Summary
The stringent response, mediated by second messenger (p)ppGpp, results in swift and massive transcriptional reprogramming under nutrient limited conditions. In this study, the role of (p)ppGpp on virulence of P. syringae pv. syringae B728a (PssB728a) was investigated. The virulence of the relA/spoT (ppGpp0) double mutant was completely impaired on bean, and bacterial growth was significantly reduced, suggesting that (p)ppGpp is required for full virulence of P. syringae. Expression of T3SS and other virulence genes was reduced in ppGpp0 mutants. In addition, ppGpp deficiency resulted in loss of swarming motility, reduction of pyoverdine production, increased sensitivity to oxidative stress and antibiotic tolerance, as well as reduced ability to utilize γ-amino butyric acid. Increased levels of ppGpp resulted in reduced cell size of PssB728a when grown in a minimal medium and on plant surfaces, while most ppGpp0 mutant cells were not viable on plant surfaces 24 h after spray inoculation, suggesting that ppGpp-mediated stringent response temporarily limits cell growth, and might control cell survival on plants by limiting their growth. These results demonstrated that ppGpp-mediated stringent response plays a central role in P. syringae virulence and survival, and indicated that ppGpp serves as a global signal for regulating various virulence traits in PssB728a.
Keywords: Alarmone, virulence, survival, T3SS, toxin, siderophore, GABA, Pseudomonas
Introduction
The plant phyllosphere represents a challenging environment for survival of microbes, as they are exposed to various stress conditions, including high doses of UV irradiation, fluctuations in temperature and relative humidity, as well as limited supply of nutrients (Beattie and Lindow, 1995). To survive such harsh environments, microbes must have undergone adaptation by regulating their physiological processes accordingly. Pseudomonas syringae, an important bacterial pathogen of many plant species and an excellent epiphyte, has developed such mechanisms for survival in hostile environments, and often establishes large populations on leaf surfaces prior to infection (Hirano and Upper, 1990, 2000).
P. syringae pv. syringae employs several strategies for causing disease, including the indispensable hrp-type III secretion systems (T3SS) and its secreted effectors, as well as phytotoxins (Bender et al., 1999; Xin and He, 2013; Yu et al., 2013). T3SS is transcriptionally regulated by a sigma factor cascade, including HrpL, a member of the extracytoplasmic function (ECF) family of alternative sigma factors, and sigma 54 (RpoN), which in turn interacts with the bacterial enhancer-binding proteins HrpS and HrpR (Alarcon-Chaidez et al., 2003; Tang et al., 2006; Thakur et al., 2013). Earlier studies have revealed that T3SS genes are induced when bacteria are in contact with plants or grown in minimal media that mimic conditions in plant apoplasts, including low nutrients, low pH, and relatively low temperature, but are repressed when grown in rich media (Anderson et al., 2014; Huynh et al., 1989; Rahme et al., 1992; Stauber et al., 2012). Despite recent efforts to better understand the critical role of T3SS, we are yet to determine the environmental host-pathogen signals that modulate T3SS and unravel the activation of sigma factor cascade.
Nucleotide second messengers, including c-di-GMP, c-di-AMP, cGMP, cAMP and (p)ppGpp, represent major signal transduction mechanisms in bacteria. These cyclic and linear nucleotides control diverse cellular processes in response to environmental cues for an organism to survive. Among these, two unusual linear nucleotides, guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp) (collectively known as (p)ppGpp, thereafter referred to as ppGpp), have been first discovered as bacterial “alarmone” compounds produced in response to nutrient starvation (e.g. lack of amino acids, phosphates, fatty acids, carbon, or iron) (Dalebroux and Swanson, 2012). Stress responses coordinated by ppGpp ultimately redirect the global transcriptional capacity of a cell from those genes responsible for growth and reproduction towards those responsible for survival, therefore referred to as the “stringent response” (Dalebroux and Swanson, 2012; Potrykus and Cashel, 2008).
In proteobacteria, synthesis and degradation of ppGpp are controlled by RelA and SpoT homologue (RSH) proteins. The ribosome-associated RelA protein synthesizes ppGpp using ATP and GTP in response to amino acid starvation, sensed by the presence of uncharged tRNA molecules in the A site of a ribosome (Haseltine and Block, 1973; Magnusson et al., 2005). The cytoplasmic SpoT is activated in response to lack of fatty acids, carbon, phosphorous, and iron, as well as to hyper-osmotic shock and oxidative stresses. In addition, SpoT also has a ppGpp hydrolase activity that mediates ppGpp turnover (Magnusson et al., 2005; Xiao et al., 1991). In Escherichia coli, global changes of cellular transcription during the stringent response result from direct interactions among RNA polymerase (RNAP), ppGpp, and its partner transcription factor DksA, that lead to down-regulation of highly expressed stable RNA, DNA replication, ribosome and protein synthesis, and simultaneous up-regulation of stress and starvation genes, as well as virulence gene expression.
In general, ppGpp directly inhibits transcription of genes involved in cell growth and cell division, and activates transcription of genes for amino acid biosynthesis by directly binding to RNAP (Barker et al., 2001; Potrykus and Cashel, 2008; Ross et al., 2013). In addition, ppGpp activates expression of many stress response genes, including oxidative and osmotic stress genes, via an indirect mechanism, known as σ-factor competition (Dalebroux and Swanson, 2012; Potrykus and Cashel, 2008). During a stringent response, ppGpp inhibits binding of RNAP to σ70-dependent stringent promoters, thus allowing more RNAP to bind to alternative σ-factors, such as RpoN, and promotes expression of alternative σ-factor-dependent genes. Importantly, inhibition of bacterial growth by ppGpp is transient and reversible, thus enabling rapid and reversible control of stress response processes (Dalebroux and Swanson, 2012).
Many studies have demonstrated that ppGpp is central to many processes related to survival and virulence (Dalebroux et al., 2010; Dalebroux and Swanson, 2012; Kalia et al., 2013). In Salmonella enterica, accumulated ppGpp induces HilA, a master regulator of Salmonella pathogenicity island 1 (SPI1). Furthermore, ppGpp directly interacts with SlyA, a transcriptional activator of pathogenicity island 2 (SPI2) to facilitate the intracellular virulence program of S. enterica (Pizarro-Cerdá and Tedin, 2004; Ramachandran et al., 2014). In E. coli, accumulation of ppGpp activates LEE gene expression and increases bacterial adherence (Nakanishi et al., 2006). For plant-associated microorganisms, ppGpp is required for nodulation, antibiotic production, Ti plasmid transfer, quorum sensing signal molecule and cell-wall degrading enzyme production (Bowden et al., 2013; Erickson et al., 2004; Khakimova et al., 2013; Moris et al., 2005; Schafhauser et al., 2014; Takeuchi et al., 2012; Vogt et al., 2011; Wang et al., 2007). However, it remains unknown as to whether or not ppGpp regulates T3SS in plant pathogenic bacteria.
The goals of this study were to determine whether ppGpp controls T3SS gene expression in P. syringae pv. syringae B728a (PssB728a), investigate the effects of ppGpp on other virulence traits, as well as determine whether ppGpp controls cell survival in a plant environment. Our results, for the first time, have provided strong evidence that ppGpp serves as an internal signal for regulating T3SS and other virulence factors in P. syringae.
Results
The RelA-SpoT system in Pseudomonas syringae pv. syringae B728a (PssB728a)
Based on the genome sequence of PssB728a, three genes are found to encode RelA-SpoT Homologue proteins (RSH). Among these, Psyr-3695 and Psyr-0209, annotated as relA and spoT genes, respectively, encode multi-domain ppGpp synthase and bifunctional synthase/hydrolase; while Psyr-4976 (pbcSpo2) encodes a small RSH protein, containing a single hydrolase domain (Atkinson et al., 2011). Following Blast-search, it is revealed that the majority of sequenced pseudomonads contain similar RSH genes as those found in PssB728a, except for P. syrinage pv. tomato DC3000 and pv. maculicola ES4326, both of which contain relA, spoT, and a gene encoding a small RSH protein with a single synthase domain (Chatnaparat et al., unpublished).
To investigate the role of ppGpp in virulence of PssB728a, relA, spoT and relA/spoT double mutants of PssB728a were constructed using overlap extension mutagenesis. However, to construct deletion mutant of pbcSpo2 was unsuccessful. In E. coli, a spoT deletion mutant has been reported to be lethal due to accumulation of high levels of ppGpp (Xiao et al., 1991). In this study, a single deletion mutant of spoT was obtained as reported in P. fluorescens, which also contains pbcSpo2 (Takeuchi et al., 2012). The recovery of such a mutant could be attributed to the presence of pbcSpo2, thus suggesting that pbcSpo2 might serve as a hydrolase of ppGpp.
When intracellular ppGpp levels were determined by LC/MS/MS, it was found that ppGpp in PssB728a wild type (WT) strain was 6.13 nmol per optical density at 600nm per ml in HMM medium at 30 min post incubation (Table 1). Levels of ppGpp in the relA mutant were 40% lower than those in the WT strain; while these were more than two-fold higher in spoT mutant (Table 1). Levels of ppGpp in the relA/spoT double mutant (thereafter referred to as ppGpp0) were undetectable (Table 1). These findings indicate that both RelA and SpoT are required for ppGpp synthesis under HMM conditions, and also suggest that SpoT is the main hydrolase responsible for degradation of ppGpp in PssB728a, while pbcSpo2 might also play a minor role in ppGpp degradation.
Table 1.
Levels of ppGpp and avrPto promoter activities in WT and mutant strains
| Strains | ppGpp concentration (nmol/OD600/ml) | Geometric mean of GFP intensity of the avrPto promoter |
|---|---|---|
| B728a | 6.13 ± 0.28 | 4.77 ± 0.15 |
| ΔrelA | 3.84 ± 0.22 | 3.4 ± 0.23 |
| ΔspoT | 17.12 ± 0.48 | 3.97 ± 0.49 |
| ΔrelA/spoT | ND | 2.73 ± 0.27 |
The bacterial alarmone ppGpp is required for full virulence of P. syringae B728a on bean
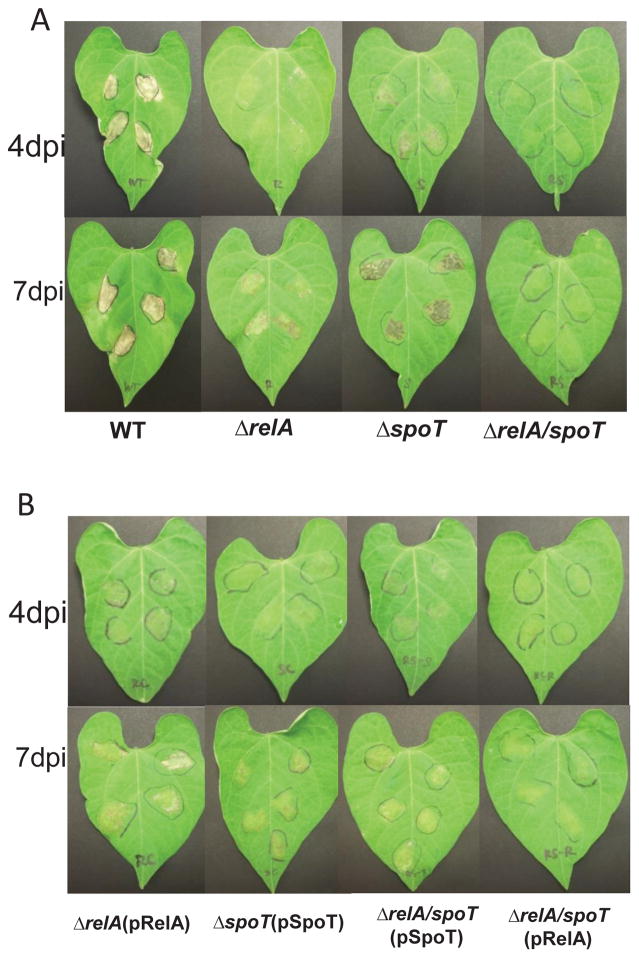
To determine the effect of ppGpp on virulence of PssB728a on bean plants, WT, relA, spoT and ppGpp0 mutant were inoculated into bean leaves using syringe infiltration (Hirano et al., 1999). The WT strain showed full necrotic lesions on bean leaves four days post-inoculation (Fig. 1A). In contrast, the ppGpp0 mutant was unable to produce any disease symptoms on bean plants at both four and seven days post inoculation. Both relA and spoT mutants were capable of causing disease, but with reduced severity when compared to the WT strain (Fig. 1A). Virulence of the ppGpp0 mutants could be partially restored by complementation with relA or spoT gene in trans, despite the fact that complementation strains remained less virulent than those of the WT, relA, and spoT mutants (Fig. 1B). These results suggest that ppGpp synthesis by both RelA and SpoT are essential for eliciting full virulence of PssB728a on bean.
