Abstract
In the present study, the molecular characterization of HPV variants 16, 18, 31, 58, 6 and 11 within the MY06/MY11 L1 genomic region was performed in 128 sequences. For HPV 16, all of the sequences analyzed had a 3 nucleotide insertion resulting in the insertion of serine in the L1 protein sequence; and 4 sequences had at least one single nucleotide polymorphism (SNP). Twelve base substitutions were detected in HPV 58, 6 SNPs produced amino acid changes, and the other SNPs detected were found to be silent mutations. For HPV 31, 25 SNPs were detected as silent mutations. Of the 8 SNPs detected on HPV 18, three produced amino acid changes, the remaining SNPs detected were silent mutations. For HPV 6, 10 SNPs were detected and none of them produced amino acid changes. From the 16 sequences analyzed for HPV 11, two SNPs were detected and neither of them produced amino acid substitutions. Phylogenetic trees were constructed for HPV 16, HPV 18, HPV 31, HPV 58, HPV 6 and HPV 11. In the current study 8 new variants were identified based on sequencing of the L1 region. Changes in the L1 region of the HPV genome may be important for discriminating the infectious potential of different variants, as well as in defining epitopes relevant to vaccine design. The findings of this study indicate that there are new variants of HPV circulating in Argentina, which need to be confirmed by further analyses of the complete HPV genome.
Keywords: HPV, Phylogenetic, Maximum-likelihood, L1 gene
Introduction
Human papillomavirus (HPV) is a heterogeneous group of viruses with circular double-stranded DNA genomes of about 8 kb size comprising three general regions: an upstream regulatory region (URR) contains sequences that control transcription and replication, an early region contains genes (E6, E7, E1, E2, E4, and E5) involved primarily in enzymatic activities, and a structural region that produces capsid proteins (L1 and L2) [7].
The persistent infection with specific types of genital human papillomavirus is the main cause of cervical cancer and its precursor, cervical intraepithelial neoplasia (CIN). Cervical cancer is the most common gynecologic malignancy and one of the leading causes of mortality in women worldwide. Approximately 90 % of anal cancers and a smaller proportion (<50 %) of other cancers such as oropharyngeal, penile, vaginal and vulvar, are attributed to HPV [19].
Over 160 HPV types have been fully characterized. They are classified according to the frequency with which they are associated with premalignant and malignant lesions and have been designated as high risk (HR), intermediate (IR) and low risk (LR) HPV types.
The recent development of prophylactic vaccines directed against the most relevant disease-causing HPV types has helped to prevent diseases related to HPV. The bivalent HPV virus-like particle vaccine against HPV types 16 and 18 was efficacious against related HPV infections and against cervical dysplasia, and the quadrivalent HPV virus-like particle vaccine against types 6, 11, 16 and 18 was efficacious against related infections and against cervical, vaginal, vulvar, and anal dysplasia and against condyloma caused by HPV 6 and 11. These vaccines are expected to prevent about 70 % of cervical cancers worldwide [14]. Recently, the U.S. Food and Drug Administration has approved a 9-valent vaccine for the prevention of certain diseases caused by nine types of HPV (HPV 6, 11, 16, 18, 31, 33, 45, 52, and 58). This vaccine offers the potential to increase the overall prevention of cervical cancer from approximately 70 % to approximately 90 % [11].
A previous analysis of HPV sequence databases showed that the L1 gene, which encodes the major capsid protein, is conserved, thus being particularly suitable for to be used as a family taxonomic criterion.
A distinct human papillomavirus “type” is established when the nucleotide sequence of the region of the L1 open reading frame (ORF) of the clonal viral genome differs from that of any other type characterized by at least 10 % [2]. Isolates of a type whose L1 genes differ by 2–10 % are referred to as “subtypes”. Isolates of the same type are referred to as “variants” when the nucleotide sequences of their L1 genes differ by less than 2 % [7]. According to previous studies, the taxonomic grouping and naming of different lineages and sub-lineages is based on the comparison of the full genome sequence variants for each HPV type. To define distinct variant lineages, researchers have used a nucleotide sequence difference of approximately 1 % between two or more variants of the same type. Similarly, differences across the genome of 0.5–1 % have been used to designate sub-lineages (i.e.: A1, A2, etc.). Each major lineage is designated using an alphanumeric code, with the “A” clade always containing the reference genome for each type [4, 5].
