Abstract
Here, we report the extensive bioinformatic and functional analyses of the unusual pLOCK 0919, a plasmid originating from the probiotic Lactobacillus casei LOCK 0919 strain. This plasmid is atypical because it harbors the spaCBA-srtC gene cluster encoding SpaCBA pili. We show that all other spaCBA-srtC sequences of the Lactobacillus genus that have been previously described and deposited in GenBank are present in the chromosomal DNA. Another important observation for pLOCK 0919 is that the spaCBA-srtC gene cluster and its surrounding genes are highly similar to the respective DNA region that is present in the most well-known and active SpaCBA pili producer, the probiotic Lactobacillus rhamnosus GG strain. Our results demonstrate that the spaCBA-srtC clusters of pLOCK 0919 and L. rhamnosus GG are genealogically similar, located in DNA regions that are rich in transposase genes and are poorly conserved among the publicly available sequences of Lactobacillus sp. In contrast to chromosomally localized pilus gene clusters from L. casei and Lactobacillus paracasei, the plasmidic spaC of L. casei LOCK 0919 is expressed and undergoes a slight glucose-induced repression. Moreover, results of series of in vitro tests demonstrate that L. casei LOCK 0919 has an adhesion potential, which is largely determined by the presence of the pLOCK 0919 plasmid. In particular, the plasmid occurrence positively influenced the hydrophobicity and aggregation abilities of L. casei LOCK 0919. Moreover, in vivo studies indicate that among the three Lactobacillus strains used to colonize the gastrointestinal tract of germ-free mice, already after 2 days of colonization, L. casei LOCK 0919 became the dominant strain and persisted there for at least 48 days.
Keywords: adhesion, spaCBA pilus cluster, Lactobacillus, plasmid, in silico analyses, probiotic properties
Introduction
The Gram-positive lactobacilli are commensal inhabitants of the gastrointestinal (GI) tract and based on their health-promoting effects on human and animal hosts, these bacteria are commonly marketed as probiotics (Saxelin et al. 2005; Bernardeau et al. 2006; Felis and Dellaglio 2007). One postulated feature that is considered indispensable for probiotic lactobacilli by the FAO-WHO (2006) guidelines for the evaluation of probiotics for human food applications is their adherence to human intestinal tissues. Bacterial adhesion to the host GI tract may promote a variety of specific interactions with the host in terms of colonization, persistence, and potential signaling (Mack et al. 2003; Vesterlund et al. 2006; Lebeer et al. 2008). Probiotic bacteria by adhering to the host GI epithelium may influence the indigenous microorganisms, enhance the intestinal epithelial barrier, stimulate the host’s immune system, and prevent infections of pathogens (Jankowska et al. 2008; Lim and Ahn 2012). It is postulated that highly adhesive bacteria have the greatest beneficial effects on the host’s health (Liu et al. 2010). The cell surface structures important for the adherence of lactobacilli to intestinal epithelial extracellular matrix (ECM) components include exopolysaccharides, teichoic acids and surface proteins such as the S-layer and LPXTG-like proteins (Lebeer et al. 2008). Among them, a major driver of adhesion to the intestinal mucosa and biofilm formation are the mucus-binding pili as those encoded by the spaCBA-srtC gene cluster in L. rhamnosus GG (Kankainen et al. 2009; von Ossowski et al. 2010; Reunanen et al. 2012). These pili are protein fibers that consist of multimers of SpaA covered by the mucus-binding protein SpaC and covalently linked to the bacterial peptidoglycan through SpaB (Reunanen et al. 2012). In lactobacilli, the genes encoding the three pilin subunits (SpaCBA) and the pilin-specific sortase (SrtC) are always present at the same chromosomal locus as a gene cluster. To date, comparative genome analyses have shown that L. rhamnosus, L. casei and L. paracasei genomes are highly related (Kant et al. 2011), as supported also by examinations of their spaCBA-srtC gene clusters (Douillard, Ribbera, Järvinen, et al. 2013). However, regarding the presence of SpaCBA pilus structures in L. rhamnosus and L. casei strains assessed by immunoblotting analysis, electron microscopy, and mucus-binding assays, only L. rhamnosus strains display functional pili that correlate with their mucus-binding abilities. The data in the literature indicate that L. rhamnosus strains remain piliated under different stress conditions, and in contrast, no expression of pili genes in L. casei has been observed under the tested conditions (Douillard, Ribbera, Järvinen, et al. 2013; Douillard, Ribbera, Kant, et al. 2013). The identification of the transcriptional start site of the spaCBA-strC operon suggested that its expression is triggered by the insertion of an insertion sequence (IS) element in the L. rhamnosus strains, in contrast to the L. casei strains. The transposition of an IS element upstream of the spaC gene appeared to have a significant impact on the evolution of L. rhamnosus species by conferring a beneficial trait that permitted colonization and persistence in mucosal-associated niches such as the human GI tract (Douillard, Ribbera, Järvinen, et al. 2013).
The DNA sequence of pLOCK 0919 plasmid that harbors the spaCBA-srtC operon was reported by us elsewhere (Koryszewska-Bagińska et al. 2013). Here, based on in silico analyses, we provide the extended data on the nucleotide sequence of this unusual pLOCK 0919, a plasmid originating from the probiotic L. casei LOCK 0919 strain (formerly L. paracasei LOCK 0919) (Cukrowska et al. 2009, 2010; Kozakova et al. 2015), with a particular focus on the plasmidic spaCBA-srtC pilus gene cluster. We found that in contrast to the chromosomal localization of the other lactobacilli pilus clusters described to date one copy of the spaCBA-srtC operon of L. casei LOCK 0919 is plasmid-localized. Moreover, the results of the comparative analyses indicated that a DNA region containing the spaCBA-srtC cluster from pLOCK 0919 might be more related to that of L. rhamnosus GG than to that of the highly conserved, albeit likely nonfunctional, spaCBA-srtC of L. casei and L. paracasei. In addition to above-mentioned in silico analyses, we also provide results of the in vitro and in vivo studies confirming that L. casei LOCK 0919 is a highly adhesive strain and this feature might be indeed plasmid-localized.
Materials and Methods
Data Collection and Phylogenetic and Sequence Analysis
Hypothetical protein sequences were analyzed using the HHpred program (Hildebrand et al. 2009). Protein sequences of SpaABC and the SrtC sortase were collected using pLOCK 0919 as the query in the PSI-BLAST (Basic Local Alignment Search Tool) program with default parameters (Altschul et al. 1997). The analysis was limited to the genus Lactobacillus. The collected protein sequences were analyzed using CLANS, a tool that enables the clustering and visualization of sequence similarities, with default parameters (Frickey and Lupas 2004).
The spaCBA-srtC nucleotide sequences were retrieved from GenBank (http://www.ncbi.nlm.nih.gov, last accessed December 17, 2015) by performing BLAST searches (Altschul et al. 1990) using the pilus cluster from pLOCK 0919 as a reference. A graphical representation of BLAST sequence comparison of pLOCK 0919 with other plasmids and trimmed chromosomes was developed using the Circos package v0.64 (Krzywinski et al. 2009). The local, shared syntenies of the spaCBA-srtC clusters and genes in terms of overlapping DNA regions were determined and visualized using the Easyfig tool (Sullivan et al. 2011).
Multiple DNA sequence alignments of the trimmed spaCBA-srtC clusters at the spaC ATG start codon and the srtC TAA stop codon were performed using the multiple sequences alignment software MAFFT v7 (Katoh and Standley 2013). The colored text of the multiple DNA sequence alignments of the spaCBA-srtC clusters and their upstream DNA regions trimmed above the spaC start codon was performed with MultAlin software (Corpet 1988).
The spaCBA-srtC sequences were subjected to molecular phylogenetic tree inferences using the phylogeny.fr online tool (Dereeper et al. 2008). Sequences were aligned with MUSCLE (v3.7) configured for highest accuracy (MUSCLE with default settings) (Edgar 2004). After alignment, ambiguous regions (i.e., containing gaps and/or poorly aligned) were removed with Gblocks (v0.91b) (Castresana 2000) using the following parameters:
minimum length of a block after gap cleaning: 10,
no gap positions were allowed in the final alignment,
all segments with contiguous nonconserved positions bigger than 8 were rejected,
minimum number of sequences for a flank position: 85%.
The phylogenetic tree was reconstructed using the maximum-likelihood (ML) method implemented in the PhyML program (v3.0) (Guindon et al. 2010). The HKY85 substitution model was selected assuming an estimated proportion of invariant sites and four gamma-distributed rate categories to account for rate heterogeneity across sites. The gamma shape parameter was estimated directly from the data. Reliability for internal branch was assessed using the approximate likelihood Shimodaira–Hasegawa ratio test (SH-like). Graphical visualization and edition of the phylogenetic tree were performed with TreeDyn (v198.3) (Chevenet et al. 2006).
Bacterial Strains, Media, and Plasmid Removal
L. rhamnosus LOCK 0900 (Aleksandrzak-Piekarczyk et al. 2013), L. rhamnosus LOCK 0908 (Koryszewska-Bagińska et al. 2014), and L. casei LOCK 0919 (Koryszewska-Bagińska et al. 2013) were obtained from the Pure Culture Collection of the Technical University of Lodz (Poland). Bacteria were cultivated anaerobically at 37 °C in the MRS broth (Oxoid) or in MRS w/o dextrose (USBiological) supplemented with 1% glucose (G-MRS) or 1% lactose (L-MRS).
