Abstract
Recent genome sequencing efforts have led to the rapid accumulation of uncharacterized or “orphaned” secondary metabolic biosynthesis gene clusters (BGCs) in public databases. This increase in DNA-sequenced big data has given rise to significant challenges in the applied field of natural product genome mining, including (i) how to prioritize the characterization of orphan BGCs, and (ii) how to rapidly connect genes to biosynthesized small molecules. Here we show that by correlating putative antibiotic resistance genes that encode target-modified proteins with orphan BGCs, we predict the biological function of pathway specific small molecules before they have been revealed in a process we call target-directed genome mining. By querying the pan-genome of 86 Salinispora bacterial genomes for duplicated house-keeping genes co-localized with natural product BGCs, we prioritized an orphan polyketide synthase-nonribosomal peptide synthetase hybrid BGC (tlm) with a putative fatty acid synthase resistance gene. We employed a new synthetic double-stranded DNA-mediated cloning strategy based on transformation-associated recombination to efficiently capture tlm and the related ttm BGCs directly from genomic DNA and to heterologously express them in Streptomyces hosts. We show the production of a group of unusual thiotetronic acid natural products, including the well-known fatty acid synthase inhibitor thiolactomycin that was first described over 30 years ago, yet never at the genetic level in regards to biosynthesis and auto-resistance. This finding not only validates the target-directed genome mining strategy for the discovery of antibiotic producing gene clusters without a priori knowledge of the molecule synthesized, but also paves the way for the investigation of novel enzymology involved in thiotetronic acid natural product biosynthesis.
Keywords: Orphan biosynthetic gene cluster, Drug discovery, Fatty acid synthase inhibitor, Resistance
Microbial natural products are of paramount biomedical importance, serving as antibiotics against a variety of pathogenic bacteria.1, 2 The profound emergence of bacterial resistance over the last several decades has marginalized many important antibiotics, thereby necessitating the need for new antibiotic discovery.3 Antibiotics from microbes are directly linked to clusters of genes that code for proteins associated with biosynthesis, resistance, regulation and transport. The ability to connect natural antibiotics to gene clusters and vice versa, along with ever-increasing knowledge of biosynthetic machineries, has spawned a new field of natural product genome mining for the rational discovery of new chemical entities.4-6 At the same time, DNA sequence data from a great variety of microbial genomes and environmental metagenomes has rapidly accumulated in public databases through sophisticated sequencing technologies, with more than 4,000 complete and 35,000 draft sequences of prokaryotic genomes in the NCBI database as of August 2015. Recently, an in silico bioinformatic analysis of 1,154 prokaryotic genome sequences predicted a total of 33,351 putative natural product biosynthetic gene clusters (BGCs),7 of which the vast majority could be considered “orphan” in that they could not be bioinformatically linked to the small molecules they produce. Thus, there is now considerable interest in prioritizing and developing new methods to study the products of these orphan BGCs, especially those with antibiotic activity.5
One of the significant challenges in the field of natural product genome mining is how to prioritize BGCs when mining for desired bioactivity, without prior knowledge of the biological targets of the compounds produced.8 Current genome mining strategies are often based on established biosynthetic information for known microbial metabolites,9-12 limiting the potential for linking chemistry with biology. Once potential orphan BGCs are prioritized for further investigation, rapidly connecting genes to molecules becomes another challenging task.8 Current strategies fall into two categories: (i) metabolic profiling coupled with mutagenesis of orphan genes13, 14 and (ii) heterologous expression of whole biosynthetic pathways in a well-established host.15, 16 The first approach relies heavily on the ability to genetically manipulate individual microbial genera, and is often not applicable to genera without amenable genetic approaches or in microbes that presently cannot be cultured. In contrast, the heterologous expression of gene clusters, usually in a well-investigated and genetically amenable host, is often more facile and practical. However, this demands cloning of the entire BGC into suitable expression vectors by laborious traditional cloning methods.8
Genes encoding self-resistance mechanisms are a characteristic trait associated with antibiotic-producing bacteria.17, 18 Self-resistance features are generally less favorable for host growth and survival, and are thus only expressed concurrently with antibiotic biosynthesis.19 The most efficient way for bacteria to link these partners, and to ensure efficient co-horizontal gene transfer, is to include the resistance gene within or adjacent to the corresponding antibiotic BGC.17 Host bacteria have evolved several resistance strategies to avoid self-toxicity, including product modification, binding and export, and target modification.20 Of these mechanisms, target modification uniquely correlates an antibiotic to its mode of action. The antibiotics novobiocin (gyrase B),21 platensin (FabB/F),22 and griselimycin (DnaN),23 for instance, represent a few examples in which target-duplicated resistance genes are co-clustered with BGCs (Supporting Information Figure 1). With the notion in mind that antibiotic-producing bacteria often duplicate and mutate genes encoding targeted proteins to confer resistance, we reasoned that identifying putative resistance genes within BGCs would provide insight to the molecular targets of BGC chemical products prior to their isolation and structure elucidation.
The marine actinomycete genus Salinispora has proven to be a remarkably prolific source of structurally diverse and biologically active secondary metabolites.24 These compounds span virtually all known biosynthetic classes, including the beta-lactone proteasome inhibitor salinosporamide A25 and the polyketide cytotoxin lomaiviticin A.26 Recently, high quality draft genomes of 75 Salinispora strains revealed 124 discrete nonribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) BGCs that unmasked fundamental information about the evolution and distribution of secondary metabolite gene clusters in bacteria.27 Here we use Salinispora as a model organism to showcase target-directed genome mining for prioritizing orphan antibiotic BGCs, allowing us, for the first time, to systematically link two unusual PKS-NRPS hybrid BGCs to a series of structurally rare thiotetronic acid fatty acid synthase inhibitors.
