Abstract
The 15-kDa selenoprotein (Sep15) has been implicated in etiology of some types of cancer. Herein, inducible RNAi cell lines were established and cell morphology and motility were analyzed. The majority of Sep15-deficient cells (>95%) formed membrane blebs in a dynamic manner. Blebbing cells transformed cell morphology from a normal flat spindle shape to a spherical morphology. In blebbing cells, actin fibers moved to the cell periphery, covering and obscuring visualization of α-tubulin. Bleb formation was suppressed by the inhibitors of Rho-associated protein kinase (ROCK), RhoA or myosin light chain (MLC), restoring blebbing cells to wild-type morphology. RhoA activation and phosphorylation of myosin phosphatase target subunit 1 was induced by Sep15 knockdown. Sep15-deficient cells were non-apoptotic, and displayed a distinct relative localization of F-actin and α-tubulin from typical apoptotic blebbing cells. Our data suggest that Sep15 in Chang Liver cells regulates the pathway that antagonizes RhoA/ROCK/MLC-dependent non-apoptotic bleb formation.
Keywords: 15-kDa Selenoprotein, Membrane blebbing, Apoptosis, Cytoskeletal protein, Selenium
Introduction
In humans and other mammals, selenium has been reported to provide many health benefits when it is obtained from the diet in adequate amounts. For example, selenium is known to play roles in preventing cancer, delaying aging, augmenting the immune system, preventing heart disease, and supporting male reproduction and development [1–5]. Many of the health benefits of selenium are mediated by selenoproteins, which contain selenocysteine (Sec) as a selenium-containing amino acid at their active site [3,5]. One of the 25 genes encoding for human selenoproteins, the 15-kDa selenoprotein (Sep15) was initially identified as a strongly [75Se]-labeled selenoprotein in human T cells, and shares homology and compartment characteristics with selenoprotein M [6,7]. Similar to all other eukaryotic selenoprotein mRNAs, Sep15 mRNA contains a UGA codon, which is recoded as a Sec codon, and a Sec insertion sequence (SECIS) element in the 3'-untranslated region [8,9]. Sep15 is evolutionarily conserved in most animals and plants; however, only the vertebrate Sep15 homologs contain Sec. Like many other selenoproteins, Sep15 has been shown to exhibit redox activity and belongs to the thiol-disulfide oxidoreductase class of proteins [8,10,11]. Although its mechanistic role is not yet elucidated, Sep15 has been implicated in cancer development. The human SEP15 gene is located at the 1p31 locus, a locus where mutations and deletions have been observed in various human cancer cells [6,12]. Sep15 expression is decreased in liver, prostate, and lung cancers [8], and in several human malignant mesothelioma cell lines [13]. Since the discovery of two single nucleotide polymorphisms (SNPs) at nucleotides 811 (C/T) and 1125 (G/A), wherein the latter polymorphism occurs in the SECIS element of Sep15 [6], specific alleles have been associated with various cancers, including colorectal cancer [14,15], malignant mesothelioma [13], and lung cancer [16].
Recently, it has been reported that the targeted reduction or loss of Sep15 in in vitro and in vivo models of colon carcinogenesis reversed the cancer phenotype. The knockdown of Sep15 mRNA in a colon cancer cell line led to the inhibition of colony formation, tumor growth, and lung metastasis [17,18]. SEP15 knockout in mice prevented chemically induced aberrant crypt formation presumably by regulating guanylate binding protein-1 [19].
To obtain insight into the molecular function of Sep15 in human cells, we constructed a Chang liver cell line that inducibly expressed short hairpin RNA (shRNA) targeting Sep15 mRNA, and analyzed the effect of Sep15-deficiency on cell morphology. Sep15 deficiency formed non-apoptotic membrane blebs by remodeling cytoskeletal proteins such as α-tubulin and F-actin through activation of the RhoA/ROCK/MLC pathway.
Materials and Methods
Materials
Chang liver and T-REx-HeLa were purchased from ATCC (#CCL-13), and Invitrogen (R714-07), respectively. G418 sulfate was purchased from AG Scientific, doxycycline from BD Biosciences, and anti-MYPT1, anti-pMYPT1, anti-RhoA antibody were purchased from Cell Signaling. RhoA Pull-down Activation Assay Biochem kit was obtained from Cytoskeleton, and Alexa Fluor 488 goat anti-mouse IgG antibody, blasticidin, pcDNA6/TR vector and TRIZOL reagent were purchased from Invitrogen. Rhodamine phalloidin was obtained from Life Technologies, pSuperior.neo vector from OligoEngine, and Mo-MuLV reverse transcriptase from Promega. In Situ Cell Death Detection Kit was purchased from Roche. Annexin V-FITC Apoptosis Detection Kit, anti-α-tubulin antibody, blebbistatin, cycloheximide, 4′,6-Diamidino-2-phenylindole dihydrochloride (DAPI), HRP-conjugated goat anti-rabbit IgG antibody, HRP-conjugated goat anti-mouse IgG antibody, human tumor necrosis factor-α (hTNF-α), phosphatase inhibitor cocktail II, III, propidium iodide, protease inhibitor mixture, RNase A and Y-27632 were purchased from Sigma. DNAs were synthesized from Cosmogenetech (Korea). The His-tagged Tat-C3 transferase exoenzyme (pHis-Tat-C3) expression vector was provided by Jae Bong Park and the recombinant C3 transferase was prepared as previously described [20].
