SUMMARY
Biological diversity among mammals is remarkable. Mammalian body weights range seven orders of magnitude and lifespans differ more than 100-fold among species. While genetic, dietary, and pharmacological interventions can be used to modulate these traits in model organisms, it is unknown how they are determined by natural selection. By profiling metabolites in brain, heart, kidney, and liver tissues of 26 mammalian species representing ten taxonomical orders, we report metabolite patterns characteristic of organs, lineages, and species longevity. Our data suggest different rates of metabolite divergence across organs and reveal patterns representing organ-specific functions and lineage-specific physiologies. We identified metabolites that correlated with species lifespan, some of which were previously implicated in longevity control. We also compared the results with metabolite changes in five long-lived mouse models and observed some similar patterns. Overall, this study describes adjustments of the mammalian metabolome according to lifespan, phylogeny, and organ and lineage specialization.
Graphical Abstract
INTRODUCTION
All modern mammals descend from a common ancestor that lived ~210 million years ago and have since undergone remarkable diversification in morphology, life history, and other characteristics. Their body parts, such as tongues, ears, fingers, and feet, have been modified for numerous functions including nectar-feeding, echolocating, swimming, flying, and digging; their body weights range from under 2 g (Etruscan shrew, Suncus etruscus) to over 150 tons (blue whale, Balaenoptera musculus); and their maximum lifespans differ by more than 100-fold (Tacutu et al., 2013). Many of the traits affecting development, body weight, and lifespan (i.e., the life history traits) are often correlated. Longer-lived species tend to be bigger, produce fewer offspring, grow more slowly, and have lower mass-specific metabolic rates (Peters, 1986; Sacher, 1959; Western, 1979), indicative of modulation by the same underlying evolutionary forces. Certain lineages, such as bats (Seim et al., 2013) and primates, have evolved to live longer as a whole, whereas other instances of exceptional longevity have emerged sporadically among short-lived taxonomic relatives such as the naked mole rat, Heterocephalus glaber, which lives ten times longer than other rodents of comparable size (Buffenstein, 2008; Fang et al., 2014; Kim et al., 2011).
Longevity is elastic and can vary along a continuum, but the underlying factors are only starting to be characterized. Research in model organisms revealed several important molecular players, such as insulin-like growth factor 1 (IGF-1) (Friedman and Johnson, 1988; Holzenberger et al., 2003; Tatar et al., 2001), mechanistic target of rapamycin (mTOR) (Kenyon, 2010; Vellai et al., 2003), and sirtuins (Lin et al., 2000; Tissenbaum and Guarente, 2001). Dietary and pharmacological interventions can also extend lifespan in diverse organisms (Harrison et al., 2009; McCay et al., 1935; Weindruch et al., 1986). In particular, the lifespan of laboratory mice can be increased by restriction of food or methionine (Flurkey et al., 2010; Sun et al., 2009), administration of rapamycin (Harrison et al., 2009; Miller et al., 2014) or acarbose (Harrison et al., 2014), or certain genetic mutations (Ladiges et al., 2009). Rapamycin, an inhibitor of mTORC1, leads to a 23%–26% increase in the median lifespan of mice (Miller et al., 2014). Acarbose inhibits glycoside hydrolases (the enzymes that digest complex carbohydrates to absorbable sugars in the gastrointestinal tract) and is used clinically to blunt post-prandial glucose surges in diabetic patients. It seems plausible that limiting peak glucose concentrations may explain its longevity benefits. Mutant strains such as Snell dwarf (defective in anterior pituitary development) (Flurkey et al., 2001) and growth hormone receptor knockout (GHRKO) (Coschigano et al., 2003) are also long-lived, due to altered signaling in growth hormone (GH) itself or through GH-stimulated production of IGF-1.
How longevity is modulated during evolution to produce both long-lived and fit animals, however, is still unclear. Lifespan is an inherent characteristic of a species and remains relatively stable through generations, but it can also change in either direction over time. In order to vary lifespan on an evolutionary timescale, a number of biological pathways may need to be altered, rewired, or reprogrammed. Omics-scale comparative studies across multiple species are instrumental in understanding the evolution of mammalian genomes and gene expression (Brawand et al., 2011; Lindblad-Toh et al., 2011). To gain insights into the metabolic basis of mammalian diversity and longevity, we quantified metabolite levels in brain, heart, kidney, and liver tissues of 26 species of mammals and identified metabolites with organ-, lineage-, and trait-specific patterns. We described the metabolite divergence and distribution in different organs, linked the lineage-specific metabolic patterns to lineage-specific physiologies, and identified metabolites with positive or negative correlation to longevity traits. In addition, we profiled the metabolites in brain and liver of five long-lived mouse models (caloric restriction, rapamycin treatment, acarbose treatment, GHRKO, and Snell dwarf) and compared the observed changes with the mammalian longevity signatures. Our study provides the first glimpse into how metabolism may have been altered to modulate mammalian lifespan.
RESULTS
Metabolite Conservation and Divergence among Organs
We applied targeted metabolite profiling to quantify the metabolite levels in brain, heart, kidney, and liver tissues of 26 species of mammals, representing ten taxonomical orders and covering a wide range of longevity-associated traits (Figure 1; Table S1A). The species were matched by biological age (all young adults) and sex (all were males, except for horse and vervet). Biological replicates (i.e., samples from multiple individuals of a species) were collected for most of the species (Table S1A). In total, 162 water-soluble metabolites and 100 lipids were reliably detected across 235 samples. Data quality was assessed graphically (Figure S1).
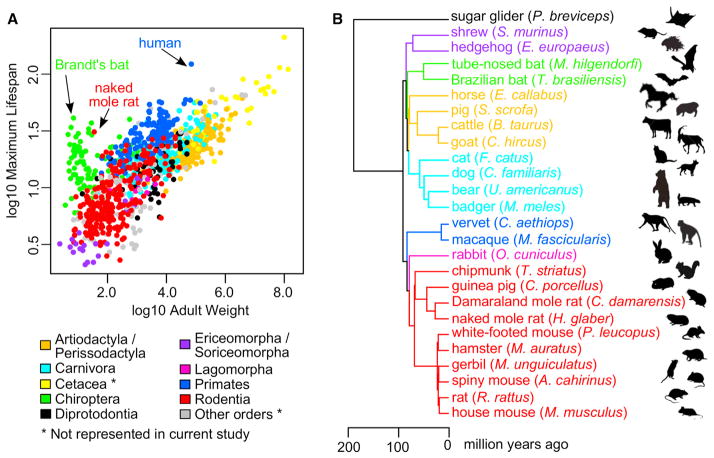
Figure 1. Diversification of Mammals.
(A) Maximum lifespan correlates positively with body mass. Maximum lifespan (years) was plotted against adult weight (g) on log10 scale for 995 mammalian species from AnAge database (Tacutu et al., 2013), color-coded by taxonomical orders. Human, the naked mole rat, and the Brandt’s bat are highlighted. To simplify the color scheme, Artiodactyla and Perissodactyla were grouped together, and Ericomorpha and Soriceomorpha were grouped together.
(B) Phylogeny of the mammals examined in the current study. Branches and tips are colored according to taxonomical orders (same color scheme as in A). Divergence times were based on previous estimates (Fushan et al., 2015; Meredith et al., 2011). Animal silhouettes are for illustration only.
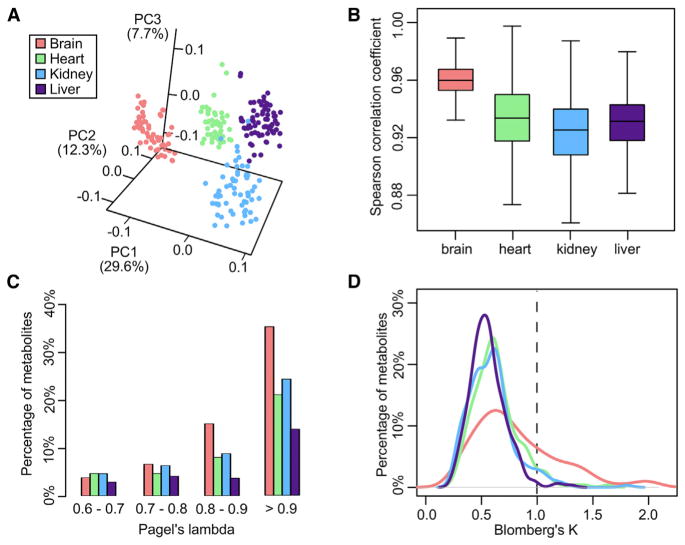
Principal-component analysis revealed the samples segregated predominantly by organ origin, with most of the replicates clustering together and the first three principal components accounting for ~50% of the total variance (Figure 2A). ANOVA confirmed that organ and species accounted for over 80% of the variation in individual metabolite levels, whereas the differences between replicates were much smaller than the differences between species (Figure S1). The clustering pattern agreed with those based on mammalian gene expression profiles (Brawand et al., 2011; Fushan et al., 2015), suggesting that metabolite levels and organ-specific metabolism were generally well conserved across the mammals.
