Abstract
High-density linkage maps are important tools for genome biology and evolutionary genetics by quantifying the extent of recombination, linkage disequilibrium, and chromosomal rearrangements across chromosomes, sexes, and populations. They provide one of the best ways to validate and refine de novo genome assemblies, with the power to identify errors in assemblies increasing with marker density. However, assembly of high-density linkage maps is still challenging due to software limitations. We describe Lep-MAP2, a software for ultradense genome-wide linkage map construction. Lep-MAP2 can handle various family structures and can account for achiasmatic meiosis to gain linkage map accuracy. Simulations show that Lep-MAP2 outperforms other available mapping software both in computational efficiency and accuracy. When applied to two large F2-generation recombinant crosses between two nine-spined stickleback (Pungitius pungitius) populations, it produced two high-density (∼6 markers/cM) linkage maps containing 18,691 and 20,054 single nucleotide polymorphisms. The two maps showed a high degree of synteny, but female maps were 1.5–2 times longer than male maps in all linkage groups, suggesting genome-wide recombination suppression in males. Comparison with the genome sequence of the three-spined stickleback (Gasterosteus aculeatus) revealed a high degree of interspecific synteny with a low frequency (<5%) of interchromosomal rearrangements. However, a fairly large (ca. 10 Mb) translocation from autosome to sex chromosome was detected in both maps. These results illustrate the utility and novel features of Lep-MAP2 in assembling high-density linkage maps, and their usefulness in revealing evolutionarily interesting properties of genomes, such as strong genome-wide sex bias in recombination rates.
Keywords: linkage map, Lep-MAP2, Pungitius pungitius, RAD-tag, recombination, SNP
Introduction
Recombination and linkage disequilibrium are two inextricably bound facets of the forces driving haplotype formation, on which natural selection can work (Hill and Robertson 1966; Feldman et al. 1996; Gessler and Xu 2000; Otto and Lenormand 2002; Posada et al. 2002). Thus, assessing the extent of recombination and linkage in the genome of an organism is important for understanding the structural, functional, and evolutionary characteristics of the genome (Wang et al. 2009; Hohenlohe et al. 2011; Kai et al. 2011). A genetic linkage map provides not only the relative order of the markers, but also a direct measure of the extent of recombination and linkage disequilibrium across chromosomes. In sexually reproducing organisms, it also allows identification of the role of each sex in creating novel haplotypes (Broman et al. 1998; Sakamoto et al. 2000; Lenormand 2003; Hedrick 2007). From a structural genomic standpoint, a linkage map provides the data necessary to analyze the presence, location, and relative size of chromosomal rearrangements, such as inversions (Tanksley et al. 1992; Agresti et al. 2000; Bansal et al. 2007). As such, linkage maps can also facilitate de novo genome assembly and validation by enabling the identification of chimeric scaffold constructs (Rastas et al. 2013; Fierst 2015). Furthermore, because haplotype length and persistence are strongly influenced by natural selection, comparisons of the levels of linkage surrounding a locus across multiple populations can provide clues to understand the evolutionary history of the locus in examination (Birky and Walsh 1988; Kreitman and Hudson 1991).
Apart from providing insights into the genomes of target species and populations, high-density linkage maps of nonmodel species can set a strong foundation for comparative genomics and the analysis of synteny across species, providing vital clues for our understanding of genome evolution and speciation (Kulathinal et al. 2009; Larkin et al. 2009; Michalak de Jimenez et al. 2013; Zhang et al. 2014). Additionally, these maps would allow linking phenotypes to specific regions of the genotype (through quantitative trait locus or association mapping analyses), and thus hold the key to understand the genetics of complex phenotypic traits (Paterson et al. 1988; Flint and Mackay 2009; Goddard and Hayes 2009).
Modern development in high-throughput sequencing technologies, such as restriction site associated DNA tags (RAD-tags; Miller et al. 2007), allow a cost-effective detection of several thousands of single nucleotide polymorphism (SNP) markers in the genome of nonmodel organisms. Compared with microsatellite markers, SNPs have the potential to substantially simplify the creation of linkage maps because they can be potentially genotyped with a greater accuracy and genome coverage than microsatellites (Kruglyak 1997; Slate et al. 2009). However, by substantially increasing the sample space containing the true marker order, large marker data sets increase the computational burden involved with linkage map construction. A number of different approaches have been devised to tackle this problem (van Os, Plet, et al. 2005; van Os, Stam, et al. 2005; Margarido et al. 2007; Tong et al. 2010; Van Ooijen 2011), but effective solutions are still few (Rastas et al. 2013; Liu et al. 2014). For instance, although the linkage mapping program Lep-MAP (Rastas et al. 2013) is in principle capable of creating linkage maps containing tens of thousands of markers, the computational time required for mapping a very large number of markers (≥2,000 per chromosome) becomes unfeasible, as it increases cubically with the number of markers per chromosome. Likewise, Lep-MAP does not allow modeling sex-specific recombination rates. Hence, there is a need for improved linkage-mapping software to make efficient use of the high-throughput data provided by new sequencing technologies.
The main aims of this study were 2-fold. First, to introduce and benchmark a substantially improved version of the Lep-MAP (Rastas et al. 2013) software (henceforth Lep-MAP2) capable of creating ultra–high-density linkage maps. Second, to use Lep-MAP2 to construct two high-density linkage maps for nine-spined sticklebacks (Pungitius pungitius) based on large SNP panels obtained using a RAD sequencing approach. The nine-spined stickleback is a nonmodel teleost, closely related (Kawahara et al. 2009; Guo et al. 2013) to the three-spined stickleback (Gasterosteus aculeatus) whose genome has been sequenced (Kingsley and Peichel 2007). Both these species are important models for an increasing amount of evolutionary biology and genetics research (Bell and Foster 1994; McKinnon and Rundle 2002; Kingsley and Peichel 2007; Wootton 2009; Merilä 2013), including the study of sex chromosome evolution (Peichel et al. 2004; Kitano et al. 2009; Ross et al. 2009; Shikano, Herczeg, et al. 2011; Shikano, Natri, et al. 2011; Natri et al. 2013). Hence, we were interested in comparing the degree of synteny and collinearity between nine-spined stickleback linkage maps and the three-spined stickleback genome in order to infer the frequency of inverted and transposed genomic regions, which are suspected to play an important role in both speciation (Flaxman et al. 2014) and local adaptation (Yeaman 2013). Specifically, we were interested in exploring possible heterogeneity in sex-specific recombination rates across the different linkage groups, as well as identifying possible structural rearrangements and recombination heterogeneity in the sex chromosomes.
Materials and Methods
A Brief Description of Lep-MAP2
Lep-MAP2 software for constructing ultradense linkage maps is based on Lep-MAP (Rastas et al. 2013) with the following novel features and improvements: 1) It takes into account achiasmatic meiosis (recombination in one sex only) and models sex-specific recombination rates, 2) the marker ordering algorithm scales to a much larger number of markers than that in Lep-MAP, 3) it can utilize and gain speed using multicore processors, and 4) the data analyzing pipeline has been improved to ease the map construction. Furthermore, it is largely automated and requires minimal user interaction. It can analyze multiple outbred families simultaneously as well as typical inbred crosses, and can handle all types of genetic marker data (e.g., SNPs, microsatellites).
The input of Lep-MAP2 consists of genotypes of one or several full-sib families (parents and their offspring), given in pre-makeped LINKAGE (Lathrop et al. 1984) pedigree format. The format gives the pedigree information on columns 1–6 and genotypes starting on column 7 onward. Only full-sib type pedigree structure is supported, but data from several types of crosses (e.g., backcrosses) can be treated as full-sibs (supplementary file S1, Supplementary Material online).
The data workflow of Lep-MAP2 with descriptions of five modules included into the program are given in supplementary file S1, Supplementary Material online. Lep-MAP2 software is publicly available together with its source and documentation at http://sourceforge.net/projects/lepmap2/ (last accessed December 22, 2015).
Simulated Data
Simulations were used to compare the performance of Lep-MAP2 with that of TMAP (Cartwright et al. 2007), JoinMap (Van Ooijen 2011), and HighMap (Liu et al. 2014). To this end, the accuracy (i.e., ability of the software to recover the correct marker order and map length) as well as computational time taken by different programs for one, two, and five family data sets with three different genotyping error rates (0%, 1%, and 5%) were assed. However, we did not evaluate the influence of genotyping errors on the five-family data sets as the run time of TMAP and JoinMap became too prohibitive. Simulated data were created as explained in supplementary file S2, Supplementary Material online. In short, it consisted of 100 individuals from a full-sib family with 10 chromosomes and 300 markers per chromosome. The recombination probability between adjacent markers was set to 0.333% and 0.167% for the father and mother, respectively. The parents were informative (heterozygous) with a probability of 0.5.
