Abstract
A safe, effective, and inexpensive vaccine against typhoid and other Salmonella diseases is urgently needed. In order to address this need, we are developing a novel vaccine platform employing buoyant, self-adjuvanting gas vesicle nanoparticles (GVNPs) from the halophilic archaeon Halobacterium sp. NRC-1, bioengineered to display highly conserved Salmonella enterica antigens. As the initial antigen for testing, we selected SopB, a secreted inosine phosphate effector protein injected by pathogenic S. enterica bacteria during infection into the host cells. Two highly conserved sopB gene segments near the 3′-region, named sopB4 and sopB5, were each fused to the gvpC gene, and resulting SopB-GVNPs were purified by centrifugally accelerated flotation. Display of SopB4 and SopB5 antigenic epitopes on GVNPs was established by Western blotting analysis using antisera raised against short synthetic peptides of SopB. Immunostimulatory activities of the SopB4 and B5 nanoparticles were tested by intraperitoneal administration of SopB-GVNPs to BALB/c mice which had been immunized with S. enterica serovar Typhimurium 14028 ΔpmrG-HM-D (DV-STM-07), a live attenuated vaccine strain. Proinflammatory cytokines IFN-γ, IL-2, and IL-9 were significantly induced in mice boosted with SopB5-GVNPs, consistent with a robust Th1 response. After challenge with virulent S. enterica serovar Typhimurium 14028, bacterial burden was found to be diminished in spleen of mice boosted with SopB4-GVNPs and absent or significantly diminished in liver, mesenteric lymph node, and spleen of mice boosted with SopB5-GVNPs, indicating that the C-terminal portions of SopB displayed on GVNPs elicit a protective response to Salmonella infection in mice. SopB antigen-GVNPs were also found to be stable at elevated temperatures for extended periods without refrigeration. The results show that bioengineered GVNPs are likely to represent a valuable platform for antigen delivery and development of improved vaccines against Salmonella and other diseases.
Keywords: Salmonella, Halobacterium, vaccine, nanoparticle, gas vesicle
1. Introduction
1.1. Salmonella diseases
Salmonella enterica, a Gram-negative intracellular pathogenic bacterium, infects humans and many warm blooded animals. Salmonellosis remains a serious problem in most developing countries and outbreaks are regularly seen in developed and industrialized countries1–4. Typhoid fever is caused by S. Typhi or S. Paratyphi, with global incidence of 21.7 million cases of Typhi and an additional 5.4 million Paratyphi cases resulting in 217,000 deaths per year5. Treatment is becoming more challenging due to increased prevalence of antibiotic resistance6–8. While two licensed vaccines for typhoid fever are commercially available, they have limited protection and/or may not be administered to immunocompromised people or children and infants. As a result, there is a critical need to develop more effective Salmonella vaccine candidates, which would also be safe and easily scalable, and inexpensive to produce and deliver. A versatile, particulate antigen delivery system, gas vesicle nanoparticles (GVNPs), which can be bioengineered to express multiple Salmonella antigens is likely to offer significant advantages over currently available vaccines, due to reduced risk and improved effectiveness.
1.2. Salmonella pathogenesis and vaccine status
Salmonella enterica includes 2500 serovars infecting humans, and several are of public health importance, including S. Typhi and S. Paratyphi, the causative agents of typhoid and paratyphoid fever2,9,10. Transmission occurs through the fecal-oral route, upon ingestion of contaminated water and food. Occurrence of the disease may be confirmed by isolation of the pathogen, detection of antibodies against Salmonella specific O (somatic) and H (flagellar) antigens in the serum, or most sensitively, PCR based methods which utilize specific primers designed against unique regions of their genomes8. Treatment however is becoming more challenging, due to increased prevalence of multiple drug resistant (MDR) strains6,7. The need for improved vaccines against typhoid fever has been amply underscored by recent WHO reports3,4.
Development of effective vaccines against Salmonella diseases is challenging due to its complex lifecycle11. The facultative intracellular pathogen enters the host by crossing intestinal epithelial cells via Peyer’s patches and infecting monocytes, macrophages, and dendritic cells. The bacteria can then travel to the mesenteric lymph nodes (MLN), spleen, and liver via circulating phagocytes. The host defense mechanisms for clearing Salmonella involve stimulation of both the adaptive and innate immune systems12. The importance of Th1 cells was shown by depletion of IFN-γ13,14 and CD4+ T cells have been shown to play a significant role in immunity induced by Salmonella flagellin15,16 and live attenuated Salmonella17. B cell depletion studies and immunization studies in Ig-deficient mice also indicate the importance of B cells for immunity against Salmonella18,19. Salmonella during its intracellular life in macrophages also induces a modification of lipopolysaccharides (LPS) recognized by TLR-4 and triggers a downstream signaling cascade to evoke host immune response. As a result of these complexities, it is clear that potent antigens as well as adjuvants are important in potentiating the immune response and modulating its quality.
