SUMMARY
Dengue is the most common vector-borne viral disease, causing nearly 400 million infections yearly. Currently there are no approved therapies. Antibody epitopes that elicit weak humoral responses may not be accessible by conventional B cell panning methods. To demonstrate an alternative strategy to generating a therapeutic antibody, we employed a non-immunodominant, but functionally relevant, epitope in domain III of the E protein, and engineered by structure-guided methods an antibody directed to it. The resulting antibody, Ab513, exhibits high-affinity binding to, and broadly neutralizes, multiple genotypes within all four serotypes. To assess therapeutic relevance of Ab513, activity against important human clinical features of dengue was investigated. Ab513 mitigates thrombocytopenia in a humanized mouse model, resolves vascular leakage, reduces viremia to nearly undetectable levels, and protects mice in a maternal transfer model of lethal antibody-mediated enhancement. The results demonstrate that Ab513 may reduce the public health burden from dengue.
Graphical Abstract
In Brief
A structure-based approach allows for the development of a monoclonal antibody that targets a nonimmunodominant epitope to effectively neutralize all four serotypes of the dengue virus. This antibody treats several symptoms of severe infection in animal models and may provide strategies for treatment in humans.
INTRODUCTION
Dengue is the most important mosquito-borne viral disease affecting humans. Half of the world population lives in areas at risk for dengue, resulting in an estimated 390 million infections per year globally (Bhatt et al., 2013). Dengue is a self-limiting, systemic illness caused by any of four dengue virus serotypes, DENV-1 through DENV-4, which share only 60%–75% identity in amino acid sequence. Infection results in life-long protection to the infecting serotype but only transient protection to heterologous serotypes. Currently, there is no specific treatment available, and the leading vaccine candidate recently demonstrated limited efficacy, estimated to be between 30%–60%, with limited to no significant protection against DENV-2 (Capeding et al., 2014; Sabchareon et al., 2012; Villar et al., 2015).
Passive immunotherapy with monoclonal antibodies represents a potentially important approach to the treatment of dengue. Treatment with monoclonal antibodies has been shown to rapidly and substantially reduce viral titers in several instances, including influenza (Ramos et al., 2015) and HIV (Caskey et al., 2015). Therapeutically viable antibodies to infectious diseases must have a broad coverage of genetically diverse strains. Such antibodies are typically identified by large-scale panning exercises of B cells from infected individuals. These approaches are inherently biased by the native humoral immune response, and as such, may be limited in accessing epitopes that elicit no or little humoral response but may yet be functionally important target epitopes. Moreover, antibody therapy to immunodominant regions has the potential to cause immune interference, such as by masking important epitopes for eliciting a memory protective response (Siber et al., 1993; Siegrist et al., 1998; Zhang et al., 2007).
Utilizing panning of B cells derived from infected patients or challenged mice, a range of DENV-neutralizing antibodies have been identified, including those with reactivity to multiple serotypes (Beltramello et al., 2010; Brien et al., 2010; de Alwis et al., 2011; Lai et al., 2013; Smith et al., 2013). Studies characterizing the human humoral response to DENV infection have found that it is dominated by antibodies to prM and domain I and II (DI/II) of the envelope (E) glycoprotein (Beltramello et al., 2010; Dejnirattisai et al., 2010; Lai et al., 2008). More recent studies have indicated that antibodies which bind complex, quaternary E protein epitopes on the virus surface (de Alwis et al., 2012; Fibriansah et al., 2014; Teoh et al., 2012), notably the hinge region connecting EDI and EDII, appear to contribute the majority of the human humoral DENV neutralizing activity and may mediate long-term protection, albeit in a serotype-specific manner (de Alwis et al., 2012). In contrast, anti-EDIII antibodies have been shown to constitute a minor proportion of the overall human humoral response and also contribute little of the anti-DENV neutralizing activity (Dejnirattisai et al., 2010; Wahala et al., 2012; Wahala et al., 2009; Williams et al., 2012). Additionally, there have been recent reports of potent antibodies that bridge E monomers (EDE-directed antibodies) (Dejnirattisai et al., 2015).
As EDIII-specific antibodies have been shown to constitute a minor component of the overall human humoral response but have high potency, we investigated whether targeting EDIII might represent an important strategy for immunotherapy. However, existing EDIII-directed antibodies are not fully cross-reactive; while they typically exhibit high potency due in part to greater antibody accessibility, none have been shown to neutralize diverse genotypes among all four serotypes. We recently described the derivation of antibody 4E5A, which was engineered from 4E11, a mouse antibody directed to an accessible EDIII epitope, without the assistance of crystal structure information through a structural-physicochemical statistical approach (Tharakaraman et al., 2013).
Herein, we describe a structural framework developed to characterize the epitope-paratope interface on domain III, thereby enabling us to engineer an optimized antibody candidate, Ab513. Ab513 was extensively characterized in vitro and in vivo to investigate its potential to alter dengue pathogenesis. Ab513 represents an alternative, complementary approach to identification of broad-spectrum antibodies, and illustrates an effective strategy to target non-immunodominant but functionally relevant epitopes.
RESULTS
Structure-Guided Redesign of 4E5A
Recently, we reported the engineering of antibody 4E5A from 4E11 with improved binding, especially to DENV-4; however, affinity to DENV-4 was still modest (100 nM) compared to the other serotypes. Analysis of genetic variability in epitope regions confirms that the region targeted by 4E5A is far more conserved compared to the EDI/II hinge epitope region. Therefore, to develop a broad-spectrum potent antibody for dengue that targets a non-immunodominant epitope, we redesigned 4E5A for improved affinity to DENV-4, leveraging analysis of the epitope-paratope interface. Adapting from network (graph) theory, we developed a framework to compute the inter-residue atomic interactions between interacting amino acid pairs of an antigen-antibody interface. As such, we define the inter-residue interactions between a CDR residue and its neighboring epitope residues, rendered in a 2D graph format, as the epitope-paratope connectivity (EPC) network (Experimental Procedures). CDR mutations or positions (when mutated) that contribute to more favorable epitope contacts, as evaluated by EPC network analysis, were investigated experimentally. To wit, absent in the identification of 4E5A was interrogation of amino acids within CDR-H1 which, unlike CDR-H3, is positioned at the Ab-Ag interface periphery, thereby potentially allowing for subtle changes to the binding interface. Therefore, 4E5A CDR-H1 was inspected as a potential region to engineer, to test the accuracy of our epitope-paratope description and to develop an optimized immunotherapy candidate targeting a specific epitope.
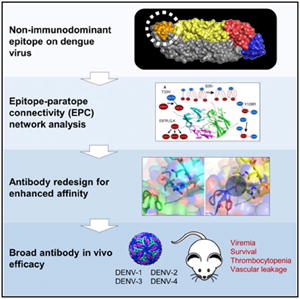
First, we hypothesized that subtle differences in the CDR backbone and/or residue side-chain conformation between the 4E11 model developed earlier (Tharakaraman et al., 2013) and the crystal structure, published recently (Cockburn et al., 2012), could have led us to underestimate the significance of certain CDR residues in our previous design calculations. While the placement of the majority of the backbone and side-chain atoms were highly similar (pairwise Cα RMSD of the six CDR loops, CDR-H1, -H2, -H3, -L1, -L2, -L3, varies from 0.176 to 0.407 Å), one notable exception was Thr33 of CDR-H1, which is observed to be in close contact with Lys310, Lys323, and Val364 of DENV-4 EDIII (Figure S1). In all the DENV-1 to -4 co-crystal structures (3UZE, 3UYP, 3UZQ, 3UZV), the CG2 carbon of Thr is proximal to the CG2 carbon of Val364 filling a void between VH and EDIII, whereas in the homology model, the CG2 carbon points away from Val364 disrupting this hydrophobic contact. In view of the different side-chain orientation of Thr33, we hypothesized that a Val in place of Thr at 33 would form a stronger hydrophobic contact with the Val at 364 of EDIII. This is evident from the EPC network analysis (Figure 1A). Furthermore, the fact that position 364 is hydrophobic in all DENV-1 to -4 suggests that a Thr33Val mutation would not be detrimental toward DENV-1 to 3 binding.
Figure 1. Epitope Paratope Connectivity Network of Putative Affinity-Enhancing Mutations and Structural Impact of Asn360Tyr Mutation (EDIII) on Antigen-Antibody Interaction.