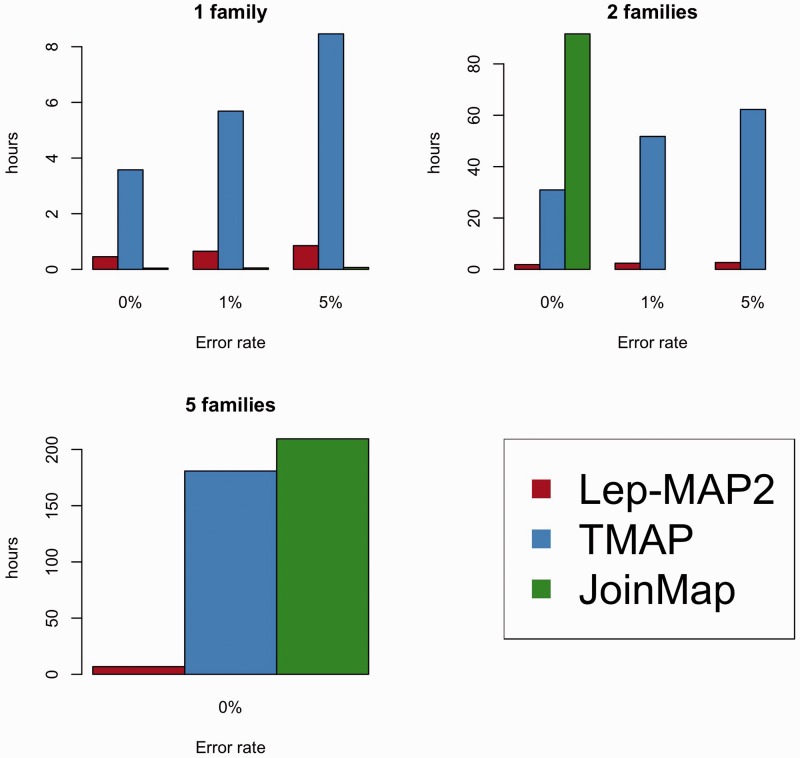
Lep-MAP2 was run ten times on each simulated data set, and both the results of the first run (LM1) and the run with the highest likelihood (LM10) are reported. For other software, only one run was conducted. For each run, we computed the Kendall tau (Kendall 1938) between the found and the correct order on the subset of informative markers with detectable recombinations. We also measured the time of each run using the Linux command “time.” The timings of LepMAP2 and TMAP were measured with a desktop computer running Linux and having 24 GB of memory and four Intel Core i7-4790 central processing units (CPUs) running at 3.60 GHz frequency. JoinMap was run on a Windows 7 Enterprise computer with 128 GB memory and dual Xeon E5-2640 v3 CPUs running at 2.60 GHz frequency. HighMap was run by the developers of the program itself, because at the time of this study, HighMap was not available for general use. One specific limitation of JoinMap was that it is a 32 bit binary with an obligatory graphical front end. Thus it is difficult to efficiently run multiple jobs or to time them, and the need for direct user input proved to be quite high. Hence, run time comparisons between JoinMap and other software were limited to single-family comparisons and multiple-family comparisons with zero error rate. HighMap runtimes could not be clocked (see above).
Stickleback F2 Recombinant Crosses
Adult nine-spined sticklebacks were collected from a marine population in Southern Finland (Helsinki, 60°13′N, 25°11′E) and from two pond populations in northeastern Finland (Rytilampi 66°23′N, 29°19′E and Pyöreälampi 66°15′N, 29°26′E) in 2006 and 2011, respectively. Two F1 hybrid generations were created by mating a marine female to a pond male from each pond population, and the F2 generations were generated from the repeated mating of a single full-sib F1 pair for each hybrid cross. In the case of the first cross (Helsinki-Rytilampi, henceforth HR cross), the resulting 283 F2 offspring are the same as used in Shikano et al. (2013) and Laine et al. (2013). In the case of the second cross (Helsinki-Pyöreälampi, henceforth HP cross), 284 F2 offspring were obtained for the purpose of this study. More details about crossing and rearing procedures used to create HR cross can be found from Shikano et al. (2013). The procedures for setting up and rearing the HP cross were mostly identical to those used for the HR cross. Sex of all the F2 offspring in both crosses was identified by genotyping all individuals for a sex-linked microsatellite marker (Stn19) as detailed in Shikano, Herczeg, et al. (2011).
This study did not involve human subjects, and our experimental protocols were approved by the National Animal Experiment Board, Finland (permission numbers: ESLH-STSTH223A and STH037A).
DNA Extraction and RAD Library Construction
Genomic DNA was extracted from ethanol-preserved fin clips using the phenol–chloroform method (Taggart et al. 1992). RAD library construction and sequencing were performed by BGI HONGKONG CO., Ltd. Briefly, DNA was fragmented by the restriction enzyme PstI, and DNA fragments of 300–500 bp were gel purified. Illumina sequencing adaptors and library-specific barcodes were ligated to the digested DNA fragments, and barcoded RAD samples were then pooled and sequenced on the Illumina HiSeq2000 platform with 45-bp single-end strategy. Twenty-four lanes were used for the HR cross and 30 for the HP cross sequencing. For each cross, grandparents and parents were sequenced in one lane (i.e., four individuals per lane) to increase their sequence coverage, and thereby also the number of mappable SNPs. Adapters and barcodes were eliminated from reads and quality was checked using FastQC (Andrews S. FastQC, http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/, last accessed December 22, 2015).
RAD-Tag Sequencing and Genotyping
An in-house pipeline was used to obtain the genotype calls from the raw single-end reads for each individual as follows. As the read length varied between 41 and 45 bp, reads were cropped by keeping only the first 41 bp. The reads of parental individuals without any missing nucleotides in both populations were pooled and identical reads were grouped together. These grouped reads were processed in the descending order of the number of occurrence. Each processed read was added to the sequence list and all its neighbors within edit (Levenshtein) distance of two were removed from the order. Only sequences occurring between 10 and 1,500 times were kept and taken as reference sequences: The sequences occurring less than 10 times were considered sequencing errors and sequences with >1,500 occurrences were likely repeat regions, thus these sequences were discarded. In the end, a total of 712,005 reference sequences, each 41 bp in length, were recorded. We verified that the number of reference sequences agreed with the number of restriction sites in the genome.
All raw reads (cropped to 41 bp) were mapped against the reference sequences with BWA (Li and Durbin 2009), together with SAMtools (Li et al. 2009), producing a single bam file for each individual.
Individual genotype posterior probabilities, taking into account the read and mapping qualities, were obtained by in-house scripts (Kvist et al. 2015, their Appendix S1) from the output of SAMtools (Li et al. 2009) mpileup on the bam files giving multiple alignment of reads for each reference position. Only positions with 2 or more alleles and no more than 20 indels among all individuals were considered. Furthermore, minimum read coverage of 3 was required for more than 158 individuals, which assured that for most (>60%) individuals it would be possible to call genotypes (or at least give some informative posterior) on the remaining markers. The parental genotypes were called by maximizing the likelihood of offspring and parent genotypes using module ParentCall, and then each offspring was called with respect to its called parental genotypes. Parental genotypes were called only if their likelihood was 100 times higher than the second best parental genotype combination. The offspring genotypes were called similarly as the parents.
Genotype calling thus identified 41,730 potential SNP markers shared between the 2 crosses. These potential SNPs were then independently quality checked for linkage map construction and low-quality SNPs were discarded.
Additional genotype data on 226 microsatellite markers were added to the HR data set. These are a subset of markers used in Shikano et al. (2013), but for the purposes of this study 28 markers (listed in supplementary table S1, Supplementary Material online) were omitted from linkage map construction because of high segregation skew or high error estimates (see section on linkage map construction and supplementary table S1, Supplementary Material online).
As a validation of the quality of the RAD-generated SNPs, and also to confirm that sample identities (including sex identity) had not been mixed between sampling and sequencing, for each cross we re-extracted DNA from the original tissue samples and genotyped all individuals for a subset of SNPs (64 for HP and 33 for HR) using the Sequenom (San Diego, USA) platform at the Finnish Institute for Molecular Medicine (FIMM). After discarding SNPs that failed to amplify, we were able to directly compare 58 SNPs for HP and 29 for HR cross between RAD and FIMM calls. These comparisons revealed that no mixing of samples had occurred between sampling and sequencing.
Linkage Map Construction with Lep-MAP2
Linkage group (LG) assignment was obtained using Lep-MAP2 software. First, the SeparateChromosomes module was executed with an logarithm of odds (LOD) score limit of 20 and minimum LG size of 10. Second, singular markers were added to the found LGs using the JoinSingles module with an LOD score limit of 10. Markers with more than 40 (about 14%) missing genotypes were removed from the LGs. Over 6,000 markers in LG12, which corresponds to the sex chromosomes (Shikano et al. 2013), were informative only for the paternal side in both crosses. To reduce the computational burden, only a common subset of 1,548 of these (paternally informative) markers with at most 10 missing values were kept in this chromosome.
Lep-MAP2 filters out markers by comparing the offspring genotype distribution and the expected Mendelian proportions (segregation distortion test). The default value of dataTolerance = 0.01 was used to filter out highly segregated markers (χ2 test, P < 0.01), thus 1 out of 100 markers should be removed by chance alone. This filtering removed 2,238 and 1,601 SNPs from HP and HP data sets, respectively, and also 28 microsatellite markers from the HR data set as described above. Marker order was determined by allowing different recombination probabilities in both sexes. Ten independent runs were conducted and the marker order with the best likelihood was kept. Only one of the exactly identical markers was used in marker ordering. Furthermore, if there were two markers with identical genotypes but one had more missing genotypes, only the one with less missing genotypes was kept. Lep-MAP2 marks unused markers as duplicated markers and takes their position from the corresponding nonduplicate marker. The final map also included an estimate for genotype error for each marker. The error parameters correspond to the hidden Markov model (HMM) used to model recombinant haplotypes in Lep-MAP2. The recombination rates correspond to the transition parameters in the HMM, whereas the emission probabilities define the error parameters (supplementary file S1, Supplementary Material online). Finally, markers with genotype error rate estimate >0.1 were removed. Few (<0.1%) markers from the ends of LGs were also removed with the criteria that 1) they contributed over 10 cM (per marker) to map length and 2) the parental coverage from their corresponding sequence was above 500 (likely repeat) or below 20 (likely haplotype or sequencing error) on all markers to be removed. The number of removed markers in this last step was 179 SNPs for HP and 174 SNPs plus 8 microsatellites for HR. The HR map was re-evaluated without microsatellites by running Lep-MAP2 on the final order as the initial marker order.