RESULTS AND DISCUSSION
Mining Salinispora BGCs for Putative Antibiotic Resistance Genes
We queried the genomes of 86 Salinispora strains (10 S. tropica, 43 S. arenicola and 33 S. pacifica) for putative target modifying resistance genes associated with natural product BGCs. First, we identified groups of related protein coding genes, or orthologous groups (OGs), using the program OrthoMCL,28 which revealed a total of 12,372 OGs within the Salinispora pan-genome (Figure 1a). Of these, 2707 OGs were conserved across all Salinispora strains analyzed and thus comprise the Salinispora core-genome. These core (house-keeping) genes could be further delineated into 1390 unique clusters of orthologous groups (COGs), each of which could be assigned a generalized function. We next searched the pan-genome for additional OGs with the same COG numbers as those found in the core-genome, which suggests they have been duplicated or were acquired by horizontal gene transfer. We detected 2393 duplicate OGs and, among these, 912 (~38%) were associated with BGCs previously identified in the Salinispora strains.27 These OGs were sorted based on COG categories to assess their potential role in resistance and to identify BGCs that may encode small molecules that act on specific targets of interest (Figure 1b and Supporting Information Methods). As proof of principle, this approach correctly identified the duplicated 20S proteasome β-subunit gene within the sal BGC, which confers resistance to the anticancer agent salinosporamide A in S. tropica.29
Figure 1.
Bioinformatic identification of candidate resistance genes. (a) The workflow for identification of duplicated orthologous groups (OGs) associated with biosynthetic gene cluster (BGC) from 86 Salinispora strains: 1. Application of OrthoMCL to 86 Salinispora genomes resulted in the identification of 12,372 OGs, representing the pan-genome of the strains in the present study; 2. From the 12,372 OGs, application of custom python scripts identified 2707 OGs that are shared among all 86 strains (core genome). The core genome is comprised of 1390 unique COG numbers. Python scripts matched these unique COG numbers to identical COG numbers in the pan-genome, identifying a total of 2393 OGs (duplicate OGs). 3. Application of PHP scripts for identifying duplicated OGs associated with predicted BGCs, resulted in the identification of 912 OGs from 20 COG categories being associated with BGCs. 4. 103 of the 912 OGs were affiliated with COG category “I” (lipid transport and metabolism). These OGs were annotated by BlastP searches, identifying one OG with homology to FabB/F, a known platensimycin and platencin resistance gene. This OG is located within the BGC PKS44 (tlm). (b) Distribution of the total number of duplicated OGs (yellow) and the OGs located within predicted BGC boundaries (green) among the COG categories.
Among the COG categories assigned to the duplicate OGs, we were particularly focused on identifying BGCs with the potential to produce compound(s) that inhibit bacterial fatty acid synthesis. The bacterial fatty acid synthase (FASII) is considered an attractive target for antibacterial drug discovery,30-32 as bacterial FASs are significantly distinct from the large, multifunctional type I synthases of mammals. Thus, we focused our attention on analyzing the BGCs linked to duplicate OGs assigned to COG category “I” of lipid transport and metabolism. BLAST analysis of the 103 “I” category OGs identified 44 related to core enzymes involved in bacterial fatty acid synthesis, including 18 similar to fabG (ketone reduction), 12 similar to fabB/F (elongation), 6 similar to fabI (enone reduction), 5 similar to fabH (chain initiation), and 3 similar to fabD (malonyl transacylation) (Supporting Information Table 1). We reasoned that while many of these genes likely encode biosynthetic enzymes important for the construction of lipid components to natural products, others could be associated with antibiotic resistance. In particular, Salin8269 (from S. pacifica CNS-863), a homolog of FabB/F, notably showed very high amino acid sequence similarity to the characterized self-resistant proteins PtmP3 (65%/79%, identity/similarity-) and PtnP3 (65%/79%, identity/similarity) from BGCs associated with the hybrid diterpenoid FASII inhibitors platensimycin and platencin,22 respectively (Supporting Information Figure 2). Thus, we suspected that Salin8269 could serve a role in antibiotic resistance rather than natural product biosynthesis. The associated BGC, PKS44, is classified as a non-canonical hybrid PKS-NRPS and not linked to diterpeniod production, as is the case for platensimycin and platencin,33 which prompted us to investigate this orphan BGC.
Discovery of the Thiolactomycin (tlm) Gene Cluster by Direct Cloning and Heterologous Expression
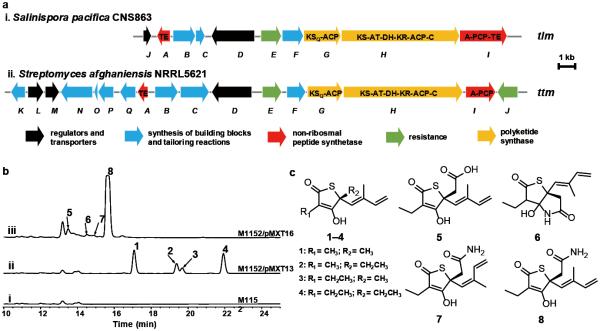
The PKS44 gene cluster was detected in four S. pacifica strains isolated from Fiji (CNS-863, CNS-996, CNT-045 and CNS-860)27 (Supporting Information Figure 3a). Closer inspection of the gene cluster (renamed tlm) from S. pacifica CNS-863 revealed a ~22 kb PKS-NRPS hybrid gene cluster of ten open reading frames (Figure 2a). In addition to the Salin8269 FabB/F homolog TlmE, the putative resistance gene, the core region of the tlm gene cluster codes for three PKS-NRPS proteins collectively containing 11 enzymatic domains (TlmG–TlmI) and a cytochrome P450 oxidoreductase (TlmF) (Figure 2a and Supporting Information Table 2). We additionally annotated five genes upstream of the tlmE–I operon encoding a type II thioesterase (TE) (TlmA), two enzymes for generating ethylmalonyl-CoA (TlmB and TlmC), and two transcriptional regulators (TlmD and TlmJ) (Figure 2a and Supporting Information Table 2). Surprisingly, we also identified two additional S. pacifica strains (CNT-084 and CNT-609) in which tlmF-I were specifically lost from the tlm locus (Supporting Information Figure 3), suggesting these strains do not produce the tlm product but maintain the resistance phenotype putatively conferred by tlmE.
Figure 2.
Identification of orphan BGCs with an extra copy of the fabB/F housekeeping gene and their metabolites. (a) Orphan gene cluster annotation. (b) HPLC profile of extracts from (i) S. coelicolor M1152, (ii) S. coelicolor M1152/pMXT13 (tlm), and (iii) S. coelicolor M1152/pMXT16 (ttm). Detection at 239 nm. (c) Structures of thiolactomycin (TLM, 1) and analogues identified in this work.
