Abstract
To understand environmental causes of disease, unbiased methods are needed to characterize the human exposome, which represents all toxicants to which people are exposed from both exogenous and endogenous sources. Because they directly modify DNA and important proteins, reactive electrophiles are probably the most important constituents of the exposome. Exposures to reactive electrophiles can be characterized by measuring adducts from reactions between circulating electrophiles and blood nucleophiles. We define an ‘adductome’ as the totality of such adducts with a given nucleophilic target. Because of their greater abundance and residence times in human blood, adducts of hemoglobin (Hb) and human serum albumin (HSA) are preferable to those of DNA and glutathione for characterizing adductomes. In fact, the nucleophilic hotspot represented by the only free sulfhydryl group in HSA (HSA-Cys34) offers particular advantages for adductomic experiments. Although targeted adducts of HSA-Cys34 have been monitored for decades, an unbiased method has only recently been reported for visualizing the HSA-Cys34 ‘subadductome’. The method relies upon a novel mass spectrometry application, termed fixed-step selected reaction monitoring (FS-SRM), to profile Cys34 adducts in tryptic digests of HSA. Here, we selectively review the literature regarding the potential of adductomics to partially elucidate the human exposome, with particular attention to the HSA-Cys34 subadductome.
Keywords: Adductome, Exposome, Reactive electrophiles, HSA, Cys34, Selected reaction monitoring
1. Environmental exposures and disease
Investigations of human twins and results from more than 400 genome-wide association studies (GWAS)1 indicate that genetic factors contribute about 10–30% of the risks of cancer and cardiovascular diseases (Lichtenstein et al., 2000; Manolio et al., 2009). Thus, it appears that non-genetic factors (i.e. ‘the environment’) are the major causes of chronic diseases in human populations. Yet the things that people generally associate with the environment, namely, air and water pollution, hygiene and sanitation, smoke from fuel combustion, and occupation, collectively contribute only about 7–10% to the burden of chronic diseases (Rodgers et al., 2004; Saracci and Vineis, 2007). Rather, the major environmental risk factors identified thus far are smoking, overweight, and the diet (Peto, 2001; Willett, 2002) all of which represent the combined effects of many toxicants. For example, cigarette smoke contains a multitude of likely human carcinogens (Hecht, 2003; Smith et al., 2003) as do many foods (Ames, 1983). However, with few exceptions, the identities of major environmental toxicants and their roles in causing chronic diseases have not been addressed.
Given the poor state of knowledge about health-impairing environmental exposures, epidemiologists pursue narrow hypotheses that largely skirt disease etiology in favor of known environmental risk factors, even when the attributable risks are small. Although such hypothesis-driven studies confirm some environmental sources of disease, they offer only fragments to our understanding of the major causes and mechanisms of chronic diseases. In fact, the current state of environmental epidemiology is reminiscent of genetic epidemiology 30 years ago, when an investigator would test for association between a single genetic polymorphism and a particular health outcome. With completion of the human genome project and subsequent technological advances leading to GWAS, such an approach would not be used today.
1.1. The exposome and biomonitoring
It has recently been argued that unbiased approaches, analogous to GWAS, are needed to investigate health effects arising from all environmental exposures, known collectively as the ‘exposome’ (Rappaport, 2011; Rappaport and Smith, 2010; Smith and Rappaport, 2009; Wild, 2005). In developing ‘environment-wide association studies’, it is essential to recognize that disease pathologies are mediated through chemicals that affect critical molecules, cells, and systems inside the body. Thus, exposures cannot be restricted to xenobiotic chemicals arising from air, water, smoking, etc., but must also include dietary constituents as well as toxicants produced endogenously by inflammation, stress, lipid peroxidation, infections, etc. In fact, blood concentrations of some toxic natural products and endogenous chemicals are much greater than those arising from polluted air and water, and are probably important contributors to cancer and cardiovascular disease (Ames, 1983; Dalle-Donne et al., 2006; Liebler, 2008). The need to include both exogenous and endogenous toxicants highlights the importance of a top-down strategy based upon biomonitoring to characterize exposures (using blood, say) rather than a bottom-up strategy using samples of air, water and food (Rappaport, 2011; Rappaport and Smith, 2010).
Another important consideration is the variability of the exposome in space and time. Indeed, knowledge that immigrant populations adopt the disease patterns of their host countries (Armstrong and Doll, 1975; Kato et al., 1973) emphasizes the importance of varying diets and lifestyles on health risks. Furthermore, whereas a person’s genome is essentially fixed at conception, his or her internal chemical environment varies during life due to changes in exogenous and endogenous exposures, age, exercise, infections, lifestyle and psychosocial factors. This variability of toxicant sources and levels places a premium upon obtaining repeated biospecimens to generate ‘snapshots’ of the exposome during critical stages of individuals’ lives, notably during gestation, early childhood, puberty, and the reproductive years (Rappaport, 2011). By investigating series of archived biospecimens from longitudinal cohort studies, it should be possible to organize such snapshots into individual exposomes, each highlighting the particular mix of environmental factors that a person experienced during life.
1.2. Candidate exposure studies
Because it is not currently feasible to measure all chemicals in the blood, we should focus upon the classes of toxicants that are likely to contribute to disease processes, namely reactive electrophiles, hormones and hormone-like substances, receptor-binding agents, and metals (Rappaport, 2011; Rappaport and Smith, 2010). Methods are currently available for measuring many such agents in human biospecimens. For example, several hundred analytes, including metals, hormone-like substances, and persistent organic compounds, have been detected in random samples of blood and urine from the U.S. population as part of the National Health and Nutrition Examination Survey (NHANES) (CDC, 2009). Using an initial list of 266 candidate exposures from the NHANES database, Patel et al. (2010) recently reported strong associations between the risk of type-2 diabetes and blood or urine levels of heptachlor epoxide, γ-tocopherol, β-carotenes, and polychlorinated biphenyls (PCBs). Note that these environmental factors include two exogenous toxicants (heptachlor epoxide and PCBs), both of which increased the risk of type-2 diabetes, as well as a vitamin (γ-tocopherol) which increased disease risk and a class of micronutrients (β-carotenes) which decreased disease risk. Interestingly, effect sizes from this candidate exposure study were comparable to the strongest loci ever reported in GWAS for any health endpoint. Although Patel >et al. identified possible environmental causes of type-2 diabetes, their study was biased in favor of a set of exogenous chemicals, vitamins, and micronutrients that had been selected a priori by NHANES investigators for various reasons. Thus, it is important for future studies to apply unbiased approaches to detect a larger cross section of toxic chemicals arising from all sources (Rappaport, 2011; Rappaport et al., 2010).