The removal of the pLOCK 0919 plasmid from L. casei LOCK 0919 was performed by the slightly modified method described by Ruiz-Barba et al. (1991) involving the use of sodium dodecyl sulfate (SDS). Prior to plasmid curing, the sublethal concentration of SDS (defined as the highest concentration allowing for the visible growth) for L. casei LOCK 0919 was determined as 0.2 mg/ml. An overnight L. casei LOCK 0919 culture in G-MRS was employed as an inoculum (102 colony forming units [CFU]/ml) for fresh G-MRS containing 0.2 mg/ml SDS, incubated and subcultured into the same medium every 72 h. At appropriate intervals (after 3, 7, 14, and 21 days of cultivation), the culture was serially diluted and plated on G-MRS solidified with 1.5% agar. After 48 h of incubation, visible colonies were randomly selected for further plasmid analysis. The absence or presence of pLOCK 0919 was assessed by PCR with primers specific for the plasmidic DNA. The resulting strain devoid of pLOCK 0919, which was selected after 21 days of cultivation in G-MRS with 0.2 mg/ml SDS, was assigned as L. casei LOCK 0919Δp.
Aggregation, Hydrophobicity, and Adherence Tests
First, to maintain the biological activity of LOCK strains, 24-h cultures in MRS were frozen at −20 °C with the addition of 20% glycerol. Before application, the bacteria were activated twice in liquid MRS broth (3% inoculum, v/v) and incubated for 24 h at 37 °C and the resultant stock cultures were stored at 4 °C. All bacterial cultures were centrifuged (3,800 × g, 15 min, 4 °C) and suspended in phosphate-buffered saline (PBS) (pH 7.4). Before experiments the final optical density of the suspensions was measured in a spectrophotometer (Beckman DU640) at 570, 600, or 630 nm (depending on the method) and was adjusted to approximately 1.0 by dilution in PBS.
For hydrophobicity tests, bacterial suspensions in PBS were mixed for 2 min with hexadecane (Sigma-Aldrich) (1:5, v/v) in order to form the emulsion. The mixtures were incubated for 60 min at ambient temperature until the hexadecane formed a separated layer above the PBS. Absorbance of the bottom layer (PBS) was measured at 600 nm. The percentage of hydrophobicity was calculated as follows: Hydrophobicity (%) = (A0 − A/A0) × 100, where A0 and A is the absorbance before and after the extraction with hexadecane, respectively.
For aggregation tests, bacterial suspensions (in three repeats each) were put in stabile, safe place at ambient temperature. After 2 and 24 h the upper layer was collected for measurement of the optical density. The percentage of aggregation was calculated as follows: Aggregation (%) = [1 − (A1/A0)] × 100, where A0 is the initial absorbance and A1 the absorbance after 2 and 24 h.
The adherence to collagen, glass, and polystyrene was tested according to the method described by Zeraik and Nitschke (2012) with some modifications. Bacterial suspensions in PBS were added into 96-well polystyrene plate (in eight repeats each), 6-well plate (in three repeats each) with a cover glass on the bottom of the well or 96-well plate coated with collagen (BD) (in eight repeats each). They were incubated for 2 h at 37 °C, rinsed with water, fixed with 80% methanol (15 min) and stained with 0.1% crystal violet (15 min, 120 rpm). Next wells were washed out with water and incubated with 33% glacial acetic acid (15 min, 120 rpm). The absorbance was measured at 630 nm with microplate reader (Tristar2 LB 942; Berthold Technologies). To calculate the adhesion ratio of bacteria to collagen/glass/polystyrene, the value of absorbance of bacteria was divided by the absorbance of the control sample (collagen/glass/polystyrene only).
The adherence of bacteria to gelatine and mucous was determined by crystal violet staining method according to Vesterlund et al. (2005) with some modifications. 96-Well plate was coated (60 min, 37 °C) with 1% sterile-filtered gelatine (Sigma-Aldrich). In case of mucous, 24-well plate was coated with mucous from porcine stomach (150 mg/ml in PBS; pH 7.2) (Type II, Sigma-Aldrich) (72 h, 4 °C). The unattached gelatine or mucous was removed and washed with PBS (pH 7.4). Next, bacterial suspensions in PBS were added to each well (in eight and four repeats in case of collagen and gelatine, respectively) and allowed to adhere (2 h, 37 °C). Nonadherent bacteria were washed with PBS and the remaining were fixed (20 min, 60 °C). Bacteria were stained with 0.1% crystal violet for 15 min, washed three times with PBS and citrate buffer (20 mM/l; pH 4.3) was added to each well (45 min, 120 rpm). The absorbance was determined at 570 nm. To calculate the adhesion ratio of bacteria to gelatine/mucous, the value of bacterial absorbance was divided by the absorbance of the control sample (gelatine/mucous only).
Bacteria were characterized as strongly adherent (A > 3), moderately adherent (3 > A > 2), weakly adherent (2 > A > 1), and nonadherent (A ≤ 1).
To determine the statistical significance of observed differences, the analysis of variance was applied to the adherence data using statistical software (version 12.0.1 for Windows, SPSS Inc., USA). P value lower than 0.05 was considered as significant.
The Quantitative PCR
Quantitative PCR (QPCR) assays were performed both to assess the spaC expression level as well as to quantify L. rhamnosus LOCK 0900, L. rhamnosus LOCK 0908, and L. casei LOCK 0919 in mice feces.
For the spaC mRNA quantification, total RNA was isolated with the use of TRI Reagent (Sigma) from three independent 20 ml L. casei LOCK 0919 cultures grown in G-MRS or L-MRS and harvested in midexponential phase (OD600 = 0.6). First-strand cDNA was synthesized from DNAse I (Sigma)-treated 2 µg RNA samples. The synthesis of cDNA was performed by the use of the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems) according to manufacturer’s instructions.
The genomic DNA of LOCK strains derived from the study of Kozakova et al. (2015) and shortly was obtained as follows: 8-week old germ-free (GF) mice (n = 6) were colonized by intragastric (i.g.) tubing with 2 × 108 CFU of equal parts of overnight cultures of L. rhamnosus LOCK 0900, L. rhamnosus LOCK 0908, and L. casei LOCK 0919 in 0.2 ml sterile PBS. Subsequently, DNA was isolated from mice feces collected on the days 2, 14, 35, and 48 after the bacterial colonization.
QPCR assays on cDNA and genomic DNA from the two abovementioned experiments were carried out on the 7500 Real Time PCR System (Applied Biosystems) and following the previously described methodology (Kozakova et al. 2015). Pairs of primers specific for chromosomal or plasmidic spaC and six unique (two per each strain) genes harbored by the respective LOCK strains are presented in the table 1. The results were normalized by the use of the reference genes, which code for the elongation factor TU and DNA-directed RNA polymerase alpha subunit.
Table 1.
Protein Details for Lactobacillus casei pLOCK 0919
| GeneID | Gene Name | Description of the Encoded Proteina |
|---|---|---|
| 16110888 | p01(repA) | Replication protein |
| 16110857 | p02 | Putative ATP synthase |
| 16110858 | p03 | Hypothetical protein |
| 16110859 | p04 | Hypothetical protein |
| 16110860 | p05 | Hypothetical protein |
| 16110861 | p06 | Hypothetical protein |
| 16110862 | p07 | Hypothetical protein containing DUF1381 and Exonuc_VII_S domains |
| 16110863 | p08 | Putative C39-like peptidase |
| 16110864 | p09 | Putative GTPase-like guanine nucleotide exchange factor containing Ecp6 and C39-like peptidase domains |
| 16110865 | p10 | Collagen adhesion protein |
| 16110866 | p11 | Transposase |
| 16110867 | p12 | IS30 family transposase |
| 16110868 | p13 (spaC) | Cell wall surface anchor family protein (ancillary protein involved in mucus-adhesion) |
| 16110869 | p14 (spaB) | Pilus specific protein (minor backbone protein) |
| 16110870 | p15 (spaA) | Fimbriae subunit (major backbone protein) |
| 16110871 | p16 (srtC) | LPXTG specific sortase A |
| 16110872 | p17 | Putative pyridoxine 5′-phosphate oxidase V related flavin-nucleotide-binding protein |
| 16110873 | p18 | Putative metallothionein containing Cu(I)-thiolate, metal binding protein and apicomplexan Apetala2 domains |
| 16110874 | p19 | Putative cytosolic protein BRCT (Replication factor C subunit 1) |
| 16110875 | p20 | DNA integration/recombination/inversion protein |
| 16110876 | p21 | Ribonucleotide reduction protein NrdI |
| 16110877 | p22 | Ribonucleotide reductase of class Ib, beta subunit |
| 16110878 | p23 | Ribonucleotide reductase of class Ib, alpha subunit |
| 16110879 | p24 | Transposase |
| 16110880 | p25 | Transposase |
| 16110881 | p26 | Transposase |
| 16110882 | p27 | Hypothetical protein PhoU |
| 16110883 | p28 | Hypothetical protein YolD |
| 16110884 | p29 (umuC) | DNA-repair protein SOS response UmuC-like protein |
| 16110885 | p30 | Putative secretin-like protein |
| 16110886 | p31 | Putative ParG protein |
| 16110887 | p32 (parA) | Putative ParA protein |
Protein functions listed here are significantly updated in comparison with the protein functions assigned basing on the L. casei pLOCK 0919 genome annotation and deposition in GenBank.
Results
General Features of L. casei pLOCK 0919
We have already reported the sequencing of pLOCK 0919 (GenBank accession number CP005487), which consists of 29,768 bp with an average GC content of 43.9% and 32 open-reading frames (ORFs) (Koryszewska-Bagińska et al. 2013). The average GC content of pLOCK 0919 is among the highest of the available complete L. casei plasmids (42.2 ± 1.9%, n = 13 by 2015) and only slightly lower than the LOCK 0919 chromosomal GC value (46.2%).