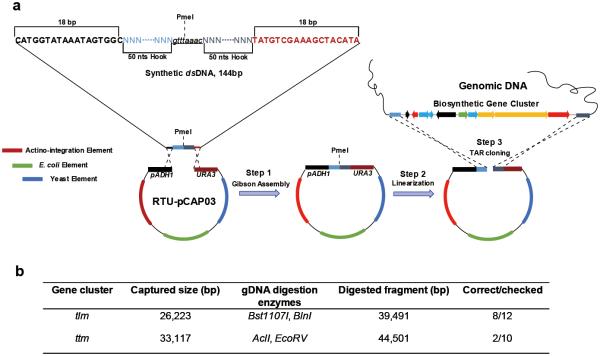
In order to identify the product(s) of the tlm gene cluster, we directly cloned an encompassing 26 kb genomic region using a synthetic double-stranded DNA-mediated (dsDNA-mediated) cloning strategy based on transformation-associated recombination (TAR) in the yeast Saccharomyces cerevisia. Although we previously developed a TAR-based platform for the direct capture of natural product BGCs from genomic DNA for targeted expression,16 the original protocol demands construction of a cluster-specific capture vector by laborious traditional cloning methods. Furthermore, we observed high levels of plasmid re-circularization due to non-homologous end joining (NHEJ), repeatedly resulting in capture rates below 2%. This inefficiency necessitated extensive screening of hundreds of colonies. To overcome these drawbacks, we revised the approach to eliminate the need for PCR for capture vector construction while introducing a counter-selectable marker to improve efficiency. We first introduced the URA3 gene under the strong promoter of the Schizosaccharomyces pombe ADH1 gene (pADH1) into pCAP01 as a counter selectable marker, to produce pCAP03 (Supporting Information Figure 4). pADH1 can tolerate an insertion of up to 130 bp between the TATA box and the transcription initiation site.34 This method allows the use of shorter capture arms and conveniently selects against NHEJ in the presence of 5-fluoroorotic acid (5-FOA).35 We then designed a synthetic dsDNA fragment that could be directly inserted into the ready-to-use pCAP03 (RTU-pCAP03, Figure 3a and Supporting Information Figure 4) using Gibson Assembly (GA).36 The 144 bp dsDNA contained two 18 bp overlapping fragments with the vector backbone for GA, two 50 bp capture hooks for recombination, and an 8 bp PmeI blunt end restriction site for linearization (Figure 3a). This approach not only greatly simplifies the procedure for constructing the capture vector, but also significantly increased the percentage of positive yeast transformants. In the capture of the tlm gene cluster, we identified eight positive clones after the screening of twelve transformants by colony PCR, and four of them were transferred into E. coli and further confirmed by restriction digestion (Figure 3b, Supporting Information Figures 5 and 6).
Figure 3.
High-efficiency TAR based direct cloning strategy for capture of natural product gene clusters. (a) The high-efficiency TAR cloning involves three steps. In step 1, the capture vector is constructed in one step by Gibson Assembly using 144 bp synthetic dsDNA. In step 2, the capture vector is linearized by the restriction enzyme PmeI. In step 3, recombination in yeast. (b) Tabulation of direct cloning parameters of two orphan gene clusters.
We introduced the resulting plasmid pMXT13 into S. coelicolor M1152 by triparental intergeneric conjugation and selected three kanamycin resistant clones. The transformed strains were cultured and extracted for HPLC analyses. The results showed that all of the S. coelicolor M1152/pMXT13 strains produced at least four new UV absorbent peaks in comparison with the untransformed S. coelicolor M1152 host (Figure 2b). These molecules were purified via preparative HPLC (Supporting Information Methods). Structural characterization by high-resolution mass spectrometry and extensive NMR spectroscopy identified the compounds as a group of previously described FASII inhibitors,37-40 including thiolactomycin (TLM, 1) and three analogues (2–4),41, 42 with 3 being isolated as a new natural product (Figure. 2c, Supporting Information Table 4 and Supporting Information Figures 7, 12, and 14–29). The production of 1–4 was additionally identified in the native producer S. pacifica CNS-863, which was aided with the results from the heterologous expression experiments (Supporting Information Figure 8).
TLM and its analogues share an unusual thiotetronic acid unit that is rare amongst natural products. The biosynthetic origin of TLM was previously reported to derive from a polyketide pathway involving acetate and three propionate building blocks and the sulfur atom from cysteine.43 Inspection of the tlm locus suggests several unconventional biosynthetic features to account for these observed metabolic substrates. An AT-independent KS-ACP (TlmG) di-domain appears to initiate polyketide chain priming with acetate and a potential iterative PKS module TlmH (KS-AT-DH-KR-ACP-C) may elongate with three branched chain malonates to construct the carbon backbone of TLM. The incorporation of sulfur from cysteine appears to originate from the NRPS TlmI (A-PCP-TE) by an unknown mechanism. Recently, Shen and co-workers established cysteine as the precursor of the sulfur atom in leinamycin and characterized two new and unrelated PKS domains involved in the transformation.44 We thus assume that the sulfur atom of the TlmI-activated cysteine residue is oxidatively removed and added to the TlmH polyketide intermediate to construct the thiotetronic acid pharmacophore.
Discovery of the Thiotetroamide (ttm) Gene Cluster by PCR-independent Cloning and Heterologous Expression
Following the successful capture and heterologous expression of the tlm gene cluster, we identified a related gene cluster (ttm) from Streptomyces afghaniensis that contained two FabB/F homologs by BLAST search (Figure 2a and Supporting Information Table 3). Like the thiolactomycin TlmE, these two proteins were similarly related to PtmP3 (TtmE, 65%/78%, identity/similarity; TtmJ, 85%/91%, identity/similarity) and PtnP3 (TtmE, 66%/78%, identity/similarity; TtmJ, 83%/91%, identity/similarity).22 The detection of two copies of the putative resistance gene led to the hypothesis that the homologous S. afghaniensis pathway may produce a more potent series of FASII antibiotics. The ttm locus is similar to the tlm gene cluster in the core structure, except that the NPRS TtmI lacks the terminal thioesterase (TE) domain in comparison to TlmI (Figure 2a and Supporting Information Table 3). Furthermore, the 17 open reading frame ttm gene cluster contains five additional biosynthetic enzymes, including a short-chain dehydrogenase (TtmK), an aminotransferase (TtmN), a ferredoxin (TtmO), an additional cytochrome p450 oxidoreductase (TtmP) and a ketoacyl-acyl carrier protein synthase III (TtmQ) (Figure 2a and Supporting Information Table 3).