Figure 2. Metabolite Divergence in Mammalian Organs.
(A) Samples segregate predominantly according to organ origin. Values in parenthesis indicate the percentage of variance explained by each of the first three principal components (PCs). Biological replicates were treated as individual points.
(B) Brain samples show highest Spearman correlation coefficients. The box plot shows the pairwise correlation among the samples in each organ. Wilcoxon rank sum test p value < 2 × 10−16 for brain against each of the other organs.
(C and D) Brain has the largest percentage of metabolites with high phylogenetic signals. In (C), only Pagel’s lambda > 0.6 are shown. In (D), the dotted line indicates Blomberg’s K = 1.0.
The phylogenetic relationship of many mammals has been established based on fossil and molecular evidence (Figure 1B) (Fushan et al., 2015; Meredith et al., 2011). To determine if their metabolite levels recapitulate this relationship, we constructed phylograms using the metabolite levels in each organ and found them largely consistent with the reference phylogeny (Figure S2A). The brain phylogram had the shortest tip-to-root branch lengths (Figures S2B and S2C), indicative of small degree of metabolite divergence (Brawand et al., 2011). The brain samples also showed highest Spearman correlation coefficients (Figure 2B) and had the largest proportion of metabolites with high phylogenetic signals (i.e., Pagel’s lambda > 0.9; Pagel, 1999; and Blomberg’s K > 1; Blomberg et al., 2003; Figures 2C and 2D), suggesting that brain metabolites are most conserved among the four organs and have evolved largely according to the phylogeny. In contrast, the metabolites in the other examined organs diverge to a much greater extent, possibly due to stronger environmental influence or other selection pressures.
Metabolite Profiles Reflect Organ Functions
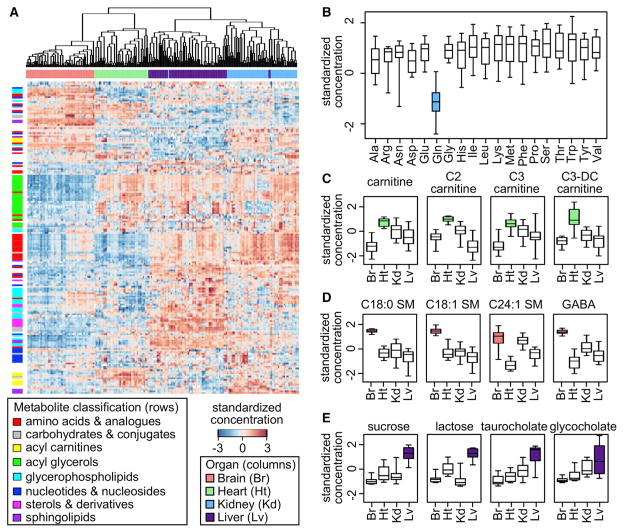
The metabolite profile of an organ is expected to reflect its biological functions. We visualized the distribution of metabolites on a heatmap (Figure 3A) and used the Wilcoxon rank sum test to identify metabolites selectively enriched or depleted in a particular organ (in comparison with at least two other organs; Table S2A).
Figure 3. Distribution of Metabolites across the Organs.
(A) The overall pattern visualized on a heat map. Hierarchical clustering was performed on standardized concentrations using average linkage. Each row represents one metabolite, and each column represents one biological sample. Selected classes of metabolites are highlighted.
(B) Kidney is depleted of glutamine. Each box represents the range of standardized concentrations for a particular amino acid in kidney across the mammals.
(C) Heart is enriched with carnitine and short-chain acylcarnitines. The alternative names are acetylcarnitine (C2 carnitine), propionylcarnitine (C3 carnitine), and malonylcarnitine (C3-DC carnitine).
(D) Brain is enriched with sphingomyelins (SM) and the neurotransmitter gamma-aminobutyric acid (GABA).
(E) Liver has high levels of sucrose, lactose, and bile acid components.
18 out of the 19 proteinogenic amino acids measured (cysteine was not quantified) were found at moderate to high levels in kidney relative to the other organs (Table S2B), likely due to reabsorption at the renal proximal tubule. One exception was glutamine (Figure 3B), which is routinely metabolized by the kidney for nitrogen disposal and acid-base balance. Glutamine is broken down to ammonia and glutamate, helping to remove excessive protons and generate bicarbonate ions (van de Poll et al., 2004).
The metabolite profile of the heart largely reflected its energy demand. Heart tissue was depleted of amino acids and many other metabolites but enriched with acylcarnitines (especially short-chain acylcarnitines; Figure 3C) and triacylglycerols (TAGs). Acylcarnitines help transport fatty acids across mitochondrial inner membranes (Vaz and Wanders, 2002), whereas carnitine acts as an acetyl group acceptor, buffering the cellular pool of coenzyme A (CoA) and preventing inhibition of pyruvate dehydrogenase, especially in tissues dependent on beta-oxidation (Hoppel, 2003).
In contrast, the brain normally relies on glucose for fuel and contains relatively few TAGs. However, it had high concentrations of glycerophospholipids and a number of sphinogmyelins (Figure 3D), which are both key constituents of animal cell membranes. In particular, sphingomyelins are mainly found in the myelin sheaths surrounding nerve cell axons. The neurotransmitters gamma-aminobutyric acid (GABA) and glutamate were also present at high levels.
Liver was enriched with a wide range of metabolites, including amino acids, glycerophospholipids, carbohydrates, and steroids (Table S2B). Some of them were significantly higher than in the other organs, likely indicative of liver-specific pathways. For example, sucrose and lactose were found at very high concentrations in liver (Figure 3E), as these sugars are not routinely utilized by the other organs. Bile acid components such as glycocholate, taurocholate, taurodeoxycholate, and taurocheno-deoxycholate were restricted mostly to liver (Figure 3E), since primary bile acids are synthesized by liver cells from cholesterol.
We grouped lipids according to LIPID MAPS Classification System (Experimental Procedures) (Fahy et al., 2007). Within each category, we compared the relative percentage abundance of individual lipids in our study with those previously reported in human plasma (Quehenberger et al., 2010). Significant correlations were observed for TAG, lysophosphatidylcholine (LPC), phosphatidylcholine (PC), and cholesteryl ester (CE) (see Experimental Procedures for lipid classification and abbreviations; Table S2D), suggesting that the overall lipid composition was conserved across mammals.
Metabolites with Lineage-Specific Changes
Since a particular lineage of mammals often exhibits biological and physiological features distinctive from the others, we grouped the species by taxonomic orders or families and applied phylogenetic ANOVA (Garland et al., 1993) to determine if the concentration of a metabolite in one group was significantly different from the other groups (Table S3).
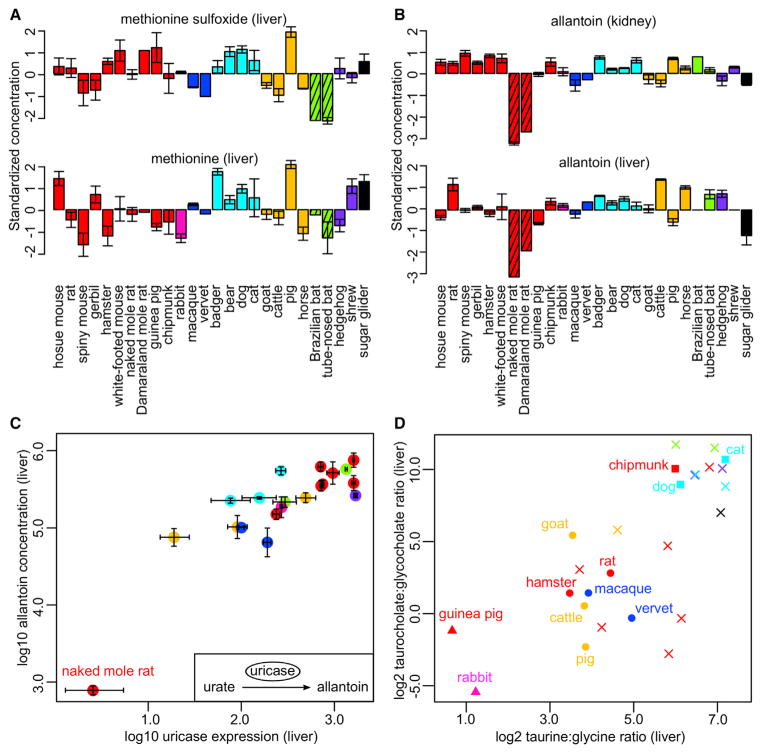
Bats (order Chiroptera) showed significantly reduced levels of methionine sulfoxide in both kidney and liver (phylogenetic ANOVA p value = 0.003 in kidney and 0.002 in liver; Table S3A), while their methionine levels were typical of other mammals (Figure 4A). Methionine sulfoxide is produced by oxidation of methionine by reactive oxygen species (ROS), and in most species, its level increases during aging or oxidative stress (Berlett and Stadtman, 1997). Bats are the longest-lived mammalian order after controlling for the effect of body size, and there is evidence that they produce less ROS and are more resistant to oxidative stress. For example, cave Myotis bats and Mexican free-tailed bats (both with maximum lifespan potential of 12 years) show lower protein carbonylation and ubiquitination in liver than mice, and their cells are more resistant to protein oxidation (Salmon et al., 2009; Shi et al., 2010). Mitochondria from bat heart also produce less hydrogen peroxide than those from shrew and white-footed mouse (Brunet-Rossinni, 2004), although the differences are less than the divergence in their maximum lifespans (Buffenstein et al., 2008). Hence, low methionine sulfoxide levels are consistent with reduced oxidative stress generally observed in bats.