Early efforts to develop an effective vaccine against Salmonella began with whole, killed cells, which were shown to be moderately effective in field trials in the 1960s, but most countries have eliminated its use due to undesired side effects. Currently, two licensed commercial vaccines for typhoid fever, a subunit (Vi polysaccharide or Vi PS) and a live attenuated S. Typhi strain (Ty21a), are commercially available. However, the use of these vaccines is limited because of the short period of protection and lack of effectiveness in small children. The Vi PS vaccine given in a single dose provides protection for only 3 years, while the live oral vaccine Ty21a requires 3–4 doses for ~7 years of protection. For Ty21a, the limitations include requirement of large numbers (109) of bacteria, its inactivation by stomach acidity, and limited period of protection. The Ty21a vaccine cannot be used by children under the age of 6 or immunocompromised individuals. The Vi PS does not induce a switched antibody response, requires frequent boosts, and also cannot be used to immunize infants under the age of 2. Additionally, there are no licensed vaccines against S. Paratyphi A or B.
In order to address these limitations, we are developing an improved vaccine utilizing an innovative new adjuvant and antigen delivery system, gas vesicle nanoparticles (GVNPs)20,21. To select antigens for display, we conducted bioinformatic analysis of secreted proteins of S. enterica serovar Typhimurium (S. typhimurium) and S. enterica serovar Typhi (S. typhi) in pathogenicity islands 1 and 2 to map MHC-I and MHC-II binding epitopes22. In initial studies, a secreted inositol phosphate phosphatase protein, SopB, was selected and displayed on GVNPs and elicited strong immunogenic responses, partially protecting animals challenged with virulent Salmonella23.
1.3. Gas Vesicle Nanoparticles
GVNPs offer a novel antigen delivery system with powerful adjuvanting properties that is likely to enhance the prospect of an improved Salmonella vaccine. The ability of GVNPs to deliver multiple antigens, and their stability, scalability and safety, represent significant innovations. These nanoparticles are buoyant, hollow, and lemon-shaped, and are naturally produced by salt-loving nonpathogenic microorganisms, called Haloarchaea, a group of the Archaea (3rd Domain of life) lacking lipopolysaccharides (LPS)24. A large gene cluster (gvp) coding these nanoparticles has been cloned and extensively characterized20,25–29. In previous work, antigenic proteins and peptides have been successfully displayed on GVNPs, and in all cases, they were highly immunogenic in mice, with no toxic effects at either at the site of administration or systemically23,30–33. GVNPs are taken up by macrophages and processed slowly, producing strong long-lived humoral and cellular immune responses. GVNPs act as adjuvants when administered intraperitoneally or subcutaneously in mice. The nanoparticles are easily purified as a result of their buoyancy after simple osmotic cell lysis. GVNPs have a very long shelf life at room temperatures due to their high material strength, and the cost of producing large quantities of bioengineered GVNPs is low. As a result, GVNPs have great potential as an inexpensive, safe, and innovative antigen delivery and vaccine development system.
1.3.1. Properties of gas vesicle nanoparticles (GVNPs)
Our approach employs GVNPs (Figure 1) from the salt-loving haloarchaeal microbe, Halobacterium sp. NRC-121,28,34–36. GVNPs consist of a thin (20 Å) lipid-free, rigid protein membrane surrounding a gas-filled space37. GVNPs are lemon-shaped, about 300 nm long, and 150 nm in diameter, and grow from small bicones to progressively larger structures. During this process, water is excluded by hydrophobicity at the inner surface of the membrane, while ambient gases accumulate through passive diffusion. GVNPs are released by osmotic cell lysis with hypotonic conditions and may be easily purified in large quantities by centrifugally accelerated flotation (Figure 1)25. Extreme resistance of the GVNP membrane to solubilization by detergents and chaotropic agents has precluded detailed biochemical studies38. However, GVNPs have been extensively studied using genetic, proteomic and immunological approaches20,26,29,39–43.
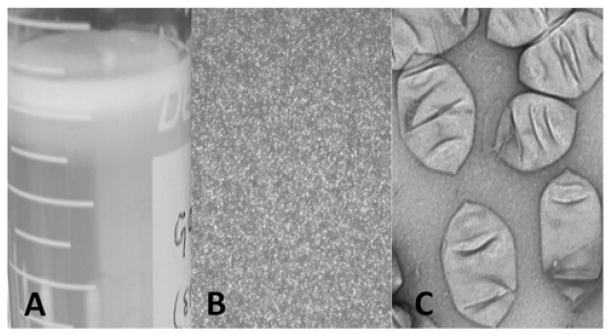
Figure 1.
Halobacterium sp. GVNPs. A. Purified by flotation; B. Observed as phase bright particles in light microscopy; C. Observed in electron microscope after negative staining20,23.
GVNPs have ideal properties as an adjuvant and antigen delivery system. They are extremely stable, yet non-toxic, and genetically engineerable for displaying proteins on their surface. Their protein composition has been studied in considerable detail using immunological probes and proteomic studies, which has shown that at least 7 proteins are present in the nanoparticles25,29,44–46. Genetic analysis has shown the involvement of 14 genes (gvpMLKJIHGFEDACNO) in GVNP biosynthesis21,27,36. Within the gvp gene cluster is the gvpC gene, which codes for the second-most abundant GVNP protein, displayed on the external surface of the nanoparticles. GvpC protein was observed in GVNPs by immunoblotting26 and gvpC mutants produced smaller GVNPs than wild-type20,27, indicating their function in strengthening the nanoparticles44,47 (our unpublished data). The NRC-1 GvpC protein contains a novel protein motif which is repeated 8 times (Figure 2), and it is likely that GvpC functions in binding to the major GvpA protein on the GVNP surface. Insertions of sequences coding ≥ 250 amino acids after the 6th GvpC repeat are tolerated without disrupting GVNP formation, providing a site for insertion of foreign antigenic sequences20,23,30–31.