In order to identify the product(s) of the homologous ttm gene cluster, we cloned a 33 kb genomic region containing the 29 kb gene cluster using the PCR-independent cloning approach, resulting in two positive colonies out of ten transformants (Figure 3b). The resulting vector pMXT16 was verified (Supporting Information Figure 6) and heterologously expressed in S. coelicolor M1152. HPLC analysis of the heterologous expression system led to the isolation and identification of a new series of TLM analogues (5–8) (Figure 2b, Supporting Information Table 5 and Supporting Information Figures 7, 13, and 30–45). Among this series, 6 and 7 are new compounds, while 8 was previously reported as Tü 301045 and 5 was generated through alkaline hydrolysis of 845 (Figure 2c). Compounds 6–8 each contain a terminal amide functionality presumably derived from the pathway specific tailoring enzymes TtmK, TtmP, and TtmN, and were accorded the trivial names thiotetroamides (TTM) A–C. In contrast, 5 contains a carboxylic acid group and is proposed as an intermediate before terminal amination, hence it was assigned the name prethiotetroamide C.
Confirmation of ttmE and ttmJ as Self-Resistance Elements
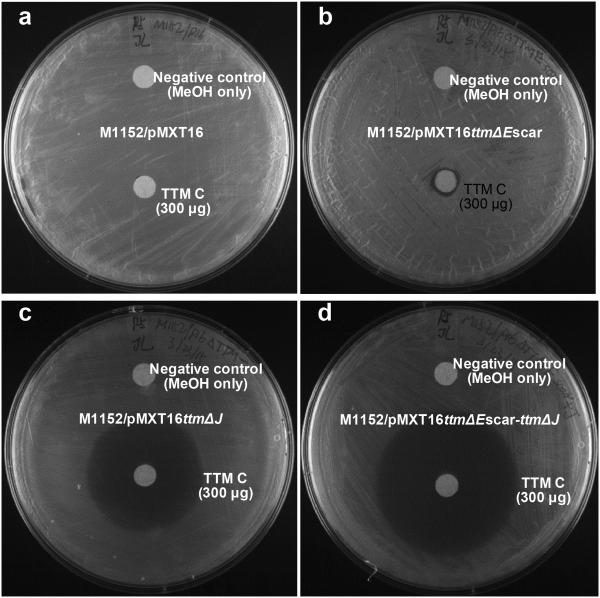
In order to test the hypothesis that the tlm and ttm-associated FabB/F homologs are associated with TLM/TTM resistance and not biosynthesis, we focused on the ttm gene cluster since it contained two FabB/F encoding genes – one shared with the tlm locus (ttmE and tlmE) and the second being unique (ttmJ). We first evaluated common Streptomyces heterologous host strains and found that S. coelicolor M1152 was susceptible to TTM C (8) with a minimum inhibitory concentration (MIC) of 12 μg/ml (Supporting Information Figure 9). S. coelicolor M1152 harboring the complete ttm BGC (S. coelicolor M1152/pMXT16), however, was resistant to 300 μg of TTM C on agar diffusion assays (Figure 4 and Supporting Information Figure 10).
Figure 4.
Representative disk diffusion assay plates for TTM C (300 μg) against S. coelicolor M1152 integrated with (a) the ttm gene cluster (designated as M1152/pMXT16) and (b-d) three mutants: (b) the ttmE deleted mutant S. coelicolor M1152/pMXT16ΔttmEscar, (c) the ttmJ deleted mutant S. coelicolor M1152/pMXT16ΔttmJ, and (d) the ttmE - ttmJ double mutant S. coelicolor M1152/pMXT16ΔttmEscar-ΔttmJ.
We generated three mutants in which the FabB/F homologous genes were individually (S. coelicolor M1152/pMXT16ΔttmEscar and S. coelicolor M1152/pMXT16ΔttmJ) and collectively (S. coelicolor M1152/pMXT16ΔttmEscar-ΔttmJ) inactivated. All three mutants retained the ability to heterologously produce the TTM compounds in same distribution (Supporting Information Figure 11), thereby establishing that the FabB/F genes ttmE and ttmJ are not required for biosynthesis. We also observed that deletion of the resistance genes did not compromise the production of TTMs. This may be attributed to the low yield of TTM C in the heterologous host (around 0.5 μg/ml), which is much lower than the MIC (12 μg/ml). The susceptibility of these recombinant S. coelicolor strains to TTM C was next determined using a disk diffusion assay. When a high concentration of TTM C (300 μg) was tested, the ttmE deletion strain S. coelicolor M1152/pMXT16ΔttmEscar, the ttmJ knockout strain S. coelicolor M1152/pMXT16ΔttmJ and the ttmE and ttmJ double-deletion strain S. coelicolor M1152/pMXT16ΔttmEscar-ΔttmJ showed inhibition zones of the sizes 0.9 ± 0.08 cm, 3.2 ± 0.3 cm and 4.4 ± 0.2 cm (Figure 4), respectively. Smaller inhibition zones were observed when lower concentrations of TTM C (50-200 μg) were applied to the ttmJ deletion strains, while the ttmE deletion strain showed complete resistance at these concentrations (Supporting Information Figure 10). These data confirm that ttmE and ttmJ provide inherent resistance to TTM C, the major compound produced by the ttm BGC. Since deletion of ttmE did not compromise resistance of the strain at these concentrations, we suggest that TtmE plays a relatively minor role in resistance. Rather the deletion experiments indicate that ttmJ is the major resistance element for TTM biosynthesis, as all ttmJ deletion mutants exhibited high susceptibility to TTM C. In line with our hypothesis, previously reported anti-bacterial activity tests showed that the MIC of TMM C is 16-32 fold lower than TLM and its analogues,30 supporting the molecular logic that the ttm pathway requires a second resistance gene. Previous studies have shown various resistance mechanisms against TLM in pathogenic bacteria, including the emrAB encoded multidrug-resistant efflux pump in E. coli,46 FabB point mutation in E. coli,47 and FabH point mutations in Staphylococcus aureus.37 Our findings reveal a distinct resistance mechanism associated with TLM/TTM producing strains.
