Abstract
Increasing evidence has demonstrated that the tumor suppressor gene Hath1 is implicated in the development and progression of tumors and is verified to be downregulated in several types of tumor. However, the roles and precise molecular mechanisms of Hath1 in cutaneous squamous cell carcinoma (SCC) remain to be elucidated. In the present study, two approaches were used to investigate the tumor-suppressing effect of Hath1 in cutaneous SCC. Firstly, the effect of inhibiting Hath1 expression with short hairpin RNA (shRNA) on tumor growth and apoptosis was investigated. KUMA5 cells were stably transfected with a plasmid expressing Hath1 shRNA (pGenesil-1-Hath1). Secondly, the anti-tumor effect of Hath1 was investigated in KUMA5 cells following transfection with pcDNA3.1-Hath1. The mRNA and protein expression of Hath1 was detected by reverse transcription quantitative polymerase chain reaction and western blot analysis, respectively. Cell proliferation in vitro was assessed using an MTT assay. Flow cytometry was used to detect cell apoptosis. The results demonstrated that compared with the control groups, the expression of Hath1 was significantly reduced in the KUMA5/pGenesil-1-Hath1 cells and markedly increased in the KUMA5/pcDNA3.1-Hath1 cells. Cell proliferation was markedly increased in the KUMA5/pGen-esil-1-Hath1 cells in a time-dependent manner; however, it was markedly inhibited in the KUMA5/pcDNA3.1-Hath1 cells. Flow cytometry revealed that apoptosis decreased in KUMA5/pGenesil-1-Hath1 cells and increased in KUMA5/pcDNA3.1-Hath1 cells. Downregulation of Hath1 expression promoted the proliferation and reduced the apoptosis of KUMA5 cells. By contrast, overexpression of Hath1 inhibited proliferation and induced the apoptosis of KUMA5 cells. These findings provide possible new strategies and therapeutic targets for the treatment and diagnosis of cutaneous SCC.
Keywords: Hath1, shRNA, overexpression, KUMA5 cell, proliferation, apoptosis
Introduction
Non-melanoma skin carcinoma is one of the most frequent types of tumor worldwide and includes two histological types: Basal cell carcinoma and squamous cell carcinoma (SCC) (1–3). Cutaneous SCC is the second most common cutaneous carcinoma and accounts for 20% of all types of cutaneous tumor (4–6). Previous studies have made significant progress in understanding the mechanisms underlying the tumorigenesis and development of cutaneous SCC (7–12). However, the incidence rates of patients with cutaneous SCC have been increasing rapidly each year, due to the lack of molecular targeted therapies and reliable molecular markers in early diagnosis. Therefore, novel molecular biomarkers and targeted therapeutic approaches for the diagnosis and treatment of patients with cutaneous SCC are urgently required.
Hath1, also termed Math1, Atoh1 and Cath1 in mouse, drosophila and chicken, respectively, is a basic helix-loop-helix (bHLH) transcription factor (13). Hath1 was initially identified as a tumor suppressor gene in colon cancer (14). However, increasing evidence has revealed that Hath1 is not only a tumor suppressor in colon cancer (14–24), but is also downregulated and involved in the tumorigenesis of various other types of human cancer, including medulloblastoma (25,26), gastric carcinoma (27,28), intestinal neuroendocrine tumors (29), lung cancer (30,31) and Merkel cell carcinoma (32). Inherited or acquired defective mutants of Atoh were initially identified in patients with Merkel cell carcinoma by Leonard in 2002 (32). Deletion of Hath1 using retrovirus vectors led to the disruption of Sonic hedgehog signaling in the developing cerebellum and finally resulted in medulloblastoma formation (26). Bossuyt et al confirmed that the incidence of colonic polyps in mice lacking the Hath1 gene was 100% compared with that in wild type mice (16). Zhu et al found that Hath1 expression is down-regulated in non-mucinous adenocarcinomas (19–21) and that Hath1 inhibits the proliferation of colon cancer cells possibly through upregulating the expression of Muc2 and p27 and downregulating the expression of cyclin D1 (24). Collectively, these studies highlight the important role of Hath1 as a tumor suppressor gene in these types of tumor and further investigation of the functions of Hath1 may provide potential molecular targets for the treatment of these tumors.