2. Reactive electrophiles and their adducts
Electrophiles, including reactive oxygen and nitrogen species, aldehydes, oxiranes and quinones, have long been suspected of causing cancer and other chronic diseases because they directly damage DNA and proteins (Brodie et al., 1971; Dalle-Donne et al., 2006; Liebler, 2008; Miller and Miller, 1966). Electrophiles enter the blood from absorption in the lungs or gut (e.g., inhalation of ethylene oxide) or, more typically, via metabolism of xenobiotics in the liver or other tissues (e.g., production of benzene oxide and acetaldehyde from metabolism of benzene and ethanol, respectively), from oxidation of lipids and other natural molecules [producing acrolein, 4-hydroxy-2(E)nonenal (HNE) and other aldehydes] and from inflammation (producing reactive oxygen and nitrogen species) associated with ionizing radiation, infections and preexisting diseases. Once in the blood, electrophiles react with all available nucleophiles to form adducts via SN1 and SN2 substitution, 1,4-addition, Schiff-base formation, and radical-mediated reactions (Liebler, 2008; Tornqvist et al., 2002).
Because they are reactive, it is difficult to measure electrophiles directly in blood. However, one can measure adducts of electrophiles resulting from reactions with DNA (in nucleated cells), reactions with prominent blood proteins, such as hemoglobin (Hb) and human serum albumin (HSA), and reactions with the ubiquitous tripeptide antioxidant, glutathione [reviewed by Blair, 2006; Rubino et al., 2009; Tornqvist et al., 2002]. Reactions of electrophiles with DNA occur primarily at nucleophilic nitrogen atoms of particular bases (especially guanine); reactions with Hb and HSA are observed primarily at the free thiol groups of Cys and amine groups of His, Trp, Lys and the N-termini; and reactions with glutathione occur primarily at the free Cys thiol. Since protein adducts are not repaired and are much more abundant than DNA adducts in blood (1 ml of blood contains about 150 mg Hb, 30 mg of HSA, and 0.003–0.008 mg of DNA) (Tornqvist et al., 2002), they are more useful measures of internal dose than DNA adducts, which have paradoxically received far more attention in this regard. And because protein adducts are also much longer lived than glutathione adducts—mean residence times are 28 d and 63 d for adducts of HSA and Hb, respectively, in humans (Furne et al., 2003; Granath et al., 1992; Troester et al., 2002), compared to hours for glutathione adducts (Wagner et al., 2007)—they provide more stable measures of exposure to electrophilic precursors. Archived blood and serum or plasma, which are biospecimens of choice in cohort studies such as NHANES or the U.K. Biobank (Ollier et al., 2005) are important sources of Hb and/or HSA for investigating adducts.
2.1. Targeted protein adducts
As indicated in Section 2, adducts of Hb and HSA are the most promising candidates for investigating exposures to reactive electrophiles in human populations. Levels of targeted Hb and/or HSA adducts have been studied in humans for several toxicants, notably acrylamide, aflatoxin B1, aldehydes, aminobiphenyl and other aromatic amines, benzene, 1,3-butadiene, ethylene oxide, and polycyclic aromatic hydrocarbons (reviewed in Rubino et al., 2009). Most of these studies relied upon older gas chromatography–mass spectrometry techniques to selectively cleave adducts from Cys residues of Hb and HSA or from the N-terminal Val residue of Hb, while newer studies have employed liquid-chromatography tandem mass spectrometry (LC–MS/MS) to investigate adducted peptides in protein digests.
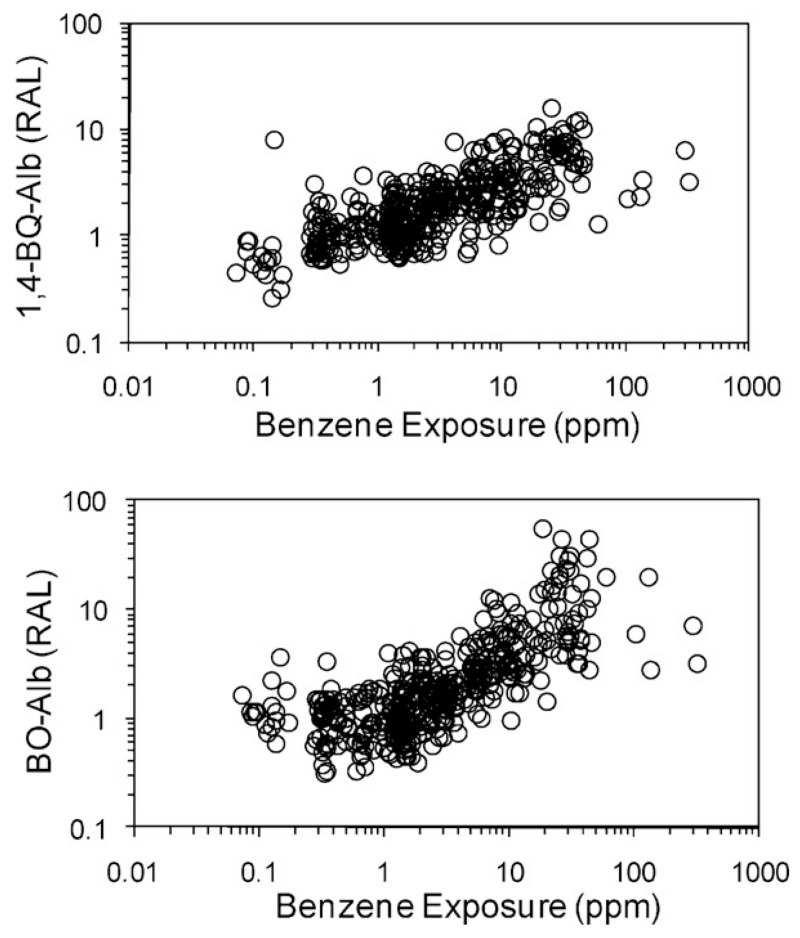
Some human studies showed strong correlations between levels of Hb and/or HSA adducts and the corresponding environmental exposures. For example, Fig. 1 depicts log-scale scatter plots of levels of cysteinyl HSA adducts of two electrophilic metabolites of benzene, i.e. benzene oxide and 1,4-benzoquinone (designated BO-Alb and 1,4-BQ-Alb), versus benzene exposure, combined from three studies of Chinese workers conducted in our laboratory (Lin et al., 2007; Rappaport et al., 2002; Yeowell-O’Connell et al., 1998). Because background levels of these adducts differed across the three populations, adduct concentrations are shown as relative adduct levels (RALs), representing ratios of the individual adduct concentrations to median values in concurrent controls. Both plots show increasing mean trends of adduct levels with increasing benzene exposure. At a given benzene exposure, the 5-fold to 10-fold ranges in adduct levels represent the combined effects of assay errors and interindividual differences in physiological, genetic, and lifestyle factors (including diet), that might have influenced production and elimination of adducts. Also, mean values of RALs were close to unity for subjects exposed to less than 1 ppm benzene, indicating that workers exposed at lower benzene concentrations had adduct levels similar to those of control subjects.