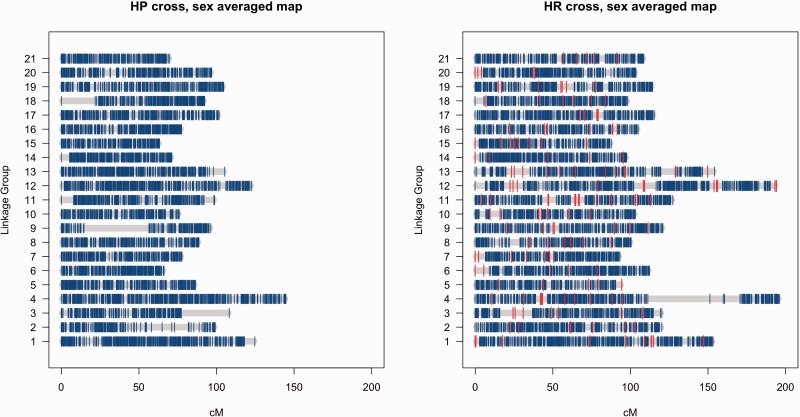
The number of initial LGs found was 23 for HP and 21 for HR. By comparing the linkage maps between HR and HP crosses, it became clear that the two smallest LGs in the HP cross were parts of LGs 2 and 9 in the HR cross. These groups were added to the corresponding LGs and the maps were re-evaluated. The reason why these parts were initially separated is in the large gaps shown in figure 2. However, it was also clear that part of HR LG4 was missing from the maps. This part was found by inspecting (SeparateChromosomes with dataTolerance = 0.0001) the markers filtered out based on the segregation distortion test. This part was also separated by a large gap from other markers in LG4, visible in figure 2, and added to the map as described above. Finally, all the maps (and also HR without microsatellites) were polished by running ten independent runs of Lep-MAP2 using the found marker order as the initial marker order.
Fig. 2.—
Ideograms of the sex-averaged linkage maps for HP and HR crosses of nine-spined sticklebacks. The position of microsatellite loci in the HR cross maps are indicated in red.
Comparisons with Three-Spined Stickleback Genome
To compare genomic synteny between nine- and three-spined sticklebacks, the reference sequences with SNP makers in the linkage map were mapped onto the three-spined stickleback genome (Ensembl release-75) by BLAST (Altschul et al. 1997) with an e-value cut-off at 1 × 10−5 by considering sequence divergence between nine- and three-spined sticklebacks (Guo et al. 2013). In order to infer whether interchromosome rearrangements have occurred in the nine- or three-spined stickleback lineage, we conducted a BLAST search against the genome sequences of medaka (Oryzias latipes), which is the closest ancestor of sticklebacks with a sequenced genome. The genomic synteny was visualized using CIRCOS (Krzywinski et al. 2009).
Statistical Analyses
LG lengths were log10 transformed to achieve normality and homoscedasticity between groups. To partition variance in LG lengths to effects of cross and sex, we used the R package lme4 to perform a two-way random-effects ANOVA with interaction. Cross, sex, and their interaction were also used as fixed terms in an ANOVA to determine the significance of their effect on the log10 of the LG lengths. Correlations between LG lengths were estimated using nonparametric Kendall’s rank correlation (tau) to account for the nonnormal distribution of LG lengths. Fisher’s exact test was used to test for associations between marker genotypes and sex. Because the two only terms in these analyses were sex and marker genotype, Fisher’s exact test is equivalent to a logistic regression. The large number of markers tested makes the use of a P value threshold not useful, because a large number of tests would exceed it purely by chance. Thus, the significance of association between marker genotypes and sex was assessed by comparing the distribution of the observed P value with the P value expected under a null hypothesis of no association.
Results
Lep-MAP2 Performance
Based on the simulations, it is clear that Lep-MAP2 can produce very accurate linkage maps. In single-family simulations, Lep-MAP2 outperformed all other software, both in recovering the correct marker order and the actual map length (150 cM; table 1). Although JoinMap was equally good as Lep-MAP2 in finding the correct marker order, it appeared to be sensitive to map-length inflation due to genotyping errors (table 1). In terms of computational time, LEP-MAP2 was substantially faster than TMAP at all genotyping error rates in single-family comparisons, but slower than JoinMap (fig. 1). For finding the correct marker order in multiple-family mapping simulations, the differences among Lep-MAP2, TMAP, and JoinMap were negligible: The different programs produced maps of roughly equally high quality (table 1). However, Lep-MAP2 was much faster than TMAP or JoinMap (fig. 1). Moreover, the speedup obtained by utilizing multicore processors to run Lep-MAP2 was very closely linear to the number of cores used (1 core: 5 h 13 min; 2 cores: 2 h 37 min; 4 cores: 1 h 21 min). Using 4 cores and the same desktop computer as used in the simulations, linkage map construction for 5,000 markers and 16 families was completed in 5 days and 12 h (parameters filterWindow = 10 and polishWindow = 100 were used for extra speedup). The accuracy (|tau|) of obtained solution was 0.999. Based on this, we estimate that the maximum data set size for Lep-MAP2 to analyze in 1 week is about 10,000 markers and 10,000 individuals on a fast computer (with 32 or more cores). The computational time in Lep-MAP2 scales linearly with the number of individuals, and quadratically with the number of markers. Based on the single-family runs, the performance of HighMap was similar to that of TMAP in terms of map order, but better in terms of map length (table 1). However, its performance in comparison with Lep-MAP2 and JoinMap was poor on all fronts (table 1).
Table 1.
Comparison of Performance of Lep-MAP2 (LM2) with Other Linkage Mapping Software in Terms of Map Order (Kendall tau correlation between expected and observed marker order) and Map Length for Different Error Rates and Number of Mapping Families
| Map Order (Kendall tau) |
Map Length (cM) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Family no. | Error rate | LM2 1st run | LM2 best of 10 runs | TMAP | JoinMAP | HighMap | LM2 1st run | LM2 best of 10 runs | TMAP | JoinMAP | HighMap |
| 1 | 0 | 0.99 | 0.99 | 0.76 | 0.99 | 0.72 | 150.5 | 148.4 | 228.1 | 155.0 | 186.3 |
| 0.01 | 0.99 | 0.99 | 0.64 | 0.99 | 0.69 | 156.4 | 151.3 | 216.2 | 266.9 | 193.9 | |
| 0.05 | 0.98 | 0.99 | 0.64 | 0.98 | 0.67 | 170.4 | 159.0 | 229.7 | 666.8 | 193.6 | |
| 2 | 0 | 0.99 | ∼1.00 | ∼1.00 | 0.98 | NA | 148.7 | 148.7 | 146.1 | 126.3 | NA |
| 0.01 | 0.99 | 0.99 | 0.97 | NA | NA | 152.3 | 152.2 | 154.1 | NA | NA | |
| 0.05 | 0.99 | 0.99 | 0.99 | NA | NA | 156.9 | 156.7 | 153.3 | NA | NA | |
| 5 | 0 | ∼1.00 | ∼1.00 | ∼1.00 | ∼0.98 | NA | 149.2 | 149.2 | 147.0 | 124.4 | NA |
Note.—NA indicates missing data when running the simulation proved impractical (see Methods).
Fig. 1.—
Performance of Lep-MAP2 linkage mapping software in comparison with two other (TMAP and JoinMap) programs in terms of computational time. The results are based on simulated data at different genotyping error rates and with different numbers of mapping families (see text for details).
Sex-Averaged Linkage Maps for Nine-Spined Sticklebacks
After quality control (QC) thinning we identified an excess of 15,000 markers in both crosses. In the HP cross, we identified 20,054 markers, of which 13,060 (65%) were uniquely informative in building the linkage map (table 2). In the HR cross, 18,691 markers were identified, of which 14,998 (80%) were uniquely informative in building the linkage map (table 2). Overall, 22,761 markers were mapped, 15,984 of which were common to both crosses. The difference in marker numbers is due to the fact that even though all markers were initially chosen from the RAD calls as common between crosses, some markers successfully mapped in only one cross due to a high rate of missing genotypes or a questionable segregation pattern in the other cross. Lep-MAP2 identified 21 LGs in both maps (table 2), a number that matches the expected number of chromosomes in the nine-spined stickleback (2n = 42; Ocalewicz et al. 2008). After applying the LG size corrections suggested by Tripathi et al. (2009), the sex-averaged HP and HR cross maps spanned a total of 1980.74 and 2528.96 cM, respectively (table 2). The average genome-wide marker density for the sex-averaged HP and HR cross maps were 6.73 and 5.99 markers/cM, respectively (table 2 and fig. 2). The actual marker positions in the linkage maps for both sexes in both crosses are given in supplementary table S2, Supplementary Material online.
Table 2.