However, to the best of our knowledge, no studies on the biological role of Hath1 in cutaneous SCC have been reported to date. Therefore, the current study predominantly focused on the biological function of the Hath1 gene. In order to examine the effects of the Hath1 gene on the growth, proliferation and apoptosis of cutaneous SCC, Hath1 was alternately silenced with short hairpin RNA (shRNA), or overexpressed using a recombinant eukaryotic expression vector carrying the Hath1 gene. KUMA5 human cutaneous squamous carcinoma cells were infected with pcDNA3.1-Hath1 or pGenesil-1-Hath1. Subsequently, cell proliferation and apoptosis were examined by MTT assay and flow cytometry to provide further insight into the potential use of Hath1 for the targeted therapy of cutaneous SCC. Collectively, these data suggest that Hath1 may be a novel molecular target for the treatment of cutaneous SCC.
Materials and methods
Construction of the pcDNA3.1-Hath1 vector
The present study was approved by an ethical review committee of the Shanghai First People's Hospital (Shanghai, China), and written informed consent was obtained from all participants at the Shanghai First People's Hospital Affiliated to Shanghai Jiao Tong University School of Medicine (Shanghai, China). Total RNA was extracted from normal human cutaneous tissue using 1 ml of TRIzol reagent (cat. no. 15596-018; Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions. cDNA was then synthesized using an ABI TaqMan one-step RT-PCR Master Mix Reagents kit (cat. no. 4309109; Applied Biosystems, Foster City, CA, USA). Based on the GenBank sequence (NC_000004.12), upstream and downstream primers were synthesized for Hath1 gene amplification. The restriction enzyme sites for BamHI and HindIII were added to the 5′ end of each primer. Subsequently, the full-length Hath1 gene was amplified by quantitative polymerase chain reaction (qPCR) from the cDNA template with the primers containing the BamHI and HindIII restriction sites. The PCR product was then ligated into the pMD18-T vector (cat. no. 6011; Takara Bio., Inc., Dalian, China), transformed and screened for positive clones. Following sequence verification, the correct sequence was cloned into the pcDNA3.1 expression vector (cat. no. V790-20; Invitrogen Life Technologies) to construct the pcDNA3.1-Hath1 recombinant expression vector. The Hath1 primer was synthesized by Sangon Biotech (Shanghai) Co., Ltd. (Shanghai, China).
shRNA design and expression plasmid vector construction
The following sequence was used to design the shRNA: Hath1, AAACGACAAGAAGCTGTCCAAATAGTGAAGCCACAGATGTATTTGGACAGCTTCTTGTCGTTG. The DNA sequence targeting the Hath1 gene was cloned into the pGenesil-1 (cat. no. VRG0358; Wuhan Genesil Biotechnology Co., Ltd., Wuhan, China), a vector expressing shRNA and enhanced green fluorescent protein in mammalian cells. Recombinant plasmid pGenesil-1-Hath1 and pGenesil-1 scramble control were constructed and verified by Wuhan Genesil Biotechnology Co., Ltd.
Cell culture and transfection
The KUMA5 human cutaneous squamous carcinoma cell line was purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and cultured in Dulbecco's modified Eagle's medium (DMEM)/F12 medium (Gibco-BRL, Carlsbad, CA, USA: nutrient mixture F12; cat. no. 11039-021) supplemented with 10% fetal bovine serum (Gibco-BRL; cat. no. 12483-020) in a humidified incubator containing 5% CO2 at 37° C. When cultured to exponential phase, KUMA5 cells were replated at 5×104 cells/well in six-well plates. Transient transfection was performed using Lipofectamine 2000 transfection reagent (cat. no. 11668-019; Invitrogen Life Technologies) according to the manufacturer's instructions. Screening of stably transfected cells was performed as follows: The cells were transfected with either the pcDNA3.1-Hath1 or pGenesil-1-Hath1 eukaryotic expression vectors and then cultured in DMEM/F12 containing 800 µg/ml G418 (cat. no. sc-29065; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) for 2 weeks. Positive clones were screened and verified by reverse transcription (RT)-qPCR and western blot analysis.