Fig. 1.
Levels of HSA adducts of benzene oxide (BO-Alb) and 1,4-benzoquinone (1,4-BQ-Alb) versus exposure to benzene for 439 Chinese workers (Lin et al., 2007; Rappaport et al., 2002; Yeowell-O’Connell et al., 1998). Relative adduct levels (RALs) represent ratios of individual adduct levels to median levels in concurrent controls.
3. The adductome
Although relationships from adduct-specific studies, such as those shown in Fig. 1, reinforce the idea that protein adducts can be useful measures of exposure to particular electrophiles, they provide little information about the universe of electrophiles to which humans are exposed. Also, such adduct-specific approaches tend to employ methods that cannot be easily multiplexed for detection of large numbers of unknown adducts.
Recently, the concept of an ‘adductome’, representing the totality of covalent adducts bound to tissue nucleophiles, has been offered as an avenue for characterizing essentially all reactive electrophiles in biospecimens (Kanaly et al., 2006; Merrick, 2008; Rappaport, 2011; Rubino et al., 2009). Kanaly et al. (2006, 2007) considered the ‘DNA adductome’ as the set of all adducts detectable in digests of DNA from lung and esophageal tissues. Similarly, Vöelkel et al. (Scholz et al., 2005; Wagner et al., 2006, 2007) and Blair (2006, 2010) described the universe of urinary glutathione adducts as a subset of the metabolome, an entity equivalent to the ‘glutathione adductome’. Merrick (2008) and Rubino et al. (2009) defined the ‘protein adductome’ in terms of all adducts or post-translational modifications (PTMs) involving a relevant subset of the human proteome. Since conventional proteomic investigations of PTMs focus upon discrete classes of modifications, such as phosphorylation of serine, threonine and tyrosine (Olsen et al., 2006), lysine acetylation (Zhang et al., 2009), arginine methylation (Ong et al., 2004), and tyrosine nitration (Zhan and Desiderio, 2006), they cover only a small portion of the protein adductome in much the same way that targeted adducts in exposure-specific studies represent small fractions of all adducts bound to a given nucleophile. Indeed, the protein adductome is staggeringly complex and cannot be fully characterized with current adduct- or PTM-focused methods.
Given the inherent advantages of Hb and HSA as measures of internal dose (abundance, lack of repair, relatively long residence times, simple kinetics as discussed in Section 2), it makes sense to focus on these two blood proteins for adductomic experiments. Thus, an incremental approach for characterizing the protein adductome would be to consider the totality of adducts bound to a nucleophilic hotspot on Hb or HSA. In describing such an approach, we will use the term ‘subadductome’ to refer to the set of modifications present at a given nucleophilic locus on a protein.
3.1. The HSA-Cys34 subadductome
Of the many loci available for substitution reactions with electrophiles, free thiols in Cys residues are attractive candidates because of their strong nucleophilicity. Since Cys subadductomes also offer the opportunity for selective enrichment via thiol-affinity chromatography (discussed in Section 3.2), they are particularly well suited for adductomic experiments. Interestingly, human Hb and HSA each have only one free thiol available for adduction, i.e. Hb-Cys93β and HSA-Cys34; all other Cys residues are either joined together by disulfide bonds (in HSA) or are buried so deep within the protein as to be inaccessible for adduction (in Hb). Binding of electrophiles to Cys residues is enhanced when the thiol exists in thiolate (–S−) form (Tornqvist et al., 2002). Thus, it is relevant that HSA-Cys34 has an unusually low pKa value (6.55 compared to about 8.0–8.5 for thiols in most other proteins and peptides) (Aldini et al., 2008; Stewart et al., 2005), and therefore contains about 88% –S− at physiological pH (7.4) compared to 20% –S− for a Cys with pKa = 8.0. The low pKa for HSA-Cys34 probably explains the 30-fold greater reactivity of HSA-Cys34 towards benzene oxide in human blood at 37 °C compared to Hb-Cys93β (0.684 M−1 s−1 versus 1.93 × 10−2 M−1 s−1) (Lindstrom et al., 1998). Thus, even though HSA has a shorter mean residence time (28 d versus 63 d) and is present at about 1/5 the blood concentration of Hb (30 versus150 mg/ml), HSA-Cys34 accumulates about 14 times more benzene oxide adducts in human blood than Hb-Cys93β at steady state. For comparison, Aldini et al. (2008) reported a rate constant for the reaction between HNE and HSA-Cys34 at 37 °C of 29.37 M−1 s−1 which is 52 times greater than that observed for benzene oxide. This indicates that HSA-Cys34 should bind to electrophilic species exhibiting a wide range of reactivities.
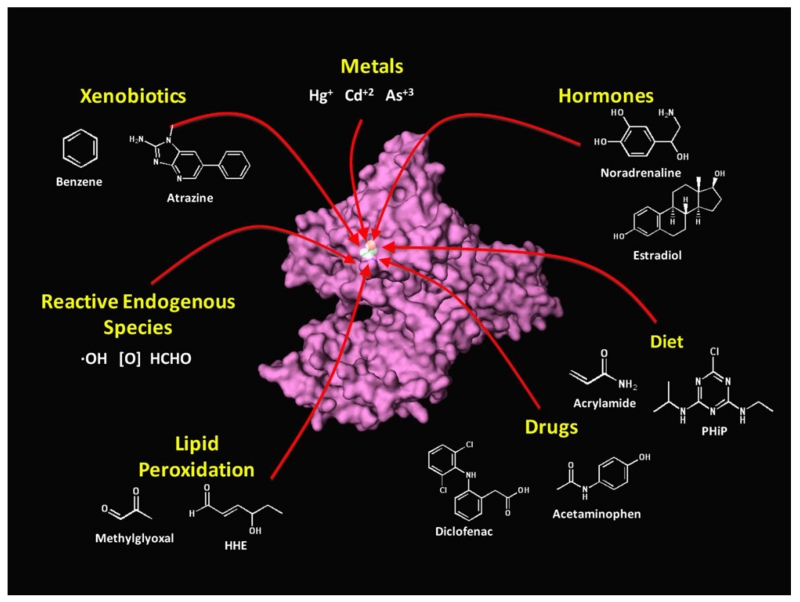
X-ray crystallography and NMR studies have shown that HSA-Cys34 resides in a small cleft of the HSA molecule (Christodoulou et al., 1995; He and Carter, 1992) and, thus, might not be accessible to some bulky electrophiles. However, the thiolate form of HSA-Cys34 exists in equilibrium between buried and exposed states, the latter of which is less prone to steric effects in forming adducts (Sengupta et al., 2001; Stewart et al., 2005). In any case, HSA-Cys34 adducts have been unambiguously detected from reactions with aldehydes, nitrogen mustards, oxiranes, quinones, metal ions, and a host of drugs (reviewed in Rubino et al., 2009) and can be oxidized to sulfenic-, sulfinic-, and sulfonic acids (Carballal et al., 2007, 2003; Turell et al., 2008). In fact, HSA-Cys34 is a well known scavenger of electrophiles that accounts for about 80% of all free thiols in human serum (Aldini et al., 2006, 2008). This indicates that a wide range of electrophiles binds to HSA-Cys34. The notion that the HSA-Cys34 subadductome can be used to characterize adducts of circulating electrophiles is illustrated in Fig. 2, which shows the location of the Cys34 hotspot on the HSA protein, and lists several classes of exogenous and endogenous toxicants that are known or expected to form adducts at this locus.