Summary of the Pungitius pungitius F2 Cross Linkage Maps
| Male | Female | Sex Average | |
|---|---|---|---|
| HP cross | |||
| No. of F2 individuals | 127 | 157 | 284 |
| No. of linkage groups | 21 | ||
| No. of markers | 20,054 | ||
| No. of unique markers (overall %) | 13,060 (65%) | ||
| Summary unique marker number/LG | Minimum = 364, mean = 621.9, maximum = 952 | ||
| Map length (cM) | 1,584.26 | 2,335.72 | 1,980.74 |
| Average LG length (cM) | 75.19 | 110.90 | 94.01 |
| Average unique marker spacing (cM) | 0.13 | 0.18 | 0.16 |
| Maximum unique marker spacing (cM) | 26.93 | 71.54 | 41.93 |
| Average unique marker density (no. of markers/cM) | 8.43 | 5.83 | 6.73 |
| HR cross | |||
| No. of F2 individuals | 141 | 142 | 283 |
| No. of linkage groups | 21 | ||
| No. of markers | 18,691 | ||
| No. of unique markers (overall %) | 14,998 (80%) | ||
| Summary unique marker number/LG | Minimum = 505, mean = 714.2, maximum = 1,209 | ||
| Map length (cM) | 1,724.27 | 3,344.09 | 2,528.96 |
| Average LG length (cM) | 81.88 | 158.80 | 120.10 |
| Average unique marker spacing (cM) | 0.11 | 0.22 | 0.17 |
| Maximum unique marker spacing (cM) | 10.87 | 43.80 | 40.07 |
| Average unique marker density (no. of markers/cM) | 8.80 | 4.48 | 5.99 |
Sex-Specific Linkage Maps
Males and females exhibited a substantial difference in map lengths, with female map length exceeding that of males by a factor of 1.5 (HP) to 1.9 (HR; table 2). The average male map lengths for HP and HR crosses were 1,584.26 and 1,724.27 cM, whereas those for females were 2,335.72 and 3,344.09 cM, respectively (table 2). Accordingly, the marker densities in both male maps (HP 8.43 markers/cM; HR 8.80 markers/cM) were 1.5–2 times higher than those in female maps (HP: 5.83 markers/cM; HR: 4.48 markers/cM; table 2). There were no significant differences in the number and distribution of sex informative markers in the two sexes in either cross (HP paired t-test, t20 = −1.25, p = 0.17; HR paired t-test, t20 = −0.06, P = 0.55), indicating that the map length differences between sexes are not explainable by differences in the number of male only versus female only informative markers. In terms of the relative importance of sex and cross identity, sex explained 58% of variance in LG length (ANOVA: F1,80 = 97.6, P < 10−14) whereas the cross effect was much smaller (7% of variance explained; F1,80 = 18.3, P < 10−4). The sex-by-cross interaction was also significant (F1,80 = 7.4, P = 0.008), but it explained only 8% variance in the data. Hence, in spite of the clear similarities in length of the different LGs across the two crosses, there were also some differences (fig. 2, table 2, and supplementary table S3, Supplementary Material online). In males, the LG length was uncorrelated between the two crosses (τ19 = 0.24, P = 0.14), whereas in females the similarity was higher (τ19 = 0.44, P = 0.005), indicating that in females the LG length order in one cross provides a reasonable approximation of the LG length order in the other cross. For the sex-averaged maps the concordance in the LG length distribution was good, albeit far from perfect (τ19 = 0.57, P < 0.001).
Association between Markers and Sex
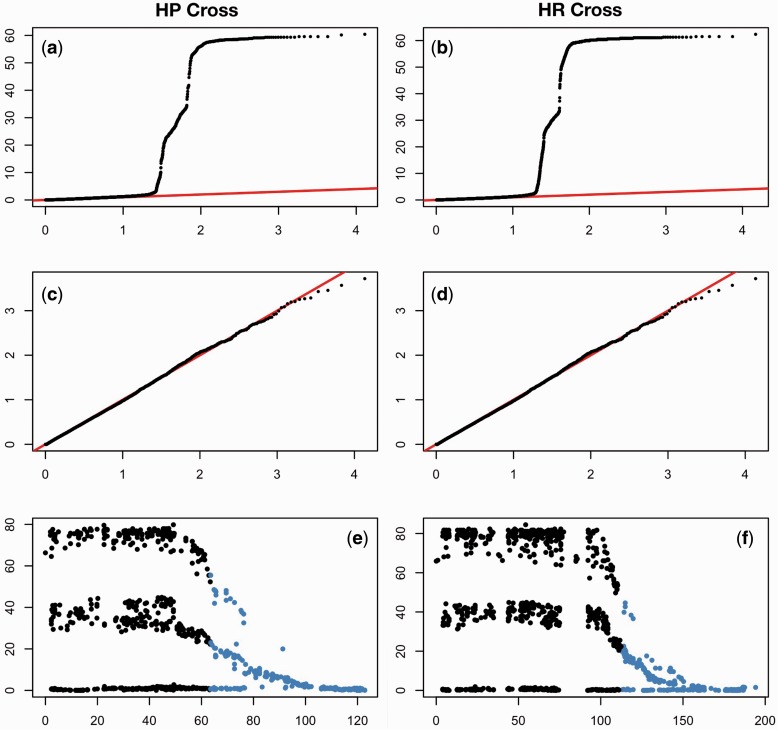
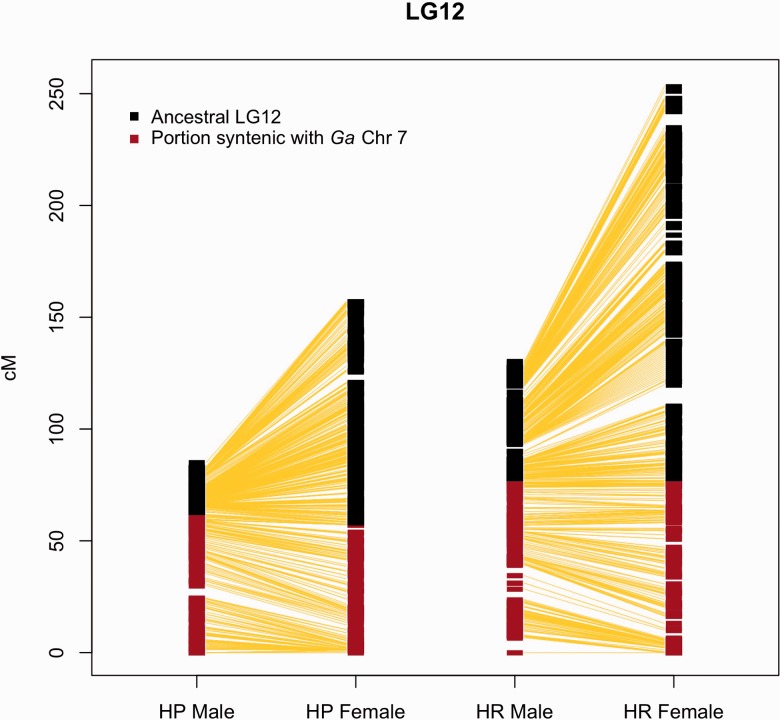
Sex determination in Gasterosteidae varies across species, with evidence of recent evolution of sex chromosomes (Ross et al. 2009). Association analyses allowed assessment of evidence for sex-associated loci outside the sex chromosomes (LG12). After determining each marker/sex association P value, LogQQ plots were used to visualize the results at the genome-wide level (fig. 3a–d). Although a plot of the P values including all LGs shows an impressive deviation from the null hypothesis of no association in both crosses (fig. 3a and b), removal of LG12 totally erases any signature of sex-marker associations (fig. 3c and d). These results suggest that although the markers on LG12 are strongly associated with sex, none of the markers on any of the other LGs are sex linked. Of particular note is the pattern of association between markers and sex on LG12 in both crosses (fig. 3e and f). When plotting the −log10 of the P value of association with given SNP and sex against map position, two interesting observations can be made. The first is that the pattern of distribution of the P values segregates at three different levels, shown in figure 3e and f as “lines.” These lines correspond to the three possible SNP genotype arrangements in the F1 parents: The “0 line” corresponds to the loci where the F1 male had the same allele on the X and the Y chromosome, and thus male and female F2 are equally divided across the genotypes; the “middle line” corresponds to the loci where both F1 parents are heterozygotes, and thus 50% of the male and 50% of the female F2 are identified by their genotype; and the “top line” corresponds to the loci where the female is a homozygote and the male is a heterozygote with a distinctive allele on the Y, giving a unique genotype to the males and to the females in F2. Thus the F1 genotype completely explains the P value pattern we observe on LG12. The second observation is that, despite a strong association between sex and genotype, the part of LG12 that is syntenic with the three-spined stickleback Chr7 (see below) does not show an association between markers and sex in its distal part. This finding suggests that while the translocated part is still recombining, recombination in the ancestral part has almost (but not entirely) ceased (fig. 3e and f). This inference is supported by comparing linkage map lengths and marker numbers (a proxy of physical size of the chromosomes) between the ancestral and translocated parts of LG12: In spite of being shorter in terms of linkage map length in males (fig. 4), the ancestral part of the chromosome has a much higher number of markers (HP male 2,077 markers; HR male 785) than the novel translocation syntenic with the three-spined stickleback Chr7 (HP male 483 markers, HR male 424 markers). Accordingly, the marker density on the ancestral part of LG12 is much higher than that in the translocated part (HP males 93.8 vs. 7.7 markers/cM, HR males 30.2 vs. 5.4 markers/cM). Note that recombination on the ancestral portion of LG12 is not completely suppressed, because this region does not collapse into one single fixed haplotype. Interestingly, recombination in this region is lower than in the novel part in females (marker density: HP 21.1 vs. 8.2 marker/cM; HR 9.7 vs. 5.4 marker/cM), despite the fact that the ancestral part of the LG is actually bigger (in terms of recombination) than the novel part (fig. 4).
Fig. 3.—
Association between sex and genotypes in the two nine-spined stickleback crosses as illustrated by LogQQ plots for (a, b) all data and (c, d) with LG12 markers removed. (e, f) Manhattan plots of the distribution of P values for association between sex and genotype across LG12. Blue depicts the parts of nine-spined stickleback LG12 syntenic to markers in three-spined stickleback chromosome 7.
Fig. 4.—
Direct comparison of marker position between male and female sex chromosome maps (LG12) in the two crosses: In males, the ancestral part of the linkage group shows much less recombination than that in the females, whereas recombination in both maps is almost identical in the new portion synthetic with G. aculeatus (Ga) chromosome 7.