Fig. 1. Pathogenicity of Pseudomonas syringae pv. syringae B728a wild type (PssB728a), relA/spoT mutants and complementation strains.
(A) Disease symptoms caused by PssB728a, relA, spoT, and relA/spoT mutants in susceptible beans. (B) Symptoms caused by complementation strains of relA, spoT, and relA/spoT mutants in susceptible bean. Beans were inoculated using needleless syringe with bacterial suspensions at 106 CFU/ml. DPI, days post-inoculation. The experiment was repeated at least three times and similar results were obtained.
To assess whether ppGpp is required for a hypersensitive response (HR) in tobacco, it was found that bacterial cells of WT and all three mutants infiltrated at higher concentrations (OD600 = 0.1) elicited typical HR in tobacco leaves. However, when bacterial cells were infiltrated at lower concentrations (OD600 = 0.01), only the ppGpp0mutant failed to elicit HR symptoms (Fig. S1A), suggesting that ppGpp may be required for efficiently eliciting HR.
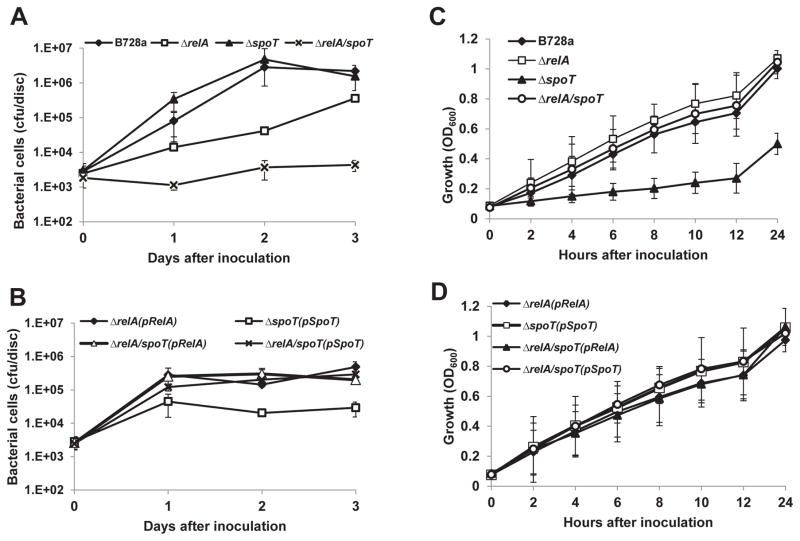
To determine whether attenuation of disease symptoms was correlated with reduced bacterial growth in plant tissues, growth of relA, spoT and ppGpp0 mutants in bean leaves was compared with that of the WT. It was observed that growth of WT and the spoT mutant was similar, reaching 107 CFU/disc two days post-inoculation (DPI) (Fig. 2A). The relA mutant appeared to grow slower, but eventually reached a similarly high population size as the WT strain (Fig. 2A). In contrast, growth of the ppGpp0mutant was about 1000fold lower than that of the WT in bean plants at all three time points (Fig. 2A). Moreover, growth of the complementation strains was partially recovered in bean plants, particularly for the ppGpp0 mutant (Fig. 2B).
Fig. 2. Growth of Pseudomonas syringae pv. syringae B728a (PssB728a), relA/spoT mutants and complementation strains.
(A) Growth of PssB728a, relA, spoT, and relA/spoT mutants in bean leaves. (B) Growth of relA, spoT, and relA/spoT mutant complementation strains in bean leaves. Bacterial growth was monitored at 0, 1, 2 and 3 days post-infiltration. (C) Growth of PssB728a, relA, spoT, and relA/spoT mutants in King’s medium B (KB). (D) Growth of relA, spoT, and relA/spoT mutant complementation strains in KB. OD600nm = optical density at 600 nm. Similar results were obtained in biologically repeated independent experiments. Vertical bars are standard deviations of means.
A classical stringent response mediated by ppGpp leads to growth arrest, resulting in lethality in spoT single mutant in E. coli (Xiao et al., 1991). To rule out the likelihood that slow growth of mutants in plants is due to bacterial growth defect, growth of relA, spoT and ppGpp0 mutants in M9 minimal medium and in KB rich medium was determined. It was found that there were no differences for all strains in M9 medium (data not shown). However, growth of the spoT mutant was highly reduced in KB medium compared to those of other strains, including relA and ppGpp0 mutants, which showed similar growth as that of the WT (Fig. 2C). Complementation of the spoT mutant with a plasmid expressing spoT gene in trans resulted in recovery of its growth (Fig. 2D). Taken together, these above results suggest that even under nutrient rich conditions, bacterial cells of the spoT mutant might accumulate inhibitory levels of ppGpp. This further indicates that SpoT is a major hydrolase responsible for ppGpp degradation, despite the presence of pbcSpo2, which may play a minor role in ppGpp degradation.
The bacterial alarmone ppGpp regulates T3SS gene expression
As the ppGpp0 mutant of PssB728a was incapable of causing disease, the effects of ppGpp on T3SS gene expression was investigated. When promoter activities of the effector avrPto gene were determined for WT and mutants carrying avrPto promoter fused to a promoterless GFP in HMM medium by flow cytometry, the geometric means of GFP intensities of the avrPto promoter in relA, spoT single mutants, and WT were 3.4, 3.97 and 4.77, respectively (Table 1); whereas the avrPto promoter activity was barely detectable in the ppGpp0 mutant (GFP intensity of 2.73 as compared to the vector control (GFP intensity of 2.54). When the WT and ppGpp0 mutant were spray-inoculated on bean leaves, only WT, but not the ppGpp0 mutant, demonstrated GFP expression at 72 h post inoculation (Fig. S1B). These results indicate that ppGpp is required for avrPto gene expression both in vitro and in vivo.
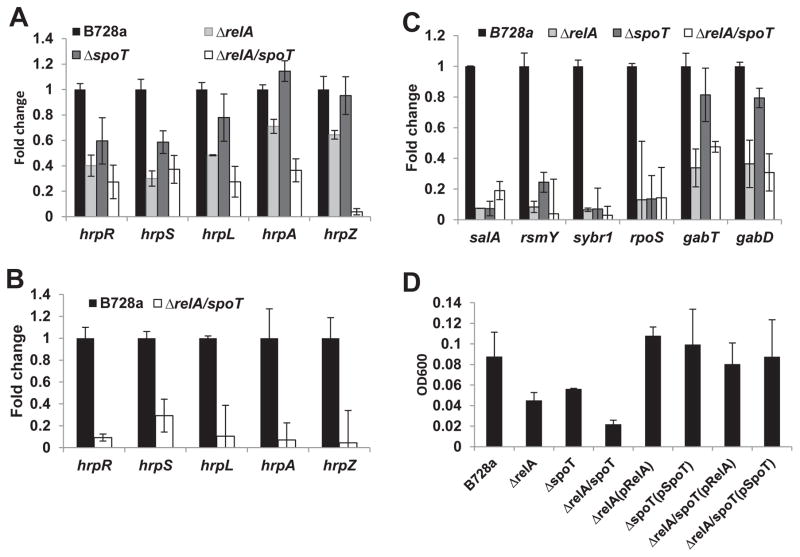
When expression of selected T3SS genes were monitored using qRT-PCR, it was found that expression of hrpR, hrpS, hrpL, hrpA and hrpZ in the ppGpp0 mutant was down-regulated more than three-fold compared tothat of the WT in HMM medium (Fig. 3B). In the relA mutant, expression of hrpL, hrpR and hrpS, but not of hrpA and hrpZ, was about two-fold lower than that of WT. In contrast, expression of hrpR and hrpS, but not of hrpL, hrpA and hrpZ, in the spoT mutant was also slightly lower than those of WT (Fig. 3A). The latter findings were consistent with symptoms observed in bean plants (Fig. 1A). In bean plants, expression of hrpR, hrpS, hrpL, hrpA and hrpZ in the ppGpp0mutant was down-regulated more than five-fold as compared with those of the WT (Fig. 3B). These results indicate that ppGpp is required for virulence in PssB728a by activating T3SS and effector genes.
Fig. 3. Virulence gene expression of Pseudomonas syringae pv. syringae B728a (PssB728a) and relA/spoT mutants.
(A) Expression of T3SS genes in relA, spoT, and relA/spoT mutant strains as compared to PssB728a WT strain grown in hrp-inducing medium 3 h post inoculation determined by qRT-PCR. (B) Expression of T3SS genes in relA/spoT mutant strain as compared to PssB728a WT 18 h post infiltration into bean leaves determined by qRT-PCR. (C) Expression of salA, rsmY, syrB, rpo, gabT and gabD in relA/spoT mutant strain compared to PssB728a 18 h post infiltration into bean leaves by qRT-PCR. The rpoD gene was used as a control. Similar results were obtained in biologically repeated independent experiments. (D) Growth of PssB728a, relA/spoT mutants and complementation strains in minimal medium supplemented with 10mM GABA at 28 °C for 24 h. Vertical bars are standard deviations of means.
The bacterial alarmone ppGpp regulates small RNA and other virulence gene expression in P. syringae B728a
In P. syringae, the Gac/Rsm system is required for lesion formation, phytotoxin production, siderophore biosynthesis, and swarming motility (Chatterjee et al., 2003; Kinscherf and Willis, 1999; Rich et al., 1994). It has also been suggested that the Gac/Rsm system and ppGpp may be involved in a reciprocal regulation (Bowden et al., 2013; Takeuchi et al., 2012). To determine the effects of ppGpp on the functionality of the Gac/Rsm system, transcriptional expression of selected genes, including small RNA rsmY and rsmZ sRNA, salA (encodes a regulator downstream of Gac regulon), syrB1, rpoS, and gabTD were determined. It was observed that expression of rsmY, salA, syrB1, and rpoS, but not of rsmZ sRNA, was down-regulated five- to ten-fold in all mutants when compared to those of the WT (Fig. 3C).
