Abstract
The PE and PPE protein family are unique to mycobacteria. Though the complete genome sequences for over 500 M. tuberculosis strains and mycobacterial species are available, few PE and PPE proteins have been structurally and functionally characterized. We have therefore used bioinformatics tools to characterize the structure and function of these proteins. We selected representative members of the PE and PPE protein family by phylogeny analysis and using structure-based sequence annotation identified ten well-characterized protein domains of known function. Some of these domains were observed to be common to all mycobacterial species and some were species specific.
Introduction
Tuberculosis (TB) caused by Mycobacterium tuberculosis (Mtb), remains a major global health problem and one of the main causes of death around the world [1]. About one third of the world’s population has latent TB infection. TB kills about two million people annually and is the second leading cause of death from an infectious disease worldwide, after the human immunodeficiency virus (HIV) [2,3]. As a result of reduced immunity in HIV patients, there is a greater risk of infection with TB [4,5] and significant increase in number of deaths. Despite the availability of effective short course chemotherapy, Directly Observed Treatment Short (DOTS) and the Mycobacterium bovis Bacille de Calmette et Guérin (BCG) vaccine, the tubercle bacillus continues to be naturally resistant to many antibiotics, making the treatment difficult [6,7]. Patients develop drug resistance resulting in the resurgence of multiple drug resistance (MDR) and extreme drug resistant (XDR) TB [8].
The complete nucleotide sequence of Mtb H37Rv strain comprising ~4,000 genes, contains two new gene families; PE and PPE, accounting for ~10% of the total genome [9]. These proteins are characterized by highly conserved N-terminal domains with approximately 110 and 180 amino acid residues, respectively. The names PE and PPE for these proteins are due to the presence of amino acid sequence motifs Pro-Glu and Pro-Pro-Glu, respectively towards the N-terminus. These proteins are proposed to be a source of antigenic variation and responsible for virulence of pathogen. Subsequently, the complete sequencing of several strains of mycobacterial genomes [10,11,12,13,14,15] identified the presence of variable numbers of PE and PPE genes. Single nucleotide polymorphisms were observed to be greater in these genes compared to the non-PE and non-PPE genes. These genes were proposed as possible vaccine candidates [9,16,17]. The PE and PPE genes were mostly arranged in a unique regulon with the PE genes located upstream to the PPE genes and scattered throughout the genome [18]. The PE and PPE genes are known to be present in pathogenic and non-pathogenic mycobacteria but have not yet been identified in non-mycobacterial species [19,20,21]. A strong evolutionary selection for PE and PPE proteins in the pathogenic mycobacteria has been reported since their expansion is linked to the ESAT-6 gene clusters that has role in immuno-pathogenesis [22].
The functions of only few PE and PPE proteins are so far known. The PE and PPE proteins are highly polymorphic and localized to the cell wall and have immunological role. Comparative analysis of the PE and PPE families in Mtb H37Rv (virulent) and H37Ra (avirulent strain) revealed genetic differences in several single nucleotide variations, insertions and deletions [23]. Comparative genomics of the M. avium complex members revealed several polymorphisms in PE and PPE family members and the presence of some unique members in PPE family that have been implicated for applications in diagnostics [24].
Among functional characterization of some of the PE and PPE proteins, it has been shown that the N-terminal PE domain of PE_PGRS33 (Rv1809) is necessary for protein localization to the cell wall in M. marinum and M. tuberculosis [25,26]. In LipY and PE_PGRS30 (Rv1651c), the C-terminus encodes the lipase activity of the protein. Both PE and PPE domains contain a signal required for secretion of LipY by the ESX-5 system and these domains are proteolytically removed upon translocation [27,28]. Studies on the enzymatic role of the PE domain in LipY revealed that PE domain down-regulates the enzyme activity of LipY, but does not effect its thermal stability [29]. Studies on the antigenic properties of operonic PE25 (Rv2431), PPE41 (Rv2430) and the complex PE25/PPE41 indicated that the PPE41 and the PE25/PPE41 complex induced significant B cell response compared to the PE25 protein [18,30]. The up-regulation of PPE32 (Rv1808) in many conditions defines its role in the host innate immune response [31,32]. The PE_PGRS63 (Rv3097c) gene is highly expressed 24 hours post-infection in murine macrophage cell lines [33] and higher expression of PE/PPE genes Rv0977, Rv1361c and Rv1840c in human macrophages upon infection have been reported [34]. Functional studies of PE and PPE family members in M. tuberculosis have reported their localization in cell wall, cytosol and membrane, and their functions have been implicated in cell wall, virulence, detoxification, adaptation, insertion sequences, lipid metabolism, intermediary metabolism, respiration and cell processes [35].
Despite the availability of the complete Mtb genome sequence very few PE, PPE proteins have been structurally and functionally characterized. Computational methods may precede the selection of proteins for wet-lab experimental validation of their structure and activity. Structure-based functional annotation of proteins has been more useful than sequence-based comparisons alone [36,37], as the protein fold and conservation of active site residues are determinants of molecular function. Therefore, in the absence of experimental structures, computer-based protein modeling methods may be employed to predict the structure and possible function [37,38,39].
Large proteins often contain domains (comprising usually >50 amino acid residues) that are known to be independent folding units, irrespective of their location along the protein sequence [40]. Our previous studies on the sequence analysis of PE and PPE proteins from Mtb H37Rv strain identified a 225 amino acid residue conserved domain. This PE-PPE domain (Pfam ID: PF08237) was observed in the C-terminus of some PE and PPE proteins [41]. Bioinformatics analyses identified a serine α/β hydrolase fold with a pentapeptide sequence motif GxSxG/S for this domain and conserved Ser, Asp and His catalytic triad residues characteristic of lipase, esterase and cutinase activities [42]. Subsequent experiments confirmed that the PE-PPE domain of PE16 (Rv1430) exhibited esterase activity [43]. Recently, Bharathy and Suguna identified and solved the three-dimensional crystal structure of an aspartic proteinase-like domain observed in the C-terminal region of the PE_PGRS16 (Rv0977) protein [44].
In the present work, we have analyzed the sequences of PE and PPE proteins from several mycobacterial species. Our results provide clues for the fold and possible functions for ten well-characterized domains predicted in some PE and PPE proteins that provide the rationale for experimental validation.
Materials and Methods
Sequence searches—PSI-BLAST
The amino acid sequences corresponding to the PE and PPE proteins from Mtb H37Rv strain were obtained from the NCBI databank (http://www.ncbi.nlm.nih.gov/). Iterative and reciprocal searches corresponding to the PE and PPE regions in these proteins were performed using the PSI-BLAST program (www.ncbi.nlm.nih.gov/BLAST/) against the protein sequences from 60 mycobacterial species. The PSI-BLAST program detects related proteins by deriving a position specific scoring matrix (PSSM) from multiple sequence alignment of proteins that are detected above a given threshold score [45]. The results obtained were manually inspected to confirm the protein family.
Selection of non-redundant proteins—CD-HIT
The large dataset of mycobacterial proteins obtained from the homology searches had a high percentage of redundant sequences. We used the CD-HIT program [46] that can efficiently handle huge datasets containing millions of protein sequences, in order to remove redundant proteins and short-list set of protein sequences based on user-defined percentage sequence identity cut-off value (http://weizhong-lab.ucsd.edu/cdhit_suite/cgi-bin/index.cgi).
Multiple sequence alignment—ClustalX
The mycobacterial PE and PPE protein sequences obtained earlier were aligned separately using the multiple sequence alignment program ClustalX 2.1. It uses a heuristic pairwise progressive sequence alignment method to generate a dendrogram. The dendrogram was used to construct the multiple sequence alignment [47]. The parameters used for multiple sequence alignment were; “10” for gap opening penalty, “0.2” for gap extension and "Gonnet Series" was chosen for protein weight matrix.