Genomic Synteny between Nine- and Three-Spined Sticklebacks
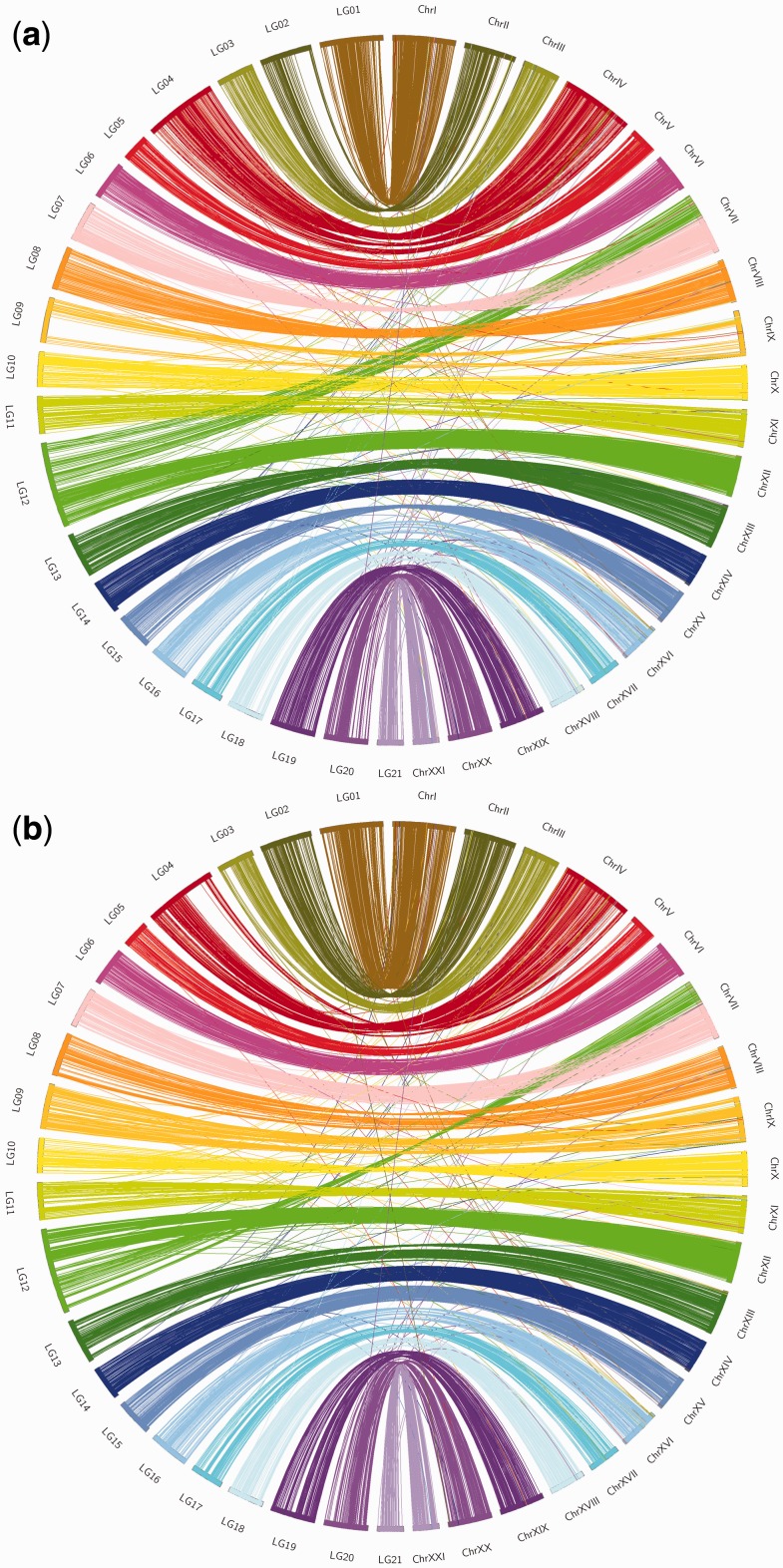
BLAST searches of the 34,015 nine-spined stickleback reference sequences with 41,730 SNPs against the three-spined stickleback genome sequence indicated a high degree of genomic synteny. A total of 11,030 of the 34,015 nine-spined stickleback reference sequences yielded high-scoring BLAST hits and 10,320 of these had unique high-scoring BLAST hits on the three-spined stickleback genome, with 6,506 located in coding regions. In all further comparisons, only these 10,320 sequences were utilized.
In the sex-average linkage map of the HP cross, 5,229 reference sequences with 5,732 SNPs mapped to the three-spined stickleback genome (table 3) indicating a high degree of genomic synteny between nine- and three-spined sticklebacks (fig. 5a). Although most of the 5,229 reference sequences were located in the syntenic LG pairs, 244 (4.3%) showed interchromosomal rearrangements between the species (table 3). For example, 120 reference sequences located on three-spined stickleback Chr7 spanning around 10 Mb (range 125,492–10,364,802 bp) were mapped to LG12 in the nine-spined stickleback, together with 601 reference sequences that mapped on Chr12 in the three-spined stickleback genome (fig. 5a).
Table 3.
Interchromosomal Rearrangements between Three- and Nine-Spined Sticklebacks
| HP Cross |
HR Cross |
||||||
|---|---|---|---|---|---|---|---|
| Nine-spined LG | Three-spined chromosome | Contigs | SNPs | Interchromosome rearrangement contigs | Contigs | SNPs | Interchromosome rearrangement contigs |
| LG 1 | Chr I | 335 | 364 | 7 | 293 | 318 | 4 |
| LG 2 | Chr II | 143 | 148 | 0 | 248 | 264 | 4 |
| LG 3 | Chr III | 191 | 214 | 4 | 170 | 190 | 4 |
| LG 4 | Chr IV | 308 | 336 | 6 | 261 | 283 | 6 |
| LG 5 | Chr V | 171 | 185 | 4 | 118 | 125 | 2 |
| LG 6 | Chr VI | 286 | 301 | 4 | 237 | 247 | 2 |
| LG 7 | Chr VII | 259 | 282 | 8 | 217 | 233 | 5 |
| LG 8 | Chr VIII | 190 | 201 | 3 | 146 | 154 | 3 |
| LG 9 | Chr IX | 79 | 84 | 3 | 194 | 207 | 8 |
| LG 10 | Chr X | 193 | 209 | 10 | 165 | 178 | 7 |
| LG 11 | Chr XI | 251 | 271 | 10 | 204 | 217 | 8 |
| LG 12 | Chr VII | 820 | 971 | 120(VII) | 784 | 932 | 103(VII) |
| + Chr XII | +15a | +11a | |||||
| LG 13 | Chr XIII | 278 | 305 | 6 | 252 | 276 | 6 |
| LG 14 | Chr XIV | 256 | 275 | 7 | 214 | 227 | 7 |
| LG 15 | Chr XV | 216 | 224 | 5 | 181 | 189 | 2 |
| LG 16 | Chr XVI | 175 | 190 | 7 | 153 | 169 | 5 |
| LG 17 | Chr XVII | 218 | 239 | 2 | 186 | 202 | 2 |
| LG 18 | Chr XVIII | 161 | 172 | 3 | 145 | 154 | 3 |
| LG 19 | Chr XIX | 231 | 252 | 6 | 217 | 237 | 3 |
| LG 20 | Chr XX | 261 | 288 | 9 | 208 | 227 | 7 |
| LG 21 | Chr XXI | 207 | 221 | 5 | 198 | 212 | 4 |
| 5,229 | 5,732 | 244 | 4,791 | 5,241 | 206 | ||
a120(VII)+15: 120 reference contigs from Group VII and 15 from other groups.
Fig. 5.—
Comparisons of synteny between (a) HP and (b) HR cross linkage maps and three-spined stickleback genome. Note the few interchromosomal rearrangements. LG = nine-spined stickleback linkage group, Chr = three-spined stickleback chromosome.
The sex-average nine-spined stickleback linkage map of the HR cross was very similar to that of the HP cross in respect to the syntenic relationships with the three-spined stickleback genome (fig. 5b). For instance, 4,791 reference sequences with 5,241 SNPs mapped to the three-spined stickleback genome mostly in perfect synteny, and only 206 (4.3%) reference sequences showed interchromosomal rearrangements between nine- and three-spined sticklebacks. As in the case of the HR cross, half (103) of these reference sequences were located on three-spined stickleback Chr7 covering a 10 Mb (range 86,906–10,364,802 bp) region of this chromosome.
Of the reference sequences showing interchromosomal rearrangements, 174 were found in both HP and HR maps, and 96 (55%) were located on Chr7 in the three-spined stickleback genome while mapping on LG12 in both nine-spined stickleback linkage maps. In addition, we found that both HP and HR maps harbored sequences which remain unassembled in the three-spined stickleback genome. The HP map included 103 such scaffolds, and the HR map included 96 similar scaffolds (supplementary table S4, Supplementary Material online). Considering the high genomic synteny between nine- and three-spined sticklebacks, these findings might help to improve the three-spined stickleback genome assembly.