It has been reported that genes involved in the GABA metabolism of PssB728a are strongly induced in planta (Yu et al., 2013). Expression of gabTD genes, which encode proteins involved in non-protein amino acid Gamma-amino butyric acid (GABA) metabolism, was also down-regulated in all mutants, and their expression patterns were highly similar to those of T3SS genes (Fig. 3B), suggesting that ppGpp is required for GABA metabolism. Furthermore, when ppGpp mutants and WT strain were grown in minimal medium containing 10 mM GABA as sole carbon or nitrogen source, bacterial growth of WT cells reached to OD600 about 0.1 at 24h; whereas cells of the ppGpp0 mutant failed to grow. Under similar conditions, growth of relA and spoT mutants were much slower than those of the WT (Fig. 3D). Complementation of mutants with corresponding genes introduced in trans restored their growth (Fig. 3D). These results suggest that P. syringae might require ppGpp for utilizing GABA as the sole carbon and nitrogen source.
The bacterial alarmone ppGpp is involved in pyoverdine production and confers resistance to hydrogen peroxide and rifampicin
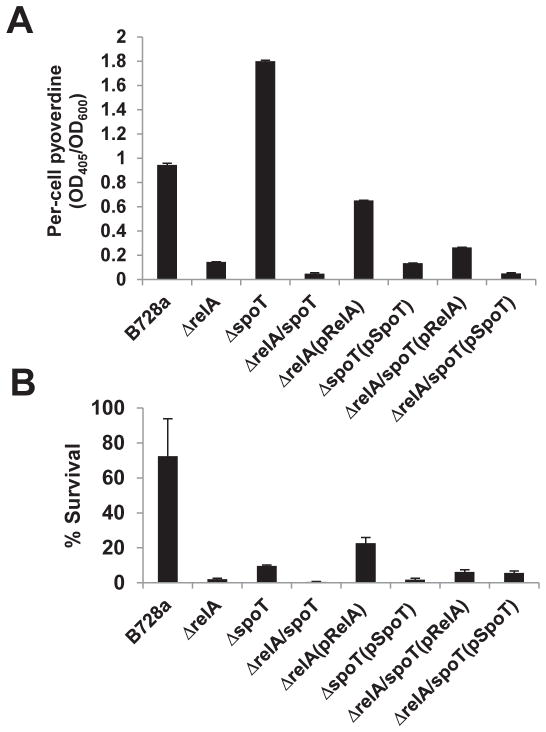
When grown on KB plates, relA and ppGpp0 mutants produced markedly less yellow-green fluorescent pigments (data not shown). This observation prompted evaluating the effects of ppGpp on production of pyoverdine, a fluorescent siderophore secreted by P. syringae. It was found that under low iron conditions, pyoverdine production was strongly reduced in both relA and the ppGpp0 mutants (Fig. 4A), while elevated levels of pyoverdine were produced in the spoT mutant as compared to the WT. Complementation strains restored the levels of pyoverdine (Fig. 4A). These results suggest that ppGpp might act as a secondary contextual signal to control pyoverdine production, possibly in response to availability of iron.
Fig. 4. ppGpp regulates pyoverdine production and oxidative stress tolerance.
(A) Pyoverdine production by Pseudomonas syringae pv. syringae B728a (PssB728a), relA, spoT, and relA/spoT mutant strains as well as complementation strains of relA, spoT, and relA/spoT mutants grown in Mannitol–Glutamate (MG) medium at 28 °C for 24 h. Pyoverdine levels were determined by measuring the absorbance at 405 nm of cell-free culture supernatants in MG medium normalized by the absorbance at 600 nm of the bacterial culture. Data was presented as relative fluorescence levels (A405/A600). The experiments were repeated three times with similar results. (B) Survival of PssB728a, relA, spoT, and relA/spoT mutant strains as well as complementation strains after exposed to 1mM hydrogen peroxide for 15 min. The vertical bars correspond to the standard deviations of means. Similar results were obtained in biologically repeated independent experiments. Vertical bars are standard deviations of means.
The stringent response is required for hydrogen peroxide and antibiotic tolerance (Braeken et al., 2006; Khakimova et al., 2013). Therefore, the survival of ppGpp mutants following exposure to hydrogen peroxide and rifampicin was determined. After exposure to 1 mM H2O2, survival of relA, spoT and ppGpp0 mutants was drastically reduced, with 2.02, 9.6, and 0.5 % survival, respectively, as compared to 72.3 % for the WT (Fig. 4B). Complementation of relA and ppGpp0 mutants partially recovered their abilities to survive following 1mM H2O2 treatment. However, complementation of the spoT mutant with a plasmid carrying the spoT gene in trans resulted in high sensitivity to1 mM H2O2 treatment. In addition, relA and ppGpp0 mutants were more sensitive to rifampicin with minimum inhibition concentration (MIC) at 312.5 and 156.25 μg/ml, respectively; whereas both WT and spoT mutant survived at 625 μg/ml(MIC) (Table 2). The reduced levels of rifampicin resistance could be complemented by expression of relA and spoT genes in trans in corresponding mutants. These results suggest that ppGpp may be required for resistance to oxidative stress and involved in antibiotic tolerance.
Table 2.
Susceptibility of P. syringae pv. syringae strains to rifampicin
| Strains | MIC (μg/ml)a |
|---|---|
| B728a | >625 |
| ΔrelA | 312.5 |
| ΔspoT | 625 |
| ΔrelA/spoT | 156.25 |
| ΔrelA(pRelA) | 625 |
| ΔspoT(pSpoT) | 312.5 |
| ΔrelA/spoT(pRelA) | 312.5 |
| ΔrelA/spoT(pSpoT) | 312.5 |
Minimal inhibition concentrations (MIC) determination by the microdilution assay in King’s medium B. The experiment was repeated at least three times and similar results were obtained.
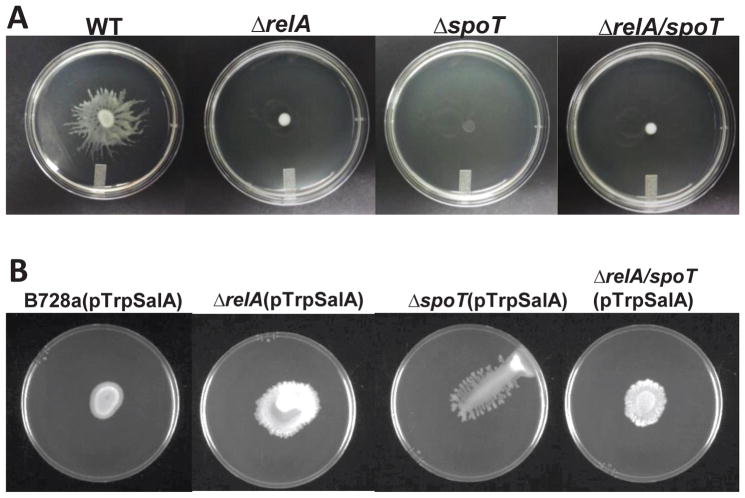
Overexpression of SalA restores swarming motility in ppGpp mutants
When the swarming motility of ppGpp mutants was assessed on a semisolid agar plates, all three mutants failed to swarm (Fig. 5A). Surprisingly, this phenotype could not be complemented by expressing corresponding genes in trans (data not shown). It has been reported that SalA positively regulates SyfA and SyfR (LuxR-type regulators), required for the production of surfactant syringafactin, which is essential for swarming motility (Berti et al., 2007; Burch et al., 2010; Hockett et al., 2013). We hypothesized that ppGpp might control swarming motility through SalA (Fig. 3C). To test this hypothesis, SalA was overexpressed in mutants as well as WT background through constitutive expression of the salA gene. The ability to swarm in ppGpp mutants expressing salA was partially restored (Fig. 5B); whereas the swarming motility of the WT overexpressing salA was reduced when compared to that of the parental strain. However, the morphology of swarming in the salA overexpression strains was different from that of the WT (Fig. 5), suggesting that ppGpp may regulate swarming through multiple pathways, including SalA.
Fig. 5. ppGpp controls swarming motility.
(A) Swarming motility of P. syringae pv. syringae B728a (PssB728a), relA, spoT, and relA/spoT mutant strains on swarming plates (KB containing 0.4% agar) 24 h post inoculation. (B) Swarming motility of PssB728a, relA, spoT, and relA/spoT mutant strains constitutively over-expressing salA gene on KB swarming plates for 24 h. The experiments were repeated at least three times with similar results.
The bacterial alarmone ppGpp controls bacterial cell size and survival
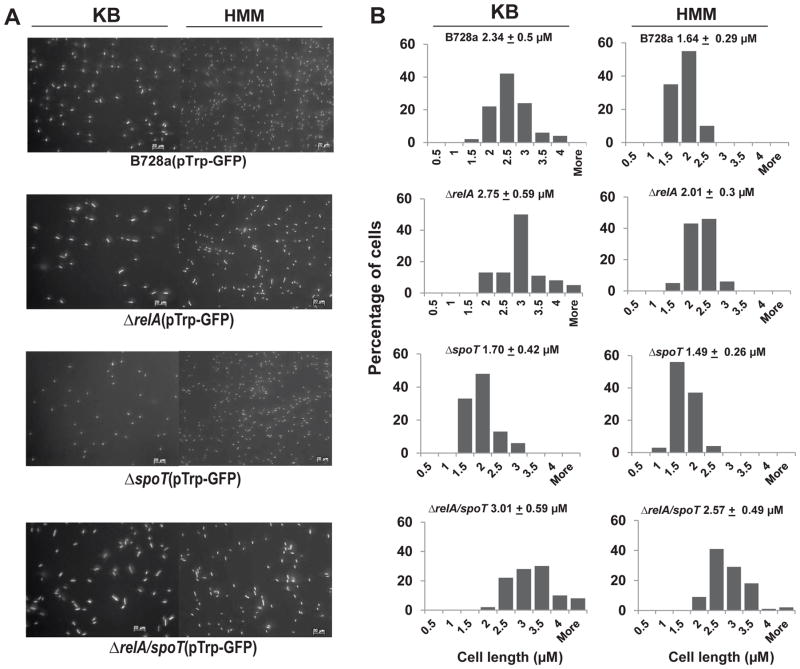
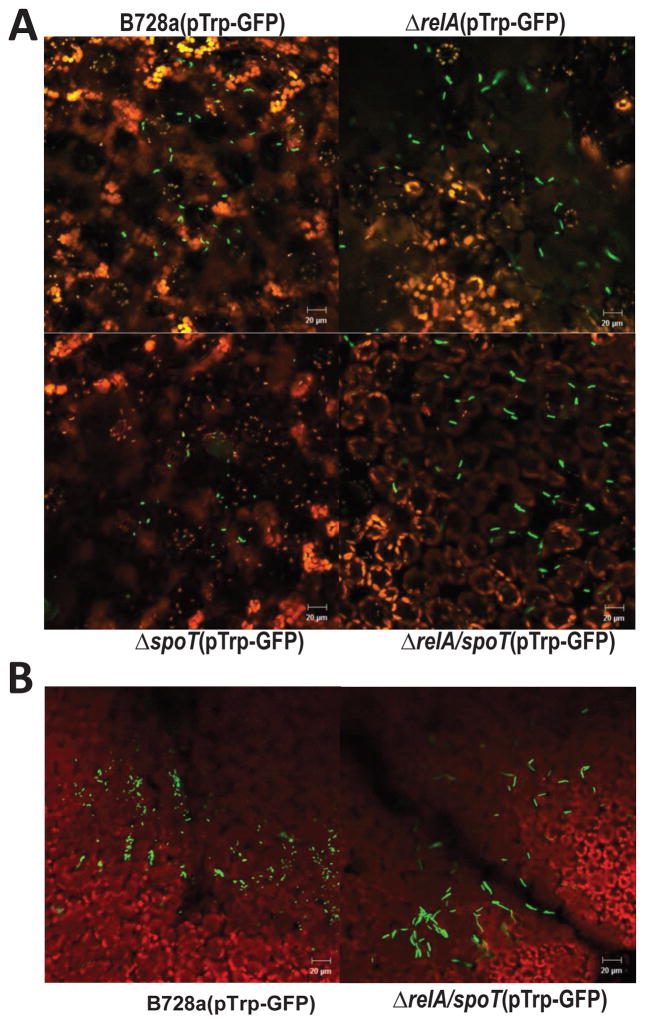
Under nutrient limitation conditions, bacterial stringent response directly inhibits ribosome and DNA replication (Dalebroux and Swanson, 2012; Potrykus et al., 2011). When grown in nutrient rich KB medium, the average length of WT cells was about 2.34 μm; whereas, the lengths of relA and ppGpp0 mutant cells were slightly longer, reaching 2.75μm and 3.0 μm, respectively, measured using epifluorescence microscopy. In contrast, the length of spoT mutant was 1.7 μm (Fig. 6). On the other hand, when grown in nutrient limited HMM medium, WT cells were shorter than those grown in KB medium (Fig. 6). The average length of WT cells was about 1.64 μm; while the lengths of relA and ppGpp0 mutant cells reached 2.01μm and 2.57 μm, respectively. In contrast, the length of spoT mutant measured only 1.49 μm (Fig. 6). Using confocal microscopy, similar change in cell size was observed when bacterial cells were spray inoculated onto bean leaves (Fig. 7A). At 4 h post inoculation, ppGpp0 cells were longer than those of relA mutant and WT, and lengths of spoT mutant cells were the shortest. Similar results were obtained for WT and ppGpp0 cells at 24 h post inoculation (Fig. 7B). These findings indicate that reduced lengths of spoT mutant cells and increased lengths of relA and ppGpp0 cells might be due to the accumulated amounts of ppGpp in these cells, thus suggesting that ppGpp controls cell size in response to nutrient availability in medium and on plant surfaces.