The analysis of HPV variants diversity worldwide is of great importance as the HPV sequence data bases, are useful for epidemiological and evolutionary studies, for the development of accurate diagnostic tests and for the design of effective vaccines. The aim of this study was to analyze the intra-type variability of 128 sequences from MY09/MY11 L1 region. These HPV types are natural genotypes and variants found in different locations of Buenos Aires. The samples were identified as different genotypes by sequence analysis [12]. These HPV types and there variants were selected because they were most frequent HPV types found in our laboratory (HPV 16, 58, 31, 18, 11 and 6).
Materials and methods
Samples
A total of 128 sequences from a specific region of the HPV genome, L1 gene (MY09: CGTCCMARRGGAWACTGATC and MY11: GCMCAGGGWCATAAYAATGG), were studied. The sequences obtained were analyzed in MANLAB—Laboratory of Genomic Medicine from samples obtained in different localities of Argentina in the period June 2011–November 2013. Patient data were anonymized prior to this study.
Sequence analysis
Sequences were analyzed using Sequencing Analysis software v5.4 (Applied Biosystems). Genotyping of HPV samples was performed by alignment sequences obtained with sequences present in the GenBank database using BLASTN software (http://www.ncbi.nlm.nih.gov/blast/htlm). The following HPV high and low risk genotypes were included in the study: HPV16 (n = 23), HPV18 (n = 9), HPV31 (n = 16) and HPV58 (n = 17) and HPV6 (n = 47) and HPV 11 (n = 16), respectively.
HPV sequence alignments were carried out using the DNA CLC Workbench 5.6 software.
Isolates were defined by detecting differences in the studied region in one or more nucleotides when compared to the reference sequence for each genotype.
The sequences of L1 (MY09/MY11) gene that has not been previously described have been deposited in GenBank under the following accession numbers: HPV16 V4 = ijc-arg-HPV16 (KT304773), HPV 58 V3 = ijc-arg-HPV58 (KT304774), HPV31 V2 = ijc-arg-1-HPV31 (KT304775), HPV31 V6 = ijc-arg-2-HPV31 (KT304776), HPV18 V1 = ijc-arg-1-HPV18 (KT304777), HPV 18 V3 = ijc-arg-2-HPV18 (KT304778), HPV 6 V5 = ijc-arg-HPV6 (KT304779) and HPV 11 V1 = ijc-arg-HPV11 (KT304780).
Phylogenetic analysis
The phylogenetic analysis was performed using the Molecular Evolutionary Genetics Analysis (MEGA) version 6 software [16]. The neighbour-joining and maximum-likelihood method based on the Jukes–Cantor’s model were used to construct the phylogenetic trees, with Rhesus monkey papillomavirus type 1 (GenBank accession number M60184) used as outgroup in the analysis of HPV sequences. A bootstrap test with 1000 replicates was used to estimate the confidence of branching patterns in the trees.
Results
Types of variation
The results revealed that nucleotide changes in the sequences were present in the form of single nucleotide polymorphisms (SNP), and only in one case, an insertion of three nucleotides was detected. Single nucleotide polymorphisms were further characterized as either silent mutations (in which amino acid change does not occur), or missense mutations (non-silent) which produce an amino acid substitution. Phylogenetic trees were constructed for HPV 16, HPV 18, HPV 31, HPV 58, HPV 6 and HPV 11 (Fig. 1a–f).
Fig. 1.