(A) The EPC networks observed in 4E11 and mutant antibodies are shown for examples of putative affinity enhancing positions. Each of these EPCs networks is shown as a 2D graph: nodes represent amino acids (antibody: red; antigen: blue) and edges represent inter-residue non-covalent interactions (black: hydrophobic bonds; red: hydrogen bonds; yellow: cation pi; green: ionic). For each position of interest, the EPC network is given before and after mutation (with the arrow pointing from the WT to the modeled structure).
(B) Sequence alignment of EDIII domain of representative DENV-4 strains from genotypes I, IIA, and IIB. 4E5A epitope residues are highlighted in red. The column corresponding to the Asn360Tyr mutation, observed in the genotype I strain H241 (Philippines/1956), is highlighted by bold letters.
(C) Close-up view of CDR-H1 and the DE-loop (residues 358–365) as observed in the 4E11-EDIII (DENV-4) co-crystal structure (PDB: 3UYP): VH: green; VL: not shown; EDIII: cyan. Asn360 forms a water-mediated hydrogen bond (dotted arrows) with Gly27 of the CDR-H1 loop of 4E11.
(D) Alternate conformers of Tyr360 generated by modeling are rendered in stick format. The side chains of these conformers are colored according to potential steric clashes: yellow-favorable; red-unfavorable.
(E) Energetic calculations as carried out using Discovery Studio.
Structural analyses coupled with EPC network analysis identified sites 27–28 and 31–33 of heavy chain as being positioned to potentially mediate new or improved contacts. To interrogate these sites, including Thr33Val, site-saturation combinatorial libraries were generated (Supplemental Experimental Procedures), combined, transformed into yeast, and assessed for binding to DENV-4 EDIII by scFv surface display using flow cytometry. Relative to 4E5A, the combined library exhibited a population with greater binding, supporting predictions that these regions are hotspots for improving DENV-4 affinity. After three rounds of sorting in which the top binders to DENV-4 EDIII were selected, the FACS profile of the population showed significant enrichment of cells with enhanced DENV-4 EDIII binding. Sequencing of 50 clones from this enriched population revealed four unique variants (Table S1). Interestingly, for the enriched triple-site variants, all contained a Thr33Val substitution and no variation was observed at position 32, despite sequencing of the unsorted library demonstrating that variation at this position was intact.
We tested the mutations identified by the EPC network analysis in the context of full-length IgG and found the variants exhibited ~10- to 20-fold enhanced affinity to DENV-4 EDIII relative to 4E5A (Table 1, rows 1–4). In this process, we noted that the Lys31Ser variant introduces a putative N-glycosylation site at position 29, therefore, this variant was not further pursued. The single mutation Thr33Val demonstrated the best binding profile overall (Table 1, row 2), consistent with our EPC network analysis. Notably, all variants demonstrated enhanced or equivalent binding to DENV-1 to -3.
Table 1.
Binding of 4E5A and Variants to EDIII of DENV-1 to −4 Determined by Competition ELISA
| Variant (chain) | EDIII-DENV-1
|
EDIII-DENV-2
|
EDIII-DENV-3
|
EDIII-DENV-4
|
||||
|---|---|---|---|---|---|---|---|---|
| KD (nM) | Relative Affinity | KD (nM) | Relative Affinity | KD (nM) | Relative Affinity | KD (nM) | Relative Affinity | |
| 4E5A | 0.27 | 0.10 | 15.8 | 94.08 | ||||
|
| ||||||||
| T33V (HC) | <0.1 | >2.7 | <0.1 | >1.0 | 0.7 | 22.6 | 3.45 | 27.3 |
|
| ||||||||
| K31Q, T33V | <0.1 | >2.7 | <0.1 | >1.0 | 1.1 | 14.4 | 4.13 | 22.8 |
|
| ||||||||
| G27Y, F28W | <0.1 | >2.7 | <0.1 | >1.0 | 14.5 | 1.1 | 5.35 | 17.6 |
|
| ||||||||
| A25Δ (HC) | 1.27 | 0.21 | <0.3 | >0.33 | 19.9 | 0.79 | 297.00 | 0.32 |
|
| ||||||||
| S26Δ (HC) | <0.3 | >0.90 | <0.3 | >0.33 | 5.5 | 2.87 | 13.80 | 6.82 |
|
| ||||||||
| G27Δ (HC) | <0.3 | >0.90 | <0.3 | >0.33 | 9.6 | 1.65 | 25.76 | 3.65 |
|
| ||||||||
| G27P (HC) | 4.58 | 0.06 | <0.3 | >0.33 | 37.4 | 0.42 | 1936.00 | 0.05 |
|
| ||||||||
| G27A (HC) | <0.3 | >0.90 | <0.3 | >0.33 | 22.3 | 0.71 | 44.70 | 2.10 |
|
| ||||||||
| Y106R (HC) | <0.3 | >0.90 | <0.3 | >0.33 | 31.3 | 0.50 | 318.50 | 0.30 |
|
| ||||||||
| E97R (LC) | 0.36 | 0.75 | <0.3 | >0.33 | 31.6 | 0.50 | 120.55 | 0.78 |
|
| ||||||||
| E97K (LC) | 0.37 | 0.73 | <0.3 | >0.33 | 29.1 | 0.54 | 112.20 | 0.84 |
|
| ||||||||
| E97Q (LC) | 0.45 | 0.60 | <0.3 | >0.33 | 26.1 | 0.61 | 123.25 | 0.76 |
|
| ||||||||
| Humanized 4E5A | <0.1 | >2.70 | <0.1 | >1.0 | 5.8 | 2.72 | 55.08 | 1.71 |
|
| ||||||||
| Ab513 | <0.1 | >2.7 | <0.1 | >1.0 | 1.2 | 13.2 | 4.32 | 21.8 |
Next, we analyzed the conservation of the epitope residues across serotypes to identify potential DENV-4 “signature” residues, as these might contribute to the relatively weaker affinity of 4E5A to DENV-4. Critically, four out of the seven epitope residues unique to DENV-4 were localized to a region in the loop between the “D” and “E” β strands (residues 358–365), suggesting that differences in this region could be partly responsible for the lower affinity binding of 4E5A against DENV-4 strains. Closer examination of this region indicated that shortening the length of the proximal antibody CDR-H1 loop via residue deletion would result in the removal of a “elbow” region (25ASGF28) in the CDR loop, resulting in roughly an 8% increase in shape complementarity (Lawrence and Colman, 1993) between the interacting surfaces (estimated shape complementarity “Sc” of 4E11 and CDR deletion mutant are 0.65 and 0.71, respectively). In addition to aiding DENV-4 EDIII binding, our analysis also suggested that deletion of a residue in the region 25–28 of CDR-H1, notably Ala25, Ser26, or Gly27 of CDR-H1, would permit the antibody to more efficiently engage DENV-1 to -3 by virtue of electrostatic interactions between the positively charged surface on the antibody VH created by Arg99 of CDR-H3 and Lys3 of FR1 and the negatively charged residues 360–363 of EDIII (D360, E362 on DENV-1; E360 and D362 on DENV-2; E362 and E363 on DENV-3) (Figure 1A and Figure S1B). Additionally, a fourth mutation—Gly27Pro—was also predicted to have a similar effect as the deletion since introduction of a Pro residue might introduce a bend in the loop backbone.
Experimental testing of these variants indicated that two of the deletion mutants, Ser26Δ and Gly27Δ, demonstrated ~7-fold and 3.6-fold greater binding to EDIII of DENV-4 (strain BC287/97, having Asn at position 360), without being detrimental to DENV-1 and DENV-2 EDIII affinity (Table 1, rows 5–9). In agreement with the structural predictions, the two deletion mutants also improved affinity to DENV-3 albeit to a lesser extent. The Ser26Δ mutant improved affinity to DENV-4 strain H241 EDIII (containing Tyr at 360) to 240.9 nM, a 19-fold improvement when compared to the parent 4E5A antibody. Notably, other putative affinity-enhancing amino acid substitutions that were predicted from a standard structure-based rational design, including Glu97Arg/Lys and Tyr106Arg, were found to have marginal or no improvement based on EPC network analysis (Figure 1)—a result that was verified experimentally (Table 1, rows 10–13). The final engineered antibody, Ab513, differs from 4E11, the starting antibody, through introduction of six affinityenhancing point mutations and an affinity-enhancing deletion at position 26 (VH) and amino acid changes to humanize the candidate. Relative to 4E5A, Ab513 exhibits a 13- and 22-fold affinity improvement to DENV-3 and DENV-4, respectively, while showing smaller gains to DENV-1 and DENV-2 (Table 1).