Conclusion
We report here a new strategy for mining orphan biosynthetic pathways, which we refer to as target-directed genome mining. By using Salinispora as a model, the present study demonstrates a systematic approach to identify BGCs with duplicated housekeeping genes that may be candidates for conferring resistance against the product of the BGCs. Combined with the synthetic dsDNA-mediated cloning method, we showed that putative fabB/F resistance genes associated with orphan BGCs in Salinispora and Streptomyces bacteria are associated with FAS inhibitors of the thiotetronic acid family, antibiotics that were first described over 30 years ago yet never at the genetic level. Our work also paves the way for investigating the biosynthesis of the structurally unique thiotetronic acid natural products, a topic that we are actively pursuing and will report elsewhere. Significantly, this work shows the first demonstration of target-directed genome mining strategy for the discovery of antibiotic producing gene clusters, without a priori knowledge of the molecule synthesized. Our comparative genomic analysis provides an indication of the frequency that housekeeping genes are involved in secondary metabolic processes. Based on the rapid increase in genome sequence data in public databases, we envision that this strategy may be widely applicable to genomics-driven natural product discovery and innovate the manner in which antibiotics are discovered, offering an efficient, hypothesis-driven genome mining platform for the development of new antibacterial drug candidates.
METHODS
Cultivation of the Strains
Seventy five Salinispora genome sequences were obtained as previously described.27 A highly transformable S. cerevisiae strain VL6-48N (MAT α, his3-D200, trp1-Δ1, ura3-Δ1, lys2, ade2-101, met14, psi+cirO)35 was used as a host for gene cluster direct cloning experiments. The yeast cells were grown in liquid YPD medium (Yeast extract Peptone Dextrose medium; 2% D-glucose, 1% yeast extract, and 2% peptone (w/v)) supplemented with 100 mg/L adenine and used for spheroplasting prior to transformation-associated recombination (TAR). Yeast transformants were selected on synthetic tryptophan drop-out agar (SD-Trp agar) containing 5-fluoroorotic acid (5-FOA) (SD-Trp-5-FOA agar) consisting of 0.17% yeast nitrogen base without amino acids and ammonium sulfate (Sigma), 0.19% yeast synthetic drop-out medium supplements without tryptophan (Sigma), 1 M sorbitol, 2% D-glucose, 0.5% ammonium sulfate, 100 mg/l adenine, 2% agar and 0.001% 5-FOA (w/v)(Zymo Research). S. coelicolor M1152 (SCP1−, SCP2−, Δact, Δred, Δcpk, Δcda, and rpoB[C1298T])48 and their respective derivatives were maintained and grown on either MS agar (2% soy flour, 2% mannitol, 2% agar (w/v for all)); components purchased from BD Biosciences) or Tryptic Soy Broth (TSB) medium (BD Biosciences). Salinispora pacifica CNS-863 was maintained on A1 agar as previously reported,49 and Streptomyces afghaniensis NRRL 5621 (accession no. NZ_AOPY01000000) was maintained on MS agar. E. coli strains were cultivated in LB medium (components purchased from BD Biosciences or Fisher Scientific) supplemented with the appropriate antibiotics. DNA isolation and manipulations were carried out according to standard methods for E. coli and Streptomyces.
Construction of the Counter-selectable Capture Vector pCAP03
pADH and URA3 were amplified from pARS-VN 35 by using the primer pairs ADH-fw/ADH-rev and URA3-fw/URA3-rev (Table S6), respectively. The two PCR amplified fragments were then combined and assembled into a single piece by PCR with primers ADH-fw and URA3-rev. The assembled fragment was digested with SpeI and KpnI and introduced into the same sites of pCAP01 16 to obtain pCAP03. In order to easy linearize pCAP03, the 1369 bp apramycin resistance cassette (acc(3)IV) was amplified from pIJ773 50 by using the primers acc(3)IV-pCAP03-fw and acc(3)IV-pCAP03-rev (Supporting Information Table 6). The resulting PCR product was digested by XhoI and NdeI and cloned into the same restriction sites of pCAP03 to obtain pCAP03-acc(3)IV. The circular construct pCAP03-acc(3)IV was digested with NdeI and XhoI and agarose gel purified, as ready-to-use pCAP03 (RTU-pCAP03) for assembly of cluster-specific capture vectors.
Construction of Specific Capture Vectors
144 bp dsDNA fragments were designed as shown in Figure 3 and synthesized by Integrated DNA Technologies, Incorporation, La Jolla, US. The 100 ng tlm-hooks and ttm-hooks (Supporting Information Table 6) were assembled with 50 ng of the RTU-pCAP03 following the instructions for the Gibson Assembly Kit (New England BioLabs Inc) to generate the capture vectors pMTZ01 and pMTZ02, respectively. Prior to direct TAR cloning, the capture vectors were digested by PmeI.
Preparation of Genomic DNA Fragments for TAR Capturing
Genomic DNA was isolated from mid-log phase cells by standard procedures.51 Approximately 200 μg of genomic DNA was digested with appropriate restriction enzymes (Figure 3b), in an overnight reaction at 37 °C. The digested genomic DNA fragments were precipitated with isopropanol and washed with 70% ethanol. The resulting DNA pellet was dissolved in 100 μL of Tris buffer (10 mM Tris·HCl, pH 8.0).