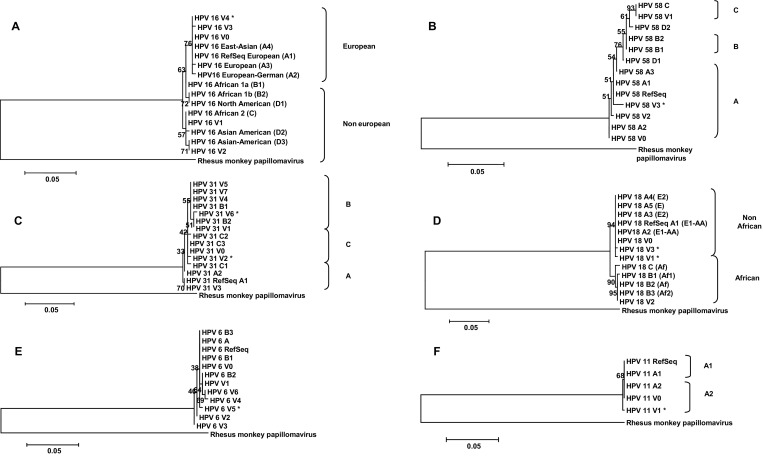
Phylogenetic analysis (Maximum-likelihood) of the L1 gene (region: MY09/MY11). Rhesus monkey papillomavirus (M60184) was used as outgroup. The asterisk indicates a new variant. a Tree of isolates of HPV 16 found in this study (HPV 16 V0, V1, V2, V3 and V4), along with HPV reference sequence [NC_001526.2 (RefSeq European HPV16/A1)] and variants of HPV 16 retrieved from GenBank: (East-Asian (EA) (A4): AF534061.1, European (A3): HQ644236.1, European-German (A2): AF536179.1, African 1a (B): AF536180.1, African 1b (B2): HQ644298.1, North American (B1): HQ644298.1, African 2 (C): AF472509.1, Asian-American (D2): AY686579.1 and Asian-American (D3): AF402678.1). b Tree of isolates of HPV 58 found in this study (HPV 58 V0, V1, V2 and V3), along with HPV reference sequence [D90400.1 A1 (RefSeq HPV 58)] and variants of HPV 58 retrieved from GenBank: lineage A1: D90400.1 A1, A2: AB819278.1, A3: AB819279.1, B1: HQ537763.1, B2: HQ537765.1, C: HQ537772.1, D1: D2 and HQ537767.1: HQ537769.1. c Tree of isolates of HPV 31 found in this study (HPV 31 V0, V1, V2, V3, V4, V5, V6 and V7), along with HPV reference sequence [J04353.1 (HPV 31 RefSeq A1)] and variants of HPV 31 retrieved from GenBank: A2: HQ537675.1, B1: HQ537676.1, B2: HQ537680.1, C1: HQ537682.1, C2 and C3 HQ537684.1: HQ537685.1. d Tree of isolates of HPV 18 found in this study (HPV 18 V0, V1, V2 and V3), along with HPV reference sequence [AY262282.1 (RefSeq HPV 18)] and variants of HPV 18 retrieved from GenBank: A1 [European 1-Asian-American (E1-AA): AY262282.1; A2 (E1-AA): EF202146.1; A3 (European 2 (E2)]: EF202147.1; A4 (E2): EF202151.1; A5 (E): GQ180787.1; B1 (African 1 (Af1)): EF202155.1; B2 (Af): KC470225. 1; B3 (Af2): EF202152.1 and C (Af): KC470229.1). e Tree of isolates of HPV 6 found in this study (HPV 6 V0, V1, V2, V3, V4, V5 and V6), along with HPV reference sequence [AF092932.1 (HPV 6 RefSeq)] and variants of HPV 6 retrieved from GenBank: A: X00203.1, B1: FR751337.1, B2 and B3 FR751328.1: L41216.1. f Tree of isolates of HPV 11 found in this study (HPV 11 V0 and V1), along with HPV reference sequence [FR872717.1 (HPV 11 RefSeq)] and variants of HPV 6 retrieved from GenBank: A1 and A2 M14119.1: FN907962.1
Variation in HPV type 16
For HPV 16, 23 sequences were analyzed for the L1 gene (region MY09/MY11) (330 bp). Out of the 23 isolates of HPV 16 sequenced, 19 were identical and 4 had at least one single nucleotide variation in comparison to other isolates of the same type. The majority of isolates were European and only 2 sequences were classified as non-European (Fig. 1a). None of the sequences analyzed was identical to the reference (NC_001526.2). From the 330 bp analyzed for L1 region, 10 base substitutions were detected, 2 of which resulted in amino acid changes (T379P and S415T). In addition, 23 samples had a three-nucleotide insertion relative to the reference sequence, resulting in the insertion of a serine (S) in the protein sequence of the HPV L1 gene (Table 1). The V4 isolate had not been previously described and according to phylogenetic analysis it can be classified as a European variant (Fig. 1a).
Table 1.