The Structure of the Ab513-EDIII Complex
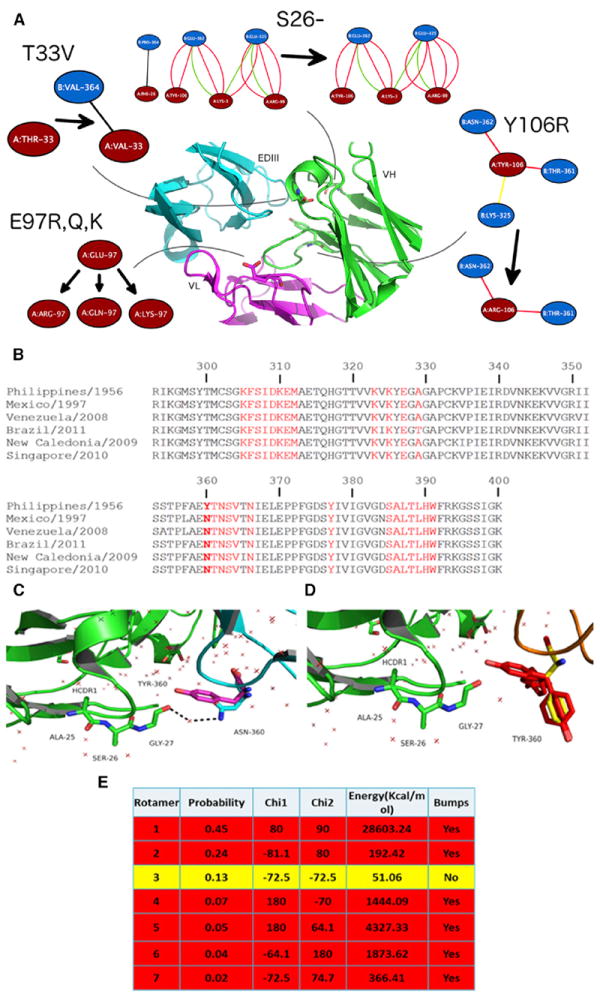
To verify the structure-based predictions of affinity-enhancing mutations and to cross-compare 4E11 against Ab513, we solved the crystal structure of Ab513 (reformatted as a scFv) bound to EDIII of DENV-4. Two crystal forms were obtained providing a total of eight independent views of the complex (Table S2). The crystal asymmetric unit of form I contains six scFv-EDIII complexes arranged as three dimers (Figure 2A). The scFvs in each dimer are related by a non-crystallographic dyad (Figure 2C) adopting a “swapped” configuration. Crystal form II (Figure 2B), however, comprises two independent monomeric mAb513scFv-DIII complexes (related by a non-crystallographic dyad). Notably, the temperature factors for the antigen in both crystal forms (temperature factors of DIII in form I and II are 80.5 and 120.4 Å2 respectively) exceed that of the scFv moiety (form I, II: 47.5, 70.4 Å2), indicating greater flexibility. Nevertheless, the eight complexes do not differ significantly from each other (average pairwise rmsd of 0.457 Å).
Figure 2. scFv Ab513-EDIII (DENV-4, BC287/97 (Mexico/1997)) Complex Structure.
(A–D) The asymmetric unit of crystal forms I (A) and II (B) contain six and two Ab513-EDIII complexes, respectively. The scFvs of form I are rendered in solvent accessible surface format and the EDIII domains are rendered in cartoon format. The interface formed by the heavy chains of the two ScFvs in the dimer is shown in C. (D) Comparison of the antibody-EDIII interface for Ab513 (left) and 4E11 (right) demonstrating that deletion of Ser26 results in higher surface complementarity due to removal of the “elbow” present in 4E11.
As expected, Ab513 recognizes the A-strand epitope on EDIII, with the heavy and light chain domains contacting the A and G β strands, respectively. The overall scFv-EDIII complex structure is similar to the 4E11-EDIII complex (PDB: 3UYP) with a root-mean-square deviation (rmsd) of 0.463 Å, indicating that the epitopes recognized by these two antibodies are nearly superimposable. Examination of the epitope-paratope interface reveals that the side chains of the six affinity-enhancing substitutions (Thr33Val and Ala55Glu of VH; Arg31Lys, Asn57Glu, Glu59Gln and Ser60Trp of VL) make the predicted contacts, with no significant deviations observed at any of the contact positions (Figure S2). Further, it is observed that the deletion of Ser26 results in higher surface complementarity with the antigen due to removal of the “elbow” present in 4E11, as predicted (Figure 2D). It should be noted that the deletion does not alter the canonical conformation (Chothia et al., 1989) of the H-CDR1 loop (Chothia type 1). A total of 898 Å2 of accessible surface area of Ab513 is buried in the Ab513-EDIII interface with the VH and VL making contact surface areas of 480 and 418 Å2, respectively. Twenty H-bonds and 13 salt bridge interactions are found across the Ab513-EDIII interface, whereas 16 H-bonds and 8 salt bridges are found across the 4E11-EDIII interface, indicating improved contacts as the principal reason of affinity enhancement.
Ab513 Neutralizes a Wide Range of DENVs
To assess the breadth of binding of Ab513, the antibody was tested against a panel of 21 EDIII proteins, which represent a set of diverse challenge strains selected, in part, for having diversity within the epitope region. Ab513 was able to bind all EDIII proteins and demonstrated affinity improvement relative to 4E5A by as much as 40-fold against DENV-3 and DENV-4 strains while marginally increasing the affinity against DENV-1 and DENV-2 strains (Table 2). Consistent with its strong binding, Ab513 demonstrated strong in vitro neutralization of DENV-1 to -4, with observed EC50 values of <200 ng/ml for all four serotypes (Table 2), a substantial enhancement compared to 4E11 (Figure S3).
Table 2.
Ab513 Breadth of Binding to Diverse Genotypes Determined by Competition ELISA and SPR, with In Vitro Neutralization of Select Strains
| Serotype | EDIII strain | ELISA KD (nM) | Fold increase from 4E5A | SPR KD (nM) | EC50 (ng/ml)a |
|---|---|---|---|---|---|
| I | Hawaii/1944 | <0.1 | NDb | 0.041 | 67 ± 12 |
| Vietnam/2008 | <0.1 | ND | 0.039 | ||
| Malaysia/2005 | <0.1 | ND | 0.064 | ||
| Mexico/2007 | <0.1 | ND | 0.071 | ||
| II | New Guinea/1944 (NGC) | <0.1 | ND | 0.015 | 190 ± 55 |
| Singapore/2008 | <0.1 | ND | 0.024 | ||
| Peru/1995 | <0.1 | ND | 0.012 | ||
| Vietnam/2007 | <0.1 | ND | 0.014 | ||
| Venezuela/2007 | <0.1 | ND | 0.029 | ||
| IIII | Philippines/1956 (H87) | 1.24 | 12.8 | 1.0 | 97 ± 20 |
| Singapore/2009 | 0.35 | 18.3 | 0.6 | ||
| Nicaragua/2010 | 3.36 | 9.5 | 4.9 | ||
| Puerto Rico/1977 | 1.36 | 9.3 | 1.4 | ||
| Cambodia/2008 | 1.32 | 10.1 | 2.5 | ||
| IV | Mexico/1997 (BC287/97) | 4.32 | 21.8 | 3.8 | 79 ± 30 |
| Singapore/2010 | 8.43 | 14.4 | 6.1 | ||
| New Caledonia/2009 | 10.81 | 15.9 | 8.9 | ||
| Philippines/1956 (H241) | 113.10 | 40.7 | 118.5 | 2,300 ± 1,500 | |
| Brazil/2011 | 14.68 | 18.2 | 7.9 | ||
| Venezuela/2008 | 4.50 | 11.9 | 3.0 | ||
| Thailand/1997 | 611.60 | 6.6 | >300c |
Average ± SEM, from three independent experiments.
ND, not determined.
Affinity estimate based on curve.