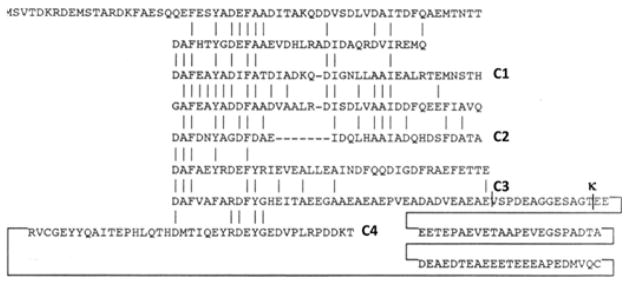
Figure 2.
The amino acid sequence of GvpC of Halobacterium sp. NRC-1 is shown with conserved residues (vertical bars) in the eight imperfect repeats. The GvpC segments used in this study are labelled C1, C2, C3 and C4 at the C-terminal end. The position of the κ site is labelled ‘κ’23.
1.3.2. Bioengineered GVNPs as adjuvant and antigen delivery system
GVNPs have advantages over live-attenuated vectors, due to reduced risk, and over conventional purified antigens, through enhanced protection48. Bioengineered GVNPs function as both adjuvant and delivery system, via presentation of antigens to the antigen presentation cells (APCs), and as immunopotentiators, increasing immunogenicity without adverse reactogenicity49. Known desirable qualities of particulate delivery systems, including the size, charge, hydrophobicity, and shape of particles, all favor GVNPs as an effective adjuvant50. The GVNPs are in the nanometer range (virus-sized), found to be ideal for cytotoxic T cell responses, including cross-presentation of vaccine antigens. GVNPs have surface charges and hydrophobic properties which have been shown to increase adjuvanting properties due to better interactions with APCs and phagocytes. The non-spherical lemon-shape of GVNPs is also expected to result in more effective adjuvanting properties. The ability to control the structure and accessibility of the GVNP surface are also desirable properties for antigen display and vaccine development.
The effectiveness of GVNPs as an adjuvant and antigen delivery system is also favored by their ability for administration via multiple routes. While traditional human vaccines have been administered by needle injection, parenteral delivery mainly induces circulating antibodies49,50. The ability to administer GVNPs via alternate administration routes, particularly mucosal administration, may reduce cost and improve compliance, while simultaneously increasing mucosal immunity without reducing systemic immunity. In the case of preventing illness resulting from the Salmonella pathogen, oral administration employing GVNPs seems highly desirable from both the convenience and effectiveness standpoints.
2. Experimental approach and findings
2.1. Salmonella antigen display on GVNPs and immunological testing
The Salmonella enterica antigen SopB was selected for GVNP-display and testing8,20,23,51. For this work, SopB4 and B5 (amino acids 300-400 and 395-561, respectively) were synthesized as codon optimized-gene fragments, using information from the fully sequenced Halobacterium sp. NRC-1 genome to replace rare codons with frequently used codons52. The synthetic fragments were cloned in the expression plasmid (pSD104) into a unique AfeI site in the plasmid gvpC gene25,30. The resulting plasmids, pSDsopB4 and pSDsopB5, contained the respective SopB coding regions inserted in-frame into the gvpC gene (Figure 3). These plasmids were transformed into Halobacterium strain SD109, which has the entire gvp gene cluster deleted53.
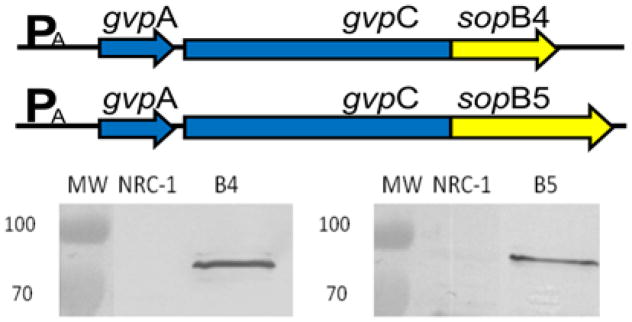
Figure 3.
(Upper panel) Salmonella SopB4 and B5-gvpC3 fusion genes used for expression in Halobacterium. Western blotting detection of GvpC-SopB fusion proteins in GVNPs purified from wild-type NRC-1, SD109 (pSDsopB4), and SD109 (pSDsopB5) probed with antisera against SopB4 (lower left panel) and SopB5 (lower right panel)23.
To determine if the gvpC-sopB4 and gvpC-sopB5 fusion genes were expressed in Halobacterium, transformed strains were grown under mevinolin selection on agar plates, and colonies examined for GVNP production. Transformants containing pSDsopB4 or pSDsopB5 were lysed by exposure to hypotonic conditions, and GVNPs were isolated by centrifugally accelerated flotation. The nanoparticle proteins were fractionated by SDS-PAGE and antigenic proteins were identified by Western blotting analysis using rabbit polyclonal antisera prepared against synthetic peptides corresponding to SopB4 and B5 (Figure 3) or GvpC8,23,29. Proteins of expected sizes were observed in the Western blots of purified GVNPs from the Halobacterium expression strains, SD109 (pSDsopB4) and SD109 (pSDsopB5), but were absent in GVNPs from NRC-1, as expected, confirming production of the fusion proteins and their presence on the GVNPs.
