Abstract
Body size, one of the most important quantitative traits under evolutionary scrutiny, varies considerably among species and among populations within species. Revealing the genetic basis underlying this variation is very important, particularly in humans where there is a close relationship with diseases and in domestic animals as the selective patterns are associated with improvements in production traits. The Debao pony is a horse breed with small body size that is unique to China; however, it is unknown whether the size-related candidate genes identified in Western breeds also account for the small body size of the Debao pony. Here, we compared individual horses from the Debao population with other two Chinese horse populations using single nucleotide polymorphisms (SNPs) identified with the Equine SNP 65 Bead Chip. The previously reported size-related candidate gene HMGA2 showed a significant signature for selection, consistent with its role observed in human populations. More interestingly, we found a candidate gene TBX3, which had not been observed in previous studies on horse body size that displayed the highest differentiation and most significant association, and thus likely is the dominating factor for the small stature of the Debao pony. Further comparison between the Debao pony and other breeds of horses from around the world demonstrated that TBX3 was selected independently in the Debao pony, suggesting that there were multiple origins of small stature in the horse.
Keywords: selective signature, di, XP-EHH, genetic differentiation, association, TBX3
Introduction
Body size, one of the most important quantitative traits under evolutionary scrutiny, varies considerably at both the inter- and intraspecific levels (Blanckenhorn et al. 1999; Kraushaar and Blanckenhorn 2002). Understanding the genetic foundation of body size variation is particularly important, especially in humans for studies on the genetic mechanisms underlying associated diseases and in domesticated animals for understanding the patterns of artificial selection and for improving production traits. Compared with populations that only experience natural selection, tremendous variation in body size is observed in domesticated species, such as dogs and chickens, which have experienced strong artificial selective pressures aimed at this trait for improvements or for exaggeration. Numerous genes that have small phenotypic effect on overall human body size have been identified, whereas the reverse pattern (few genes with large effect) has been described in domesticated animals, with a subset of genes convergently identified from both lists (Sutter et al. 2007; Boyko et al. 2010; Lango Allen et al. 2010; Makvandi-Nejad et al. 2012). For example, IGF1 together with almost 700 other genes explains only approximately 16% of the height variation observed in humans (Lango Allen et al. 2010), but a single mutation within IGF1 is the dominant explanation for small body size in several different breeds of dogs (Sutter et al. 2007), and the majority of the variation in the average body mass of dogs from different breeds can be explained by only six loci (Boyko et al. 2010; Rimbault et al. 2013).
The official definition of a pony is a horse that measures less than 14.2 hands (58 inches, 147 cm) at the withers (Mattern 2010). Guangxi Mountains of Southern China is of particular interest due to its extremely small size (withers height less than 106 cm) (China National Commission of Animal Genetic Resources 2011) (fig. 1A). It is thought that this breed was selected for its small size to facilitate work in mountainous regions. Similar small stature horse breeds (e.g., Shetland and Caspian ponies) are found in other location in the world, where they serve a variety of functions (e.g., transport in mines and carrying children). Given the restricted geographic distributions of these different breeds, it is possible that small stature was independently selected across these distinct horse breed lineages. Although previous studies have identified quantitative trait loci on equine chromosomes (e.g., horse chromosomes 3, 6, 9, and 11) that contribute to size variation in breeds developed in Europe, America and the Middle East, where frequent migration occurred during the breeding process, it remains unknown if these loci account for the size variation observed in all horse breeds (Makvandi-Nejad et al. 2012).
