Scholar]
- Mayr E. 1963. Animal species and evolution. Cambridge (MA): Harvard University Press. [Google Scholar]
- Min XJ, Butler G, Storms R, Tsang A. 2005. OrfPredictor: predicting protein-coding regions in EST-derived sequences. Nucleic Acids Res. 33:W677–W680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner JB. 2005. Organizational behavior 2: essential theories of process and structure. Armonk (NY): M.E. Sharpe. [Google Scholar]
- Mokievsky VO, Filippova KA, Chesunov AV. 2005. Nematode fauna associated with detached kelp accumulations in the subtidal zone of the White Sea. Oceanology 45:689–697. [Google Scholar]
- Mombaerts P. 1999. Seven-transmembrane proteins as odorant and chemosensory receptors. Science 286:707–711. [DOI] [PubMed] [Google Scholar]
- Morgan M., et al. 2009. ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics 25(19):2607–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T. 2008. Comparative sequence analysis of myosin heavy chain proteins from congeneric shallow- and deep-living rattail fish (genus Coryphaenoides). J Exp Biol. 211:1362–1367. [DOI] [PubMed] [Google Scholar]
- Morita T. 2010. High-pressure adaptation of muscle proteins from deep-sea fishes, Coryphaenoides yaquinae and C. armatus. Ann N Y Acad Sci. 1189:91–94. [DOI] [PubMed] [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35:W182–W185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura A, Savageau MA, Alves R. 2013. Relative amino acid composition signatures of organisms and environments. PLoS One 8(10):e77319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musto H, et al. 2006. Genomic GC level, optimal growth temperature, and genome size in prokaryotes. Biochem Biophys Res Commun. 347:1–3. [DOI] [PubMed] [Google Scholar]
- Mustroph A, et al. 2010. Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol. 152:1484–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New AM, et al. 2014. Different levels of catabolite repression optimize growth in stable and variable environments. PLoS Biol. 12(1):e1001764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NRC (National Research Council). 1979. Subcomittee on Hydrogen Sulfide. Baltimore: University Park Press. [Google Scholar]
- Oliver JL, Marin A. 1996. A relationship between GC content and coding-sequence length. J Mol Evol. 43:216–223. [DOI] [PubMed] [Google Scholar]
- Olson-Manning CF, Wagner MR, Mitchell-Olds T. 2012. Adaptive evolution: evaluating empirical support for theoretical predictions. Nat Rev Genet. 13:867–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbi SR. 1994. Genetic-divergence, reproductive isolation, and marine speciation. Annu Rev Ecol Syst. 25:547–572. [Google Scholar]
- Pfennig DW, et al. 2010. Phenotypic plasticity's impacts on diversification and speciation. Trends Ecol Evol. 25:459–467. [DOI] [PubMed] [Google Scholar]
- Pond DW, Tarling GA, Mayor DJ. 2014. Hydrostatic pressure and temperature effects on the membranes of a seasonally migrating marine copepod. PLoS One 9:e111043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnova D, Haflidason H, Todt C. 2010. Nematode species distribution patterns at the Nyegga pockmarks. Proceedings of the 14th International Meiofauna Conference (FourtIMCo). Ghent (Belgium): Ghent University. p. 174.
- Powell MA, Somero GN. 1986. Hydrogen sulfide oxidation is coupled to oxidative phosphorylation in mitochondria of Solemya reidi. Science 233:563–566. [DOI] [PubMed] [Google Scholar]
- Pradel N, et al. 2013. The first genomic and proteomic characterization of a deep-sea sulfate reducer: insights into the piezophilic Lifestyle of Desulfovibrio piezophilus. PLoS One 8: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C, Zhou SK, Xiao YB, Chen L. 2014. Erythropoietin enhances mitochondrial biogenesis in cardiomyocytes exposed to chronic hypoxia through Akt/eNOS signalling pathway. Cell Biol Int. 38:335–689. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2013. R: a language and environment for statistical computing. [Internet]. Vienna (Austria): R Foundation for statistical computing; Available from: http://www.R-project.org/. [Google Scholar]
- Ramani AK, et al. 2011. Genome-wide analysis of alternative splicing in Caenorhabditis elegans. Genome Res. 21:342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisner K. 2012. Physiological roles of fatty acids and Endocannabinoids in the Nematode Caenorhabditis elegans. Kuopio (Finland): Publications of the University of Easten Finland. [Dissertations in Health Sciences 100]. 100 p.