Cell culture and establishment of cell lines
Cell culture and transfection of cells were carried out as described previously [21]. An inducible Sep15 knockdown cell line was constructed in two steps. The TetR expression cell line (TetR) was constructed by transfecting pcDNA6/TR into Chang liver cells and selecting with blasticidin (5 µg/ml). To construct the doxycycline (Dox)-inducible Sep15 shRNA vector (pSuperior.neo.Sep15i), the hairpin sequence (GTATGTCCGTGGTTCAGACTTCAAGAG AGTCTGAACCACGGACATACTTTTTT) was inserted into the BglII and BamHI restriction sites of pSuperior.neo, which was subsequently transfected into the TetR cell line. To select the cells harboring pSuperior.neo.Sep15i, G418 (4 mg/ml) was added to the medium and the surviving colonies were selected. Single cell clones were isolated by diluting cells from each colony in a 96-well plate. Vector insertion into the host genome was confirmed by Southern blot analysis.
Measurement of knockdown efficiency
After induction of shRNA expression by 10 µg/ml of Dox treatment, total RNA was isolated from pSuperior.neo.Sep15i containing cells and subjected to realtime PCR using the ABI 7300 Real-time PCR system (Applied Biosystems) following the manufacturer’s instructions.
Immunocytochemistry and Western blot analysis
Immunocytochemistry was carried out as described previously [22] with minor modifications. After the cells were fixed, they were permeabilized, blocked, and incubated with anti-α-tubulin antibody (1:1000) and subsequently incubated with Alexa 488 anti-mouse IgG (1:1000). For F-actin and nuclear counterstaining, cells were stained with rhodamine phalloidin (1 µM) and DAPI (10 µg/ml), respectively. Cells were imaged with an LSM 700 confocal microscope (Carl Zeiss). Western analysis was carried out as described previously [23] using anti-pMYPT antibody (1: 500). The same membrane was used for another western analysis with anti-MYPT and anti-α-tubulin antibodies used as controls, after stripping with stripping buffer (62.5 mM Tris/HCl, pH6.8, 2% SDS and 100 mM β-mercaptoethanol) at 50°C for 10 min.
Live cell imaging
To examine the effect of the inhibitors of proteins that participate in RhoA/ROCK pathway on the morphology of Sep15 deficient cells, blebbistatin (10 µM), Y27632 (5 µM) or C3 transferase (2.5 µg/ml) was added to the medium after mounting the cells on the stage of a time-lapse microscope (Deltavision, Applied Precision). Cells were incubated at 37°C with 5% CO2 and images acquired with 10 sec time intervals for initial 2 hours and then 1 min intervals up to 12 hours.
Binding assays for cytoskeleton-regulating proteins
RhoA activity was measured using a RhoA Pull-down Activation Assay Biochem kit following the manufacturer’s instructions with slight modifications. When necessary, C3 transferase was added overnight. Cells were harvested with cell lysis buffer and clarified. The lysates were then mixed with GST-rhotekin-RBD beads (50 µg) for RhoA binding at 4°C for 2 h and the beads washed with wash buffer. Bound proteins were eluted by boiling in Laemmli sample buffer and subjected to western blot analysis using anti-RhoA (1:1000).
Detection of apoptosis
TUNEL assay
A terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assay was carried out using the In Situ Cell Death Detection Kit following the manufacturer’s instructions. As a positive control, cells were treated with 50 µg/ml of cycloheximide and 10 ng/ml of hTNF-α for 6 h. Cells were also incubated for 10 min in DNase I (50 units/ml) as TUNEL positive controls.
Annexin V and propidium iodide (PI) staining
Annexin V and PI staining was performed using the Annexin V-FITC Apoptosis Detection Kit following the manufacturer’s instructions. After staining, cells were viewed under a confocal microscope.
Statistical analysis
Each experiment was performed in biological triplicate for statistical analysis. Statistical significance was tested by the Student’s t-test followed by an f-test to ensure the equality of the 2 variances.