To determine the effect of Salmonella SopB4 and B5 antigens displayed on GVNPs, we immunized mice intraperitoneally (i.p.) with 103 colony forming units (cfu) of the S. Typhimurium attenuated strain followed by two boosts with 100 μg each of SopB4, B5, or wild-type NRC-1 GVNPs respectively, 1 and 2 weeks post immunization. Cytokine responses were determined for each cohort of mice, and the most notable changes found were found for mice immunized with SopB5-GVNPs. Serum levels of IFN-γ, GM-CSF, and IL-2, all of which are Th1 cytokines, were found to be significantly increased in post-SopB5-GVNP-immunization sera compared to pre-immunization sera, indicating that the Th1 pathway involved in response to intracellular bacterial pathogens is induced by these GVNPs23 (Figure 4A).
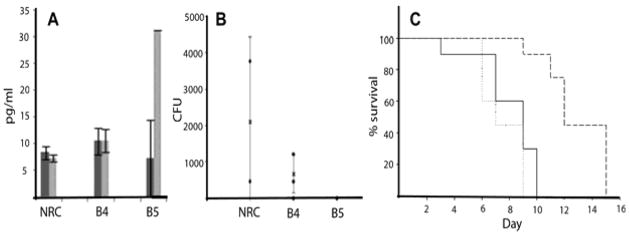
Figure 4.
Effectiveness of Salmonella SopB4 and B5-GVNPs for immunogenicity and protection in a mouse model. A. Serum IFN-γ levels in pre- (dark gray) and post- (light gray) immunized animals; B: Bacterial load in spleen of GVNP-immunized and pathogen challenged animals, and C. Survival after pathogen challenge (PBS-solid line, NRC-1 GVNPs-dotted line and SopB5-GVNPs-dashed line)23.
Following immunizations and boosting, mice were challenged orally with 107 cfu of wild-type Salmonella, and the MLN, liver, and spleen were isolated one week post-challenge. Bacterial loads were determined for each organ by plating dilutions and counting cfu. In each of the three organs, bacterial cfu were lower by at least two orders of magnitude for mice immunized with SopB5-GVNP23 (Figure 4B). Bacterial load in the spleen was also significantly reduced in SopB4-GVNP immunized mice. CD4+ T-cells were also found to be elevated by 2–4-fold in the spleen of mice immunized with SopB5-GVNPs compared to wild-type23. Further challenge experiments were conducted to determine protection and mice immunized with SopB5-GVNP survived 3–5 days longer than those immunized with wild-type NRC-1 (Figure 4C).
2.2. Stability of GVNPs in Salmonella vaccine producing strains
Halobacterium strains containing SopB-GVNPs were tested over several months for stability of the GvpC-SopB proteins using Western blotting assays. The recombinant GVNPs were found to be stable over several months at room temperature, as well as when exposed to high temperatures of 50°C, with little to no degradation of the Salmonella antigen displayed on the nanoparticles23 (Figure 5). Thus, our results clearly showed that the GVNP-antigen display system is shelf-stable without the need for refrigeration, even under tropical summer conditions.
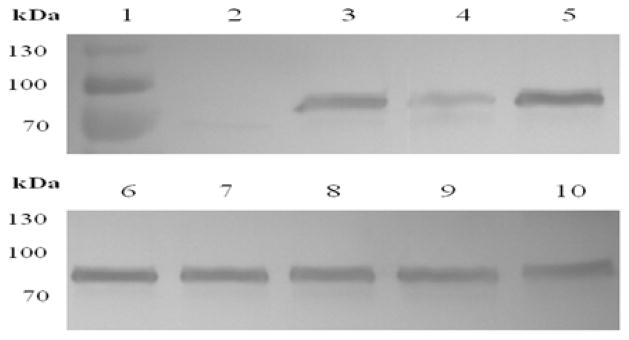
Figure 5.
NRC-1, lanes 2–10: Halobacterium sp. SD109 (SopB4). Top: A culture at room temperature over time, Bottom: Lysates exposed to 50°C over time; Lane 1: Molecular weight marker, 2: 8 months, 3: 8 months, 4: 4 months, 5: 7 days, 6: 6h, 7: 12h, 8: 24h, 9: 72h, 10: 120h.
2.3. Improvement of the GVNP bioengineering system
The genetic system used for bioengineering of SopB-GVNPs employed a relatively large plasmid, pSD104, containing the entire gvp gene28. In order to facilitate future bioengineering of nanoparticles, we constructed a series of smaller, more versatile plasmid expression vectors containing gvpC, as well as a gvpC deletion host strain20(Figure 6). The Halobacterium sp. NRC-1Δura3ΔgvpC deletion strain, constructed using the ura3-based gene deletion method54, showed a partially gas vesicle-deficient phenotype with small, mainly lemon-shaped gas vesicles. We next constructed a gvpC expression vector series using pMC2, an expression plasmid with the cold-inducible cspD2 promoter, and pDRK, an expression plasmid incorporating the higher-level gvpA promoter55,56. A series of PCR amplified gvpC gene fragments (C-series) were then inserted into the expression plasmids to test their ability to bind to GVNPs and complement nanoparticle production. The plasmids were transformed into the ΔgvpC deletion strain and the results showed correlation between progressively longer gvpC fragments and larger size GVNPs20.