- Russell DV, Patricia AM, Barbara J, John OB. 1989. Reduced sulfur compounds in the marine environment In: Biogenic sulfur in the environment: American Chemical Society; p. 243–261. [Google Scholar]
- Seehausen O, et al. 2014. Genomics and the origin of species. Nat Rev Genet. 15:176–192. [DOI] [PubMed] [Google Scholar]
- Shannon P, et al. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer GAC, Hickey DA. 2000. Nucleotide bias causes a genomewide bias in the amino acid composition of proteins. Mol Biol Evol. 17:1581–1588. [DOI] [PubMed] [Google Scholar]
- Singh R, Valcarcel J, Green MR. 1995. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268:1173–1176. [DOI] [PubMed] [Google Scholar]
- Somero G, Childress J, Anderson A. 1989. Transport, metabolism, and detoxification of hydrogen sulfide in animals from sulfide-rich marine environments. Rev Aqu Sci. 1(4):591–614. [Google Scholar]
- Stamm S. 2002. Signals and their transduction pathways regulating alternative splicing: a new dimension of the human genome. Hum Mol Genet. 11:2409–2416. [DOI] [PubMed] [Google Scholar]
- Stanhope MJ, Connelly MM, Hartwick B. 1992. Evolution of a crustacean chemical communication channel—behavioral and ecological genetic-evidence for a habitat-modified, race-specific pheromone. J Chem Ecol. 18:1871–1887. [DOI] [PubMed] [Google Scholar]
- Sueoka N. 1988. Directional mutation pressure and neutral molecular evolution. Proc Natl Acad Sci U S A. 85:2653–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MA, et al. 1993. The inheritance of germline-specific epigenetic modifications during development. Philos Trans Soc Lond Ser B Biol Sci. 339:165–172. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchesunov AV, Portnova DA, Van Campenhout J. 2015. Description of two free-living nemaode species of Halomonhystera disjuncta complex (Nematoda: Monhysterida) from two peculiar habitats in the seat. Helgoland Mar Res. 1–29. [Google Scholar]
- Theron M, Guerrero F, Sebert P. 2000. Improvement in the efficiency of oxidative phosphorylation in the freshwater eel acclimated to 10.1 MPa hydrostatic pressure. J Exp Biol. 203:3019–3023. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toffaletti DL, Del Poeta M, Rude TH, Dietrich F, Perfect JR. 2003. Regulation of cytochrome c oxidase subunit 1 (COX1) expression in Cryptococcus neoformans by temperature and host environment. Microbiology 149:1041–1049. [DOI] [PubMed] [Google Scholar]
- Tokuda S, Miyazaki H, Nakajima KI, Yamada T, Marunaka Y. 2009. Hydrostatic pressure regulates tight junctions, actin cytoskeleton and transcellular ion transport. Biochem Biophys Res Commun. 390:1315–1321. [DOI] [PubMed] [Google Scholar]
- Tondera D, et al. 2009. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 28:1589–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter D, Webster JM. 1983. Distribution and abundance of marine nematodes on the kelp Macrocystis-Integrifolia. Mar Biol. 78:39–43. [Google Scholar]
- Van Campenhout J, Derycke S, Moens T, Vanreusel A. 2014. Differences in life-histories refute ecological equivalence of cryptic species and provide clues to the origin of bathyal Halomonhystera (Nematoda). PLos One 9:e111889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Campenhout J, Derycke S, Tchesunov A, Portnova D, Vanreusel A. 2013. The Halomonhystera disjuncta population is homogeneous across the Håkon Mosby mud volcano (Barents Sea) but is genetically differentiated from its shallow-water relatives. J Zool Syst Evol Res. 52:203–216. [Google Scholar]