Phylogeny analysis—MEGA5
Phylogenetic trees were generated for PE and PPE proteins using the draw tree clustering option in ClustalX based on the neighbor joining clustering algorithm. The phylogenetic tree was viewed and analyzed using Mega 5.0. MEGA5 is a collection of maximum likelihood (ML) analyses for inferring evolutionary trees, selecting best-fit substitution models for both nucleotides or amino acids, inferring ancestral states and estimating evolutionary rates [48]. The representative proteins based on these phylogenetic trees were selected for further analysis.
Protein fold recognition—Phyre2
The non-PE and non-PPE regions in the PE and PPE proteins were selected in order to identify functional domains. Phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2) comprises a suite of tools that can predict and analyze protein structure, function and mutations [49]. Phyre2 is particularly useful to recognize the protein folds of distantly related sequences and uses advanced methods for constructing three-dimensional model. The results of the model obtained for a given protein sequence can be interpreted based on percentage confidence, percentage sequence coverage, sequence alignment with the target structure, secondary and tertiary structure of the models, domain composition, conservation of the active site and model quality.
Results and Discussion
Mycobacterial species are known to comprise variable numbers of PE and PPE family proteins. Several variations in their protein sequences have been attributed to synonymous and non-synonymous single nucleotide polymorphism (SNPs), in-frame deletions and insertions, resulting in their altered physico-chemical properties [23,24,50]. These features indicate the observed differences between the mycobacterial species. The N-terminal PE and PPE domains, and the Gly-rich regions in these families of proteins provide limited clues on their possible structure and function. The structure-based annotation of the protein folds are more useful, which have therefore been applied in the present work using Phyre2 program that has also been useful to assign a possible function. The list of mycobacterial species for the analysis of structure and function corresponding to the PE and PPE protein families studied in this work are shown in the supplementary data (S1 Table). The protein fold templates identified with ‘high’ confidence and described as "certain" according to the program and corresponding to significant sequence coverage over whole length of the protein were selected as the probable fold. The three-dimensional models based on the templates identified and the alignments of the sequence to each of the templates produced by Phyre2 were manually inspected to examine whether the catalytic residues and the cofactor binding residues were also conserved. In situations where such conservation was confirmed, the corresponding domains were assigned as the protein fold. We discuss below the different domains that were identified in the PE and PPE proteins.
Hydrolase domain
The PE and PPE proteins from several mycobacterial species (M. tuberculosis, M. bovis, M. africanum, M. canettii, M. marinum, M. kansasii, M. liflandi, M. heckeshornense, M. bovis BCG strain, M. asiaticum, M. caprae, M. nebraskense, M. gordonae, M. haemophilum, M. lentiflavum, M. simiae, M. triplex, M. sinense, M. arupense, M. heraklionense, M. neworleansense) that were predicted to comprise a conserved C-terminal domain with α/β hydrolase fold are shown in the supplementary data (S1 Appendix). This domain was modelled on the crystal structures of two templates comprising msmeg_6394 from M. smegmatis str. MC2 155 (PDB_ID: 3AJA:A) and a putative esterase from Staphylococcus aureus (PDB_ID: 3D7R:B). The template structures represent hydrolase family which exhibit an overall α/β hydrolase fold with central β-sheet flanked by α-helices on either side of the sheet and has the functional characterization of a lipase. Most hydolase family proteins are characterized by a beta-sheet core made up of five to eight β-strands connected by α-helices forming an α/β/α sandwich with a conserved pentapeptide sequence motif GxSxG, and Ser, Asp and His as the catalytic residues [51].
We previously identified this domain in the C-terminal region of 8 PE and PPE proteins (Rv0151c, Rv0152c, Rv0159c, Rv0160c, Rv1430, Rv1800, Rv2608 and Rv3539) in M. tuberculosis H37Rv strain and was termed ‘PE-PPE domain’ [41]. Subsequently, we modelled this domain, characterized the active site, lid insertion close to the active site, the oxyanion hole required for function and predicted that this domain would specifically possess esterase/lipase/cutinase activity [42]. Further, we carried out wet-lab experimental studies using biochemical assays and mutational analyses and confirmed that the purified full-length Rv1430 protein and its ‘PE-PPE domain’ possesses esterase activity and hydrolyses short to medium chain fatty acid esters with highest specific activity for p-nitrophenyl caproate [43].
Aspartic proteinase domain
The PE proteins from mycobacterial species (M. tuberculosis, M. bovis, M. caprae, M. africanum, M. caprae, M. orygis, M. canettii, M. gordonae) predicted to comprise the aspartic proteinase domain are shown in the supplementary data (S2 Appendix). This domain was reported in the PE_PGRS16 (Rv0977) protein and its three-dimensional crystal structure was solved (PDB_ID: 4EHC) [44]. The aspartic proteinase domain has low overall sequence similarity to HIV proteinase with a characteristic pepsin-fold and catalytic site architecture. The overall fold comprises a six stranded β-sheet located at the centre formed by the contribution of 3 strands each from N- and C-terminal domains. On either side of the central region, β-sheet rich regions and two α-helices connected by loops that harbor the conserved DTG motifs were located. These motifs are essential for the peptide hydrolysis function of this enzyme. In this work, the fold for some of the PE proteins, for instance, WP_015355774.1, WP_036418539.1 and WP_031667305.1 (refer S2 Appendix for detailed list) comprising a C-terminal 280 amino acid domain was predicted as the aspartic proteinase domain. This domain in the mycobacterial PE proteins was modeled on the crystal structure of PDB_ID: 4EHC_A. The models comprise the two conserved DT(S)G motifs characteristic of aspartic proteinases. In the peptide hydrolysis reaction, one of the aspartic acid residues from the conserved motifs acts as a general base and the other acts as a general acid followed by the nucleophilic attack of a catalytic water molecule [52]. Typically aspartic proteinases hydrolyse a peptide bond between hydrophobic residues. For example, renin is an aspartic proteinase that cleaves angiotensiogen to a decapetide angiotensin with high specific cleavage between Leu-Leu bond [53]. Several of these proteins are drug targets, for example, HIV proteinase in AIDS and renin in hypertension [54].
Glucosyl-3-phosphoglycerate phosphatase domain
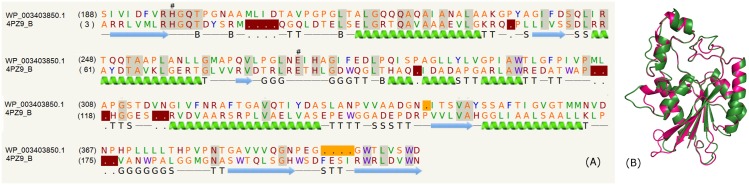
The list of PE proteins from mycobacterial species (M. tuberculosis, M. bohemicum, M. haemophilum, M. canettii, M. kansasii, M. africanum, M. bovis, M. marinum, M. liflandii, M. xenopi, M. heckeshornense, M. gordonae) comprising the glucosyl-3-phosphoglycerate phosphatase domain is shown in the supplementary data (S3 Appendix). Some of these PE proteins, for instance, WP_003403850.1, WP_015357328.1 and CPR12073.1 recognized the template PDB_ID: 4PZ9:B corresponding to the mycobacterial glucosyl-3-phosphoglycerate phosphatase Rv2419c [55]. This enzyme consists of a single domain made up of a central β-sheet flanked by α-helices on either side and is known to catalyze the second step in the biosynthesis of methylglucose lipopolysaccharides (MGLPs) pathway. The synthesis of mycolic acids, which forms an important lipid component of the mycobacterium cell wall, is regulated by MGLPs. The alignment of the sequence to the template (~21% identity) is shown in Fig 1A and highlights the regions of secondary structure. The corresponding structure alignment of the model and template is shown in Fig 1B and suggests the overall similarity in the protein fold.