Figure 4. Metabolite Patterns Reflect Species Physiology.
(A) Bats have low levels of methionine sulfoxide in liver. The error bars indicate SEs (only for those with biological replicates). The species are colored according to taxonomical orders (same color scheme as in Figure 1A). The bars representing the bats are shaded.
(B) African mole rats have low levels of allantoin in kidney and liver. The bars representing the naked mole rat and Damaraland mole rat are shaded.
(C) Liver allantoin levels correlate positively with uricase expression. The error bars indicate SEs in gene expression measurements (horizontal direction) or in metabolite measurements (vertical direction). The correlation relationship is robust (correlation coefficients using all points: Pearson = 0.86, Spearman = 0.78; excluding naked mole rat: Pearson = 0.76, Spearman = 0.74). Gene expression data were not available for Damaraland mole rat.
(D) Use of taurine and glycine for bile acid conjugation among the mammals. The species with known conjugated strategy are highlighted. Square (■), taurine-conjugation only; triangle (▲), glycine-conjugation only; circle (●); both taurine- and glycine-conjugation; cross (×), conjugation strategy unknown.
Several genetic and physiological features of African mole rats (family Bathyergidae) are distinct from those of other rodents (Fang et al., 2014; Kim et al., 2011), so we compared Bathyergidae against the other examined species (Table S3B), as well as against the other rodents in this study (Table S3C). Several metabolites were detected in both comparisons across multiple organs, including enrichment of acetylglycine (in heart and liver), enrichment of trimethylamine N-oxide (in brain and heart), and depletion of allantoin (in brain, heart, kidney, and liver) (Figure 4B). This depletion of allantoin in the Bathyergidae is particularly striking, since other African rodents (in particular the Cricetidae) excrete high levels of allantoin (Buffenstein et al., 1985). Using the gene expression data for some of these species (Fushan et al., 2015), we confirmed the positive correlation between uricase expression and allantoin level in liver (Pearson correlation coefficient = 0.86, Spearman correlation coefficient = 0.78; Figure 4C), with particularly low expression in the naked mole rat. In mammals, degradation of purine produces urate, which is then converted to allantoin by the enzyme uricase and excreted in urine (Buffenstein et al., 1985; Ngo and Assimos, 2007). In humans and other higher primates, the gene coding for uricase is a pseudogene and urate is excreted instead. However, these genetic changes were not found in the African mole rat enzymes, so the low uricase expression appears to be achieved by a different mechanism.
Since the mammalian species used in this study include carnivores, insectivores, omnivores, and herbivores, we wondered if the dietary preferences would also be reflected in the metabolic profiles, especially in terms of different bile acid conjugates. Bile acids can be conjugated with either taurine or glycine, depending on their concentrations in liver and affinities for the enzyme bile acid CoA:amino acid N-acyltransferase. Most animals conjugate exclusively with taurine, whereas glycine conjugation is limited to certain placental mammals and herbivores (Huxtable, 2002; Vessey, 1978). Indeed, in liver, the taurocholate:glycocholate ratio correlated positively with the taurine:glycine ratio (Pearson correlation coefficient = 0.74; Figure 4D). Rabbit and guinea pig are known to employ glycine-conjugation only and had low values for both ratios, while cat, being an obligate carnivore, was high in both (Figure 4D). Based on the clustering pattern, hedgehog and shrew (predominantly insectivores), as well as animals belonging to order Carnivora, probably use taurine conjugation only, whereas most rodents of the family Muridae and animals of orders Artiodactyla and Perissodactyla use both taurine and glycine conjugation (Figure 4D).
Metabolome Remodeling and Longevity Variation
Next, we examined the general trend in longevity and body mass across these species. We obtained the traits data from public databases (Carey and Judge, 2000; Tacutu et al., 2013) and focused primarily on adult weight (AW) and the longevity metrics average lifespan (AL), maximum lifespan (ML), female time to maturity (FTM), as well as their body-mass-adjusted residuals (i.e., ALres, MLres, and FTMres, respectively) (Table S1A). While AL and ML are most closely related to the concept of longevity, FTM can be measured more easily and may be less prone to reporting bias. They also correlated strongly with one another (Pearson correlation coefficient = 0.91 between AL and ML; 0.87 between AL and FTM; 0.84 between ML and FTM). Since AW correlates positively with lifespan (Figure 1A), the longevity residuals were computed to remove the body mass influence. To account for the evolutionary relationship of the species, we performed regression by phylogenetic generalized least squares (Felsenstein, 1985; Grafen, 1989) (Table S4). Different models of trait evolution were tested and within-species variations were incorporated in the calculation (Supplemental Experimental Procedures). To assess the result robustness, we applied a two-step verification procedure (Experimental Procedures). The results were also adjusted for false discovery rate (“q value”) and tabulated across the organs and traits (Table S4).
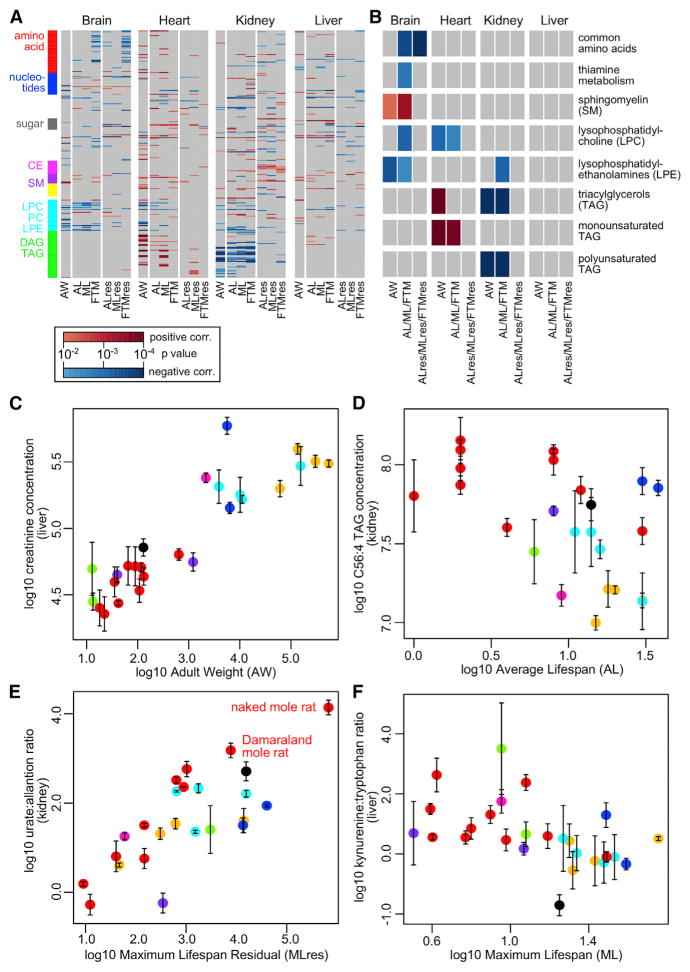
When the results were visualized on a heatmap (Figure 5A), a few observations became apparent. Within each organ, the correlations with the longevity metrics were largely consistent. Although the reported lifespan data were obtained from different databases and might not be entirely accurate, they did not significantly affect the calculated correlation, suggesting the results were robust to sample variation or slight measurement inaccuracy. On the other hand, the patterns were rather distinct across the four organs, suggesting the metabolites in different organs may follow different trajectories during evolution. By pooling the top hits (p value.robust < 0.01) of the two sets of longevity metrics (i.e., combining AL, ML, and FTM as one set; ALres, MLres, and FTMres as the other set), a number of positively and negatively correlating pathways were found to be enriched in each organ (Figure 5B).