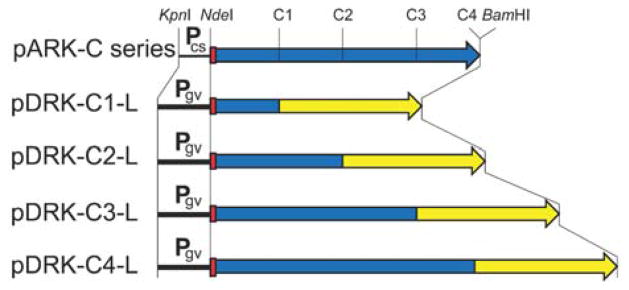
Figure 6.
The upper map (pARK-C series) displays the KpnI-BamHI region of the pARK-C series plasmids, with the cspD2 promoter labelled Pcs, His-tag shown as red box, and gvpC gene (with C1, C2, C3, and C4 regions marked as in Figure 2) shown as blue arrow. The four lower maps (labelled pDRK-C1-L to C4-L) show the KpnI-BamHI regions of pDRK-C-L plasmid series containing the gvpA promoter (labelled Pgv), His-tag (red box), C1, C2, C3, and C4 regions of gvpC (blue boxes), and codon optimized Gaussia princeps luciferase gene (yellow arrow). The corresponding sites of KpnI, NdeI, and BamHI cleavage are indicated, while the AfeI sites at the GvpC-luciferase gene boundaries are not shown20.
Next, a codon-optimized synthetic gene coding luciferase from the marine copepod Gaussia princeps was fused to the gvpC gene fragments on the expression plasmid and resulted in production of the chemiluminescence protein on GVNPs20 (Figure 7). When inserted together with a wild-type gvpC gene, we obtained GVNPs displaying both of GvpC and GvpC-luciferase fusion proteins. This finding confirmed that multiple GvpC protein types may be expressed simultaneously on the surface of GVNPs, providing the capability to express multiple antigenic proteins and delivering multivalent GVNP vaccines in a single strain. This system also opens the potential for expression of other therapeutic proteins on GVNPs for applications to drug delivery.
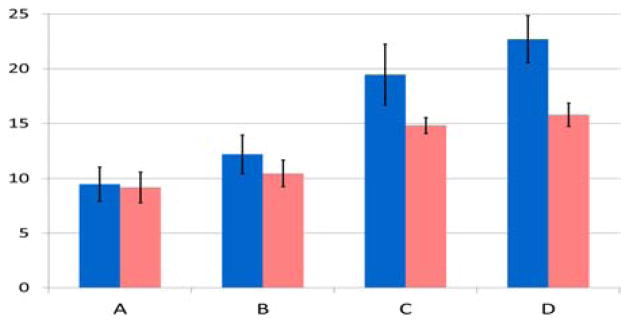
Figure 7.
Luciferase activity in purified GVNPs from Halobacterium sp. NRC-1Δura3ΔgvpC and Halobacterium sp. NRC-1 strains containing gvpC and luciferase expression plasmids. Percent luciferase activity, chemiluminescence activity detected in gas vesicles compared to total activity observed in cell lysates, is plotted on vertical axis for pDRK-C-L plasmid series. Values plotted are the average of triplicate experiments, and standard deviation is shown with error bars. A: pDRK-C1-L, B: pDRK-C2-L, C: pDRK-C3-L, D: pDRK-C4-L in either Halobacterium sp. NRC-1Δura3ΔgvpC (blue), or in wild-type Halobacterium sp. NRC-1 (pink) strain20.
3. Conclusion
Salmonella pathogens remain important causes of morbidity and mortality due to the lack of an effective, long-lasting vaccine. Novel gas vesicle nanoparticles (GVNPs) produced by extremophilic Halobacterium sp. NRC-1 are being used to develop an improved vaccine against Salmonella pathogens that is both inexpensive and effective57. When administered to animals, GVNPs have previously been shown to produce strong long-lived humoral and cellular immune responses against surface-displayed antigens. In this study, a secreted protein conserved in Salmonella enterica pathogens, SopB, was displayed by genetic fusion to GvpC, a GVNP surface proteins, and the bioengineered SopB-GVNPs were found to be highly immunogenic in mice. Our results showed that animals boosted with SopB-GVNPs stimulated IgGs, proinflammatory cytokines, and CD4+ T cells, and resulted in reduced bacterial load in key organs. Additionally, the vaccine-producing strains were found to be shelf-stable over months when stored at room temperature and at elevated temperatures. We also developed an improved GVNP expression host and plasmid series and demonstrated its potential by displaying the Gaussia princeps luciferase reporter. This work opens the possibility of delivery of therapeutic proteins displayed on GVNPs.
Acknowledgments
This work was supported by National Institutes of Health grant R03 AI107634 and Bill & Melinda Gates Foundation grant OPP1061509 to S.D., and by the Director of the Indian Institute of Science, Bangalore, India and the Department of Biotechnology (DBT 311, DBT197 and DBT 172), Life Science Research Board (LSRB0008) and DBT-IISc partnership program for advanced research in biological sciences and bioengineering to D.C.
