Abstract
Fifth-generation cephalosporins, ceftobiprole and ceftaroline, are promising drugs for treatment of bacterial infections from methicillin-resistant Staphylococcus aureus (MRSA). These antibiotics are able to bind native PBP2a, the penicillin-binding protein encoded by the mecA resistance determinant that mediates broad class resistance to nearly all other beta-lactam antibiotics, at clinically achievable concentrations. Mechanisms of resistance to ceftaroline based on mecA mutations have been previously described. Here we compare the genomes of 11 total parent-daughter strains of Staphylococcus aureus for which specific selection by serial passaging with ceftaroline or ceftobiprole was used to identify novel non-mecA mechanisms of resistance. All 5 ceftaroline-resistant strains, derived from 5 different parental strains, contained mutations directly upstream of the pbp4 gene (coding for the PBP4 protein), including four with the same thymidine insertion located 377 nucleotides upstream of the promoter site. In 4 of 5 independent ceftaroline-driven selections, we also isolated mutations to the same residue (Asn138) in PBP4. In addition, mutations in additional candidate genes such as ClpX endopeptidase, PP2C protein phosphatase and transcription terminator Rho, previously undescribed in the context of resistance to ceftaroline or ceftobiprole, were detected in multiple selections. These genomic findings suggest that non-mecA mechanisms, while yet to be encountered in the clinical setting, may also be important in mediating resistance to 5th-generation cephalosporins.
Introduction
Multi-drug resistant Staphyloccocus aureus (MRSA) is a ubiquitous problem in hospitals and in the community [1, 2]. The United States Centers for Disease Control and Prevention (CDC) estimates that there are over 75,000 MRSA infections annually [3], with the vast majority occurring in older persons in healthcare-associated settings. Ceftaroline is a fifth-generation cephalosporin with broad-spectrum activity against gram-negative and gram-positive bacteria, including MRSA [4], and is FDA-approved for skin and soft tissue infections, including those caused by MRSA [5, 6]. To date, a number of moderately ceftaroline-resistant strains (MIC 4 μg/ml) of MRSA have been described clinically. Moderate ceftaroline resistance has recently been shown to be particularly high among MRSA strains in China and Thailand. High-level ceftaroline resistance is rare, with only one clinical strain to date demonstrating resistance of MIC > 32 μg/ml [7–9]. Whole-genome and candidate gene sequencing have identified Y446N and E447K mutations in the mecA / pbp2a gene to be associated with high-level ceftaroline resistance [10, 11]. However, whether ceftaroline-associated resistance mutations exist outside of the mecA / pbp2a locus remains to be determined.
To discover potential mecA-independent resistance mechanisms, we sequenced the genomes of 7 total parent-daughter strains of S. aureus from which the mecA locus had been removed prior to passaging of ceftaroline or ceftobiprole in vitro. In addition, we sequenced and analyzed the full genome of a ceftaroline-resistant mecA positive strain (CRT) reported to exhibit non-mecA mediated ceftaroline resistance [10] in order to elucidate its mechanisms of resistance. Our data reveal a number of mutations in key genes within the S. aureus genome that are associated with the establishment of high-level resistance to ceftobiprole and ceftaroline.
Materials and Methods
Construction of strains
The parental strains for this study include Coln (a mecA positive strain) and Colnex and SF8300ex (strains from which the mecA gene has been excised) (Table 1). These parental strains were passaged daily 28 days, as previously described [12]. Briefly, 10 ml preparations of trypticase soy broth (TSB) containing various concentrations of antibiotic (ceftaroline or ceftobiprole) were inoculated at a 1:100 dilution with overnight cultures containing 109 CFU ml-1. The drug concentration was doubled at each passage as tolerated until bacterial growth was observed in at least 128 μg ml-1 of the drug specified. At the end of passaging, a single clone was chosen randomly for further studies.
Table 1. Strains used in this study and mutations detected in penicillin binding proteins, gdpP and acrB genes.
| Strains | MecA | Driver for selection | MIC to CFTR* | MIC to AMP** | MIC to NAF*** | PBP1 | PBP2 | PBP3 | pbp4 promoter | PBP4 | GdpP | AcrB |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Colna | (+) | 1 S | 16 R | 128 R | ||||||||
| CRT# | (+) | CFTR | >64 R | 256 R | >256 R | G631S | 724602_724603insT (-377); 724500_724590del (-275) | N138K | ||||
| Colnexb# | (-) | <0.25 R | 0.25 S | 0.5 S | ||||||||
| CmTcc# | (-) | CFTR | >64 R | >256 R | >256 R | D156N | 724602_724603insT (-377); 724624T>G (-399) | T201A; F241L | H443Y | |||
| SF8300exd# | (-) | 0.25 S | 0.25 S | 0.5 S | ||||||||
| SRT# | (-) | CFTR | >64 R | >256 R | 64 R | 716955_716965del (-301) | N138K; H270L | Y306X | ||||
| SRB# | (-) | CFBP | 4 R | 4 S | 8 S | H499R; E567K | Y437C; V445L; Q453R; M559I | W228X | E183V; F241R | T509A | ||
| Sgape | (-) | 0.5 S | <0.25 S | 0.5 S | E183A; F241R | N182K | I960V | |||||
| SgapT# | (-) | CFTR | 64 R | >256 R | >256 R | H499R | 717031_717032insT (-377) | N138I; E183A; R200L; F241R | N182K | I960V | ||
| Spf | (-) | 0.25 S | 0.25 S | 1 S | E183A; F241R | |||||||
| SpT# | (-) | CFTR | >64 R | >256 R | >256 R | G581D | 716675del (-21); 717031_717032insT (-377) | N138I; E183V; T201A; F241R | N214del |
*CFTR = ceftaroline; CLSI breakpoints are ≤1 S, 2 I, ≥4 R (CLSI document M100-S23; ISBN 1-56238-865-7)
**AMP = ampicillin; CLSI breakpoints are ≤8 S, ≥16 R (CLSI document M100-S23; ISBN 1-56238-865-7)
***NAF = nafcillin; CLSI breakpoints are ≤8 S, ≥16 R (CLSI document M100-S23; ISBN 1-56238-865-7)
#Strains sequenced in this study
aParental strain of CRT
bCol strain with mecA excised
cColnex strain containing exogenous mecA plasmid (pYK20); mecA plasmid was evicted after ceftaroline selection (Chan L 2015 AAC)
dSF8300 strain with mecA excised, parental strain of SRT and SRB
eSF8300ex strain in which PBP4 (E183A, F241R), GdpP (N182K) and AcrB (I960V) mutations analogous to those in the CRB strain (Banerjee, et al. 2010 AAC) were introduced
fSF8300ex strain in which PBP4 (E183A, F241R) mutations analogous to those in the CRB strain (Banerjee, et al. 2010 AAC) were introduced
Abbreviations: MIC = minimal inhibitory concentration; R = resistant; S = susceptible, susc = susceptibility; CFTR = ceftaroline; CFBP = ceftobiprole.