Figure 5. Metabolites Correlating with Body Mass and Longevity.
(A) Overview of correlation with body mass and longevity. The grids represent the robust regression p value (“p value.robust”) between metabolite levels in each organ and the indicated traits (AW, adult weight; AL, average lifespan; ML, maximum lifespan; FTM, female time to maturity; ALres, average lifespan residual; MLres, maximum lifespan residual; FTMres, female time to maturity residual). Only p value.robust < 0.01 are shown in color, with positive correlation in red and negative correlation in blue. Selected classes of metabolites are highlighted by rows (same color scheme as in Figure 3A). See Table S4H for more details.
(B) Top pathways correlating with body mass and longevity. The grids represent the pathway enrichment analysis p values (only p values < 0.01 are shown in color), with positive correlation in red and negative correlation in blue. For the purpose of enrichment analysis, the top hits (p value.robust < 0.01) in AL, ML, and FTM were pooled together, and the top hits in ALres, MLres, and FTMres were also pooled together. “Monounsaturated TAG” refers to TAG with at most two double bonds in total. “Polyunsaturated TAG” refers to TAG with three or more double bonds. See Table S4J for more details.
(C) Liver creatinine level correlates positively with AW. The vertical error bars indicate SE. The points are colored according to taxonomical orders (same color scheme as in Figure 1A). Regression p value.robust = 1.01 × 10−10; p value.max = 4.20 × 10−10.
(D) Kidney C56:4 TAG level correlates negatively with average lifespan. Regression p value.robust = 9.75 × 10−3; p value.max = 3.70 × 10−2.
(E) Kidney urate:allantoin ratio correlates positively with maximum lifespan residual. The points representing the naked mole rat and the Damaraland mole rat are indicated. Regression p value.robust = 8.41 × 10−6; p value.max = 1.60 × 10−4.
(F) Liver kynurenine:tryptophan ratio correlates negatively with maximum lifespan. Regression p value.robust = 7.23 × 10−3; p value.max = 1.89 × 10−2.
Body Mass and Longevity Signatures
With respect to AW, creatinine (Crn) showed significant positive correlation in all four organs (p value.robust <10−8; Figure 5C). A related metabolite, creatine (Cr), also emerged as a top hit in heart and liver (Table S4). It is well known that urinary and serum Crn levels increase with body mass (especially lean body mass) (Forbes and Bruining, 1976), as most Crn is derived from Cr in skeletal muscles and larger animals tend to have greater muscle mass. On the other hand, several glycerophospholipids (e.g., C16:0 LPE, C22:6 LPE, C18:0 LPC, and C22:6 LPC) negatively correlated with body mass, especially in brain and heart (Figure 5B; Table S4). A number of TAGs showed significant but opposite trends in heart (positive correlation) and kidney (negative correlation) (Figure 5B).
In terms of the longevity traits, a negative correlation was observed for amino acids, LPC, lysophosphatidylethanolamine (LPE), and metabolites involved in thiamine metabolism, whereas a positive correlation was observed mainly for sphingomyelin (SM) (Figure 5B). LPC and LPE are generated by phospholipase-dependent hydrolysis of PC and phosphatidyl-ethanolamine (PE), respectively. Phospholipase A2 (PLA2) activity releases fatty acids such as arachidonic acid from the sn-2 position of the glycerol backbone of phospholipids and is commonly associated with inflammatory signaling in mammalian tissues. For example, elevated circulating lipoprotein-associated PLA2 activity is linked to coronary artery disease in humans (Rosenson and Stafforini, 2012), supporting a potential inverse relationship between phospholipase activities (and hence LPC and LPE levels) and longevity.
Similar to the situation with body mass, TAG as a whole showed opposing trends to longevity in heart (positive) and kidney (negative). Closer examination revealed that the negative correlations in kidney were largely attributed to TAG with polyunsaturated fatty acid (PUFA) side chains (i.e., multiple double bonds; Figure 5D), whereas the positive correlations in heart were due to TAG with saturated or monounsaturated fatty acid (MUFA) side chains. A recent study on human plasma lipidomes of middle-aged offspring of nonagenarians revealed a signature of 19 lipid species associating with female familial longevity, including high levels of SM and low levels of PUFA TAG (Gonzalez-Covarrubias et al., 2013). Analysis of phospholipids in heart of a number of mammals also revealed a negative correlation between double-bond content and ML (Pamplona et al., 2000). Naked mole rat tissues contain much lower levels of docosahex-aenoic-acid-containing (with six double bonds) phospholipids compared to mouse (Mitchell et al., 2007). Since PUFAs are particularly sensitive to peroxidation damage, the reduced level of polyunsaturated TAG in long-lived species may reflect their enhanced resistance to oxidative stress.
Allantoin correlated negatively with longevity in brain, kidney, and liver, whereas urate showed some moderate positive correlation (Table S4H). Furthermore, the urate:allantoin ratio showed significant positive correlation with ML, ALres, and MLres in kidney (p value.robust < 10−3; Figure 5E), indicating that long-lived mammals had higher urate and lower allantoin levels. The ranges of p values in kidney remain significant even when each species was left out one at a time (p value.max = 1.21 × 10−2 for ML, 1.89 × 10−3 for ALres, and 1.60 × 10−4 for MLres; Table S4H), so the observation is generally applicable across the examined mammals and does not depend on any particular species. A previous study in primate and non-primate mammals also found a significant positive correlation between ML potential and urate concentration in serum and brain per specific metabolic rate (Cutler, 1984). Interestingly, humans have the highest serum urate level and are the longest-lived primates (Cutler, 1984). The naked mole rat, the longest-lived rodent, also had very low levels of uricase expression in liver (Figure 4C). Allantoin can also be produced from urate by free radical oxidation (Kaur and Halliwell, 1990), and studies on human samples suggest high allantoin level may be a marker of oxidative stress (Kand’ár and Záková, 2008; Yardim-Akaydin et al., 2006).
Liver concentrations of two tryptophan degradation products, anthranilic acid and kynurenine, showed a robust negative correlation with longevity (Table S4H). Over 95% of free tryptophan is degraded via the kynurenine pathway, with the first rate-limiting step catalyzed by indoleamine 2,3-dioxygenase (IDO) or tryptophan 2,3-dioxygenase (TDO). Anthranilic acid is produced from enzymatic hydrolysis of kynurenine. Several studies have linked tryptophan metabolism to aging and longevity. Knockdown of tdo-2 gene in C. elegans can suppress the toxicity of aggregation-prone proteins and extend lifespan (van der Goot et al., 2012). Fruit flies with TDO deficiency live significantly longer than wild-type controls (Oxenkrug, 2010). In mammals, reducing dietary tryptophan can extend lifespan and delay age-related changes in rats and mice (De Marte and Enesco, 1986; Segall and Timiras, 1976), and the kynurenine:tryptophan ratio in humans increases with aging (Capuron et al., 2011; Frick et al., 2004). In agreement, we also observed significant negative correlation with longevity for the kynurenine:tryptophan ratio and the anthranilic acid:tryptophan ratio in liver (Figure 5F; Table S4H).
Reducing dietary amino acids levels has proved effective in lifespan extension (Grandison et al., 2009; Lee et al., 2014; Min and Tatar, 2006). Here, the amino acids in brain showed negative correlation predominantly with FTM and FTMres (Table S4H), implying that mammals that mature more slowly (and which are usually also longer-lived) tend to have lower levels of brain amino acids. The levels of branched chain amino acids such as leucine and isoleucine are also low in long-lived Ames dwarf mice (Wijeyesekera et al., 2012), which are defective in adenohypophyseal development and have stunted growth. 4-pyridoxate (catabolite of vitamin B6) in brain and thiamine (vitamin B1) in kidney and liver also negatively correlated with lifespan (Table S4H). They are required, respectively, for the synthesis of pyridoxal phosphate (PLP) and thiamine pyrophosphate (TPP), which are the essential cofactors for many enzymes involved in amino acid metabolism (Eliot and Kirsch, 2004; Lonsdale, 2006). Overall, the result is consistent with reduced rate of mass-specific metabolism in longer-lived mammals.