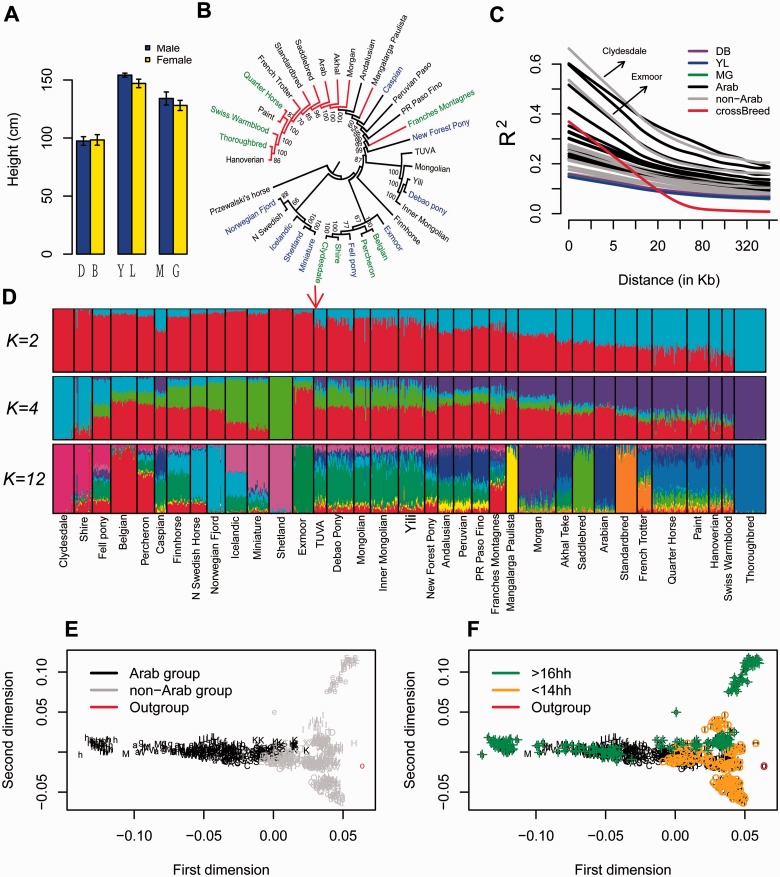
Fig. 1.—
Phylogenetic analysis of horse breeds. (A) Height of the three breeds: DB, MG, and YL. Data were retrieved from China National Commission of Animal Genetic Resources (2011). (B) Phylogenetic tree constructed by TreeMix with bootstrap analysis with the Przewalski’s horse as the outgroup. Red branches are “Arab group.” Names labeled in blue are short stature horses/pony, and green are tall horses. (C) Decay of LD among 35 breeds of horses. (D) Population structure analysis among the 35 breeds. Red arrow signs Przewalski’s horse. (E, F) Multidimensional clustering. Each letter represents a breed. A, Akhal; B, Andalusian; C, Arabian; D, Belgian; E, Caspian; F, Clydesdale; G, Debao; H, Exmoor; I, Fell pony; J, Finnhorse; K, Franches-Montagnes; L, French Trotter; M, Hanoverian; N, Icelandic; O, Inner Mongolian; P, MangalargaPaulista; Q, Miniature; R, Mongolian; S, Morgan; T, New Forest Pony; U, North Swedish Horse; V, Norwegian Fjord; W, Paint; X, Percheron; Y, Peruvian Paso; Z, Puerto Rican Paso Fino; a, Quarter Horse; b, Saddlebred; d: Shetland; e, Shire; f, Standardbred; g, Swiss Warmblood; h, Thoroughbred; i, Tuva; j, Yili. The abbreviation “hh” stands for “hands high.” One hand is equal to 4 inches or 10.16 cm.
Here, we conducted a genomic scan to search for signatures of positive selection for diminutive body size in the Debao pony (DB) and selection for the relatively higher stature of the Yili (YL) and Inner Mongolian (MG) horse breeds from northern China (fig. 1A), using a comparative approach with a world-wide sample of 730 horses. Our objectives were to 1) investigate the phylogenic relationship between the three Chinese horses and previously studied horses to estimate the origin of the DB, 2) compare the population genomic structure of the DB with those of other horses to identify genetic mechanisms underlying the small size of the DB, and 3) identify if introgression of size-related genes occurred among different horse breeds to determine whether small stature had a single or multiple origins.
Materials and Methods
A total of 96 horse individuals from three horse breeds (DB [n = 32], YL [n = 32], and MG [n = 32]) were genotyped using the GeneSeek Equine SNP 65 Bead Chip panel. Genotyped SNPs data from 32 other breeds of horses (n = 729) from previous studies (Petersen, Mickelson, Cothran, et al. 2013; Petersen, Mickelson, Rendahl, et al. 2013) were also obtained. SNPs that failed in exact tests for Hardy–Weinberg equilibrium at P < 0.001 or had more than 10% missing genotype data or had Minor Allele Frequency less than 5% were excluded using PLINK (Purcell et al. 2007). Individuals with more than 10% missing genotyped data were also removed. In total, 816 individuals with 21,740 SNPs were retrieved for the phylogenetic analysis and multidimensional clustering (Purcell et al. 2007). SNP data from a Przewalski’s horse were downloaded from a study (Orlando et al. 2013) and merged with the data from the domesticated horses to construct a maximum-likelihood tree using TreeMix (Pickrell and Pritchard 2012), based on the genome-wide allele frequency data. With a pruned data set, which discarded SNPs in linkage disequilibrium (LD) across breeds (PLINK –indep 50 5 2), we implemented a Bayesian model-based approach to assess the population relationship of the world-wide horses using the program STRUCTURE (Pritchard et al. 2000). Following the same SNP quality filtration, a new small data set including only the three Chinese breeds was created and the population relationships were analyzed after LD-based SNP pruning. To compare the diversity of the Chinese population with reported diversity of other horse breeds (Petersen, Mickelson, Cothran, et al. 2013), we pruned the data set for pairwise r2 < 0.1 considering 100 SNP windows with a step size of 25 SNPs. To compare LD decay among breeds with different sample sizes, we selected a random subset of 14 horses from each of the 35 breeds. All breeds combined LD decay were calculated with each 14 individuals for a breed (14 × 35 = 490).