Discussion
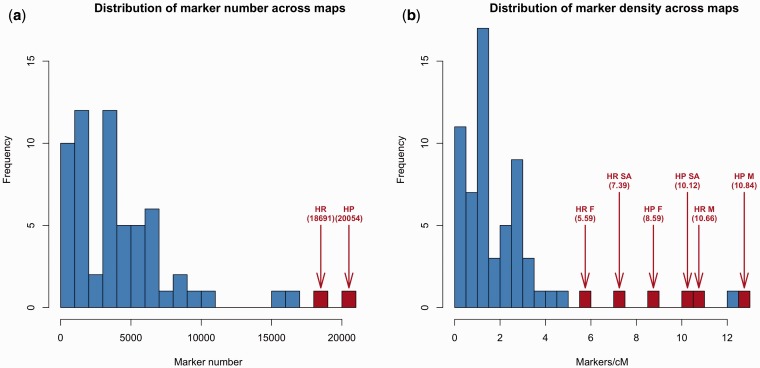
High-density linkage maps, such as those constructed here using Lep-MAP2, provide means to gain insights to genome-wide linkage and recombination patterns and thereby the structural, functional, and evolutionary characteristics of the genome itself (Wang et al. 2009; Kai et al. 2011; Hohenlohe et al. 2011). A quick survey of the current literature (supplementary table S5, Supplementary Material online) shows that although marker numbers in recent linkage mapping studies continue to increase (median = 3,677 markers), they are still modest compared with the maps we produced with the Lep-MAP2 (fig. 6a). Also in terms of marker density, our maps are among the highest published to date (fig. 6b). Apart from suggesting genome-wide recombination suppression in male nine-spined sticklebacks, these maps support the suggestion (Shikano et al. 2013) that a translocation of an autosomal chromosome arm to a sex chromosome has taken place after the nine-spined stickleback diverged from the three-spined stickleback. The results further suggest that the translocated part of this neo-sex chromosome is still recombining, whereas the ancestral part has nearly, but not entirely, ceased to do so. Furthermore, the comparative genomic analyses revealed a high degree of synteny between three- and nine-spined stickleback genomes, with some evidence of infrequent interchromosomal rearrangements. In the following, we will discuss these findings and their implications to our understanding of stickleback genome evolution, highlighting the value of these new ultra–high-density linkage maps as genomic resources of broad utility. We also discuss the utility and advantages of Lep-MAP2 in the construction of high-density linkage maps.
Fig. 6.—
Distributions of (a) marker numbers and (b) marker densities in a sample of recently (from 2000 onwards) published linkage maps (see supplementary table S5, Supplementary Material online, for data sources). Blue bars = earlier studies, red bars = this study. M = male, F = female, SA = sex average.
The two high-density second-generation SNP-based linkage maps, constructed using the RAD-seq approach, provide substantial improvements over the previously available microsatellite-based maps for nine-spined sticklebacks (Shapiro et al. 2009; Shikano et al. 2013). The basic structure of the SNP-based HR map was in agreement with the microsatellite-only HR linkage map: In both maps the microsatellites mapped to the same LGs, and their overall order was comparable between the two maps (Wilcoxon rank-sum test, 198 matched observations, W = 19,678, P = 0.34). However, the new maps increased the overall coverage of the LGs, which is not surprising in the view that the new maps contained 83–105 times more markers than the previous maps (Shikano et al. 2013), illustrating the effectiveness of the RAD-seq approach in SNP discovery, genotyping, and linkage mapping in a nonmodel organism (Etter and Johnson 2012). Hence, the high marker densities and more even distribution of markers across the different LGs in the SNP-based maps yielded a far more refined image of the genetic landscape of the nine-spined stickleback genome than that provided by first-generation microsatellite-based linkage maps. Both these features—increased marker density and coverage—helped us to not only verify the high degree of synteny in genomes of nine- and three-spined sticklebacks, but also to detect genomic rearrangements that have occurred during the evolutionary history of sticklebacks. Namely, earlier comparative genomic analyses between the three- and nine-spined sticklebacks have provided some preliminary insights into the genome evolution between these two model species (Shikano et al. 2010; Guo et al. 2013), and our results refine this picture. Based on their estimated divergence of around 13 Ma (Bell et al. 2009), a high degree of synteny was expected. This expectation was fulfilled: >5,000 SNPs were uniquely mapped onto the three-spined stickleback genome in each of the maps with a high degree of synteny. About 65% of these SNPs were located within coding regions, possibly because substitution rates are higher in noncoding than in coding regions of genomes (Guo et al. 2013). We also discovered that many scaffold sequences which remained unassembled to chromosomes in the three-spined stickleback genome assembly (Jones et al. 2012) showed strong linkage to specific LGs in both of our nine-spined stickleback linkage maps, suggesting that the utility of the high-density linkage maps in genome assembly is not limited to target species, but could also aid in assembling genomes of closely related species.
An earlier study identified a possible rearrangement between an autosome (LG7) and the sex chromosome (LG12) in the nine-spined stickleback (Shikano et al. 2013). Results of this study confirmed this finding with two independent crosses, and provided higher resolution information about the size and location of this rearrangement. Namely, it appears that a chromosomal segment corresponding to 36% (ca. 10 Mb) of Chr7 in three-spined sticklebacks has fused to LG12 in the nine-spined stickleback. Frequent chromosomal rearrangements in fish genomes are well known (Mank et al. 2006; Mank and Avise 2009), and especially sex chromosome fusions with autosomes appear to be common (Mank and Avise 2009; Kitano and Peichel 2012). Based on comparative analyses of the three-spined stickleback and medaka genomes, no interchromosomal rearrangement were found in these LGs. Therefore, it appears that the chromosome rearrangement between LG7 and LG12 occurred in the nine-spined stickleback after it diverged from the three-spined stickleback. In the linkage map from North American nine-spined sticklebacks, the segmental part of LG7 that linked to LG12 in our study did not show linkage to either LG12 or to the remaining part of LG7 (Shapiro et al. 2009). This discrepancy could be explained if this rearrangement is not present in the North American populations of nine-spined sticklebacks. Alternatively, the small number (=120) of individuals utilized in the North American study (Shapiro et al. 2009) might have rendered the power to detect the rearrangement low. Further cytogenetic analyses and/or more refined genetic maps based on larger sample sizes from North American and other nine-spined stickleback clades can clarify this issue. Irrespective of the situation in other populations, the occurrence of this rearrangement among the eastern European nine-spined stickleback populations is now an undisputed fact.
Apart from this translocation event, interchromosomal rearrangements were infrequent (<5%) in both maps. The marker order rearrangements in each LG varied substantially, from almost complete homology to cases where several inversion and translocation events between the two species were indicated to have happened. These findings align with the conjecture that although synteny among species is often conserved, the gene order within syntenic blocks is frequently changed (Woods et al. 2005; Kasahara et al. 2007). In addition, our findings support the observations that intrachromosomal rearrangements are fixed more frequently than translocations among nonhomologous chromosomes (Woods et al. 2005; Sémon and Wolfe 2007). Furthermore, studies utilizing physical genetic maps of closely related species or populations show that most detected inversions tend to be of small size (<1 kb; Kirkpatrick 2010; see also Feuk et al. 2005; Jones et al. 2012). In our study, the detected inversions were all relatively large, but this may be partly explained by our approach, which did not allow the detection of small inversions. Single SNP rearrangements are hard to interpret in our data as the linkage map is based on recombination frequencies, rather than on physical genetic positions. Therefore, a more refined estimate of inversions that are fixed among these two stickleback species cannot be performed until the whole genome sequence is available for the nine-spined stickleback. Also, we wish to emphasize that although our linkage maps were de novo assembled, all the comparative genomic analyses rely heavily on the three-spined stickleback reference genome sequence, which has been shown to contain assembly errors (Kasahara et al. 2007; Glazer et al. 2015). Although we corrected for known errors in our analyses, it is possible that some of the rearrangements we have discovered (or overlooked) might still owe to problems in the reference sequence. Although we have no a priori reason to believe that this would have biased our results—especially given that three-spined stickleback studies have found evidence for frequent inversions even at the intra-specific level (Deagle et al. 2012)—only access to the physical map of the nine-spined stickleback can help to eliminate doubts about the fixed inversions among these two species.
We discovered evidence for dramatic sexual dimorphism (SD) in recombination frequency at the genome-wide level: In both crosses, female maps were substantially (1.5–1.9 times) longer than male maps, and these differences were not confined to the sex LG but also occurred in autosomal LGs. Likewise, linkage blocks were on average shorter in females than in males, both in autosomal and sex chromosomal LGs. These observations suggest genome-wide recombination suppression in males, which are the heterogametic sex in this species. Similar observations are common in a wide variety of taxa (Burt et al. 1991; Lenormand and Dutheil 2005; Hedrick 2007; Brandvain and Coop 2012), including many fish species showing even more extreme SD in recombination frequency (Onchorynchys mykiss: 3.2, Sakamoto et al. 2000; Danio rerio: 2.7; Singer et al. 2002). Although the ultimate reasons for sex-specific recombination rates remains elusive (Lenormand and Dutheil 2005; Hedrick 2007; Brandvain and Coop 2012), the fact that it occurs implicates an important role for female meiosis in generating genetic variability in many species, including the nine-spined stickleback. The higher female recombination frequency also means that for some specific gene-mapping applications, the choice of study design with emphasis on either segregation of male or female-specific variability may be desirable (Singer et al. 2002).
Sex chromosomes in teleost fish show rapid evolution (Charlesworth 2004; Volff et al. 2007; Kondo et al 2009; Natri et al 2013; Shikano et al. 2013), to the point that closely related stickleback (Gasterosteidae) species have different sex chromosome systems (Ross et al. 2009). Our results confirmed that LG12 is the sex chromosome in the nine-spined stickleback, and that there are no sex-linked markers on any of the other LGs. Hence, sex determination in this species is caused by a well-defined sex chromosome, and not by polygenic factors spread across the genome. Of particular note is that the part of LG12 (the neo-sex chromosome) that is syntenic with the three-spined stickleback Chr7 showed a clearly different pattern of association with sex as compared with the rest of this LG. In particular, the P value pattern in this part of LG12 shows the presence of a transition between the ancestral LG12 and the syntenic portion. The gradual decay of association between sex and genotype across LG12 is clearly indicative of total recombination suppression in the ancestral part of LG12, whereas most of the syntenic part of the LG is still recombining. Hence, the data suggest that the sex chromosomes in the nine-spined stickleback are undergoing a rapid evolution making it an ideal model to understand both the evolution of sex determination systems, as well as patterns and processes occurring in early stages of sex chromosome evolution.