References
- 1.Hohmann EL. Nontyphoidal salmonellosis. Clin Infect Dis. 2001;32:263–269. doi: 10.1086/318457. [DOI] [PubMed] [Google Scholar]
- 2.Mastroeni P, Maskell D, editors. Salmonella infections. Clinical, immunological, and molecular aspects. Advances in Molecular and Cellular Microbiology. Vol. 9. Cambridge: Cambridge University Press; 2006. [Google Scholar]
- 3.WHO. Typhoid vaccines. Wkly Epidemiol Rec. 2000;75:257–264. [PubMed] [Google Scholar]
- 4.WHO. Typhoid vaccines. Wkly Epidemiol Rec. 2008;83:49–58. [PubMed] [Google Scholar]
- 5.Crump JA, Luby SP, Mintz ED. The global burden of typhoid fever. Bull World Health Organ. 2004;82:346–253. [PMC free article] [PubMed] [Google Scholar]
- 6.Helms M, Ethelberg S, Molbak K. International Salmonella Typhimurium DT104 infections, 1992–2001. Emerg Infect Dis. 2005;11:859–867. doi: 10.3201/eid1106.041017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poppe C, Smart N, Khakhria R, Johnson W, Spika J, Prescott J. Salmonella Typhimurium DT104: a virulent and drug-resistant pathogen. Can Vet J. 1998;39:559–565. [PMC free article] [PubMed] [Google Scholar]
- 8.Nagarajan AG, Karnam G, Lahiri A, Allam US, Chakravortty D. Reliable means of diagnosis and serovar determination of blood-borne Salmonella strains: quick PCR amplification of unique genomic loci by novel primer sets. J Clin Microbiol. 2009a;47:2435–2441. doi: 10.1128/JCM.00327-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Popoff MY, Bockemühl J, Gheesling LL. Supplement 2002 (no. 46) to the Kauffmann-White scheme. Res Microbiol. 2004;155:568–570. doi: 10.1016/j.resmic.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 10.Baumler AJ, Tsolis RM, Ficht TA, Adams LG. Evolution of host adaptation in Salmonella enterica. Infect Immun. 1998;66:4579–4587. doi: 10.1128/iai.66.10.4579-4587.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haraga A, Ohlson MB, Miller SI. Salmonellae interplay with host cells. Nat Rev Microbiol. 2008;6:53–66. doi: 10.1038/nrmicro1788. [DOI] [PubMed] [Google Scholar]
- 12.Lahiri A, Das P, Chakravortty D. Engagement of TLR signaling as adjuvant: towards smarter vaccine and beyond. Vaccine. 2008;26:6777–6783. doi: 10.1016/j.vaccine.2008.09.045. [DOI] [PubMed] [Google Scholar]
- 13.Nauciel C. Role of CD4+ T cells and T-independent mechanisms in acquired resistance to Salmonella typhimurium infection. J Immunol. 1990;145:1265–1269. [PubMed] [Google Scholar]
- 14.Nauciel C, Espinasse-Maes F. Role of gamma interferon and tumor necrosis factor alpha in resistance to Salmonella typhimurium infection. Infect Immun. 1992;60:450–454. doi: 10.1128/iai.60.2.450-454.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ravindran R, McSorley SJ. Tracking the dynamics of T-cell activation in response to Salmonella infection. Immunol. 2005;114:450–458. doi: 10.1111/j.1365-2567.2005.02140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraser A, Paul M, Goldberg E, Acosta CJ, Leibovici L. Typhoid fever vaccines: systematic review and meta-analysis of randomised controlled trials. Vaccine. 2007;25:7848–7857. doi: 10.1016/j.vaccine.2007.08.027. [DOI] [PubMed] [Google Scholar]
- 17.Lundin BS, Johansson C, Svennerholm AM. Oral immunization with a Salmonella enterica serovar typhi vaccine induces specific circulating mucosa-homing CD4(+) and CD8(+) T cells in humans. Infect Immun. 2002;70:5622–5627. doi: 10.1128/IAI.70.10.5622-5627.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mastroeni P, Simmons C, Fowler R, Hormaeche CE, Dougan G. Igh-6(−/−) (B cell-deficient) mice fail to mount solid acquired resistance to oral challenge with virulent Salmonella enterica serovar typhimurium and show impaired Th1 T-cell responses to Salmonella antigens. Infect Immun. 2000;68:46–53. doi: 10.1128/iai.68.1.46-53.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mittrucker HW, Raupach B, Kohler A, Kaufmann SH. Cutting edge: role of B lymphocytes in protective immunity against Salmonella typhimurium infection. J Immunol. 2000;164:1648–1652. doi: 10.4049/jimmunol.164.4.1648. [DOI] [PubMed] [Google Scholar]
- 20.DasSarma S, Karan R, DasSarma P, Barnes S, Ekulona F, Smith B. An improved genetic system for bioengineering buoyant gas vesicle nanoparticles from Haloarchaea. BMC Biotechnol. 2013;13:112. doi: 10.1186/1472-6750-13-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shively JM, Cannon GC, Heinhorst S, Bryant DA, DasSarma S, Bazylinski D, Preiss J, Steinbüchel A, Docampo R, Dahl C. Encyclopedia of Life Sciences. London: Wiley; 2011. Bacterial and Archaeal Inclusions. [DOI] [Google Scholar]