Results
Sep15 deficiency stimulates membrane bleb formation
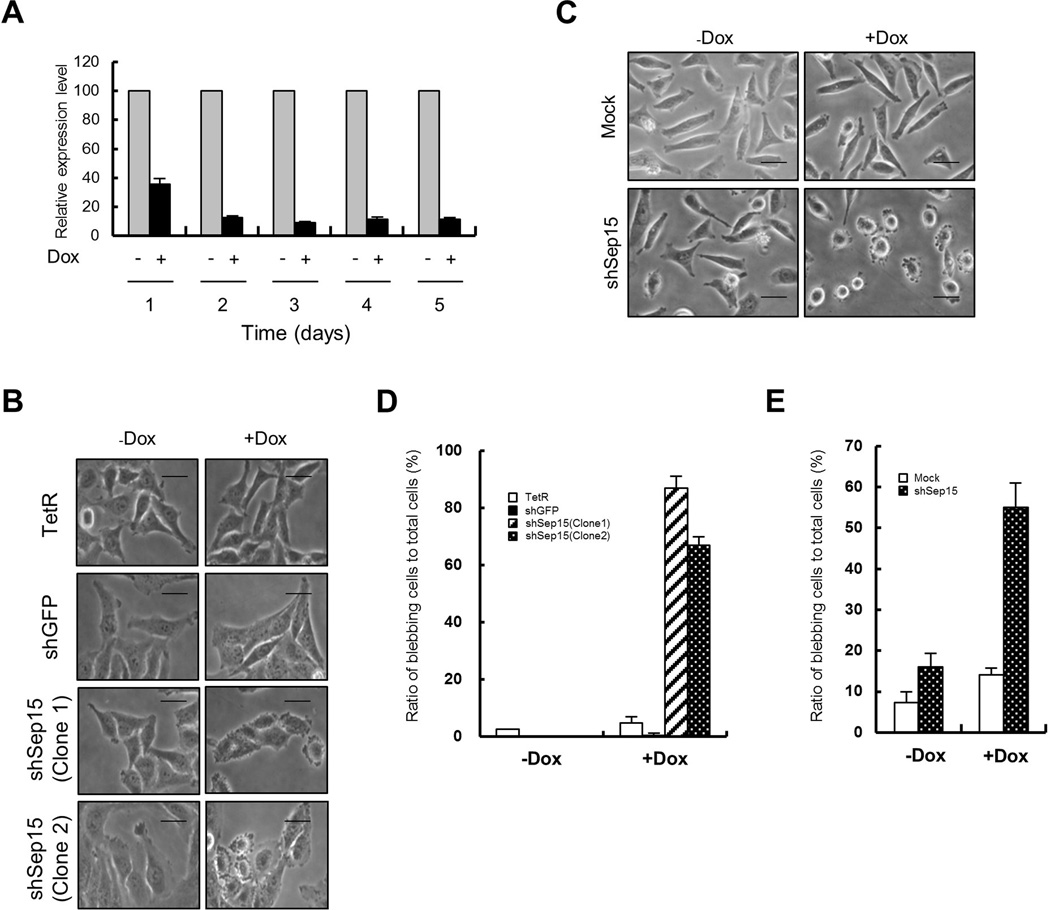
We constructed three cell lines that harbor pSuperior.neo.Sep15i inserted in different sites of chromosomes and can be induced to express Sep15-targeted shRNA. One of them (Clone 1) was chosen arbitrarily, renamed as shSep15 and used for subsequent analyses. The temporal knockdown efficiency of the shSep15 cell line was measured by quantitative RT-PCR. Sep15 expression was significantly reduced and the knockdown efficiency reached over 90% by day 2 (Fig. 1A).
Fig. 1. Sep15 knockdown leads to membrane bleb formation.
(A) Efficiency of Sep15 knockdown after the induction of shRNA expression was measured by quantitative RT-PCR GAPDH was used as an internal control. (B) The morphology of cells was examined under a light microscope. Cell names are indicated on the left of Y-axis. (C) Knockdown of Sep15 in T-Rex HeLa led to membrane bleb formation. (D) Relative ratio of blebbing cells to total cells calculated from (B). (E) Relative ratio of blebbing cells to total cells calculated from (C). Blebbing cells were counted from 3 independent experiments. Scale bars represent 10 µm.
One striking observation with the Sep15-deficient cells was that membrane blebs formed in more than 90% of these cells (Fig. 1B,D). Because Dox-treated TetR-expressing cells did not produce blebs, membrane blebbing appears to be solely attributable to Sep15 deficiency. In addition to shSep15 (Clone 1), another clone (Clone 2) that contains pSuperior.neo.Sep15i on different site of chromosome from shSep15 (Clone 1, Supplementary Fig. S1) also showed membrane blebbing after Dox treatment (Fig. 1B,D). These results suggest that insertion of the vector into a chromosome did not affect bleb formation in these cell lines. It should be noted that membrane blebbing was also induced in T-REx-HeLa, which contains TetR gene in HeLa, when Sep15 was knocked down with the same shRNA gene construct used in Chang liver cells (Fig. 1C,E). However, Dox treatment of T-Rex HeLa containing mock vector (pSuperior.neo) did not induce bleb formation. These results indicate that membrane blebbing by Sep15 deficiency is a common event in HeLa and Chang liver. Higher magnification differential interference contrast (DIC) images of the cells showed a marked change in overall cell shape upon lack of Sep15. Cells changed from an oblong to round morphology and they possessed blebs of varying sizes, suggesting that bleb formation was dynamic (Fig. 2A). The blebs disappeared after removing Dox by changing the medium that did not contain Dox (Dox wash). Live cell images showed dynamic bleb formation (Fig. 2B and Supplementary Movie 1). It should be noted that in some cases, very long extended blebs were formed and then disappeared by shrinking in shSep15+Dox cells (Supplementary Movie 2).