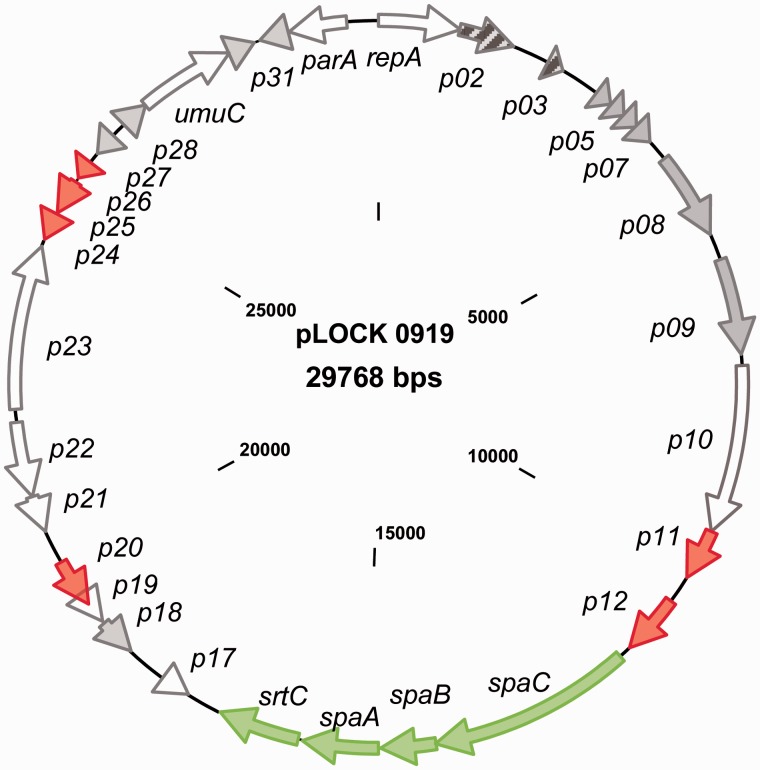
All of the identified genes encode proteins that exhibit similarities to proteins present in the public databases. Approximately half of them show similarity to proteins with hypothetical functions (table 1). Most of the genes encode proteins that are highly similar to proteins encoded in the genomes of the Lactobacillus genus. However, two genes (p02 and p03) encode proteins that are similar to those encoded in the Enterococcus genus (fig. 1). Moreover, five transposases/insertion elements (ISs) and the p20 gene encoding the DNA integration/recombination/inversion protein were identified in the pLOCK 0919 nucleotide sequence (table 1). The presence of ORFs with similarities to integrases, recombinases, and transposases suggests that pLOCK 0919 is a plasmid with a high degree of genetic plasticity and that some genes present in pLOCK 0919 might have been acquired by horizontal gene transfer (HGT).
Fig. 1.—
The genetic map of pLOCK 0919. All of the identified ORFs are indicated by arrows showing the direction of transcription. ORFs encoding proteins of hypothetical function are colored in gray, transposases and IS elements are shown in red, and ORFs encoding proteins that are homologous to bacteria other than lactobacilli are striped. The pilus spaCBA-srtC operon is depicted in green.
Comaparative analyses suggested that p32 and p01 are involved in plasmid stability and replication, respectively. The translated product of p32 shows extensive similarity to the ParA family of ATPases, which are involved in active partition mechanisms that counteract plasmid loss during cell division. The p01 gene, which is divergently transcribed with respect to parA, encodes the replication protein A (RepA). RepA includes the pfam06970 RepA_N domain in its N terminus, which is known to contain a helix-turn-helix (HTH) motif. This feature is characteristic for pLS32-type theta replication proteins (Tanaka and Ogura 1998) that represent a family of Gram-positive theta replicons, recognized as class F. This group lacks an AT-rich region that could function as a putative origin of replication and contains DNA iterons within the coding sequence of the replication protein (Cui et al. 2015). Accordingly, an inspection of the pLOCK 0919 DNA sequence revealed the presence of three discontiguous 22-bp repeats and lack of noticeable AT-rich region within and upstream of the repA gene, respectively. The further confirmation that pLOCK 0919 follows a theta model of replication is its size (∼30 kb). It has been shown that plasmids of that size found in natural isolates of Lactobacillus, typically replicate through the theta mechanism (Wang and Lee 1997).
Furthermore, pLOCK 0919 encodes two proteins that are likely involved in the SOS response to DNA damaging factors. Among them, p29 encodes the UmuC protein, and p28 encodes a hypothetical protein that is predicted to be a member of the YolD family (table 1). The YolD family members are functionally uncharacterized; however, it has been postulated that they can represent a functional equivalent of the UmuD subunit of PolV (Permina et al. 2002).
Regarding the unique functional traits that are often localized in plasmids in lactic acid bacteria, such as the hydrolysis of milk sugar and metal resistance (Cai et al. 2009), we did not identify any genes encoding lactose hydrolysis. However, we found that the metallothionein encoded by the p18 gene might be engaged in copper homeostasis (table 1). None of resistance genes was detected in pLOCK 0919.
In regard to possible elements of plasmid transmission, the inspection of the pLOCK 0919 sequence revealed the lack of neither conjugation nor mobilization genes. Moreover, no putative origin of transfer (oriT) sequence similar to those of other plasmids with mobilization genes was detected in the pLOCK 0919 plasmid sequence. The absence of all these necessary key components excludes conjugative mobilization capacity of the pLOCK 0919 plasmid.
Finally, the pilus-encoding island known as the spaCBA-srtC operon (Kankainen et al. 2009) was detected in pLOCK 0919. This pilus operon comprises three pilin subunit genes and one pilin-specific sortase gene: p13 (spaC), p14 (spaB), p15 (spaA), and p16 (srtC).
The pLOCK 0919 DNA Sequence Is Poorly Conserved among Bacterial Genomes
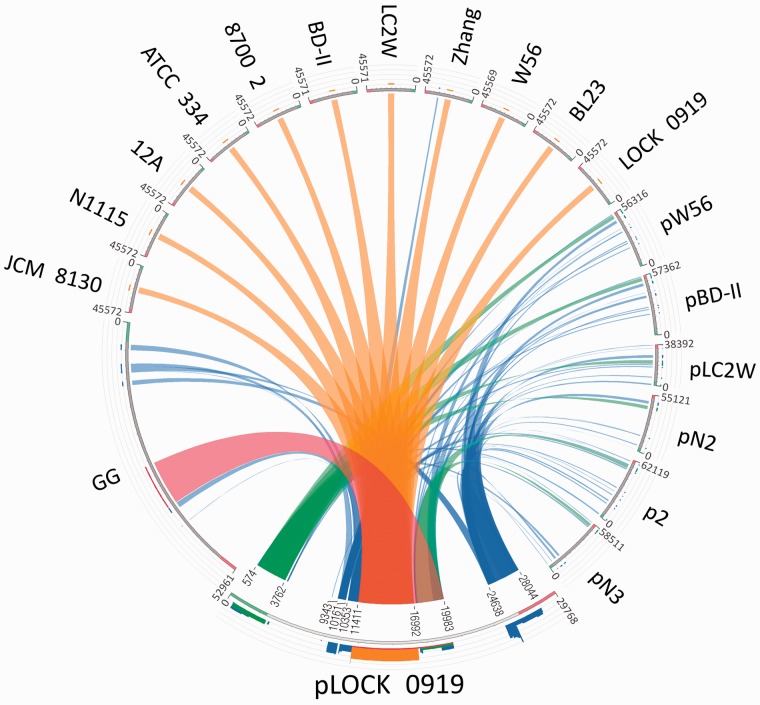
The comparative analysis of the nucleotide sequence of pLOCK 0919 indicated a low overall gene synteny with the plasmids available in GenBank. The highest similarity was detected between pLOCK 0919 and plasmids from L. casei (pW56, pBD-II, and pLC2W) and L. paracasei (pN2, pN3, and p2) (fig. 2). However, none of these plasmids has an analogous pLOCK 0919 architecture, and the similar DNA regions are rather randomly dispersed. The total similarity spans 39% of the pLOCK 0919 sequence, including the DNA regions encompassing the following genes: p1–p6 (574–3,762 bp), p11 (9,343–10,161 bp), p12 (10,353–11,405 bp), p17–p20 (17,163–19,983 bp), and p25–p30 (24,638–28,044 bp) (fig. 2). Among the pLOCK 0919 genes found in other plasmids, the genes encoding hypothetical or putative proteins are most widely represented. In addition, the L. casei and L. paracasei plasmids harbor the homologs of the majority of the pLOCK 0919 transposases/ISs (p11, p12, p25, and p26). The presence of mobile sequences on plasmids is a common phenomenon and may indicate that many homologous plasmid-localized operons have a common origin that underwent recombination events within the Lactobacillus sp. Remarkably, the pLOCK 0919 DNA region encompassing the spaCBA genes is not present in any of the Lactobacillus sp. plasmid DNA sequences available in GenBank, but it resides in selected chromosomes of Lactobacillus sp. (fig. 2).