The compact yet dynamic surface structure of flaviviruses, including DENV, impacts epitope accessibility and thereby antibody neutralization activity (Lok et al., 2008; Sukupolvi-Petty et al., 2013). Therefore, extending beyond the binding studies, we performed a series of neutralization studies to characterize further the activity of Ab513. First, to validate the affinity gain observed by Ser26Δ mutation to DENV-4 strain H241 (containing a bulky Tyr at position 360) EDIII protein, as predicted by structural modeling analyses, we also tested Ab513 for in vitro neutralization against this strain. Compared to 4E5A, Ab513 exhibits a 4-fold improvement in neutralization potency to H241, with an EC50 of about 2 μg/ml. Next, to challenge Ab513 neutralization breadth, we performed a bioinformatic analysis of strains available from the World Reference Center for Emerging Viruses and Arboviruses to identify diverse isolates having sequence diversity within and near the Ab513 epitope region. This analysis resulted in the identification of 12 isolates, three from each serotype, which collectively represent a true challenge panel of viruses that are most likely to be refractory to Ab513 neutralization. Ab513 was able to fully neutralize all tested challenge viruses, with 9 of 12 viruses yielding EC50 values of <0.5 μg/ml, and the remaining three viruses neutralized at <4 μg/ml (Table S3).
To further assess and compare Ab513 with other DENV-neutralizing antibodies, we performed comparison studies of in vitro neutralization. We note that variations exist in methods of measuring in vitro neutralization of DENV, and studies have shown that even when using the same method, substantial titer/EC50 differences are often observed between laboratories with the same antibody samples (Rain-water-Lovett et al., 2012; Thomas et al., 2009). We therefore performed side-by-side direct comparisons, first against four of the most potent antibodies from the recently described EDE class of antibodies (Dejnirattisai et al., 2015). Results demonstrated that Ab513 exhibits similar or better potency than EDE mAbs of sub-class 1 and comparable activity to those of subclass 2, which are sensitive to glycosylation state of DENV (Table S4). We also directly compared Ab513 with a representative fusion loop-directed antibody (4G2) (Henchal et al., 1982), two DI/II hinge epitope-directed antibodies (14c10 and 1F4) (de Alwis et al., 2012; Teoh et al., 2012), and a potent human cross-reactive antibody directed to DIII (DV87.1) (Beltramello et al., 2010). Ab513 showed greater potency than the fusion-loop mAb, comparable activity to the potent DIII-directed mAb and one DI/II hinge-directed mAb and slightly lower potency than the other DI/II hinge-directed mAb (Figure S4). Collectively, these results demonstrate that Ab513 is able to efficiently neutralize a broad panel of challenge viruses which contain sequence diversity within the epitope region and which represent genotypic and geographical diversity of DENV. Additionally, Ab513 is able to neutralize virus more strongly than fusion loop-directed antibodies and with similar or better potency than the most potent EDE antibodies.
Ab513 Neutralizes DENV Despite Fc Receptor-Mediated Phagocytosis
Secondary infection with a heterologous DENV serotype has been associated with more severe illness; one mechanism that has been posited to explain this observation is antibody-mediated enhancement of virus uptake through the Fc receptor upon virus binding with either a non-neutralizing antibody, for example to prM, or binding of a neutralizing antibody at sub-neutralizing concentrations. Therefore, we investigated the ability of Ab513 to enhance virus uptake in the context of an ex vivo model.
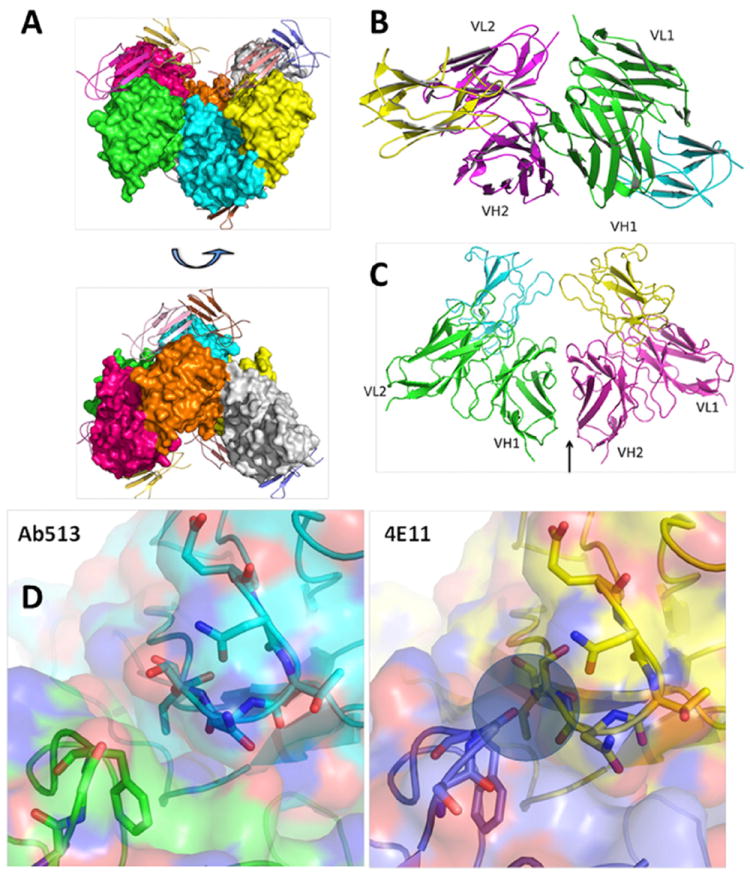
We compared the extent of enhanced virus replication with Ab513 and a chimeric version of the fusion loop-directed antibody, 4G2, in Fc-receptor bearing cells. Significantly lower levels of enhancement were observed with Ab513 as compared to 4G2 against all four DENV serotypes (Figure 3A). Since neutralization of the homologous DENV serotype can occur in the presence of Fc receptor-mediated phagocytosis, whereas heterologous DENV neutralizes by inhibiting uptake (Chan et al., 2011), we assessed the ability of Ab513 to neutralize the four DENV in the presence of cellular uptake. We examined the fate of fluores-cently labeled DENV with the highest dilution of antibodies that resulted in complete virus neutralization in THP-1 cells. Then, the localization of DENV-immune complexes was visualized by immunofluorescence. Ab513 neutralized all four DENV serotypes in the presence of uptake, with DENV-immune complexes trafficked to the LAMP-1 compartment (Figure 3B). This is in contrast to chimeric 4G2 (and 4E5A, data not shown) where DENV was neutralized by inhibition of initial virus uptake (Figure 3C). Collectively, these results demonstrate that Ab513 can neutralize all DENV serotypes in the presence of phagocytosis, which has been previously observed exclusively when convalescent serum samples were reacted with the homologous but not heterologous DENV serotypes (Chan et al., 2011; Wu et al., 2012).
Figure 3. Effects of Antibody on DENV Uptake in a Monocytic Cell Line.
(A) Enhanced virus infection in THP-1.2S cells with addition of either chimeric 4G2 or Ab513.
(B) Analysis of Ab513 and DENV localization in THP-1.2S cells. The late endosomal and lysosomal compartments of cells were stained by LAMP-1.
(C) Analysis identical to (B) except chimeric 4G2 is used as antibody.
Ab513 Demonstrates Activity in Multiple Mouse Models Capturing Key Clinical Features of Disease
Severe dengue infection is associated with increases in vascular permeability, which can lead to life-threatening hypovolemic shock. The increased permeability is often accompanied by thrombocytopenia. Currently, there are no specific therapies for treating dengue and management consists of supportive care only. Therefore, development of a therapeutic strategy that attenuates the duration and severity of symptoms and/or reduces the incidence of these major complications is of clinical importance (Simmons et al., 2012). To test the hypothesis of whether an immunotherapy can reduce clinical signs and symptoms of DENV infection, we deployed Ab513 in a set of animal models having multiple relevant endpoints: (1) viremia, (2) throm-bocytopenia, (3) vascular leak/permeability, and (4) antibody-enhanced disease.
To test the ability of Ab513 to reduce viremia, we administered Ab513 to 7- to 10-week-old AG129 mice infected with DENV-2. While the AG129 mouse model has potential limitations, we were particularly interested in whether we could prevent virus from migrating to the CNS. In this model, using DENV-2 strain NGC as the infective agent, administration of 25 mg/kg of an irrelevant isotype-matched antibody (“Ctl. mAb”) 24 hr prior to virus injection had no effect on CNS-related migration of virus, paralysis, and death. In contrast, a single prophylactic administration of Ab513 at 5 mg/kg resulted in survival of 6/10 animals (p < 0.0001) out to day 31 post-challenge, indicating elimination of most virus, little to no migration of DENV-2 to the CNS, and protection against CNS-related symptoms, such as paralysis (Figures 4A and 4B). This effect was even more pronounced at 25 mg/kg, where 9/10 animals survived. This increase in survival was also reflected in measurement of viremia levels at day 3 post-infection, the day of peak viremia in this model (Figure 4A). Administration of 5 mg/kg of Ab513 resulted in a 1.7 log10 reduction in viral titer; a 2.4 log10 mean reduction was observed in mice treated with 25 mg/kg Ab513, with three of the animals in this group having titers below the limit of detection. We note that the typical read-outs of this model are viremia at day 3 or following animals for 14–21 days, prior to evidence of CNS-related symptoms. However, as demonstrated here, a single dose of Ab513 is able to effectively neutralize the virus and prevent migration of virus to the CNS.