Fig 1. Glucosyl-3-phosphoglycerate phosphatase domain.
(A) Sequence alignment of model WP_003403850.1 and template PDB_ID: 4PZ9:B indicating the catalytic residues (#). In all the pair-wise sequence alignments, identical residues are indicated in gray shaded regions, deletions in the template sequence is indicated as dots in brown shaded region, deletions in the query sequence is indicated as dots in yellow shaded region. (B) Structure alignment of model (green) and template (pink).
In the crystal structure (PDB_ID: 4QIH), the active site is located in a positively charged cleft situated above a central β-sheet. Unambiguous electron density for a vanadate ion covalently bound to His11 (numbering according to PDB_ID: 4PZ9) mimicking the phosphohistidine intermediate and acetate ion was observed. In WP_003403850.1 too, the catalytic residues His11 and Glu84 are conserved (Fig 1A) and most of the ligand binding residues close to the acetate and vanadate, for instance, Arg10, His11, Asn17, Gln23, Arg60, Glu84, His159 and Leu209 which are important for enzymatic activity are conserved, except Gln23 which was replaced by Thr and His159 by Tyr. According to the structure analyses, most residues being conserved, especially residues close to vanadate, suggests that the protein function is also likely to be conserved.
Laminaripentaose-producing beta-1,3-glucanase domain
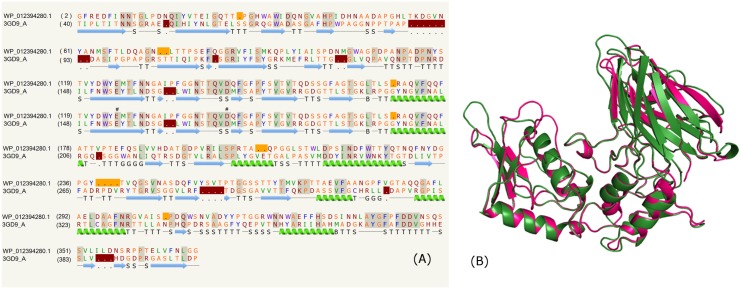
The list of PE proteins from mycobacterial species (M. marinum, Mycobacterium sp. 012931, M. ulcerans str. Harvey) comprising the laminaripentaose-producing beta-1,3-glucanase (LPHase) domain is shown in the supplementary data (S4 Appendix). The PE protein, for instance, WP_012394280.1 from M. marinum C-terminal region recognized the template PDB_ID: 3GD9:A corresponding to the crystal structure of LPHase in complex with laminaritetraose [56]. Glycoside hydrolases have been classified into families based on sequence similarity and have been further grouped into clans based on the similarity of their overall fold, active site architecture and catalytic mechanism [57,58,59]. LPHase is a glycoside hydrolase family 64 protein which cleaves a long chain polysaccharide β-1,3-glucan into specific pentasaccharide oligomers. The structure consists of a crescent-like fold; a barrel domain and a mixed (α/β) domain forming a wide-open groove between the two domains. The sequence alignment highlighting the secondary structures predicted is shown in Fig 2A and has ~24% identity. The structural overlay of the model and template is shown in Fig 2B that suggests the protein has overall similar fold and certain variable loop regions. The glycoside hydrolases are known to catalyse the hydrolysis of the glycosidic bond between two or more carbohydrates or between a carbohydrate and non-carbohydrate moiety [60]. Depending on the nature of the organism, these enzymes are associated with a variety of roles, such as degradation of biomass by cellulases, pathogenesis during the activity of influenza virus neuraminidase [61], normal cellular metabolic processes that involves the formation and breakage of glycosidic bonds [62]. Our model suggests the conservation of catalytic residues; Glu154 and Asp170 (numbering according to the PDB_ID: 3GD9) as shown in Fig 2A. Between the N and C-terminal domains, the model contains an electronegatively charged wide groove comprising several conserved residues that include the above catalytic residues and four amino acid residues; Thr156, Asn158, Trp163 and Thr167 involved in sugar binding that accommodate the laminaritetraose molecule. According to the crystal structure of LPHase (PDB_ID:3GD9), the enzyme uses a direct displacement mechanism involving Glu154 and Asp170 via acid-base catalysis to cleave β-1,3-glucan into specific α-pentasaccharide oligomer. The side chains of Thr156, Asn158 and Trp163 are known to demarcate the subsite +5 in the active site.
Fig 2. Laminaripentaose-producing beta-1,3-glucanase domain.
(A) Sequence alignment of model WP_012394280.1 and template PDB _ID: 3GD9:A indicating the catalytic residues (#). (B) Structure alignment of model (green) and template (pink).
Chitinase domain
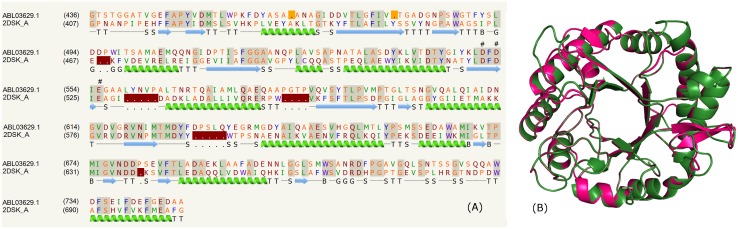
The list of PE proteins from mycobacterial species (M. liflandii, M. marinum, Mycobacterium sp. 012931, M. ulcerans str. Harvey, M. gastri, M. gordonae) comprising the chitinase domain is shown in the supplementary data (S5 Appendix). Some of these PE proteins, for instance, ABL03629.1, WP_015355330.1, WP_023367572.1, WP_036414736.1 and WP_012395848.1 recognize the template PDB_ID:2DSK:A corresponding to the chitinase domain. The catalytic site residues; Asp522, Asp524 and Glu526 form the characteristic DxDxE motif observed in the crystal structure were conserved in these PE proteins as shown for one of the illustrative examples in Fig 3A that shares ~37% identity. The model was constructed on the crystal structure of the catalytic domain of chitinase from Pyrococcus furiosus (PDB _ID: 2DSK:A). The overall structure of this domain comprises a TIM-barrel fold with a tunnel-like active site, a common feature of family 18 chitinases. The high degree of the overall structural similarity is shown in Fig 3B. The chitinases hydrolyze chitin, a polymer of β-1,4-linked N-acetylglucosamine (GlcNAc) and classified into two families (families 18 and 19 in the CAZy database; http://www.cazy.org/) according to amino acid sequence similarity [63].
Fig 3. Chitinase domain.
(A) Sequence alignment of model ABL03629.1 and template PDB _ID: 2DSK:A indicating the catalytic residues (#). (B) Structure alignment of model (green) and template (pink).