We used a total of 52,244 SNPs to calculate the population differentiation among the DB, YL, and MG horses as described in Akey et al. (2002). To retrieve candidate SNPs under selection in the DB, di values were also calculated for each SNP as described in Akey et al. (2010). Haplotypes were phased by the fastPHASE program (Scheet and Stephens et al. 2006). XP-EHH values of each SNP in the Debao compared with the YL and the MG were calculated by the xpehh program (http://hgdp.uchicago.edu/Software/, last accessed December 16, 2015). The two values of XP-EHH (DB vs. YL) and XP-EHH (DB vs. MG) were merged using where P is the percentile rank value of the XP-EHH values of each SNP.
All SNPs within this small data set were also considered in scans for association (Pickrell and Pritchard 2012). A potential difference in association with stature variation between DB and YL was ignored after testing the “quantitative trait interaction” (G×E). Although the influence of sex was not significant in the multiple regression analysis, we still considered sex when performing the association analysis. We also performed FDR adjustment and “adaptive” model-based permutation using PLINK (Pickrell and Pritchard 2012). Candidate genes were identified whether they are located within 150 kb of an SNP of interest. Gene ontology (GO) enrichment analysis was performed on g: Profiler (http://biit.cs.ut.ee/gprofiler). The STRING database was queried for the protein–protein interaction (http://string-db.org/) (Szklarczyk et al. 2011).
Results and Discussion
Phylogenetic Analysis of the DB
The GeneSeek Equine SNP (single nucleotide polymorphism) 65 Bead Chip panel was used to genotype 32 individuals each from the DB, YL, and MG breeds of horse. To assess the phylogenetic relationships of the Chinese horses to other horse breeds, we obtained SNP genotype data from 32 different breeds from a previous study (Petersen, Mickelson, Rendahl, et al. 2013), which yielded a total of 21,740 SNPs that passed our quality filtration (see Materials and Methods). After merging with the SNP data from a Przewalski’s horse (Orlando et al. 2013), a total of 14,959 SNPs were used to infer a maximum-likelihood tree of the horse populations using the genome-wide allele frequency from the Przewalski’s horse as the outgroup (fig. 1B). As expected, all three Chinese breeds genotyped here clustered with the Mongolian horses (fig. 1B), consistent with their close geographic distribution. The relationships of the ponies, Scandinavian breeds, heavy draft horses, recent breeds derived from the Thoroughbred, modern US breeds, and the trotting breeds (fig. 1B) were in agreement with that described in a previous study (Petersen, Mickelson, Cothran, et al. 2013). Landrace breeds, which originated geographically, ranged freely and experience lesser degrees of management, located basally in the tree with close relationship with the wild horse, and many of the modern breeds clustered together (fig. 1B). Considering the fact that most modern breeds were recently created by infusing Arabian, and/or Thoroughbred, which is also inherited majorly from Arabian, we simply grouped these breeds into an “Arab group” and the remaining into a “non-Arab group” (Materials and Methods). Ancient breeds located basally in the tree with close relationship with the wild horse, and many of the modern breeds clustered together (fig. 1B). Breeds with short stature were located at disperse locations in the phylogenetic trees, suggesting independent domestication of these short stature breeds. However, when the TreeMix program (Pickrell and Pritchard 2012) was used to infer migration events, the tree topology frequently changed with changes in the defined number of migration events (supplementary fig. S1, Supplementary Material online), suggesting a complex history for horse domestication with genetic admixtures occurring among breeds. The data also suggest that the exact phylogenetic relationship might be difficult to reveal based only on allele frequency data, particularly here, since genetic admixture occurred and only one wild Przewalski’s horse was available. This is illustrated by comparing our results with those of previous studies. Petersen, Mickelson, Cothran, et al. (2013) found that the Mangalarga Paulista was located basally in a phylogenetic tree constructed by the parsimony method, whereas in contrast, McCue et al. (2012) identified the Norwegian Fjord and Icelandic as being basal in their phylogenetic tree, and here, we have the Norwegian Fjord, N Swedish, and Shetland horses appearing close to the Przewalski’s horse (fig. 1B); however, we also detected migrations from the Przewalski’s horse to these three breeds (supplementary fig. S1, Supplementary Material online).To better analyze the phylogenetic relationships, we next implemented a Bayesian model-based approach to assess the individual assignment using the program STRUCTURE (Pritchard et al. 2000) (fig. 1D). A gradient from “non-Arab group” to “Arab group” was found whether we assumed that there were two ancestral populations (K = 2). Some European draft (Clydesdale, Shire) and pony breeds (Icelandic, Miniature, Shetland) separated when four ancestral populations (K = 4) were assumed. Horses from Mongolia, China and Siberia grouped into a distinct cluster when K = 12, suggesting a much closer relationship among these horses from the middle part of the Eurasian continent. When K = 12, Przewalski’s horse showed closer relationship and clustered with the Chinese breeds. Consistently, the neighbor-joining tree based on all individuals showed that the Chinese breeds had the closest relationship to the Przewalski’s horse (supplementary fig. S2, Supplementary Material online), which contrasts with the phylogenetic tree inferred by whole population allele frequency data (fig. 1B). These results also suggest that the phylogenetic relationships are difficult to reveal based only on allele frequency data. Genetic migrations between the Przewalski’s horse and the Chinese horse breeds were not detected by our TreeMix analysis, which potentially suggests that there is a true closer relationship between the Przewalski’s horse and the Chinese breeds (supplementary fig. S1, Supplementary Material online).