Fig. 2. Fine structure of blebbing cells.
Three days after induction of knockdown, cell morphology was examined. (A) Light microscope images of cells. (B) Live cell transmission images obtained using a time-lapse microscope. Arrowheads in the inbox indicate the position of a newly generated bleb on the cell membrane. (C) Cells were stained for α-tubulin (green), F-actin (red) and nuclei (blue). Images obtained by confocal microscopy. (D) Three-dimensional images obtained by stacking two-dimensional images from confocal sections taken at 0.45 µm intervals. Scale bars represent 10 µm.
We stained cells for F-actin and α-tubulin and examined their localization using confocal microscopy. As shown in Fig. 2C, the majority of F-actin became localized to both the plasma and bleb membranes in Sep15 knockdown cells (+Dox). In the control cells (−Dox), F-actin was usually enriched at the lamellipodia tips and in filopodia. Neither filopodia nor lamellipodia could be detected in Sep15-deficient cells. In these cells, the majority of α-tubulin became localized to near the nuclear membrane and to the inside of the blebs. The addition of Dox to shGFP cells did not lead to the formation of blebs and the positions of F-actin and α-tubulin were not changed (Supplementary Fig. S2A) suggesting membrane blebbing in Sep15 knockdown cells is dependent on Sep15 deficiency. A three-dimensional reconstruction shows the reorganization of the actin and microtubule cytoskeletons more clearly (Fig. 2D). The F-actin was so abundant at the periphery of shSep15+Dox cells that α-tubulin, which is normally enriched there, was nearly undetectable. We measured the thickness of the blebbing cells and found that it was increased approximately 2-fold relative to control cells (Fig. 2D, side view). In contrast, the relative positions of F-actin and α-tubulin were not changed by Dox addition in shGFP cells (Supplementary Fig. S2B). These results support the notion that the cytoskeleton is contractile during bleb formation. Based on these data, we propose that Sep15 deficiency in Chang liver cells leads to the formation of membrane blebs through the reorganization of cytoskeletal proteins, changing their positions in the cell and making them more contractile. To our knowledge, this is the first evidence demonstrating that the positions of the cytoskeletal proteins, F-actin and α-tubulin, are reversed during membrane bleb formation.
Membrane blebs are formed through the RhoA/ROCK/MLCK pathway
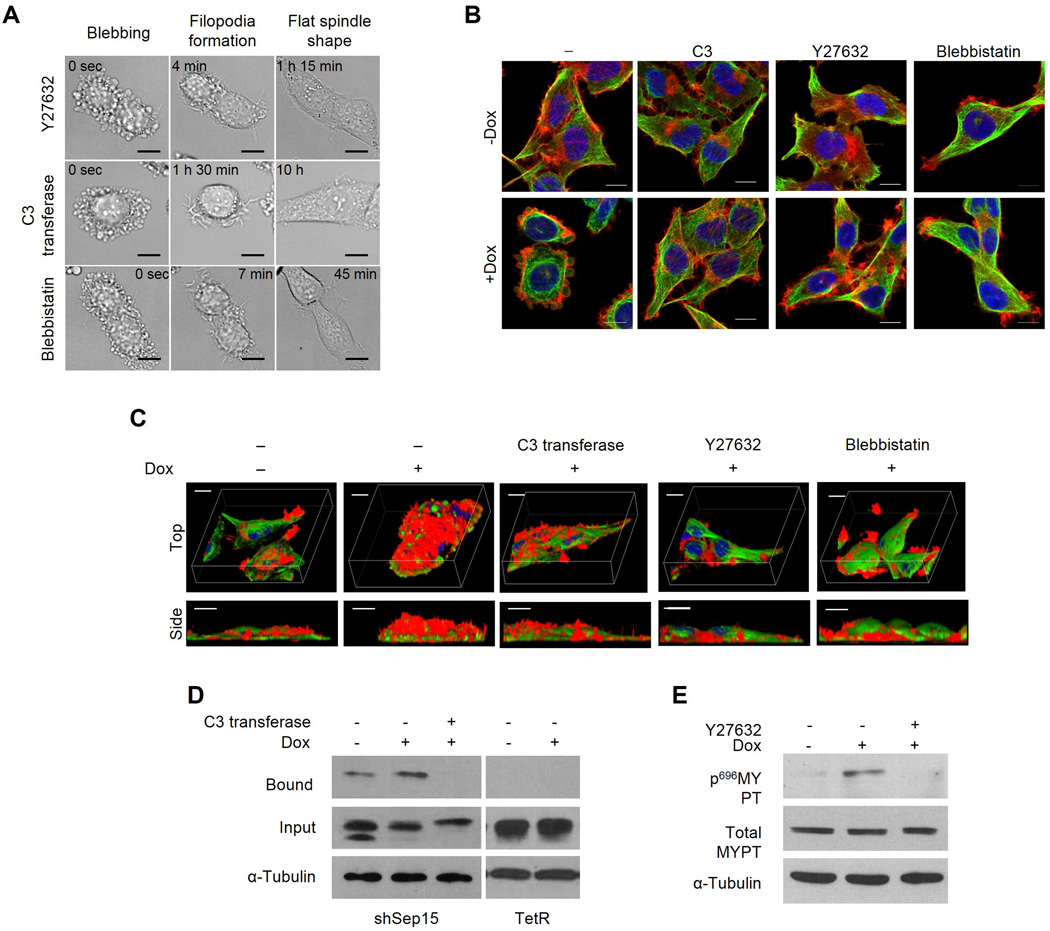