Insights from the Analysis of Long-Lived Mouse Models
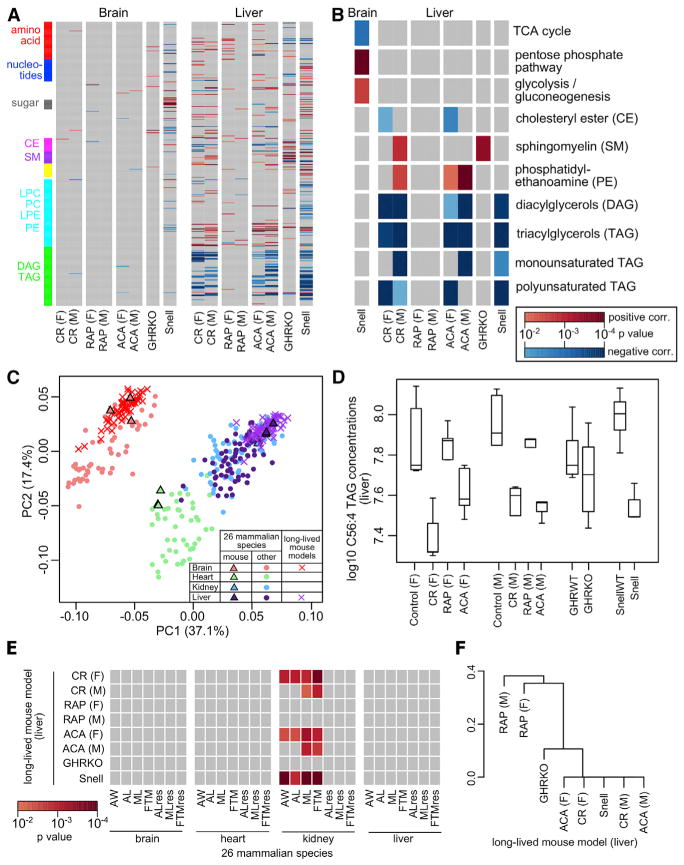
To compare our results with established long-lived animal models, we performed metabolite profiling on brain and liver tissues of mice under caloric restriction (CR), rapamycin treatment (RAP), and acarbose treatment (ACA), as well as GHRKO and Snell dwarf mice (Snell), against their respective wild-type controls under control diets (Table S1B). Five age-matched (~1 year old when sacrificed) biological replicates were collected for each condition, with both males and females for CR, RAP, and ACA and males only for GHRKO and Snell (Table S1B). In total, 358 metabolites were reliably quantified across the 120 samples, and 241 of these metabolites overlapped with the mammalian dataset (Figure S3).
We identified the metabolites differentially distributed between the long-lived mouse models and the corresponding controls and performed pathway enrichment analysis (Figures 6A and 6B; Table S5). The long-lived mouse model dataset clustered with the mouse data in the mammalian dataset (Figure 6C), indicating the overall metabolic signatures inherent in the species were well preserved. Interestingly, while a significant number of top hits were found in liver, the brain metabolite levels did not change much between the treatment and control (Figure 6A), and they were more conserved than those in liver (Figure S3B). The blood-brain barrier may help keep the brain metabolism in tight homeostasis and refractory to external modulations. The only exception was the Snell mice, which are defective in anterior pituitary development. Compared to control, the brain of Snell mice exhibits a shift from oxidative phosphorylation toward glycolysis (Figure 6B).
Figure 6. Metabolites Differentially Distributed in Long-Lived Mouse Models.
(A) Overview of metabolite differential distribution. The grids represent the linear model p values for differential distribution in the indicated long-lived mouse models with respect to their corresponding controls in brain and liver (CR, caloric restriction; RAP, rapamycin treatment; ACA, acarbose treatment; GHRKO, growth hormone receptor knockout; Snell, Snell dwarf mouse; F, female; M, male). Only p values < 0.01 are shown in color, with positive correlation in red and negative correlation in blue. Selected classes of metabolites are highlighted by rows (same color scheme as in Figure 3A). See Table S5C for more details.
(B) Top enriched pathways. The grids represent the pathway enrichment analysis p values (only p values < 0.01 are shown in color), with positive correlation in red and negative correlation in blue. For brain, only Snell is shown. See Table S5F for more details.
(C) Long-lived mouse model data cluster well with mammalian species data. Values in parenthesis indicate the percentage of variance explained by each principal component (PC). Biological replicates were treated as individual points.
(D) Liver C56:4 TAG level across the long-lived mouse models. C56:4 TAG levels were significant lower in CR(F) (p value = 2.21 × 10−5), CR(M) (p value = 8.17 × 10−3), ACA(F) (p value = 7.22 × 10−3), and Snell (p value = 9.07 × 10−5), compared to their respective controls.
(E) Overlap among longevity signatures. Between each pair of comparison, the numbers of metabolites with matching and opposite direction of correlation to longevity were calculated. p value was based on binomial statistics, assuming equal probability of getting a match or a mismatch by chance. Only p values < 0.01 are shown in color. See Table S5H for more details.
(F) Hierarchical clustering of the long-lived mouse models. The distance matrix is based on the similarity among the longevity signatures (i.e., pairwise binomial p values; Table S5H). Only the liver data are shown. The mouse models are as shown above.
In liver, CR, ACA, and Snell mice produced very similar metabolic shifts, and these patterns were observed in both males and females (Figures 6A and 6B). Remarkably, there was extensive reduction in PUFA TAG levels across all these three models (Figures 6B and 6D; Table S5C), which was consistent with the longevity signature we identified across the mammalian species and may indicate reduced susceptibility to peroxidation damage and oxidative stress in the long-lived mice. While the low PUFA TAG levels might be partly explained by the lower body weights of these long-lived mice, this signature was not observed in GHRKO dwarf mice or in RAP mice. There were no significant differences in body weight among CR, RAP, and ACA in either gender (Figure S3C). In addition, the long-lived mouse models exhibited elevated levels of SM (in particular C14:0 SM, C16:0 SM, C18:0 SM and C18:1 SM), which also showed positive correlation in longevity in the mammalian species dataset. Previously, SM levels were reported to be low in old mice but at normal levels in those under chronic CR (De Guzman et al., 2013) and were found to be high in the serum of centenarians (Montoliu et al., 2014). High SM levels are also associated with human female familial longevity (Gonzalez-Covarrubias et al., 2013). Sphingosine-linked fatty acids like ceramides are often regarded as “damage-associated molecular patterns” and may cause inflammatory damage by activating Nlrp3 inflammasome (De Guzman et al., 2013; Vandanmagsar et al., 2011). Elevated SM levels may also reflect reduced turnover to ceramides.
Other similarities as well as differences exist between our two datasets and those in the literature. For example, methionine is found at high levels in long-lived Ames dwarf mice, which may represent an increased methionine flux to transsulfuration and improved oxidative stress resistance (Wijeyesekera et al., 2012). Methionine level is also high in brain of male CR mice and liver of female ACA mice (Table S5). LPC levels were previously found to decrease with age but maintained in CR mice (De Guzman et al., 2013); in both our datasets they were low in long-lived animals. Furthermore, the mammalian dataset signatures of high urate:allantoin ratio and low kynurenine:tryptophan ratio were either insignificant or showed the opposite trends in the mouse models (Table S5).
To quantify the similarity between the longevity signatures from our two datasets, we counted the number of top hits in both datasets that had the same direction of correlation to longevity and compared that with the probability of getting similar results by chance (Tables S5G–S5I). The liver signatures of Snell, CR, and ACA mice matched very well to those based on AL, ML, and FTM in kidney of the mammalian dataset (Figure 6E; Tables S5H and S5I). In addition, these liver signatures also clustered together (Figure 6F), suggesting lifespan extension by CR, acarbose treatment, and in Snell mutants may affect certain common pathways, where rapamycin treatment and growth hormone receptor knockout may achieve lifespan extension via different mechanisms.