Fig. 6. ppGpp controls cell size in medium.
(A) Microscopy images of P. syringae pv. syringae B728a (PssB728a), relA, spoT, and relA/spoT mutant strains constitutively expressing green fluorescent protein (GFP) observed by epifluorescence microscope after grown in KB and in HMM medium for 4 h. Pictures were taken at 200 X magnification with a scale bar of 20 μm. (B) Distribution of bacterial sizes and average bacterial cell lengths of PssB728a, relA, spoT, and relA/spoT mutant strains constitutively expressing GFP in KB and HMM media. The experiments were repeated at least two times with similar results.
Fig. 7. ppGpp controls cell size survival on plants.
(A)Microscopy images of P. syringae pv. syringae B728a (PssB728a), relA, spoT, and relA/spoT mutant strains constitutively expressing green fluorescent protein (GFP) observed by confocal laser scanning microscopy 3 h following spray inoculation on bean leaf surfaces; (B) Microscopy images of PssB728a and relA/spoT mutant strain constitutively expressing GFP observed by confocal laser scanning microscopy 24 h post spray inoculation on bean leaf surface. GFP-labeled bacterial cells are green, and leaf cells are either red or black in color. Pictures were taken at 200x magnification with a scale bar of 20 μm. The experiments were repeated three times with similar results.
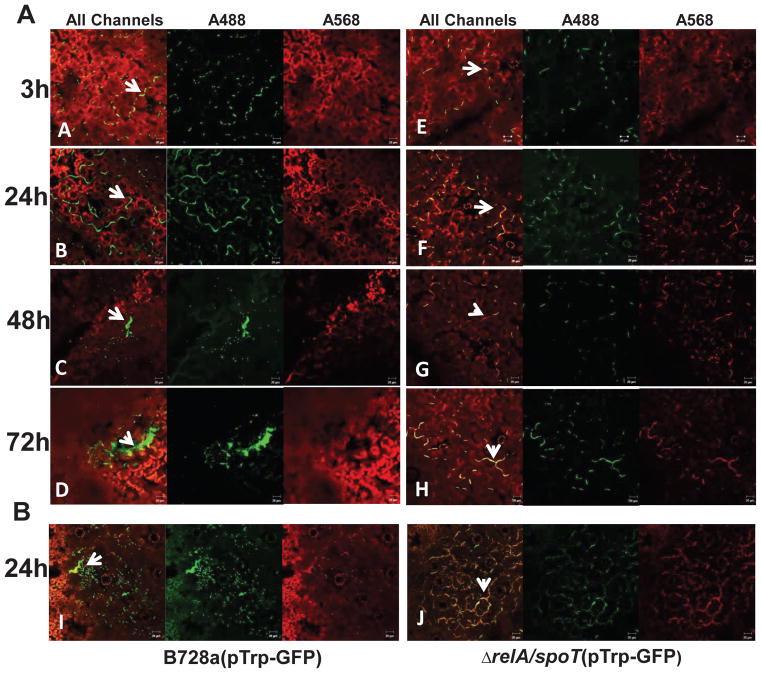
To determine whether ppGpp influences bacterial cell survival on the leaf surfaces, WT and ppGpp0 mutants on both host (bean) and non-host (tobacco) plants were compared. Similarly, strains constitutively expressing GFP were spray-inoculated onto leaf surfaces and observed by confocal microscopy. Propidium iodide was used to distinguish membrane-compromised cells or dead cell (stained in red) from viable cells (retained in green). At 3 h post inoculation, WT and ppGpp0cells were found as individual cells and in small groups along epidermal cell surfaces (Fig. 8A, E). At 24 h, most WT cells were observed as small aggregates along epidermal cell surfaces, particularly in grooves along veins and cell junctions (Fig. 8B). After 48 h, WT cells observed on leaf surface were alive and formed large aggregates around stomatal areas and these aggregates became larger at 72 h (Fig. 8C, D). In contrast, most ppGpp0 mutant cells were found to be dead on epidermal cells and cell junctions by 24 h, and no aggregates were observed at 48 and 72 h (Fig. 8F, G, H). In addition, on non-host tobacco leaf surfaces, the ppGpp0 mutant could not survive, and the numbers of dead cells were higher than those of WT, which were smaller in size (Fig. 8I, J). These results indicate that ppGpp controls survival of PssB728a on both host and non-host leaf surfaces.
Fig. 8. ppGpp mediates survival on plants.
(A)Microscopy images of P. syringae pv. syringae B728a (PssB728a) (A–D) and relA/spoT mutant strains (E–H) constitutively expressing green fluorescent protein (GFP) observed by confocal laser scanning microscopy at 3, 24, 48 and 72 h post spray inoculation on bean. (B) Microscopy images of PssB728a (I) and relA/spoT mutant strain (J) constitutively expressing GFP and observed by confocal laser scanning microscopy 24 h post spray inoculation on tobacco leaf surface. GFP-labeled bacterial cells were green, dead bacterial cells stained with propidium iodide were red, and leaf cells were either red or black in color. White arrow indicates GFP-labeled bacteria. Pictures were taken at 200 X magnification with a scale bar of 20 μm.
Discussion
As one of the most studied strains of P. syringae, PssB782a requires an epiphytic phase on leaf surfaces before causing disease on beans. This process is also dependent on the onset of T3SS and other virulence factors to suppress host defense responses and survive the hostile leaf surface environment, which is also nutrient limited (Lee et al., 2012). Earlier studies have determined that a sigma factor cascade (HrpL and RpoN) is critical for T3SS and virulence in P. syringae (Alarcón-Chaidez et al., 2003; Ferreira et al., 2006; Hendrickson et al., 2000; Jovanovic et al., 2011; Rahme et al., 1992; Tang et al., 2006). In this study, it has been demonstrated that ppGpp-mediated stringent response is essential for T3SS gene expression and virulence in PssB728a, possibly through activation of the sigma factor cascade. Furthermore, ppGpp controls bacterial cell size and survival both in vitro and on plant surfaces. These findings demonstrate that Pss728a perceives environmental/host signals in order to activate T3SS via synthesis of the intracellular ppGpp and regulation of bacterial pathogenesis, suggesting that signals triggering ppGpp biosynthesis are most likely to be responsible for activation of T3SS.
In many bacterial systems containing only RelA/SpoT, the null mutation of the spoT gene in the relA+ background is found to be lethal, thus suggesting that high levels of ppGpp may be toxic (Bergman et al., 2014; Hernandez and Bremer, 1990; Spira et al., 2008; Sarubbi et al., 1988). In this study, a spoT null mutant was successfully generated in PssB728a, which accumulated higher levels of ppGpp with reduced growth in rich medium, suggesting that this could be due to the presence of a small mono-hydrolase domain protein, pbcSpo2. These results likely indicated that pbcSpo2 is functional in PssB728a as a hydrolase, though a deletion mutant could not be obtained. This finding also suggests that SpoT in PssB728a may function mainly as a hydrolase under nutrient rich conditions. In contrast, growth of the ppGpp0 mutant in bean was completely impaired and the mutant was unable to cause disease, suggesting that lack of ppGpp strongly influences the growth of P. syringae in bean plants. While growth of the spoT mutant was similar to that of the WT in plants, growth of relA mutant was reduced, thus suggesting that accumulation of ppGpp, which may depend on both RelA and SpoT within plants, is required for bacterial multiplication in the plant’s apoplast environment. Although accumulation of ppGpp was higher in the spoT mutant and lower in the relA mutant, when compared to that of WT in vitro, both mutants caused reduced disease symptoms, suggesting that both RelA and SpoT are required for full virulence of PssB728a in beans. These findings indicate that lesion formation in PssB728a might require certain levels of ppGpp.
Previous studies have pointed out that availability of nutrients has an adverse effect on cell size (Abranches et al., 2009; Hill et al., 2013; Liu et al., 2014; Monahan et al., 2014; Traxler et al., 2008; Yao et al., 2012). It has also been reported that cells of smaller sizes become increasingly resistant to abiotic stresses, including acid and oxidative stresses, thus enhancing their ability to survive under harsh environmental conditions (Crompton et al., 2014; Hase, 1999; Jubair et al., 2012; Steinberger et al., 2002). Monier and Lindow (2003b) have reported that lengths of PssB728a cells were rapidly shortened upon inoculation on bean leaf surfaces, suggesting that leaf surfaces offer limited sources of nutrients (Monier and Lindow, 2003b). When E. coli ppGpp mutant cells were grown under isoleucine starvation conditions, they were found to be considerably longer than WT cells (Traxler et al., 2008; 2011). On the other hand, bacterial stringent response are required for tolerance to hydrogen peroxide, antibiotics, as well as for other biotic stresses such as osmotic stress (Abranches et al., 2009; Khakimova et al., 2013; Vercruysse et al., 2011). In this study, it has been demonstrated that cell size reduction and survival on leaf surfaces was primarily controlled by ppGpp in response to nutrient availability. It was found that ppGpp0 cells were unable to adjust their size, and most larger/longer cells were not viable within 24 h after inoculation onto both bean and tobacco leaf surfaces. Furthermore, lengths of spoT mutant cells were significantly shorter than those of WT cells, suggesting that spoT mutant cells have smaller cell size and accumulate higher amount of ppGpp. Furthermore, all ppGpp mutants of PssB728a were more susceptible/sensitive to oxidative stress, suggesting that reduced cell size is related to tolerance to abiotic stresses (therefore survival), and may both be under positive regulation of ppGpp. In addition, since both rifampicin and ppGpp target RNAP, it will be interesting to investigate why high level of ppGpp leads to increased resistance to rifampicin in the future.