Because it has been reported that membrane blebbing occurs through a ROCK-dependent pathway [24], we examined whether membrane blebbing in Sep15-deficient cells also occurred through a ROCK pathway. Y-27632 is known to be a potent ROCK inhibitor. The addition of Y-27632 (5 µM) to the culture medium reversed the blebbing cells to their normal morphology within 1.5 h (Fig. 3A and Supplementary Movie 3). Cells became flattened and the lamellipodia and filopodia were regenerated. ROCK can be activated by either caspase-3 or RhoA [24]. Therefore, we determined whether ROCK is activated through the RhoA-dependent pathway by treating blebbing cells with recombinant C3 transferase, a RhoA inhibitor. Addition of C3 transferase (2.5 µg/ml) to the medium also restored cells to normal morphology within 10 h (Fig. 3A and Supplementary Movie 4). Usually myosin light chain kinase (MLCK) that activates myo II is the target of ROCK. Blebbistatin (10 µM), a MLCK inhibitor, also reversed the blebbing cell morphology within 10 min (see also Supplementary Movie 5). Normal relative localization of these cytoskeletal components and the formation of both lamellipodia and filopodia were restored by the treatment of C3 transferase, Y27632 and blebbistatin (Fig. 3B). A three-dimensional reconstruction clearly shows that the cell thickness decreased to that of normal cells (see side view of Fig. 3C). These data strongly suggest that Sep15 deficiency activates RhoA, ROCK, and then MLCK. The activation of myoII might then become a cue for membrane blebbing. Since the above data suggest that RhoA is activated by Sep15 knockdown, we carried out a GST-rhotekin pull-down assay to obtain direct evidence that RhoA is activated by Sep15 knockdown. The amount of rhotekin-bound RhoA was significantly increased by Dox treatment in shSep15 cells (Fig. 3D), but the binding was blocked by treatment of C3 transferase. We also examined whether the phosphorylation of MYPT was stimulated by Sep15 knockdown and found that Sep15 deficiency increased the levels of phosphorylated MYPT1, while Y27632 reduced the phosphorylated form of MYPT1 significantly (Fig. 3E).
Fig. 3. The effect of C3 transferase, Y27632 and blebbistatin on membrane bleb formation and the activation of RhoA and ROCK in Sep15-deficient cells.
Cells were prepared as described in Materials and Methods. (A) Live cell images of cells treated with C3 transferase, Y27632 or blebbistatin. Characteristic morphological changes are indicated on the top. The varying times to reach each specific cell morphology are indicated on the inside of each panel. The two-dimensional (B) and three-dimensional (C) images obtained by confocal microscopy. Cells were stained for α-tubulin (red), F-actin (green) and nucleus (blue), and observed using a confocal microscope. (D) Measuring active forms of RhoA by pull-down assay. Films were exposed for 30 sec after antibody binding. (E) Detection of phospho-MYPT by western blotting. α-tubulin was used as an internal control of input proteins (30 µg). Scale bars represent 10 µm.