DISCUSSION
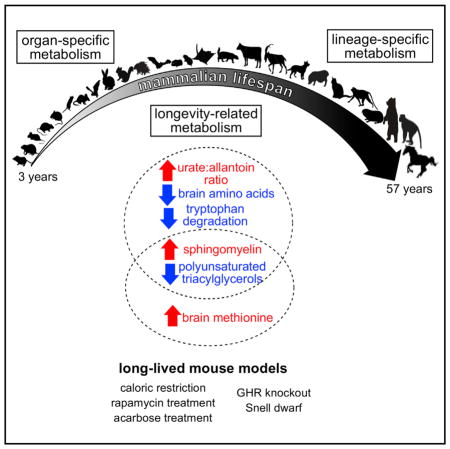
Mammals have diversified dramatically over the tens of millions of years of evolution with remarkably different longevity profiles. How are their lifespans modulated by evolution while preserving competitiveness within their ecological niches? Which metabolites are involved and, more generally, how is metabolism adjusted in order to increase lifespan? While most of the research on the control of lifespan was performed on single-model organisms, our study addressed these questions by analyzing metabolite levels in several organs across the class of Mammalia. We found that metabolites in brain diverged less than in the other examined organs and the organ-differential distribution of metabolites represented their respective biological functions. The lineage-specific metabolite features we identified reflect known physiology of animals (e.g., low oxidative stress in bats) and also offer some new insights (e.g., bile acid conjugation strategies among mammals and diminished conversion of urate to allantoin in African mole rats). With regard to the longevity traits, we identified metabolites previously implicated in lifespan control as well as several new candidates. In particular, long-lived mammals were associated with low polyunsaturated TAGs, low tryptophan degradation products, and low brain amino acids, as well as high sphingomyelin levels and a high urate:allantoin ratio. Comparison of our signatures with the metabolite changes in long-lived mouse models indicated some overlap with mice under CR, mice treated with acarbose, and Snell dwarf mice, especially for decrease in polyunsaturated TAGs and increase in sphingomyelin. Similar changes were also previously reported in studies on human centenarians and other long-lived animal models. Furthermore, these three mouse models produced metabolite signatures distinct from those observed in rapamycin treatment and GHRKO mice, so the lifespan extension effects may have been achieved via different mechanisms.
Our study also reveals some unexpected complexities in analyzing metabolites and longevity. While some metabolites show consistent correlation with longevity traits across multiple organs, many patterns seem to be organ specific. In the long-lived mouse models, many liver metabolites change significantly compared to the controls, but the brain exhibits very little perturbation. Furthermore, the longevity signatures in liver of the long-lived mouse models matched with the kidney (but not liver) signature across the mammalian species, suggesting certain aspects of the mammalian longevity signatures may be distinct from the long-lived mouse models. In addition, the molecular mechanisms underlying the lifespan extension in these mouse models are not yet well delineated, and differences among various long-lived mouse models have been previously reported. For example, in heart, kidney, and liver tissues, Snell and GHRKO mice showed different levels of chaperone mRNAs (Swindell et al., 2009). A low-calorie diet is beneficial to Ames dwarf mice (Bartke et al., 2001), but not to GHRKO mice (Bonkowski et al., 2009). Expression of genes related to xenobiotic detoxification in liver are distinctly different among rapamycin-treated mice, CR mice (Miller et al., 2014), and GHR-deletion mice (Li et al., 2013). CR mice also differ from rapamycin-treated mice in terms of leptin, FGF-21, and glucose tolerance (Lamming et al., 2013; Miller et al., 2014).
Compared with research that focuses on a single species, the current study benefited from the large effects of trait differences. While various factors such as feeding status, circadian cycle, gender, and body weight differences can introduce additional noise, ANOVA suggests that the variation between different species is generally much greater than the variation among replicates of the same species. Even with the ablation of GHR or anterior pituitary, the brain and liver profiles of the long-lived mice still clustered well with the mouse data in the mammalian dataset, and very similar longevity signatures were also obtained from both males and females of the same long-lived model. However, our study also suffers from a number of limitations. The current study does not prove causality between the metabolites and longevity traits, as the metabolite levels may influence and also be influenced by longevity. The number of metabolites quantified here only represents a fraction of the entire metabolome space and potentially important candidates may have been missed by our targeted approach. Many metabolites correlated strongly among one another and can inflate the signals observed. The metabolic fluxes through pathways and the metabolic changes during aging would not be reflected in our data either. While the biological implications of many metabolites identified here are far from fully understood, our study provides the first report of metabolite signatures of longevity across the mammalian spectrum, from which future studies should benefit.
EXPERIMENTAL PROCEDURES
Targeted metabolite profiling using three liquid chromatography-mass spectrometry (LC-MS) methods (Townsend et al., 2013) was applied to characterize metabolites and lipids in brain, heart, kidney, and liver of 26 mammalian species, as well as brain and liver tissues of five long-lived mouse models (Table S1). For the mammalian species, guinea pig, rabbit, hamster, gerbil, and rat were purchased from Charles River; naked mole rat, Damaraland mole rat, mouse, white-footed mouse, shrew, primates, and bats were from our laboratories; and other species were as described previously (Fushan et al., 2015). The samples were homogenates of freshly frozen tissues of sacrificed animals, matched by age and sex; biological replicates (i.e., samples from multiple individuals of a species) were obtained for most species. All long-lived mouse models as well as genotype and diet matched controls were from the colonies at University of Michigan Medical School. Liver and brain cortex samples were taken at 12 months of age from male and female mice treated from 4 months of age with rapamycin (14.7 ppm, as in Miller et al., 2014) or acarbose (1,000 ppm, as in Harrison et al., 2014), from mice subjected to 40% dietary restriction, or from untreated littermate control mice of the genetically heterogeneous stock UM-HET3. Liver and brain cortex samples from Snell dwarf (Flurkey et al., 2001) and GHRKO (Coschigano et al., 2003) males, and their corresponding littermate controls, were taken from young adults aged 4–6 months. Stringent filtering, processing, and normalization procedures were applied to generate the metabolome datasets (Data S1 and S2). Data quality was assessed graphically (Figures S1 and S3).
Most of the phylogenetic and statistical analysis was performed using R packages “phytools” (Revell, 2012) and “phylolm” (Ho and Ané, 2014). Based on LIPID MAPS Classification System (Fahy et al., 2007), we grouped the lipids as acylglycerols (diacylglycerol [DAG] and TAG), glycerophospholipids (PC, PE, LPC, and LPE), sphingolipids (SM), and sterols (CE). Pathway enrichment statistics were based on hypergeometric distribution and a 5,000-time bootstrap procedure. For organ-differential distribution, we required a metabolite to reach statistical significance in at least 2 organ pairs to qualify as a hit. To identify lineage-specific distribution, a chosen group (based on taxonomical order or family) was compared against all other species (unless otherwise stated) by phylogenetic ANOVA.
Regression by generalized least-squares method was used to identify correlation between longevity traits and metabolite levels and test four models of trait evolution (Supplemental Experimental Procedures). We evaluated the robustness of our results using a two-step verification procedure. First, regression was repeated by excluding the point with largest residue error (“p value.robust”), so that the overall relationship was not skewed by a potential outlier. Next, each species was left out, one at a time, and regression was performed on the remaining species to calculate the maximal (i.e., least significant) p value (“p value.max”), ensuring that the correlation was generalizable and did not depend on a single species. For the long-lived mouse models, differentially distributed metabolites between treatment and the corresponding control samples were identified. Overlap among the longevity signatures was assessed by binomial statistics and a 5,000-times bootstrap. Detailed experimental procedures can be found in Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Metabolic patterns reflect organ functions and lineage-specific physiologies
Long-lived mammals show distinctive signatures in amino acid and lipid levels
Some overlapping metabolite changes can be observed in long-lived mouse models
Rapamycin and caloric restriction extend lifespan by different mechanisms
Acknowledgments
Supported by NIH grants AG047745, AG021518, AG047200, and GM109312 and by funds from the Glenn Foundation for Medical Research.
Footnotes
AUTHOR CONTRIBUTIONS
V.N.G. coordinated the study. S.M. carried out data analyses. S.H.Y. carried out preliminary analyses. C.B.C. supervised metabolite profiling. S.H.Y., S.G.L., E.B.K., S.R.L., K.T.C., R.B., K.N.L., T.J.P., and R.A.M. provided, collected, and prepared samples. All authors contributed to data interpretation. S.M. and V.N.G. wrote the paper with input from all authors.
Supplemental Information includes Supplemental Experimental Procedures, three figures, five tables, and two datasets and can be found with this article online at http://dx.doi.org/10.1016/j.cmet.2015.07.005.
