Abstract
Being driven by non-covalent interactions, the formation of functional assemblies (or aggregates) of small molecules at nanoscale is a more common process in water than one would think. While most efforts on self-assembly in cellular environment concentrate on the assemblies of proteins (e.g., microtubules or amyloid fibers), nanoscale assemblies of small molecules are emerging functional entities that exhibit important biological function in cellular environments. This review describes the increasing efforts on the exploration of nanoscale assemblies of small molecules that largely originate from the serendipitous observations in research fields other than nanoscience and technology. Specifically, we describe that nanoscale assemblies of small molecules exhibit unique biological functions in extracellular and intracellular environment, thus inducing various cellular responses, like causing cell death or promoting cell proliferation. We first survey certain common feature of nanoscale molecular assemblies, then discuss several specific examples, such as, nanoscale assemblies of small peptides accumulated in the cells for selectively inhibiting cancer cells via promiscuous interactions with proteins, and nanoscale assemblies of a glycoconjugate for promoting the proliferation of stem cells or for suppressing immune responses. Subsequently, we emphasize the spatiotemporal control of nanoscale assemblies for controlling the cell fate, particularly illustrate a paradigm-shifting approach—enzyme-instructed self-assembly (EISA), that is, the integration of enzymatic reaction and self-assembly—for generating nanoscale assemblies from innocuous monomers for selectively inhibiting cancer cells. Moreover, we introduce a convenient assay for proteomic study of the proteins that interact with nanoscale assemblies of small molecules in cellular environment. Furthermore, we introduce the use of ligand-receptor interaction to catalyze the formation of nanoscale assemblies. By illustrating these experimental strategies for controlling the formation of nanoscale assemblies of small molecules and for identifying their corresponding protein targets, we aim to highlight that, though not being defined at the genetic level, nanoscale assemblies of small molecules are able to perform many critical biological functions. We envision that nanoscale assemblies of small molecules are a new frontier at the intersection of nanoscience and cell biology and biomedicine. In addition, we discuss the challenges and perspectives of relevant potential biomedical applications of nanoscale assemblies of small molecules.
Keywords: Nanoscale, assemblies, small molecule, cell fate, self-assembly, cancer, stem cells, EISA
1. Introduction
Aggregation of molecules or molecular self-assembly—the spontaneous, non-covalent association of molecules to give nanoscale assemblies with emergent properties—is ubiquitous in biology. In this article, we use aggregates to describe the less ordered nanostructures and assemblies to refer more ordered nanostructures. However, their distinction becomes blurred in cellular environment, which is highly dynamic. Self-assembly of proteins generates assemblies that either are crucial for cellular functions (e.g., actin filaments and microtubules for cell dynamics,[1, 2] ribosomes for protein production,[3, 4] apoptosome[5] for program cell death, and inflammasome[6] for immune response) or associate with certain diseases (e.g., fibrillar assemblies of aberrant proteins as hallmarks of neurodegenerative diseases[7, 8]). Self-assembly of lipids forms cell membranes that compartmentalize subcellular organelles and control the substances in and out cells.[9] Besides leading to intensive studies on the assemblies of proteins for elucidating cellular functions and mechanisms of diseases[10] and on the assemblies of lipids for understanding the origin of life,[11] these fundamental facts have raised a fundamental question about biological functions of nanoscale assemblies of small molecules, a largely ignored phenomenon that is scientifically intriguing and increasingly significant in biology and medicine.
Emerging evidence from several unrelated fields (e.g., biomaterials,[12] high throughput drug screening,[13] and neurodegenerative diseases[14–16]) over the last decade has highlighted the significance of nanoscale assemblies (or aggregates) of small molecules in biology and medicine. Being able to sequester enzymes or unfold proteins,[17] activate enzymes,[18, 19] or inhibit cell growth,[20] recruit and retain mRNAs to form cell-free RNA granules,[21] nanoscale assemblies of small molecules, undoubtedly, can act as functional molecular entities in cellular environment. The development of small molecule hydrogels (e.g., lanreotide autogel) for treating acromegaly[12] and the surprising high potency of non-nucleoside reverse transcriptase inhibitors (NNRTIs) in cell culture[22] clearly validates applications of nanoscale assemblies of small molecules in medicine. Moreover, these results support the notion that nanoscale assemblies of small molecules behave drastically differently from their un-assembled components and exhibit specific biological functions (Shown in Fig. 1).[23–29] Despite their significance and potentials, nanoscale assemblies of small molecules, however, are not defined at the genetic level, thus, conventional biochemical and genetic methods are inadequate to control their formation and to identify their corresponding protein targets. The intention of this review is to illustrate the methodology for exploring nanoscale assemblies of small molecules and to highlight their importance and promise.
Figure 1.
Schematic illustration of self-assemblies of small molecules to control cell fates.
In this article, we first summarize a common feature of the molecular assemblies derived from the analysis of relatively large number of reports on the cytotoxicity of nanoscale molecular assemblies, then we discuss several specific examples, such as, intracellular nanoscale assemblies of ultrashort dipeptide for selectively inhibiting cancer cells via promiscuous interactions with proteins, and nanoscale assemblies of a glycoconjugate for promoting the proliferation of stem cells or suppressing immune responses. Subsequently, we highlight the spatiotemporal control of nanoscale assemblies for controlling the cell fate, particularly illustrate a paradigm-shifting approach—enzyme-instructed self-assembly (EISA), that is, the integration of enzymatic reaction and self-assembly—for controlling the spatiotemporal profiles of nanoscale assemblies of innocuous molecules for selectively inhibiting cancer cells. After that, we introduce a convenient assay for identifying the proteins that interact with nanoscale assemblies of small molecules in cellular environment. Furthermore, we introduce the formation of nanoscale assemblies of small molecules catalyzed by ligand-receptor interactions. By illustrating the experimental strategies for generating nanoscale assemblies of small molecules and for identifying their corresponding protein targets, we aim to demonstrate that, though not being defined at the genetic level, nanoscale assemblies of small molecules are able to perform many critical biological functions. Finally, we discuss the challenges and perspectives of relevant potential biomedical applications of nanoscale assemblies of small molecules. We hope that the results briefly outlined in this progress review will stimulate the research attentions on nanoscale assemblies of small molecules, an intriguing type of molecular entities that reside at the intersections of chemistry, biology, nanoscience, and nanotechnology.
2. Nanoscale molecular assemblies and biological functions
Common feature of nanoscale assemblies
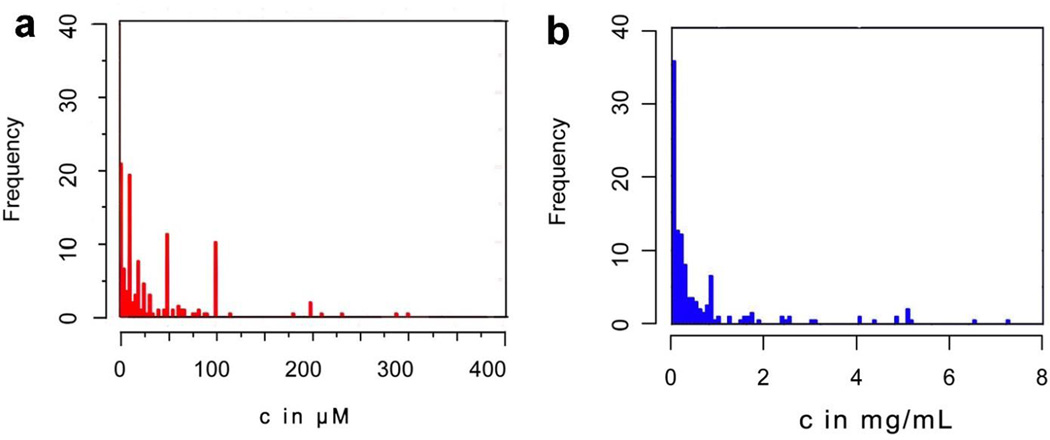
The most intriguing case of nanoscale molecular assemblies would be the nanofibrils of β-amyloid (Aβ) peptides, which associate with Alzheimer’s disease.[30] Recently studies on aberrant proteins or peptides,[31–33] as well as non-pathogenic proteins,[34, 35] have suggested a common, yet unknown mechanism for the cytotoxicity exhibited by nanoscale assemblies of proteins or peptides. To understand such common mechanism, we used meta-analysis to evaluate the cytotoxicity of these nanoscale assemblies reported in 628 studies. As shown in Fig. 2, our analysis indicates that the cytotoxicity of the nanoscale assemblies converges in a narrower range in the mass concentrations (with the mean value at about 240 µg/mL) than in the molar concentrations, suggesting that the volume of the assemblies rather than the numbers of the individual molecules dictates the cytotoxicity. This new perspective implies that these assemblies likely have less-specific interaction with one or two key receptors, but rather have promiscuous interactions with many cellular proteins to cause cell death. Moreover, the cytotoxicities of the assemblies converge to a narrow range, suggesting that the assemblies may crowd biomacromolecules to disrupt multiple cellular processes.[36] More importantly, this analysis further supports the notion that the biological functions of nanoscale molecular assemblies can differ drastically from the un-assembled individual molecules.
Figure 2.
Quantification of cytotoxicities (a) in mole concentration and (b) in weight concentration. The histogram shows how cytotoxicity data spread out in certain concentration ranges, the data was obtained from 628 studies.[37].