RNA isolation, RT-qPCR and western blot analysis
mRNA transcript expression was quantified via RT-qPCR and normalized to the expression of β-actin. Total RNA from the KUMA5 cells that had been transfected with either the pcDNA3.1-Hath1, pGenesil-1-Hath1, pGenesil-1 scramble control or blank control was isolated using TRIzol reagent according to the manufacturer's instructions and the quality of the RNA was confirmed (i.e., A260/A280>1.8). qPCR was performed using an ABI 7900HT Fast Real-Time PCR System (cat. no. 4329003; Applied Biosystems). Detailed sequences of the primers are as follows: β-actin, forward 5′-ATGATATCGCCGCGCTCGTCGTC-3′ and reverse 5′-CGCTCGGCCGTGGTGGTGAA-3′; Hath1, forward 5′-CGCCGCCCAGTATTTGCTA-3′ and reverse 5′-ATTCACCTGTTTGCTGGAAGG-3′. Total protein from cell lines was extracted using the ReadyPrep Protein Extraction kit (cat. no. 163-2087; Bio-Rad Laboratories, Hercules, CA, USA). The protein concentration was determined using a BCA Protein Assay kit (cat. no. 23227; Pierce Biotechnology, Inc., Rockford, IL, USA). Protein lysates were loaded onto a 10% sodium dodecyl sulfate polyacryl-amide gel. Separated protein bands were electrotransferred onto polyvinylidene fluoride (PVDF) membranes. The PVDF membranes were then blocked in Tris-buffered saline (TBS; 10 mM Tris-HCl, pH 7.5 and 150 mM NaCl) containing 5% non-fat dry milk at room temperature for 1 h and incubated at 4° C overnight with anti-Hath1 monoclonal antibody (1:400; cat. no. sc-136173; Santa Cruz Biotechnology, Inc.) and anti-β-actin monoclonal antibody (1:1,000; cat. no. sc-47778; Santa Cruz Biotechnology, Inc.). Following primary antibody incubation, the membranes were washed in TBS with Tween 20 (10 mM Tris-HCl, pH 7.5, 150 mM NaCl and 0.05% Tween-20) and a rabbit anti-mouse IgG-horseradish peroxidase antibody (1:2,000; cat. no. sc-358920; Santa Cruz Biotechnology, Inc.) was added. The membrane was then incubated at 37° C for 2 h. ECL Plus Western Blotting Substrate (cat. no. 32132; Pierce Biotechnology, Inc.) was used to visualize the immunoreactive bands. Relative protein level was normalized to β-actin concentration. Three separate experiments were performed in duplicate for each treatment.
Examination of cell viability
Cell viability was determined using an MTT assay (cat. no. M2128; Sigma-Aldrich, St. Louis, MO, USA). Cells were cultured in 24-well plates at a concentration of 5×104 and allowed to adhere. Following treatment for different time intervals, 100 µl MTT (0.5 mg/ml) was added into the cells and the mixture was incubated for 4 h at 37° C. Subsequently, the supernatant was removed and dimethyl sulfoxide (cat. no. D2650; Sigma-Aldrich) was used to dissolve the resultant formazan crystals. The absorbance was read at 570 nm (EL309 Automated Microplate Reader; Bio-Tek Instruments, lnc., Winooski, VT, USA). Six wells were measured for each group and the experiment was repeated three times.
Flow cytometric analysis of cell apoptosis
To detect cell apoptosis, the proliferating phase KUMA5 cells (5×104 clones) were trypsinized, washed with cold phosphate-buffered saline and resuspended in binding buffer according to the manufacturer's instructions of the Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) Apoptosis Detection kit (cat. no. KGA106; Nanjing KeyGen Biotech Co., Ltd., Nanjing, China). FITC-Annexin V and PI were added to the fixed cells for 20 min in darkness at room temperature. Subsequently, Annexin V binding buffer was added to the mixture prior to measuring fluorescence using a FACSort flow cytometer (BD Biosciences, Mountain View, CA, USA). Cell apoptosis was analyzed using Cell Quest 3.0 software (Becton Dickinson, Franklin Lakes, NJ, USA).
Statistical analysis
SPSS 13.0 (SPSS, Inc., Chicago, IL, USA) was used for data analysis. All experiments were repeated three times and data are presented as the mean ± standard deviation. Differences between groups were analyzed using one-way analysis of variance. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of Hath1 in KUMA5 cells
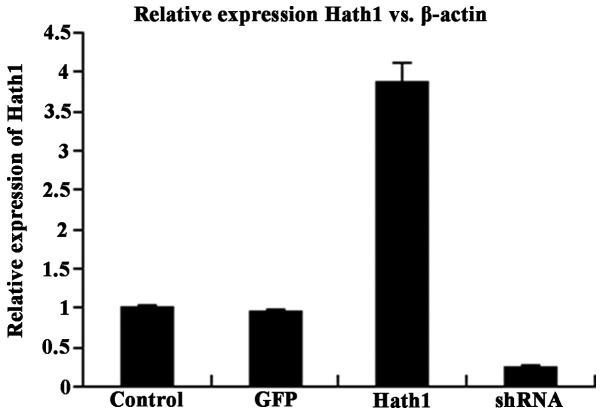
Total cellular RNA and protein were extracted from KUMA5 cells that had been transfected with the pcDNA3.1-Hath1, pGenesil-1-Hath1, pGenesil-1 scramble control or a blank control. RT-qPCR and western blot analysis were performed to determine the mRNA and protein expression of Hath1, respectively. The results demonstrated that the expression of Hath1 in KUMA5 cells transfected with pcDNA3.1-Hath1 was nearly four times higher than in the control groups (Fig. 1). The efficiency of gene silencing through pGenesil-1-Hath1 transfection was 78% (Fig. 1).