Fig. 2.
Humans are exposed to reactive and potentially toxic electrophiles from both exogenous and endogenous sources. The HSA molecule contains a nucleophilic hotspot (HSA-Cys34) that forms adducts with electrophiles in the blood and can be used to characterize human exposures to reactive electrophiles (or their precursors) during the month prior to blood collection.
3.2. Enrichment of HSA-Cys34 adducts
Despite its high reactivity with metals, thiols, and electrophilic species, most freshly isolated HSA remains in the free thiol form (mercaptalbumin), while the bulk of bound thiols (about 20–30%) is represented by disulfides with cysteine, glutathione, and other small Cys-containing peptides (Beck et al., 2004; Carballal et al., 2003). While these HSA-Cys34 disulfides are readily reduced with dithiothreitol (DTT), the remaining HSA-Cys34 covalent adducts are not reducible with DTT (Carballal et al., 2003).
Although the fraction of HSA-Cys34 represented by covalent adducts has not been extensively studied, Aldini et al. (2008) reported that a fresh sample of fully reduced HSA contained 96% mercaptalbumin, suggesting that about 4% of HSA-Cys34 was covalently modified with unknown adducts. Since this pool of covalent modifications to HSA-Cys34 is rather small, and probably contains hundreds of individual adducts, it is beneficial to remove mercaptalbumin prior to adductomic experiments. To accomplish this, Funk et al. (2010) incubated DTT-reduced samples of fresh and archived HSA with a thiol-affinity resin to remove mercaptalbumin. After removal of free thiols, a mass corresponding to mercaptalbumin (66,436 Da) was not observed in the enriched samples. Comparing HSA concentrations before and after enrichment, the mass fraction of modified HSA-Cys34 in a sample of fresh HSA was 2.9%, a value similar to the estimate of 4% derived from Aldini’s experiment with fresh HSA (Aldini et al., 2008). In contrast, the mean fraction of HSA-Cys34 adducts in 8 samples of archived HSA was 32.3% (SD = 5.9%), suggesting that the adduct fraction had been increased due to either isolation of HSA or storage for 11 years at −80 °C.
4. Profiling adducts by fixed-step selected reaction monitoring
Investigators have recently used triple-quadrupole MS to obtain profiles of unknown adducts of glutathione (mercapturic acids) in urine (Scholz et al., 2005; Wagner et al., 2006, 2007), DNA adducts in autopsy tissues (Kanaly et al., 2006, 2007), and HSA-Cys34 adducts in archived samples of HSA (Li et al., 2011). We refer to this general methodology as ‘fixed-step selected reaction monitoring’ or FS-SRM (Li et al., 2011). Unlike conventional SRM, which searches MS/MS spectra for parent and product ions with masses corresponding to those of targeted analytes, FS-SRM employs sequential lists of theoretical parent and product ions, each separated by a fixed m/z, and thereby detects essentially all modifications of the targeted nucleophile within a specified range of mass increases (adduct masses). Although each of the cited applications of FS-SRM employed triple quadrupole MS/MS for analysis, the characteristics of the biospecimens, adducts, and methodology varied across studies.
4.1. FS-SRM of glutathione and DNA adducts
Experiments which profiled glutathione and DNA adducts employed LC–MS/MS with electrospray ionization (ESI) to detect mass spectra displaying constant neutral losses, characteristic of the parent nucleophiles. The human studies of glutathione adducts by Vöelkel’s laboratory (Wagner et al., 2006, 2007) employed 1 ml of urine that had been acidified and then purified by solid-phase extraction as part of on-line LC–MS/MS. Glutathione adducts were detected as mercapturic acids by LC–ESI-MS/MS in SRM mode by monitoring a constant neutral loss of glutamate [m/z → (m/z – 129)]− Da, using a fixed-step m/z of 1 Da over a range of parent ion m/z between 200 and 450 (251 theoretical transitions), corresponding to added masses between 52 and 302 Da. Quantitation employed isotopic internal standards of synthetic adducts. Principal components of adduct profiles were distinguishable between male and female subjects (Wagner et al., 2007). Adducts of a metabolite of the drug acetaminophen were also unambiguously detected in volunteer subjects following a single administration of either 50 or 500 mg acetaminophen (Wagner et al., 2006).
Kanaly et al. (2007) enzymatically digested 100 μg portions of purified DNA in post mortem samples of lung and esophageal tissues from a female subject. Adducts were detected as modified DNA nucleosides by LC–ESI-MS/MS in SRM mode via a constant neutral loss of 2′-deoxyribose – corresponding to [M+H]+ → [M+H–116]+ Da – using a fixed-step m/z of 1 Da over a range→of parent ion m/z between 228.8 and 602.8 (374 theoretical transitions). Twelve repeated LC–MS/MS runs were required to obtain sufficient data for analysis of the full transition list. Quantitation employed isotopic internal standards of synthetic adducts. Adduct profiles were compared across tissue samples obtained at different locations in the lung, the esophagus, and calf-thymus DNA (control samples). Of the hundreds of putative adducts detected, 7 DNA adducts representing addition of ethyl-, etheno-, and propano-moieties to guanosine and adenosine were tentatively identified by comparison of masses and retention times with synthetic standards.