Endoglucanase domain
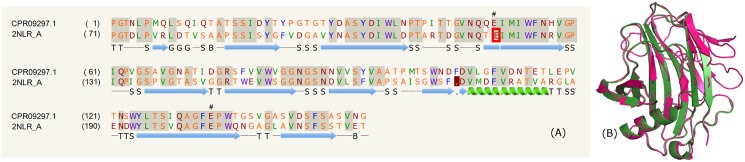
The list of PE proteins from mycobacterial species (M. kansasii, M. gastri, M. gordonae, M. bohemicum DSM 44277, M. asiaticum) comprising the endoglucanase domain is shown in the supplementary data (S6 Appendix). The PE proteins, for example, MGAST_01715 and CPR09297.1 have a conserved C-terminal region. The fold for these proteins corresponds to the endoglucanases (PDB_IDs: 1OA4:A and 2NLR). These glycoside hydrolase clan GH-C group endoglucanases comprise the family 11 xylanases and family 12 cellulases, which share a jelly-roll topology. The two predominantly anti-parallel β-sheets form a long substrate-binding cleft. The catalysis of these enzymes is via a double-displacement mechanism in which a covalent glycosyl enzyme intermediate is formed and subsequently hydrolyzed with acid-base assistance, via oxocarbenium ion transition states and performs the catalysis with net retention of anomeric configuration [64,65]. Fig 4A shows the sequence alignment (~58% identity) along with the catalytic residues and the secondary structural information. The structural comparison is shown in Fig 4B. The concave surface of the larger β-sheet produces a wide substrate-binding cleft across one face of the enzyme [66]. Further, the cleft has two invariant catalytic residues Glu120 and Glu203 (amino acid numbering according to PDB_IDs: 2NLR) that point into the active site cleft from opposite sides. A long loop crossing the substrate-binding groove terminates at the reducing end, known as the “cord” and that is a common feature of all family 11 and family 12 structures. A number of residues located in the loop, importantly, Pro133 is conserved throughout clan GH-C members. A possible loop movement upon substrate binding has been speculated [66].
Fig 4. Endoglucanase domain.
(A) Sequence alignment of model CPR09297.1 and template PDB_ID: 2NLR:A indicating the catalytic residues (#, red box). (B) Structure alignment of model (green) and template (pink).
Carbohydrate binding domain
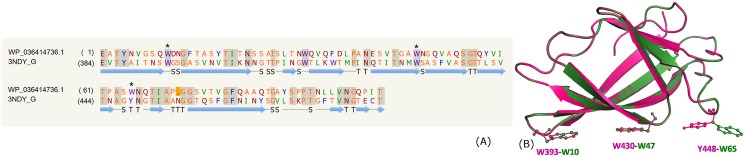
The list of PE proteins from mycobacterial species (M. kansasii, M. gastri, Mycobacterium sp. 012931, M. bohemicum DSM 44277) comprising the carbohydrate binding domain is shown in the supplementary data (S7 Appendix). In the cellulose degradation process, the binding of the cellulolytic enzyme is mediated by carbohydrate binding domain (CBD) which typically comprises ~100 amino acid residues [67]. CBDs are separated from the catalytic domain by a short amino acid linker region and the enzymatic degradation is carried out by the cellulolytic domain [68,69]. From our analysis, we observed that the CPR09297.1 comprising an endoglucanase domain described above has one CBD, while ETW25608.1 has two consecutive CBDs separated by 87 amino acids. The two proteins with CBD domains are linked with C-terminal glycosyl hydrolase 12 (GH12) family domains discussed above. Likewise, another PE protein; WP_036414736.1, comprises a CBD that is linked to a glycosyl hydrolase 18 (GH18) family domain. The three-dimensional structures of these CBDs were modelled on the CBD domain present in the endoglucanase D from Clostridium cellulovorans (PDB_ID: 3NDY). The sequence alignment (~35% identity) is shown in Fig 5A. The alignment demonstrates that the secondary structure is mainly comprised of beta-strands. The structural overlay of model with template is shown in Fig 5B which reveal eight major beta-strands that fold into a beta-sheet structure. Three conserved hydrophobic amino acids (Trp, Trp, Trp/Tyr) ‘strip’ are located on loops connecting the beta-strands in the model and the template as shown in Fig 5B. The pi-electron dense aromatic rings of the ‘strip’ contact the cellulose hydrophobic region and drive the enzymes to perform their catalytic functions [70,71].
Fig 5. Carbohydrate binding domain.
(A) Sequence alignment of model WP_036414736.1 and template PDB_ID: 3NDY:G indicating aromatic residues (*) from hydrophobic strip. (B) Structure alignment of model (green) and template (pink).
Cytochrome P450 domain
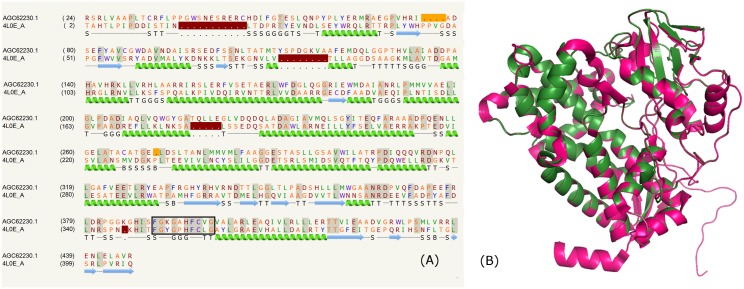
The 1761 amino acid PE protein AGC62230.1 from M. liflandii 128FXT comprises a 500 amino acid C-terminus domain. The fold for this domain was recognized as the cytochrome P450 from Streptomyces sp. Acta 2897 (PDB_ID:4L0E) [72]. Cytochrome P450s are a class of heme cofactor binding proteins and found in all domains of life. The high diversity in sequences and functions resulted in an expanded family of cytochrome P450s. They catalyse a variety of reactions, for instance, carbon heteroatom oxygenation, dealkylation, epoxidation, aromatic hydroxylation, reduction and dehalogenation [73]. A unique consensus sequence motif; ‘FXXGXXXCXG’ is present in all cytochrome P450s and located between helices K and L that forms a heme binding decapeptide loop [74]. This motif was also observed in the PE protein AGC62230.1 from Mycobacterium liflandii 128FXT (Fig 6A).
Fig 6. Cytochrome P450 domain.
(A) Sequence alignment of model AGC62230.1 and template PDB _ID: 4L0E:A indicating consensus heme binding decapeptide motif in box. (B) Structure alignment of model (green) and template (pink).
The non-ribosomal peptide synthetases (NRPSs) are involved in the synthesis of diverse peptides known as non-ribosomally synthesised peptides (NRPs) [75]. One of the prominent modifications found in NRPs is the β-hydroxylation of various amino acid residues including the hydroxylation of non-activated C-H bonds [76] that are catalysed by cytochrome P450 enzymes. The cytochrome P450 encoded (sky32) is associated with the skyllamycin biosynthesis gene cluster. The cyclodepsipeptide skyllamycin A isolated from streptomyces is an inhibitor of the platelet derived growth factor signaling pathway [77]. The crystal structure of sky32 (PDB_ID:4L0E) is responsible for the β-hydroxylation of three separate amino acids at positions 5 (β-hydroxyphenylalanine), 7 (β-hydroxy-OMe-tyrosine), and 11 (β-hydroxyleucine). The sequence alignment (~23% identity) corresponding to the cytochrome P450 domain in the above PE protein and sky32 mainly comprising helices is shown in Fig 6A. The comparison of the overall fold shown in Fig 6B reveals the high degree of structural similarity.