References
- Bartke A, Wright JC, Mattison JA, Ingram DK, Miller RA, Roth GS. Extending the lifespan of long-lived mice. Nature. 2001;414:412. doi: 10.1038/35106646. [DOI] [PubMed] [Google Scholar]
- Berlett BS, Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- Blomberg SP, Garland T, Jr, Ives AR. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution. 2003;57:717–745. doi: 10.1111/j.0014-3820.2003.tb00285.x. [DOI] [PubMed] [Google Scholar]
- Bonkowski MS, Dominici FP, Arum O, Rocha JS, Al Regaiey KA, Westbrook R, Spong A, Panici J, Masternak MM, Kopchick JJ, Bartke A. Disruption of growth hormone receptor prevents calorie restriction from improving insulin action and longevity. PLoS ONE. 2009;4:e4567. doi: 10.1371/journal.pone.0004567. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Brawand D, Soumillon M, Necsulea A, Julien P, Csárdi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, Kircher M, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- Brunet-Rossinni AK. Reduced free-radical production and extreme longevity in the little brown bat (Myotis lucifugus) versus two non-flying mammals. Mech Ageing Dev. 2004;125:11–20. doi: 10.1016/j.mad.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Buffenstein R. Negligible senescence in the longest living rodent, the naked mole-rat: insights from a successfully aging species. J Comp Physiol B. 2008;178:439–445. doi: 10.1007/s00360-007-0237-5. [DOI] [PubMed] [Google Scholar]
- Buffenstein R, Campbell WE, Jarvis JU. Identification of crystalline allantoin in the urine of African Cricetidae (Rodentia) and its role in their water economy. J Comp Physiol B. 1985;155:493–499. doi: 10.1007/BF00684680. [DOI] [PubMed] [Google Scholar]
- Buffenstein R, Edrey YH, Yang T, Mele J. The oxidative stress theory of aging: embattled or invincible? Insights from non-traditional model organisms. Age (Dordr) 2008;30:99–109. doi: 10.1007/s11357-008-9058-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capuron L, Schroecksnadel S, Féart C, Aubert A, Higueret D, Barberger-Gateau P, Layé S, Fuchs D. Chronic low-grade inflammation in elderly persons is associated with altered tryptophan and tyrosine metabolism: role in neuropsychiatric symptoms. Biol Psychiatry. 2011;70:175–182. doi: 10.1016/j.biopsych.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Carey JR, Judge DS. Longevity Records: Life Spans of Mammals, Birds, Amphibians, Reptiles, and Fish. Odense: Odense University Press; 2000. [Google Scholar]
- Coschigano KT, Holland AN, Riders ME, List EO, Flyvbjerg A, Kopchick JJ. Deletion, but not antagonism, of the mouse growth hormone receptor results in severely decreased body weights, insulin, and insulin-like growth factor I levels and increased life span. Endocrinology. 2003;144:3799–3810. doi: 10.1210/en.2003-0374. [DOI] [PubMed] [Google Scholar]
- Cutler RG. Urate and ascorbate: their possible roles as antioxidants in determining longevity of mammalian species. Arch Gerontol Geriatr. 1984;3:321–348. doi: 10.1016/0167-4943(84)90033-5. [DOI] [PubMed] [Google Scholar]
- De Guzman JM, Ku G, Fahey R, Youm YH, Kass I, Ingram DK, Dixit VD, Kheterpal I. Chronic caloric restriction partially protects against age-related alteration in serum metabolome. Age (Dordr) 2013;35:1091–1104. doi: 10.1007/s11357-012-9430-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marte ML, Enesco HE. Influence of low tryptophan diet on survival and organ growth in mice. Mech Ageing Dev. 1986;36:161–171. doi: 10.1016/0047-6374(86)90017-5. [DOI] [PubMed] [Google Scholar]
- Eliot AC, Kirsch JF. Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annu Rev Biochem. 2004;73:383–415. doi: 10.1146/annurev.biochem.73.011303.074021. [DOI] [PubMed] [Google Scholar]
- Fahy E, Sud M, Cotter D, Subramaniam S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007;35:W606–W612. doi: 10.1093/nar/gkm324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Seim I, Huang Z, Gerashchenko MV, Xiong Z, Turanov AA, Zhu Y, Lobanov AV, Fan D, Yim SH, et al. Adaptations to a subterranean environment and longevity revealed by the analysis of mole rat genomes. Cell Rep. 2014;8:1354–1364. doi: 10.1016/j.celrep.2014.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1–15. [Google Scholar]
- Flurkey K, Papaconstantinou J, Miller RA, Harrison DE. Lifespan extension and delayed immune and collagen aging in mutant mice with defects in growth hormone production. Proc Natl Acad Sci USA. 2001;98:6736–6741. doi: 10.1073/pnas.111158898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flurkey K, Astle CM, Harrison DE. Life extension by diet restriction and N-acetyl-L-cysteine in genetically heterogeneous mice. J Gerontol A Biol Sci Med Sci. 2010;65:1275–1284. doi: 10.1093/gerona/glq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes GB, Bruining GJ. Urinary creatinine excretion and lean body mass. Am J Clin Nutr. 1976;29:1359–1366. doi: 10.1093/ajcn/29.12.1359. [DOI] [PubMed] [Google Scholar]
- Frick B, Schroecksnadel K, Neurauter G, Leblhuber F, Fuchs D. Increasing production of homocysteine and neopterin and degradation of tryptophan with older age. Clin Biochem. 2004;37:684–687. doi: 10.1016/j.clinbiochem.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Friedman DB, Johnson TE. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushan AA, Turanov AA, Lee SG, Kim EB, Lobanov AV, Yim SH, Buffenstein R, Lee SR, Chang KT, Rhee H, et al. Gene expression defines natural changes in mammalian lifespan. Aging Cell. 2015;14:352–365. doi: 10.1111/acel.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland T, Dickerman AW, Janis CM, Jones JA. Phylogenetic analysis of covariance by computer simulation. Syst Biol. 1993;42:265–292. [Google Scholar]
- Gonzalez-Covarrubias V, Beekman M, Uh HW, Dane A, Troost J, Paliukhovich I, van der Kloet FM, Houwing-Duistermaat J, Vreeken RJ, Hankemeier T, Slagboom EP. Lipidomics of familial longevity. Aging Cell. 2013;12:426–434. doi: 10.1111/acel.12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafen A. The phylogenetic regression. Philos Trans R Soc Lond B Biol Sci. 1989;326:119–157. doi: 10.1098/rstb.1989.0106. [DOI] [PubMed] [Google Scholar]
- Grandison RC, Piper MD, Partridge L. Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature. 2009;462:1061–1064. doi: 10.1038/nature08619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Allison DB, Ames BN, Astle CM, Atamna H, Fernandez E, Flurkey K, Javors MA, Nadon NL, et al. Acarbose, 17-α-estradiol, and nordihydroguaiaretic acid extend mouse lifespan preferentially in males. Aging Cell. 2014;13:273–282. doi: 10.1111/acel.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Ls, Ané C. A linear-time algorithm for Gaussian and non-Gaussian trait evolution models. Syst Biol. 2014;63:397–408. doi: 10.1093/sysbio/syu005. [DOI] [PubMed] [Google Scholar]
- Holzenberger M, Dupont J, Ducos B, Leneuve P, Géloën A, Even PC, Cervera P, Le Bouc Y. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- Hoppel C. The role of carnitine in normal and altered fatty acid metabolism. Am J Kidney Dis. 2003;41:S4–S12. doi: 10.1016/s0272-6386(03)00112-4. [DOI] [PubMed] [Google Scholar]
- Huxtable RJ. Expanding the circle 1975–1999: sulfur biochemistry and insights on the biological functions of taurins. In: Corte L, Huxtable RJ, Sgaragli G, Tipton KF, editors. Taurine 4: Taurine and Excitable Tissues. Springer; 2002. pp. 1–25. [Google Scholar]
- Kand’ár R, Záková P. Allantoin as a marker of oxidative stress in human erythrocytes. Clin Chem Lab Med. 2008;46:1270–1274. doi: 10.1515/CCLM.2008.244. [DOI] [PubMed] [Google Scholar]
- Kaur H, Halliwell B. Action of biologically-relevant oxidizing species upon uric acid. Identification of uric acid oxidation products. Chem Biol Interact. 1990;73:235–247. doi: 10.1016/0009-2797(90)90006-9. [DOI] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Kim EB, Fang X, Fushan AA, Huang Z, Lobanov AV, Han L, Marino SM, Sun X, Turanov AA, Yang P, et al. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature. 2011;479:223–227. doi: 10.1038/nature10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladiges W, Van Remmen H, Strong R, Ikeno Y, Treuting P, Rabinovitch P, Richardson A. Lifespan extension in genetically modified mice. Aging Cell. 2009;8:346–352. doi: 10.1111/j.1474-9726.2009.00491.x. [DOI] [PubMed] [Google Scholar]