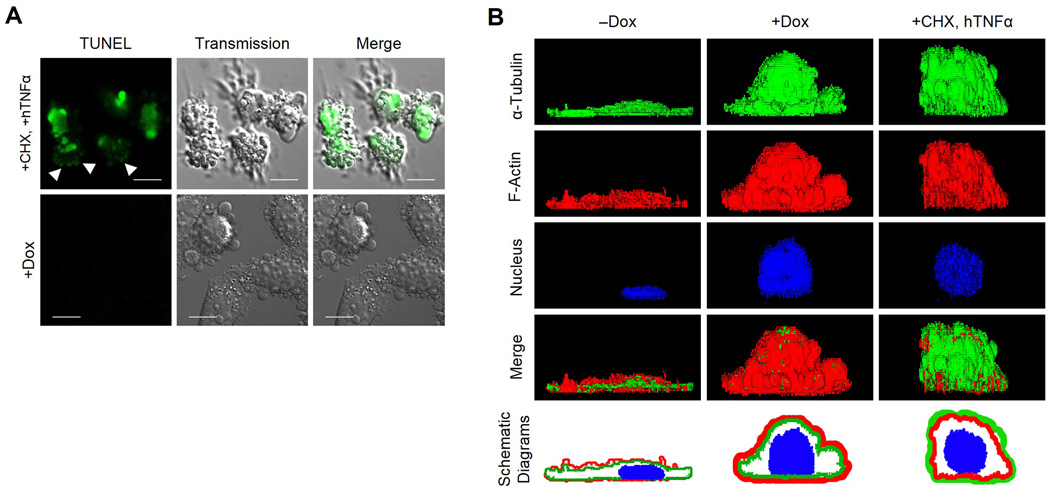
Blebbing Sep15-deficient cells are non-apoptotic and have a unique cytoskeletal organization distinct from apoptotic cells
We performed 3 assays (annexin V, TUNEL, and PI) to determine whether the Sep15-deficient blebbing cells were undergoing apoptosis. Unexpectedly, the Sep15-deficient cells were not stained by any of the 3 methods used (Supplementary Fig. S3). Notably, the TUNEL-positive cells resulting from both cycloheximide and hTNFα treatment floated in the culture medium; however, the Sep15-deficient blebbing cells remained attached to the bottom of the culture plate (Fig. 4A). The TUNEL-positive cells showed both the nucleus and membrane blebs were stained, although the signal intensity in the blebs was lower than in the nucleus suggesting that the nuclear membrane was ruptured allowing the DNA to move into the blebs (arrows in the Fig. 4A). In contrast, there was no TUNEL signal in either the nucleus or the blebs of Sep15-deficient cells. These data indicate that, unlike most cells undergoing ROCK-dependent membrane blebbing, Sep15-deficient cells do not undergo apoptosis. As mentioned previously, the side view of Sep15-deficient cells clearly shows that the actin fibers moved to the periphery and α-tubulin to the interior of the cell, while both cytoskeletal proteins could be detected clearly on the surface of normal cells (Fig. 4B). In Sep15-deficient cells, the intensity of the α-tubulin signal was not reduced. However, the image size of α-tubulin was smaller than F-actin in Sep15-deficient cells (+Dox) and the F-actin image covered the α-tubulin image when they were overlaid (see the schematic diagrams of +Dox). However, the relative localization of F-actin and α-tubulin was reversed in cycloheximide/hTNFα derived apoptotic cells (Fig. 4B). In these cells, the merged image showed that α-tubulins cover actin fibers (see merge and schematic diagrams of +CHX, hTNFα). The nuclear image of apoptotic cell shows many pores suggesting that fragmentation of the nucleus occurs in this cell. These data indicate that the relative positions of cytoskeletal proteins, α-tubulin and F-actin, are opposite between non-apoptotic and apoptotic blebbing cells.
Fig. 4. Blebbing Sep15-deficient cells show unique characteristics distinct from apoptotic cells.
(A) TUNEL assays for Sep15 knockdown cells. Scale bars represent 10 µm. Images of positive control cells were obtained by focusing on the floating blebbing cells (upper panels). The adherent Sep15 deficient cells (+Dox) were negative for TUNEL signals (lower panels). (B) Three dimensional images of apoptotic and non-apoptotic cells. Cells were stained for α-tubulin (green), F-actin (red) and nucleus (blue) before observation. Schematic diagrams were produced by merging the images of α-tubulin, β-actin and nucleus after redrawing each image with the same thickness using Photoshop software. Scale bars represent 10 µm.
Discussion
Sep15 has been implicated in cancer development [6,8,12,13,18,19]. However, the mechanistic role of Sep15 during cancer development is not fully understood. In this study, we constructed inducible Sep15 knockdown cell lines and examined the phenotypic changes that occurred after the induction of Sep15 shRNA. Membrane blebs were formed by Sep15 knockdown in both HeLa and Chang liver cells and the blebbing cells were non-apoptotic unlike most of other blebbing cells as described in the literature.
Blebs are round cellular protruding structures formed when the plasma membrane separates from the cell cortex. Blebs reportedly form during apoptosis, cytokinesis, cell division and migration, and even during virus uptake [25,26]. Obviously, blebs formed during cell division and migration should be non-apoptotic. Two different hypotheses have been proposed on blebbing during cell division; a side effect of tension build-up in the cortex during cell division [27], and generation of additional cortex and/or membrane surface area during cytokinesis [28]. During cell migration, blebbing occurs for a short time before lamellipodia formation [29,30]. Apoptotic blebbing is known to occur as a result of over-activation of myosin II [31]. And it can occur through either caspase-dependent or caspase-independent pathways [31,32]. Although many blebbing cells become apoptotic and MLCK was activated when Sep15 was knocked down, we could not obtain any evidence indicating that Sep15-deficient cells underwent apoptosis. Our data, therefore, indicate that the blebs observed in Sep15-deficient cells were generated through a novel mechanism that is distinct from the one responsible for general non-apoptotic blebbing. We found two characteristics that separate non-apoptotic blebs induced by Sep15 deficiency from those of typical apoptotic blebs: one is that Sep15-deficient cells are more adhesive than apoptotic bleb cells and the other is that the relative localization of actin/tubulin is opposite in the two groups (see Fig. 4A,B). Further studies are required to determine whether these characteristics are general features that can be used to distinguish apoptotic blebs from non-apoptotic ones.