- 22.Nagarajan AG, Balasundaram SV, Janice J, Karnam G, Eswarappa SM, Chakravortty D. sopB of Salmonella enterica serovar Typhimurium is a potential DNA vaccine candidate in conjugation with live attenuated bacteria. Vaccine. 2009b;27:2804–2811. doi: 10.1016/j.vaccine.2009.02.092. [DOI] [PubMed] [Google Scholar]
- 23.DasSarma P, Negi VD, Balakrishnan A, Karan R, Barnes S, Ekulona F, Chakravortty D, DasSarma S. Haloarchaeal gas vesicle nanoparticles displaying Salmonella SopB antigen reduce bacterial burden when administered with live attenuated bacteria. Vaccine. 2014;32:4543–4549. doi: 10.1016/j.vaccine.2014.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DasSarma S, Coker JC, DasSarma P. Archaea-overview. In: Schaechter M, editor. Desk encyclopedia of microbiology. 2. San Diego, CA: Academic Press; 2010. pp. 118–139. [Google Scholar]
- 25.Halladay JT, Ng W-L, DasSarma S. Genetic transformation of a halophilic archaebacterium with a gas vesicle gene cluster restores its ability to float. Gene. 1992;119:131–136. doi: 10.1016/0378-1119(92)90078-4. [DOI] [PubMed] [Google Scholar]
- 26.Halladay JT, Jones JG, Lin F, MacDonald AB, DasSarma S. The rightward gas vesicle operon in Halobacterium halobium plasmid pNRC-100: identification of the gvpA and gvpC gene products by use of antibody probes and genetic analysis of the region downstream of gvpC. J Bacteriol. 1993;175:684–692. doi: 10.1128/jb.175.3.684-692.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DasSarma S, Arora P, Lin F, Molinari E, Yin LR. Wild-type gas vesicle formation requires at least ten genes in the gvp gene cluster of Halobacterium halobium plasmid pNRC100. J Bacteriol. 1994;176:7646–52. doi: 10.1128/jb.176.24.7646-7652.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DasSarma S, Arora P. Genetic analysis of the gas vesicle gene cluster in haloarchaea. FEMS Microbiol Lett. 1997;153:1–10. [Google Scholar]
- 29.Shukla HD, DasSarma S. Complexity of gas vesicle biogenesis in Halobacterium sp. strain NRC-1: identification of five new proteins. J Bacteriol. 2004;186:3182–3186. doi: 10.1128/JB.186.10.3182-3186.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stuart ES, Morshed F, Sremac M, DasSarma S. Antigen presentation using novel particulate organelles from halophilic archaea. J Biotechnol. 2001;88:119–128. doi: 10.1016/s0168-1656(01)00267-x. [DOI] [PubMed] [Google Scholar]
- 31.Stuart ES, Morshed F, Sremac M, DasSarma S. Cassette-based presentation of SIV epitopes with recombinant gas vesicles from halophilic archaea. J Biotechnol. 2004;114:225–237. doi: 10.1016/j.jbiotec.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 32.Sremac M, Stuart ES. Recombinant gas vesicles from Halobacterium sp. displaying SIV peptides demonstrate biotechnology potential as a pathogen peptide delivery vehicle. BMC Biotechnol. 2008;8:9. doi: 10.1186/1472-6750-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sremac M, Stuart ES. SIVsm Tat, Rev, and Nef1: functional characteristics of r-GV internalization on isotypes, cytokines, and intracellular degradation. BMC Biotechnol. 2010;10:54. doi: 10.1186/1472-6750-10-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DasSarma S, DasSarma P. Encyclopedia of Life Sciences. London: Wiley; 2012. Halophiles. [DOI] [Google Scholar]
- 35.Walsby AE. Gas vesicles. Micro Rev. 1994;58:94–144. doi: 10.1128/mr.58.1.94-144.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfeifer F. Distribution, formation, and regulation of gas vesicles. Nature Rev Microbiol. 2012;10:705. doi: 10.1038/nrmicro2834. [DOI] [PubMed] [Google Scholar]
- 37.McMaster TJ, Miles MJ, Walsby AE. Direct observation of proteins secondary structure in gas vesicles by atomic force microscopy. Biophys J. 1996;70:2432–2436. doi: 10.1016/S0006-3495(96)79813-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Belenky M, Meyers R, Herzfeld J. Subunit structure of gas vesicles: a MALDI-TOF mass spectrometry study. Biophys J. 2004;86:499–505. doi: 10.1016/S0006-3495(04)74128-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DasSarma S, Fleischmann EM, editors. Archaea: a laboratory manual – Halophiles. NY: Cold Spring Harbor press; 1995. [Google Scholar]
- 40.Ng W-L, Ciufo SA, Smith TM, Bumgardner RE, Baskin D, Faust J, Hall B, Loretz C, Seto J, Slagel J, Hood L, DasSarma S. Snapshot of a large dynamic replicon from a halophilic Archaeon: Megaplasmid or minichromosome? Genome Res. 1998;8:1131–1141. doi: 10.1101/gr.8.11.1131. [DOI] [PubMed] [Google Scholar]