To exclude the possibility that the estimated population structures were influenced by the specific algorithmic strategy, we also used principal component analysis (PCA) to examine the population relationships (supplementary fig. S3, Supplementary Material online). Similar to the above analysis, the first dimension moderately distinguished the “Arab group” and “non-Arab group” (fig. 1E), or the ancient and modern breeds (supplementary fig. S4, Supplementary Material online), whereas the second dimension moderately separated horses with differing statures (fig. 1F). Horses from the Middle East and Asia region clustered together in the central part of the multidimensional scaling plot (fig. 1E and F, supplementary fig. S5, Supplementary Material online). Consistent with the above, the Przewalski’s horse is close to the ancient breeds and non-Arab breeds (fig. 1E and supplementary fig. S4B, Supplementary Material online), but in two dimensions we could not distinguish clearly the Przewalski’s horse from the domestic breeds. The individual closest to the Przewalski’s horse in the PCA was the Exmoor, but the TreeMix analysis found strong evidence of admixture between the Exmoor and the Przewalski’s horse (supplementary fig. S1, Supplementary Material online).
Consistent with the phylogenetic tree and the structural analysis, the Chinese breeds were located within the ancient breeds in the PCA analysis (supplementary fig. S4B, Supplementary Material online), and closer than most breeds to the Przewalski’s horse (supplementary fig. S6, Supplementary Material online). The Chinese breeds also showed higher levels of observed heterozygosity (supplementary fig. S7, Supplementary Material online), which were comparable to the expected level of heterozygosity (Petersen, Mickelson, Cothran, et al. 2013). In addition, LD, which can be informative for population demography, found a similar decay in each of the three Chinese breeds, with their low levels of LD similar to those seen within other landrace breeds (fig. 1C). There were two exceptions: Clydesdale and Exmoor, which have small population sizes compared with other landrace breeds (Petersen, Mickelson, Cothran, et al. 2013) and therefore showed relative long linked segments (fig. 1C). Thus, the higher levels of genetic diversity, the closer relationship to the ancient breeds, and the lower levels of LD in the DB than in other recent breeds suggest an ancient origin for the DB, consistent with its antiquity record dating to at least 2,000 years ago (Hendricks 2007). We also quantified runs of homozygosity (ROH) to assess recent inbreeding. The three Chinese breeds showed similar ROH means, with the peaks of the ROH distributions near the same lengths (supplementary fig. S8A and B, Supplementary Material online), indicative of similar contemporary population sizes and mating systems. Taken together, these results indicate that the three Chinese breeds have an ancient origin, which is consistent with their anecdotal records of breed history (Wentong 1990; China National Commission of Animal Genetic Resources 2011). Perhaps surprisingly, LD across whole breeds did not rapidly decay, a pattern that is very different from that observed in dogs (Boyko et al. 2010) (fig. 1C), which may indicate that numerous IBD (identity by descent) segments are shared across multiple breeds and/or the shared segments are quite large. This pattern is consistent with horse breeding history as many modern breeds originated, at least in part, from the Arabian horses, in a relatively short period of time.
As the DB individuals could not be assigned to a single cluster using the SNPs detected by all horses, we next assessed the populations of the three Chinese breeds as a group using the 52,244 SNPs passed after quality filtration. The results from both STRUCTURE and PCA clearly distinguished the DB from the other two populations (supplementary fig. S8C and D, Supplementary Material online), which may indicate a differentiation between South China and North China horse populations.