Figure 1.
Relative mRNA expression of Hath1 in KUMA5 cells. Control, blank control group (Dulbecco's modified Eagle's medium/F12); GFP, pGen-esil-1 scramble control group; Hath1, pcDNA3.1-Hath1-transfected group; shRNA, pGenesil-1-Hath1-transfected group. GFP, green fluorescent protein; shRNA, short hairpin RNA.
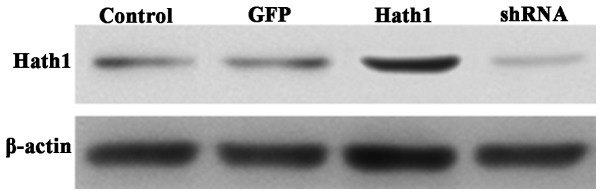
The protein expression of Hath1 was high in the pcDNA3.1-Hath1-transfected group, low in the control group, and barely detectable in the pGenesil-1-Hath1-transfected group (Fig. 2). Therefore, it was concluded that KUMA5 strains with stable overexpression or depletion of the Hath1 gene had been successfully created.
Figure 2.
Relative protein expression of Hath1 in KUMA5 cells. Control, blank control group (Dulbecco's modified Eagle's medium/F12); GFP, pGen-esil-1 scramble control group; Hath1, pcDNA3.1-Hath1-transfected group; shRNA, pGenesil-1-Hath1-transfected group. GFP, green fluorescent protein; shRNA, short hairpin RNA.
Effect of Hath1 on KUMA5 cell proliferation
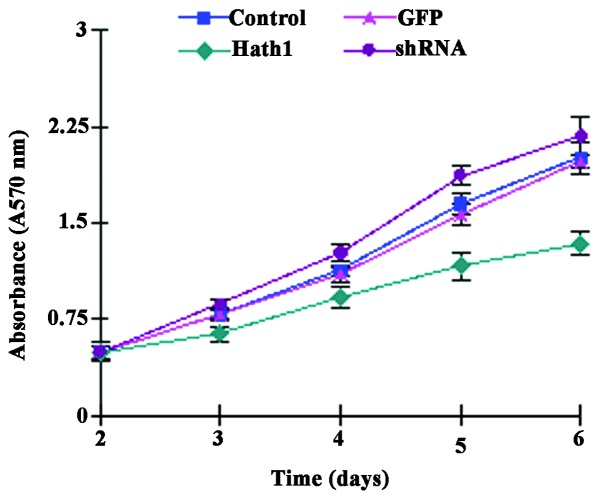
It was hypothesized that cell proliferation may be affected by the level of Hath1 in cutaneous SCC cells. To examine the effect of Hath1 on cell proliferation, an equal number of KUMA5 cells transfected with either the pcDNA3.1-Hath1, pGenesil-1-Hath1, pGenesil-1 scramble control or a blank control were seeded and analyzed using an MTT assay. The present study found that Hath1 overexpression attenuated the proliferation of KUMA5 cells and Hath1 gene RNA interference enhanced the proliferation of KUMA5 cells (Fig. 3). Taken together, Hath1 clearly inhibited the proliferation and growth of SCC cells, indicating that Hath1 may act as a potential tumor suppressor in cutaneous SCC and that Hath1 may represent a novel target for the treatment of SCC.
Figure 3.
Effect of Hath1 on KUMA5 cell growth. Control, blank control group (Dulbecco's modified Eagle's medium/F12); GFP, pGenesil-1 scramble control group; Hath1, pcDNA3.1-Hath1-transfected group; shRNA, pGen-esil-1-Hath1-transfected group. GFP, green fluorescent protein; shRNA, short hairpin RNA.