- 41.Ng WV, Kennedy SP, Mahairas GG, Berquist B, Pan M, Shukla HD, Lasky SR, Baliga N, Thorsson V, Sbrogna J, Swartzell S, Weir D, Hall J, Dahl TA, Welti R, Goo YA, Leithauser B, Keller K, Cruz R, Danson MJ, Hough DW, Maddocks DG, Jablonski PE, Krebs MP, Angevine CM, Dale H, Isenbarger TA, Peck RF, Pohlschroder M, Spudich JL, Jung K-H, Alam M, Freitas T, Hou S, Daniels CJ, Dennis PP, Omer AD, Ebhardt H, Lowe TM, Liang P, Riley M, Hood L, DasSarma S. Genome sequence of Halobacterium species NRC-1. Proc Natl Acad Sci USA. 2000;97:12176–12181. doi: 10.1073/pnas.190337797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DasSarma S. Genome sequence of an extremely halophilic archaeon. In: Fraser C, Read T, Nelson KE, editors. Microbial Genomes. Totowa, NJ: Humana Press; 2004. pp. 383–399. [Google Scholar]
- 43.Berquist BR, Müller JA, DasSarma S. Genetic Systems for Halophilic Archaea. Meth Microbio. 2006;35:649–680. [Google Scholar]
- 44.Walsby AE, Hayes PK. The minor cyanobacterial gas vesicle protein. GVPc, is attached to the outer surface of the gas vesicle. J Gen Microbiol. 1988;134:2647–2657. [Google Scholar]
- 45.Chu LJ, Chen MC, Setter J, Tsai YS, Yang H, Fang X, Ting YS, Shaffer SA, Taylor GK, von Haller PD, Goodlett DR, Ng WV. New structural proteins of Halobacterium salinarum gas vesicle revealed by comparative proteomics analysis. J Proteome Res. 2011;10:1170–1178. doi: 10.1021/pr1009383. [DOI] [PubMed] [Google Scholar]
- 46.Xu B-Y, Dai Y-N, Zhou K, Liu Y-T, Sun Q, Ren Y-M, Chen Y, Zhou C-Z. Structure of the gas vesicle protein GvpF from the cyanobacterium Microcystis aeruginosa. Acta Cryst. 2014;D70:3013–3022. doi: 10.1107/S1399004714021312. [DOI] [PubMed] [Google Scholar]
- 47.Kinsman R, Walsby AE, Hayes PK. GvpCs with reduced numbers of repeating sequence elements bind to and strengthen cyanobacterial gas vesicles. Mol Microbiol. 1995;17:147–154. doi: 10.1111/j.1365-2958.1995.mmi_17010147.x. [DOI] [PubMed] [Google Scholar]
- 48.Perrie Y, Mohammed AR, Kirby DJ, McNeil SE, Bramwell VW. Vaccine adjuvant systems: Enhancing the efficacy of sub-unit protein antigens. Int J Pharm. 2008;364:272–280. doi: 10.1016/j.ijpharm.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 49.Storni T, Kqndig TM, Senti G, Johansen P. Immunity in response to particulate antigen-delivery systems. Adv Drug Deliv Rev. 2005;57:333–355. doi: 10.1016/j.addr.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 50.Foged C. Subunit vaccines of the future: the need for safe, customized and optimized particulate delivery systems. Ther Del. 2011;2:1057–1077. doi: 10.4155/tde.11.68. [DOI] [PubMed] [Google Scholar]
- 51.Negi VD, Singhamahapatra S, Chakravortty D. Salmonella enterica serovar Typhimurium strain lacking pmrG-HM-D provides excellent protection against salmonellosis in murine typhoid model. Vaccine. 2007;25:5315–5323. doi: 10.1016/j.vaccine.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 52.Kennedy SP, Ng WV, Salzberg SL, Hood L, DasSarma S. Understanding the adaptation of Halobacterium species NRC-1 to its extreme environment through computational analysis of its genome sequence. Genome Res. 2001;11:1641–1650. doi: 10.1101/gr.190201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ng W-L, Arora P, DasSarma S. Large deletions in class III gas vesicle-deficient mutants of Halobacterium halobium. Syst Appl Microbiol. 1994;16:560–568. [Google Scholar]
- 54.Peck RF, DasSarma S, Krebs MP. Homologous gene knockout in the archaeon Halobacterium salinarum with ura 3 as a counterselectable marker. Mol Microbiol. 2000;35:667–676. doi: 10.1046/j.1365-2958.2000.01739.x. [DOI] [PubMed] [Google Scholar]
- 55.Karan R, Capes MD, DasSarma P, DasSarma S. Cloning, overexpression, purification, and characterization of a polyextremophilic β-galactosidase from the Antarctic haloarchaeaon Halorubrum lacusprofundi. BMC Biotechnol. 2013;13:3. doi: 10.1186/1472-6750-13-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karan R, DasSarma P, Balcer-Kubiczek E, Weng RR, Liao C-C, Goodlett DR, Ng WV, DasSarma S. Bioengineering radioresistance by overproduction of RPA, a mammalian-type single-stranded DNA-binding protein, in a halophilic archaeon. Appl Microbio Biotech. 2014;98:1737–1747. doi: 10.1007/s00253-013-5368-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DasSarma S, Berquist BR, Coker JA, DasSarma P, Muller JA. Post-genomics of the model haloarchaeon Halobacterium sp. NRC-1 Saline Systems. 2006;2:3. doi: 10.1186/1746-1448-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]