Nucleotide and amino acid sequence variation in sequences of L1 gene (region: MY09/MY11) of HPV 16 (23 samples)
| L1 nucleotide position | 6666 | 6694 | 6720 | 6802 | 6853 | 6864 | 6902–6904 |
|---|---|---|---|---|---|---|---|
| Non-synonymous mutations | – | T379P | – | S415T | – | – | Ins S |
| HPV 16 Ref (NC_001526.2) | A | A | G | T | C | C | – |
| V0 (19) | – | – | – | – | – | – | ATC |
| V1 (1) | – | C | A | – | T | T | ATC |
| V2 (1) | – | C | A | A | T | T | ATC |
| V3 (1) | C | – | – | – | – | – | ATC |
| V4 (1) | – | – | – | – | – | – | GTC |
The coordinates refer to the first nucleotide of each specific HPV reference sequence used. Isolates found in this study were designated as HPV 16 V0, V1, V2, V3 and V4. The number of isolates with identical sequences is indicated in parentheses
Variation in HPV type 58
For HPV 58, 17 sequences (356 bp) were analyzed, 14 of which were identical and 3 had at least one nucleotide variation between them (Table 2). None of the sequences analyzed was identical to the reference (D90400.1). Twelve base substitutions were detected, 6 SNPs belonging to the sequence isolate V1 which produce amino acid changes (T375N, G379D, D383N, I412V, D420N and N422D). The other isolates detected were silent mutations. One of the isolates found has not been previously described (V3) so far.
Table 2.
Nucleotide and amino acid sequence variation in sequences of L1 gene (region: MY09/MY11) of HPV 58 (17 samples)
| L1 nucleotide position | 6641 | 6682 | 6689 | 6691 | 6697 | 6711 | 6719 | 6798 | 6822 | 6827 | 6828 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Non-synonymous mutations | – | – | – | T375N | G379D | D383N | – | I412V | D420N | – | N422D |
| HPV 58 Ref (D90400.1) | G | A | C | G | G | G | T | A | G | C | A |
| V0 (14) | A | – | – | – | – | – | – | – | – | – | – |
| V1 (1) | – | – | – | A | A | A | – | G | A | A | G |
| V2 (1) | C | – | – | – | – | – | – | – | – | – | – |
| V3 (1) | – | G | A | – | – | – | C | – | – | – | – |
The coordinates refer to the first nucleotide of each specific HPV reference sequence used. Isolates found in this study were designated as HPV 58 V0, V1, V2 and V3. The number of isolates with identical sequences is indicated in parentheses
The phylogenetic analysis shows that most of the isolates (V0, V2 and V3) were within the lineage A corresponding to the most prevalent worldwide. The V1 isolate is within the lineage C which is predominant in Africa (Fig. 1b).
Variation in HPV type 31
Among the 16 sequences corresponding to HPV 31 (349 bp), 9 were identical (see Table 3). None of the sequences analyzed was identical to the reference (J04353.1). Twenty-five SNPs were detected, producing no amino acid changes. Two isolates (V2 and V6) found have not been previously described so far.
Table 3.
Nucleotide and amino acid sequence variation in sequences of L1 gene (region: MY09/MY11) of HPV 31 (16 samples)
| L1 nucleotide position | 6568 | 6586 | 6742 | 6772 | 6778 | 6796 | 6817 |
|---|---|---|---|---|---|---|---|
| Non-synonymous mutations | – | – | – | – | – | – | – |
| HPV 31 Ref (J04353.1) | T | T | T | G | A | G | A |
| V0 (9) | – | G | – | – | – | A | C |
| V1 (1) | C | – | – | A | – | A | C |
| V2 (1) | – | G | – | – | G | A | C |
| V3 (1) | – | – | – | – | – | – | – |
| V4 (1) | – | – | – | A | – | A | C |
| V5 (1) | – | – | – | A | – | A | C |
| V6 (1) | C | – | C | A | – | A | C |
| V7 (1) | – | – | – | A | – | A | C |
The coordinates refer to the first nucleotide of each specific HPV reference sequence used. Isolates found in this study were designated as HPV 31 V0, V1, V2, V3, V4, V5, V6 and V7. The number of isolates with identical sequences is indicated in parentheses
The phylogenetic tree shows that most of the isolates (62.5 %) are included within the lineage C including isolate V2. However, isolate V6 is within the lineage B (Fig. 1c).
Variation in HPV type 18
For HPV 18, 9 sequences (374 bp) were analyzed. Six sequences were identical to the reference sequence (AY262282.1). Eight SNPs were detected, 3 of them produce amino acid changes (V384I and P412S). The remaining SNPs detected were silent mutations (Table 4). Two of the isolates found have not been previously described (V1 and V3). The phylogenetic analysis showed that most of the isolates are within the non-African lineage, including isolates V3 and V1. However, one of the isolates (V2) was within the African lineage (Fig. 1d).