Fig. 2.—
Relationship between the complete sequence of pLOCK 0919 and respective chromosome fragments and plasmids of other lactobacilli genomes. The DNA sequence of L. rhamnosus GG is inverted. The sequences are placed around a circle in a counterclockwise direction, with the query sequence located at the bottom. The genomic nucleotide sequences are not fully drawn to scale—for better visibility the sequences of pLOCK 0919 and L. rhamnosus GG have been disproportionately enlarged in relation to the other genomes. The ribbons represent the local alignments produced by BLAST (with a normal E value of 10−10), the width shows the alignment length, and the colors blue, green, orange, and red correspond to the alignment bit scores in the four quartiles. Blue represents the lowest values of the maximum bit score (i.e., a bit score below 25%), green—the next 25%, orange—the third quartile, and red ribbons—the best relative bit scores of 75–100% of the maximum bit score. The ribbons are inverted if the local alignments are inverted. The coordinates of homologous regions between query sequence and other genomes are indicated on ribbons from pLOCK 0919. The names above the sequences represent the respective chromosome or plasmid deriving from the respective organism (chromosomes: L. casei LOCK 0919 GenBank accession no. NC_021722.1; L. rhamnosus GG GenBank accession no. NC_013198.1; L. paracasei subsp. paracasei 8700:2 GenBank accession no. NC_022112.1; L. casei ATCC 334 GenBank accession no. NC_008526.1; L. casei str. Zhang GenBank accession no. NC_014334.1; L. paracasei N1115 GenBank accession no. CP007122.1; L. casei LOCK 0919 GenBank accession no. NC_021721.1; L. casei LC2W GenBank accession no. NC_017473.1; L. casei BL23 GenBank accession no. NC_010999.1; L. casei W56 GenBank accession no. NC_018641.1; L. casei BD-II GenBank accession no. NC_017474.1; plasmids: pLOCK 0919 GenBank accession no. NC_021722.1 from L. casei LOCK 0919, pW56 GenBank accession no. NC_020057.1 from L. casei W56, pBD-II GenBank accession no. NC_017476.1 from L. casei BD-II, pLC2W GenBank accession no. NC_017475.1 from L. casei LC2W, p2 GenBank accession no. NC_022123.1 from L. paracasei subsp. paracasei 8700:2, pN2 and pN3 from L. paracasei N1115).
The pLOCK 0919 spaCBA-srtC Pilus Cluster Is Highly Conserved among Some Species in the L. casei Taxonomic Group
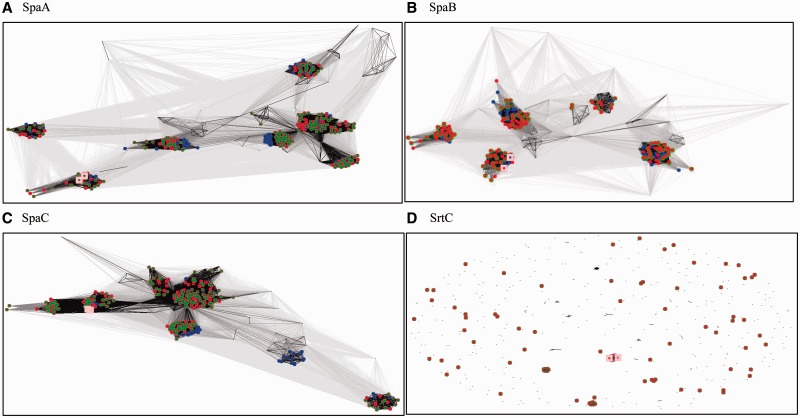
Although studies investigating the spaCBA pilus cluster in Lactobacillus sp. are restricted mainly to L. rhamnosus GG, thorough PSI-BLAST searches indicated that homologs of pLOCK 0919 SpaCBA are widely represented among the Lactobacillus genus. Moreover, the results of a CLANS analysis clearly showed a large variety of clans for all SpaCBA proteins from the different species in this genus (fig. 3A–C). Among them, SpaCBA was present in an overwhelming number of strains belonging to the related so-called L. casei taxonomic group. This group contains four species of the Lactobacillus genus (L. casei, L. paracasei, L. rhamnosus, and L. zeae) and is evolutionarily distant from other lactobacilli (Felis and Dellaglio 2007). Among these species, the SpaCBA proteins were found to be most abundant in L. casei, occurred less frequently in L. paracasei, and were the least represented in L. rhamnosus (fig. 3A–C). The SpaCBA proteins from these species can be found in most clans produced by the CLANS program. In contrast, L. gasseri, L. johnsonii, L. plantarum, L. ruminus, and L. brevis are conserved and produce single clans. Whether this effect results from a large quantity of sequenced genomes in the L. casei taxonomic group and a smaller quantity in the other Lactobacillus species depends on the timing and presence of new sequenced genomes. A completely different result can be observed for the Lactobacillus sp. sortase family, which is scattered, mostly dissimilar and comprises only a few clans (fig. 3D). This dissimilarity suggests the presence of very little evolutionary pressure on the sortase family or, in contrast, a strong pressure to diversify. In each of these cases, these proteins of L. casei pLOCK 0919 and L. rhamnsosus GG are positioned in common clans in close proximity to one another, indicating a high level of similarity (fig. 3A–D).
Fig. 3.—
CLANS clustering of SpaCBA and sortase protein sequences from the Lactobacillus genus. The color coding of the species is as follows: red—L. casei, green—L. paracasei, and navy blue—L. rhamnosus. Proteins of other species present on panels, but not colored, include L. plantarum, L. brevis, L. gasseri, L. johnsonii, L. delbrueckii, L. crispatus, and L. ruminis. Proteins encoded by L. rhamnosus GG and in pLOCK 0919 are indicated by salmon-colored larger squares. The intensity of the connecting lines reflects the level of sequence similarity. In cases of L. casei and L. paracasei, SpaB and SrtC are overlaid because of identical protein sequences. The sortase sequences do not form any well-defined groups excluding a few L. casei/L. paracasei clans.
At the time this report was written, 13 complete genomic sequences in 12 Lactobacillus strains carrying the spaCBA-srtC pilus cluster were deposited in GenBank. Among them, we identified its presence only in certain strains of the L. casei taxonomic group. Among these strains, the spaCBA-srtC pilus cluster was found to be most prevalent in L. casei (eight strains), occurred much less frequently in L. paracasei (three strains), and was represented only once in L. rhamnosus (fig. 2). It is worth mentioning that only strain L. casei LOCK 0919 assessed herein carried the spaCBA-srtC gene cluster in a plasmid. All of the other cases demonstrated a chromosomal location. Interestingly, in L. casei LOCK 0919, the pilus cluster was also present in the chromosome (fig. 2), and thus, this strain was identified as the only host carrying two copies of spaCBA-srtC.
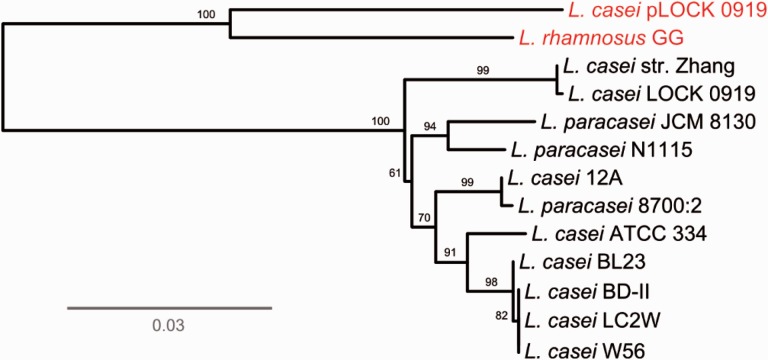
In addition, the nucleotide BLAST comparison of the pLOCK 0919 spaCBA-srtC sequence with other pilus gene clusters derived from the fully sequenced strains of L. casei taxonomic group revealed a high level of overall sequence identity (>97%), which indicated that they were likely inherited from a common ancestor. To analyze the phylogenetic relationships between them, we constructed an ML tree for the approximately 5,552 bp of sequence spanning the DNA region of the four spaCBA-srtC loci from 12 strains in the L. casei taxonomic group (fig. 4). The tree revealed that the spaCBA-srtC clusters of pLOCK 0919 and L. rhamnosus GG are closely related to one another and the most different among all of other ten strains, as evidenced by their clustering in independent branches. This observation indicated that spaCBA-srtC of pLOCK 0919 and L. rhamnosus GG are genealogically similar and distinct from the pilus clusters of other lactobacilli strains, which is indicative of their ancient divergence from a common ancestor.
Fig. 4.—
Genome-based unrooted phylogenetic tree of the spaCBA-srtC clusters. Phylogenetic relationships between the 13 nucleotide sequences of the spaCBA-srtC clusters from 12 sequenced lactobacilli strains. The scale bar represents the evolutionary distance. The bootstrap values (%) are indicated at the nodes.
The highest sequence similarity was observed in the DNA region encompassing the spaB, spaA and srtC genes, with only very few differences consisting of nucleotide changes. The similarity of the spaC genes was much less pronounced because, in comparison to the L. casei and L. paracasei strains, these genes in pLOCK 0919 and L. rhamnosus GG exhibited three regions with 3–15 nt deletions (supplementary fig. S1, Supplementary Material online).