Although marker numbers and densities in our maps were very high in comparison with most earlier linkage maps—even in comparison with those created with RAD-seq approach (e.g., 8,257 SNPs, Gonen et al. 2014; 1,622 SNPs, 1.16 markers/cM, Kakioka et al. 2013; 755 SNPs, 0.5 markers/cM, Recknagel et al. 2013; fig. 6)—we did not observe a perfectly uniform distribution of markers across maps in either of the crosses or sexes. The presence of large intermarker gaps observed in the maps is especially interesting because the marker densities in our maps were higher than what could be fully resolved with the number of recombination events associated with an F2-cross, and many markers mapped to the same linkage positions despite the fact that these markers were individually resolved in the sequence assembly. Hence, one possible explanation for these large gaps is that they signal the presence of recombination hotspots across the P. pungitius genome. This is not implausible because recombination is known to be uneven across the genome and to be mostly affected by recombination hotspots (Auton and McVean 2007). When compared with an exponential distribution these intermarker gaps do not seem to arise by chance alone (HP female map: D = 0.072, p = 0.003; HP sex-averaged map: D = 0.0838, p = 0.0002; HR sex-averaged map: D = 0.0644, p = 0.002), but at present, given the lack of a physical map, it is difficult to determine whether recombination or another cause, such as a localized absence of RAD-seq derived loci, is the cause of these large intermarker gaps. Because Lep-MAP2 removes markers that show a significant deviation from the Mendelian proportions expected in an F2, and because we were able to select a high number of markers that fulfilled this condition for our maps, our data are not suited to study segregation distortion. We note though that for three LGs (LG2 and LG9 in the HP cross and LG4 in the HR cross) the markers on one end of the LG, while mapping together, mapped almost completely independently compared with the rest of the LG and were placed on maps through multiple mapping iterations. We cannot currently offer an explanation for the difficulties in mapping markers at these particular LGs.
Finally, being a modern implementation of a linkage mapping software applicable to sequencing-based data sets, Lep-MAP2 aims to directly tackle the software limitations which are holding back our ability to create ultradense maps from high-throughput data sets. The increased data output by genotyping-by-sequencing technologies have rendered many linkage mapping software obsolete, and there is an increasing demand for high-density linkage maps as tools for genome assembly validation (Fierst 2015). In addition to its ability to deal with large data sets, our simulations on smaller data show that Lep-MAP2 produces results that are both highly reliable and precise, and much more so than those of other available linkage mapping software. Because Lep-MAP2 is implemented fully in JAVA, it is truly machine independent, and thus compatible with any computing choices or needs of the user. It can utilize multiple cores of typical CPUs and is easily run on a computing cluster without any direct user intervention. These features together with its ability to handle high-throughput data—such as the one analyzed in this article—place Lep-MAP2 among the top state-of-the-art linkage mapping software currently available.
In conclusion, apart from giving novel insights into the genomic architecture of the nine-spined stickleback, the ultra–high-density linkage maps described in this study illustrate the power and utility of Lep-MAP2 software in handling large marker data sets, as well as in modeling sex-specific recombination. The homology analyses revealed a high degree of interchromosomal synteny between three- and nine-spined sticklebacks, but also that inversions have frequently occurred during their divergence. The results also confirmed (Shikano et al. 2013) the presence of an interchromosomal rearrangement that has led to the formation of a neo-sex chromosome in the nine-spined stickleback, as well as fairly strong genome-wide recombination suppression in male nine-spined sticklebacks. The constructed maps should also provide useful resources for further QTL mapping and comparative genomic analyses, as well as aid in the assembly of the nine-spined stickleback genome sequence. We note that our maps can also prove to be a valuable resource for improving the three-spined stickleback genome assembly: Given the high degree of synteny between the genomes of these two species, our finding of unmapped scaffolds in the three-spined stickleback assembly suggests their likely location in the three-spined stickleback genome. Hence, we envision that the results, insights, and resources created in this study will not only be useful for future genomic studies of nine-spined sticklebacks, but also for those of other closely related taxa.
Supplementary Material
Supplementary files S1 and S2 and tables S1–S5 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Chris Eberlein, Abigel Gonda, Gabor Herczeg, Sami Karja, Heini M. Natri, Sara Negazzi, Yukinori Shimada, Mirva Turtianen, and Jing Yang for help in fish rearing, and Kirsi Kähkönen for help in laboratory. Special thanks are due to Gabor Herczeg for access to the samples from the HR cross, two anonymous reviewers’ comments that improved the earlier version of this article, and Jacquelin DeFaveri for linguistic corrections. The parental fish from Kuusamo were collected with license from Metsähallitus, and the fish rearing was performed under license (STH211A) from Finnish National Animal Experiment Board. This work was supported by the Academy of Finland (grant numbers 129662, 134728, 218343 to J.M.).
Literature Cited
- Agresti JJ, et al. 2000. Breeding new strains of tilapia: development of an artificial center of origin and linkage map based on AFLP and microsatellite loci. Aquaculture 185:43–56. [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, McVean G. 2007. Recombination rate estimation in the presence of hotspots. Genome Res. 17:1219–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal V, Bashir A, Bafna V. 2007. Evidence for large inversion polymorphisms in the human genome from HapMap data. Genome Res. 17:219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell MA, Foster SA. 1994. The evolutionary biology of the threespine stickleback Oxford: Oxford University Press. [Google Scholar]
- Bell MA, Stewart JD, Park PJ. 2009. The world's oldest fossil threespine stickleback fish. Copeia 2009:256–265. [Google Scholar]
- Birky CW, Walsh JB. 1988. Effects of linkage on rates of molecular evolution. Proc Natl Acad Sci U S A. 85:6414–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandvain Y, Coop G. 2012. Scrambling eggs: meiotic drive and the evolution of female recombination rates. Genetics 190:709–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. 1998. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet. 63:861–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A, Bell G, Harvey PH. 1991. Sex differences in recombination. J Evol Biol. 4:259–277. [Google Scholar]
- Cartwright DA, Troggio M, Velasco R, Gutin A. 2007. Genetic mapping in the presence of genotyping errors. Genetics 176:2521–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. 2004. Sex determination: primitive Y chromosomes in fish. Curr Biol. 14:R745–R747. [DOI] [PubMed] [Google Scholar]
- Deagle BE, et al. 2012. Population genomics of parallel phenotypic evolution in stickleback across stream–lake ecological transitions. Proc R Soc B Biol Sci. 279:1277–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etter PD, Johnson E. 2012. RAD paired-end sequencing for local de novo assembly and SNP discovery in non-model organisms. Methods Mol Biol. 888:135–151. [DOI] [PubMed] [Google Scholar]
- Feldman MW, Otto SP, Christiansen FB. 1996. Population genetic perspectives on the evolution of recombination. Annu Rev Genet. 30:261–295. [DOI] [PubMed] [Google Scholar]
- Feuk L, et al. 2005. Discovery of human inversion polymorphisms by comparative analysis of human and chimpanzee DNA sequence assemblies. PLoS Genet. 1:e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierst JL. 2015. Using linkage maps to correct and scaffold de novo genome assemblies: methods, challenges, and computational tools. Front Genet. 6:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaxman SM, Wacholder AC, Feder JL, Nosil P. 2014. Theoretical models of the influence of genomic architecture on the dynamics of speciation. Mol Ecol. 23:4074–4088. [DOI] [PubMed] [Google Scholar]
- Flint J, Mackay TFC. 2009. Genetic architecture of quantitative traits in mice, flies, and humans. Genome Res. 19:723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessler DD, Xu S. 2000. Meiosis and the evolution of recombination at low mutation rates. Genetics 156:449–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazer AM, Killingbeck EE, Mitros T, Rokhsar DS, Miller CT. 2015. Genome assembly improvement and mapping convergently evolved skeletal traits in sticklebacks with Genotyping-by-Sequencing. G3 (Bethesda) 5:1463–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard ME, Hayes BJ. 2009. Mapping genes for complex traits in domestic animals and their use in breeding programmes. Nat Rev Genet. 10:381–391. [DOI] [PubMed] [Google Scholar]
- Guo B, Chain FJJ, Bornberg-Bauer E, Leder EH, Merilä J. 2013. Genomic divergence between nine- and three-spined sticklebacks. BMC Genomics 14:756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick PW. 2007. Sex: differences in mutation, recombination, selection, gene flow, and genetic drift. Evolution 61:2750–2771. [DOI] [PubMed] [Google Scholar]
- Hill WG, Robertson A. 1966. Effect of linkage on limits to artificial selection. Genet Res. 8:269–294. [PubMed] [Google Scholar]
- Hohenlohe PA, Bassham S, Currey M, Cresko WA. 2011. Extensive linkage disequilibrium and parallel adaptive divergence across threespine stickleback genomes. Philos Trans R Soc Lond B Biol Sci. 367:395–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FC, et al. 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai W, Kikuchi K, Tohari S, Chew AK, Tay A, et al. 2011. Integration of the genetic map and genome assembly of fugu facilitates insights into distinct features of genome evolution in teleosts and mammals. Genome Biol Evol. 3:424–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakioka R, Kokita T, Kumada H, Watanabe K, Okuda N. 2013. A RAD-based linkage map and comparative genomics in the gudgeons (genus Gnathopogon, Cyprinidae). BMC Genomics 14:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M, et al. 2007. The medaka draft genome and insights into vertebrate genome evolution. Nature 447:714–719. [DOI] [PubMed] [Google Scholar]