Supplementary Material
Acknowledgements
This work was supported by the Priority Research Centers Program (Grant No. 2010-0029694) and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (Grant No. 2011-0012947), the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science, ICT & Future Planning (2012M3A9D1054622) to BJL, and in part by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research to DLH. JB, MJ, JHH, JN and MS were supported by the Brain Korea 21 Research Fellowship from the Korea Ministry of Education and Human Resources Development.
Abbreviations
- Sep15
15-kDa Selenoprotein
- ROCK
Rho-associated protein kinase
- MLC
myosin light chain
- MYPT
Myosin phosphatase targeting protein
- Sec
Selenocysteine
- SECIS
Sec insertion sequence
- Dox
Doxycycline
References
- 1.Boosalis MG. The role of selenium in chronic disease. Nutr Clin Pract. 2008;23:152–160. doi: 10.1177/0884533608314532. [DOI] [PubMed] [Google Scholar]
- 2.Flohe L. Selenium in mammalian spermiogenesis. Biol Chem. 2007;388:987–995. doi: 10.1515/BC.2007.112. [DOI] [PubMed] [Google Scholar]
- 3.Hatfield DL, Gladyshev VN. How selenium has altered our understanding of the genetic code. Mol Cell Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papp LV, Lu J, Holmgren A, Khanna KK. From selenium to selenoproteins: synthesis, identity, and their role in human health. Antioxid Redox Signal. 2007;9:775–806. doi: 10.1089/ars.2007.1528. [DOI] [PubMed] [Google Scholar]
- 5.Roman M, Jitaru P, Barbante C. Selenium biochemistry and its role for human health. Metallomics. 2013 doi: 10.1039/c3mt00185g. [DOI] [PubMed] [Google Scholar]
- 6.Gladyshev VN, Jeang KT, Wootton JC, Hatfield DL. A new human selenium-containing protein. Purification, characterization, and cDNA sequence. J Biol Chem. 1998;273:8910–8915. doi: 10.1074/jbc.273.15.8910. [DOI] [PubMed] [Google Scholar]
- 7.Korotkov KV, Novoselov SV, Hatfield DL, Gladyshev VN. Mammalian selenoprotein in which selenocysteine (Sec) incorporation is supported by a new form of Sec insertion sequence element. Mol Cell Biol. 2002;22:1402–1411. doi: 10.1128/mcb.22.5.1402-1411.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumaraswamy E, Malykh A, Korotkov KV, Kozyavkin S, Hu Y, Kwon SY, Moustafa ME, Carlson BA, Berry MJ, Lee BJ, Hatfield DL, Diamond AM, Gladyshev VN. Structure-expression relationships of the 15-kDa selenoprotein gene. Possible role of the protein in cancer etiology. J Biol Chem. 2000;275:35540–35547. doi: 10.1074/jbc.M004014200. [DOI] [PubMed] [Google Scholar]
- 9.Low SC, Berry MJ. Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends Biochem Sci. 1996;21:203–208. [PubMed] [Google Scholar]
- 10.Ferguson AD, Labunskyy VM, Fomenko DE, Arac D, Chelliah Y, Amezcua CA, Rizo J, Gladyshev VN, Deisenhofer J. NMR structures of the selenoproteins Sep15 and SelM reveal redox activity of a new thioredoxin-like family. J Biol Chem. 2006;281:3536–3543. doi: 10.1074/jbc.M511386200. [DOI] [PubMed] [Google Scholar]
- 11.Labunskyy VM, Hatfield DL, Gladyshev VN. The Sep15 protein family: roles in disulfide bond formation and quality control in the endoplasmic reticulum. IUBMB Life. 2007;59:1–5. doi: 10.1080/15216540601126694. [DOI] [PubMed] [Google Scholar]
- 12.Nasr MA, Hu YJ, Diamond AM. Allelic loss at the Sep15 locus in breast cancer. Cancer Therapy. 2013;1:293–298. [Google Scholar]
- 13.Apostolou S, Klein JO, Mitsuuchi Y, Shetler JN, Poulikakos PI, Jhanwar SC, Kruger WD, Testa JR. Growth inhibition and induction of apoptosis in mesothelioma cells by selenium and dependence on selenoprotein SEP15 genotype. Oncogene. 2004;23:5032–5040. doi: 10.1038/sj.onc.1207683. [DOI] [PubMed] [Google Scholar]
- 14.Davis CD, Tsuji PA, Milner JA. Selenoproteins and cancer prevention. Annu Rev Nutr. 2012;32:73–95. doi: 10.1146/annurev-nutr-071811-150740. [DOI] [PubMed] [Google Scholar]
- 15.Sutherland A, Kim DH, Relton C, Ahn YO, Hesketh J. Polymorphisms in the selenoprotein S and 15-kDa selenoprotein genes are associated with altered susceptibility to colorectal cancer. Genes Nutr. 2010;5:215–223. doi: 10.1007/s12263-010-0176-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jablonska E, Gromadzinska J, Sobala W, Reszka E, Wasowicz W. Lung cancer risk associated with selenium status is modified in smoking individuals by Sep15 polymorphism. Eur J Nutr. 2008;47:47–54. doi: 10.1007/s00394-008-0696-9. [DOI] [PubMed] [Google Scholar]