4.2. FS-SRM of HSA-Cys34 adducts
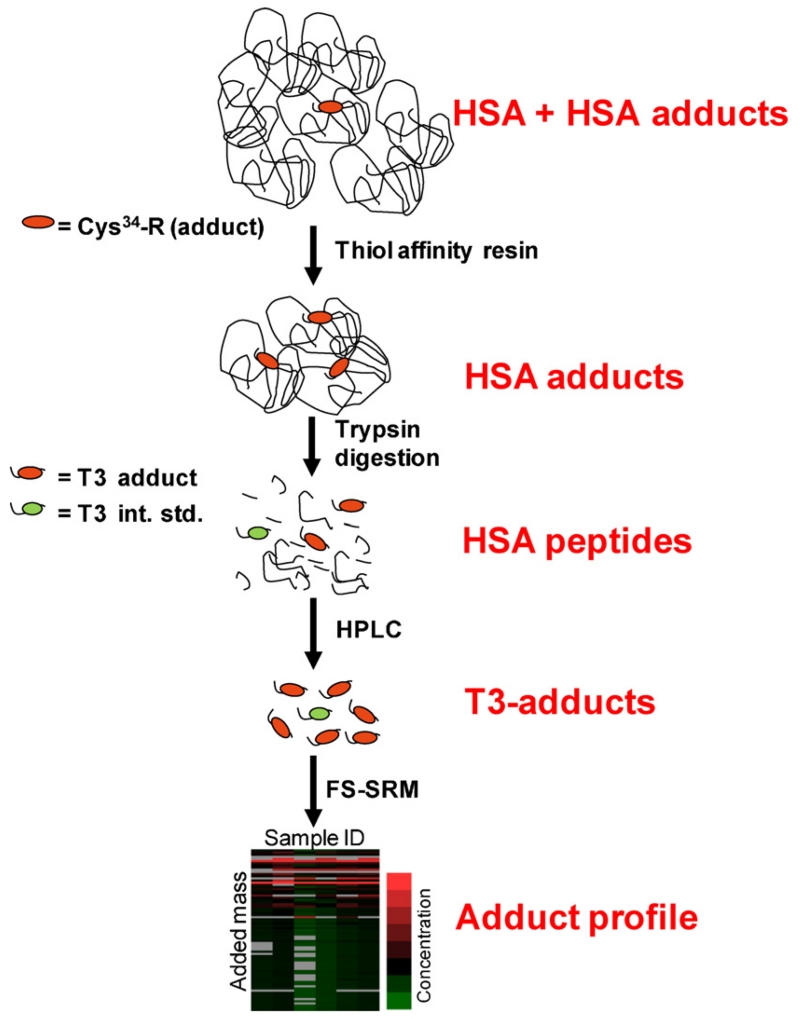
After tryptic digestion of HSA, all HSA-Cys34 adducts are represented by modifications to a 21-mer peptide, designated as the T3 peptide (the 3rd largest tryptic peptide of HSA with sequence ALVLIAFAQYLQQC34PFEDHVK and a mass of 2432 Da). Fig. 3 shows the analytical scheme used in our laboratory to profile HSA-Cys34 adducts as modified T3 peptides (Li et al., 2011). After removing mercaptalbumin from HSA (see Section 3.2), adduct-enriched HSA is digested with trypsin, and an isotopically labeled T3 adduct of iodoacetamide (IAA) is added as internal standard (IAA-iT3) for quantification of putative adducts. Modified T3 peptides are then purified by off-line high performance liquid chromatography (HPLC) and analyzed by FS-SRM. Results of FS-SRM are displayed in profiles of putative adduct concentrations vs. adduct masses.
Fig. 3.
Scheme for profiling HSA-Cys34 adducts (Li et al., 2011). Thiol-affinity resins are used to remove mercaptalbumin (i.e. HSA containing free Cys34). Enriched HSA-Cys34 adducts are purified by high-performance liquid chromatography (HPLC), detected as modified T3 peptides by fixed-step selected reaction monitoring (FS-SRM), and displayed as a profile of adduct concentration vs. added mass.
To perform FS-SRM, purified T3-adduct fractions are statically infused into the nano-ESI source of the MS, and sequential lists of 312 theoretical triply charged parent ions and four production transitions (b4 +, y15 2+ , y16 2+ and y17 2+ for each parent ion) are scanned 200 times over a range of parent ion m/z between 815 and 929 (Li et al., 2011). In parallel, FS-SRM is performed with a negative control consisting of the analogous T3 peptide from bovine serum albumin which also contains a Cys34 but with a different amino acid sequence (GLVLIAFSQYLQQC34FFDEHVK). With a fixed-step m/z of 1.5, each triply-charged parent ion represents an adduct-mass bin of 4.6 Da between 9.1 Da and 351.1 Da (77 total bins) and a single bin can contain multiple adducts of similar mass. Quantitation of putative adducts is performed by comparing the y16 2+ production response with the corresponding response for IAA-iT3 (internal standard). To minimize false positives, a set of structured screening rules, based upon synthetic T3 adducts, is applied to classify putative adducts.
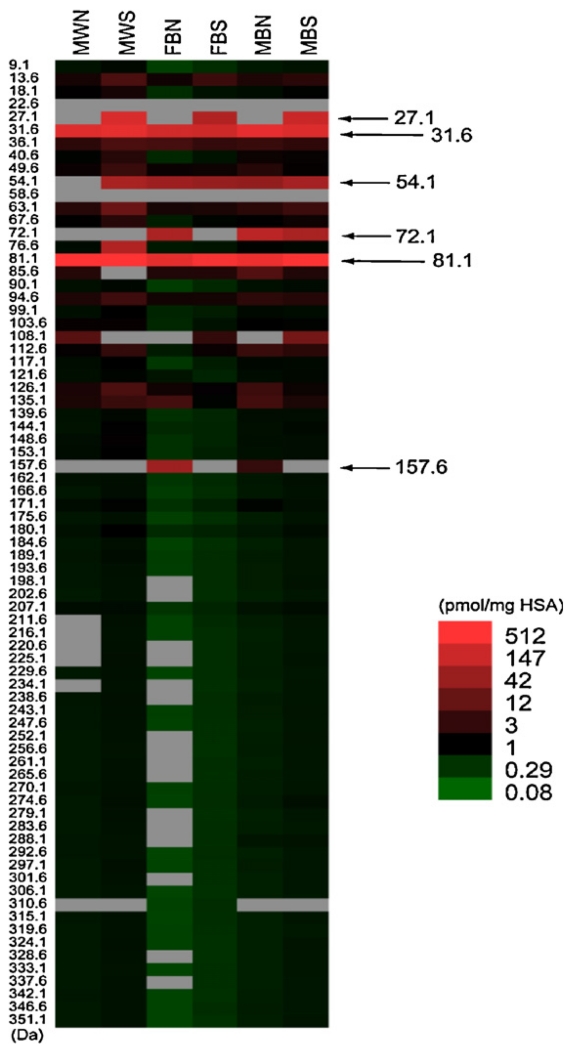
Adduct profiles from FS-SRM of 6 archived specimens of pooled HSA are shown in Fig. 4, which uses the same heat-map format that is commonly employed for gene-expression profiling. Each of the 6 pooled specimens in Fig. 4 is identified at the top of a column representing 77 possible HSA-Cys34 adduct-mass bins with midpoints shown at the left (Li et al., 2011). The intensity of each adduct hit is indicated by its color. An average of 66 adduct hits (out of 77 possible) were observed in these samples, with smokers having somewhat more hits (median = 71) than nonsmokers (median = 63). The adduct-mass range of the putative adducts extended from 9.1 to 351.1 Da, with more abundant adducts concentrated below 100 Da. (Note that the term ‘adduct mass’ refers to any chemical modification of HSA-Cys34 which results in a net mass increase and not necessarily to a simple addition product).