Figure 4. In Vivo Activity of Ab513 in AG129 and Humanized Mice.
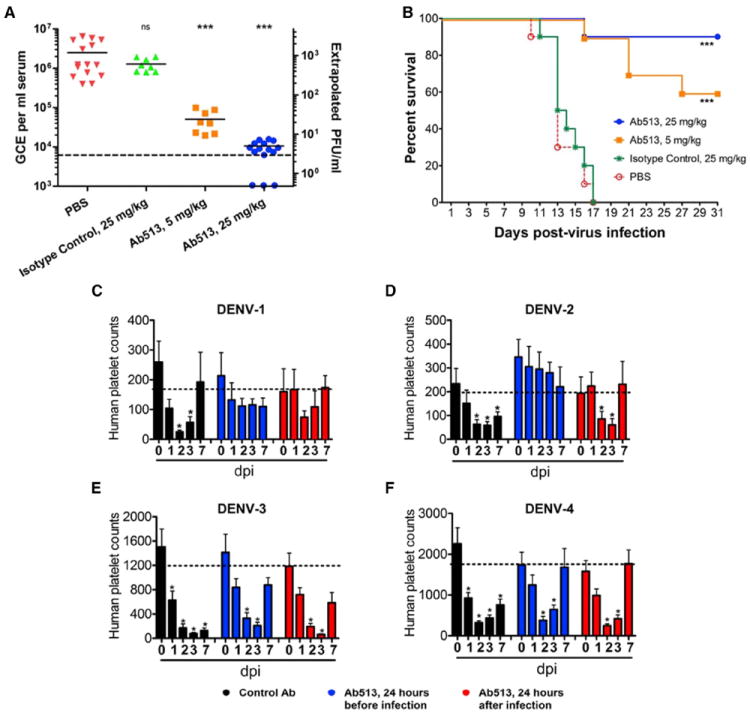
(A) AG129 mice (n = 10) were treated with Ab513, an irrelevant IgG1 mAb (“Isotype control”), or PBS prior to challenge with 106.4 CCID50 of DENV-2. Viremia from serum 3 days post-infection (dpi) was measured by qRT-PCR. Dotted line represents limit of detection. The three dots on the x axis represent values (samples) below the lower limit of detection.
(B) A separate cohort of animals were monitored for survival. ***p < 0.0001, as compared with PBS controls.
(C–F) Comparison of human platelet levels in uninfected and infected humanized mice (humice) without treatment or treated with an isotype control Ab (“Control Ab”) or Ab513. The dashed line indicates the average of human platelet counts in uninfected humice. Results are shown as the average counts of human platelets per microliter of blood at different days post-infection (n = 5–7). **p < 0.05. Log-rank (Mantel-Cox) statistical test was performed to assess for significance. Error bars denote SD.
Next, we directly tested the prophylactic and therapeutic potential of Ab513 in the context of platelet loss upon DENV infection. To this end, we adapted a recently reported humanized mice (humice) dengue model developed using DENV-2. Humice were reconstituted with human blood lineage cells, leading to production of a significant level of human platelets. We have shown previously that DENV infection in humice reproduces some key features of dengue in human, most notably thrombocytopenia (Sridharan et al., 2013). Critically, the reduction in platelet count occurs with human but not mouse platelets, thus allowing us to evaluate if Ab513 can specifically prevent human thrombocytopenia, in vivo. We applied the same infection approach with clinical isolates representing all four DENV serotypes for evaluation of Ab513, without any adaptation. Humice challenged with virus only or virus with isotype control antibody display a sharp reduction of human platelets after virus infection, with the nadir typically observed 2–3 days post challenge followed by a gradual recovery (Figures 4C–4F). Humice receiving a single administration of Ab513 (25 mg/kg) either 24 hr prior to or after virus challenge demonstrated a significantly accelerated recovery of human platelet levels (Figures 4C–4F), with a more dramatic impact on humice challenged with DENV-1 and DENV-2 (Figures 4C and 4D). In contrast, mouse platelet levels were not affected by virus infection or by Ab513 administration (Figure S5). Quantification of virus levels in sera (as determined by plaque assay) from humice challenged with DENV-1 and DENV-2 showed a significant reduction in viral load by administration of Ab513 prior to or post-infection. DENV-4 viremia could not be detected by plaque assay or qRT-PCR and DENV-3 viremia could only be detected by qRT-PCR but mostly near the limit of detection, precluding a robust analysis (Figure S5). Despite the current limitations of this model in the context of the DENV-3 and DENV-4 clinical isolates, our data indicate that treatment with Ab513 shortened the duration of human thrombocytopenia. Additionally, levels of IFN-γ and IL-10 were markedly reduced in DENV-2 challenged humice treated with Ab513 in comparison to those that received no Ab or the control serotype-specific mAb. A significant reduction in IFN-γ was also observed in DENV-4 challenged humice in response to administration of Ab513. These results demonstrate that a single dose of Ab513 administered before or after infection is able to effectively prevent thrombocytopenia or accelerate recovery of human platelets to normal levels in humanized mice across all four DENV serotypes. Additionally, in the more robust models of DENV-1 and -2, Ab513 causes a significant reduction in viremia in humanized mice, consistent with the data generated in the AG129 mouse.
In addition to thrombocytopenia, another crucial aspect to address in use of an immunotherapy for dengue treatment is whether it can mitigate vascular leakage. Therefore, we assessed the extent of vascular leakage in Ab513-treated DENV2-infected mice by Evans Blue assay on day 6 post-infection, the time point at which the isotype control mice were moribund and expected to display significant increased vascular permeability (Ng et al., 2014). Evans’ blue dye binds strongly to albumin present in the blood and the amount of the dye detected in perfused organs is proportional to the extent of vascular leakage. In the AG129 ADE model, significant vascular leakage can be detected in the liver, intestine, spleen, and kidney of infected mice born to immune mothers (Ng et al., 2014). In this model, a significant increase in vascular leakage was observed in the isotype antibody-treated mice compared to control mice as measured by elevated Evans Blue content in their livers, intestines, spleen, and kidneys (Figure 5A). Upon treatment with Ab513 on day 1 post-infection, the extent of vascular leakage detected for all the organs was significantly reduced (Figure 5A). Taken together, this data indicates that Ab513 treatment significantly limited the extent of vascular leak in key organs.
Figure 5. Ab513 Protects Animals from Enhanced Disease.
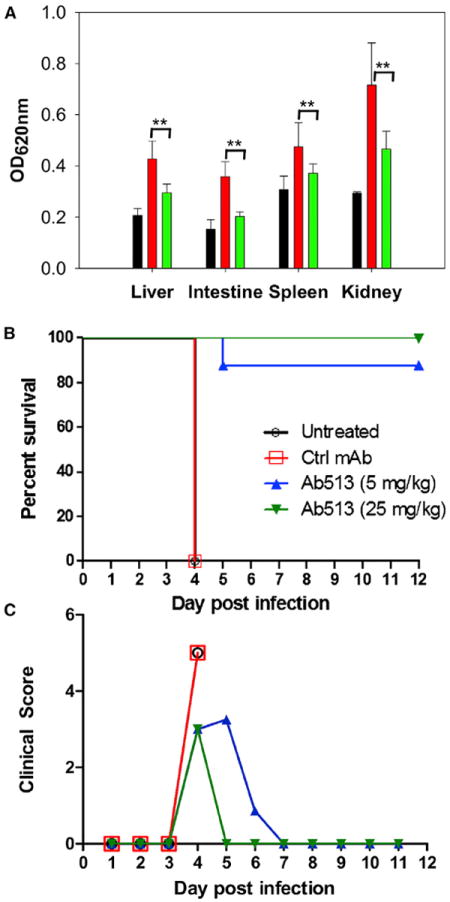
(A) Five- to six-week-old AG129 mice born to DENV-1 immune mothers were infected with DENV-2; included was an uninfected control (black). On day 1 post-infection, infected mice were either treated with 25 mg/kg of Ab513 (green) or isotype control antibody (red). On day 6 post-infection, the extent of vascular leakage in the mice were assessed by Evans blue assay. **p < 0.01 based on the Mann-Whitney test with reference to the isotype control.