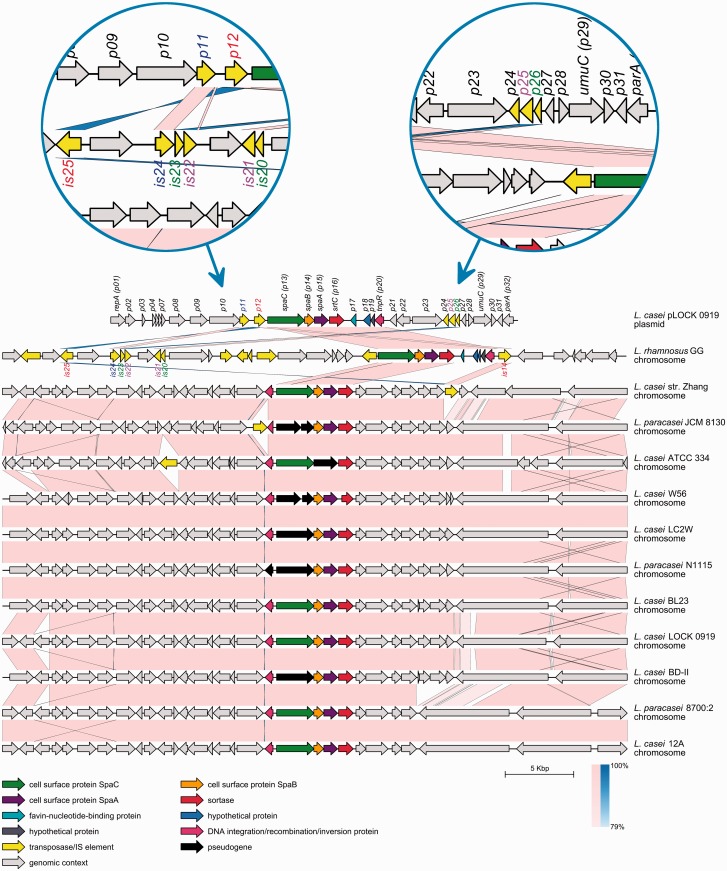
Moreover, a comparison of the layout of the spaCBA-srtC genes among the publicly available fully sequenced genomes of lactobacilli revealed a high degree of synteny (fig. 5), suggesting a strong evolutionary conservation. Despite the high level of sequence conservation, certain strains (L. casei ATCC 334, L. casei LC2W, L. casei BD-II, L. casei W56, L. paracasei JCM 8130, and L. paracasei N1115) appeared to carry some remains of the pili genes in a degenerated form (fig. 5), which indicated that these clusters are not evolutionarily conserved and may be the result of progressive pseudogenization. In contrast, the pLOCK 0919 spaCBA-srtC genes did not appear to contain any critically mutated coding sequences (fig. 5).
Fig. 5.—
Synteny between the complete sequence of pLOCK 0919 and the respective DNA fragments of other lactobacilli genomes. The DNA sequence of L. rhamnosus GG is inverted. The respective genome designations are indicated on the right side of each genome line. The predicted ORFs are shown in their respective orientation as boxes with arrows. Conserved genes between pLOCK 0919 and Lactobacillus sp. genomes are depicted in the same color. The local genomic maps are drawn to scale. The direct nucleotide similarity between individual sequences of neighboring genomes is shown as pale pink bands, and inverted similarities are indicated as blue bands. The color shading indicates the level of similarity, with greater saturation of a similarity band indicating more conservation of two ORFs pairs. For better visibility, two IS-reach regions of pLOCK 0919 and L. rhamnosus GG are enlarged in blue circles. The names of homologous ISs from pLOCK 0919 and L. rhamnosus GG are shown in the same colors.
The DNA Regions and Transposable Elements Overlapping spaCBA-srtC Are Conserved between pLOCK 0919 and L. rhamnosus GG
Comparing the pLOCK 0919-localized spaCBA-strC up- and downstream DNA regions with the respective DNA sequences of L. rhamnosus GG and other strains in the L. casei taxonomic group, a number of similarities in their genetic organization are apparent between pLOCK 0919 and GG. First, the downstream region of the pLOCK 0919-localized spaCBA-srtC shares common DNA regions (on pLOCK 0919, encompassing the p17–p20 genes) with L. rhamnosus GG (figs. 2 and 5). No other Lactobacillus sp. genome carries homologs located in close proximity to the pilus cluster (fig. 5). This unique similarity suggests that these whole DNA regions (on pLOCK 0919, consisting of the spaCBA-srtC,p17, p18, p19, and p20 genes) of pLOCK 0919 and L. rhamnosus GG may have originated from a common ancestor that was distinct to the ancestor(s) from other Lactobacillus sp. Another similarity between the spaCBA-srtC pili clusters of pLOCK 0919 and L. rhamnosus GG is that they are flanked by numerous IS elements/transposase genes (fig. 5). Their presence indicates that these IS element-rich DNA regions might be subjected to genetic recombination events within species, such as, for example, the events that led to the acquisition of the spaCBA-strC gene cluster by HGT in both strains. Some of the ISs/transposase genes found in pLOCK 0919 are located adjacent to spaCBA-srtC as in the case of p11 and p12, which are inserted directly upstream of this gene cluster. Transposition events involve both recombination and replication processes, which frequently generate two daughter copies of the original transposable elements. One copy remains at the parent site, and another is present at the target site (Mahillon and Chandler 1998). Interestingly, we found no copies of the p11 and p12 ISs/transposase genes in the chromosome of the L. casei LOCK 0919 strain. Their homologs, however, commonly reside in the plasmids and chromosomes of many other species of the Lactobacillus genus (fig. 2), paradigmatically in L. rhamnosus GG, in which they are particularly widely represented. In this bacterium, the p11 and p12 homologs resided in close but not direct proximity to the spaCBA-srtC cluster and were assigned as follows: is14, is24, and is25 (fig. 5). Moreover, two other pLOCK 0919 ISs/transposase genes (p25 and p26) residing at a greater distance downstream from spaCBA-strC also had counterparts in the vicinity of the pilus gene cluster in the genome of L. rhamnosus GG. In this bacterium, these two genes occurred in duplicated form and were designated as follows: is20, is21, is22, and is23 (fig. 5). The scattering of ISs/transposase genes in the genomes of other Lactobacillus sp. and their absence in the L. casei LOCK 0919 chromosome suggest that pLOCK 0919 is not a permanent, long-term resident of L. casei LOCK 0919 and that the p11 and p12 genes might jump into this host plasmid from the genome of another species. However, we cannot exclude the possibility of a conservative way of the p11 and p12 transposition, by which they might be transposed from the chromosome of L. casei LOCK 0919 to its pLOCK 0919 plasmid.
Further analysis of pLOCK 0919 mobile elements indicated that the majority of them likely represents the remnants of ISs that were once active. An indication of the lack of p24, p25 and p26 functionality is provided by their lengths (372, 471, and 261 bp, respectively), which are considerably too short in comparison to the average length of the bacterial insertion element (0.7–2.7 kb; Siguier et al. 2006). Similarly, the pLOCK 0919 p11 gene also appears to be truncated because it lacks the HTH motif that is necessary for the recognition and binding of terminal inverted repeats. The degeneration of the majority of pLOCK 0919 ISs/transposase genes may indicate that their insertion in the plasmid is evolutionarily more ancient when compared with nontruncated is14 and is15 of L. rhamnosus GG.
In contrast to pLOCK 0919 and L. rhamnosus GG, the pilus gene clusters detected in other Lactobacillus sp. are located in chromosomal regions that are devoid of ISs/transposase genes or in which these genes occur only occasionally (fig. 5). This finding suggests that spaCBA-srtC of L. casei and L. paracasei species were not acquired by HGT but rather originated from an ancient common ancestor. Therefore, it can be suggested that the spaCBA-srtC operon hitchhiked from stably inherited genomes to the plasmid sequences.
The pLOCK 0919 spaCBA-srtC Upstream Noncoding DNA Sequence Differs from Relatives of L. casei Taxonomic Group Species
A comparison of the pLOCK 0919 noncoding sequence upstream of spaCBA-srtC with the respective sequences of other Lactobacillus strains revealed differences in their similarity. Along their entire length, these sequences are almost identical among all of the Lactobacillus strains excluding pLOCK 0919 and L. rhamnosus GG (fig. 6). In these two strains, when compared with other lactobacilli, there is a variable degree of similarity that ranges from very high similarity (26 nucleotides directly upstream of the spaC gene start codon), to low similarity in the subsequent nucleotides, and to virtually no similarity upstream of this region (fig. 6). The observed differences in the spaCBA-srtC noncoding upstream sequence between pLOCK 0919, L. rhamnosus GG, and other Lactobacillus strains consist of mismatches, insertions or deletions of single or strings of nucleotides (fig. 6). This strong conservation of the 26-nt region upstream of the spaC gene across species may indicate its significant role in, for example, the spaCBA-srtC cluster expression. In fact, a survey reported by Douillard, Ribbera, Järvinen, et al. (2013) demonstrated that in L. rhamnosus GG, the transcription start site (TSS) mapped to 23 nucleotides upstream of the start of spaC translation, which coincides with the end of the region with a high similarity (fig. 6). Therefore, we can assume that the putative start of spaC transcription in pLOCK 0919 is coordinated similarly to that in L. rhamnosus GG and, thus, that the highly conserved DNA region spans the beginning of the spaC mRNA (fig. 6). The possibility of spaC expression is strengthened by the presence of a putative transcriptional promoter located approximately ten nucleotides upstream of the region with a high level of similarity (fig. 6 and section below).
Fig. 6.—
Multiple sequence alignment of the spaC gene upstream region in selected strains of the L. casei taxonomic group. The complete consensus nucleotides are marked in red, and nucleotides with a low level of consensus are marked in either blue (majority) or black (minority). The DNA regions with a variable similarity are separated from one another by three rectangles. Yellow-shaded sequences highlight the potential −10/−35 promoter regions upstream of pLOCK 0919 spaC and p12 (IS30). The putative ribosome binding sites (RBSs) are shaded in red, and the start and stop codons are shaded in green. The position of the TSS and the −10 and −35 regions in L. rhamnosus GG (as determined previously by Douillard, Ribbera, Järvinen, et al. 2013) are highlighted in yellow and dark gray, respectively.