Promiscuity of nanoscale assemblies of small molecules
Like certain aggregates of proteins, the nanoscale assemblies of small molecule also play important role in biological activity.[18, 26, 38] Encouraged by the results from our earlier exploration of the intracellular self-assembly of small molecules,[25] we evaluated the steepcytotoxicity of a dipeptide (naphthalene-phenylalanine-phenylalanine (Nap-FF), 1 (Fig. 3a)). 1 suddenly exhibits cytotoxicity at about 400 µM, implying the formation of aggregates, which is the case: A series of assays proved that the molecules of 1 self-assemble in water to form nanofibrils.[39] Unexpectedly, the nanoscale assemblies of 1 slectively inhibit glioblastoma cells over neuronal cells and effectively inhibit xenograft tumors in a mouse model.[40]
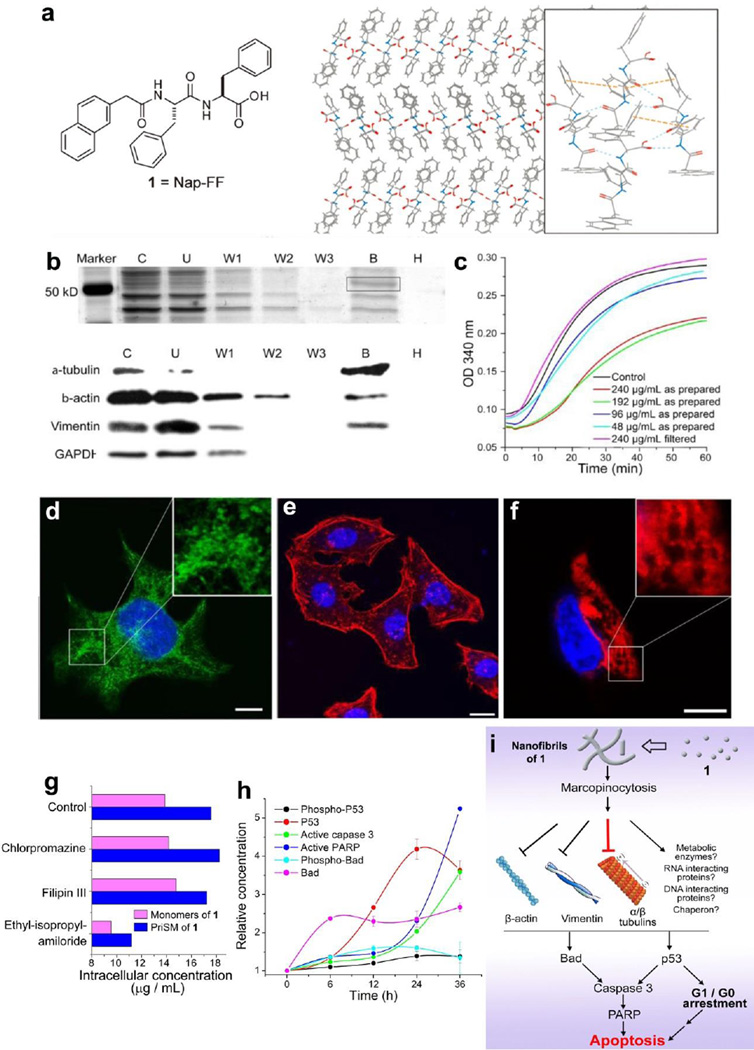
Figure 3.
(a) The chemical structure of 1 and a possible molecular arrangement in the nanofibers of 1 based on its crystal structure. (b) Molecular hydrogel protein binding (MHPB) assay. Upper panel: Silver stain of SDS-PAGE shows a major protein band at ~ 55 kDa in lane B. lower panel: Western blot indicates cytoskeletal proteins as the primary molecular targets. (c) Tubulin polymerization assays with 1. (d, e, f) Confocal images show the assemblies of 1 impede the dynamic of cytoskeletal proteins. (g) Cellular uptake of 1 in HeLa cells treated by endocytosis inhibitors. (h) Time dependent activation of apoptotic proteins of HeLa cells treated with 1. (i) The mechanism of the selective cytotoxicity of 1 towards cancer cell.[38, 44] (Print with the permission from © 2013 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim)
To understand the mechanism that how assemblies of 1 inhibit the proliferation of cells, we developed a molecular hydrogel protein binding (MHPB) assay[41, 42] to investigate the interaction between assemblies of 1 and cytosol proteins. The results of the MHBP assay (upper panel of Fig. 3b) and mass protein spectrometry show that assemblies of 1 promiscuously interact with different proteins (e.g. cytoskeleton proteins such as tubulins, vimentin, and actins). This result agrees with the Western-blot (lane B, lower panel of Fig. 3b) analysis. Moreover, as indicated by the lack of GAPDH in the lane B (lower panel of Fig. 3b), nanoscale assemblies of 1, indeed, bind to protein in a more specific manner than random, albeit promiscuously. Furthermore, the significant reduction of the polymerization rate in tubulin polymerization assay (Fig. 3c), together with confocal images (Fig. 3d), confirms that assemblies of 1 impede the formation of microtubules.[43] Meanwhile, confocal images (Fig. 3e, f) and the result of MHBP assay imply that assemblies of 1 also disrupt the dynamics of actin filaments and intermediate filaments of vimentins in cells. By measuring the intracellular concentrations of 1 (Fig. 3g) during the inhibition of endocytosis processes, we inferred that both the assemblies of 1 and the monomers of 1 enter the cell via macropinocytosis. The changes of several key signalling molecules of apoptosis (Fig. 3h) indicate that the assemblies of 1 activate Bad and p53, which later initiate the caspase cascade and downstream activation of PARP for the apoptosis in HeLa cells. These results allow us to give partial mechanism of the functions of nanoscale assemblies of 1 (Fig. 3i): after entering the cell via macropinocytosis, assemblies of 1 promiscuously interact with proteins and eventually lead the apoptosis via the intrinsic cell death pathway. As the first case of nanoscale assemblies of small molecule to impede multiple cytoskeletal proteins, this work not only reveals a mechanism for inherent cytotoxicity of hydrophobic, nanoscale molecular assemblies, but also implies a new paradigm for ultilizing nanoscale assemblies of small molecules for a variety of biological functions, including serving as a fundamental new type of anticancer agents.
Nanoscale assemblies of small glycoconjugates as potential mimics of glycoproteins
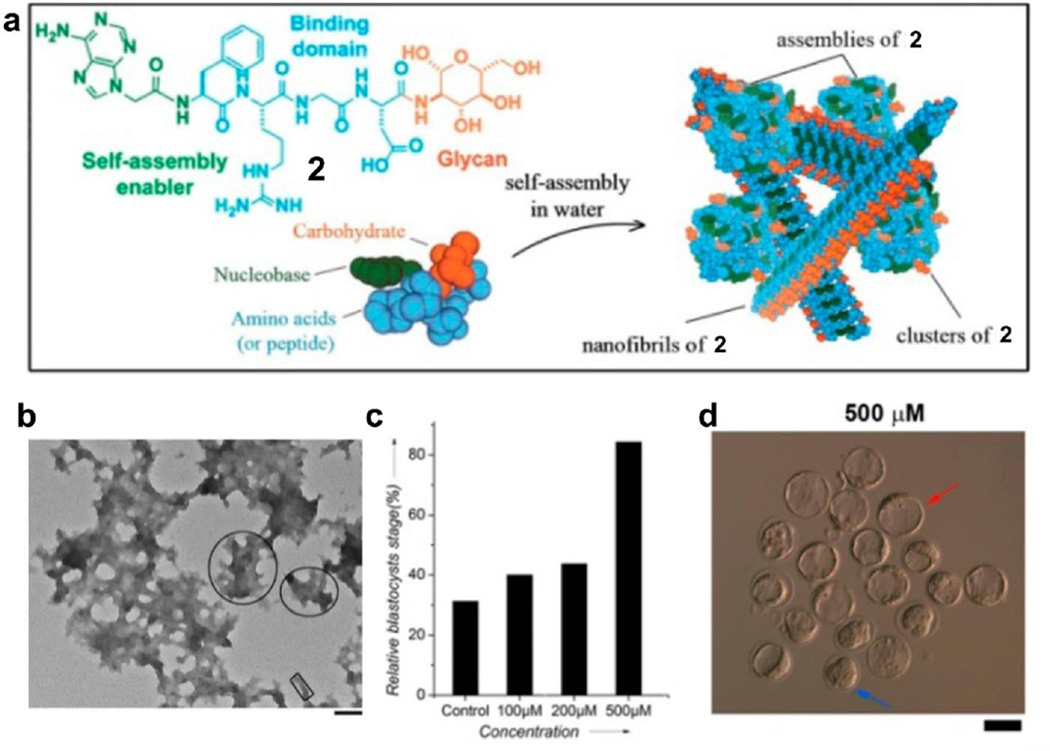
Due to the synthetic challenge in glycobiology and glycochemistry, the development of glycobiomaterials for biomedical applications remains a challenge. To address this issue, we explored the use of the nanoscale assemblies of simple glycocojugates (2) to mimic the functions of glycoproteins/proteoglycans. We designed and synthesized a glycoconjugate, 2, which consist of nucleobases, Arg-Gly-Asp (RGD) domain, and saccharides (Fig. 4a). Besides that 2 is significantly more stable than RGD[45], we found that the assemblies of 2 are able to promote the proliferation of murine embryonic stem (mES) cells.[46] As shown in Fig. 4a, the molecules of 2 self-assemble in water to form nanofibrils and nanoparticles (Fig. 4b) via aromatic-aromatic interactions and hydrogen-bonding. Moreover, assemblies of 2 promote the early development of mouse zygotes. Fig. 4c shows that 84% of zygotes become blastocysts (red arrows in Fig. 4d) after 4 days incubation with 2 at 500 µM, which is higher than control (only 31%). This result further confirms that the assemblies of 2 promote the proliferation of mES cells. Molecular engineering underscores the critical role of each component (e.g. glucosamine, RGD, adenine), and also reveal that assemblies of 2 bind with integrin to cluster integrin, thus mediate the cell proliferation.[47] As the first report of the use of self-assemblies of small glycoconjugates to control cellular response, this work illustrates a promising way to mimic the function of glycoproteins and proteoglycans.
Figure 4.