(B and C) A group of A129 infected pups was followed daily. (B) Survival curves of four cohorts (n = 8): untreated, receiving a control antibody (Ctrl mAb), or 5 mg/kg or 25 mg/kg Ab513. Statistical inferences were made by pairwise comparisons for each treatment group to the control group, considering the survival of both. Ab513 at all doses had a significant effect on survival, log-rank (Mantel-Cox) test, p < 0.0001). (C). Assessment of mean clinical score of the three treatment cohorts.
Finally, for Ab513 to be useful therapeutically to treat dengue, this antibody must be able to compete with heterologous antibodies that could enhance DENV infection of Fc-receptor bearing cells. This is particularly relevant since severe dengue is more common in patients having secondary infection with a DENV serotype heterologous to initial infection. To test whether our antibody can be effective under such circumstances, we examined whether Ab513 could mitigate enhanced disease caused by heterotypic antibodies. Consistent with previous reports (Ng et al., 2014), a sublethal challenge with 106 PFU of DENV-2 of A129 pups from DENV naive mothers resulted in a transient infection with 100% survival. In contrast, 100% of pups from DENV-1 immune mothers reached moribund state on day 4 post-DENV-2 infection (Figure 5B), indicating that these animals underwent enhanced disease severity mediated by maternally acquired heterologous DENV antibodies. The protection efficacy of Ab513 in the presence of these heterologous enhancing antibodies was then determined by treating the infected pups from DENV-1 immune mother with 25 mg/kg or 5 mg/kg of Ab513 on day 1 post-infection. Ab513 treatment was able to efficiently prevent disease enhancement in these infected pups with 100% and 88% survival rate, respectively (Figure 5B). In sharp contrast, administration of an isotype Ab control resulted in 100% mortality on day 4 post-infection. Notably, the mice from the treatment groups displayed mild diarrhea but were still very active on day 4 post-infection. In contrast, infected animals administered an isotype control displayed ruffled fur, severe diarrhea, hunched backs and lethargy (Figure 5C). A dose response was observed for Ab513, with more rapid recovery time associated with the higher antibody dose.
DISCUSSION
In this study, we sought to address the question of whether an engineered anti-DENV antibody targeted to a non-immundominant—but functionally relevant—epitope could be used for immunotherapy. Importantly, development of such a strategy to treat dengue, unlike most other infectious agents such as influenza virus or HIV, faces a unique set of challenges arising from the fact that DENV antibodies potentially have the capacity to mediate protection or exacerbate disease. Additionally, recent discovery of antibodies that neutralize DENV-1 to -4 and bind epitopes that span two adjacent E monomers within a single dimer (EDE-directed antibodies) raises interesting and important questions with regards to the human immunological response (Dejnirattisai et al., 2015). While EDE antibodies exhibit potent pan-serotype neutralizing activity, they have features which may limit their potential as effective immunotherapies. First, they have long CDR-H3 loops (15–27 amino acids) with a high level of Tyr residues. Both these features have been linked to antibody polyreactivity (Mouquet and Nussenzweig, 2012; Wardemann et al., 2003). Additionally, activity of many EDE antibodies is dependent on glycosylation state of the virus, thereby limiting their breadth and providing potential viable escape mechanisms (Dejnirattisai et al., 2015).
Our structure-based EPC network analysis enabled the successful prediction of a CDR-proximal deletion to enhance epitope-paratope complementarity. Engineering of Ab513 validates the EPC network approach, enabling a structural understanding of antibody diversification with regards to antigen engagement, thereby providing a complementary tool to existing genetic approaches that aim to trace the development of broadly neutralizing antibodies from germline (Lingwood et al., 2012; Pappas et al., 2014).
Ab513 exhibits broad binding and neutralization, regardless of virus genotype, neutralizes all four DENV serotypes even in the presence of FcR-mediated uptake, and demonstrates in vivo efficacy against all four DENV serotypes. Taken together, these data demonstrate that an immunotherapy has the potential to effectively control viremia and disease in humans.
EXPERIMENTAL PROCEDURES
Materials
Cell culture and virus propagation were carried out as previously described (Tharakaraman et al., 2013). Briefly, viruses were procured from ATCC or BEI Resources and propagated in C6/36 or Vero cells using an MOI of approximately 0.1, and harvested after 4–7 days, depending on the strain. Aliquots were stored at −80°C. For breadth of neutralization studies, the 12 viruses were obtained from the UTMB World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) repository.
Computation of Epitope-Paratope Connectivity Network of Dengue EDIII-Antibody Complexes
The X-ray co-crystal structures of 4E11 in complex with the EDIII antigen (PDB: 3UYP [DENV4], 3UZE [DENV3], 3UZV [DENV2], 3UZQ [DENV1]) were used to determine the various inter-residue inter-atomic contacts across the antigen-antibody interface, including putative hydrogen bonds (including water-bridged ones), disulfide bonds, pi-bonds, polar interactions, salt bridges, and Van der Waals interactions (non-hydrogen) as described previously (Soundararajan et al., 2011). The interactions between a CDR residue and its neighboring epitope residues are represented by a 2D network graph as a visual aid, where nodes represent amino acids and the edges represent inter-residue non-covalent interactions (black: hydrophobic bonds; red: hydrogen bonds; yellow: cation pi; green: ionic).
Structure Determination, Refinement, and Analysis
Data were collected at a wavelength of 1.00 Å at the Swiss Light Source beam-line PXIII using a Pilatus 6M detector (Dectris, Baden, Switzerland). Indexing, integration, and merging of the intensities were carried out with program XDS (Kabsch, 2010) and scaling was performed using program SCALA (Evans, 2006). Data collection statistics are summarized in Table S2. The structure of form I crystals was determined by molecular replacement using scFvE11 and DIII DENV4 as individual search probes (Cockburn et al., 2012). The molecular model was rebuilt using COOT and refined with REFMAC (Winn et al., 2011). Subsequently, a second crystal form (form II) diffracting to higher resolution (2.49 Å) was obtained in the monoclinic space group P21 with two scFv513-DIII complexes per asymmetric unit. The data were collected by exposing two different regions of the prismatic crystals and processed using the same packages. The structure was solved by molecular replacement using the refined coordinates obtained from crystal form I and the structure was refined using REFMAC. Figures were prepared in PyMOL (http://pymol.sourceforge.net) and the structure was validated using the Molprobity web server (http://molprobity.biochem.duke.edu). Structural superimpositions, buried surface areas and inter-molecular contacts were calculated using programs LSQKAB, AREAIMOL, SC, and NCONT from the CCP4 package (Winn et al., 2011).
Assessment and Visualization of DENV Fate in the Presence of Antibody
THP-1.2S was subcloned from THP-1 by limiting dilution (Chan et al., 2014). DENV-1 (06K2402DK1), DENV-2 (ST), DENV-3 (05K863DK1), and DENV-4 (06K2270DK1) are clinical isolates. For visualization of DENV immune complexes, DiD-labeled DENV was incubated with media, antibodies, or serum for 1 hr at 37°C before adding to cells at MOI 10. Uptake was assessed as described previously (Chan et al., 2011).
Administration of Ab513 in a Humanized Mouse Model
All experiments were performed in compliance with the guidelines of the institutional committees at the National University of Singapore and Massachusetts Institute of Technology. Humanized mice were generated as previously described (Sridharan et al., 2013). Experiments were initiated 1 week before infection with the i.v. administration of 300 mg/kg human immune globulins (IVIG, GAMMAGARD [BAXTER]) twice a week to ensure normal levels of circulating IgG. Humice were infected by tail vein injection of 1 × 107 PFU of virus (DENV2 strain 05K3295) in 200 μl of RPMI 1640 medium. IVIG administration continued during the infection period. Uninfected humice reconstituted with the same batch of fetal liver cells were injected with 200 μl of RPMI medium. Humice were prophylactically (24 hr before DENV infection) or therapeutically (24 hr after infection) administered with 25 mg/kg of Ab513 or an isotype control antibody (control Ab) intravenously (i.v). Plaque assay and platelet counts were performed as described (Sridharan et al., 2013). Results are shown as means ± SEM except for viremia which presented as a median. Differences were compared by using ANOVA followed by Student-Newman-Keuls post hoc analysis. Results with a p < 0.05 were considered significant. All calculations to examine differences between cohorts were completed using GraphPad Prism v5.0 (GraphPad Software).