Direct Capture of the tlm and ttm Gene Clusters
Direct TAR cloning of the tlm and ttm gene clusters from genomic DNA were carried out using a previously reported protocol with minor modifications.16, 52 S. cerevisiae strain VL6-48N was grown in 50 ml YPD medium supplemented with adenine (100 mg/L) at 30°C with shaking until OD600 of 0.7-1.0 was reached. The cells were harvested and washed with ice-cold water and osmotically stabilized in 1 M sorbitol at 4 °C for overnight prior to spheroplasting. Preparation of spheroplast cells was carried out using a lytic enzyme (Zymolyase-20T, MP Biomedicals, US) in final concentration of 0.1 mg/ml, with 30-40 min incubation. 2.5 to 3 μg of genomic DNA fragments and 0.5 μg linearized specific capture vector were added to spheroplast cells, and the transformation was mediated by PEG8000 (Sigma, US). The transformed spheroplasts were mixed with 8 ml synthetic tryptophan drop-out (SD-Trp) top agar (containing 3% agar) at 55 °C and overlaid on SD-Trp-5-FOA agar. The plates were incubated at 30 °C for 4-5 days. Normally, around 10-20 transformants appeared per plate and were picked and transferred onto new SD-Trp agar plates, and incubated for 2 days at 30 °C. Cells were lysed using Zymolyase-20T at 37 °C for 2 h and subsequently boiled at 98 °C for 5 min. The captured tlm and ttm gene clusters were screened by the primer pairs of tlm-test-fw/tlm-test-rev and ttm-test-fw/ttm-test-rev, respectively (Supporting Information Table 6). Plasmids were extracted from PCR positive clones and then transferred into E. coli Top10 cells by electroporation. The plasmids were purified from kanamycin resistant E. coli clones, and the resulting constructs were confirmed by restriction analysis. The vector containing tlm gene cluster and ttm gene cluster were designated as pMXT13 and pMXT16, respectively.
Heterologous Expression of the tlm and ttm Gene Clusters
pMXT13 and pMXT16 were transferred into E. coli ET12567 and introduced into S. coelicolor M1152 by triparental intergeneric conjugation with the help of E. coli ET12567/pUB307.53 Kanamycin resistant clones were selected, confirmed by PCR and designated as S. coelicolor M1152/pMXT13 and S. coelicolor M1152/pMXT16. Three milliliters of TSB medium were inoculated with spore suspension of S. coelicolor M1152 or a derivative thereof. The cultures were incubated for 2 d at 30 °C at 220 r.p.m. For the production of the compounds, 1 ml of preculture was inoculated into 50 ml of the production medium containing 1% soytone, 1% soluble starch and 2% D-maltose (w/v) adjusted to pH 6.7 (components purchased from BD Biosciences, US). The cultures were incubated for 7 d at 30 °C with 220 rpm shaking. The culture supernatant was adjusted to pH 4 with acetic acid and subsequently extracted with an equal volume of ethyl acetate. The organic phase was evaporated and extracts were dissolved in 1 ml methanol. Each extract was monitored at 239 nm during separation by HPLC using a Luna 100A-C18 column (5 μm, 250 × 4.6 mm; Phenomenex, US) as follows: 0-23 min, 35%-70% B; 24-28 min, 70%-100% B; 29-33 min, 100% B; 34-35 min, 100%-35% B; 36-40 min, 35% B (solvent A: water/trifluoroacetic acid (999:1); solvent B: acetonitrile/trifluoroacetic acid (999:1)).
Other Methods
Other methods, including the identification of orthologous groups, BGC annotation, disk diffusion assays, isolation and structural elucidation of thiolactomycins and thiotetroamides, are described in Supporting Information Methods.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank V. Larionov (National Cancer Institute, NIH) for providing S. cerevisiae strain VL6-48N and plasmid pARS-VN, Mervyn Bibb (John Innes Centre, UK) for providing Streptomyces coelicolor M1152, the Agricultural Research Service (ARS) culture collection for providing Streptomyces afghaniensis NRRL 5621 strain, and N. Ziemert (Scripps Institution of Oceanography, UCSD) and M. Wietz (ICBM, University of Oldeburg) for valuable discussions. This work was supported by US National Institutes of Health grants R01-GM085770 and U19-TW007401, the Roddenberry Foundation, and graduate fellowships from the NSF (to J.J.Z.) and Consejo Nacional de Ciencia y Tecnología (CONACyT-213497 to N.M-A.). Genome sequencing was conducted by the U.S. Department of Energy Joint Genome Institute and supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
ABBREVIATIONS
- PKS
polyketide synthase
- NRPS
nonribosomal peptide synthetase
- TAR
transformation-associated recombination
- dsDNA
double-stranded DNA
- NHEJ
non-homologous end joining
Footnotes
Supporting Information
This material is available free of charge via the Internet at http://pubs.acs.org.
Accession Codes:
Eleven genome sequences reported in this paper have been deposited in the Joint Genome Institute’s Integrated Microbial Genomes (IMG) database, http://img.jgi.doe.gov/(accession nos. 2540341193, 2524614530, 2540341192, 2528311034, 2524614561, 2524023246, 2526164509, 2528311033, 2524614529, 2524614515, 2521172655). The nucleotide sequences of the thilactomycin gene cluster from Salinispora pacifica CNS-863 and the thiotetroamide gene cluster from Streptomyces afghaniensis NRRL 5621 have been deposited in the GenBank database (accession nos. KT282100 and KT282101, respectively).