Effect of Hath1 on the apoptosis of KUMA5 cells
To further confirm the inhibitory effect of Hath1 on KUMA5 cells, the rate of apoptosis was examined by Annexin V and PI staining of KUMA5 cells transfected with the pcDNA3.1-Hath1, pGenesil-1-Hath1, pGenesil-1 scramble control or a blank control. FITC-Annexin V and PI were added to the fixed cells for 20 min in darkness at room temperature. Subsequently, Annexin V binding buffer was added to the mixture prior to measuring the fluorescence using a FACSort flow cytometer (BD Biosciences). Cell apoptosis was analyzed using Cell Quest 3.0 software (Becton Dickinson). The results demonstrated that a significantly higher rate of apoptosis was present in the KUMA5 cells overexpressing Hath1 compared with that of the control groups (Fig. 4). In addition, a significantly lower rate of apoptosis was observed in the KUMA5 cells transfected with pGenesil-1-Hath1 compared with that of the control groups (Fig. 4). Therefore, these results suggest that overexpression of Hath1 may promote apoptosis in KUMA5 cells.
Figure 4.
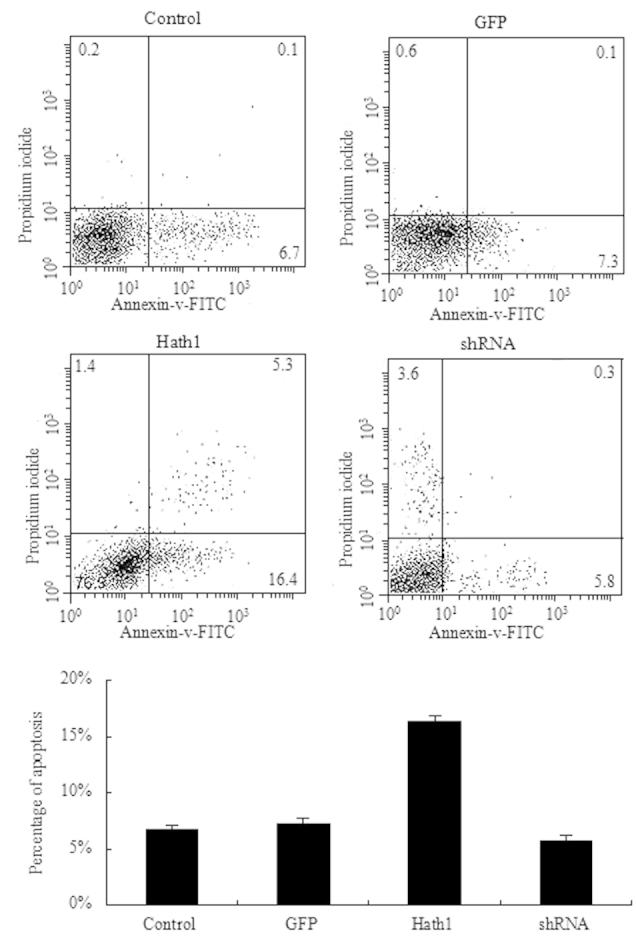
Effect of Hath1 on the apoptosis of KUMA5 cells. Control, blank control group (Dulbecco's modified Eagle's medium/F12); GFP, pGenesil-1 scramble control group; Hath1, pcDNA3.1-Hath1-transfected group; shRNA, pGenesil-1-Hath1-transfected group. GFP, green fluorescent protein; FITC, fluorescein isothiocyanate; shRNA, short hairpin RNA.
Discussion
Hath1, located on the chromosomal region 4q22, is the mammalian homologue of Drosophila atonal, a bHLH transcription factor. It is known to be important in the differentiation of various cells, including cerebellar granule cells (33), inner hair cells in the auditory system (34), epidermal Merkel cells (32), intestinal secretory cells (35,36) and hindbrain neurons (37). Previous studies have demonstrated that Hath1 acts as a tumor suppressor gene in various types of cancer, including colon cancer (14-24), medulloblastoma (25,26), gastric carcinoma (27,28), intestinal neuroendocrine tumors (29), lung cancer (30,31) and Merkel cell carcinoma (32).
However, to date, no studies on the biological role of Hath1 in cutaneous SCC have been reported. The present study demonstrated for the first time, to the best of our knowledge, the anti-proliferative and pro-apoptotic activity of Hath1 in cutaneous SCC, using a variety of techniques, including overexpression of Hath1 and shRNA-mediated Hath1 silencing. As shown in Figs. 1 and 2, Hath1 was successfully silenced or overexpressed. The expression level of Hath1 in pGenesil-1-Hath1-transfected cells was significantly decreased to 22% of the normal levels (P<0.05). Cells transfected with pcDNA3.1-Hath1 had significantly higher levels of Hath1 expression, which was ~3.9 times higher than the control groups (P<0.05). MTT assays demonstrated that the proliferation of KUMA5/pGenesil-1-Hath1-transfected cells was promoted in a time-dependent manner, however, proliferation was inhibited in KUMA5/pcDNA3.1-Hath1-infected cells. Furthermore, the apoptotic rate of the pGenesil-1-Hath1-trans-fected group significantly decreased. By contrast, the apoptotic rate of the KUMA5/pcDNA3.1-Hath1-infected group significantly increased. These experimental results demonstrated that overexpression of Hath1 could repress the proliferation of KUMA5 cells and promote apoptosis. However, shRNA-mediated Hath1 silencing enhanced the growth of KUMA5 cells and inhibited the apoptosis of KUMA5 cells.