- 17.Irons R, Tsuji PA, Carlson BA, Ouyang P, Yoo MH, Xu XM, Hatfield DL, Gladyshev VN, Davis CD. Deficiency in the 15-kDa selenoprotein inhibits tumorigenicity and metastasis of colon cancer cells. Cancer Prev Res (Phila) 2010;3:630–639. doi: 10.1158/1940-6207.CAPR-10-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsuji PA, Naranjo-Suarez S, Carlson BA, Tobe R, Yoo MH, Davis CD. Deficiency in the 15 kDa selenoprotein inhibits human colon cancer cell growth. Nutrients. 2011;3:805–817. doi: 10.3390/nu3090805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsuji PA, Carlson BA, Naranjo-Suarez S, Yoo MH, Xu XM, Fomenko DE, Gladyshev VN, Hatfield DL, Davis CD. Knockout of the 15 kDa selenoprotein protects against chemically-induced aberrant crypt formation in mice. PLoS One. 2012;7:e50574. doi: 10.1371/journal.pone.0050574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park J, Kim JS, Jung KC, Lee HJ, Kim JI, Kim J, Lee JY, Park JB, Choi SY. Exoenzyme Tat-C3 inhibits association of zymosan particles, phagocytosis, adhesion, and complement binding in macrophage cells. Mol Cells. 2003;16:216–223. [PubMed] [Google Scholar]
- 21.Kim JY, Lee KH, Shim MS, Shin H, Xu XM, Carlson BA, Hatfield DL, Lee BJ. Human selenophosphate synthetase 1 has five splice variants with unique interactions, subcellular localizations and expression patterns. Biochem Biophys Res Commun. 2010;397:53–58. doi: 10.1016/j.bbrc.2010.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shim MS, Kim JY, Jung HK, Lee KH, Xu XM, Carlson BA, Kim KW, Kim IY, Hatfield DL, Lee BJ. Elevation of glutamine level by selenophosphate synthetase 1 knockdown induces megamitochondrial formation in Drosophila cells. J Biol Chem. 2009;284:32881–32894. doi: 10.1074/jbc.M109.026492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim M, Chen Z, Shim MS, Lee MS, Kim JE, Kwon YE, Yoo TJ, Kim JY, Bang JY, Carlson BA, Seol JH, Hatfield DL, Lee BJ. SUMO modification of NZFP mediates transcriptional repression through TBP binding. Mol Cells. 2013;35:70–78. doi: 10.1007/s10059-013-2281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leverrier Y, Ridley AJ. Apoptosis: caspases orchestrate the ROCK 'n' bleb. Nat Cell Biol. 2001;3:E91–93. doi: 10.1038/35070151. [DOI] [PubMed] [Google Scholar]
- 25.Charras G, Paluch E. Blebs lead the way: how to migrate without lamellipodia. Nat Rev Mol Cell Biol. 2008;9:730–736. doi: 10.1038/nrm2453. [DOI] [PubMed] [Google Scholar]
- 26.Hickson GR, Echard A, O'Farrell PH. Rho-kinase controls cell shape changes during cytokinesis. Curr Biol. 2006;16:359–370. doi: 10.1016/j.cub.2005.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paluch E, Sykes C, Prost J, Bornens M. Dynamic modes of the cortical actomyosin gel during cell locomotion and division. Trends Cell Biol. 2006;16:5–10. doi: 10.1016/j.tcb.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 28.Charras GT, Hu CK, Coughlin M, Mitchison TJ. Reassembly of contractile actin cortex in cell blebs. J Cell Biol. 2006;175:477–490. doi: 10.1083/jcb.200602085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bereiter-Hahn J, Luck M, Miebach T, Stelzer HK, Voth M. Spreading of trypsinized cells: cytoskeletal dynamics and energy requirements. J Cell Sci. 1990;96(Pt 1):171–188. doi: 10.1242/jcs.96.1.171. [DOI] [PubMed] [Google Scholar]
- 30.Bergert M, Chandradoss SD, Desai RA, Paluch E. Cell mechanics control rapid transitions between blebs and lamellipodia during migration. Proc Natl Acad Sci U S A. 2012;109:14434–14439. doi: 10.1073/pnas.1207968109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sebbagh M, Renvoize C, Hamelin J, Riche N, Bertoglio J, Breard J. Caspase-3-mediated cleavage of ROCK I induces MLC phosphorylation and apoptotic membrane blebbing. Nat Cell Biol. 2001;3:346–352. doi: 10.1038/35070019. [DOI] [PubMed] [Google Scholar]
- 32.Sebbagh M, Hamelin J, Bertoglio J, Solary E, Breard J. Direct cleavage of ROCK II by granzyme B induces target cell membrane blebbing in a caspase-independent manner. J Exp Med. 2005;201:465–471. doi: 10.1084/jem.20031877. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.