Beta-propeller
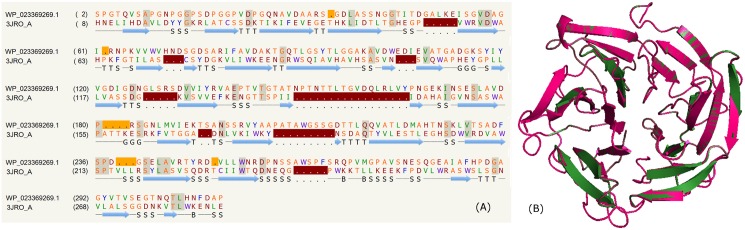
We earlier reported certain PE family proteins to comprise YVTN repeats [41]. These repeats contain 40–45 amino acid residues present in tandem along the protein sequence and located towards the C-terminus. In this work, we have identified several PE proteins from mycobacterial species (M. tuberculosis, M. bovis, M. africanum MAL020173, M. caprae, M. orygis 112400015, M. canettii CIPT 140070010, M. haemophilum, M. marinum) that contain the above repeat as shown in the supplementary data (S8 Appendix). Some of these PE proteins, for example, WP_023369269.1, CCP43730.1, WP_013988789.1 and WP_013988787.1 were modelled on the crystal structures of nitrous oxide reductase from Pseudomonas nautical (PDB_ID: 1QNI:E) [78], nitrous oxide reductase from P. denitrificans (PDB_ID: 1FWX:B) [79], cytochrome cd1 nitrite reductase (PDB_ID: 1GQ1:B) [80] and nup84-nup145c-sec13 (PDB_ID: 3JRO:A) [81]. These diverse proteins comprise a 6–8 bladed beta-propeller fold. Typically, beta- propellers contain 4–8 blades that are arranged circularly around a central axis [82]. Several other diverse ~40–45 amino acid repeats, such as, WD, YWTD, etc., are known to be present in tandem and fold as beta-propellers [83]. The proteins containing the beta-propellers are associated with diverse functions such as transport, hydrolases, transferases, sugar and cofactor binding proteins, cell surface proteins, lyases and isomerases [83]. In some cases, the active site is present in the loops that connect the tandem blades. For example, in the crystal structure of influenza neuraminidase (PDB_ID: 1BJI), the active site is located in the region connected by several loops [84]. The sequence alignment (~13% identity) corresponding to the PE protein; CMB12570.1 and its template is shown in Fig 7A. The structural comparison of the model and template is shown in Fig 7B. This protein represents a 6 bladed beta-propeller.
Fig 7. Beta-propeller.
(A) Sequence alignment of model WP_023369269.1 and template PDB_ID: 3JRO:A. (B) Structure alignment of model (green) and template (pink).
Beta-helix
The beta-helix predicted for the PPE proteins from mycobacterial species (M. tuberculosis, M. kansasii, M. asiaticum, M. gastri, Mycobacterium sp. 012931, M. marinum, M. canettii, M. gordonae, M. ulcerans, M. bovis, M. orygis, M. liflandii) is shown in the supplementary data (S9 Appendix). Several PPE proteins are characterized by Gly-rich pentapeptide sequence repeats. Some of the PPE proteins (WP_049959014.1, WP_049959014.1 and WP_012396836.1) were modelled on the N-terminal domain of a ubiquitin ligase (PDB _ID: 3NB2:B) as the template. The template structure contains two structural domains: an N-terminal four stranded beta-helix domain made up of penta-peptide sequence repeats and a C-terminal α-helical catalytic domain [85,86]. Beta-helices were initially identified in the crystal structure of pectate lyase [87] and their functions, such as, polysaccharide lyases, cellulose or acid sugar binding lyases have been reviewed [88]. The beta-helix forms a helical pattern due to the hydrogen bonds between parallel beta sheets and can form two/three/four beta-stranded helices. The sequence alignment (~15% identity) predicted to mainly comprise the beta-strands is shown in Fig 8A and structure superposition on the template for this domain predicted in WP_012396836.1 is shown in Fig 8B.
Fig 8. Beta-helix.
(A) Sequence alignment of model WP_012396836.1 and template PDB_ID: 3NB2:B. (B) Structure alignment of model (green) and template (pink).
Acetyl hydrolase / cutinase domain
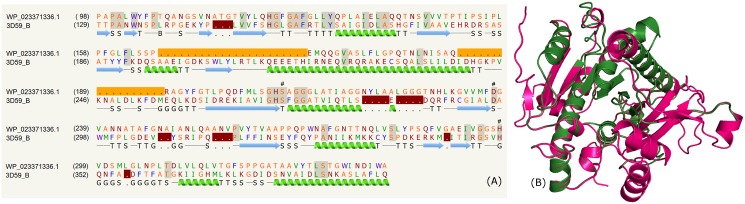
The list of PE proteins from mycobacterial species (M. bohemicum DSM 44277, M. marinum, M. liflandii, Mycobacterium sp. 012931, M. kansasii, M. asiaticum, M. gordonae) predicted to contain the acetyl hydrolase / cutinase domain is shown in the supplementary data (S10 Appendix). Some of these proteins were modeled, for instance, WP_023371336.1, WP_015356298.1, WP_036353055.1 and CPR09862.1. The crystal structures of human plasma platelet activating factor acetyl hydrolase (PDB_IDs: 3D5E, 3D59:B) were used as templates in the modeling procedure. These structures have a α/β-hydrolase fold containing a catalytic triad of Ser, His and Asp. The alignment of the sequences (~15% identity) is shown in Fig 9A. The structural comparison for the PE protein; WP_023371336.1 with template is shown in Fig 9B. The location of the catalytic residues Ser273, Asp296 and His351 (numbering according to PDB_ID: 3D59) are shown in Fig 9A.
Fig 9. Acetyl hydrolase/cutinase.
(A) Sequence alignment of model WP_023371336.1 and template PDB _ID: 3D59:B indicating the catalytic residues (#). (B) Structure alignment of model (green) and template (pink).
Transmembrane domain
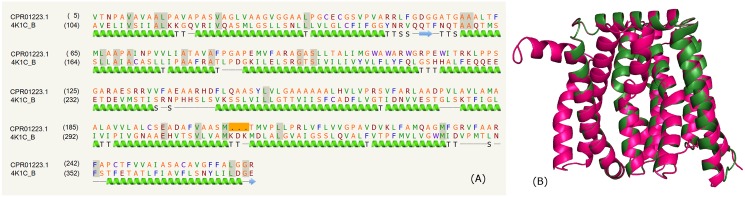
Some PPE family proteins; WP_036437315.1, WP_036444818.1 and CPR01223.1 have been predicted to comprise transmembrane helices with large stretches of intervening sequences suggesting these as membrane proteins. We modeled, one of the above proteins, CPR01223.1, a M. bohemicum protein on the crystal structure of a eukaryotic calcium/proton exchanger (PDB_ID: 4K1C). The sequence alignment that shares ~9% identity and predicted secondary structure is shown in Fig 10A, suggesting this protein is mainly composed of helices. The three-dimensional model superimposed on the template is shown in Fig 10B.
Fig 10. Transmembrane domain.
(A) Sequence alignment of model CPR01223.1 and template PDB _ID: 4K1C:B. (B) Structure alignment of model (green) and template (pink).
In this work, we have identified certain well-characterized domains present in the PE and PPE proteins of mycobacteria. These domains are known to be associated with a variety of functions, such as, hydrolysis of lipids (α/β hydrolase fold and acetyl hydrolase), hydrolysis of carbohydrates (chitinase, endoglucanase and laminaripentaose-producing beta-1,3-glucanase domain) and hydrolysis of proteins (aspartic proteinase). Glucosyl-3-phosphoglycerate phosphatase plays an important role in the synthesis of mycobacterial cell wall components and cytochrome P450 domain is implicated in metabolic activity. The transmembrane domains, beta-propellers, CBD and beta-helices are regulators of protein function. This suggests that some of the PE and PPE family proteins may be associated with enzymatic and regulatory roles.