Maternal Transfer Model and Measurement of Vascular Leakage
Five- to six-week-old AG129 pups that were born to DENV1-immune mothers were infected via the subcutaneous route with 103 PFU of DENV2 (D2Y98P-PP1) diluted in 0.1 ml of sterile PBS. On day 1 post-infection, mice received the respective treatments, either control antibody or Ab513, via the intravenous (i.v.) route. The mice were then monitored twice daily for their survival and clinical score for a period of 27 days (0: healthy; 1: ruffled fur; 2: hunched back; 3: severe diarrhea; 4: lethargic; 5: moribund). In addition, on day 6 post-infection (the time point at which mice from the isotype antibody-treated group became moribund), Evans blue assay was performed on 5 mice per group (infected control, isotype control, 5 mg/kg and 25 mg/kg group) as previously described (Ng et al., 2014). An uninfected control group, comprising age-matched AG129 mice, was included to obtain the baseline absorbance readings.
Supplementary Material
Highlights.
Structure-guided affinity enhancement of a cross-reactive dengue antibody
mAb neutralizes all four serotypes with a low level of viral-enhancing activity
Antibody demonstrates in vivo ability to resolve symptoms of severe dengue infection
Crystal structure of antibody-antigen validates the predicted designs
Acknowledgments
This work was funded in part by NIH Award (1R01AI111395 to R.S.), NIEHS training grant (2T32ES007020 to L.N.R.), NIH Merit Award (R37 GM057073-13 to R.S.), and National Research Foundation supported Interdisciplinary Research group in Infectious Diseases of SMART (Singapore MIT alliance for Research and Technology). The authors would like to acknowledge the work of Wilton DePina and Andrew Wollacott in designing and testing of E proteins and dengue strains. L.N.R. is an employee and shareholder of Visterra, Inc.; K.J.R. is an employee and shareholder of Visterra, Inc.; T.K. is an employee and shareholder of Visterra, Inc.; K.V. is an employee and shareholder of Visterra, Inc.; H.T. is an employee and shareholder of Visterra, Inc.; B.R. is an employee and shareholder of Visterra, Inc.; J.R.M. is an employee and shareholder of Visterra, Inc.; G.J.B.; Z.S. is an employee and shareholder of Visterra, Inc.; and R.S. is a founder, shareholder, and member of the board of Visterra, Inc.
Footnotes
AUTHOR CONTRIBUTIONS
L.N.R., K.T., V.S., J.R.M., G.J.B., Z.S. and R.S. conceptualized and designed the study. K.T., V.S., and R.S. designed the EPCN approach. D.C. performed structural modeling and sequence analysis. L.N.R., J.R.M., G.J.B., K.V., Z.S. and R.S. designed the biochemical, in vitro, and in vivo experiments. L.N.R., K.J.R., T.K., R.K., and H.T. performed the biochemical and in vitro experiments. K.R.C., H.C.T., and E.E.O. performed monocyte-based in vitro studies. V.V.C., J.C. and E.E.O. performed the humice studies. L.C.O and S.A. carried out the A129 and AG129 mouse experiments. Y.H.W., C.W.L., B.R., and J.L. determined the co-crystal structure of Ab513 and Domain III and completed structural analysis. L.N.R., E.E.O., K.T., Z.S., V.S. and R.S. wrote the manuscript.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, five figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2015.06.057.
ACCESSION NUMBERS
The accession numbers for the atomic coordinates and structure factors for ScFv Ab513-EDIII (DENV-4) co-crystal structures reported in this paper are Protein Data Bank: 4UDZ (form I) and 4UD3 (form II).
References
- Beltramello M, Williams KL, Simmons CP, Macagno A, Simonelli L, Quyen NT, Sukupolvi-Petty S, Navarro-Sanchez E, Young PR, de Silva AM, et al. The human immune response to Dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe. 2010;8:271–283. doi: 10.1016/j.chom.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, et al. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brien JD, Austin SK, Sukupolvi-Petty S, O’Brien KM, Johnson S, Fremont DH, Diamond MS. Genotype-specific neutralization and protection by antibodies against dengue virus type 3. J Virol. 2010;84:10630–10643. doi: 10.1128/JVI.01190-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capeding MR, Tran NH, Hadinegoro SR, Ismail HI, Chotpitayasunondh T, Chua MN, Luong CQ, Rusmil K, Wirawan DN, Nallusamy R, et al. CYD14 Study Group. Clinical efficacy and safety of a novel tetravalent dengue vaccine in healthy children in Asia: a phase 3, randomised, observer-masked, placebo-controlled trial. Lancet. 2014;384:1358–1365. doi: 10.1016/S0140-6736(14)61060-6. [DOI] [PubMed] [Google Scholar]
- Caskey M, Klein F, Lorenzi JC, Seaman MS, West AP, Jr, Buckley N, Kremer G, Nogueira L, Braunschweig M, Scheid JF, et al. Virae-mia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature. 2015;522:487–491. doi: 10.1038/nature14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KR, Zhang SL, Tan HC, Chan YK, Chow A, Lim AP, Vasudevan SG, Hanson BJ, Ooi EE. Ligation of Fc gamma receptor IIB inhibits antibody-dependent enhancement of dengue virus infection. Proc Natl Acad Sci USA. 2011;108:12479–12484. doi: 10.1073/pnas.1106568108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KR, Ong EZ, Tan HC, Zhang SL, Zhang Q, Tang KF, Kaliaperumal N, Lim AP, Hibberd ML, Chan SH, et al. Leukocyte immunoglobulin-like receptor B1 is critical for antibody-dependent dengue. Proc Natl Acad Sci USA. 2014;111:2722–2727. doi: 10.1073/pnas.1317454111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C, Lesk AM, Tramontano A, Levitt M, Smith-Gill SJ, Air G, Sheriff S, Padlan EA, Davies D, Tulip WR, et al. Conformations of immunoglobulin hypervariable regions. Nature. 1989;342:877–883. doi: 10.1038/342877a0. [DOI] [PubMed] [Google Scholar]
- Cockburn JJ, Navarro Sanchez ME, Fretes N, Urvoas A, Staropoli I, Kikuti CM, Coffey LL, Arenzana Seisdedos F, Bedouelle H, Rey FA. Mechanism of dengue virus broad cross-neutralization by a monoclonal antibody. Structure. 2012;20:303–314. doi: 10.1016/j.str.2012.01.001. [DOI] [PubMed] [Google Scholar]
- de Alwis R, Beltramello M, Messer WB, Sukupolvi-Petty S, Wahala WM, Kraus A, Olivarez NP, Pham Q, Brien JD, Tsai WY, et al. In-depth analysis of the antibody response of individuals exposed to primary dengue virus infection. PLoS Negl Trop Dis. 2011;5:e1188. doi: 10.1371/journal.pntd.0001188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alwis R, Smith SA, Olivarez NP, Messer WB, Huynh JP, Wahala WM, White LJ, Diamond MS, Baric RS, Crowe JE, Jr, de Silva AM. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc Natl Acad Sci USA. 2012;109:7439–7444. doi: 10.1073/pnas.1200566109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W, Jumnainsong A, Onsirisakul N, Fitton P, Vasanawathana S, Limpitikul W, Puttikhunt C, Edwards C, Duangchinda T, Supasa S, et al. Cross-reacting antibodies enhance dengue virus infection in humans. Science. 2010;328:745–748. doi: 10.1126/science.1185181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W, Wongwiwat W, Supasa S, Zhang X, Dai X, Rouvinski A, Jumnainsong A, Edwards C, Quyen NT, Duangchinda T, et al. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat Immunol. 2015;16:170–177. doi: 10.1038/ni.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Fibriansah G, Tan JL, Smith SA, de Alwis AR, Ng TS, Kostyuchenko VA, Ibarra KD, Wang J, Harris E, de Silva A, et al. A potent anti-dengue human antibody preferentially recognizes the conformation of E protein monomers assembled on the virus surface. EMBO Mol Med. 2014;6:358–371. doi: 10.1002/emmm.201303404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henchal EA, Gentry MK, McCown JM, Brandt WE. Dengue virus-specific and flavivirus group determinants identified with monoclonal antibodies by indirect immunofluorescence. Am J Trop Med Hyg. 1982;31:830–836. doi: 10.4269/ajtmh.1982.31.830. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CY, Tsai WY, Lin SR, Kao CL, Hu HP, King CC, Wu HC, Chang GJ, Wang WK. Antibodies to envelope glycoprotein of dengue virus during the natural course of infection are predominantly cross-reactive and recognize epitopes containing highly conserved residues at the fusion loop of domain II. J Virol. 2008;82:6631–6643. doi: 10.1128/JVI.00316-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CY, Williams KL, Wu YC, Knight S, Balmaseda A, Harris E, Wang WK. Analysis of cross-reactive antibodies recognizing the fusion loop of envelope protein and correlation with neutralizing antibody titers in Nicaraguan dengue cases. PLoS Negl Trop Dis. 2013;7:e2451. doi: 10.1371/journal.pntd.0002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- Lingwood D, McTamney PM, Yassine HM, Whittle JR, Guo X, Boyington JC, Wei CJ, Nabel GJ. Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature. 2012;489:566–570. doi: 10.1038/nature11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok SM, Kostyuchenko V, Nybakken GE, Holdaway HA, Battisti AJ, Sukupolvi-Petty S, Sedlak D, Fremont DH, Chipman PR, Roehrig JT, et al. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat Struct Mol Biol. 2008;15:312–317. doi: 10.1038/nsmb.1382. [DOI] [PubMed] [Google Scholar]
- Mouquet H, Nussenzweig MC. Polyreactive antibodies in adaptive immune responses to viruses. Cell Mol Life Sci. 2012;69:1435–1445. doi: 10.1007/s00018-011-0872-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng JK, Zhang SL, Tan HC, Yan B, Martinez JM, Tan WY, Lam JH, Tan GK, Ooi EE, Alonso S. First experimental in vivo model of enhanced dengue disease severity through maternally acquired heterotypic dengue antibodies. PLoS Pathog. 2014;10:e1004031. doi: 10.1371/journal.ppat.1004031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas L, Foglierini M, Piccoli L, Kallewaard NL, Turrini F, Silacci C, Fernandez-Rodriguez B, Agatic G, Giacchetto-Sasselli I, Pellicciotta G, et al. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature. 2014;516:418–422. doi: 10.1038/nature13764. [DOI] [PubMed] [Google Scholar]
- Rainwater-Lovett K, Rodriguez-Barraquer I, Cummings DA, Lessler J. Variation in dengue virus plaque reduction neutralization testing: systematic review and pooled analysis. BMC Infect Dis. 2012;12:233. doi: 10.1186/1471-2334-12-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos EL, Mitcham JL, Koller TD, Bonavia A, Usner DW, Balaratnam G, Fredlund P, Swiderek KM. Efficacy and safety of treatment with an anti-m2e monoclonal antibody in experimental human influenza. J Infect Dis. 2015;211:1038–1044. doi: 10.1093/infdis/jiu539. [DOI] [PubMed] [Google Scholar]
- Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, Suvannadabba S, Jiwariyavej V, Dulyachai W, Pengsaa K, Wartel TA, et al. Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. Lancet. 2012;380:1559–1567. doi: 10.1016/S0140-6736(12)61428-7. [DOI] [PubMed] [Google Scholar]
- Siber GR, Werner BG, Halsey NA, Reid R, Almeido-Hill J, Garrett SC, Thompson C, Santosham M. Interference of immune globulin with measles and rubella immunization. J Pediatr. 1993;122:204–211. doi: 10.1016/s0022-3476(06)80114-9. [DOI] [PubMed] [Google Scholar]
- Siegrist CA, Córdova M, Brandt C, Barrios C, Berney M, Tougne C, Kovarik J, Lambert PH. Determinants of infant responses to vaccines in presence of maternal antibodies. Vaccine. 1998;16:1409–1414. doi: 10.1016/s0264-410x(98)00100-5. [DOI] [PubMed] [Google Scholar]
- Simmons CP, Wolbers M, Nguyen MN, Whitehorn J, Shi PY, Young P, Petric R, Nguyen VV, Farrar J, Wills B. Therapeutics for dengue: recommendations for design and conduct of early-phase clinical trials. PLoS Negl Trop Dis. 2012;6:e1752. doi: 10.1371/journal.pntd.0001752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, de Alwis AR, Kose N, Harris E, Ibarra KD, Kahle KM, Pfaff JM, Xiang X, Doranz BJ, de Silva AM, et al. The potent and broadly neutralizing human dengue virus-specific monoclonal antibody 1C19 reveals a unique cross-reactive epitope on the bc loop of domain II of the envelope protein. MBio. 2013;4:e00873, e13. doi: 10.1128/mBio.00873-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundararajan V, Zheng S, Patel N, Warnock K, Raman R, Wilson IA, Raguram S, Sasisekharan V, Sasisekharan R. Networks link antigenic and receptor-binding sites of influenza hemagglutinin: mechanistic insight into fitter strain propagation. Sci Rep. 2011;1:200. doi: 10.1038/srep00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan A, Chen Q, Tang KF, Ooi EE, Hibberd ML, Chen J. Inhibition of megakaryocyte development in the bone marrow underlies dengue virus-induced thrombocytopenia in humanized mice. J Virol. 2013;87:11648–11658. doi: 10.1128/JVI.01156-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukupolvi-Petty S, Brien JD, Austin SK, Shrestha B, Swayne S, Kahle K, Doranz BJ, Johnson S, Pierson TC, Fremont DH, Diamond MS. Functional analysis of antibodies against dengue virus type 4 reveals strain-dependent epitope exposure that impacts neutralization and protection. J Virol. 2013;87:8826–8842. doi: 10.1128/JVI.01314-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoh EP, Kukkaro P, Teo EW, Lim AP, Tan TT, Yip A, Schul W, Aung M, Kostyuchenko VA, Leo YS, et al. The structural basis for serotype-specific neutralization of dengue virus by a human antibody. Sci Transl Med. 2012;4:139ra183. doi: 10.1126/scitranslmed.3003888. [DOI] [PubMed] [Google Scholar]
- Tharakaraman K, Robinson LN, Hatas A, Chen YL, Siyue L, Raguram S, Sasisekharan V, Wogan GN, Sasisekharan R. Redesign of a cross-reactive antibody to dengue virus with broad-spectrum activity and increased in vivo potency. Proc Natl Acad Sci USA. 2013;110:E1555–E1564. doi: 10.1073/pnas.1303645110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SJ, Nisalak A, Anderson KB, Libraty DH, Kalayanarooj S, Vaughn DW, Putnak R, Gibbons RV, Jarman R, Endy TP. Dengue plaque reduction neutralization test (PRNT) in primary and secondary dengue virus infections: How alterations in assay conditions impact performance. Am J Trop Med Hyg. 2009;81:825–833. doi: 10.4269/ajtmh.2009.08-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar L, Dayan GH, Arredondo-García JL, Rivera DM, Cunha R, Deseda C, Reynales H, Costa MS, Morales-Ramírez JO, Carrasquilla G, et al. CYD15 Study Group. Efficacy of a tetravalent dengue vaccine in children in Latin America. N Engl J Med. 2015;372:113–123. doi: 10.1056/NEJMoa1411037. [DOI] [PubMed] [Google Scholar]
- Wahala WM, Kraus AA, Haymore LB, Accavitti-Loper MA, de Silva AM. Dengue virus neutralization by human immune sera: role of envelope protein domain III-reactive antibody. Virology. 2009;392:103–113. doi: 10.1016/j.virol.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahala WM, Huang C, Butrapet S, White LJ, de Silva AM. Recombinant dengue type 2 viruses with altered e protein domain III epitopes are efficiently neutralized by human immune sera. J Virol. 2012;86:4019–4023. doi: 10.1128/JVI.06871-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- Williams KL, Wahala WM, Orozco S, de Silva AM, Harris E. Antibodies targeting dengue virus envelope domain III are not required for serotype-specific protection or prevention of enhancement in vivo. Virology. 2012;429:12–20. doi: 10.1016/j.virol.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RS, Chan KR, Tan HC, Chow A, Allen JC, Jr, Ooi EE. Neutralization of dengue virus in the presence of Fc receptor-mediated phagocytosis distinguishes serotype-specific from cross-neutralizing antibodies. Antiviral Res. 2012;96:340–343. doi: 10.1016/j.antiviral.2012.09.018. [DOI] [PubMed] [Google Scholar]
- Zhang P, Wu CG, Mihalik K, Virata-Theimer ML, Yu MY, Alter HJ, Feinstone SM. Hepatitis C virus epitope-specific neutralizing antibodies in Igs prepared from human plasma. Proc Natl Acad Sci USA. 2007;104:8449–8454. doi: 10.1073/pnas.0703039104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