(a) The chemical structure of 2 and its self-assemblies. (b) TEM images of 2 at 500 µM. (c) Percentage of zygotes reaching the blastocyst stage after incubation without 2 and with 2 at different concentrations. (d) Photomicrographs of embryos incubated with 2. Red arrows point to blastocysts, and blue arrows to morulas.[46] (Print with the permission from © 2014 American chemical society)
Nanoscale assemblies of small glycoconjugates for immunomodulation
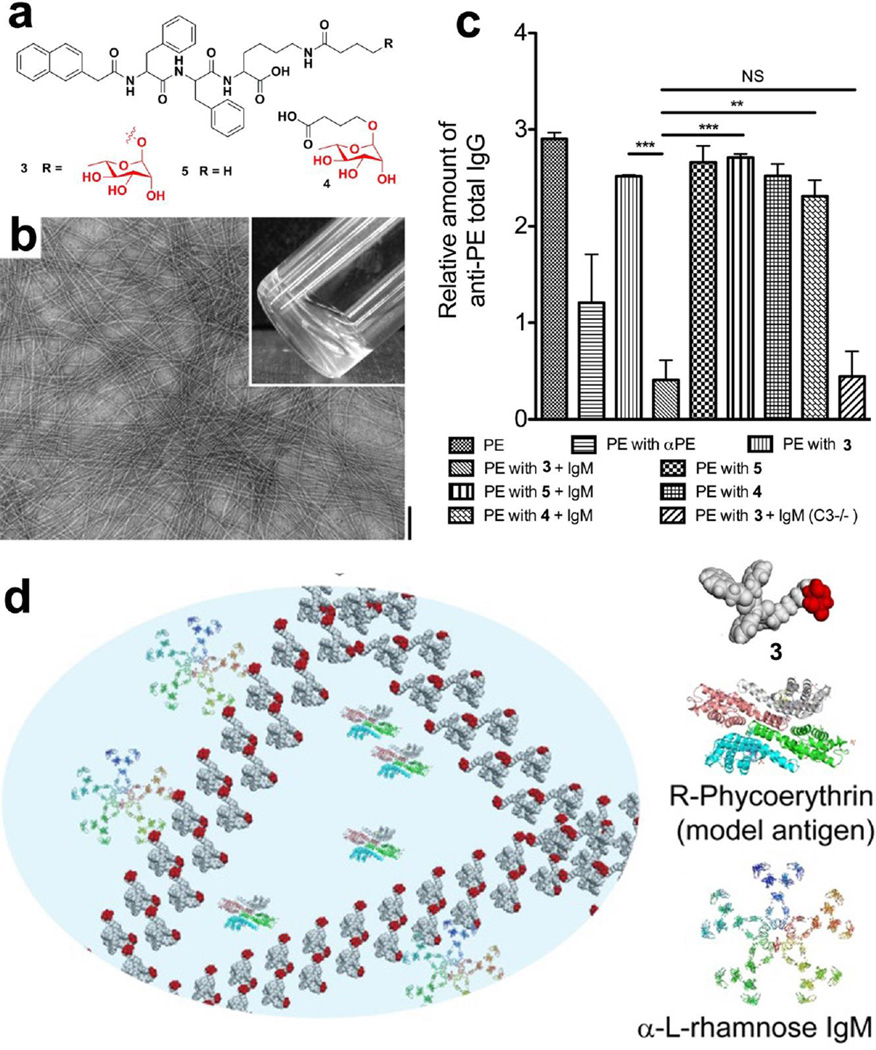
Inspired by the achievement of self-assembling small glycopeptides[46] and the advancement of immunology, we introduced a natural antigenic saccharide[48, 49] into nanoscale assemblies for modulating immune responses. We designed and synthesized a conjugates (3) containing L-rhamnose and a self-assembling motif (Fig. 5a).[50] TEM image shows that 3 self-assembles to form nanofibrils and result in a hydrogel at a concentration of 0.4 wt% (Fig. 5b). The resulting hydrogel is able to encapsulate a fluorescent model antigen, R-phycoerythrin (PE, Fig. 5d). After intraperitoneal (i.p.) injection of PE encapsulated by assemblies of 3 into mice that were pre-injected with purified human IgM, we observed a dramatic reduction in IgG against PE in the murine model. Meanwhile, this reduction is absent in i.p. immunization of PE inside assemblies of 5 (“PE with 5+IgM” in Fig. 5c), strongly suggesting that the presence of L-rhamnose is essential for mediating the immunosuppression. However, monomeric L-rhamnose (4) also is unable to modulate the immunosuppression at same condition (“PE with 4+IgM” in Fig. 5c), indicating that assemblies of L-rhamnose exhibit a dramatically different immune response from monomeric L-rhamnose. As the first example of nanoscale assemblies of a saccharide to suppress immunity, this work illustrates an approach of molecular nanofibers to modulate immune response. Furthermore, the understanding of the mechanisms behind this work will certainly offer a new way for developing immunomodulatory materials.
Figure 5.
(a) Molecular structures of 3, 4, and 5. (b) TEM image and optical image (inset) of 3 at the concentration of 0.4 wt%.(scale bar: 100 nm). (c) Total immunoglobulin G (lgG) levels against PE in serum of mice 28 days after intraperitoneal immunization by the indicated samples. (d) Illustration of the interaction of L-rhamnose in molecular nanofibrils with natural antibodies (α-L-rhamnose IgM) in mediating the immune response.[50] (Print with the permission from © 2014 Royal Society of Chemistry with permission)
3. Spatiotemporal control of nanoscale assemblies and the biological applications and implications
Intracellular nanoscale assemblies of small molecules
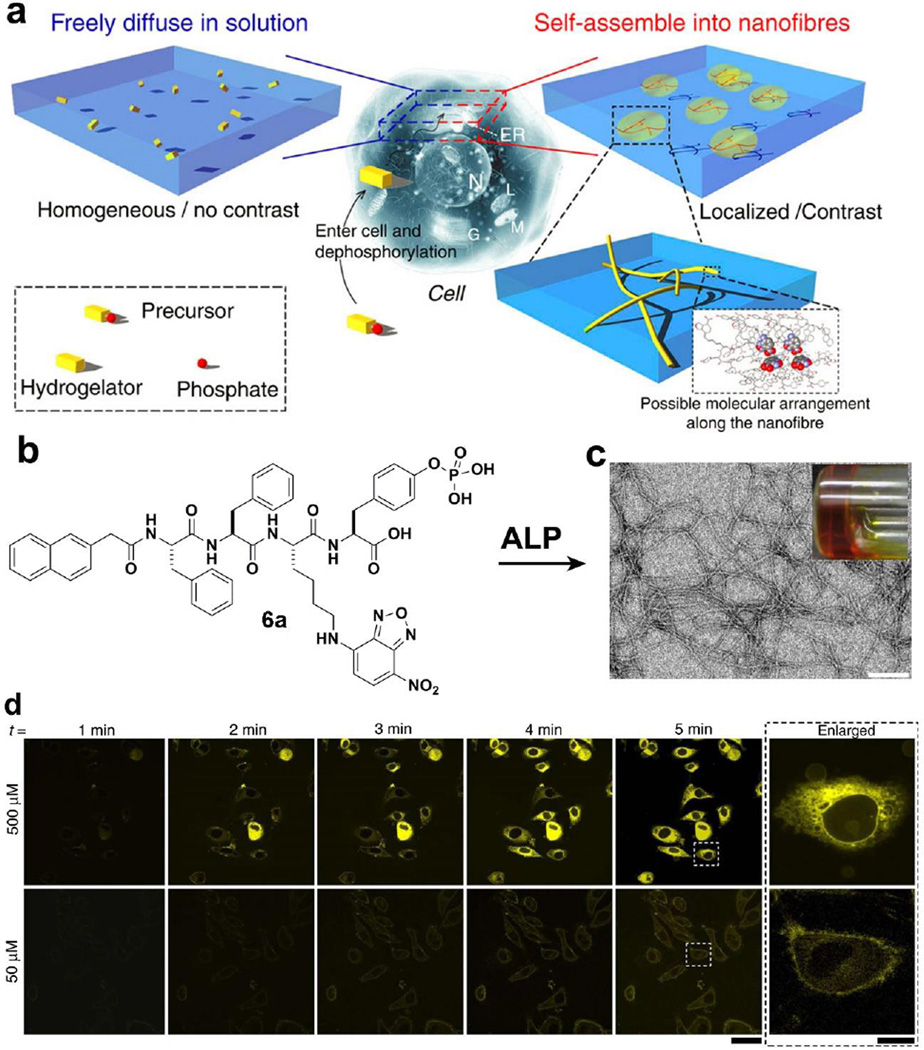
Because it is critical to control the self-assembly of small molecules in cellular environments for exploring the biological functions of assemblies of small molecules, we have been developing EISA of small molecules as a multistep process to produce nanoscale assemblies in cellular environment. In addition, it is necessary to evaluate the spatiotemporal profile[51] of nanoscale assemblies of small molecules in the cellular environment for understanding how nanoscale assemblies to control cell fates. Thus, we designed and synthesized a precursor (6a) by linking a fluorophore (4-nitro-2,1,3-benzoxadiazole(NBD)) to a self-assembly motif.[28] In particular, as shown in Fig. 6a, 6a consists of a phosphate group that not only acts as the enzyme trigger, but also makes the precursor dissolve well in water. The homogeneous distribution of the fluorescence of 6a ensures a uniform background and low fluorescence. After dephosphorylation by ALP, the resulting hydrogelators (6b: without phosphate group in 6a) self-assemble to form nanofibers at certain concentrations (e.g., above the critical aggregation concentration) (Fig. 6b, c). Because the nanofibers consist of aggregated fluorophores, they provide a high contrast for imaging the self-assembly of the hydrogelator. As shown in Fig. 6d, the incubation of the cells with 500 µM of 6a for 5 min, the cells glow brighter in the center of cytoplasm than the outer membrane of cells, indicating the formation of intracellular nanoscale assemblies. Cells incubated with 6a at a concentration (50 µM) below the threshold concentration of self-assembly of 6b, hardly exhibit any fluorescence, because there are too few molecules 6b to result in nanofibers for imaging. Furthermore, the co-incubation of cells with 6a and an inhibitor of phosphatase (CinnGEL 2Me, 25µM) is able to delay the appearance of fluorescence. This observation, together with the result of cell fraction experiments, indicates that the dephosphorylation of 6a and subsequently self-assembly of 6b occur in the outer membrane of the endoplasmic reticulum. This work provides important insights for understanding the nanoscale assemblies of small molecules in cellular environments.
Figure 6.