The α/β hydrolase domain is present in both PE and PPE family proteins, whereas aspartic proteinase, chitinase, endoglucanase and beta-propeller domains were only detected for PE proteins. The beta-helix has been observed in PPE proteins. It was also observed that some domains such as α/β hydrolase were present in all mycobacterial species, whereas some domains, such as, chitinase and endoglucanase were specific only to certain mycobacterial species. These findings suggest that the PE and PPE family proteins co-ordinate diverse roles that are mycobacterial species dependent.
Out of several hundred diverse sequences that were analysed, we were able to predict the fold with ‘high’ confidence for ~30% proteins. The structure and function predicted for the PE and PPE proteins discussed in this work provide the rationale for validation by experimental studies.
Conclusions
The bioinformatics analyses of several PE and PPE proteins from a number of mycobacterial species allowed us to identify the following well-characterized domains; hydrolase, aspartic proteinase, glucosyl-3-phosphoglycerate phosphatase, laminaripentaose-producing beta-1,3-glucanase, chitinase, endoglucanase, carbohydrate binding, cytochrome P450, beta-propeller, beta-helix, acetyl hydrolase/cutinase and transmembrane domains. Some of these domains have enzymatic roles and hydrolyse substrates, such as, proteins, lipids and carbohydrates, while some domains have a regulatory role. Further, some domains were observed to be common to several mycobacterial species, while some were present only in few mycobacterial species. Our work sheds new light on the structural and functional aspects of these important classes of mycobacterial proteins.
Supporting Information
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(DOC)
Acknowledgments
LGP thanks DBT for Fast Track fellowship and University with Potential for Excellence-2, University of Hyderabad for research facilities. RS thanks Council of Scientific and Industrial Research for Senior Research Fellowship. KT thanks DSK Kothari, UGC for post-doctoral fellowship. The authors also thank the anonymous reviewers and the Academic Editor for very useful comments to improve the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
LGP thanks DBT for the Fast Track Fellowship and University with Potential for Excellence-2, University of Hyderabad for research facilities. RS thanks the Council of Scientific and Industrial Research for the Senior Research Fellowship. KT thanks DSK Kothari, UGC for the post-doctoral fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Zaman K (2010) Tuberculosis: a global health problem. Journal of health, population, and nutrition 28: 111–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaufmann SHE, Rubin E, editor (2008) Handbook of Tuberculosis: Clinics, Diagnostics, Therapy and Epidemiology. [Google Scholar]
- 3.Ehebauer MT, Wilmanns M (2011) The progress made in determining the Mycobacterium tuberculosis structural proteome. Proteomics 11: 3128–3133. 10.1002/pmic.201000787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narita M, Ashkin D, Hollender ES, Pitchenik AE (1998) Paradoxical worsening of tuberculosis following antiretroviral therapy in patients with AIDS. American journal of respiratory and critical care medicine 158: 157–161. [DOI] [PubMed] [Google Scholar]
- 5.Harries AD, Zachariah R, Corbett EL, Lawn SD, Santos-Filho ET, Chimzizi R, et al. (2010) The HIV-associated tuberculosis epidemic—when will we act? Lancet 375: 1906–1919. 10.1016/S0140-6736(10)60409-6 [DOI] [PubMed] [Google Scholar]
- 6.Snider DE Jr, La Montagne JR (1994) The neglected global tuberculosis problem: a report of the 1992 World Congress on Tuberculosis. The Journal of infectious diseases 169: 1189–1196. [DOI] [PubMed] [Google Scholar]
- 7.Cole ST, Telenti A (1995) Drug resistance in Mycobacterium tuberculosis. The European respiratory journal Supplement 20: 701s–713s. [PubMed] [Google Scholar]
- 8.WHO (2006) Guidelines for the programmatic management of drug-resistant tuberculosis.
- 9.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393: 537–544. [DOI] [PubMed] [Google Scholar]
- 10.Vissa VD, Brennan PJ (2001) The genome of Mycobacterium leprae: a minimal mycobacterial gene set. Genome biology 2: REVIEWS1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li L, Bannantine JP, Zhang Q, Amonsin A, May BJ, Alt D, et al. (2005) The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proceedings of the National Academy of Sciences of the United States of America 102: 12344–12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim BJ, Choi BS, Lim JS, Choi IY, Lee JH, Chun J, et al. (2012) Complete genome sequence of Mycobacterium intracellulare strain ATCC 13950(T). Journal of bacteriology 194: 2750 10.1128/JB.00295-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garnier T, Eiglmeier K, Camus JC, Medina N, Mansoor H, Pryor M, et al. (2003) The complete genome sequence of Mycobacterium bovis. Proceedings of the National Academy of Sciences of the United States of America 100: 7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bentley SD, Comas I, Bryant JM, Walker D, Smith NH, Harris SR, et al. (2012) The genome of Mycobacterium africanum West African 2 reveals a lineage-specific locus and genome erosion common to the M. tuberculosis complex. PLoS neglected tropical diseases 6: e1552 10.1371/journal.pntd.0001552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stinear TP, Seemann T, Harrison PF, Jenkin GA, Davies JK, Johnson PD, et al. (2008) Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome research 18: 729–741. 10.1101/gr.075069.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akhter Y, Ehebauer MT, Mukhopadhyay S, Hasnain SE (2012) The PE/PPE multigene family codes for virulence factors and is a possible source of mycobacterial antigenic variation: perhaps more? Biochimie 94: 110–116. 10.1016/j.biochi.2011.09.026 [DOI] [PubMed] [Google Scholar]
- 17.Sampson SL (2011) Mycobacterial PE/PPE proteins at the host-pathogen interface. Clinical & developmental immunology 2011: 497203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tundup S, Akhter Y, Thiagarajan D, Hasnain SE (2006) Clusters of PE and PPE genes of Mycobacterium tuberculosis are organized in operons: evidence that PE Rv2431c is co-transcribed with PPE Rv2430c and their gene products interact with each other. FEBS letters 580: 1285–1293. [DOI] [PubMed] [Google Scholar]
- 19.Ishikawa J, Yamashita A, Mikami Y, Hoshino Y, Kurita H, Hotta K, et al. (2004) The complete genomic sequence of Nocardia farcinica IFM 10152. Proceedings of the National Academy of Sciences of the United States of America 101: 14925–14930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nair S (2014) Immunomodulatory role of Mycobacterial PE/PPE family of proteins. Proc Indian Natn Sci Acad 80: 1055–1072. [Google Scholar]
- 21.Abdallah AM, Verboom T, Weerdenburg EM, Gey van Pittius NC, Mahasha PW, Jiménez C, et al. (2009) PPE and PE_PGRS proteins of Mycobacterium marinum are transported via the type VII secretion system ESX-5. Molecular microbiology 73: 329–340. 10.1111/j.1365-2958.2009.06783.x [DOI] [PubMed] [Google Scholar]