At Least Two Putative Promoters May Drive the Expression of the pLOCK 0919 Pilus spaCBA Genes Cluster
Data reported in the literature show that in spite of the high level of conservation and sequence identity of the spaCBA-srtC pilus gene cluster, some L. rhamnosus strains, but none of the L. casei strains, produce pili (Douillard, Ribbera, Järvinen, et al. 2013;Douillard, Ribbera, Kant, et al. 2013). It has been postulated that this deficiency may be due to a loss of the −35 and −10 consensus regions and the TSSs in L. casei, whereas integration of the IS30 element provided a promoter that allowed the expression of the pili genes in L. rhamnosus GG (Douillard, Ribbera, Järvinen, et al. 2013). This transcriptional alteration of gene expression by the introduction of IS elements is not an unusual phenomenon (Reynolds et al. 1986; Poirel et al. 2003). Interestingly, the DNA region of the pilus cluster identified in pLOCK 0919 differs significantly from the sequences present in other L. casei and L. paracasei strains, both with respect to the noncoding spaCBA-srtC upstream DNA region (fig. 6) and the presence of transposases (fig. 5). Thus, it is tempting to speculate that the integration of transposases upstream of the plasmid spaC gene in L. casei LOCK 0919 might also affect the activation of pilus gene expression in this strain. To determine whether the insertions might result in the acquisition of the −35 and −10 regions consensus regions, we inspected the upstream DNA sequence of the pLOCK 0919 spaCBA-srtC pilus gene cluster. The pLOCK 0919 spaCBA-srtC pilus gene cluster appeared to be preceded by at least two putative promoters. The first (P1; −35 AAGCCT/−10 TATTAT) was found in close proximity to spaC because its −10 region was located 58 nucleotides upstream of the spaC start codon (fig. 6). A comparison of the pLOCK 0919 P1 region with the corresponding chromosomal DNA of L. rhamnosus GG revealed that both regions contain similarly positioned promoters, although the spacing between the spaC start codon and predicted −10 promoter region was found to be 8 nt longer in pLOCK 0919. Moreover, although the pLOCK 0919 upstream sequence of spaCBA-srtC lacks a triplet of adenines, which has been designated as the transcriptional start site in L. rhamnosus GG (Douillard, Ribbera, Järvinen, et al. 2013), it contains instead a stretch of other nucleotides that may serve as a TSS (fig. 6). Further inspection of the more distant noncoding DNA region situated between p11 and p12 revealed the presence of the second possible promoter (P2; −35 GTGAAA/−10 TAAAAT) located 42 bp upstream of the p12 start codon (fig. 6). Its presence and the absence of a clear terminator of transcription between p12 and spaC suggest that the spaCBA-srtC gene cluster in pLOCK 0919 may be cotranscribed with the p12 gene encoding a transposase. Although neither promoter fully resembles the canonical promoters defined as TATAAT and TTGACA (Browning and Busby 2004), they may still drive high levels of pilus gene expression, as observed for the noncanonical promoter that drives the efficient expression of the pilus cluster in L. rhamnosus GG (Douillard, Ribbera, Järvinen, et al. 2013).
Lactobacilluscasei LOCK 0919 Strongly Adheres to Different Surfaces and Colonize Mice Intestine
For the experimental confirmation of L. casei LOCK 0919 adhesive potential and evaluation whether this potential may reside on its plasmid, series of in vitro and in vivo assays were performed on L. casei LOCK 0919, L. casei LOCK 0919Δp, L. rhamnosus LOCK 0900, and/or L. rhamnosus LOCK 0908 strains. For these experiments, LOCK 0900 and LOCK 0908 strains were used as negative controls as no spaCBA clusters were detected in their genomes (Aleksandrzak-Piekarczyk et al. 2013; Koryszewska-Bagińska et al. 2014).
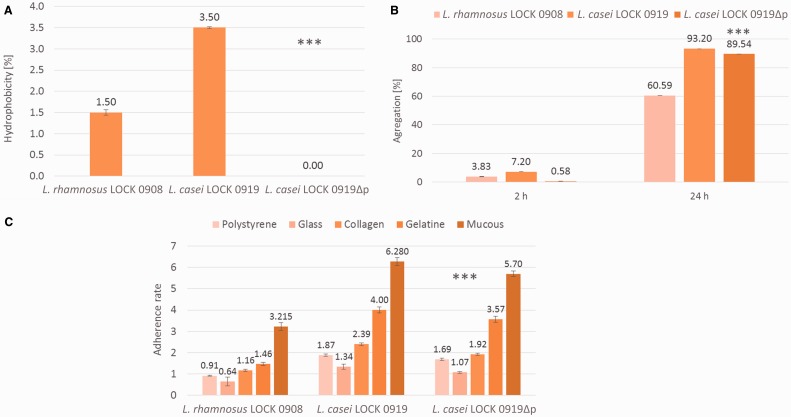
In vitro tests included the evaluation of strains’ hydrophobic properties, aggregation, adherence to polystyrene, glass, mucous, collagen, and gelatin. The percentage of hydrophobicity varied between strains. The highest value was observed for L. casei LOCK 0919 (3.50%), whereas the same strain after removing of its plasmid (L. casei LOCK 0919Δp) was not hydrophobic at all (fig. 7A). Similarly, the percentage of the aggregation was strain-dependent and after 2 h L. casei LOCK 0919 aggregated to highest extent (7.2%), whereas L. casei LOCK 0919Δp was the least aggregative (0.58%). After 24 h both L. casei LOCK 0919 and L. casei LOCK 0919Δp strongly aggregated and the percentage of aggregated cells was 93.2% and 89.5%, respectively (fig. 7B). In both of experiments, L. rhamnosus LOCK 0908 was less hydrophobic and aggregative than L. casei LOCK 0919 but more hydrophobic and aggregative (after 2 h, only) than L. casei LOCK 0919Δp (fig.7A and B). Also, the adherence ability of tested bacteria varied significantly between strains. L. casei LOCK 0919 showed the highest adherence to all surfaces, whereas the lack of pLOCK919 led in all cases to statistically significant decrease of the L. casei LOCK 0919Δp adherence (fig. 7C). Among the different strains and surfaces tested, L. casei LOCK 0919 and L. casei LOCK 0919Δp were classified as strongly adhesive to mucous and gelatine; L. casei LOCK 0919 was moderately adhesive to collagen. L. rhamnosus LOCK 0908 was classified as nonadherent to polystyrene and glass and weakly adherent to gelatine and collagen.
Fig. 7.—
Hydrophobicity (A), aggregation after 2 and 24 h (B) and adherence to abiotic (polystyrene, glass) and biotic (collagen, gelatin, mucous) surfaces (C) of Lactobacillus strains. ***Results statistically different from L. casei LOCK 0919 (P < 0.05).
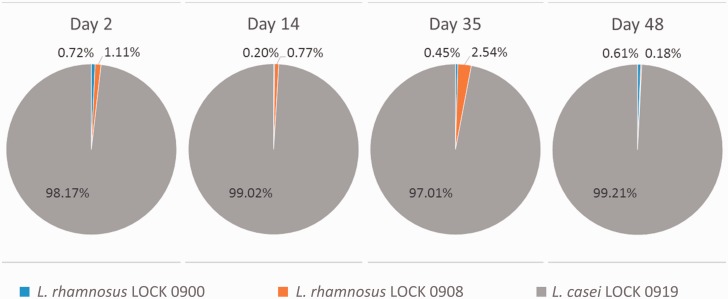
For an in vivo assessment of the L. casei LOCK 0919 persistence in the gut, we performed a strain-specific QPCR on the DNA isolated from feces of GF mice, whom three Lactobacillus strains were administrated. As shown in figure 8, starting from the second day after bacterial application and throughout of the study, the amount of L. casei LOCK 0919 in stools was overwhelming ranging from 97% to 99%, whereas the L. rhamnosus LOCK 0900 and LOCK 0908 strains were barely detected.
Fig. 8.—
Representation of the Lactoctobacillus LOCK strains isolated from mice feces at indicated time points. Data are shown as the percentage of each strain from all detected Lactobacillus LOCK strains at the given day after GF mice colonization.
Plasmid-Encoded SpaC Pilin Is Expressed in L. casei LOCK 0919 and Undergoes Glucose-Induced Repression
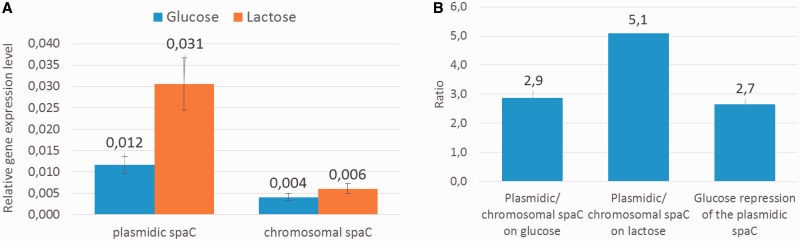
Minor differences between the DNA sequences of the two paralogues spaC genes from L. casei LOCK 0919 (supplementary fig. S1, Supplementary Material online) allowed us to design specific set of primers (table 2) and to perform the quantification of the genes expression. In the presence of any of the sugars tested, the level of the chromosomal spaC expression was on the threshold of detection (fig. 9A), whereas the expression of the plasmidic spaC was significantly 2.9- or 5.1-fold higher, respectively on glucose or lactose (fig. 9B). Moreover, the use of two sugars, glucose or lactose, allowed us to test whether the expression of the spaC genes may be under the control of the global catabolic repression system. The main example of such a general regulator is catabolite control protein A (CcpA) (Hueck and Hillen 1995) that promotes, in the repressive conditions (mostly in the presence of glucose), carbon catabolite regulation by binding to 14-nt cis-acting DNA target sites known as cre (catabolite responsive element) (Miwa et al. 2000). A thorough analysis of the DNA sequences upstream of the plasmidic and chromosomal spaC genes revealed no obvious cres. Despite the lack of clear cre boxes, spaC expression could be still regulated by CcpA in an indirect manner (Ludwig et al. 2002). Indeed, we found the plasmidic spaC expression to be slightly sugar-dependent as it had 2.9-fold elevated mRNA level on lactose in comparison to glucose (fig. 9B).