Table 4.
Nucleotide and amino acid sequence variation in sequences of L1 gene (region: MY09/MY11) of HPV 18 (9 samples)
| L1 nucleotide position | 6579 | 6581 | 6626 | 6663 | 6719 | 6749 | 6845 | 6917 |
|---|---|---|---|---|---|---|---|---|
| Non-synonymous mutations | V384I | V384I | – | P412S | – | – | – | – |
| HPV 18 Ref (AY262282.1) | G | T | C | C | G | G | A | G |
| V0 (6) | – | – | – | – | – | – | – | – |
| V1 (1) | – | – | – | – | – | – | T | – |
| V2 (1) | A | C | T | – | A | A | – | A |
| V3 (1) | – | – | – | T | – | – | – | – |
The coordinates refer to the first nucleotide of each specific HPV reference sequence used. Isolates found in this study were designated as HPV 18 V0, V1, V2 and V3. The number of isolates with identical sequences is indicated in parentheses
Variation in HPV type 6
For HPV 6, 47 sequences (377 bp) were analyzed of which 28 were identical to each other and equal to the reference sequence (AF092932.1). Ten SNPs were detected and none of them were found to produce an amino acid change (Table 5). One isolate has not been described in previous works (V5). When performing the phylogenetic analysis of the isolates it was found that they do not group according to the lineages and sub lineages described previously (Fig. 1e).
Table 5.
Nucleotide and amino acid sequence variation in sequences of L1 gene (region: MY09/MY11) of HPV 6 (47 samples)
| L1 nucleotide position | 6830 | 6974 | 7001 | 7077 | 7078 | 7100 |
|---|---|---|---|---|---|---|
| Non-synonymous mutations | – | – | – | K430Q | E431Q | – |
| HPV 6 Ref (AF092932.1) | T | A | C | A | G | G |
| V0 (28) | – | – | – | – | – | – |
| V1 (7) | – | – | – | – | C | – |
| V2 (7) | – | – | – | – | – | A |
| V3 (2) | – | G | – | – | – | A |
| V4 (1) | G | – | – | C | C | – |
| V5 (1) | – | – | T | – | – | – |
| V6 (1) | – | – | – | C | C | – |
The coordinates refer to the first nucleotide of each specific HPV reference sequence used. Isolates found in this study were designated as HPV 6 V0, V1, V2, V3, V4, V5 and V6. The number of isolates with identical sequences is indicated in parentheses
Variation in HPV type 11
From the 16 sequences analyzed for HPV 11 (380 bp), 15 were identical to the reference sequence (AF092932.1). Limited variation was detected and from the 380 bp aligned, 2 SNPs were detected and neither of them produced an amino acid substitution (Table 6). The isolate V1 has not been previously described. Taken all together, the phylogenetic studies allow concluding that all the sequences are within the A2 sub-lineage (Fig. 1f).
Table 6.
Nucleotide and amino acid sequence variation in sequences of L1 gene (region: MY09/MY11) of HPV 11 (16 samples)
| L1 nucleotide position | 6763 |
|---|---|
| Non-synonymous mutations | – |
| HPV 11 Ref (FR872717.1) | A |
| V0 (15) | – |
| V1 (1) | G |
The coordinates refer to the first nucleotide of each specific HPV reference sequence used. Isolates found in this study were designated as HPV 11 V0 and V1. The number of isolates with identical sequences is indicated in parentheses
Discussion
In the present study the molecular characterization of HPV 16, 18, 31, 58, 6 and 11 variants within the MY06/MY11 L1 genomic region was performed in 128 sequences.
Based on literature data, phylogenetic tree branches of HPV 16 were constructed to visualize their geographical origin. Such variants were classified as European (E) and non-European the latter including the following sub-lineages: African 1 (Af1), African 2 (Af2) and Asian American (AA) (1, 5). This tree reflects the evolution and worldwide migration of the human host and suggests that certain variants diverged at approximately the same time when the major human ethnic groups formed [9].
The results of this study indicate that 91 % of the sequences of HPV 16 are grouped in the European lineage. Only 2 isolates (9 %) were grouped in the non-European lineage (Table 1; Fig. 1a). This result agrees with previous studies carried out in different regions of Argentina [1, 17]. HPV 16 variants of the European lineage are the most prevalent worldwide; while African and Asian-American variants are less prevalent, but they feature different biological and epidemiological properties which result, in an increased carcinogenesis potential.