Signatures of Positive Selection in the DB
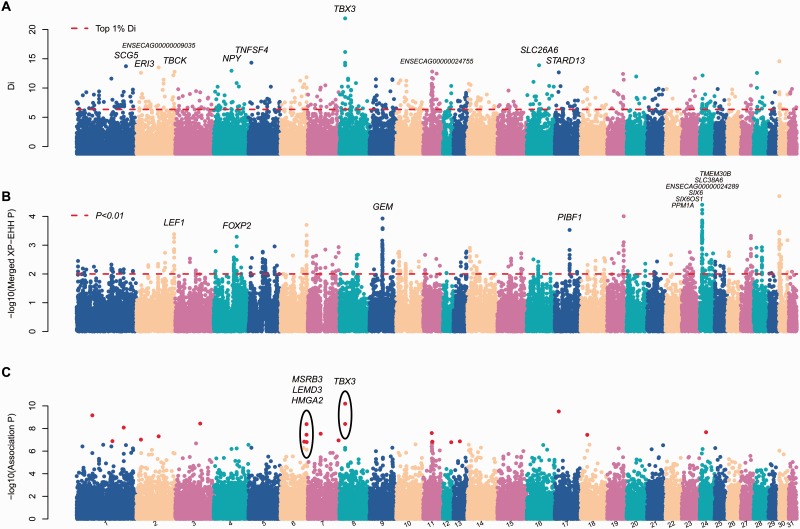
Given that the DB possesses a significantly low stature and their population characteristics imply an ancient history, we sought to identify candidate loci that regulate this phenotype and estimate the origin of this phenotype. Population structure analysis implied a much closer relationship among the DB, YL, and MG breeds; therefore, we first evaluated the population differentiation of DB from the other two Chinese breeds to search for loci that show evidence of positive selection specific to the DB, using the statistic di, as described in Akey et al. (2010). We first extracted the top SNPs using a cutoff of 0.5%, and found 262 SNPs, which mapped to 153 genes (within 150 kb of the SNPs). Gene enrichment analysis found some categories associated with development of the skeletal system, such as “Abnormal morphology of forearm bone,” “Abnormality of the ischium,” “Dumbbell-shaped long bone,” and “Aplasia/Hypoplasia involving bones of the feet,” but each had very few genes (supplementary table S1, Supplementary Material online). When a cutoff of 1% was used, a total of 523 SNPs (di > 6.34) were identified (fig. 2A). Gene enrichment analysis of the 338 genes closest to these SNP found a significant overrepresentation in developmental process, for example, 87 genes involved in “anatomical structure development” (GO:0048856), 17 genes involved in “skeletal system development” (GO:0001501), 10 genes in “cartilage development” (GO:0051216), 11 genes (HAND2, LEF1, ALX3, COL2A1, ZBTB16, TBX3, SULF1, ASPH, EN1, PCSK5, CHST11) involved in “limb development” (GO:0060173), and “limb morphogenesis” (GO:0035108) (table 1). We also extracted the top SNPs using cutoffs of 0.1%, and obtained similar results for enrichment of categories associated development of skeletal system (supplementary tables S2, Supplementary Material online). However, due to ascertainment bias and the small number of genotyped SNPs, it is difficult to identify the number of genes that have evolved under positive selection in the Debao. Whole-genome sequences from multiple individuals would be necessary for validating this.
Fig. 2.—
Positive selection analysis of the DB. Genome-wide distribution of (A) di, (B) the P values of the merged XP-EHH values, (C) and Manhattan plot presenting the association P value across the genome in the DB.
Table 1.
Overrepresented GO Categories among Genes Showing High di Value in DB
| Term ID | Description | P Value | Gene Number |
|---|---|---|---|
| GO:0009653 | Anatomical structure morphogenesis | 2.36E-05 | 57 |
| GO:0009888 | Tissue development | 4.33E-05 | 42 |
| GO:0007275 | Multicellular organismal development | 5.62E-05 | 82 |
| GO:0030902 | Hindbrain development | 5.80E-05 | 11 |
| GO:0009790 | Embryo development | 6.85E-05 | 35 |
| GO:0048856 | Anatomical structure development | 7.07E-05 | 87 |
| GO:0044767 | Single-organism developmental process | 1.29E-04 | 90 |
| GO:0032502 | Developmental process | 1.53E-04 | 90 |
| GO:0043009 | Chordate embryonic development | 1.89E-04 | 26 |
| GO:0009792 | Embryo development ending in birth or egg hatching | 2.23E-04 | 26 |
| GO:0002062 | Chondrocyte differentiation | 2.51E-04 | 9 |
| GO:0048731 | System development | 3.29E-04 | 74 |
| GO:0048513 | Organ development | 6.02E-04 | 60 |
| GO:0072089 | Stem cell proliferation | 1.12E-03 | 10 |
| GO:0035107 | Appendage morphogenesis | 1.35E-03 | 11 |
| GO:0035108 | Limb morphogenesis | 1.35E-03 | 11 |
| GO:0061448 | Connective tissue development | 2.07E-03 | 12 |
| GO:0048736 | Appendage development | 2.64E-03 | 11 |
| GO:0060173 | Limb development | 2.64E-03 | 11 |
| GO:0001944 | Vasculature development | 2.72E-03 | 20 |
| GO:0001501 | Skeletal system development | 2.91E-03 | 17 |
| GO:0072359 | Circulatory system development | 4.14E-03 | 26 |
| GO:0072358 | Cardiovascular system development | 4.14E-03 | 26 |
| GO:0001701 | In utero embryonic development | 4.93E-03 | 17 |
| GO:0007417 | Central nervous system development | 5.05E-03 | 22 |
| GO:0061061 | Muscle structure development | 5.08E-03 | 18 |
| GO:0006029 | Proteoglycan metabolic process | 6.30E-03 | 6 |
| GO:0048468 | Cell development | 7.98E-03 | 38 |
| GO:0051216 | Cartilage development | 9.05E-03 | 10 |
| GO:0007420 | Brain development | 9.85E-03 | 18 |
| GO:0048568 | Embryonic organ development | 1.03E-02 | 17 |
| GO:0072091 | Regulation of stem cell proliferation | 1.04E-02 | 7 |
| GO:0009887 | Organ morphogenesis | 1.13E-02 | 26 |