- 22.Gey van Pittius NC, Sampson SL, Lee H, Kim Y, van Helden PD, Warren RM (2006) Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC evolutionary biology 6: 95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohli S, Singh Y, Sharma K, Mittal A, Ehtesham NZ, Hasnain SE (2012) Comparative genomic and proteomic analyses of PE/PPE multigene family of Mycobacterium tuberculosis H(3)(7)Rv and H(3)(7)Ra reveal novel and interesting differences with implications in virulence. Nucleic acids research 40: 7113–7122. 10.1093/nar/gks465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mackenzie N, Alexander DC, Turenne CY, Behr MA, De Buck JM (2009) Genomic comparison of PE and PPE genes in the Mycobacterium avium complex. Journal of clinical microbiology 47: 1002–1011. 10.1128/JCM.01313-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cascioferro A, Daleke MH, Ventura M, Dona V, Delogu G, Palù G, et al. (2011) Functional dissection of the PE domain responsible for translocation of PE_PGRS33 across the mycobacterial cell wall. PloS one 6: e27713 10.1371/journal.pone.0027713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zumbo A, Palucci I, Cascioferro A, Sali M, Ventura M, D'Alfonso P, et al. (2013) Functional dissection of protein domains involved in the immunomodulatory properties of PE_PGRS33 of Mycobacterium tuberculosis. Pathogens and disease 69: 232–239. 10.1111/2049-632X.12096 [DOI] [PubMed] [Google Scholar]
- 27.Daleke MH, Cascioferro A, de Punder K, Ummels R, Abdallah AM, van der Wel N, et al. (2011) Conserved Pro-Glu (PE) and Pro-Pro-Glu (PPE) protein domains target LipY lipases of pathogenic mycobacteria to the cell surface via the ESX-5 pathway. The Journal of biological chemistry 286: 19024–19034. 10.1074/jbc.M110.204966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iantomasi R, Sali M, Cascioferro A, Palucci I, Zumbo A, Soldini S, et al. (2012) PE_PGRS30 is required for the full virulence of Mycobacterium tuberculosis. Cellular microbiology 14: 356–367. 10.1111/j.1462-5822.2011.01721.x [DOI] [PubMed] [Google Scholar]
- 29.Garrett CK, Broadwell LJ, Hayne CK, Neher SB (2015) Modulation of the Activity of Mycobacterium tuberculosis LipY by Its PE Domain. PloS one 10: e0135447 10.1371/journal.pone.0135447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tundup S, Pathak N, Ramanadham M, Mukhopadhyay S, Murthy KJ, Ehtesham NZ, et al. (2008) The co-operonic PE25/PPE41 protein complex of Mycobacterium tuberculosis elicits increased humoral and cell mediated immune response. PloS one 3: e3586 10.1371/journal.pone.0003586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cascioferro A, Delogu G, Colone M, Sali M, Stringaro A, Arancia G, et al. (2007) PE is a functional domain responsible for protein translocation and localization on mycobacterial cell wall. Molecular microbiology 66: 1536–1547. [DOI] [PubMed] [Google Scholar]
- 32.Deng W, Li W, Zeng J, Zhao Q, Li C, Zhao Y, et al. (2014) Mycobacterium tuberculosis PPE family protein Rv1808 manipulates cytokines profile via co-activation of MAPK and NF-kappaB signaling pathways. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology 33: 273–288. [DOI] [PubMed] [Google Scholar]
- 33.Srivastava V, Rouanet C, Srivastava R, Ramalingam B, Locht C, Srivastava BS (2007) Macrophage-specific Mycobacterium tuberculosis genes: identification by green fluorescent protein and kanamycin resistance selection. Microbiology 153: 659–666. [DOI] [PubMed] [Google Scholar]
- 34.Dubnau E, Fontan P, Manganelli R, Soares-Appel S, Smith I (2002) Mycobacterium tuberculosis genes induced during infection of human macrophages. Infection and immunity 70: 2787–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fishbein S, van Wyk N, Warren RM, Sampson SL (2015) Phylogeny to function: PE/PPE protein evolution and impact on Mycobacterium tuberculosis pathogenicity. Molecular microbiology 96: 901–916. 10.1111/mmi.12981 [DOI] [PubMed] [Google Scholar]
- 36.Anand P, Sankaran S, Mukherjee S, Yeturu K, Laskowski R, Bhardwaj A, et al. (2011) Structural annotation of Mycobacterium tuberculosis proteome. PloS one 6: e27044 10.1371/journal.pone.0027044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh S, Guttula PK, Guruprasad L (2014) Structure based annotation of Helicobacter pylori strain 26695 proteome. PloS one 9: e115020 10.1371/journal.pone.0115020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pearson WR (2013) An introduction to sequence similarity ("homology") searching Current protocols in bioinformatics / editoral board, Baxevanis Andreas D [et al. ] Chapter 3: Unit3 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pieper U, Webb BM, Barkan DT, Schneidman-Duhovny D, Schlessinger A, et al. (2011) ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic acids research 39: D465–474. 10.1093/nar/gkq1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adindla S, Sultana R, Tanneeru K, Singh S, Guruprasad L (2013) To make the most of a protein sequence. Special issue Bioinformatics, Proceedings of Andhra Pradesh of Akademic of Sciences 125–130. [Google Scholar]
- 41.Adindla S, Guruprasad L (2003) Sequence analysis corresponding to the PPE and PE proteins in Mycobacterium tuberculosis and other genomes. Journal of biosciences 28: 169–179. [DOI] [PubMed] [Google Scholar]
- 42.Sultana R, Tanneeru K, Guruprasad L (2011) The PE-PPE domain in mycobacterium reveals a serine alpha/beta hydrolase fold and function: an in-silico analysis. PloS one 6: e16745 10.1371/journal.pone.0016745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sultana R, Vemula MH, Banerjee S, Guruprasad L (2013) The PE16 (Rv1430) of Mycobacterium tuberculosis is an esterase belonging to serine hydrolase superfamily of proteins. PloS one 8: e55320 10.1371/journal.pone.0055320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barathy DV, Suguna K (2013) Crystal structure of a putative aspartic proteinase domain of the Mycobacterium tuberculosis cell surface antigen PE_PGRS16. FEBS open bio 3: 256–262. 10.1016/j.fob.2013.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic acids research 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659. [DOI] [PubMed] [Google Scholar]
- 47.Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ (1998) Multiple sequence alignment with Clustal X. Trends in biochemical sciences 23: 403–405. [DOI] [PubMed] [Google Scholar]
- 48.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution 28: 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nature protocols 10: 845–858. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McEvoy CR, Cloete R, Muller B, Schurch AC, van Helden PD, Gagneux S, et al. (2012) Comparative analysis of Mycobacterium tuberculosis pe and ppe genes reveals high sequence variation and an apparent absence of selective constraints. PloS one 7: e30593 10.1371/journal.pone.0030593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hotelier T, Renault L, Cousin X, Negre V, Marchot P, Chatonnet A (2004) ESTHER, the database of the alpha/beta-hydrolase fold superfamily of proteins. Nucleic acids research 32: D145–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dunn B (1989) "Determination of protease mechanism". Proteolitic Enzymes a practical approach. eds. Beynon R. and Bond J.. oxford: Oxford University Press. [Google Scholar]
- 53.Poulsen K, Haber E, Burton J (1976) On the specificity of human renin. Studies with peptide inhibitors. Biochimica et biophysica acta 452: 533–537. [DOI] [PubMed] [Google Scholar]
- 54.Cooper JB (2002) Aspartic proteinases in disease: a structural perspective. Current drug targets 3: 155–173. [DOI] [PubMed] [Google Scholar]
- 55.Zheng Q, Jiang D, Zhang W, Zhang Q, Zhao Q, Jin J, et al. (2014) Mechanism of dephosphorylation of glucosyl-3-phosphoglycerate by a histidine phosphatase. The Journal of biological chemistry 289: 21242–21251. 10.1074/jbc.M114.569913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu HM, Liu SW, Hsu MT, Hung CL, Lai CC, Cheng WC, et al. (2009) Structure, mechanistic action, and essential residues of a GH-64 enzyme, laminaripentaose-producing beta-1,3-glucanase. The Journal of biological chemistry 284: 26708–26715. 10.1074/jbc.M109.010983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Henrissat B, Callebaut I, Fabrega S, Lehn P, Mornon JP, Davies G (1995) Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases. Proceedings of the National Academy of Sciences of the United States of America 92: 7090–7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Henrissat B, Davies G (1997) Structural and sequence-based classification of glycoside hydrolases. Current opinion in structural biology 7: 637–644. [DOI] [PubMed] [Google Scholar]