Although signals that activate T3SS in P. syringae remains unknown, it has been reported that T3SS of PssB728a was induced on leaf surfaces and required for epiphytic survival and growth (Lee et al., 2012). In this study, it was found that ppGpp positively regulates the expression of hrpRS, hrpL, hrpZ and avrPto both in vitro and in planta, suggesting that ppGpp is required for T3SS expression on leaves. As ppGpp indirectly controls transcription via sigma factor competition (Dalebroux et al., 2010; Dalebroux and Swanson, 2012; Magnusson et al., 2005), it is proposed that ppGpp accumulation leads to increased transcription of genes regulated by alternative sigma factors, such as σ54 (RpoN), which in turn activate hrpL gene expression, thus suggesting that ppGpp could serve as an internal signal for T3SS activation (Tang et al., 2006). In E. coli, transcription of relA is activated by NtrC during nitrogen starvation (Brown et al., 2014). Similar to HrpS and HrpR, NtrC is a bacterial enhancer binding protein, required for RpoN-dependent gene expression, including nitrogen metabolism genes. Therefore, it is likely that P. syringae employs similar mechanism as well. However, further studies are needed to support this finding.
Besides T3SS, ppGpp regulates other virulence-associated traits, including toxin, oxidative stress response, and siderophore production. In this study, transcription levels of salA, rsmY, and sybr1 were found to be five-folds lower in ppGpp0 mutant than in the WT. Furthermore, SalA, a LuxR family regulator protein, acts downstream of the GacS/GacA signal transduction system and is required for pathogenicity, as well as syringomycin and syringopeptin production (Kitten et al., 1998). It has been reported that SalA positively regulates expression of the syringomycin synthetase gene syrB1, syrF, and rsmY (Lu et al., 2002, 2005; Wang et al., 2006). In addition, salA also positively regulates syfR and syfA, which controls swarming motility of P. syringae by regulating syringafactin production (Hockett et al., 2013). Interestingly, when over-expression salA, the swarming deficient phenotype was partially restored in all ppGpp mutants, indicating that ppGpp may regulate swarming motility partly via SalA. Recent studies demonstrated that there is a reciprocal regulation between ppGpp and the GacS/GacA system. In E. coli, ppGpp activates transcription of csrB and csrC (Edwards et al., 2011). Furthermore, CsrA/RsmA and ppGpp are known to exhibit opposite effects on expression of specific genes, such as flhDC. In P. fluorescens, levels of ppGpp were higher in a gacA mutant, but were lower in a rsmA/E and retS mutant (Takeuchi et al., 2012). In both P. aeroginosa and P. syringae, RetS and LadS reciprocally regulate T3SS and interact with the Gac/Rsm pathway (Records and Gross, 2010; Ventre et al., 2006). Our results showed that ppGpp positively regulates rsmY expression, suggesting that ppGpp likely interacts with the GacS/LadS/RetS multisensory signaling network; however, this requires further investigation.
In E. coli, expression of genes in the RpoS regulon is dependent on ppGpp under both carbon and amino acid starvation conditions (Bougdour and Gottesman, 2007; Traxler et al., 2008; 2011). In P. aeroginosa, it has been reported that expression of catalase genes requires ppGpp through a RpoS-dependent pathway, and ppGpp is required for oxidative stress response in several Pseudomonas species (Dalebroux et al., 2010; Khakimova et al., 2013;Takeuchi et al., 2012). In PssB728a, genes involved in oxidative stress, including katEGN and sodAC genes, were found to be induced in planta, suggesting PssB728a is under oxidative stress in host plants (Yu et al., 2013). Our findings showed that rpoS gene expression was reduced in all mutant strains, and as a consequence, resistance to H2O2 was greatly diminished, particularly in the ppGpp0 strain. On the other hand, pyoverdine production was higher in the spoT mutant, but lower in relA and the ppGpp0 mutants as compared to WT, when cells were grown under low iron conditions. This finding is consistent with observation reported in P. aeroginosa, wherein a relA/spoT double mutant has exhibited reduced production of pyoverdine (Vogt et al., 2011). In contrast, It has been reported that mutation of the relA gene did not influence siderophore production in Pectobacterium carotovora subsp. atroseptica (Wang et al., 2007). Furthermore, growth of a pyoverdine-deficient PssB728a strain on leaf surfaces was highly affected (Karamanoli et al., 2011). Our results suggest that ppGpp contributes to pyoverdine production, thus playing an important role in epiphytic growth and in survival of P. syringae.
As one of the most abundant non-protein amino acid in plants, GABA could serve both as a carbon and nitrogen sources for P. syringae and also play regulatory roles in increased defenses against P. syringae (Park et al., 2010, Yu et al., 2013). In a recent study, it has been revealed that genes involved in metabolism and transport of GABA were highly induced in PssB728a cells exposed to nitrogen limitation conditions and in bean apoplasts (Yu et al., 2013), suggesting that PssB728a could utilize plant derived GABA in bean plants. In addition, increased accumulation of GABA in tomato plants inhibited growth of P. syringae pv. tomato DC3000, suppressed T3SS gene expression, and activated plant defense gene expression (Park et al., 2010). Our results demonstrated that ppGpp affects the ability of PssB728a in utilizing GABA in vitro and positively regulates expression of GABA uptake genes in planta. Thus, it is proposed that, by upregulating GABA catabolic genes, P. syringae could utilize GABA as a nutrient source by reducing GABA levels in planta, thereby promoting bacterial growth and suppressing plant defense responses mediated by GABA.
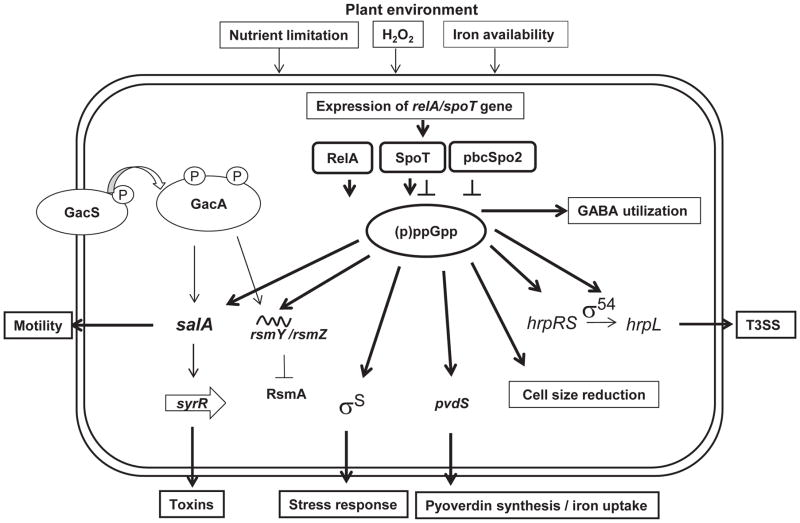
Based on the above and previous findings, the following model is proposed as to how PssB728a incorporates host and environmental signals in regulating virulence and survival (Fig. 9). Upon contact on plant surfaces, P. syringae cells are subjected to stress conditions, including low nutrients, low iron, and oxidative stress, all of which trigger activation of the RelA/SpoT system, leading to accumulation of ppGpp. Subsequently, ppGpp-mediated stringent response indirectly activates alternative sigma factor activities, such as RpoN, leading to expression of T3SS and other virulence factors, as well as stress-related genes. Most P. syringae genomes, including PssB728a, contain 14 alternative sigma factors (Oguiza et al., 2005; Thakur et al., 2013). Among these, 10 ECF sigma factors are involved in regulating T3SS gene expression (HrpL), alginate production (AlgU), iron uptake (PvdS, AcsS, Psyr_1040, Psyr_1107, and Psyr_4731), maintenance of cell shape (SigX), toluene tolerance (Psyr_0362), and cytoplasmic sensing (Psyr_0892)(Oguiza et al. 2005). In addition, the ECF sigma factor Psyr_1107 plays a role in both swarming motility and fimbrial gene expression (Thakur et al., 2013). It is likely that RpoS, global regulator SalA, and GacSA-sRNA-RsmA(RNA-binding protein) may interact with the ppGpp regulatory network, leading to changes in physiological and stress responses, including swarming motility, toxin and siderophore production, tolerance to antibiotics, oxidative stress, cell size reduction, survival, and nutrient acquisition. However, there was not always a direct linear relationship between ppGpp levels and various phenotypes observed in this study. It is possible that quantitative differences in the intracellular ppGpp level determine the precise expression of genes controlling various phenotypes in different environments, including in planta and in medium, which requires further investigation (Traxler et al., 2011). Future studies should also investigate the global effects of ppGpp, both in vitro and in planta, and those signals that activate ppGpp accumulation. Given that the ppGpp0 strain is unable to survive and cause disease in host plants, further studies are needed to target ppGpp in plant pathogenic bacteria for developing control strategies.
Fig. 9. A working model illustrated the role of ppGpp in Pseudomonas syringae pv. syringae B728a (PssB728a) in response to plant and environmental stimuli.
This model derives from findings in this study as well as data reported previously (Hockett et al. 2013; Lu et al. 2005; Records and Gross 2010; Takeuchi et al. 2012). Symbols: ↓, positive effect; ⊥, negative effect; bold lines, results from this study.
Experimental procedures
Bacterial strains, plasmids, and recombinant techniques
Bacterial strains and plasmids used in this study are listed in Table S1. P. syringae pv. syringae strains were cultured on either King’s medium B (KB) or Mannitol–Glutamate (MG) medium at 28°C (Park et al. 2010) or a hrp-inducing minimal medium (HMM) at 22 °C (Huynh et al. 1989). E. coli strains were routinely grown on LB medium at 37°C. Bacterial growth was monitored by measuring absorbance of cell suspensions at 600 nm. All primers used in this study are listed in Table S2. Antibiotics were added to medium at the following concentrations: kanamycin, 50μg/ml; rifampicin, 100μg/ml; tetracycline, 15μg/ml; and spectinomycin, 100μg/ml. All DNA manipulations, including DNA isolation, plasmid extraction, restriction digestion, ligation, and gel electrophoresis were performed as described previously (Sambrook et al., 1989).
Construction of relA, spoT and relA/spoT double mutants
Deletion mutation of relA, spoT and relA/spoT were generated using splice overlap extension mutagenesis (Heckman and Pease, 2007). Briefly, approximately 1-kb upstream and downstream fragments of relA and spoT in P. syringae pv. syringae strain B728a were amplified using primers unique to these genes (Table S2); One primer also contained an extension sequence complementary to a kanamycin resistance cassette flanked with FLP recombination target (FRT) sites from pKD13. The two PCR products contained ends overlapping with those of the kanamycin resistance cassette (Datsenko and Wanner, 2000). The two amplicons were then joined with the kanamycin resistance cassette using overlap extension PCR. The final fragment was then cloned into the pTok2 vector, resulting in pTok2::ΔrelA and pTok2:: ΔspoT, respectively. The resulting plasmids were electroporated into the mobilizing strain E. coli S17-1 λpir. Then, E. coli S17-1 λpir and P. syringae were allowed to mate overnight by biparental mating. Sunsequently, transformants were plated on KB plates containing rifampicin and kanamycin, and the resulting kanamycin resistant, but tetracycline sensitive mutants were selected. Gene disruption was confirmed using PCR, with primers specific to the sequences flanking each of the genes. To generate markerless mutant strains, plasmid pFLP2-omega, expressing FLP recombinase, was electroporated into the mutant strains. Transformants were plated on KB plates containing spectinomycin to allow Flp recombinase-mediated recombination between the FRT sites, resulting in the loss of the kanamycin resistance cassette. The pFLP2-omega plasmid was cured from the mutants by replica-plating on KB plates. Markerless deletions were confirmed by PCR. In order to generate a relA/spoT double deletion mutant, plasmid pTok2::ΔspoT was transferred to the relA markerless mutant strain via conjugation.