Fig. 4.
HSA-Cys34 adducts detected in archived HSA from subjects stratified by race, gender and smoking status (Li et al., 2011). Each HSA sample was pooled from 5 subjects stratified by race (B = black, W = white), gender (F = female, M = male, and smoking status (N = nonsmoker, S = smoker). Adduct-mass hits in each HSA sample are shown with estimated concentrations expressed as pmol/mg of HSA (represented by colors). Each adduct mass represents the midpoint of a mass bin ±2.3 Da.
Although the results shown in Fig. 4 could have been influenced by artifacts due to isolation and storage of HSA, as well as modifications arising during processing, the observed concentrations appear reasonable. Levels of 398 total adduct hits in these samples ranged from 0.2 to 785 pmol/mg HSA, with median and mean values of 0.61 and 14.6 pmol/mg HSA, respectively (Li et al., 2011). These putative adduct concentrations are comparable to mean values between <0.01 and 313 pmol/mg HSA that were reported for 9 targeted HSA-Cys34 adducts measured in control populations (Rubino et al., 2009). Smokers had between 17% and 34% higher cumulative concentrations of putative adducts than nonsmokers, which is also consistent with results from the 9 control populations summarized by Rubino et al. (2009).
5. Comparing adduct profiles across populations
Returning now to the motivation for characterizing exposures to reactive electrophiles, we will consider potential epidemiologic applications of adductomics. The basic idea is to compare adduct profiles across populations of interest. Using HSA-Cys34 adducts as an example, samples of serum or plasma would be obtained from two or more populations representing different categories of disease or exposure status. For example, samples could be derived from a case control study of a particular cancer or from smokers and nonsmokers. After processing the HSA from the samples, adduct profiles would be compared within and between the populations to determine the approximate masses of abundant and discordant adducts. Using statistical tools, which have been developed for performing multiple comparisons in GWAS and metabolomics studies, adduct profiles would be compared and discordant adducts pinpointed. Then follow-up experiments would be conducted to chemically identify these important adducts (and their precursors), using a combination of high resolution MS, to obtain accurate masses and elemental compositions, and synthesis of reference compounds. Finally, validation studies would be performed to accurately and precisely measure targeted adducts, preferably with repeated serum or plasma samples obtained from subjects at different times or stages of life. Such studies would address questions about measurement errors, adduct variability and possible formation of artifacts. Archived specimens from prospective cohort studies would be well suited for such validation exercises.
6. Conclusions
Although environmental rather than genetic factors are the primary contributors to chronic diseases, investigations of ‘the environment’ have focused upon air and water pollutants of limited importance to the overall disease burden. To investigate the environmental causes of chronic diseases, unbiased methods are needed for charactering human exposures to the myriad toxicants arising from exogenous (air, water, diet, tobacco use, etc.) and endogenous (inflammation, stress, lipid peroxidation, infection, etc.) sources, which collectively comprise the human exposome. Exposures to reactive electrophiles are of particular concern because of the inherent toxicity of these substances, due to their ability to modify DNA and important proteins. Levels of reactive electrophiles in the body can be inferred from the concentrations of adducts, which arise from reactions between these electrophiles and blood nucleophiles. We define the totality of such adducts with a given nucleophilic target (e.g., DNA, protein, or glutathione) as its ‘adductome’. The subadductome of HSA-Cys34, which is the strongest and most abundant nucleophile in serum with a residence time of 28 days, is an appealing candidate for adductomic studies of human populations. Although HSA-Cys34 adducts of targeted electrophiles have been investigated for many years, an unbiased method for detecting all such adducts has only recently been reported (Li et al., 2011). As with previous human studies of glutathione and DNA adductomes (Kanaly et al., 2006, 2007; Wagner et al., 2006, 2007), this method relies upon a novel application of mass spectrometry called FS-SRM to generate adduct profiles over a given mass range. Putative HSA-Cys34 adducts in human samples covered a wide range of concentrations, were most abundant in the mass range below 100 Da (see Fig. 3), and were more abundant in smokers than nonsmokers. By comparing adduct profiles across populations representing different categories of health or exposure status, it should be possible to zero in upon particular adducts and precursor molecules of importance.
Acknowledgements
We thank Martyn Smith and Suramya Waidyanatha for thoughtful discussions and to our many collaborators who contributed to the studies of Chinese benzene-exposed workers cited in Fig. 1. This work was supported by grant U54ES016115 from the U.S. National Institute for Environmental Health Sciences (NIEHS) through the trans-National Institutes of Health (NIH) Genes, Environment, and Health Initiative and by a contract from the American Chemistry Council Long-range Research Initiative. Funding sources played no role in study design; in the collection, analysis and interpretation of data; in the writing of the manuscript; and in the decision to submit the paper for publication.