- Van Gaever S, et al. 2009. Trophic specialisation of metazoan meiofauna at the HAyenkon Mosby Mud Volcano: fatty acid biomarker isotope evidence. Mar Biol. 156:1289–1296. [Google Scholar]
- Van Gaever S, Moodley L, de Beer D, Vanreusel A. 2006. Meiobenthos at the Arctic Hakon Mosby Mud Volcano, with a parental-caring nematode thriving in sulphide-rich sediments. Mar Ecol Prog Ser. 321:143–155. [Google Scholar]
- van Megen H, et al. 2009. A phylogenetic tree of nematodes based on about 1200 full-length small subunit ribosomal DNA sequences. Nematology 11:927 S927. [Google Scholar]
- Vismann B. 1991. Sulfide tolerance: physiological mechanisms and ecological implications. Ophelia 34:1–27. [Google Scholar]
- Voet D, Voet JG, Pratt CW. 2006. Fundamentals of biochemistry: life at the molecular level. New Jersey: Wiley. [Google Scholar]
- Völkel S, Grieshaber M. 1996. Sulphide oxidation and oxidative phosphorylation in the mitochondria of the lugworm. J Exp Biol. 200:83–92. [DOI] [PubMed] [Google Scholar]
- Vranken G, Herman PMJ, Heip C. 1988. Studies of the life-history and energetics of marine and brackish-water nematodes: 1. Demography of Monhystera-Disjuncta at different temperature and feeding conditions. Oecologia 77:296–301. [DOI] [PubMed] [Google Scholar]
- Vranken G, Tire C, Heip C. 1988. The toxicity of paired metal mixtures to the nematode Monhystera-Disjuncta (Bastian, 1865). Mar Environ Res. 26:161–179. [Google Scholar]
- Vranken G, Tire C, Heip C. 1989. Effect of temperature and food on hexavalent chromium toxicity to the marine nematode Monhystera-Disjuncta. Mar Environ Res. 27:127–136. [Google Scholar]
- Vranken G, Vanderhaeghen R, Heip C. 1985. Toxicity of cadmium to free-living marine and brackish water nematodes (Monhystera-Microphthalma, Monhystera-Disjuncta, Pellioditis-Marina). Dis Aqu Organ. 1:49–58. [Google Scholar]
- Vrijenhoek RC. 2009. Cryptic species, phenotypic plasticity, and complex life histories: Assessing deep-sea faunal diversity with molecular markers. Deep-Sea Res Part II Topic Stud Oceanogr. 56:1713–1723. [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136:701–718. [DOI] [PubMed] [Google Scholar]
- Walsh SJ, Mena CF. 2013. Science and conservation in the Galapagos Islands: frameworks & perspectives. New York: Springer. [Google Scholar]
- Wang D-P, Wan H-L, Zhang S, Yu J. 2009. gamma-MYN: a new algorithm for estimating Ka and Ks with consideration of variable substitution rates. Biol Direct. 4:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zhang Y, Zhang Z, Zhu J, Yu J. 2010. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinformatics 8:77–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CM, Burdge G. 2006. Long-chain n-3 PUFA: plant v. marine sources. Proc Nutr Soc. 65:42–50. [DOI] [PubMed] [Google Scholar]
- Wilquet V, Van de Casteele M. 1999. The role of the codon first letter in the relationship between genomic GC content and protein amino acid composition. Res Microbiol. 150:21–32. [DOI] [PubMed] [Google Scholar]
- Yano Y, Nakayama A, Yoshida K. 1997. Distribution of polyunsaturated fatty acids in bacteria present in intestines of deep-sea fish and shallow-sea poikilothermic animals. Appl Environ Microbiol. 63:2572–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XQ, Zhang Z, Yan JW, Yu J. 2007. GC content variability of eubacteria is governed by the pol III alpha subunit. Biochem Biophys Res Commun. 356:20–25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.