Complementation of mutants and generation of SalA overexpression plasmid
For complementation of the relA/spoT mutants, a 2.8-kb fragment containing the native promoter and the relA gene and a 3.1-kb fragment containing native promoter and the spoT and rpoZ genes were amplified using the primer pairs RelAcom-F(BamHI) - RelAcom-R(BamHI) and SpoTcom-F(BamHI) - SpoTcom-R (BamHI), respectively. PCR fragments were digested with BamHI and cloned into the pVSP61 vector to yield plasmids pRelA and pSpoT, which were then introduced into corresponding relA/spoT markerless mutant strains by electroporation, respectively. To construct SalA overexpression plasmid, an E. coli trp operon promoter was fused to the salA gene of PssB728a using primers Ptrp-salA1, Ptrp-salA2, and Ptrp-salA-R. PCR fragment was cloned into the SmaI site of pVSP61 to yield pTrp-SalA plasmid.
Nucleotide extraction and ppGpp measurement by LC/MS/MS
Bacterial strains were grown in 100 ml KB to an OD600 ~ 1. Cells were harvested by centrifugation at 4,000 rpm for 15 min and washed with 5 ml HMM medium. Cell pellets were re-suspended in 100 ml HMM medium and incubated with shaking at 250 rpm at 22°C for 30 min. Subsequently, bacterial cells were pelleted, and cells were immediately suspended in 2 ml 100% cold methanol, vortexed for 50 sec, frozen in liquid nitrogen, and allowed to thaw on ice. Cell suspensions were then centrifuged at 4000 rpm for 10 min at 4 °C. Supernatants were transferred to a new tube and stored on ice, and another 2 ml of 100% cold methanol was added to the pellet. The freeze thaw cycle was repeated, and the methanol extract was then combined, and filtered using 0.22 μm pore size (Millipore) filter. The filtrate was freeze-dried and resuspended in 200 μl distilled water for LC/MS/MS analysis. Pure ppGpp purchased from TriLink Biotech Inc. (CA, USA) was used as a positive control.
Nucleotide-enriched samples were analyzed in a 5500 QTRAP LC/MS/MS system (AB Sciex, Foster City, CA) with a 1200 series HPLC system (Agilent Technologies, Santa Clara, CA), including a degasser, an autosampler, and a binary pump. The LC separation was performed on an Agilent SB-Aq column (4.6×50mm, 5μm) with a mobile phase A (10mM ammonia formate in water) and a mobile phase B (methanol). The flow rate was 0.3 mL/min. The linear gradient was as follows: 0–4 min, 99%A; 4.1–8.0 min, 5%A; and 8.1–13.0 min, 99%A. The autosampler was set at 5°C. The injection volume was 10 μL. Mass spectra were acquired under positive electrospray ionization (ESI) with an ion spray voltage of +5500 V. The source temperature was 450 °C. The curtain gas, ion source gas 1, and ion source gas 2 were 35, 65, and 55psi, respectively. Multiple reaction monitoring (MRM) was used for quantitation of ppGpp (m/z 604.1-->m/z 152.0). The final ppGpp concentration was normalized to OD600.
RNA isolation and quantitative RT-PCR analysis
Bacterial strains, grown overnight at 28 °C in KB medium, were harvested by centrifugation, washed with phosphate buffered saline (PBS), reinoculated in HMM, and then grown for an additional 3 h. An RNA protect reagent (Qiagen, Hilden, Germany) was added to bacterial cells and total RNA was extracted using the RNeasy Mini BacteriaProtect Kit (Qiagen, Hilden, Germany). For in planta conditions, one gram of bacterial-infected plant tissue was grounded into a powder in liquid nitrogen, and total RNA was extracted using Direct-zol RNA MiniPrep (Zymo Research) according to the manufacturer’s protocol. DNA was removed using the Turbo DNA-free kit (Ambion, Austin, TX, U.S.A.) and RNA samples were quantified using a Nano-Drop ND-100 spectrophotometer (Nano-Drop Technologies; Wilmington, DE, USA). cDNA was synthesized from 1μg of RNA using SuperScript III (Invitrogen Life Technologies, Carlsbad, CA, U.S.A.) and random hexamers. qRT-PCR was performed using 100 ng cDNA and a Power SYBR Green PCR Master Mix on a StepOnePlus Real-Time PCR System (Applied Biosystems) following the manufacturer’s specifications. Melting curve analysis was used to verify amplification of a single product. Gene expression was calculated using the relative quantification (ΔΔCt) method. The Ct values of each gene were normalized to Ct values of the housekeeping sigma factor gene rpoD.
Flow cytometry analysis
Bacterial cells were analyzed by flow cytometry (BD Biosciences, San Jose, CA, USA) for induction of the avrPto-GFP as described previously (Wang et al. 2009, Zhao et al. 2009). The WT and mutant strains containing the pAvrPto-GFP were grown overnight in KB, harvested, washed, and resuspended in HMM and grown for 16 h at 18 °C. Subsequently, bacterial cells were diluted in PBS for flow cytometry assays. Flow cytometry was performed using LSRII 10-parameter multilaser analyzer (BD Biosciences). Data were collected for a total of 100,000 events, and analyzed statistically by gating using flow cytometry software FCS Express V3 (De Novo Software, Los Angeles, CA, USA). A geometric mean was calculated for each sample, and the experiment was repeated three times.
Swarming motility, pyoverdine production, oxidative stress, and GABA utilization assay
For swarming motility assay, cells were grown overnight in KB, harvested, and washed in PBS. Cells were resuspended in PBS to a final concentration of 5.0×108 CFU/ml (OD600 = 0.3), and 10 μl was spotted onto the center of swarming plates (0.4% KB agar) and incubated for 24 h at room temperature. Motility of bacterial cells was visually examined.
For pyoverdine production, bacterial cells of overnight cultures in KB were washed, resuspended to an OD at 600 nm of 0.05 in MG medium, and incubated with shaking at 28°C for 24 h. Pyoverdine from supernatants was measured at OD405 and normalized at OD600 (Imperi et al., 2009).
For oxidative stress assay, bacterial cells were grown in KB medium, washed, and resuspended in PBS buffer with or without 1 mM H2O2 to a final concentration of ~ 1.0×108 CFU/ml. Bacterial suspensions were then incubated at room temperature for 15 min. Subsequently, serial dilutions were plated on KB agar to enumerate surviving cells. Percentage survival was calculated by dividing H2O2 treated versus untreated cells. The experiments were repeated at least three times.
For GABA utilization assays, bacterial cells of overnight cultures in KB were washed and resuspended to OD at 600 nm of 0.02 in modified MG medium by replacing mannitol and L-glutamic acid in MG medium with 10 mM GABA. Cells were grown overnight at 28°C, and growth was monitored by measuring OD at 600 nm.
Pathogenicity assays, HR test, and bacterial growth in bean
Bacterial cells were grown overnight in KB broth, harvested by centrifugation and resuspended in PBS buffer with bacterial cells adjusted to concentrations of ~106 cfu/μL (OD600 = 0.3 and then diluted 100 times) in PBS. Bacterial cells were infiltrated into leaves of two-week-old bean plants (Phaseolus vulgaris L. cv. Tender Bounty) using a needleless syringe. Infiltrated plants were maintained in a greenhouse at 25°C. Symptoms were recorded at 4 and 7 days following inoculation. These experiments were repeated at least three times with similar results. For bacterial growth, three leaves were collected from each plant at a given time point and leaf tissues at inoculation sites were excised using a #6 cork borer (12-mm diameter), and homogenized in 1 ml PBS. Serial dilutions were made in PBS, and plated on KB agar amended with appropriate antibiotics. Cell numbers were calculated after enumeration of colonies on plates incubated at 28°C for 48 h. HR assays were conducted using tobacco (Nicotiana tabacum) plants. Briefly, overnight cultures were harvested by centrifugation, cells were resuspended in PBS to OD600 = 0.1 or 0.01, and infiltrated into fully expanded third and fourth leaves using needleless syringe. Infiltrated plants were maintained in greenhouse, and HR symptoms were recorded at 24 h post infiltration.
Determination of lengths of cells by epifluorescence microscopy
Lengths of P. syringae cells were determined by epifluorescence microscopy. Bacterial strains constitutively expressing GFP were grown in KB to mid-exponential phase (OD600 = 0.7), harvested, washed, and reinoculated at OD600 0.1 in either KB broth or in HMM. After 4 h, 3 μl of bacterial suspensions was mixed with 5 μl of Aqua-Polymount (Polysciences, Warrington, Pennsylvania, USA), mounted between cover slips, and immediately observed under an Axiovert 200M fluorescence microscope (Carl Zeiss, Jena, Germany) using a FITC filter sets (Absorbance: 490–494 nm, Emission: 517 nm). Images were captured with an AxioCam® MPc digital camera (Carl Zeiss, Jena, Germany). At least 200 individual cells from 10 different images were measured using ImageJ software, available online at the National Institutes of Health (Rasband, 1997 – 2005).
Determination of cell viability by confocal microscopy
Bacterial cell viability on leaf surfaces was observed using a confocal laser scanning microscope (Zeiss). Suspensions of bacterial strains harboring pKT-trp or pAvrPto-GFP in PBS buffer (OD600 = 0.1) were sprayed onto bean (host) and tobacco (non-host) leaves and incubated under controlled conditions at 22°C. For the first 24 h after inoculation, plants were held in an enclosed plastic bag to maintain high humidity and moisture. Two plants were inoculated simultaneously, and leaves were randomly sampled from plants at each time point. Leaf discs (12 mm diameter) were mounted between cover slips (22×50-1) in Aqua-Polymount (Polysciences, Warrington, Pennsylvania, USA), and mixed with 5 μl propidium iodide (10 μg/ml). Samples were kept in the dark at room temperature for 15 min, and observed using a LSM 700 laser scanning microscope (Zeiss) using a 20× objective. At least 6 leaf discs were evaluated for each time point. Images were obtained by mixing the signals recorded in two excitation lines (488-nm laser for GFP fluorescence and 568-nm laser for chloroplastic autofluorescence and propidium iodide emission). At least 10 randomly selected fields were examined for viability of bacterial cells on leaf surfaces.
Supplementary Material
Fig. S1. (A) Hypersensitive response (HR) assays on tobacco leaves. Tobacco leaves were infiltrated using needleless syringe with bacterial cell suspensions at OD600nm 0.1 and 0.01 of Pss728a, relA, spoT, and relA/spoT mutants. (B) Green fluorescence of bacterial cells expressing GFP from pAvrPto-GFP in PssB728a WT and a relA/spoT double mutant observed by confocal laser microscopy 72 h post spray inoculation on bean leaf surfaces. Pictures were taken at 200 X magnification with a scale bar of 20 μm.