(a) Principle of imaging enzyme-instructed self-assembly inside cells. (b) Molecular structure of 6a. (c) Optical image and TEM image of the hydrogel made of 6b. (d) Fluorescent confocal microscope images show the time course of fluorescence emission inside the HeLa cells incubated with 500 or 50 µM of 6a in PBS buffer. Scale bar in Fig.6c is 100 nm, 50µm in Fig.6d.[28] (Print with the permission from © 2012 Nature Publishing group)
Imaging nanoscale assemblies of small molecules in cellular environments
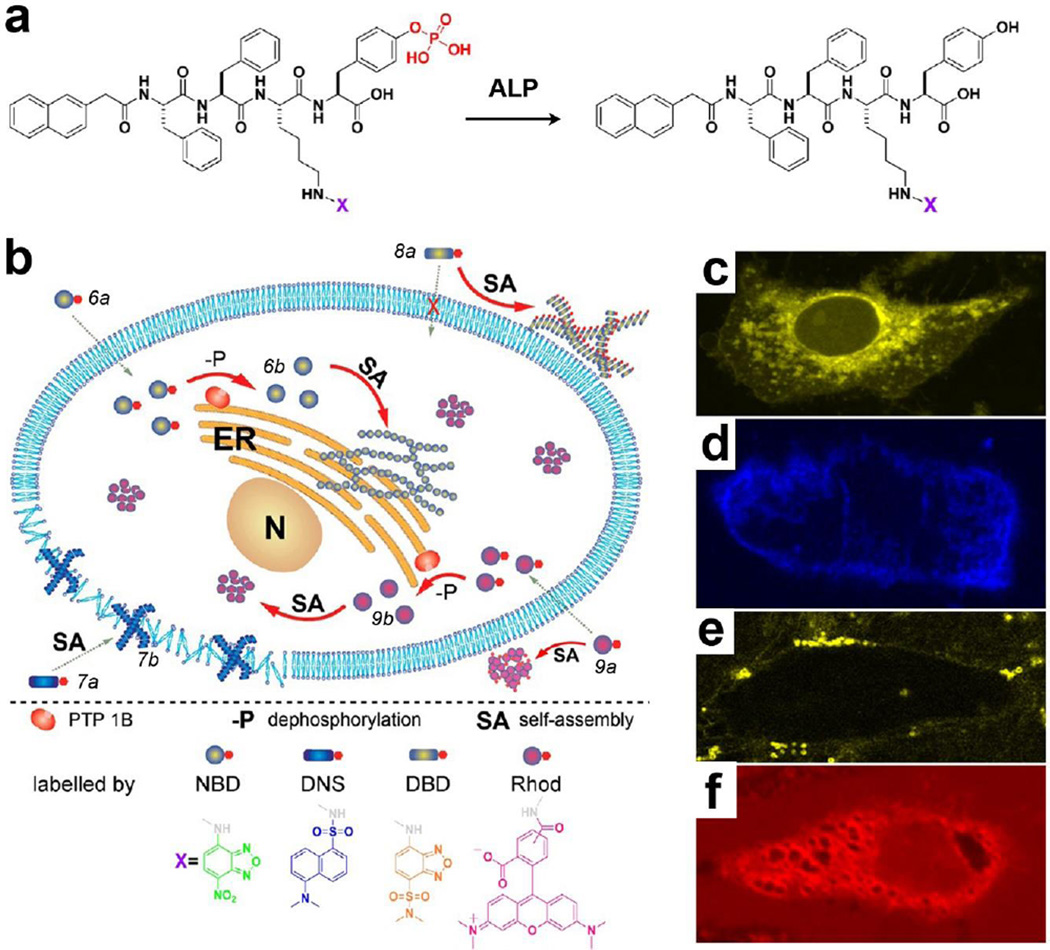
To further understand the properties of the nanoscale assemblies of small molecules in the cellular environment, we prepared a series of fluorescent precursors (e.g. 7a) via conjugating with different dyes (e.g. DBD).[52] Upon enzymatic dephosphorylation (Fig. 7a), these precursors transform to their corresponding hydrogelators and exhibit higher contrast than background for imaging. As shown in Fig. 7b, fluorescence images reveal the distribution of the assemblies. Due to the different self-assembling ability originates from their subtly different structures, these molecules exhibit distinct spatiotemporal profiles in the cells, which lead to different cellular responses. For example, the assemblies of 6b localize in the cytoplasm of cells and are innocuous for 48h even if the concentration is as high as 500 µM (Fig. 7c). The assemblies of 7b mainly adhere to the cell membrane to result in inhibition of HeLa cells (Fig. 7d). The assemblies of 8b distribute randomly outside of cells and exhibit little effect on the cells (Fig. 7e), thus it is cytocompatible. The molecules of 9b, being unable to self-assemble, show homogenous fluorescence inside and outside of cells (Fig. 7f), indicating that 9b exhibits little specific interaction with cell organelles. Thus, 9a/9b exhibit dose-dependent cytotoxicity to the cells. This work illustrates that EISA can control the fate of cells via modulating the spatiotemporal profile of the nanoscale assemblies of small molecules.
Figure 7.
(a) Molecular structures of the precursors and the hydrogelators containing different fluorophores. (b) Illustration of the distinct spatial distribution of the small molecules in a cellular environment. Fluorescent confocal images of the HeLa cells incubated with 500 µM of (c) 6a, (d) 7a, (e) 8a, and (f) 9a for 30 min.[52] (Print with the permission from © 2013 American Chemical Society)
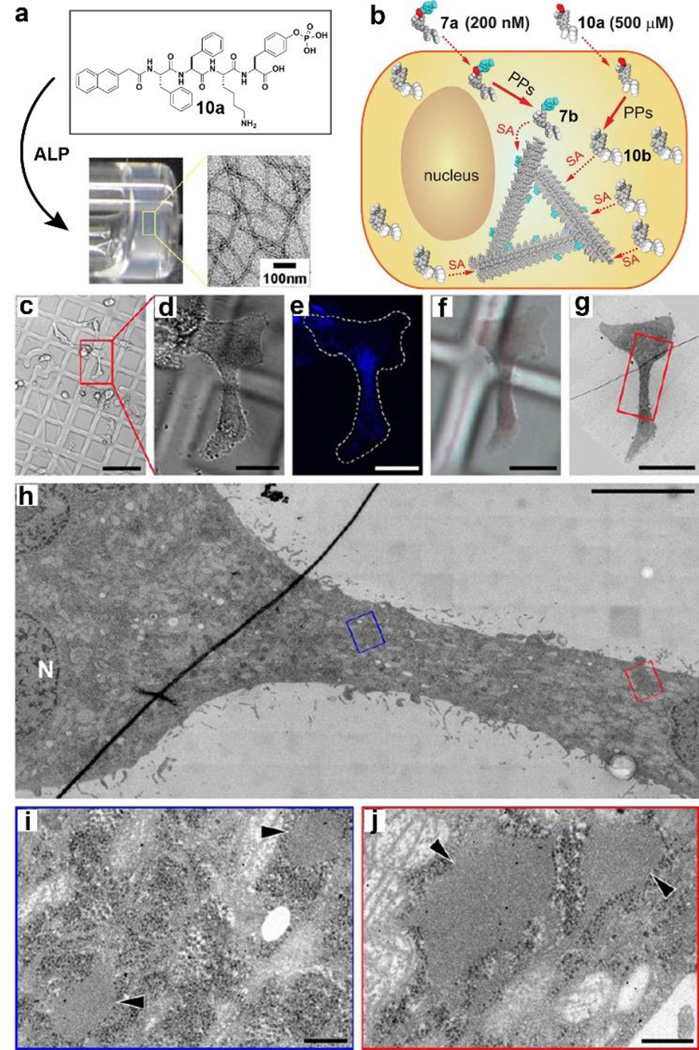
By generating heterotypic nanoscale assemblies of small molecules, we developed a new way to visualize the assemblies of non-fluorescent small molecule in cellular environment.[53] A well-designed precursor (10a) self-assembles to form nanofibers after enzymatic dephosphorylation at the concentration of 500 µM. To evaluate the spatiotemporal profile of assemblies of 10b, we chose 7a as fluorescence dopant since 7b and 10b share the same self-assembly motif. 7b is unable to form nanofibers at the concentration of 200 nM, thus it hardly shows fluorescence. However, 7b co-assembles with 10b at 500 µM to reveal the assemblies of 10b in cellular environment (Fig. 8b).[53] In addition, we used correlate light and electron microscopy (CLEM) to image cells treated with 10a and 7a. This technique allows us to correlate the fluorescence signal of co-assemblies of 10b/7b in live cells and the ultrastructural organization of the same cells. As shown in Fig. 8c–j, CLEM shows a high accumulation of vesicles with low electron-dense materials in the cytoplasmic region that also shows a high fluorescence signal. Further studies found that the molecules of 10b self-assemble and localize in the endoplasmic reticulum, also may enter the early exocytosis via the cellular secretory pathway (i.e., ER-Golgi-lysosomes/secretion). This work illustrates CLEM as a general approach to evaluate the spatiotemporal profile of nanoscale assemblies of small molecules in cellular environment. These insights should promise a new paradigm for controlling cell fates based on interactions between the nanoscale assemblies of small molecules and subcellular organelles.
Figure 8.
(a) Precursor 10a, upon the addition of ALP, become self-assembling 10b to form nanofibers/hydrogels. (b) Illustration of doping method to visualize assemblies of 10b. (c–j) Correlative light and electron microscopy (CLEM) of HeLa cells incubated for 48h with 500 µM 10a and 200 nM 7a. (c–f) DIC and fluorescence light microscopy images of treated HeLa cells growing on Aclar plastic film. (h–j) TEM images of the cell of interest shown in Fig. 8c–f.[53] (Print with the permission from © 2013 American Chemical Society)
Pericellular nanoscale assemblies of small molecules
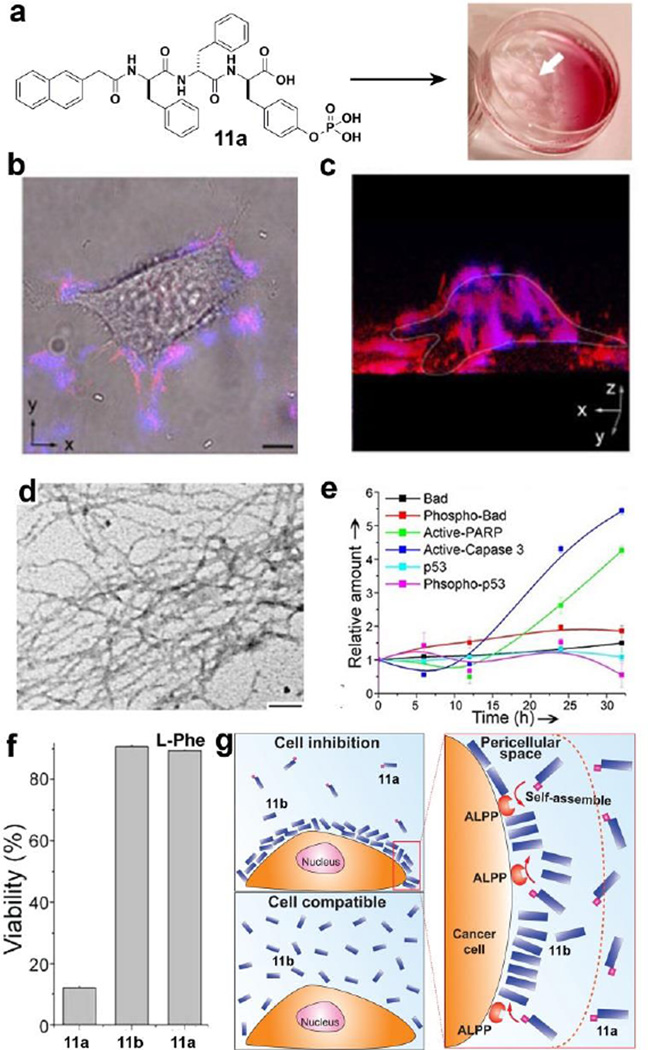
As enzymes located on the cell surface, ectoenzymes[54, 55] play important roles in cellular processes, but they have received little attention for regulating the self-assembly of small molecules. We reported the first example of using ectoenzyme to catalyse the self-assembly of small molecules, which leads to the formation of nanofibers/hydrogel in pericellular space.[56] As shown in Fig. 9a, the designed precursor 11a, based on a D-tripeptide, is able to form a hydrogel upon incubation of HeLa cells with 11a (560 µM) for 2h. TEM reveals the resulting hydrogelator 11b self-assembles to form nanofibers (Fig. 9d). After treating with 11a at 280 µM and staining with Congo red and DAPI for 12h, HeLa cells exhibits significantly red fluorescence on the cell surface, indicating the formation of nanofibers/hydrogels in the pericellular space (Fig. 9b–c). Meanwhile, DAPI (nucleus dye, blue) is unable to stain the nucleus, but co-localizes with the red fluorescence from Congo red, suggesting that the pericellular nanofibers even prevent diffusion of small molecules into the cells. We also found that the pericellular nanofibers block the cellular mass exchange between HeLa cells and their environment to inhibit the proliferation of the HeLa cells. Moreover, the pericellular nanofibers decrease migration of HeLa cells and inhibit several cancer cells, including a drug-resistance cell line (e.g. MES-SA/dx5 cells). In addition, as revealed by ELISA, active-caspase-3 and PARP show significant increase at 24 or 32 h, suggesting that the HeLa cells undergo caspase-dependent apoptosis.