Table 2.
Primers for QPCR Amplifications
| Gene or Encoded Protein | 5′–3′ DNA Sequence of Forward/Reverse Primera |
|---|---|
| Primers to quantify the plasmidic or chromosomal spaC expression level | |
| Plasmidic spaC | GGTGGCGAATCGGGT/CCATCGAAGTTGTGCGGTATCGCT |
| Chromosomal spaC | CGGCAATAAAACCACTGG/ATTTTTAAGTTCTGCGGTATAGCC |
| Primers to quantify Lactobacillus rhamnosus LOCK 0900, L. rhamnosus LOCK 0908 and Lactobacillus casei LOCK 0919 in mice feces | |
| Primers for L. casei LOCK 0900-specific genes | |
| Phage anti-repressor protein | GGGCTGCTCTACAAAGATG/AGTTGATGGATGCCAGTTAC |
| DNA repair protein RecT | CCAGTAACGCCCTCAATTC/CCCTTGTATCACCATGTTCAG |
| Primers for L. rhamnosus LOCK 0908-specific genes | |
| DNA helicase YeeB | GGCACGAGTCTATCAACATC/CCTTTCGGACACCTTGATTAG |
| YeeC-like protein | GCGAAGAAGTTGAGCTTAATG/CCATACAGAGATGAGGCAAG |
| Primers for L. rhamnosus LOCK 0919-specific genes | |
| Ornithine cyclodeaminase | GAAGATCGTATCCGTTTACCC/CTGTTCGAAGCTGAGTAAGG |
| Phospholipase A2 family enzyme | GTATGGGAATTATTGCGGTAAAG/CGTTGACGAAATTCCTGATTAC |
| Primers for reference genes | |
| RNA polymerase α subunit | GATATCGTCGCTGATCCTG/GTCAGCTGCCACATACC |
| Translation elongation factor Tu | GACCTTGGATCTTGGTGAAG/TGGATTGAACCTGGCTTTG |
All primers were designed on the basis of the L. rhamnosus LOCK 0900, L. rhamnosus LOCK 0908, and L. casei LOCK 0919 genome nucleotide sequences, which are available from NCBI (http://www.ncbi.nlm.nih.gov/genome, last accessed December 17, 2015) with the respective accession numbers CP005484, CP005485, CP005486 (chromosomes) and CP005487 (plasmid).
Fig. 9.—
The relative plasmidic and chromosomal spaC expression level (A), spaC plasmid/chromosomal ratio on glucose or lactose and glucose repression ratio (B). The relative spaC genes expression level was measured by QPCR; plasmidic/chromosomal spaC ratios were calculated as a quotient of the relative plasmidic and chromosomal spaC expressions in L. casei LOCK 0919 grown in G-MRS or in L-MRS; glucose repression ratio of the plasmidic spaC was calculated as a quotient of the relative plasmidic spaC expression in L. casei LOCK 0919 grown in L-MRS and in G-MRS.
Discussion
L. casei LOCK 0919 is a probiotic strain with an antiallergic potential, which has been proven in a number of clinical trials and animal experiments (Cukrowska et al. 2009, 2010; Kozakova et al. 2015). Among numerous desirable features of microorganisms defined as probiotics is their ability to adhere to the host GI epithelium. Adherence of microorganisms to surfaces involves numerous factors, such as cell hydrophobicity, electrostatic forces, and specific adhesins. Among the bacterial adhesion molecules, the mucus-binding SpaCBA pili are of key importance due to their ability of strong adherence to enterocytes, formation of biofilm, and dampening of IL-8 production by intestinal epithelial cells (Segers and Lebeer 2014). According to our in silico analyses, homologs of the individual SpaCBA proteins are widely represented among different Lactobacillus sp. (fig. 3); however, the encoding genes, in the order of spaCBA, are present in a limited number of species, including L. casei and L. paracasei. This observation is apparent from our analyses of the completely sequenced Lactobacillus sp. genomes (fig. 5) and their draft genome sequences (data not shown). According to the literature (Douillard, Ribbera, Kant, et al. 2013), some of L. rhamnosus strains also produce pili, but so far among them only the genome of L. rhamnosus GG has been released into the GenBank database. All of the hitherto known spaCBA sequences are carried by the chromosomes and, to the best of our knowledge, this is a first comprehensive report on the analysis of the plasmid-encoded SpaCBA pili. Our comparative analyses indicate the highest similarity of the plasmidic L. casei LOCK 0919 spaCBA region to the corresponding DNA region of the best ever known SpaCBA pili producer L. rhamnosus GG, and significantly lower similarity to the corresponding operons from L. casei and L. paracasei strains. We detected this unique homology between sequences of both strains regarding the DNA sequence downstream of spaCBA, transposable elements overlapping spaCBA and amino acid sequence of the SpaC protein. This result is even more interesting due to the fact that all so far identified spaCBA of L. casei and L. paracasei are not expressed and L. casei LOCK 0919 seems to be an exception in this group as its plasmidic spaC is expressed even in the repressive conditions. This expression may be driven by any of two potential promoters found either directly upstream of the spaCBA operon or above the p12 IS, and absent in other L. casei and L. paracasei strains. Thus, similarly to L. rhamnosus GG, in L. casei LOCK 0919 the insertion of an IS element upstream of the plasmidic spaCBA may trigger the expression of the pilus genes.
In order to confirm the results of our bioinformatic predictions implying occurrence of an adhesive potential in L. casei LOCK 0919 and to gain information on the genetic localization of structural properties that are indispensable for the adhesion, series of the in vitro assays including hydrophobicity, aggregation, and adherence to abiotic and biotic surfaces were conducted in Lactobacillus strains. The fact that a high percentage of L. casei LOCK 0919 cells adhered to a nonpolar solvent—n-hexadecane, demonstrated a hydrophobic cell surface of this strain. The removal of pLOCK 0919 from L. casei LOCK 0919 resulted in a complete abolition of the L. casei LOCK 0919Δp cells hydrophobicity indicating the plasmidic localization of genetic determinants of this phenomenon. Bacterial adhesion initially depends on nonspecific physical interactions between two surfaces following specific interactions between mostly proteinaceous adhesins and corresponding receptors. Previous studies have shown cell hydrophobicity to be positively correlated with the presence of a proteinaceous surface-coat and thus leading to the aggregation and adherence ability (Kos et al. 2003; Lukić et al. 2012; Ren et al. 2012). Indeed, our further research on cells autoaggregation confirmed this observation—the most hydrophobic (3.5%) strain L. casei LOCK 0919 was also shown to be highly aggregative (93.2%), whereas L. rhamnosus LOCK 0908 was the least hydrophobic (1.5%) and aggregative (60.6%). The removal of pLOCK 0919 resulted in a significant decrease of L. casei LOCK 0919Δp autoaggregation (0.58%) implying the plasmidic localization of genetic determinants encoding a proteinaceous surface-coat involved in this phenomenon. The fact that after 24 h of the autoaggregation experiment this effect was not pronounced anymore in L. casei LOCK 0919Δp (89.5% of aggregating cells) implies that pLOCK 0919-encoded cell surface proteins may be important factors only in the process of initial attachment and in the early stages of biofilms establishment. The further experimental indication that an adhesion system may indeed operate efficiently in L. casei LOCK 0919 was the strong adherence of this strain to collagen-, mucus-, gelatin-coated or bare polystyrene plates. In contrast, the control L. rhamnsosus LOCK 0908 strain was barely able to adhere to these surfaces. The observed lack or low level of L. rhamnsosus LOCK 0908 adhesion to abiotic and biotic surfaces seems to be reasonable as no spaCBA pilus cluster has been detected in its genome (Koryszewska-Bagińska et al. 2014). The deprivation of pLOCK 0919 caused a statistically significant decrease of L. casei LOCK 0919Δp adherence indicating the plasmidic localization of L. casei LOCK 0919 adhesive potential.
The further confirmation of L. casei LOCK 0919 adhesive potential are the results of our in vivo studies demonstrating that among the three Lactobacillus strains used to colonize GF mice, already after 2 days of i.g. bacteria application, L. casei LOCK 0919 became the dominant strain in the gut. As shown in figure 8, starting from the second day, the amount of L. casei LOCK 0919 in feces was overwhelming ranging from 97% to 99%, whereas L. rhamnosus strains were barely detected. This long-term persistence and high abundance of L. casei LOCK 0919 in mice gut may indicate that its functional adhesion system prevented its immediate elimination by peristalsis as it happened in the case of L. rhamnosus LOCK 0900 and LOCK 0908 strains that do not encode the SpaCBA system. However, we cannot exclude the possibility that the other than the lack of a functional adhesion system and yet unknown factors contributed to the weak persistence of LOCK 0900 and LOCK 0908 in mice guts.
Therefore, we determined that the plasmid-encoded L. casei LOCK 0919 cell surface proteins may be important factors in the process of the attachment and in the initial stages of biofilms establishment. Consequently, we may infer conclusions regarding the prospective, invaluable role of pLOCK 0919 in terms of its adhesive potential. The possession of such a plasmid by any bacterium greatly increases the range of inhabited environments, allows for colonization and longer persistence in the host’s gut, and provides a competitive advantage over other bacteria (including pathogens) in this tough to survive ecosystem.