Table S1. Bacterial strains and plasmids used in this study
Table S2. Primers used in this study
Acknowledgments
We would like to thank Dr. Steven Lindow at University of California, Berkeley and Dr. Jean Greenberg at University of Chicago for providing plasmids. We also thank Dr. Raymond Zielinski at University of Illinois for his help with epifluorescence microscope. This project was supported by the Agriculture and Food Research Initiative Competitive Grants Program Grant no. 2010-65110-20497 from the USDA National Institute of Food and Agriculture (YFZ) and USDA-SCRI grant AG 2009-51181-06023 (SSK). 5500 QTRAP MS is funded by NIH National Center For Research Resources grant S10RR024516 (ZL).
References
- Abranches J, Martinez AR, Kajfasz JK, Chávez V, Garsin DA, Lemos JA. The molecular alarmone (p)ppGpp mediates stress responses, vancomycin tolerance, and virulence in Enterococcus faecalis. J Bacteriol. 2009;191:2248–2256. doi: 10.1128/JB.01726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcón-Chaidez FJ, Keith L, Zhao Y, Bender CL. RpoN (σ54) is required for plasmid-encoded coronatine biosynthesis in Pseudomonas syringae. Plasmid. 2003;49:106–117. doi: 10.1016/s0147-619x(02)00155-5. [DOI] [PubMed] [Google Scholar]
- Anderson JC, Wan Y, Kim YM, Pasa-Tolic L, Metz TO, Peck SC. Decreased abundance of type III secretion system-inducing signals in Arabidopsis mkp1 enhances resistance against Pseudomonas syringae. Proc Natl Acad Sci USA. 2014;111:6846–6851. doi: 10.1073/pnas.1403248111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson GC, Tenson T, Hauryliuk V. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One. 2011;6:e23479. doi: 10.1371/journal.pone.0023479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MM, Gaal T, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J Mol Biol. 2001;305:689–702. doi: 10.1006/jmbi.2000.4328. [DOI] [PubMed] [Google Scholar]
- Beattie GA, Lindow SE. The secret life of foliar bacterial pathogens on leaves. Annu Rev Phytopathol. 1995;33:145–172. doi: 10.1146/annurev.py.33.090195.001045. [DOI] [PubMed] [Google Scholar]
- Bender CL, Alarcon-Chaidez F, Gross DC. Pseudomonas syringae Phytotoxins: Mode of Action, Regulation, and Biosynthesis by Peptide and Polyketide Synthetases. Microbiol Mol Biol Rev. 1999;63:266–292. doi: 10.1128/mmbr.63.2.266-292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman JM, Hammarlöf DL, Hughes D. Reducing ppGpp level rescues an extreme growth defect caused by mutant EF-Tu. PLoS One. 2014;9:e90486. doi: 10.1371/journal.pone.0090486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berti AD, Greve NJ, Christensen QH, Thomas MG. Identification of a biosynthetic gene cluster and the six associated lipopeptides involved in swarming motility of Pseudomonas syringae pv. tomato DC3000. J Bacteriol. 2007;189:6312–6323. doi: 10.1128/JB.00725-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougdour A, Gottesman S. ppGpp regulation of RpoS degradation via anti-adaptor protein IraP. Proc Natl Acad Sci USA. 2007;104:12896–12901. doi: 10.1073/pnas.0705561104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden SD, Eyres A, Chung JCS, Monson RE, Thompson A, Salmond GPC, Spring DR, Welch M. Virulence in Pectobacterium atrosepticum is regulated by a coincidence circuit involving quorum sensing and the stress alarmone, (p)ppGpp. Mol Microbiol. 2013;90:457–471. doi: 10.1111/mmi.12369. [DOI] [PubMed] [Google Scholar]
- Braeken K, Moris M, Daniels R, Vanderleyden J, Michiels J. New horizons for (p)ppGpp in bacterial and plant physiology. Trends Microbiol. 2006;14:45–54. doi: 10.1016/j.tim.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Brown DR, Barton G, Pan Z, Buck M, Wigneshweraraj S. Nitrogen stress response and stringent response are coupled in Escherichia coli. Nat Commun. 2014;5:4115. doi: 10.1038/ncomms5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch AY, Shimada BK, Browne PJ, Lindow SE. Novel high-throughput detection method to assess bacterial surfactant production. Appl Environ Microbiol. 2010;76:5363–5372. doi: 10.1128/AEM.00592-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Cui Y, Yang H, Collmer A, Alfano JR, Chatterjee AK. GacA, the response regulator of a two-component system, acts as a master regulator in Pseudomonas syringae pv. tomato DC3000 by controlling regulatory RNA, transcriptional activators, and alternate sigma factors. Mol Plant Microbe Interact. 2003;16:1106–1117. doi: 10.1094/MPMI.2003.16.12.1106. [DOI] [PubMed] [Google Scholar]
- Crompton MJ, Dunstan RH, Macdonald MM, Gottfries J, von Eiff C, Roberts TK. Small changes in environmental parameters lead to alterations in antibiotic resistance, cell morphology and membrane fatty acid composition in Staphylococcus lugdunensis. PLoS One. 2014;9:e92296. doi: 10.1371/journal.pone.0092296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux ZD, Svensson SL, Gaynor EC, Swanson MS. ppGpp conjures bacterial virulence. Microbiol Mol Biol Rev. 2010;74:171–199. doi: 10.1128/MMBR.00046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux ZD, Swanson MS. ppGpp: magic beyond RNA polymerase. Nat Rev Microbiol. 2012;10:203–212. doi: 10.1038/nrmicro2720. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AN, Patterson-Fortin LM, Vakulskas CA, Mercante JW, Potrykus K, Vinella D, Camacho MI, Fields JA, Thompson SA, Georgellis D, Cashel M, Babitzke P, Romeo T. Circuitry linking the Csr and stringent response global regulatory systems. Mol Microbiol. 2011;80:1561–1580. doi: 10.1111/j.1365-2958.2011.07663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson DL, Lines JL, Pesci EC, Venturi V, Storey DG. Pseudomonas aeruginosa relA contributes to virulence in Drosophila melanogaster. Infect Immun. 2004;72:5638–5645. doi: 10.1128/IAI.72.10.5638-5645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira AO, Myers CR, Gordon JS, Martin GB, Vencato M, Collmer A, Wehling MD, Alfano JR, Moreno-Hagelsieb G, Lamboy WF, DeClerck G, Schneider DJ, Cartinhour SW. Whole-genome expression profiling defines the HrpL regulon of Pseudomonas syringae pv. tomato DC3000, allows de novo reconstruction of the Hrp cis clement, and identifies novel coregulated genes. Mol Plant Microbe Interact. 2006;19:1167–1179. doi: 10.1094/MPMI-19-1167. [DOI] [PubMed] [Google Scholar]
- Hase C. Nutrient deprivation and the subsequent survival of biocontrol Pseudomonas fluorescens CHA0 in soil. Soil Biol Biochem. 1999;31:1181–1188. [Google Scholar]
- Haseltine WA, Block R. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc Natl Acad Sci USA. 1973;70:1564–1568. doi: 10.1073/pnas.70.5.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman KL, Pease LR. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc. 2007;2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- Hendrickson EL, Guevera P, Ausubel FM. The alternative sigma factor RpoN is required for hrp activity in Pseudomonas syringae pv. maculicola and acts at the level of hrpL transcription. J Bacteriol. 2000;182:3508–3516. doi: 10.1128/jb.182.12.3508-3516.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez VJ, Bremer H. Guanosine tetraphosphate (ppGpp) dependence of the growth rate control of rrnB P1 promoter activity in Escherichia coli. J Biol Chem. 1990;265:11605–11614. [PubMed] [Google Scholar]
- Hill NS, Buske PJ, Shi Y, Levin PA. A moonlighting enzyme links Escherichia coli cell size with central metabolism. PLoS Genet. 2013;9:e1003663. doi: 10.1371/journal.pgen.1003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano SS, Charkowski AO, Collmer A, Willis DK, Upper CD. Role of the Hrp type III protein secretion system in growth of Pseudomonas syringae pv. syringae B728a on host plants in the field. Proc Natl Acad Sci USA. 1999;96:9851–9856. doi: 10.1073/pnas.96.17.9851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano SS, Upper CD. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae-a pathogen, ice nucleus, and epiphyte. Microbiol Mol Biol Rev. 2000;64:624–653. doi: 10.1128/mmbr.64.3.624-653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano SS, Upper CD. Population Biology and Epidemiology of Pseudomonas Syringae. Annu Rev Phytopathol. 1990;28:155–177. [Google Scholar]
- Hockett KL, Burch AY, Lindow SE. Thermo-regulation of genes mediating motility and plant interactions in Pseudomonas syringae. PLoS One. 2013;8:e59850. doi: 10.1371/journal.pone.0059850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh TV, Dahlbeck D, Staskawicz BJ. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science. 1989;245:1374–1377. doi: 10.1126/science.2781284. [DOI] [PubMed] [Google Scholar]
- Imperi F, Tiburzi F, Visca P. Molecular basis of pyoverdine siderophore recycling in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2009;106:20440–20445. doi: 10.1073/pnas.0908760106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic M, James EH, Burrows PC, Rego FGM, Buck M, Schumacher J. Regulation of the co-evolved HrpR and HrpS AAA+ proteins required for Pseudomonas syringae pathogenicity. Nat Commun. 2011;2:177. doi: 10.1038/ncomms1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubair M, Morris JG, Ali A. Survival of Vibrio cholerae in nutrient-poor environments is associated with a novel “persister” phenotype. PLoS One. 2012;7:e45187. doi: 10.1371/journal.pone.0045187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia D, Merey G, Nakayama S, Zheng Y, Zhou J, Luo Y, Guo M, Roembke BT, Sintim HO. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem Soc Rev. 2013;42:305–341. doi: 10.1039/c2cs35206k. [DOI] [PubMed] [Google Scholar]
- Karamanoli K, Bouligaraki P, Constantinidou HIA, Lindow SE. Polyphenolic compounds on leaves limit iron availability and affect growth of epiphytic bacteria. Ann Appl Biol. 2011;159:99–108. [Google Scholar]
- Khakimova M, Ahlgren HG, Harrison JJ, English AM, Nguyen D. The stringent response controls catalases in Pseudomonas aeruginosa and is required for hydrogen peroxide and antibiotic tolerance. J Bacteriol. 2013;195:2011–2020. doi: 10.1128/JB.02061-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinscherf TG, Willis DK. Swarming by Pseudomonas syringae B728a requires gacS (lemA) and gacA but not the acyl-homoserine lactone biosynthetic gene ahlI. J Bacteriol. 1999;181:4133–6. doi: 10.1128/jb.181.13.4133-4136.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitten T, Kinscherf TG, McEvoy JL, Willis DK. A newly identified regulator is required for virulence and toxin production in Pseudomonas syringae. Mol Microbiol. 1998;28:917–929. doi: 10.1046/j.1365-2958.1998.00842.x. [DOI] [PubMed] [Google Scholar]