Figure 9.
(a) HeLa cells incubated with precursor 11a (560 µM) for 2h afford a hydrogel on the cells. (b) Overlaid images and (c) 3D stacked z-scan images of Congo red and DAPI stained HeLa cells after the incubation of the HeLa cells with 11a for 12h. (d) TEM images of the pericellular nanofibers on the HeLa cells treated by 11a (280 µM). (e) Change of relative amount of apoptosis signal molecules over time in HeLa cells treated by 11a (280 µM). (f) Cell viability of HeLa cells incubated with 11a (280 µM), 11b (280 µM), and 11a (280 µM) plus L-Phe (1.0 mM). (g) Enzyme-instructed self-assembly to form pericellular nanofibers/hydrogel and selectively induce cell death.[56, 57] (Print with the permission from © 2004 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim)
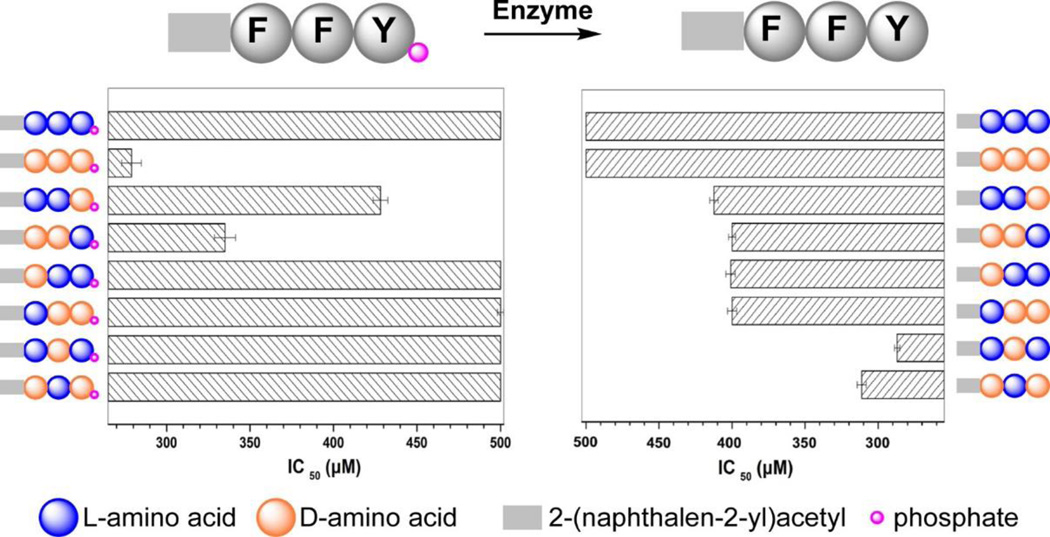
Furthermore, as the enantiomer of 11a, L-peptide derivative (12a) hardly inhibits HeLa cells because 12a undergoes proteolysis. Based on this observation, we used D-amino acids systematically replacing the corresponding L-amino acids in the tripeptidic precursor 12a and hydrogelator 12b.[57] As shown in Fig. 10, the enantiomer pair of the precursors inhibits HeLa cells requiring quite different dosage, while the enantiomer pair of the hydrogelators exhibits similar inhibitory activities. Except 12a, other precursors are able to resist proteolytic hydrolysis due to the incorporation of D-amino acid(s). Moreover, the precursors that undergo higher rate of dephosphorylation exhibit higher cytotoxicities towards HeLa cells, indicating that the kinetics of phosphorylation determine the rate of the formation of nanoscale assemblies of the hydrogelators on the HeLa cells, thus, resulting in different cellular response. We found that the use of L-Phe, an uncompetitive inhibitor of placental alkaline phosphatase (ALPP), to treat HeLa cells abrogates the cytotoxicity of 11a, confirming that the overexpressed ALPP on HeLa cells contributes to EISA (Fig. 9f) resulted inhibition of the cells. In addition, the hydrogelator, 11b, is innocuous to HeLa cells even at 500 µM. This observation indicates that ALPP, as the ectoenzyme, catalyzes the dephosphorylation of 11a, results in the accumulation of 11b on the cell surface, and leads to in-situ formation of the nanofibers of 11b. Thus, the pericellular nanofibers selectively form on cancer cells to inhibit the cancer cells. This work illustrates EISA as a fundamentally new approach to modulate the spatiotemporal profiles of nanoscale assemblies in cellular microenvironment to control the fate of cells.
Figure 10.
IC50 values of precursors (e.g., 12a, the first one in left panel) and its hydrogelators (e.g.,12b, the first one in right panel) on HeLa cells. F and Y indicate phenylalanine and tyrosine.[57] (Print with the permission from © 2014 American Chemical Society)
Recently, Pires and Ulijn et al. designed a simple and innovative carbohydrate amphiphile that is able to self-assemble into nanofibers and result in a hydrogel on the surface of SaO2 rather than ATDC5 cells. The EISA of the carbohydrate amphiphile results in the formation of pericellular nanofibers, which selectively inhibit the proliferation of the SaO2 cancer cells.[58] Based on the EISA process, Maruyama et al. reported that matrix metalloproteinase-7 (MMP-7) hydrolyze a well-designed lipid-peptide to afford a hydrogelator. The resulting hydrogelator enters the cells and self-assembly to form nanofibrils that impair cellular functions, which lead to selectively inhibition to cancer cells.[59] In addition, we reported that the CD73, an ectoenzyme that catalyzes the dephosphorylation of a nucleopeptide forms a hydrogelator. The assemblies of the resulting hydrogelators are able to inhibit the HepG2 cells that express CD73.[60] In another experiment, we decorated the magnetic nanoparticles with D-tyrosine phosphate. The resulting nanoparticles selectively adhere to cancer cells in co-culture after dephosphorylation, and selectively inhibit the cancer cells.[61, 62]
4. Molecular Hydrogel Protein Binding (MHPB) Assay
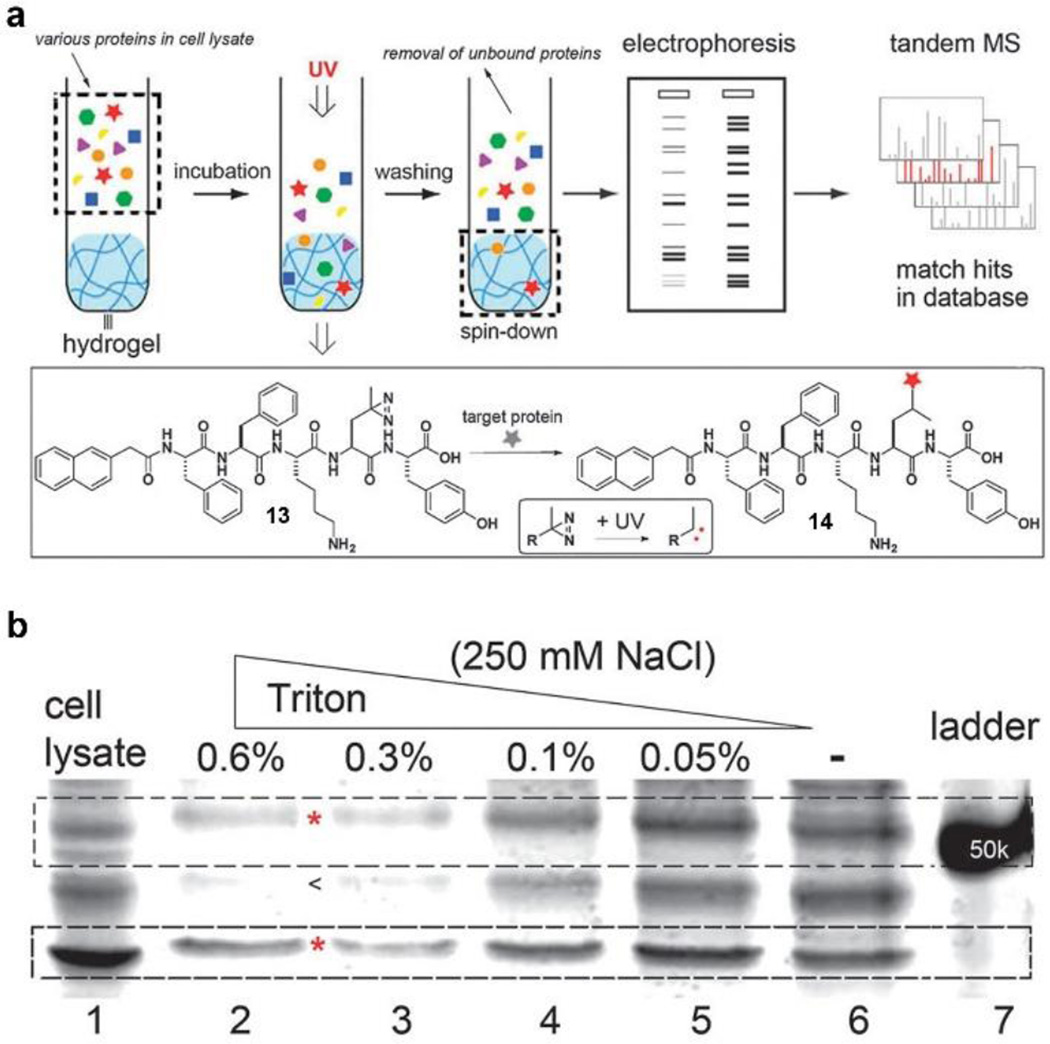
To provide a general assay for evaluating the interaction between nanoscale assemblies of small molecules and cellular components, we, based on the resemblance of the cellular environment and hydrogels, used supramolecular nanofibers in hydrogel to discover the target proteins of the nanoscale assemblies of small molecules. As shown in Fig. 11a, the hydrogel is able to retain the proteins that bind with the nanofibers of small molecules. SDS-Page separates the bound proteins for silver staining, and tandem MS identifies the composition of the proteins.[41] Specifically, we designed a hydrogelator (13) containing photoleucine. 13 self-assembles to form nanofibers and afford a hydrogel at physiological conditions. Being incubated with the cell lysate of HeLa cells, the resulting hydrogel retains the target proteins, which are fixed by photo activation. We used a series of concentration of a Triton surfactant together with NaCl solution to wash the hydrogel for removing non-specifically bound proteins. After that, SDS-Page separates the proteins released from the hydrogel. As shown in Fig. 11b, the intensities of the bands of the nonspecific-absorbed protein decrease with the increase in Triton concentration, the intensities of bands in lane 2 and 3 remain constant, indicating the protein in those bands has specific interactions with nanoscale assemblies of 13. Tandem MS and Western-blot analysis suggest that nanoscale assemblies of 13 interact with tubulins, actins, and several other proteins in the cells. This work provides a facile approach to study the interaction of nanoscale assemblies of small molecules and cellular proteins. Using this facile assay, we explored the interactions of cytosol proteins with morphologically different nanoscale assemblies formed by the same molecule (1).[63] MHPB assay shows the nanofibers of 1 bind to more proteins than precipitates of 1 do, confirming that the morphology of nanoscale assemblies of small molecules plays a critical role in interacting with cellular proteins. The correlation of morphological control of the nanoscale assemblies and their biological functions will contribute to the understanding of the biological, as well as pharmacological, properties of the assemblies of small molecules.