The expression level of the Hath1 gene was frequently found to significantly affect cell proliferation and apoptosis (14,16,20–22,24). Zhu et al (20,21,24) and Leow et al (14) identified that overexpression of Hath1 inhibits proliferation of HT29 colon cancer cells through downregulation of cyclin D1 and upregulation of p27 and MUC2, a goblet cell differentiation marker. In a previous study, knockout of Hath1 was demonstrated to promote tumorigenesis in colorectal mouse models with intestine-specific deletion of Hath1 (18). The anti-tumor effects of Hath1, including the inhibition of colon cancer cell proliferation and enhancement of apoptosis in vitro and in vivo, may be achieved by activation of the Jun N-terminal kinase signaling pathway (16). According to Zhao et al (22), Hath1 can effectively inhibit the proliferation of four colon cancer cell lines, namely HT-29, LS17T, SW480 and SW620 with weak or absent expression of Hath1. In addition, the inhibition of cell proliferation by Hath1 was inversely proportional to the expression of Hath1 in these four colon cancer cell lines. However, instead of inducing apoptosis, an increase in tumor cell sensitivity to the chemotherapeutic drug 5-fluorouracil was observed in one of the four colon cancer cell lines. By contrast, shRNA-mediated Hath1 gene silencing using retroviral vectors could significantly accelerate the growth rate of the normal rat intestinal epithelial cell line IEC6 with suppressed growth by high expression of the Hath1 gene, which also confirmed the tumor suppressor activity of Hath1 (22). A previous study (38) demonstrated that retroviral-mediated Hath1 expression can significantly reduce human esophageal keratinocyte cell proliferation and induce marked expression of Barrett's esophagus markers Mucin-2 and Keratin-20. The results of the present study, which build on earlier studies (14,16,20–22,24), confirm the anti-proliferative and pro-apoptotic activities of Hath1.
In conclusion, the present results verify the anti-tumor effects of Hath1 on the proliferation and apoptosis of cutaneous SCC. Hath1 may be a potential therapeutic target for the inhibition of growth and progression of cutaneous SCC. These studies provide insights into the tumorigenesis and development of cutaneous SCC and provide new strategies and targets for the treatment of cutaneous SCC. However, further studies are required to clarify the associated molecular mechanisms and signal transduction of Hath1.
References
- 1.Veness MJ, Morgan GJ, Palme CE, Gebski V. Surgery and adjuvant radiotherapy in patients with cutaneous head and neck squamous cell carcinoma metastatic to lymph nodes: Combined treatment should be considered best practice. Laryngoscope. 2005;115:870–875. doi: 10.1097/01.MLG.0000158349.64337.ED. [DOI] [PubMed] [Google Scholar]
- 2.Preston DS, Stern RS. Nonmelanoma cancers of the skin. N Engl J Med. 1992;327:1649–1662. doi: 10.1056/NEJM199212033272307. [DOI] [PubMed] [Google Scholar]
- 3.Euvrard S, Kanitakis J, Claudy A. Skin cancers after organ transplantation. N Engl J Med. 2003;348:1681–1691. doi: 10.1056/NEJMra022137. [DOI] [PubMed] [Google Scholar]
- 4.McGuire JF, Ge NN, Dyson S. Nonmelanoma skin cancer of the head and neck I: Histopathology and clinical behavior. Am J Otolaryngol. 2009;30:121–133. doi: 10.1016/j.amjoto.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Martinez JC, Cook JL. High-risk cutaneous squamous cell carcinoma without palpable lymphadenopathy: Is there a therapeutic role for elective neck dissection? Dermatol Surg. 2007;33:410–420. doi: 10.1111/j.1524-4725.2007.33087.x. [DOI] [PubMed] [Google Scholar]
- 6.Weinberg AS, Ogle CA, Shim EK. Metastatic cutaneous squamous cell carcinoma: An update. Dermatol Surg. 2007;33:885–899. doi: 10.1111/j.1524-4725.2007.33190.x. [DOI] [PubMed] [Google Scholar]