Footnotes
Abbreviations: BO-Alb, HSA adduct of benzene oxide; 1,4-BQ-Alb; HSA adduct of 1,4-benzoquinone; DTT, dithiothreitol; ESI, electrospray ionization; FS-SRM, fixed-step SRM; GWAS, genome-wide association studies; Hb, hemoglobin; HNE, 4-hydroxy-2(E)nonenal; HSA, human serum albumin; HPLC, high-performance liquid chromatography; IAA, iodoacetamide; IAA-iT3, internal standard consisting of the adduct of IAA with isotopically labeled T3; MS, mass spectrometry; NHANES, National Health and Nutrition Examination Survey; PCBs, polychlorinated biphenyls; PTM, post-translational modification; RAL, relative adduct level; SRM, selected reaction monitoring; T3, the third largest tryptic peptide of HSA.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
References
- Aldini G, Gamberoni L, Orioli M, Beretta G, Regazzoni L, Maffei Facino R, Carini M. Mass spectrometric characterization of covalent modification of human serum albumin by 4-hydroxy-trans-2-nonenal. J. Mass Spectrom. 2006;41:1149–1161. doi: 10.1002/jms.1067. [DOI] [PubMed] [Google Scholar]
- Aldini G, Vistoli G, Regazzoni L, Gamberoni L, Facino RM, Yamaguchi S, Uchida K, Carini M. Albumin is the main nucleophilic target of human plasma: a protective role against pro-atherogenic electrophilic reactive carbonyl species? Chem. Res. Toxicol. 2008;21:824–835. doi: 10.1021/tx700349r. [DOI] [PubMed] [Google Scholar]
- Ames BN. Dietary carcinogens and anticarcinogens, oxygen radicals and degenerative diseases. Science. 1983;221:1256–1264. doi: 10.1126/science.6351251. [DOI] [PubMed] [Google Scholar]
- Armstrong B, Doll R. Environmental factors and cancer incidence and mortality in different countries, with special reference to dietary practices. Int. J. Cancer. 1975;15:617–631. doi: 10.1002/ijc.2910150411. [DOI] [PubMed] [Google Scholar]
- Beck JL, Ambahera S, Yong SR, Sheil MM, de Jersey J, Ralph SF. Direct observation of covalent adducts with Cys34 of human serum albumin using mass spectrometry. Anal. Biochem. 2004;325:326–336. doi: 10.1016/j.ab.2003.10.041. [DOI] [PubMed] [Google Scholar]
- Blair IA. Endogenous glutathione adducts. Curr. Drug Metab. 2006;7:853–872. doi: 10.2174/138920006779010601. [DOI] [PubMed] [Google Scholar]
- Blair IA. Analysis of endogenous glutathione-adducts and their metabolites. Biomed. Chromatogr. 2010;24:29–38. doi: 10.1002/bmc.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie BB, Reid WD, Cho AK, Sipes G, Krishna G, Gillette JR. Possible mechanism of liver necrosis caused by aromatic organic compounds. Proc. Natl. Acad. Sci. U. S. A. 1971;68:160–164. doi: 10.1073/pnas.68.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballal S, Alvarez B, Turell L, Botti H, Freeman BA, Radi R. Sulfenic acid in human serum albumin. Amino Acids. 2007;32:543–551. doi: 10.1007/s00726-006-0430-y. [DOI] [PubMed] [Google Scholar]
- Carballal S, Radi R, Kirk MC, Barnes S, Freeman BA, Alvarez B. Sulfenic acid formation in human serum albumin by hydrogen peroxide and peroxynitrite. Biochemistry. 2003;42:9906–9914. doi: 10.1021/bi027434m. [DOI] [PubMed] [Google Scholar]
- CDC . Fourth National Report on Human Exposure to Environmental Chemicals. National Center for Environmental Health. Centers for Disease Control and Prevention; Atlanta, GA: 2009. [Google Scholar]
- Christodoulou J, Sadler PJ, Tucker A. 1H NMR of albumin in human blood plasma: drug binding and redox reactions at Cys34. FEBS Lett. 1995;376:1–5. doi: 10.1016/0014-5793(95)01231-2. [DOI] [PubMed] [Google Scholar]
- Dalle-Donne I, Rossi R, Colombo R, Giustarini D, Milzani A. Biomarkers of oxidative damage in human disease. Clin. Chem. 2006;52:601–623. doi: 10.1373/clinchem.2005.061408. [DOI] [PubMed] [Google Scholar]
- Furne JK, Springfield JR, Ho SB, Levitt MD. Simplification of the endalveolar carbon monoxide technique to assess erythrocyte survival. J Lab Clin Med. 2003;142:52–57. doi: 10.1016/S0022-2143(03)00086-6. [DOI] [PubMed] [Google Scholar]
- Funk WE, Li H, Iavarone AT, Williams ER, Riby J, Rappaport SM. Enrichment of cysteinyl adducts of human serum albumin. Anal. Biochem. 2010;400:61–68. doi: 10.1016/j.ab.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granath F, Ehrenberg L, Tornqvist M. Degree of alkylation of macromolecules in vivo from variable exposure. Mutat. Res. 1992;284:297–306. doi: 10.1016/0027-5107(92)90014-s. [DOI] [PubMed] [Google Scholar]
- He XM, Carter DC. Atomic structure and chemistry of human serum albumin. Nature. 1992;358:209–215. doi: 10.1038/358209a0. [DOI] [PubMed] [Google Scholar]
- Hecht SS. Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat. Rev. Cancer. 2003;3:733–744. doi: 10.1038/nrc1190. [DOI] [PubMed] [Google Scholar]
- Kanaly RA, Hanaoka T, Sugimura H, Toda H, Matsui S, Matsuda T. Development of the adductome approach to detect DNA damage in humans. Antioxid. Redox Signal. 2006;8:993–1001. doi: 10.1089/ars.2006.8.993. [DOI] [PubMed] [Google Scholar]
- Kanaly RA, Matsui S, Hanaoka T, Matsuda T. Application of the adductome approach to assess intertissue DNA damage variations in human lung and esophagus. Mutat. Res. 2007;625:83–93. doi: 10.1016/j.mrfmmm.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Kato H, Tillotson J, Nichaman MZ, Rhoads GG, Hamilton HB. Epidemiologic studies of coronary heart disease and stroke in Japanese men living in Japan, Hawaii and California. Am. J. Epidemiol. 1973;97:372–385. doi: 10.1093/oxfordjournals.aje.a121518. [DOI] [PubMed] [Google Scholar]
- Li H, Grigoryan H, Funk WE, Lu SS, Rose S, Williams ER, Rappaport SM. Profiling Cys34 adducts of human serum albumin by fixed-step selected reaction monitoring. Mol Cell Proteomics. 2011;10(3):1–14. doi: 10.1074/mcp.M110.004606. M110.004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer–analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- Liebler DC. Protein damage by reactive electrophiles: targets and consequences. Chem. Res. Toxicol. 2008;21:117–128. doi: 10.1021/tx700235t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YS, Vermeulen R, Tsai CH, Waidyanatha S, Lan Q, Rothman N, Smith MT, Zhang L, Shen M, Li G, Yin S, Kim S, Rappaport SM. Albumin adducts of electrophilic benzene metabolites in benzene-exposed and control workers. Environ. Health Perspect. 2007;115:28–34. doi: 10.1289/ehp.8948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom AB, Yeowell-O’Connell K, Waidyanatha S, McDonald TA, Golding BT, Rappaport SM. Formation of hemoglobin and albumin adducts of benzene oxide in mouse, rat, and human blood. Chem. Res. Toxicol. 1998;11:302–310. doi: 10.1021/tx9701788. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whitte-more AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick BA. The plasma proteome, adductome and idiosyncratic toxicity in toxicoproteomics research. Brief. Funct. Genomic Proteomic. 2008;7:35–49. doi: 10.1093/bfgp/eln004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EC, Miller JA. Mechanisms of chemical carcinogenesis: nature of proximate carcinogens and interactions with macromolecules. Pharmacol. Rev. 1966;18:805–838. [PubMed] [Google Scholar]