| GO:2000648 | Positive regulation of stem cell proliferation | 1.30E-02 | 6 |
| GO:0048010 | Vascular endothelial growth factor receptor signaling pathway | 1.44E-02 | 5 |
| GO:0001568 | Blood vessel development | 1.47E-02 | 18 |
| GO:0060021 | Palate development | 1.49E-02 | 7 |
| GO:0007605 | Sensory perception of sound | 1.84E-02 | 8 |
| GO:0010629 | Negative regulation of gene expression | 1.85E-02 | 27 |
| GO:0051301 | Cell division | 1.89E-02 | 17 |
| GO:1902679 | Negative regulation of RNA biosynthetic process | 2.08E-02 | 25 |
| GO:0035295 | Tube development | 2.16E-02 | 18 |
| GO:0021549 | Cerebellum development | 2.43E-02 | 6 |
| GO:0048729 | Tissue morphogenesis | 2.47E-02 | 19 |
| GO:0014706 | Striated muscle tissue development | 2.52E-02 | 13 |
| GO:0048598 | Embryonic morphogenesis | 3.21E-02 | 19 |
| GO:0051253 | Negative regulation of RNA metabolic process | 3.25E-02 | 25 |
| GO:0048869 | Cellular developmental process | 3.26E-02 | 58 |
| GO:0007399 | Nervous system development | 3.32E-02 | 35 |
| GO:0050954 | Sensory perception of mechanical stimulus | 3.33E-02 | 8 |
| GO:0030154 | Cell differentiation | 3.38E-02 | 54 |
| GO:0007049 | Cell cycle | 3.50E-02 | 28 |
| GO:0007588 | Excretion | 3.60E-02 | 4 |
| GO:2000113 | Negative regulation of cellular macromolecule biosynthetic process | 3.94E-02 | 26 |
| GO:0002009 | Morphogenesis of an epithelium | 4.04E-02 | 16 |
| GO:0001570 | Vasculogenesis | 4.22E-02 | 6 |
| GO:0060537 | Muscle tissue development | 4.37E-02 | 13 |
| GO:0016486 | Peptide hormone processing | 4.54E-02 | 3 |
Beside high population differentiation, regions under directional selection also show other signatures of variation, such as long range haplotype homozygosity (Sabeti et al. 2006). To identify alleles potentially under selection in the DB, we employed XP-EHH, which is based on long-range haplotype homozygosity and is suitable for the detection of recent positive selection and is robust to the ascertainment bias of SNPs (Sabeti et al. 2006). A total of 505 SNPs have significantly high merged XP-EHH values in the DB (P < 0.01) (fig. 2B). A total of 635 genes were found within 150 kp of these SNPs, and these genes were found to be significantly enriched in GO terms for bone and muscle variation (supplementary table S3, Supplementary Material online), such as “upper limb undergrowth,” “abnormal cortical bone morphology,” “aplasia/hypoplasia affecting bones of the axial skeleton,” “aplasia/hypoplasia involving bones of the feet,” “abnormality of the musculature of the limbs,” “limb-girdle muscle weakness,” and “foot dorsiflexor weakness.” We also retrieved 122 genes with more significance of merged XP-EHH (P < 0.005), which showed similar enrichment in the development, bone and muscle variation, such as “regulation of developmental process,” “skeletal muscle tissue development,” “spinal cord lesions,” and “abnormality of the mandible” (supplementary table S4, Supplementary Material online).
Although many candidate selected genes are involved in the development of skeletal system, the selected alleles in DB pony identified by the outlier methods cannot be directly associated with small stature in DB pony, as they might be confounded by other unique phenotypes found in the DB. To verify whether the identified loci play a role in the small stature size of the DB, we conducted association scans within the DB and YL for variation in stature (fig. 2C). After adjustment for FDR, 22 SNPs showed a significant association with stature (BONF P value < 0.01), all of which harbored significantly high di values (top 1%) and/or significant merged XP-EHH values (P < 0.01). To control for potential confounding effects of stratification and the nonindependence of related individuals that may bias the association study (although the above population structure analysis showed no legacy of stratification for each of the Chinese breeds and we tried to collect samples from different families or villages), we performed an adaptive permutation and confirmed the most significant 22 SNPs for stature variation (supplementary table S5, Supplementary Material online). Furthermore, we also excluded the possibility that there could be differences for these 22 SNPs in their association with stature variation between the DB and YL genomic backgrounds, as no significance was found in an interaction analysis (G×E) (supplementary table S5, Supplementary Material online). These 22 SNPs together explain most of stature variance among the 63 horses from the DB and YL breeds (R2 = 0.943).