- 7.Bertero T, Bourget-Ponzio I, Puissant A, et al. Tumor suppressor function of miR-483-3p on squamous cell carcinomas due to its pro-apoptotic properties. Cell Cycle. 2013;12:2183–2193. doi: 10.4161/cc.25330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu D, Feng X, Wu X, et al. Tumor suppressor in lung cancer 1 (TSLC1), a novel tumor suppressor gene, is implicated in the regulation of proliferation, invasion, cell cycle, apoptosis and tumorigenicity in cutaneous squamous cell carcinoma. Tumour Biol. 2013;34:3773–3783. doi: 10.1007/s13277-013-0961-2. [DOI] [PubMed] [Google Scholar]
- 9.Yang C, Wu D, Jia J, et al. DLC1 as a regulator of proliferation, invasion, cell cycle and apoptosis in cutaneous squamous cell carcinoma. Tumour Biol. 2013;34:2633–2643. doi: 10.1007/s13277-013-0813-0. [DOI] [PubMed] [Google Scholar]
- 10.Scortegagna M, Martin RJ, Kladney RD, Neumann RG, Arbeit JM. Hypoxia-inducible factor-1alpha suppresses squamous carcinogenic progression and epithelial-mesenchymal transition. Cancer Res. 2009;69:2638–2646. doi: 10.1158/0008-5472.CAN-08-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma A, Ramanjaneyulu A, Ray R, Rajeswari MR. Involvement of high mobility group B proteins in cisplatin-induced cytotoxicity in squamous cell carcinoma of skin. DNA Cell Biol. 2009;28:311–318. doi: 10.1089/dna.2009.0851. [DOI] [PubMed] [Google Scholar]
- 12.Bito T, Sumita N, Ashida M, et al. Inhibition of epidermal growth factor receptor and PI3K/Akt signaling suppresses cell proliferation and survival through regulation of Stat3 activation in human cutaneous squamous cell carcinoma. J Skin Cancer. 2011;2011:874571. doi: 10.1155/2011/874571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ben-Arie N, McCall AE, Berkman S, Eichele G, Bellen HJ, Zoghbi HY. Evolutionary conservation of sequence and expression of the bHLH protein Atonal suggests a conserved role in neurogenesis. Hum Mol Genet. 1996;5:1207–1216. doi: 10.1093/hmg/5.9.1207. [DOI] [PubMed] [Google Scholar]
- 14.Leow CC, Romero MS, Ross S, Polakis P, Gao WQ. Hath1, down-regulated in colon adenocarcinomas, inhibits proliferation and tumorigenesis of colon cancer cells. Cancer Res. 2004;64:6050–6057. doi: 10.1158/0008-5472.CAN-04-0290. [DOI] [PubMed] [Google Scholar]
- 15.Leow CC, Polakis P, Gao WQ. A role for Hath1, a bHLH transcription factor, in colon adenocarcinoma. Ann NY Acad Sci. 2005;1059:174–183. doi: 10.1196/annals.1339.048. [DOI] [PubMed] [Google Scholar]
- 16.Bossuyt W, Kazanjian A, De Geest N, et al. Atonal homolog 1 is a tumor suppressor gene. PLoS Biol. 2009;7:e39. doi: 10.1371/journal.pbio.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sikandar SS, Pate KT, Anderson S, et al. NOTCH signaling is required for formation and self-renewal of tumor-initiating cells and for repression of secretory cell differentiation in colon cancer. Cancer Res. 2010;70:1469–1478. doi: 10.1158/0008-5472.CAN-09-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kazanjian A, Noah T, Brown D, Burkart J, Shroyer NF. Atonal homolog 1 is required for growth and differentiation effects of notch/gamma-secretase inhibitors on normal and cancerous intestinal epithelial cells. Gastroenterology. 2010;139:918–928. doi: 10.1053/j.gastro.2010.05.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu DH, Gong J, Ren K, et al. Hath1 expression is down-regulated in colon non-mucinous adenocarcinomas compared with the expression in normal colon mucosa. Chong Qing Yi Ke Da Xue Xue Bao. 2010;35:666–669. In Chinese. [Google Scholar]
- 20.Zhu DH, Gong JP, Ren K, Sun JM, Wei SD. Hath1 gene transfer inhibits the proliferation of colonic cancer cells in vitro. Nan Fang Yi Ke Da Xue Xue Bao. 2010;30:1005–1008. In Chinese. [PubMed] [Google Scholar]