HPV 58 is considered the seventh most common high risk HPV type in cervical cancer and the sixth most common in cervical samples taken from asymptomatic patients [13]. Previous phylogenetic analyses indicate that this genotype has four distinct lineages (A, B, C and D). Lineage A predominates in all regions except Africa, where lineages A and C exist in comparable proportions. The distribution of sublinages A1, A2 and A3 also displayed geographical variation. Sublineage A2 is predominant in Africa, America and Europe. Some authors hypothesize that lineage A (probably A2) is the oldest one [6]. The data analyzed in this work are in the line with those of other authors, whose reports also indicate that the majority of the studied sequences can be grouped in the lineage A, specifically with sequence A2 sublineage (Fig. 1b).
Previous works have shown that the phylogenetic trees generated from the complete genome nucleotide sequences clustered HPV 31 variants into three distinct lineages designated A, B and C [8]. In genotype 31 we have noted that the sequences of the studied isolates were located mostly within the lineage C, unlike the reference sequence (J04353.1) which was located in the lineage A. The latter was an unexpected finding, for according to published data, lineage A is the one predominant in Caucasian women, and C is the predominant lineage among African-American women [18].
In line with literature data, the phylogenetic tree made with HPV 18 sequences revealed the presence of branches of African (A), European (E), and Asian-American (AA) origins and they can be grouped into African and Non-African origins. Likewise HPV 16 branches of phylogenetic trees show their geographical origin [15]. Most isolates found for HPV 18 were in the Non-African group. Only one isolate was grouped together with the African variants.
HPV 6 can be classified into two lineages (A and B), with the B lineage consisting of three sub-lineages (1, 2 and 3). HPV 11 variants are highly conserved and are classified in one lineage with two sublineages (A1 and A2). Previously reports indicate that no geographical association exists between variants of HPV 6 and HPV 11, and no association with different pathologies has been found [10].
Herein, a phylogenetic study was undertaken only by analyzing the region MY09/MY11 L1 gene, thus this methodology did not allow us to group the sequences of genotype 6 according to the lineages and sublineages previously described in the literature, in which complete HPV genomic sequences were employed [16]. Analogously, the grouping of sequences belonging to lineage D (D1 and D2) of HPV genotype 58 do not follow the same pattern as that published in other works where sequences of different genes (E6, E7, E2, E5, L1 and LCR) were used for the classification [6].
In the present study, 8 new variants were identified based on the sequencing of the L1 region, 6 belonged to the HPV high-risk group and two for the low-risk group. Changes in the L1 region of the HPV genome may be important for discriminating the infectious potential of different variants, as well as in defining epitopes relevant to vaccine design. The L1 protein present in HPV vaccine formulations elicits high-titres of neutralizing antibodies and confers type-specific and long-lasting protection against persistent infection and associated cervical neoplasia attributable to HPV vaccine types [3]. Changes in the nucleotide sequence of the L1 gene may induce structural changes, which modify not only its epitopes, but could also alter the viral particle assembly.
The findings of this study indicate that there are new variants of HPV circulating in the region. Further studies are needed to confirm the presence of new HPV variants and subtypes and to understand the evolution of HPV isolates in Argentina by analyzing the complete HPV genome or different regions of HPV genes such as L2, LCR, E6 and E7.