REFERENCES
- 1.Clardy J, Fischbach MA, Walsh CT. New antibiotics from bacterial natural products. Nat Biotechnol. 2006;24:1541–1550. doi: 10.1038/nbt1266. [DOI] [PubMed] [Google Scholar]
- 2.Demain AL. Antibiotics: natural products essential to human health. Med Res Rev. 2009;29:821–842. doi: 10.1002/med.20154. [DOI] [PubMed] [Google Scholar]
- 3.Cooper MA, Shlaes D. Fix the antibiotics pipeline. Nature. 2011;472:32. doi: 10.1038/472032a. [DOI] [PubMed] [Google Scholar]
- 4.Corre C, Challis GL. New natural product biosynthetic chemistry discovered by genome mining. Nat Prod Rep. 2009;26:977–986. doi: 10.1039/b713024b. [DOI] [PubMed] [Google Scholar]
- 5.Genilloud O. The re-emerging role of microbial natural products in antibiotic discovery. Antonie Van Leeuwenhoek. 2014;106:173–188. doi: 10.1007/s10482-014-0204-6. [DOI] [PubMed] [Google Scholar]
- 6.Milshteyn A, Schneider JS, Brady SF. Mining the metabiome: identifying novel natural products from microbial communities. Chem Biol. 2014;21:1211–1223. doi: 10.1016/j.chembiol.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cimermancic P, Medema MH, Claesen J, Kurita K, Wieland Brown LC, Mavrommatis K, Pati A, Godfrey PA, Koehrsen M, Clardy J, Birren BW, Takano E, Sali A, Linington RG, Fischbach MA. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158:412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rutledge PJ, Challis GL. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat Rev Microbiol. 2015 doi: 10.1038/nrmicro3496. [DOI] [PubMed] [Google Scholar]
- 9.Eustaquio AS, Nam SJ, Penn K, Lechner A, Wilson MC, Fenical W, Jensen PR, Moore BS. The discovery of salinosporamide K from the marine bacterium "Salinispora pacifica" by genome mining gives insight into pathway evolution. Chembiochem. 2011;12:61–64. doi: 10.1002/cbic.201000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaysser L, Tang X, Wemakor E, Sedding K, Hennig S, Siebenberg S, Gust B. Identification of a napsamycin biosynthesis gene cluster by genome mining. Chembiochem. 2011;12:477–487. doi: 10.1002/cbic.201000460. [DOI] [PubMed] [Google Scholar]
- 11.Kersten RD, Yang YL, Xu Y, Cimermancic P, Nam SJ, Fenical W, Fischbach MA, Moore BS, Dorrestein PC. A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nat Chem Biol. 2011;7:794–802. doi: 10.1038/nchembio.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kersten RD, Ziemert N, Gonzalez DJ, Duggan BM, Nizet V, Dorrestein PC, Moore BS. Glycogenomics as a mass spectrometry-guided genome-mining method for microbial glycosylated molecules. Proc Natl Acad Sci U S A. 2013;110:E4407–4416. doi: 10.1073/pnas.1315492110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lautru S, Deeth RJ, Bailey LM, Challis GL. Discovery of a new peptide natural product by Streptomyces coelicolor genome mining. Nat Chem Biol. 2005;1:265–269. doi: 10.1038/nchembio731. [DOI] [PubMed] [Google Scholar]
- 14.Laureti L, Song L, Huang S, Corre C, Leblond P, Challis GL, Aigle B. Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. Proc Natl Acad Sci U S A. 2011;108:6258–6263. doi: 10.1073/pnas.1019077108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu J, Bian X, Hu S, Wang H, Huang F, Seibert PM, Plaza A, Xia L, Muller R, Stewart AF, Zhang Y. Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat Biotechnol. 2012;30:440–446. doi: 10.1038/nbt.2183. [DOI] [PubMed] [Google Scholar]
- 16.Yamanaka K, Reynolds KA, Kersten RD, Ryan KS, Gonzalez DJ, Nizet V, Dorrestein PC, Moore BS. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc Natl Acad Sci U S A. 2014;111:1957–1962. doi: 10.1073/pnas.1319584111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Costa VM, McGrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006;311:374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 18.Thaker MN, Wang W, Spanogiannopoulos P, Waglechner N, King AM, Medina R, Wright GD. Identifying producers of antibacterial compounds by screening for antibiotic resistance. Nat Biotechnol. 2013;31:922–927. doi: 10.1038/nbt.2685. [DOI] [PubMed] [Google Scholar]
- 19.Andersson DI, Levin BR. The biological cost of antibiotic resistance. Curr Opin Microbiol. 1999;2:489–493. doi: 10.1016/s1369-5274(99)00005-3. [DOI] [PubMed] [Google Scholar]
- 20.Wright GD. Molecular mechanisms of antibiotic resistance. Chem Commun (Camb) 2011;47:4055–4061. doi: 10.1039/c0cc05111j. [DOI] [PubMed] [Google Scholar]
- 21.Steffensky M, Muhlenweg A, Wang ZX, Li SM, Heide L. Identification of the novobiocin biosynthetic gene cluster of Streptomyces spheroides NCIB 11891. Antimicrob Agents Chemother. 2000;44:1214–1222. doi: 10.1128/aac.44.5.1214-1222.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peterson RM, Huang T, Rudolf JD, Smanski MJ, Shen B. Mechanisms of self-resistance in the platensimycin- and platencin-producing Streptomyces platensis MA7327 and MA7339 strains. Chem Biol. 2014;21:389–397. doi: 10.1016/j.chembiol.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kling A, Lukat P, Almeida DV, Bauer A, Fontaine E, Sordello S, Zaburannyi N, Herrmann J, Wenzel SC, Konig C, Ammerman NC, Barrio MB, Borchers K, Bordon-Pallier F, Bronstrup M, Courtemanche G, Gerlitz M, Geslin M, Hammann P, Heinz DW, Hoffmann H, Klieber S, Kohlmann M, Kurz M, Lair C, Matter H, Nuermberger E, Tyagi S, Fraisse L, Grosset JH, Lagrange S, Muller R. Antibiotics. Targeting DnaN for tuberculosis therapy using novel griselimycins. Science. 2015;348:1106–1112. doi: 10.1126/science.aaa4690. [DOI] [PubMed] [Google Scholar]