For the 22 candidate SNPs, only two regions (of less than 1 Mb) contained more than one adjacent SNP. The first genomic region is located on ECA8 and contained two SNPs: ECA8.18101000 and ECA8.18120526. ECA8.18101000 shows both the most significant association and the highest di value (di = 21.90) (fig. 2A and C) and the major genotype for this SNP in the DB was “A,” with a frequency of 80.65%, whereas this genotype had a frequency of only 18.85%, with no homozygous “AA” genotype individuals observed, in the two other breeds. The proportion of the variance for height explained by a single SNP was the highest for this SNP, with a R2 = 0.506. ECA8.18120526 has the fifth highest association significance and the second highest di value, with 70.97% of the genotypes being “G” in the DB, but only 19.30% in the other two breeds, and only one “GG” homozygote was observed in the MG. Both of these SNPs are located less than 100 kb upstream of the gene TBX3, a T-box gene that plays a role in the development of the anterior/posterior axis of the tetrapod forelimb (Bamshad et al. 1997, 1999). TBX3 was not detected in previous studies for short stature (Makvandi-Nejad et al. 2012; Signer-Hasler et al. 2012; Petersen, Mickelson, Cothran, et al. 2013; Tetens et al. 2013), suggesting the possibility of independent selection for small stature in the DB.
The second region with clustered SNPs is on ECA6 and contained three SNPs with high di values, high merged XP-EHH values, and significant association P values (fig. 2A–C). Although this region contains three genes, MSRB3, LEMD3 and HMGA2, HMGA2 is the likely candidate gene as it has been previously associated with height variation in human populations (Weedon et al. 2007; Yang et al. 2010) and was studied previously, and found to be associated with stature in horse populations (Makvandi-Nejad et al. 2012; Petersen, Mickelson, Rendahl, et al. 2013).HMGA2, like IGF1, has previously been suggested to have experienced convergent evolution in horses and humans (Petersen, Mickelson, Rendahl, et al. 2013). In addition, a previous association study in dogs, where these three genes are also clustered together, found the strongest association signal with body weight to be located near HMGA2, whereas the strongest association signal with ear floppiness was near MSRB3 (Boyko et al. 2010). Thus HMGA2 is the likely target of selection for short stature in the DB. The genotypes of all five SNPs (two near TBX3 and three near HMGA2) significantly associate with variation in stature (pairwise t-test) (supplementary fig. S9, Supplementary Material online). More interestingly, although TBX3 does not interact directly with HMGA2, they both directly interact with BMP3 and CDH4, which regulate the bone synthesis. Furthermore, TBX3 and HMGA2 together explain 71.8% of the variance in height, with the explained proportion rising to 83.3% when an interaction between these two genes is added. These results therefore strongly suggest that a complex network underlies the evolution of small body size in the DB.
Nearly 1% of all human genes have been implicated in contributing to size variation (Lango Allen et al. 2010). In contrast, only a small number of genes appear to dominate size variation in domestic animals, such as dogs (Boyko et al. 2010; Rimbault et al. 2013) and horses (Makvandi-Nejad et al. 2012). Qualitative evidence points to the genetic control of size by many loci with small effect in naturally evolving species, but by few loci with large effect in domesticated species. Still, as most genes perform conserved roles in regulating size across species, this should provide opportunities to explore the genetics of size variation in humans and other mammals.
Although previous studies have identified several genes that may be influential in the determination of horse size, imaging a common genetic mechanism that accounts for all horse populations is difficult as many populations are restricted to localized environment where they were bred for different goals. Here we report that the gene TBX3 likely dominates the small stature of the DB horse, an ancient small pony specific to China that was selected for transport in mountainous regions. TBX3 has not been associated with body size in other horse populations (Makvandi-Nejad et al. 2012; Petersen, Mickelson, Rendahl, et al. 2013). Moreover, only one gene, HMGA2, from all reported horse-size candidate genes (Makvandi-Nejad et al. 2012; Petersen, Mickelson, Rendahl, et al. 2013) showed a selective signature in the DB, although we cannot address whether the selective signature found in this gene was shared by other pony breeds, as the three SNPs around HMGA2 were not genotyped in any of the other pony breeds. Therefore, it is likely that the DB independently evolved small stature. To test this hypothesis, we also evaluated the di value of DB and other ponies from a total of 35 breeds of horses. Although the two most differentiated SNPs in DB were removed from this analysis, due to a greater than 10% missing rate among all 35 breeds, the most differentiated SNP in the DB (chr8:17883095, di = 183.346) was still located approximately 300 kb upstream of TBX3. In contrast to the DB, none of the other pony breeds showed relatively high di values for this SNP. In addition, we also conducted the f3 test (Reich et al. 2009) implemented in TreeMix (Pickrell and Pritchard 2012) to investigate whether there were any potential migration events among the pony breeds. No evidence of migration between the DB and the other pony breeds was found (supplementary table S6, Supplementary Material online). Taking together, these results suggest the independent evolution of small stature size in DB. IHH, which has been reported as a candidate contributing to stature size variation in human (Weedon et al. 2008), also showed high XP-EHH specific to the DB. However, neither a signature of high diversification nor stature-associated significance was found, which may indicate that this gene has a modest effect in a subset of the DB population.