- Ollier W, Sprosen T, Peakman T. UK Biobank: from concept to reality. Pharmacogenomics. 2005;6:639–646. doi: 10.2217/14622416.6.6.639. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Ong SE, Mittler G, Mann M. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat. Methods. 2004;1:119–126. doi: 10.1038/nmeth715. [DOI] [PubMed] [Google Scholar]
- Patel CJ, Bhattacharya J, Butte AJ. An Environment-Wide Association study (EWAS) on type 2 diabetes mellitus. PLoS One. 2010;5:e10746. doi: 10.1371/journal.pone.0010746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peto J. Cancer epidemiology in the last century and the next decade. Nature. 2001;411:390–395. doi: 10.1038/35077256. [DOI] [PubMed] [Google Scholar]
- Rappaport SM. Implications of the exposome for exposure science. J. Expo. Sci. Environ. Epidemiol. 2011;21:5–9. doi: 10.1038/jes.2010.50. [DOI] [PubMed] [Google Scholar]
- Rappaport SM, Kim S, Lan Q, Li G, Vermeulen R, Waidyanatha S, Zhang L, Yin S, Smith MT, Rothman N. Human benzene metabolism following occupational and environmental exposures. Chem. Biol. Interact. 2010;184:189–195. doi: 10.1016/j.cbi.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport SM, Smith MT. Environment and disease risks. Science. 2010;330:460–461. doi: 10.1126/science.1192603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport SM, Waidyanatha S, Qu Q, Shore R, Jin X, Cohen B, Chen LC, Melikian AA, Li G, Yin S, Yan H, Xu B, Mu R, Li Y, Zhang X, Li K. Albumin adducts of benzene oxide and 1,4-benzoquinone as measures of human benzene metabolism. Cancer Res. 2002;62:1330–1337. [PubMed] [Google Scholar]
- Rodgers A, Ezzati M, Vander Hoorn S, Lopez AD, Lin RB, Murray CJ. Distribution of major health risks: findings from the global burden of disease study. PLoS Med. 2004;1:e27. doi: 10.1371/journal.pmed.0010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubino FM, Pitton M, Di Fabio D, Colombi A. Toward an “omic” physiopathology of reactive chemicals: thirty years of mass spectrometric study of the protein adducts with endogenous and xenobiotic compounds. Mass Spectrom. Rev. 2009;28:725–784. doi: 10.1002/mas.20207. [DOI] [PubMed] [Google Scholar]
- Saracci R, Vineis P. Disease proportions attributable to environment. Environ. Health. 2007;6:38. doi: 10.1186/1476-069X-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz K, Dekant W, Volkel W, Pahler A. Rapid detection and identification of N-acetyl-l-cysteine thioethers using constant neutral loss and theoretical multiple reaction monitoring combined with enhanced product-ion scans on a linear ion trap mass spectrometer. J. Am. Soc. Mass Spectrom. 2005;16:1976–1984. doi: 10.1016/j.jasms.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Chen H, Togawa T, DiBello PM, Majors AK, Budy B, Ketterer ME, Jacobsen DW. Albumin thiolate anion is an intermediate in the formation of albumin-S-S-homocysteine. J. Biol. Chem. 2001;276:30111–30117. doi: 10.1074/jbc.M104324200. [DOI] [PubMed] [Google Scholar]
- Smith CJ, Perfetti TA, Garg R, Hansch C. IARC carcinogens reported in cigarette mainstream smoke and their calculated log P values. Food Chem. Toxicol. 2003;41:807–817. doi: 10.1016/s0278-6915(03)00021-8. [DOI] [PubMed] [Google Scholar]
- Smith MT, Rappaport SM. Building exposure biology centers to put the E into “G × E” interaction studies. Environ. Health Perspect. 2009;117:A334–A335. doi: 10.1289/ehp.12812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart AJ, Blindauer CA, Berezenko S, Sleep D, Tooth D, Sadler PJ. Role of Tyr84 in controlling the reactivity of Cys34 of human albumin. FASEB J. 2005;272:353–362. doi: 10.1111/j.1742-4658.2004.04474.x. [DOI] [PubMed] [Google Scholar]
- Tornqvist M, Fred C, Haglund J, Helleberg H, Paulsson B, Rydberg P. Protein adducts: quantitative and qualitative aspects of their formation, analysis and applications. J. Chromatogr. B: Analyt. Technol. Biomed. Life Sci. 2002;778:279–308. doi: 10.1016/s1570-0232(02)00172-1. [DOI] [PubMed] [Google Scholar]
- Troester MA, Lindstrom AB, Waidyanatha S, Kupper LL, Rappaport SM. Stability of hemoglobin and albumin adducts of naphthalene oxide, 1,2-naph-thoquinone, and 1,4-naphthoquinone. Toxicol. Sci. 2002;68:314–321. doi: 10.1093/toxsci/68.2.314. [DOI] [PubMed] [Google Scholar]
- Turell L, Botti H, Carballal S, Ferrer-Sueta G, Souza JM, Duran R, Freeman BA, Radi R, Alvarez B. Reactivity of sulfenic acid in human serum albumin. Biochemistry. 2008;47:358–367. doi: 10.1021/bi701520y. [DOI] [PubMed] [Google Scholar]
- Wagner S, Scholz K, Donegan M, Burton L, Wingate J, Volkel W. Metabonomics and biomarker discovery: LC–MS metabolic profiling and constant neutral loss scanning combined with multivariate data analysis for mercapturic acid analysis. Anal. Chem. 2006;78:1296–1305. doi: 10.1021/ac051705s. [DOI] [PubMed] [Google Scholar]
- Wagner S, Scholz K, Sieber M, Kellert M, Voelkel W. Tools in metabonomics: an integrated validation approach for LC–MS metabolic profiling of mercapturic acids in human urine. Anal. Chem. 2007;79:2918–2926. doi: 10.1021/ac062153w. [DOI] [PubMed] [Google Scholar]
- Wild CP. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol. Biomarkers Prev. 2005;14:1847–1850. doi: 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- Willett WC. Balancing life-style and genomics research for disease prevention. Science. 2002;296:695–698. doi: 10.1126/science.1071055. [DOI] [PubMed] [Google Scholar]
- Yeowell-O’Connell K, Rothman N, Smith MT, Hayes RB, Li G, Waidyanatha S, Dosemeci M, Zhang L, Yin S, Titenko-Holland N, Rappaport SM. Hemoglobin and albumin adducts of benzene oxide among workers exposed to high levels of benzene. Carcinogenesis. 1998;19:1565–1571. doi: 10.1093/carcin/19.9.1565. [DOI] [PubMed] [Google Scholar]
- Zhan X, Desiderio DM. Nitroproteins from a human pituitary adenoma tissue discovered with a nitrotyrosine affinity column and tandem mass spectrometry. Anal. Biochem. 2006;354:279–289. doi: 10.1016/j.ab.2006.05.024. [DOI] [PubMed] [Google Scholar]
- Zhang J, Sprung R, Pei J, Tan X, Kim S, Zhu H, Liu CF, Grishin NV, Zhao Y. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol. Cell. Proteomics. 2009;8:215–225. doi: 10.1074/mcp.M800187-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]