- Kawahara R, Miya M, Mabuchi K, Near TJ, Nishida M. 2009. Stickleback phylogenies resolved: evidence from mitochondrial genomes and 11 nuclear genes. Mol Phylogenet Evol. 50:401–404. [DOI] [PubMed] [Google Scholar]
- Kendall M. 1938. A new measure of rank correlation. Biometrika 30:81–89. [Google Scholar]
- Kingsley DM, Peichel CL. 2007. The molecular genetics of evolutionary change in sticklebacks In: Östlund-Nilsson S, Mayer I, Huntingford FA, editors. Biology of the three-spined stickleback. Boca Raton (FL): CRC Press; p. 41–81. [Google Scholar]
- Kirkpatrick M. 2010. How and why chromosome inversions evolve. PLoS Biol. 8:e1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano J, Peichel CL. 2012. Turnover of sex chromosomes and speciation in fishes. Environ Biol Fishes. 94:549–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano J, et al. 2009. A role for a neo-sex chromosome in stickleback speciation. Nature 461:1079–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M, Nanda I, Schmid M, Schartl M. 2009. Sex determination and sex chromosome evolution: insights from medaka. Sex Dev. 3:88–98. [DOI] [PubMed] [Google Scholar]
- Kreitman M, Hudson RR. 1991. Inferring the evolutionary histories of the Adh and Adh-dup loci in Drosophila melanogaster from patterns of polymorphism and divergence. Genetics 127:565–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L. 1997. The use of genetic map of biallelic markers in linkage studies. Nat Genet. 17:21–24. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulathinal RJ, Stevison LS, Noor MAF. 2009. The genomics of speciation in Drosophila: diversity, divergence, and introgression estimated using low-coverage genome sequencing. PLoS Genet. 5:e1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvist J, et al. 2015. Flight-induced changes in gene expression in the Glanville fritillary butterfly. Mol Ecol. 24:4886–4900. [DOI] [PubMed] [Google Scholar]
- Laine VN, Shikano T, Herczeg G, Vilkki J, Merilä J. 2013. Quantitative trait loci for growth and body size in the nine-spined stickleback Pungitius pungitius L. Mol Ecol. 22:5861–5876. [DOI] [PubMed] [Google Scholar]
- Larkin DM, et al. 2009. Breakpoint regions and homologous synteny blocks in chromosomes have different evolutionary histories. Genome Res. 19:770–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop G, Lalouel J, Julier C, Ott J. 1984. Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci U S A. 81:3443–3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand T. 2003. The evolution of sex dimorphism in recombination. Genetics 163:811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand T, Dutheil J. 2005. Recombination difference between sexes: a role for haploid selection. PLoS Biol. 3:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The Sequence Alignment/Map (SAM) format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, et al. 2014. Construction and analysis of high-density linkage map using high-throughput sequencing data. PLoS One 9:e98855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE, Avise JC. 2009. Evolutionary diversity and turn-over of sex determination in teleost fishes. Sex Dev. 3:60–67. [DOI] [PubMed] [Google Scholar]
- Mank JE, Promislow DEL, Avise JC. 2006. Evolution of alternative sex-determining mechanisms in teleost fishes. Biol J Linn Soc. 87:83–93. [Google Scholar]
- Margarido GR, Souza AP, Garcia AA. 2007. OneMap: software for genetic mapping in outcrossing species. Hereditas 144:78–79. [DOI] [PubMed] [Google Scholar]
- McKinnon JS, Rundle HD. 2002. Speciation in nature: the threespine stickleback model systems. Trends Ecol E 17:480–488. [Google Scholar]
- Merilä J. 2013. Nine-spined stickleback (Pungitius pungitius): an emerging model for evolutionary biology research. Ann N Y Acad Sci. 1289:18–35. [DOI] [PubMed] [Google Scholar]
- Michalak de Jimenez MK, et al. 2013. A radiation hybrid map of chromosome 1D reveals synteny conservation at a wheat speciation locus. Funct Integr Genomics. 13:19–32. [DOI] [PubMed] [Google Scholar]
- Miller MR, Dunham JP, Amores A, Cresko WA, Johnson EA. 2007. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res. 17:240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natri H, Shikano T, Merilä J. 2013. Progressive recombination suppression and differentiation in recently evolved neo-sex chromosomes. Mol Biol Evol. 30:1131–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocalewicz K, Fopp-Bayat D, Woznicki P, Jankun M. 2008. Heteromorphic sex chromosomes in the ninespine stickleback Pungitius pungitius. J Fish Biol. 73:456–462. [Google Scholar]
- Otto SP, Lenormand T. 2002. Resolving the paradox of sex and recombination. Nat Rev Genet. 3:252–261. [DOI] [PubMed] [Google Scholar]
- Paterson AH, et al. 1988. Resolution of quantitative traits into Mendelian factors by using a complete linkage map of restriction fragment length polymorphisms. Nature 335:721–726. [DOI] [PubMed] [Google Scholar]
- Peichel CL, et al. 2004. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr Biol. 14:1416–1424. [DOI] [PubMed] [Google Scholar]
- Posada D, Keith A, Crandall KA, Holmes HC. 2002. Recombination in evolutionary genomics. Annu Rev Genet. 36:75–97. [DOI] [PubMed] [Google Scholar]
- Rastas P, Paulin L, Hanski I, Lehtonen R, Auvinen P. 2013. Lep-MAP: fast and accurate linkage map construction for large SNP datasets. Bioinformatics 29:3128–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recknagel H, Elmer KR, Meyer A. 2013. A hybrid genetic linkage map of two ecologically and morphologically divergent Midas cichlid fishes (Amphilophus spp.) obtained by massively parallel DNA sequencing (ddRADSeq). G3 (Bethesda) 3:65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JA, Urton JR, Boland J, Shapiro MD, Peichel CL. 2009. Turnover of sex chromosomes in the stickleback fishes (Gasterosteidae). PLoS Genet. 5:e1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, et al. 2000. A microsatellite linkage map of rainbow trout (Oncorhynchus mykiss) characterized by large sex-specific differences in recombination rates. Genetics 155:1331–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sémon M, Wolfe KH. 2007. Rearrangement rate following the whole-genome duplication in teleosts. Mol Biol Evol. 24:860–867. [DOI] [PubMed] [Google Scholar]
- Shapiro MD, et al. 2009. The genetic architecture of skeletal convergence and sex determination in ninespine sticklebacks. Curr Biol. 19:1140–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikano T, Herczeg G, Merilä J. 2011. Molecular sexing and population genetic inference using a sex-linked microsatellite marker in the nine-spined stickleback (Pungitius pungitius). BMC Res Notes 4:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikano T, Laine VN, Herczeg G, Vilkki J, Merilä J. 2013. Genetic architecture of parallel pelvic reduction in ninespine sticklebacks. G3 (Bethesda) 3:1833–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikano T, Natri HM, Shimada Y, Merilä J. 2011. High degree of sex chromosome differentiation in stickleback fishes. BMC Genomics 12:474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikano T, Ramadevi J, Shimada Y, Merilä J. 2010. Utility of sequenced genomes for microsatellite marker development in non-model organisms: a case study of functionally important genes in nine-spined sticklebacks (Pungitius pungitius). BMC Genomics 11:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer A, et al. 2002. Sex-specific recombination rates in zebrafish (Danio rerio). Genetics 160:649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slate J, et al. 2009. Gene mapping in the wild with SNPs: guidelines and future directions. Genetica 136:97–107. [DOI] [PubMed] [Google Scholar]
- Taggart JB, Hynes RA, Prodöuhl PA, Ferguson A. 1992. A simplified protocol for routine total DNA isolation from salmonid fishes. J Fish Biol. 40:963–965. [Google Scholar]
- Tanksley SD, et al. 1992. High density molecular linkage maps of the tomato and potato genomes. Genetics 132:1141–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong C, Zhang B, Shi J. 2010. A hidden Markov model approach to multilocus linkage analysis in a full-sib family. Tree Genet Genomes. 6:651–662. [Google Scholar]
- Tripathi N, et al. 2009. Genetic linkage map of the guppy, Poecilia reticulata, and quantitative trait loci analysis of male size and colour variation. Proc Biol Sci. 276:2195–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen J. 2011. Multipoint maximum likelihood mapping in a full-sib family of an outbreeding species. Genet Res. 93:343–349. [DOI] [PubMed] [Google Scholar]
- van Os H, Plet S, Visser RGF, van Eck HJ. 2005a. Record: a novel method for ordering loci on a genetic linkage map. Theor Appl Genet. 112:30–40. [DOI] [PubMed] [Google Scholar]
- van Os H, Stam P, Visser RGF, van Eck HJ. 2005b. SMOOTH: a statistical method for successful removal of genotyping errors from high-density genetic linkage data. Theor Appl Genet. 112:187–194. [DOI] [PubMed] [Google Scholar]
- Volff JN, Nanda I, Schmid M, Schartl M. 2007. Governing sex determination in fish: regulatory putsches and ephemeral dictators. Sex Dev. 1:85–99. [DOI] [PubMed] [Google Scholar]
- Woods IG, et al. 2005. The zebrafish gene map defines ancestral vertebrate chromosomes. Genome Res. 15:1307–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton RJ. 2009. The Darwinian stickleback Gasterosteus aculeatus: a history of evolutionary studies. J Fish Biol. 75:1919–1942. [DOI] [PubMed] [Google Scholar]
- Yeaman S. 2013. Genomic rearrangements and the evolution of clusters of locally adaptive loci. Proc Natl Acad Sci U S A. 110:E1743–E1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zhang L, Meyer E, Matz MV. 2009. Construction of a high-resolution genetic linkage map and comparative genome analysis for the reef-building coral Acropora millepora. Genome Biol. 10:R126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, et al. 2014. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346:1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.