Figure 11.
Illustration of the molecular hydrogel protein binding (MHPB) assay that the hydrogel protein pull-down coupled with electrophoresis and tandem mass spectrometry for identifying of cytosolic proteins that bind to molecular assemblies. (a) Photo reaction of hydrogelator (13) and molecular assemblies bind with proteins. (b) Silver staining of SDS-Page gel shows that different conditions alter the proteins binding on the supramolecular hydrogel.[41] (Print with the permission from © 2012 Royal Society of Chemistry)
5. Ligand-Receptor Interaction Catalyzes the Formation of Nanoscale Assemblies of Small Molecules
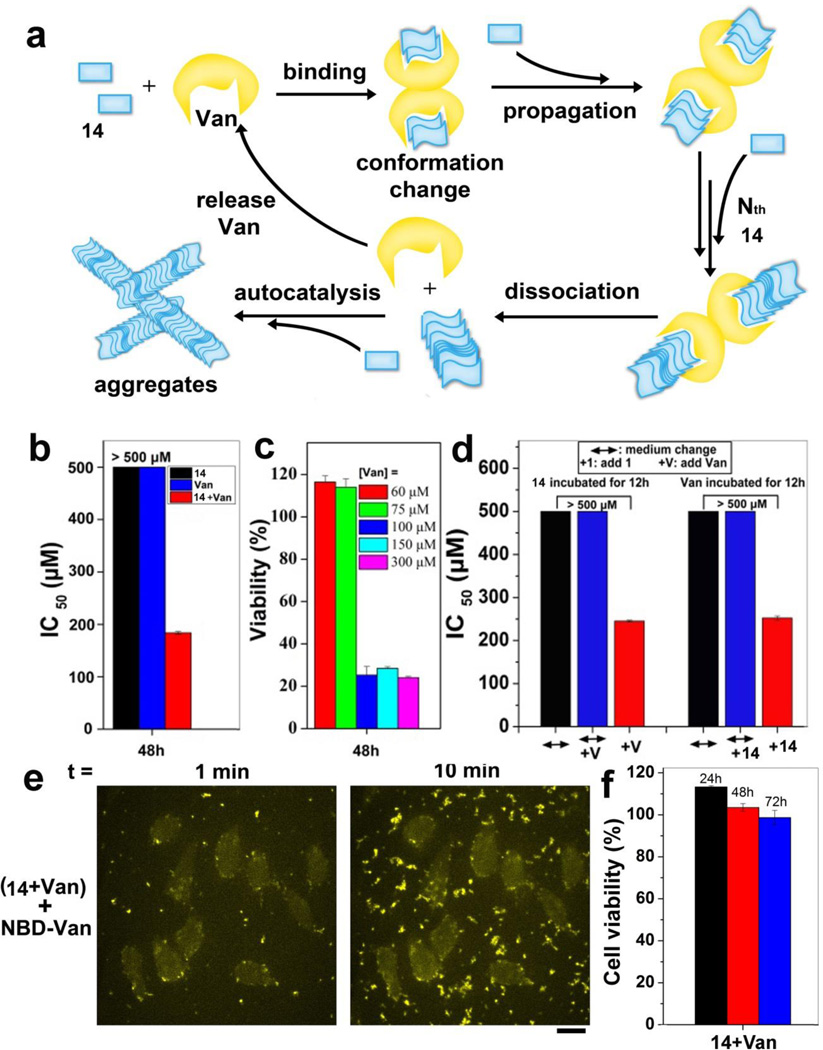
In addition to the use of EISA or the change of pH or temperature to form the nanoscale assemblies of small molecules, we recently developed a new approach that uses ligand-receptor interaction for catalyzing the formation of nanoscale assemblies of small molecules.[64] As shown in Fig. 12a, via ligand-receptor interactions, two molecules of vancomycin (Van) bind with four molecules of Fmoc-Lys-Gly-Gly-D-Ala-D-Ala (14). Such binding catalyzes the formation of assemblies of 14. Meanwhile, the Van in assemblies released to surrounding solution over time continues to catalyze the production of assemblies of 14. The resulting assemblies of 14 are able to inhibit the proliferation of HeLa cells with IC50 value of 184 µM (Fig. 12b). Moreover, when the concentrations of Van reduced from 300 µM to 100 µM, the assemblies catalyzed by Van still inhibit HeLa cells, suggested that ligand-receptor interaction, indeed, catalyzes the formation of nanoscale assemblies (Fig. 12c) that possess biological activities. As shown in Fig. 12d, the cell viability test under different incubation conditions not only confirms that ligand-receptor interaction catalyzes the assemblies of 14 and induces the cell death, but also suggests that the extracellular aggregates result in the cell death. Fluorescence images show that the assemblies of 14 increase significantly in extracellular environment, implying that the resulting assemblies of 14 cause cell death (Fig. 12e). Further experiment reveals that the cells likely undergo necroptosis. It was worth to note that the pre-formed aggregates of 14, without sufficient amount of Van, are innocuous to HeLa cells (Fig. 12f). This result further proves the essential role of Van in producing, as well as maintaining, the assemblies of 14. As the first case that the ligand-receptor interaction catalyzes the assembly of small molecules, this result shares common feature with prions[15, 65] that pathogenic prions unfold normal prion proteins to form neurotoxic aggregates. In addition, we also demonstrated the use of enzymatic transformation to regulate the ligand-receptor interaction of small molecules, in which the enzymatic reaction change the binding ratios of Van and D-Ala-D-Ala derivatives from 1:1 to 1: 2.[66] Since the integration of enzyme catalysis, ligand-receptor interactions, and self-assembly is a common feature of many cellular processes (e.g., apoptosis),[67] the use of nanoscale assemblies of small of molecules to achieve a similar feature may lead to a new way to control cell fates.
Figure 12.
(a) Illustration of ligand-receptor interaction catalyzes the formation of nanoscale molecular assemblies. (b) IC50 values of 14 without and with vancomycin ([14]0/[Van]0 = 1:1) against HeLa cells for 48 h. (c) At 48 h, the viability of HeLa cells incubated with 14 ([1]0 = 300 µM) and varying amounts of Van (from 60 µM to 300 µM). (d) IC50 values of (14+Van) against HeLa cells at different conditions. (e) Confocal images showing the time course of fluorescence emission from HeLa cells incubated with 14, Van, and NBD-Van. [14]0 = 300 µM, [Van]0 = 294 µM, [NBD-Van]0 = 6 µM. (f)Cytotoxicity of pre-formed aggregates of 14 against HeLa cells.14 and Van mixed well for 12h, separated the supernatant and aggregates by centrifugation, the obtained aggregates are pre-formed aggregates that is used to treat cells. [14]0 = [Van]0 = 500 µM. [64] (Print with the permission from © 2015 American Chemical Society)
6. Perspective and Challenge
The last decade has witnessed the emergence of nanoscale assemblies of small molecules as a new class of molecular entities to perform multiple biological functions in the cellular environment. Despite the scattering of the research of nanoscale assemblies of small molecules in a number of quite different fields, the biological significance of nanoscale assemblies of small molecules is an undeniable fact, and their promises are beginning to be realized. However, the exploration of the assemblies is just at its beginning,[68–78] and the relevant mechanistic understanding is far from complete. Despite the promiscuity of the nanoscale assemblies of small molecules in interacting with the proteins in the cells, the kinetics of the formation of the assemblies (e.g., EISA), the dynamics of their dissociation, and their stability in cellular environment offer abundant opportunities for developing selective therapies. One important application of the assemblies is to create novel nanomedicines with prescribed function. To achieve this goal, future studies likely need to focus on understanding the function of nanoscale assemblies of small molecules in the cellular environment, especially in cell-based assays.