Supplementary Material
Supplementary figure S1 is available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was partially supported by a grant awarded by the Polish National Centre for Research and Development (no. 12/P01/2010/10) and RVO 61388971 of the Czech Republic.
Literature Cited
- Aleksandrzak-Piekarczyk T, Koryszewska-Bagińska A, Bardowski J. 2013. Genome sequence of the probiotic strain Lactobacillus rhamnosus (formerly Lactobacillus casei) LOCK900. Genome Announc. 1:e00640–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Bernardeau M, Guguen M, Vernoux JP. 2006. Beneficial lactobacilli in food and feed: long-term use, biodiversity and proposals for specific and realistic safety assessments. FEMS Microbiol Rev. 30:487–513. [DOI] [PubMed] [Google Scholar]
- Browning DF, Busby SJW. 2004. The regulation of bacterial transcription initiation. Nat Rev Microbiol. 2:57–65. [DOI] [PubMed] [Google Scholar]
- Cai H, Thompson R, Budinich MF, Broadbent JR, Steele JL. 2009. Genome sequence and comparative genome analysis of Lactobacillus casei: insights into their niche-associated evolution. Genome Biol Evol. 1:239–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552. [DOI] [PubMed] [Google Scholar]
- Chevenet F, Brun C, Bañuls A-L, Jacq B, Christen R. 2006. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7:439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, et al. 2015. Plasmids from food lactic acid bacteria: diversity, similarity, and new developments. Int J Mol Sci. 16:13172–13202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukrowska B, et al. 2009. Probiotic Lactobacillus strains: in vitro and in vivo studies. Folia Microbiol. 54:533–537. [DOI] [PubMed] [Google Scholar]
- Cukrowska B, et al. 2010. Impact of heat-inactivated Lactobacillus casei and Lactobacillus paracasei strains on cytokine responses in whole blood cell cultures of children with atopic dermatitis. Folia Microbiol. 55:277–280. [DOI] [PubMed] [Google Scholar]
- Dereeper A, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douillard FP, Ribbera A, Järvinen HM, et al. 2013. Comparative genomic and functional analysis of Lactobacillus casei and Lactobacillus rhamnosus strains marketed as probiotics. Appl Environ Microbiol. 79:1923–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douillard FP, Ribbera A, Kant R, et al. 2013. Comparative genomic and functional analysis of 100 Lactobacillus rhamnosus strains and their comparison with strain GG. PLoS Genet. 9:e1003683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO-WHO. 2006. Probiotics in food. Health and nutritional properties and guidelines for evaluation. FAO Food and Nutritional Paper No. 85. doi: ISBN: 92-5-105513-0.
- Felis GE, Dellaglio F. 2007. Taxonomy of lactobacilli and bifidobacteria. Curr Issues Intest Microbiol. 8:44–61. [PubMed] [Google Scholar]
- Frickey T, Lupas A. 2004. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics 20:3702–3704. [DOI] [PubMed] [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321. [DOI] [PubMed] [Google Scholar]
- Hildebrand A, Remmert M, Biegert A, Söding J. 2009. Fast and accurate automatic structure prediction with HHpred. Proteins 77:128–132. [DOI] [PubMed] [Google Scholar]
- Hueck CJ, Hillen W. 1995. Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the Gram-positive bacteria? Mol Mikrobiol. 15:395–401. [DOI] [PubMed] [Google Scholar]
- Jankowska A, Laubitz D, Antushevich H, Zabielski R, Grzesiuk E. 2008. Competition of Lactobacillus paracasei with Salmonella enterica for adhesion to Caco-2 cells. J Biomed Biotechnol. 2008:357964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankainen M, et al. 2009. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc Natl Acad Sci U S A. 106:17193–17198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant R, Blom J, Palva A, Siezen RJ, de Vos WM. 2011. Comparative genomics of Lactobacillus. Microb Biotechnol. 4:323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koryszewska-Bagińska A, Aleksandrzak-Piekarczyk T, Bardowski J. 2013. Complete genome sequence of the probiotic strain Lactobacillus casei (formerly Lactobacillus paracasei) LOCK919. Genome Announc. 1:e00758–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koryszewska-Bagińska A, Bardowski J, Aleksandrzak-Piekarczyk T. 2014. Genome sequence of the probiotic strain Lactobacillus rhamnosus (formerly Lactobacillus casei) LOCK908. Genome Announc. 2:e00120–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos B, et al. 2003. Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J Appl Microbiol. 94:981–987. [DOI] [PubMed] [Google Scholar]
- Kozakova H, et al. 2015. Colonization of germ-free mice with a mixture of three lactobacillus strains enhances the integrity of gut mucosa and ameliorates allergic sensitization. Cell Mol Immunol. Available from: http://dx.doi.org/10.1038/cmi.2015.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeer S, Vanderleyden J, De Keersmaecker SCJ. 2008. Genes and molecules of lactobacilli supporting probiotic action. Microbiol Mol Biol Rev. 72:728–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S-M, Ahn D-H. 2012. Factors affecting adhesion of lactic acid bacteria to Caco-2 cells and inhibitory effect on infection of Salmonella typhimurium. J Microbiol Biotechnol. 22:1731–1739. [DOI] [PubMed] [Google Scholar]
- Liu C, Zhang Z-Y, Dong K, Guo X-K. 2010. Adhesion and immunomodulatory effects of Bifidobacterium lactis HN019 on intestinal epithelial cells INT-407. World J Gastroenterol. 16:2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig H, Rebhan N, Blencke H-M, Merzbacher M, Stülke J. 2002. Control of the glycolytic gapA operon by the catabolite control protein A in Bacillus subtilis: a novel mechanism of CcpA-mediated regulation. Mol Microbiol. 45:543–553. [DOI] [PubMed] [Google Scholar]
- Lukić J, et al. 2012. Different roles for lactococcal aggregation factor and mucin binding protein in adhesion to gastrointestinal mucosa. Appl Environ Microbiol. 78:7993–8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack DR, Ahrne S, Hyde L, Wei S, Hollingsworth MA. 2003. Extracellular MUC3 mucin secretion follows adherence of Lactobacillus strains to intestinal epithelial cells in vitro. Gut 52:827–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahillon J, Chandler M. 1998. Insertion sequences. Microbiol Mol Biol Rev. 62:725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa Y, Nakata A, Ogiwara A, Yamamoto M, Fujita Y. 2000. Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis. Nucleic Acids Res. 28:1206–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Permina EA, Mironov AA, Gelfand MS. 2002. Damage-repair error-prone polymerases of eubacteria: association with mobile genome elements. Gene 293:133–140. [DOI] [PubMed] [Google Scholar]
- Poirel L, Decousser J-W, Nordmann P. 2003. Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob Agents Chemother. 47:2938–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, et al. 2012. Inhibition of Staphylococcus aureus adherence to Caco-2 cells by lactobacilli and cell surface properties that influence attachment. Anaerobe 18:508–515. [DOI] [PubMed] [Google Scholar]
- Reunanen J, von Ossowski I, Hendrickx APA, Palva A, de Vos WM. 2012. Characterization of the SpaCBA pilus fibers in the probiotic Lactobacillus rhamnosus GG. Appl Environ Microbiol. 78:2337–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds AE, Mahadevan S, LeGrice SFJ, Wright A. 1986. Enhancement of bacterial gene expression by insertion elements or by mutation in a CAP-cAMP binding site. J Mol Biol. 191:85–95. [DOI] [PubMed] [Google Scholar]
- Ruiz-Barba JL, Piard JC, Jiménez-Díaz R. 1991. Plasmid profiles and curing of plasmids in Lactobacillus plantarum strains isolated from green olive fermentations. J Appl Bacteriol. 71:417–421. [DOI] [PubMed] [Google Scholar]
- Saxelin M, Tynkkynen S, Mattila-Sandholm T, de Vos WM. 2005. Probiotic and other functional microbes: from markets to mechanisms. Curr Opin Biotechnol. 16:204–211. [DOI] [PubMed] [Google Scholar]
- Segers M, Lebeer S. 2014. Towards a better understanding of Lactobacillus rhamnosus GG- host interactions. Microb Cell Fact. 13:S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siguier P, Filée J, Chandler M. 2006. Insertion sequences in prokaryotic genomes. Curr Opin Microbiol. 9:526–531. [DOI] [PubMed] [Google Scholar]
- Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Ogura M. 1998. A novel Bacillus natto plasmid pLS32 capable of replication in Bacillus subtilis. FEBS Lett. 422:243–246. [DOI] [PubMed] [Google Scholar]
- Vesterlund S, Karp M, Salminen S, Ouwehand AC. 2006. Staphylococcus aureus adheres to human intestinal mucus but can be displaced by certain lactic acid bacteria. Microbiology 152:1819–1826. [DOI] [PubMed] [Google Scholar]
- Vesterlund S, Paltta J, Karp M, Ouwehand AC. 2005. Measurement of bacterial adhesion—in vitro evaluation of different methods. J Microbiol Methods. 60:225–233. [DOI] [PubMed] [Google Scholar]
- von Ossowski I, et al. 2010. Mucosal adhesion properties of the probiotic Lactobacillus rhamnosus GG SpaCBA and SpaFED pilin subunits. Appl Environ Microbiol. 76:2049–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T-T, Lee BH. 1997. Plasmids in Lactobacillus. Crit Rev Biotechnol. 17:227–272. [DOI] [PubMed] [Google Scholar]
- Zeraik AE, Nitschke M. 2012. Influence of growth media and temperature on bacterial adhesion to polystyrene surfaces. Braz Arch Biol Technol. 55:569–576 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.