- Lamming DW, Ye L, Astle CM, Baur JA, Sabatini DM, Harrison DE. Young and old genetically heterogeneous HET3 mice on a rapamycin diet are glucose intolerant but insulin sensitive. Aging Cell. 2013;12:712–718. doi: 10.1111/acel.12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BC, Kaya A, Ma S, Kim G, Gerashchenko MV, Yim SH, Hu Z, Harshman LG, Gladyshev VN. Methionine restriction extends lifespan of Drosophila melanogaster under conditions of low amino-acid status. Nat Commun. 2014;5:3592. doi: 10.1038/ncomms4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Bartke A, Berryman DE, Funk K, Kopchick JJ, List EO, Sun L, Miller RA. Direct and indirect effects of growth hormone receptor ablation on liver expression of xenobiotic metabolizing genes. Am J Physiol Endocrinol Metab. 2013;305:E942–E950. doi: 10.1152/ajpendo.00304.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al. Whole Genome Assembly Team; Baylor College of Medicine Human Genome Sequencing Center Sequencing Team; Genome Institute at Washington University. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale D. A review of the biochemistry, metabolism and clinical benefits of thiamin(e) and its derivatives. Evid Based Complement Alternat Med. 2006;3:49–59. doi: 10.1093/ecam/nek009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCay CM, Crowell MF, Maynard LA. The effect of retarded growth upon the length of life span and upon the ultimate body size: one figure. J Nutr. 1935;10:63–79. [PubMed] [Google Scholar]
- Meredith RW, Janecka JE, Gatesy J, Ryder OA, Fisher CA, Teeling EC, Goodbla A, Eizirik E, Simão TL, Stadler T, et al. Impacts of the cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science. 2011;334:521–524. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]
- Miller RA, Harrison DE, Astle CM, Fernandez E, Flurkey K, Han M, Javors MA, Li X, Nadon NL, Nelson JF, et al. Rapamycin-mediated lifespan increase in mice is dose and sex dependent and metabolically distinct from dietary restriction. Aging Cell. 2014;13:468–477. doi: 10.1111/acel.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min KJ, Tatar M. Restriction of amino acids extends lifespan in Drosophila melanogaster. Mech Ageing Dev. 2006;127:643–646. doi: 10.1016/j.mad.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Mitchell TW, Buffenstein R, Hulbert AJ. Membrane phospholipid composition may contribute to exceptional longevity of the naked mole-rat (Heterocephalus glaber): a comparative study using shotgun lipidomics. Exp Gerontol. 2007;42:1053–1062. doi: 10.1016/j.exger.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Montoliu I, Scherer M, Beguelin F, DaSilva L, Mari D, Salvioli S, Martin FP, Capri M, Bucci L, Ostan R, et al. Serum profiling of healthy aging identifies phospho- and sphingolipid species as markers of human longevity. Aging (Albany, NY) 2014;6:9–25. doi: 10.18632/aging.100630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo TC, Assimos DG. Uric Acid nephrolithiasis: recent progress and future directions. Rev Urol. 2007;9:17–27. [PMC free article] [PubMed] [Google Scholar]
- Oxenkrug GF. The extended life span of Drosophila melanogaster eye-color (white and vermilion) mutants with impaired formation of kynurenine. J Neural Transm. 2010;117:23–26. doi: 10.1007/s00702-009-0341-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401:877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- Pamplona R, Portero-Otín M, Ruiz C, Gredilla R, Herrero A, Barja G. Double bond content of phospholipids and lipid peroxidation negatively correlate with maximum longevity in the heart of mammals. Mech Ageing Dev. 2000;112:169–183. doi: 10.1016/s0047-6374(99)00045-7. [DOI] [PubMed] [Google Scholar]
- Peters RH. The Ecological Implications of Body Size. Vol. 2. Cambridge University Press; 1986. [Google Scholar]
- Quehenberger O, Armando AM, Brown AH, Milne SB, Myers DS, Merrill AH, Bandyopadhyay S, Jones KN, Kelly S, Shaner RL, et al. Lipidomics reveals a remarkable diversity of lipids in human plasma. J Lipid Res. 2010;51:3299–3305. doi: 10.1194/jlr.M009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell LJ. phytools: an R package for phylogenetic comparative biology (and other things) Methods Ecol Evol. 2012;3:217–223. [Google Scholar]
- Rosenson RS, Stafforini DM. Modulation of oxidative stress, inflammation, and atherosclerosis by lipoprotein-associated phospholipase A2. J Lipid Res. 2012;53:1767–1782. doi: 10.1194/jlr.R024190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher GA. Relation of lifespan to brain weight and body weight in mammals. In: Wolstenholme GEW, O’Conner M, editors. Ciba Foundation Symposium: The Lifespan of Animals (Colloquia on Ageing) John Wiley & Sons; 1959. pp. 115–141. [Google Scholar]
- Salmon AB, Leonard S, Masamsetti V, Pierce A, Podlutsky AJ, Podlutskaya N, Richardson A, Austad SN, Chaudhuri AR. The long lifespan of two bat species is correlated with resistance to protein oxidation and enhanced protein homeostasis. FASEB J. 2009;23:2317–2326. doi: 10.1096/fj.08-122523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall PE, Timiras PS. Patho-physiologic findings after chronic tryptophan deficiency in rats: a model for delayed growth and aging. Mech Ageing Dev. 1976;5:109–124. doi: 10.1016/0047-6374(76)90012-9. [DOI] [PubMed] [Google Scholar]
- Seim I, Fang X, Xiong Z, Lobanov AV, Huang Z, Ma S, Feng Y, Turanov AA, Zhu Y, Lenz TL, et al. Genome analysis reveals insights into physiology and longevity of the Brandt’s bat Myotis brandtii. Nat Commun. 2013;4:2212. doi: 10.1038/ncomms3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Buffenstein R, Pulliam DA, Van Remmen H. Comparative studies of oxidative stress and mitochondrial function in aging. Integr Comp Biol. 2010;50:869–879. doi: 10.1093/icb/icq079. [DOI] [PubMed] [Google Scholar]
- Sun L, Sadighi Akha AA, Miller RA, Harper JM. Life-span extension in mice by preweaning food restriction and by methionine restriction in middle age. J Gerontol A Biol Sci Med Sci. 2009;64:711–722. doi: 10.1093/gerona/glp051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR, Masternak MM, Kopchick JJ, Conover CA, Bartke A, Miller RA. Endocrine regulation of heat shock protein mRNA levels in long-lived dwarf mice. Mech Ageing Dev. 2009;130:393–400. doi: 10.1016/j.mad.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacutu R, Craig T, Budovsky A, Wuttke D, Lehmann G, Taranukha D, Costa J, Fraifeld VE, de Magalhães JP. Human Ageing Genomic Resources: integrated databases and tools for the biology and genetics of ageing. Nucleic Acids Res. 2013;41:D1027–D1033. doi: 10.1093/nar/gks1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292:107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Townsend MK, Clish CB, Kraft P, Wu C, Souza AL, Deik AA, Tworoger SS, Wolpin BM. Reproducibility of metabolomic profiles among men and women in 2 large cohort studies. Clin Chem. 2013;59:1657–1667. doi: 10.1373/clinchem.2012.199133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Poll MC, Soeters PB, Deutz NE, Fearon KC, Dejong CH. Renal metabolism of amino acids: its role in interorgan amino acid exchange. Am J Clin Nutr. 2004;79:185–197. doi: 10.1093/ajcn/79.2.185. [DOI] [PubMed] [Google Scholar]
- van der Goot AT, Zhu W, Vázquez-Manrique RP, Seinstra RI, Dettmer K, Michels H, Farina F, Krijnen J, Melki R, Buijsman RC, et al. Delaying aging and the aging-associated decline in protein homeostasis by inhibition of tryptophan degradation. Proc Natl Acad Sci USA. 2012;109:14912–14917. doi: 10.1073/pnas.1203083109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandanmagsar B, Youm YH, Ravussin A, Galgani JE, Stadler K, Mynatt RL, Ravussin E, Stephens JM, Dixit VD. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat Med. 2011;17:179–188. doi: 10.1038/nm.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz FM, Wanders RJ. Carnitine biosynthesis in mammals. Biochem J. 2002;361:417–429. doi: 10.1042/0264-6021:3610417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Müller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- Vessey DA. The biochemical basis for the conjugation of bile acids with either glycine or taurine. Biochem J. 1978;174:621–626. doi: 10.1042/bj1740621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weindruch R, Walford RL, Fligiel S, Guthrie D. The retardation of aging in mice by dietary restriction: longevity, cancer, immunity and lifetime energy intake. J Nutr. 1986;116:641–654. doi: 10.1093/jn/116.4.641. [DOI] [PubMed] [Google Scholar]
- Western D. Size, life history and ecology in mammals. Afr J Ecol. 1979;17:185–204. [Google Scholar]
- Wijeyesekera A, Selman C, Barton RH, Holmes E, Nicholson JK, Withers DJ. Metabotyping of long-lived mice using 1H NMR spectroscopy. J Proteome Res. 2012;11:2224–2235. doi: 10.1021/pr2010154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yardim-Akaydin S, Sepici A, Ozkan Y, Simşek B, Sepici V. Evaluation of allantoin levels as a new marker of oxidative stress in Behçet’s disease. Scand J Rheumatol. 2006;35:61–64. doi: 10.1080/03009740510026878. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.