- Kitten T, Willis DK. Suppression of a sensor kinase-depedent phenotype in Pseudomonas syringae by ribosomal proteins L35 and L20. J Bacteriol. 1996;178:1548–1555. doi: 10.1128/jb.178.6.1548-1555.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Teitzel GM, Munkvold K, del Pozo O, Martin GB, Michelmore RW, Greenberg JT. Type III secretion and effectors shape the survival and growth pattern of Pseudomonas syringae on leaf surfaces. Plant Physiol. 2012;158:1803–1818. doi: 10.1104/pp.111.190686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Qimuge Hao J, Yan H, Bach T, Fan L, Morigen AspC-mediated aspartate metabolism coordinates the Escherichia coli cell cycle. PLoS One. 2014;9:e92229. doi: 10.1371/journal.pone.0092229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loper JE, Lindow SE. A biological sensor for iron available to bacteria in their habitats on plant surfaces. Appl Environ Microbiol. 1994;60:1934–1941. doi: 10.1128/aem.60.6.1934-1941.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SE, Scholz-Schroeder BK, Gross DC. Characterization of the salA, syrF, and syrG regulatory genes located at the right border of the syringomycin gene cluster of Pseudomonas syringae pv. syringae. Mol Plant Microbe Interact. 2002;15:43–53. doi: 10.1094/MPMI.2002.15.1.43. [DOI] [PubMed] [Google Scholar]
- Lu SE, Wang N, Wang J, Chen ZJ, Gross DC. Oligonucleotide microarray analysis of the salA regulon controlling phytotoxin production by Pseudomonas syringae pv. syringae. Mol Plant Microbe Interact. 2005;18:324–333. doi: 10.1094/MPMI-18-0324. [DOI] [PubMed] [Google Scholar]
- Magnusson LU, Farewell A, Nyström T. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 2005;13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Monahan LG, Hajduk IV, Blaber SP, Charles IG, Harry EJ. Coordinating bacterial cell division with nutrient availability: a role for glycolysis. MBio. 2014;5:e00935. doi: 10.1128/mBio.00935-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier JM, Lindow SE. Differential survival of solitary and aggregated bacterial cells promotes aggregate formation on leaf surfaces. Proc Natl Acad Sci USA. 2003a;100:15977–15982. doi: 10.1073/pnas.2436560100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier JM, Lindow SE. Pseudomonas syringae responds to the environment on leaves by cell size reduction. Phytopathology. 2003b;93:1209–1216. doi: 10.1094/PHYTO.2003.93.10.1209. [DOI] [PubMed] [Google Scholar]
- Moris M, Braeken K, Schoeters E, Verreth C, Beullens S, Vanderleyden J, Michiels J. Effective symbiosis between Rhizobium etli and Phaseolus vulgaris requires the alarmone ppGpp. J Bacteriol. 2005;187:5460–5469. doi: 10.1128/JB.187.15.5460-5469.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi N, Abe H, Ogura Y, Hayashi T, Tashiro K, Kuhara S, Sugimoto N, Tobe T. ppGpp with DksA controls gene expression in the locus of enterocyte effacement (LEE) pathogenicity island of enterohaemorrhagic Escherichia coli through activation of two virulence regulatory genes. Mol Microbiol. 2006;61:194–205. doi: 10.1111/j.1365-2958.2006.05217.x. [DOI] [PubMed] [Google Scholar]
- Oguiza JA, Kiil K, Ussery DW. Extracytoplasmic function sigma factors in Pseudomonas syringae. Trends Microbiol. 2005;13:565–568. doi: 10.1016/j.tim.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Park DH, Mirabella R, Bronstein PA, Preston GM, Haring MA, Lim CK, Collmer A, Schuurink RC. Mutations in γ-aminobutyric acid (GABA) transaminase genes in plants or Pseudomonas syringae reduce bacterial virulence. Plant J. 2010;64:318–330. doi: 10.1111/j.1365-313X.2010.04327.x. [DOI] [PubMed] [Google Scholar]
- Pizarro-Cerdá J, Tedin K. The bacterial signal molecule, ppGpp, regulates Salmonella virulence gene expression. Mol Microbiol. 2004;52:1827–1844. doi: 10.1111/j.1365-2958.2004.04122.x. [DOI] [PubMed] [Google Scholar]
- Potrykus K, Cashel M. Gpp: still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- Potrykus K, Murphy H, Philippe N, Cashel M. ppGpp is the major source of growth rate control in E. coli. Environ Microbiol. 2011;13:563–575. doi: 10.1111/j.1462-2920.2010.02357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahme LG, Mindrinos MN, Panopoulos NJ. Plant and environmental sensory signals control the expression of hrp genes in Pseudomonas syringae pv. phaseolicola. J Bacteriol. 1992;174:3499–3507. doi: 10.1128/jb.174.11.3499-3507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran VK, Shearer N, Thompson A. The primary transcriptome of Salmonella enterica Serovar Typhimurium and its dependence on ppGpp during late stationary phase. PLoS One. 2014;9:e92690. doi: 10.1371/journal.pone.0092690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband WS. ImageJ. U. S. National Institutes of Health; Bethesda, Maryland, USA: 1997–2005. http://rsb.info.nih.gov/ij/ [Google Scholar]
- Records AR, Gross DC. Sensor Kinases RetS and LadS Regulate Pseudomonas syringae Type VI Secretion and Virulence Factors. J Bacteriol. 2010;192:3584–3596. doi: 10.1128/JB.00114-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich JJ, Kinscherf TG, Kitten T, Willis DK. Genetic evidence that the gacA gene encodes the cognate response regulator for the lemA sensor in Pseudomonas syringae. J Bacteriol. 1994;176:7468–7475. doi: 10.1128/jb.176.24.7468-7475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W, Vrentas CE, Sanchez-Vazquez P, Gaal T, Gourse RL. The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol Cell. 2013;50:420–429. doi: 10.1016/j.molcel.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, U.S.A: 1989. [Google Scholar]
- Sarubbi E, Rudd KE, Cashel M. Basal ppGpp level adjustment shown by new spoT mutants affect steady state growth rates and rrnA ribosomal promoter regulation in Escherichia coli. MGG Mol Gen Genet. 1988;213:214–222. doi: 10.1007/BF00339584. [DOI] [PubMed] [Google Scholar]
- Schafhauser J, Lepine F, McKay G, Ahlgren HG, Khakimova M, Nguyen D. The Stringent response modulates 4-Hydroxy-2-Alkylquinoline biosynthesis and quorum-sensing hierarchy in Pseudomonas aeruginosa. J Bacteriol. 2014;196:1641–1650. doi: 10.1128/JB.01086-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat Biotechnol. 1983;1:784–791. [Google Scholar]
- Spira B, Hu X, Ferenci T. Strain variation in ppGpp concentration and RpoS levels in laboratory strains of Escherichia coli K-12. Microbiology. 2008;154:2887–2895. doi: 10.1099/mic.0.2008/018457-0. [DOI] [PubMed] [Google Scholar]
- Stauber JL, Loginicheva E, Schechter LM. Carbon source and cell density-dependent regulation of type III secretion system gene expression in Pseudomonas syringae pathovar tomato DC3000. Res Microbiol. 2012;163:531–539. doi: 10.1016/j.resmic.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Steinberger RE, Allen AR, Hansa HG, Holden PA. Elongation correlates with nutrient deprivation in Pseudomonas aeruginosa-unsaturates biofilms. Microb Ecol. 2002;43:416–423. doi: 10.1007/s00248-001-1063-z. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Yamada K, Haas D. ppGpp Controlled by the Gac/Rsm Regulatory Pathway Sustains Biocontrol Activity in Pseudomonas fluorescens CHA0. Mol Plant Microbe Interact. 2012;25:1440–1449. doi: 10.1094/MPMI-02-12-0034-R. [DOI] [PubMed] [Google Scholar]
- Tang X, Xiao Y, Zhou JM. Regulation of the type III secretion system in phytopathogenic bacteria. Mol Plant Microbe Interact. 2006;19:1159–1166. doi: 10.1094/MPMI-19-1159. [DOI] [PubMed] [Google Scholar]
- Thakur PB, Vaughn-Diaz VL, Greenwald JW, Gross DC. Characterization of five ECF sigma factors in the genome of Pseudomonas syringae pv. syringae B728a. PLoS One. 2013;8:e58846. doi: 10.1371/journal.pone.0058846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, Conway T. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxler MF, Zacharia VM, Marquardt S, Summers SM, Nguyen H, Stark SE, Conway T. Discretely calibrated regulatory loops controlled by ppGpp partition gene induction across the ‘feast to famine’ gradient in Escherichia coli. Mol Microbiol. 2011;79:830–845. doi: 10.1111/j.1365-2958.2010.07498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, Molin S, Bleves S, Lazdunski A, Lory S, Filloux A. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc Natl Acad Sci USA. 2006;103:171–176. doi: 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercruysse M, Fauvart M, Jans A, Beullens S, Braeken K, Cloots L, Engelen K, Marchal K, Michiels J. Stress response regulators identified through genome-wide transcriptome analysis of the (p)ppGpp-dependent response in Rhizobium etli. Genome Biol. 2011;12:R17. doi: 10.1186/gb-2011-12-2-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt SL, Green C, Stevens KM, Day B, Erickson DL, Woods DE, Storey DG. The stringent response is essential for Pseudomonas aeruginosa virulence in the rat lung agar bead and Drosophila melanogaster feeding models of infection. Infect Immun. 2011;79:4094–4104. doi: 10.1128/IAI.00193-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Korban SS, Zhao YF. The Rcs phosphorelay system is essential for pathogenicity in Erwinia amylovora. Mol Plant Pathol. 2009;10:277–290. doi: 10.1111/j.1364-3703.2008.00531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gardiol N, Burr T, Salmond GPC, Welch M. RelA-dependent (p)ppGpp production controls exoenzyme synthesis in Erwinia carotovora subsp. atroseptica. J Bacteriol. 2007;189:7643–7652. doi: 10.1128/JB.00920-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Lu SE, Records AR, Gross DC. Characterization of the transcriptional activators SalA and SyrF, which are required for syringomycin and syringopeptin production by Pseudomonas syringae pv. syringae. J Bacteriol. 2006;188:3290–3298. doi: 10.1128/JB.188.9.3290-3298.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Goodwin SM, Xiao Y, Sun Z, Baker D, Tang X, Jenks MA, Zhou JM. Arabidopsis CYP86A2 represses Pseudomonas syringae type III genes and is required for cuticle development. EMBO J. 2004;23:2903–2913. doi: 10.1038/sj.emboj.7600290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
- Xin XF, He SY. Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu Rev Phytopathol. 2013;51:473–498. doi: 10.1146/annurev-phyto-082712-102321. [DOI] [PubMed] [Google Scholar]
- Yao Z, Davis RM, Kishony R, Kahne D, Ruiz N. Regulation of cell size in response to nutrient availability by fatty acid biosynthesis in Escherichia coli. Proc Natl Acad Sci USA. 2012;109:2561–2568. doi: 10.1073/pnas.1209742109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Lund SP, Scott RA, Greenwald JW, Records AH, Nettleton D, Lindow SE, Gross DC, Beattie GA. Transcriptional responses of Pseudomonas syringae to growth in epiphytic versus apoplastic leaf sites. Proc Natl Acad Sci USA. 2013;110:425–434. doi: 10.1073/pnas.1221892110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YF, Wang D, Nakka S, Sundin GW, Korban SS. Systems level analysis of two-component signal transduction systems in Erwinia amylovora: role in virulence, regulation of amylovoran biosynthesis and swarming motility. BMC Genomics. 2009;10:245. doi: 10.1186/1471-2164-10-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. (A) Hypersensitive response (HR) assays on tobacco leaves. Tobacco leaves were infiltrated using needleless syringe with bacterial cell suspensions at OD600nm 0.1 and 0.01 of Pss728a, relA, spoT, and relA/spoT mutants. (B) Green fluorescence of bacterial cells expressing GFP from pAvrPto-GFP in PssB728a WT and a relA/spoT double mutant observed by confocal laser microscopy 72 h post spray inoculation on bean leaf surfaces. Pictures were taken at 200 X magnification with a scale bar of 20 μm.
Table S1. Bacterial strains and plasmids used in this study
Table S2. Primers used in this study