- 24.Jensen PR, Moore BS, Fenical W. The marine actinomycete genus Salinispora: a model organism for secondary metabolite discovery. Nat Prod Rep. 2015;32:738–751. doi: 10.1039/c4np00167b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gulder TA, Moore BS. Salinosporamide natural products: Potent 20 S proteasome inhibitors as promising cancer chemotherapeutics. Angew Chem Int Ed Engl. 2010;49:9346–9367. doi: 10.1002/anie.201000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colis LC, Woo CM, Hegan DC, Li Z, Glazer PM, Herzon SB. The cytotoxicity of (-)-lomaiviticin A arises from induction of double-strand breaks in DNA. Nat Chem. 2014;6:504–510. doi: 10.1038/nchem.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ziemert N, Lechner A, Wietz M, Millan-Aguinaga N, Chavarria KL, Jensen PR. Diversity and evolution of secondary metabolism in the marine actinomycete genus. Salinispora, Proc Natl Acad Sci U S A. 2014;111:E1130–1139. doi: 10.1073/pnas.1324161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L, Stoeckert CJ, Jr., Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kale AJ, McGlinchey RP, Lechner A, Moore BS. Bacterial self-resistance to the natural proteasome inhibitor salinosporamide A. ACS Chem Biol. 2011;6:1257–1264. doi: 10.1021/cb2002544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Young K, Jayasuriya H, Ondeyka JG, Herath K, Zhang C, Kodali S, Galgoci A, Painter R, Brown-Driver V, Yamamoto R, Silver LL, Zheng Y, Ventura JI, Sigmund J, Ha S, Basilio A, Vicente F, Tormo JR, Pelaez F, Youngman P, Cully D, Barrett JF, Schmatz D, Singh SB, Wang J. Discovery of FabH/FabF inhibitors from natural products. Antimicrob Agents Chemother. 2006;50:519–526. doi: 10.1128/AAC.50.2.519-526.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balemans W, Lounis N, Gilissen R, Guillemont J, Simmen K, Andries K, Koul A. Essentiality of FASII pathway for Staphylococcus aureus. Nature. 2010;463:E3. doi: 10.1038/nature08667. discussion E4. [DOI] [PubMed] [Google Scholar]
- 32.Yao J, Rock CO. How bacterial pathogens eat host lipids: implications for the development of fatty acid synthesis therapeutics. J Biol Chem. 2015;290:5940–5946. doi: 10.1074/jbc.R114.636241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smanski MJ, Yu Z, Casper J, Lin S, Peterson RM, Chen Y, Wendt-Pienkowski E, Rajski SR, Shen B. Dedicated ent-kaurene and ent-atiserene synthases for platensimycin and platencin biosynthesis. Proc Natl Acad Sci U S A. 2011;108:13498–13503. doi: 10.1073/pnas.1106919108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Furter-Graves EM, Hall BD. DNA sequence elements required for transcription initiation of the Schizosaccharomyces pombe ADH gene in Saccharomyces cerevisiae. Mol Gen Genet. 1990;223:407–416. doi: 10.1007/BF00264447. [DOI] [PubMed] [Google Scholar]
- 35.Noskov VN, Kouprina N, Leem SH, Ouspenski I, Barrett JC, Larionov V. A general cloning system to selectively isolate any eukaryotic or prokaryotic genomic region in yeast. BMC Genomics. 2003;4:16. doi: 10.1186/1471-2164-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 37.Parsons JB, Yao J, Frank MW, Rock CO. FabH mutations confer resistance to FabF-directed antibiotics in Staphylococcus aureus. Antimicrob Agents Chemother. 2015;59:849–858. doi: 10.1128/AAC.04179-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackowski S, Murphy CM, Cronan JE, Jr., Rock CO. Acetoacetyl-acyl carrier protein synthase. A target for the antibiotic thiolactomycin. J Biol Chem. 1989;264:7624–7629. [PubMed] [Google Scholar]
- 39.Hayashi T, Yamamoto O, Sasaki H, Kawaguchi A, Okazaki H. Mechanism of action of the antibiotic thiolactomycin inhibition of fatty acid synthesis of Escherichia coli. Biochem Biophys Res Commun. 1983;115:1108–1113. doi: 10.1016/s0006-291x(83)80050-3. [DOI] [PubMed] [Google Scholar]
- 40.Price AC, Choi KH, Heath RJ, Li Z, White SW, Rock CO. Inhibition of beta-ketoacyl-acyl carrier protein synthases by thiolactomycin and cerulenin. Structure and mechanism. J Biol Chem. 2001;276:6551–6559. doi: 10.1074/jbc.M007101200. [DOI] [PubMed] [Google Scholar]
- 41.Sasaki H, Oishi H, Hayashi T, Matsuura I, Ando K, Sawada M. Thiolactomycin, a new antibiotic. II. Structure elucidation. J Antibiot (Tokyo) 1982;35:396–400. doi: 10.7164/antibiotics.35.396. [DOI] [PubMed] [Google Scholar]
- 42.Hayashi T, Yamamoto O, Sasaki H, Okazaki H, Kawaguchi A. Inhibition of fatty acid synthesis by the antibiotic thiolactomycin. J Antibiot (Tokyo) 1984;37:1456–1461. doi: 10.7164/antibiotics.37.1456. [DOI] [PubMed] [Google Scholar]
- 43.Brown MS, Akopiants K, Resceck DM, McArthur HA, McCormick E, Reynolds KA. Biosynthetic origins of the natural product, thiolactomycin: a unique and selective inhibitor of type II dissociated fatty acid synthases. J Am Chem Soc. 2003;125:10166–10167. doi: 10.1021/ja034540i. [DOI] [PubMed] [Google Scholar]
- 44.Ma M, Lohman JR, Liu T, Shen B. C-S bond cleavage by a polyketide synthase domain. Proc Natl Acad Sci U S A. 2015;112:10359–10364. doi: 10.1073/pnas.1508437112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rapp C, Jung G, Isselhorst-Scharr C, Zähner H. A new member of the class of antibiotics with thiotetronic acid structure isolated from Streptomyces olivaceus Tü 3010. Liebigs Annalen der Chemie. 1988;1988:1043–1047. [Google Scholar]
- 46.Furukawa H, Tsay JT, Jackowski S, Takamura Y, Rock CO. Thiolactomycin resistance in Escherichia coli is associated with the multidrug resistance efflux pump encoded by emrAB. J Bacteriol. 1993;175:3723–3729. doi: 10.1128/jb.175.12.3723-3729.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jackowski S, Zhang YM, Price AC, White SW, Rock CO. A missense mutation in the fabB (beta-ketoacyl-acyl carrier protein synthase I) gene confers tiolactomycin resistance to Escherichia coli. Antimicrob Agents Chemother. 2002;46:1246–1252. doi: 10.1128/AAC.46.5.1246-1252.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gomez-Escribano JP, Bibb MJ. Streptomyces coelicolor as an expression host for heterologous gene clusters. Methods Enzymol. 2012;517:279–300. doi: 10.1016/B978-0-12-404634-4.00014-0. [DOI] [PubMed] [Google Scholar]
- 49.Jensen PR, Gontang E, Mafnas C, Mincer TJ, Fenical W. Culturable marine actinomycete diversity from tropical Pacific Ocean sediments. Environ Microbiol. 2005;7:1039–1048. doi: 10.1111/j.1462-2920.2005.00785.x. [DOI] [PubMed] [Google Scholar]
- 50.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci U S A. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics (John Innes Foundation. Norwich; UK: 2000. [Google Scholar]
- 52.Kouprina N, Larionov V. Selective isolation of genomic loci from complex genomes by transformation-associated recombination cloning in the yeast Saccharomyces cerevisiae. Nat Protoc. 2008;3:371–377. doi: 10.1038/nprot.2008.5. [DOI] [PubMed] [Google Scholar]
- 53.Flett F, Mersinias V, Smith CP. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol Lett. 1997;155:223–229. doi: 10.1111/j.1574-6968.1997.tb13882.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.