- 59.Henrissat B, Bairoch A (1996) Updating the sequence-based classification of glycosyl hydrolases. The Biochemical journal 316 (Pt 2): 695–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davies G, Henrissat B (1995) Structures and mechanisms of glycosyl hydrolases. Structure 3: 853–859. [DOI] [PubMed] [Google Scholar]
- 61.Colman PM (1994) Influenza virus neuraminidase: structure, antibodies, and inhibitors. Protein science: a publication of the Protein Society 3: 1687–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sinnott ML (1990) Catalytic mechanisms of enzymatic glycosyl transfer. Chemical Reviews 90: 1171–1202. [Google Scholar]
- 63.Henrissat B (1991) A classification of glycosyl hydrolases based on amino-acid-sequence similarities. Biochemical Journal 280: 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koshland DE (1953) Stereochemistry and the mechanism of enzymatic reactions. Biol Rev Camb Philos Soc 28: 416–436. [Google Scholar]
- 65.Davies G, Sinnott M L, Withers S G (1997) Comprehensive bBiological Catalysis. Sinnott M. L., Ed London: Academic Press; 119–209 p. [Google Scholar]
- 66.Torronen A, Harkki A, Rouvinen J (1994) Three-dimensional structure of endo-1,4-beta-xylanase II from Trichoderma reesei: two conformational states in the active site. The EMBO journal 13: 2493–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tormo J, Lamed R, Chirino AJ, Morag E, Bayer EA, Shoham Y, et al. (1996) Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose. The EMBO journal 15: 5739–5751. [PMC free article] [PubMed] [Google Scholar]
- 68.Din N GN, Tekant B, Miller RC Jr, Warren RAJ, Kilburn DG (1991) Non–hydrolytic disruption of cellulose fibres by the binding domain of a bacterial cellulase. Nature Biotechnology 9: 1096–1099. [Google Scholar]
- 69.Din N, Damude HG, Gilkes NR, Miller RC Jr, Warren RA, Kilburn DG (1994) C1-Cx revisited: intramolecular synergism in a cellulase. Proceedings of the National Academy of Sciences of the United States of America 91: 11383–11387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Doxey AC, Cheng Z, Moffatt BA, McConkey BJ (2010) Structural motif screening reveals a novel, conserved carbohydrate-binding surface in the pathogenesis-related protein PR-5d. BMC structural biology 10: 23 10.1186/1472-6807-10-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnson PE, Joshi MD, Tomme P, Kilburn DG, McIntosh LP (1996) Structure of the N-terminal cellulose-binding domain of Cellulomonas fimi CenC determined by nuclear magnetic resonance spectroscopy. Biochemistry 35: 14381–14394. [DOI] [PubMed] [Google Scholar]
- 72.Uhlmann S, Sussmuth RD, Cryle MJ (2013) Cytochrome p450sky interacts directly with the nonribosomal peptide synthetase to generate three amino acid precursors in skyllamycin biosynthesis. ACS chemical biology 8: 2586–2596. 10.1021/cb400555e [DOI] [PubMed] [Google Scholar]
- 73.Danielson PB (2002) The cytochrome P450 superfamily: biochemistry, evolution and drug metabolism in humans. Current drug metabolism 3: 561–597. [DOI] [PubMed] [Google Scholar]
- 74.Song WC, Funk CD, Brash AR (1993) Molecular cloning of an allene oxide synthase: a cytochrome P450 specialized for the metabolism of fatty acid hydroperoxides. Proceedings of the National Academy of Sciences of the United States of America 90: 8519–8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hur GH, Vickery CR, Burkart MD (2012) Explorations of catalytic domains in non-ribosomal peptide synthetase enzymology. Natural product reports 29: 1074–1098. 10.1039/c2np20025b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cryle MJ (2011) Carrier protein substrates in cytochrome P450-catalysed oxidation. Metallomics: integrated biometal science 3: 323–326. [DOI] [PubMed] [Google Scholar]
- 77.Pohle S, Appelt C, Roux M, Fiedler HP, Sussmuth RD (2011) Biosynthetic gene cluster of the non-ribosomally synthesized cyclodepsipeptide skyllamycin: deciphering unprecedented ways of unusual hydroxylation reactions. Journal of the American Chemical Society 133: 6194–6205. 10.1021/ja108971p [DOI] [PubMed] [Google Scholar]
- 78.Brown K, Tegoni M, Prudencio M, Pereira AS, Besson S, Moura JJ, et al. (2000) A novel type of catalytic copper cluster in nitrous oxide reductase. Nature structural biology 7: 191–195. [DOI] [PubMed] [Google Scholar]
- 79.Brown K, Djinovic-Carugo K, Haltia T, Cabrito I, Saraste M, Moura JJ, et al. (2000) Revisiting the catalytic CuZ cluster of nitrous oxide (N2O) reductase. Evidence of a bridging inorganic sulfur. The Journal of biological chemistry 275: 41133–41136. [DOI] [PubMed] [Google Scholar]
- 80.Gordon EH, Sjogren T, Lofqvist M, Richter CD, Allen JW, Higham CW, et al. (2003) Structure and kinetic properties of Paracoccus pantotrophus cytochrome cd1 nitrite reductase with the d1 heme active site ligand tyrosine 25 replaced by serine. The Journal of biological chemistry 278: 11773–11781. [DOI] [PubMed] [Google Scholar]
- 81.Brohawn SG, Schwartz TU (2009) Molecular architecture of the Nup84-Nup145C-Sec13 edge element in the nuclear pore complex lattice. Nature structural & molecular biology 16: 1173–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Adindla S, Inampudi KK, Guruprasad L (2007) Cell surface proteins in archaeal and bacterial genomes comprising "LVIVD", "RIVW" and "LGxL" tandem sequence repeats are predicted to fold as beta-propeller. International journal of biological macromolecules 41: 454–468. [DOI] [PubMed] [Google Scholar]
- 83.Chen CK, Chan NL, Wang AH (2011) The many blades of the beta-propeller proteins: conserved but versatile. Trends in biochemical sciences 36: 553–561. 10.1016/j.tibs.2011.07.004 [DOI] [PubMed] [Google Scholar]
- 84.Taylor NR, Cleasby A, Singh O, Skarzynski T, Wonacott AJ, Smith PW, et al. (1998) Dihydropyrancarboxamides related to zanamivir: a new series of inhibitors of influenza virus sialidases. 2. Crystallographic and molecular modeling study of complexes of 4-amino-4H-pyran-6-carboxamides and sialidase from influenza virus types A and B. Journal of medicinal chemistry 41: 798–807. [DOI] [PubMed] [Google Scholar]
- 85.Lin DY, Diao J, Zhou D, Chen J (2011) Biochemical and structural studies of a HECT-like ubiquitin ligase from Escherichia coli O157:H7. The Journal of biological chemistry 286: 441–449. 10.1074/jbc.M110.167643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin DY, Diao J, Chen J (2012) Crystal structures of two bacterial HECT-like E3 ligases in complex with a human E2 reveal atomic details of pathogen-host interactions. Proceedings of the National Academy of Sciences of the United States of America 109: 1925–1930. 10.1073/pnas.1115025109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lietzke SE, Yoder MD, Keen NT, Jurnak F (1994) The three-dimensional structure of pectate lyase E, a plant virulence factor from Erwinia chrysanthemi. Plant physiology 106: 849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mitraki A, Miller S, van Raaij MJ (2002) Review: conformation and folding of novel beta-structural elements in viral fiber proteins: the triple beta-spiral and triple beta-helix. Journal of structural biology 137: 236–247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.