- 21.Zhu DH, Wen YW, Yang H, Liang G. Anti-cancer effect of Hath1 in pathogenesis of non-mucinous colon adenocarcinoma. Ji Lin Da Xue Xue Bao (Yi Xue Ban) 2011;37:700–706. In Chinese. [Google Scholar]
- 22.Zhao LL, Liu Z, Zhang YJ. Hath1 gene inhibits colorectal cancer cell growth. Zhong Guo Yi Yao Sheng Wu Ji Shu. 2011;6:204–208. In Chinese. [Google Scholar]
- 23.Peignon G, Durand A, Cacheux W, et al. Complex interplay between β-catenin signalling and Notch effectors in intestinal tumorigenesis. Gut. 2011;60:166–176. doi: 10.1136/gut.2009.204719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu DH, Niu BL, Du HM, Ren K, Sun JM, Gong JP. Hath1 inhibits proliferation of colon cancer cells probably through up-regulating expression of Muc2 and p27 and down-regulating expression of cyclin D1. Asian Pac J Cancer Prev. 2012;13:6349–6355. doi: 10.7314/APJCP.2012.13.12.6349. [DOI] [PubMed] [Google Scholar]
- 25.Zhao H, Ayrault O, Zindy F, Kim JH, Roussel MF. Post-transcriptional down-regulation of Atoh1/Math1 by bone morphogenic proteins suppresses medulloblastoma development. Genes Dev. 2008;22:722–727. doi: 10.1101/gad.1636408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flora A, Klisch TJ, Schuster G, Zoghbi HY. Deletion of Atoh1 disrupts Sonic Hedgehog signaling in the developing cerebellum and prevents medulloblastoma. Science. 2009;326:1424–1427. doi: 10.1126/science.1181453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sekine A, Akiyama Y, Yanagihara K, Yuasa Y. Hath1 up-regulates gastric mucin gene expression in gastric cells. Biochem Biophys Res Commun. 2006;344:1166–1171. doi: 10.1016/j.bbrc.2006.03.238. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, Yu H, Yang Y, et al. SOX2 in gastric carcinoma, but not Hath1, is related to patients' clinicopathological features and prognosis. J Gastrointest Surg. 2010;14:1220–1226. doi: 10.1007/s11605-010-1246-3. [DOI] [PubMed] [Google Scholar]
- 29.Heiskala K, Arola J, Heiskala M, Andersson LC. Expression of Reg IV and Hath1 in neuroendocrine neoplasms. Histol Histopathol. 2010;25:63–72. doi: 10.14670/HH-25.63. [DOI] [PubMed] [Google Scholar]
- 30.Chen JB, Liu JH, Liu YF, et al. Expression and clinical significance of Atoh1 in lung cancer. Lin Chuang Fei Ke Za Zhi. 2011;11:1744–1746. In Chinese. [Google Scholar]
- 31.Xu HT, Xie XM, Li QC, et al. Atonal homolog 1 expression in lung cancer correlates with inhibitors of the Wnt pathway as well as the differentiation and primary tumor stage. APMIS. 2013;121:111–119. doi: 10.1111/j.1600-0463.2012.02946.x. [DOI] [PubMed] [Google Scholar]
- 32.Leonard JH, Cook AL, Van Gele M, et al. Proneural and proneuroendocrine transcription factor expression in cutaneous mechanoreceptor (Merkel) cells and Merkel cell carcinoma. Int J Cancer. 2002;101:103–110. doi: 10.1002/ijc.10554. [DOI] [PubMed] [Google Scholar]
- 33.Ben-Arie N, Bellen HJ, Armstrong DL, et al. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- 34.Bermingham NA, Hassan BA, Price SD, et al. Math1: An essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- 35.Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- 36.Shroyer NF, Helmrath MA, Wang VY, Antalffy B, Henning SJ, Zoghbi HY. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology. 2007;132:2478–2488. doi: 10.1053/j.gastro.2007.03.047. [DOI] [PubMed] [Google Scholar]
- 37.Rose MF, Ren J, Ahmad KA, et al. Math1 is essential for the development of hindbrain neurons critical for perinatal breathing. Neuron. 2009;64:341–354. doi: 10.1016/j.neuron.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kong J, Crissey MA, Sepulveda AR, Lynch JP. Math1/Atoh1 contributes to intestinalization of esophageal keratinocytes by inducing the expression of Muc2 and Keratin-20. Dig Dis Sci. 2012;57:845–857. doi: 10.1007/s10620-011-1998-y. [DOI] [PMC free article] [PubMed] [Google Scholar]