Additionally, some of fundament questions remain to be addressed. For example, what are the molecular targets of the nanoscale assemblies? How to generate nanoscale assemblies with controlled dimensions and desired functions? How do nanoscale assemblies transport in and out the cells? How to use molecular engineering to control the stability of nanoscale assemblies in the cells? How to improve the potency or activity of nanoscale assemblies of small molecules? To answer these questions, it requires the interdisciplinary collaboration among the scientist from chemistry, biological, physical, and medical science. Particularly, it is important for chemists to move beyond molecules.[79] One of the reasonable and convenient starting point of nanoscale assemblies of small molecules is to explore the biological functions of molecules that self-assemble in water.[80–94] The development of supramolecular gelators over the last two decades,[95–106] apparently, has already provided a large pool of molecular candidates for such kind of exploration. We hope that this review will stimulate increased number of scientists, who work in nanoscience and nanotechnology, to design and to explore nanoscale assemblies of small molecules for solving a wide range of problems in biomedicine as well as other relevant fields.
Acknowledgements
We are grateful for the financial support from NIH (R01CA142746), NSF MRSEC (DMR-1420382), and the W. M. Keck Foundation.
References
- 1.Mitchison T, Kirschner M. Nature. 1984;312:232–237. doi: 10.1038/312232a0. [DOI] [PubMed] [Google Scholar]
- 2.Mitchison T, Kirschner M. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 3.Brenner S. Cold Spring Harb Sym. 1961;26:101–110. doi: 10.1101/sqb.1961.026.01.015. [DOI] [PubMed] [Google Scholar]
- 4.Brenner S, Jacob F, Meselson M. Nature. 1961;190:576–581. doi: 10.1038/190576a0. [DOI] [PubMed] [Google Scholar]
- 5.Shi Y. Structure. 2002;10:285–288. doi: 10.1016/s0969-2126(02)00732-3. [DOI] [PubMed] [Google Scholar]
- 6.Martinon F, Burns K, Tschopp J. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 7.Lashuel HA, Hartley D, Petre BM, Walz T, Lansbury PT. Nature. 2002;418:291-291. doi: 10.1038/418291a. [DOI] [PubMed] [Google Scholar]
- 8.Hegde RS, Mastrianni JA, Scott MR, DeFea KA, Tremblay P, Torchia M, DeArmond SJ, Prusiner SB, Lingappa VR. Science. 1998;279:827–834. doi: 10.1126/science.279.5352.827. [DOI] [PubMed] [Google Scholar]
- 9.Mann S. Angew. Chem. Int. Ed. 2008;47:5306–5320. doi: 10.1002/anie.200705538. [DOI] [PubMed] [Google Scholar]
- 10.Glover JR, Lindquist S. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 11.Szostak JW. Phil. Trans. R. Soc. B. 2011;366:2894–2901. doi: 10.1098/rstb.2011.0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caron P, Beckers A, Cullen DR, Goth MI, Gutt B, Laurberg P, Pico AM, Valimaki M, Zgliczynski W. J. Clin. Endocrinol. Metab. 2002;87:99–104. doi: 10.1210/jcem.87.1.8153. [DOI] [PubMed] [Google Scholar]
- 13.McGovern SL, Caselli E, Grigorieff N, Shoichet BK. J. Med. Chem. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- 14.Feng BY, Toyama BH, Wille H, Colby DW, Collins SR, May BCH, Prusiner SB, Weissman J, Shoichet BK. Nat. Chem. Biol. 2008;4:197–199. doi: 10.1038/nchembio.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prusiner SB. Science. 1991;252:1515–1522. doi: 10.1126/science.1675487. [DOI] [PubMed] [Google Scholar]
- 16.Prusiner SB, McKinley MP, Bowman KA, Bolton DC, Bendheim PE, Groth DF, Glenner GG. Cell. 1983;35:349–358. doi: 10.1016/0092-8674(83)90168-x. [DOI] [PubMed] [Google Scholar]
- 17.McGovern SL, Helfand BT, Feng B, Shoichet BK. J. Med. Chem. 2003;46:4265–4272. doi: 10.1021/jm030266r. [DOI] [PubMed] [Google Scholar]
- 18.Zorn JA, Wille H, Wolan DW, Wells JA. J. Am. Chem. Soc. 2011;133:19630–19633. doi: 10.1021/ja208350u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zorn JA, Wolan DW, Agard NJ, Wells JA. J. Biol. Chem. 2012;287:33781–33795. doi: 10.1074/jbc.M112.386128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Z, Liang G, Guo Z, Xu B. Angew. Chem., Int. Ed. 2007;46:8216–8219. doi: 10.1002/anie.200701697. [DOI] [PubMed] [Google Scholar]
- 21.Kato M, Han Tina W, Xie S, Shi K, Du X, Wu Leeju C, Mirzaei H, Goldsmith Elizabeth J, Longgood J, Pei J, Grishin Nick V, Frantz Douglas E, Schneider Jay W, Chen S, Li L, Sawaya Michael R, Eisenberg D, Tycko R, McKnight Steven L. Cell. 149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frenkel YV, Clark AD, Das K, Wang YH, Lewi PJ, Janssen PAJ, Arnold E. J. Med. Chem. 2005;48:1974–1983. doi: 10.1021/jm049439i. [DOI] [PubMed] [Google Scholar]
- 23.Xing BG, Yu CW, Chow KH, Ho PL, Fu DG, Xu B. J. Am. Chem. Soc. 2002;124:14846–14847. doi: 10.1021/ja028539f. [DOI] [PubMed] [Google Scholar]
- 24.Xing BG, Ho PL, Yu CW, Chow KH, Gu HW, Xu B. Chem Commun. 2003:2224–2225. doi: 10.1039/b305886g. [DOI] [PubMed] [Google Scholar]
- 25.Yang ZM, Xu KM, Guo ZF, Guo ZH, Xu B. Adv Mater. 2007;17:3152–3156. [Google Scholar]
- 26.Yang Z, Liang G, Guo Z, Guo Z, Xu B. Angew. Chem. Int. Ed. 2007;46:8216–8219. doi: 10.1002/anie.200701697. [DOI] [PubMed] [Google Scholar]
- 27.Yang Z, Liang G, Ma M, Abbah AS, Lu WW, Xu B. Chem. Commun. 2007:843–845. doi: 10.1039/b616563j. [DOI] [PubMed] [Google Scholar]
- 28.Gao Y, Shi J, Yuan D, Xu B. Nat. Commun. 2012;3:1033. doi: 10.1038/ncomms2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li JY, Kuang Y, Gao Y, Du XW, Shi JF, Xu B. J. Am. Chem. Soc. 2013;135:542–545. doi: 10.1021/ja310019x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Querfurth HW, LaFerla FM, Engl N. J. Med. 2010;362:329–344. doi: 10.1056/NEJMra0909142. [DOI] [PubMed] [Google Scholar]
- 31.Hardy J, Selkoe DJ. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 32.Haass C, Selkoe DJ. Nat Rev Mol Cell Bio. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 33.Liu Z, Meray RK, Grammatopoulos TN, Fredenburg RA, Cookson MR, Liu Y, Logan T, Lansbury PT. Proc. Natl. Acad. Sci. U.S.A. 2009;106:4635–4640. doi: 10.1073/pnas.0806474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolognesi B, Jahn TR, Brorsson AC, Luheshi LM, Yerbury JJ, Crowther DC, Dobson CM. FEBS J. 2010;277:79–80. [Google Scholar]
- 35.Svanborg C, Agerstam H, Aronson A, Bjerkvig R, Duringer C, Fischer W, Gustafsson L, Hallgren O, Leijonhuvud I, Linse S, Mossberg AK, Nilsson H, Pettersson J, Svensson M. Adv Cancer Res. 2003;88:1–29. doi: 10.1016/s0065-230x(03)88302-1. [DOI] [PubMed] [Google Scholar]
- 36.Harada R, Tochio N, Kigawa T, Sugita Y, Feig M. J Am Chem Soc. 2013;135:3696–3701. doi: 10.1021/ja3126992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou R, Xu B. PLoS One. 2014;9:e95759. doi: 10.1371/journal.pone.0095759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuang Y, Xu B. Angew. Chem. Int. Edit. 2013;52:6944–6948. doi: 10.1002/anie.201302658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shoichet BK. J. Med. Chem. 2006;49:7274–7277. doi: 10.1021/jm061103g. [DOI] [PubMed] [Google Scholar]
- 40.Kuang Y, Du X, Zhou J, Xu B. Adv Healthcare Mater. 2014;3:1217–1221. doi: 10.1002/adhm.201300645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao Y, Long MJC, Shi J, Hedstrom L, Xu B. Chem. Commun. 2012;48:8404–8406. doi: 10.1039/c2cc33631f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuang Y, Yuan D, Zhang Y, Kao A, Du X, Xu B. RSC Adv. 2013;3:7704–7707. doi: 10.1039/C3RA41523F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clément M-J, Jourdain I, Lachkar S, Savarin P, Gigant B, Knossow M, Toma F, Sobel A, Curmi PA. Biochemistry. 2005;44:14616–14625. doi: 10.1021/bi0512492. [DOI] [PubMed] [Google Scholar]
- 44.Kuang Y, Long MJC, Zhou J, Shi J, Gao Y, Xu C, Hedstrom L, Xu B. J. Biol. Chem. 2014 doi: 10.1074/jbc.M114.600288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X, Du X, Gao Y, Shi J, Kuang Y, Xu B. Soft Matter. 2012;8:7402–7407. doi: 10.1039/C2SM25725D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du X, Zhou J, Guvench O, Sangiorgi FO, Li X, Zhou N, Xu B. Bioconjugate Chem. 2014;25:1031–1035. doi: 10.1021/bc500187m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giancotti FG, Ruoslahti E. Science. 1999;285:1028–1033. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 48.Chen W, Gu L, Zhang W, Motari E, Cai L, Styslinger TJ, Wang PG. ACS Chem. Biol. 2011;6:185–191. doi: 10.1021/cb100318z. [DOI] [PubMed] [Google Scholar]