A caveat of our study is the possible shortcomings in the genotyped SNP data, for example, SNP ascertainment bias. Although, a group of genes were identified, the relationship of the genotype and phenotype still needs additional functional evidence. Whole-genome sequences from multiple individuals would be necessary for validating the potential selective targets, which should also lead to the identification of additional key mutations responsible for the body size seen in the DB.
Supplementary Material
Supplementary figures S1–S9 and tables S1–S6 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors gratefully acknowledge Xingkui Yao and Dianxin Xu for their assistance in the sample collection. They thank Dr Bridgett M. vonHoldt for her suggestions and comments on the manuscript. This work was supported by the Agricultural Science and Technology Innovation Program of China (ASTIP-IAS01), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB13020600), the National Natural Science Foundation of China (31272403) and the Domestic Animals Sharing Platform in China.
Literature Cited
- Akey JM, et al. 2002. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 12:1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey JM, et al. 2010. Tracking footprints of artificial selection in the dog genome. Proc Natl Acad Sci U S A. 107:1160–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad M, et al. 1997. Mutations in human TBX3 alter limb, apocrine and genital development in ulnar-mammary syndrome. Nat Genet. 16:311–315. [DOI] [PubMed] [Google Scholar]
- Bamshad M, et al. 1999. The spectrum of mutations in TBX3: Genotype/Phenotype relationship in ulnar-mammary syndrome. Am J Hum Genet. 64:1550–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanckenhorn W, Morf C, Muhlhauser C, Reusch T. 1999. Spatiotemporal variation in selection on body size in the dung fly Sepsis cynipsea. J Evol Biol. 12(3):563–576. [Google Scholar]
- Boyko AR, et al. 2010. A simple genetic architecture underlies morphological variation in dogs. PLoS Biol. 8:e1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- China National Commission of Animal Genetic Resources. 2011. Animal genetic resources in China: horse, donkey, camels. China Agriculture Press. [Google Scholar]
- Hendricks BL. 2007. International encyclopedia of horse breeds. University of Oklahoma Press. [Google Scholar]
- Kraushaar U, Blanckenhorn WU. 2002. Population variation in sexual selection and its effect on size allometry in two dung fly species with contrasting sexual size dimorphism. Evolution 56:307–321. [DOI] [PubMed] [Google Scholar]
- Lango Allen H, et al. 2010. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467:832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makvandi-Nejad S, et al. 2012. Four loci explain 83% of size variation in the horse. PLoS One 7:e39929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattern J. 2010. Horses on the farm. Rourke Pub Group. [Google Scholar]
- McCue ME, et al. 2012. A high density SNP array for the domestic horse and extant Perissodactyla: utility for association mapping, genetic diversity, and phylogeny studies. PLoS Genet. 8:e1002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando L, et al. 2013. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature 499:74–78. [DOI] [PubMed] [Google Scholar]
- Petersen JL, Mickelson JR, Cothran EG, et al. 2013. Genetic diversity in the modern horse illustrated from genome-wide SNP data. PLoS One 8:e54997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen JL, Mickelson JR, Rendahl AK, et al. 2013. Genome-wide analysis reveals selection for important traits in domestic horse breeds. PLoS Genet. 9:e1003211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Pritchard JK. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8:e1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, et al. 2009. Reconstructing Indian population history. Nature 461:489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimbault M, et al. 2013. Derived variants at six genes explain nearly half of size reduction in dog breeds. Genome Res. 23:1985–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, et al. 2006. Positive natural selection in the human lineage. Science 312:1614–1620. [DOI] [PubMed] [Google Scholar]
- Scheet P, Stephens M. 2006. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 78:629–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer-Hasler H, et al. 2012. A genome-wide association study reveals loci influencing height and other conformation traits in horses. PLoS One 7:e37282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter NB, et al. 2007. A single IGF1 allele is a major determinant of small size in dogs. Science 316:112–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, et al. 2011. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39:D561–D568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetens J, Widmann P, Kuhn C, Thaller G. 2013. A genome-wide association study indicates LCORL/NCAPG as a candidate locus for withers height in German Warmblood horses. Anim Genet. 44:467–471. [DOI] [PubMed] [Google Scholar]
- Weedon MN, et al. 2007. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat Genet. 39:1245–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedon MN, et al. 2008. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 40:575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentong H. 1990. A brief analysis of the origin and development of the pony in China. J Agric Archaeol 1:064. [Google Scholar]
- Yang TL, et al. 2010. HMGA2 is confirmed to be associated with human adult height. Ann Hum Genet. 74:11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.