References
- 1.Badano I, Totaro ME, Culasso ACA, Sanabria DJ, Schurr TG, Balette IC, Roisman A, Basiletti J, Picconi MA, Campos RH, Domingo LJ. Genetic characterization and clinical implications of human papillomavirus type 16 (HPV16) variants from northeastern Argentina. Infect Genet Evol. 2015;29:103–109. doi: 10.1016/j.meegid.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 2.Bernard HU, Burk RD, Chen Z, van Doorslaer K, Hausen H, et al. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology. 2010;401:70–79. doi: 10.1016/j.virol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop B, Dasgupta J, Klein M, Garcea RL, Christensen ND, et al. Crystal structures of four types of human papillomavirus L1 capsid proteins: understanding the specificity of neutralizing monoclonal antibodies. J Biol Chem. 2007;282:31803–31811. doi: 10.1074/jbc.M706380200. [DOI] [PubMed] [Google Scholar]
- 4.Burk RD, Chen Z, Harari A, Smith BC, Kocjan BJ, et al. Classification and nomenclature system for human Alphapapillomavirus variants: general features, nucleotide landmarks and assignment of HPV6 and HPV11 isolates to variant lineages. Acta Dermatovenerol Alp Panonica Adriat. 2011;20:113–123. [PMC free article] [PubMed] [Google Scholar]
- 5.Burk RD, Harari A, Chen Z. Human papillomavirus genome variants. Virology. 2013;445:232–243. doi: 10.1016/j.virol.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan PK, Luk AC, Park JS, Smith-McCune KK, Palefsky JM, Konno R, Giovannelli L, Coutlée F, Hibbitts S, Chu TY, Settheetham-Ishida W, Picconi MA, Ferrera A, De Marco F, Woo YL, Raiol T, Piña-Sánchez P, Cheung JL, Bae JH, Chirenje MZ, Magure T, Moscicki AB, Fiander AN, Di Stefano R, Cheung TH, Yu MM, Tsui SK, Pim D, Banks L. Identification of human papillomavirus type 58 lineages and the distribution worldwide. J Infect Dis. 2011;203:1565–1573. doi: 10.1093/infdis/jir157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Z, Terai M, Fu L, Herrero R, DeSalle R, Burk RD. Diversifying selection in human papillomavirus type 16 lineages based on complete genome analyses. J Virol. 2005;79:7014–7023. doi: 10.1128/JVI.79.11.7014-7023.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Z, et al. Evolution and taxonomic classification of human papillomavirus 16 (HPV16)-related variant genomes: HPV31, HPV33, HPV35, HPV52, HPV58 and HPV67. PLoS ONE. 2011;6:e20183. doi: 10.1371/journal.pone.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Villiers EM, Fauquet C, Broker TR, Bernard HU, Zur Hausen H. Classification of papillomaviruses. Virology. 2004;324:17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 10.Heinzel PA, Chan SY, Ho L, O’Connor M, Balaram P, et al. Variation of human papillomavirus type 6 (HPV-6) and HPV-11 genomes sampled throughout the world. J Clin Microbiol. 1995;33:1746–1754. doi: 10.1128/jcm.33.7.1746-1754.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joura EA, Giuliano AR, Iversen OE, Bouchard C, et al. A 9-valent HPV vaccine against infection and intraepithelial neoplasia in women. N Engl J Med. 2015;372:711–723. doi: 10.1056/NEJMoa1405044. [DOI] [PubMed] [Google Scholar]
- 12.Lee SH, Vigliotti VS, Vigliotti JS, et al. Validation of human papillomavirus genotyping by signature DNA sequence analysis. BMC Clin Pathol. 2009;9:3. doi: 10.1186/1472-6890-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muñoz N, Bosch FX, de Sanjose S, Herrero R, Castellsagué X, Shah KV, Snijders PJF, Meijer CJLM. Epidemiological classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348:518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 14.Muñoz N, Bosch FX, Castellsague X, et al. Against which human papillomavirus types shall we vaccinate and screen? The international perspective. Int J Cancer. 2004;111:278–285. doi: 10.1002/ijc.20244. [DOI] [PubMed] [Google Scholar]
- 15.Ong CK, Chan SY, Campo MS, Fujinaga K, Mavromara-Nazos P, Labropoulou V, Pfister H, Tay SK, ter Meulen J, Villa LL, et al. Evolution of human papillomavirus type 18: an ancient phylogenetic root in Africa and intratype diversity reflect coevolution with human ethnic groups. J Virol. 1993;67:6424–6431. doi: 10.1128/jvi.67.11.6424-6431.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA 6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tonon SA, Basiletti J, Badano I, Alonio LV, Villa LL, Teyssie AR, Picconi MA. Human papillomavirus type 16 molecular variants in Guarani Indian women from Misiones, Argentina. Int J Infect Dis. 2007;11:76–81. doi: 10.1016/j.ijid.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Xi LF, Schiffman M, Koutsky LA, He Z, Winer RL, Hulbert A, Lee SK, Ke Y, Kiviat NB. Persistence of newly detected human papillomavirus type 31 infection, stratified by variant lineage. Int J Cancer. 2013;132:549–555. doi: 10.1002/ijc.27689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zur Hausen H. Papillomavirus infections—a major cause of human cancers. Biochim Biophys Acta. 1996;1288:F55–F78. doi: 10.1016/0304-419x(96)00020-0. [DOI] [PubMed] [Google Scholar]