- 49.Sarkar S, Lombardo SA, Herner DN, Talan RS, Wall KA, Sucheck SJ. J. Am. Chem. Soc. 2010;132:17236–17246. doi: 10.1021/ja107029z. [DOI] [PubMed] [Google Scholar]
- 50.Zhao F, Heesters BA, Chiu I, Gao Y, Shi J, Zhou N, Carroll MC, Xu B. Org. Biomol. Chem. 2014;12:6816–6819. doi: 10.1039/c4ob01362j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams RJ, Smith AM, Collins R, Hodson N, Das AK, Ulijn RV. Nat Nano. 2009;4:19–24. doi: 10.1038/nnano.2008.378. [DOI] [PubMed] [Google Scholar]
- 52.Gao Y, Kuang Y, Du X, Zhou J, Chandran P, Horkay F, Xu B. Langmuir. 2013;29:15191–15200. doi: 10.1021/la403457c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao Y, Berciu C, Kuang Y, Shi J, Nicastro D, Xu B. ACS Nano. 2013;7:9055–9063. doi: 10.1021/nn403664n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fishman WH, Inglis NR, Green S, Anstiss CL, Gosh NK, Reif AE, Rustigian R, Krant MJ, Stolbach LL. Nature. 1968;219:697–699. doi: 10.1038/219697a0. [DOI] [PubMed] [Google Scholar]
- 55.Huang Y, Shi J, Yuan D, Zhou N, Xu B. Peptide Science. 2013;100:790–795. doi: 10.1002/bip.22240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuang Y, Shi J, Li J, Yuan D, Alberti KA, Xu Q, Xu B. Angew. Chem. Int. Ed. 2014;53:8104–8107. doi: 10.1002/anie.201402216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi J, Du X, Yuan D, Zhou J, Zhou N, Huang Y, Xu B. Biomacromolecules. 2014;15:3559–3568. doi: 10.1021/bm5010355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pires RA, Abul-Haija YM, Costa DS, Novoa-Carballal R, Reis RL, Ulijn RV, Pashkuleva I. J. Am. Chem. Soc. 2015;137:576–579. doi: 10.1021/ja5111893. [DOI] [PubMed] [Google Scholar]
- 59.Tanaka A, Fukuoka Y, Morimoto Y, Honjo T, Koda D, Goto M, Maruyama T. J. Am. Chem. Soc. 2015;137:770–775. doi: 10.1021/ja510156v. [DOI] [PubMed] [Google Scholar]
- 60.Wu D, Du X, Shi J, Zhou J, Zhou N, Xu B. J. Colloid Interface Sci. 2015;447:269–272. doi: 10.1016/j.jcis.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Du X, Zhou J, Wu L, Sun S, Xu B. Bioconjugate Chem. 2014;25:2129–2133. doi: 10.1021/bc500516g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Du X, Zhou J, Xu B. J. Colloid Interface Sci. 2015;447:273–277. doi: 10.1016/j.jcis.2014.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuang Y, Yuan D, Zhang Y, Kao A, Du XW, Xu B. RSC Adv. 2013;3:7704–7707. doi: 10.1039/C3RA41523F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi J, Du X, Huang Y, Zhou J, Yuan D, Wu D, Zhang Y, Haburcak R, Epstein IR, Xu B. J. Am. Chem. Soc. 2015;137:26–29. doi: 10.1021/ja5100417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prusiner S. Science. 1991;252:1515–1522. doi: 10.1126/science.1675487. [DOI] [PubMed] [Google Scholar]
- 66.Shi J, Du X, Yuan D, Haburcak R, Wu D, Zhou N, Xu B. Chem. Commun. 2015;51:4899–4901. doi: 10.1039/c4cc10374b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou J, Xu B. Bioconjug Chem. 2015;26:987–999. doi: 10.1021/acs.bioconjchem.5b00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yuan D, Zhou R, Shi J, Du X, Li X, Xu B. RSC Adv. 2014;4:26487–26490. doi: 10.1039/C4RA04765F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Julien O, Kampmann M, Bassik MC, Zorn JA, Venditto VJ, Shimbo K, Agard NJ, Shimada K, Rheingold AL, Stockwell BR, Weissman JS, Wells JA. Nat. Chem. Biol. 2014;10:969–976. doi: 10.1038/nchembio.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang H, Wei J, Yang C, Zhao H, Li D, Yin Z, Yang Z. Biomaterials. 2012;33:5848–5853. doi: 10.1016/j.biomaterials.2012.04.047. [DOI] [PubMed] [Google Scholar]
- 71.Liu J, Liu J, Chu L, Zhang Y, Xu H, Kong D, Yang Z, Yang C, Ding D. ACS Appl. Mater. Interfaces. 2014;6:5558–5565. doi: 10.1021/am406007g. [DOI] [PubMed] [Google Scholar]
- 72.Li J, Gao Y, Kuang Y, Shi J, Du X, Zhou J, Wang H, Yang Z, Xu B. J. Am. Chem. Soc. 2013;135:9907–9914. doi: 10.1021/ja404215g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu H, Li Y, Lyu Z, Wan Y, Li X, Chen H, Chen H, Li X. J Mater Chem. B. 2014;2:8303–8309. doi: 10.1039/c4tb01563k. [DOI] [PubMed] [Google Scholar]
- 74.Martin AD, Robinson AB, Mason AF, Wojciechowski JP, Thordarson P. Chem. Commun. 2014;50:15541–15544. doi: 10.1039/c4cc07941h. [DOI] [PubMed] [Google Scholar]
- 75.Tang A-M, Wang W-J, Mei B, Hu W-L, Wu M, Liang G-L. Sci. Rep. 2013;3 doi: 10.1038/srep01848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu X-D, Liang L, Cheng H, Wang X-H, Jiang F-G, Zhuo R-X, Zhang X-Z. J. Mater. Chem. 2012;22:18164–18171. [Google Scholar]
- 77.Standley SM, Toft DJ, Cheng H, Soukasene S, Chen J, Raja SM, Band V, Band H, Cryns VL, Stupp SI. Cancer Res. 2010;70:3020–3026. doi: 10.1158/0008-5472.CAN-09-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zheng L-H, Wang Y-J, Sheng J, Wang F, Zheng Y, Lin X-K, Sun M. Mar. Drugs. 2011;9:1840–1859. doi: 10.3390/md9101840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Whitesides GM. Angew. Chem. Int. Ed. 2015;54:3196–3209. doi: 10.1002/anie.201410884. [DOI] [PubMed] [Google Scholar]
- 80.Ulijn RV, Bibi N, Jayawarna V, Thornton PD, Todd SJ, Mart RJ, Smith AM, Gough JE. Mat Today. 2007;10:40–48. [Google Scholar]
- 81.Branco MC, Schneider JP. Acta Biomater. 2009;5:817–831. doi: 10.1016/j.actbio.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang SG. Biotechnol. Adv. 2002;20:321–339. doi: 10.1016/s0734-9750(02)00026-5. [DOI] [PubMed] [Google Scholar]
- 83.Estroff LA, Hamilton AD. Chem. Rev. 2004;104:1201–1217. doi: 10.1021/cr0302049. [DOI] [PubMed] [Google Scholar]
- 84.Rajagopal K, Schneider JP. Curr. Opin. Struc. Biol. 2004;14:480–486. doi: 10.1016/j.sbi.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 85.Palmer LC, Stupp SI. Acc. Chem. Res. 2008;41:1674–1684. doi: 10.1021/ar8000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Alivisatos AP, Barbara PF, Castleman AW, Chang J, Dixon DA, Klein ML, McLendon GL, Miller JS, Ratner MA, Rossky PJ, Stupp SI, Thompson ME. Adv Mater. 1998;10:1297–1336. [Google Scholar]
- 87.Zhao X, Pan F, Xu H, Yaseen M, Shan H, Hauser CAE, Zhang S, Lu JR. Chem. Soc. Rev. 2010;39:3480–3498. doi: 10.1039/b915923c. [DOI] [PubMed] [Google Scholar]
- 88.Zhang SG. Nat. Biotechnol. 2003;21:1171–1178. doi: 10.1038/nbt874. [DOI] [PubMed] [Google Scholar]
- 89.Zelzer M, Ulijn RV. Chem. Soc. Rev. 2010;39:3351–3357. doi: 10.1039/c0cs00035c. [DOI] [PubMed] [Google Scholar]
- 90.Ulijn RV, Smith AM. Chem. Soc. Rev. 2008;37:664–675. doi: 10.1039/b609047h. [DOI] [PubMed] [Google Scholar]
- 91.Zhao F, Ma ML, Xu B. Chem. Soc. Rev. 2009;38:883–891. doi: 10.1039/b806410p. [DOI] [PubMed] [Google Scholar]
- 92.Yang Z, Liang G, Xu B. Acc. Chem. Res. 2008;41:315–326. doi: 10.1021/ar7001914. [DOI] [PubMed] [Google Scholar]
- 93.Gao Y, Yang Z, Kuang Y, Ma M-L, Li J, Zhao F, Xu B. Biopolymers. 2010;94:19–31. doi: 10.1002/bip.21321. [DOI] [PubMed] [Google Scholar]
- 94.Zhang Y, Kuang Y, Gao Y, Xu B. Langmuir. 2011;27:529–537. doi: 10.1021/la1020324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tam AY-Y, Yam VW-W. Chem. Soc. Rev. 2013;42:1540–1567. doi: 10.1039/c2cs35354g. [DOI] [PubMed] [Google Scholar]
- 96.Sangeetha NM, Maitra U. Chem. Soc. Rev. 2005;34:821–836. doi: 10.1039/b417081b. [DOI] [PubMed] [Google Scholar]
- 97.Dastidar P. Chem. Soc. Rev. 2008;37:2699–2715. doi: 10.1039/b807346e. [DOI] [PubMed] [Google Scholar]
- 98.Suzuki M, Hanabusa K. Chem. Soc. Rev. 2009;38:967–975. doi: 10.1039/b816192e. [DOI] [PubMed] [Google Scholar]
- 99.Buerkle LE, Rowan SJ. Chem. Soc. Rev. 2012;41:6089–6102. doi: 10.1039/c2cs35106d. [DOI] [PubMed] [Google Scholar]
- 100.Kawase T, Kurata H. Chem. Rev. 2006;106:5250–5273. doi: 10.1021/cr0509657. [DOI] [PubMed] [Google Scholar]
- 101.Yu T, Cristiano R, Weiss RG. Chem. Soc. Rev. 2010;39:1435–1447. doi: 10.1039/b821320h. [DOI] [PubMed] [Google Scholar]
- 102.Terech P, Weiss RG. Chem. Rev. 1997;97:3133–3160. doi: 10.1021/cr9700282. [DOI] [PubMed] [Google Scholar]
- 103.van Esch JH, Feringa BL. Angew. Chem. Int. Edit. 2000;39:2263–2266. doi: 10.1002/1521-3773(20000703)39:13<2263::aid-anie2263>3.3.co;2-m. [DOI] [PubMed] [Google Scholar]
- 104.de Loos M, Feringa BL, van Esch JH. Eur. J. Org. Chem. 2005:3615–3631. [Google Scholar]
- 105.Smith DK, Diederich F. Chem-eur J. 1998;4:1353–1361. [Google Scholar]
- 106.Banerjee S, Das RK, Maitra U. J. Mater. Chem. 2009;19:6649–6687. [Google Scholar]