The passaged daughter strains include (a) CRT, a ceftaroline-passaged derivative of the Coln strain [10], (b) CmTc, a strain in which a mecA-containing plasmid (pYK20) was re-introduced into Colnex before passaging in ceftaroline (CmTc) [10], and (c) SRT and SRB, two newly generated strains derived from passaging of SF8300ex in ceftaroline and ceftobiprole, respectively (Table 1). The mutant strain Sgap was created by introducing into SF8300ex select mutations from the pbp4, acrB, and gdpP genes that were identified via whole genome sequencing of a S. aureus strain selected for high-level ceftobiprole-resistance (CRB) [13]. Daughter strain SgapT was created by passaging the Sgap strain in ceftaroline. Finally, strain SpT was created by passaging in ceftaroline Sp, a mutant SF8300ex strain in which only pbp4 mutations from strain CRB had been introduced.
Sgap and Sp were included in the study to determine if there were any differences in baseline ceftaroline resistance between a wild-type strain with only pbp4 mutations (Sp) and a strain with mutations in pbp4, gdpP, and acrB (Sgap). Furthermore, passing of the Sp and Sgap strains were carried out in ceftaroline and subsequently genome sequenced to determine what changes were present in the resistant SpT and SgapT strains.
Genomic DNA extraction
Bacterial cultures incubated in broth overnight (1 mL) were collected by centrifugation and re-suspended in 500 μL buffer containing 50 mM Tris-Cl, pH 8.0, 10 mM EDTA and 100 μg/mL RNase A. Bacterial suspensions were then transferred to Lysing matrix B column (MP Biomedicals) to lyse the bacteria. Bacterial lysates were incubated in ice for 5 min and centrifuged at 14,000g for 15 min. Supernatants containing the genomic DNA were transferred to a fresh tube, precipitated using ethanol and re-suspended in sterile water.
Sequencing library preparation
One ng of genomic DNA was used as input for the Nextera XT kit (Illumina), followed by sample barcoding and amplification with 12 cycles of PCR. Libraries were quantified on the Bioanalyzer (Agilent Technologies) and combined in an equimolar mixture. Next-generation sequencing (NGS) libraries were sequenced on a single run on the Illumina MiSeq instrument (300 bp paired-end reads).
Sequencing analysis
Reads were adapter- and quality-trimmed (Q30 cutoff; minimum length >20 nucleotides) using cutadapt [14], followed by re-pairing using pairfq [15]. Paired-end reads were mapped to the reference genomes of Staphylococcus aureus COL (NC_002951) or USA300 (CP000730) using Geneious v8.0 [16]. More than 97% of paired-end reads mapped to the reference genomes, with coverage ranging from 138X−287X. High-confidence variants were called using a minimum coverage of 25X and minimum variant frequency of 90%. All called variants were manually reviewed to correct large deletions that were erroneously called as SNPs. Remaining unmapped reads were de novo assembled to find genetic elements not present in the reference sequence. Of note, all USA300-based strains de novo assembled a ~3,100-bp contig with >99% identity to the Staphylococcus aureus SAP046B plasmid (GQ900404), which did not comprise part of the sequenced core genome of Staphylococcus aureus USA300 in the National Center for Biotechnology Information (NCBI) GenBank reference database.
Accession Numbers
All genomic data from this study have been deposited publicly in NCBI under BioProject PRJNA293093.
Results
Four independent selections in ceftaroline result in similar pbp4 coding and promoter mutations
To identify key genes associated with non-mecA mediated ceftaroline resistance in MRSA, we recovered the whole-genome sequences of 2 parental strains and 6 resistant daughter strains that had been passaged in either ceftaroline (n = 5) or ceftobiprole (n = 1) (Table 1). We first examined 6 genes that had been previously implicated in beta-lactam resistance in Staphylococcus aureus (Table 1), including pbp1, pbp2, pbp3, pbp4, pbp4 promoter, gdpP, and acrB [10, 17, 18].
In the SRT strain, selected in ceftaroline directly from parental strain SF8300ex, 2 coding mutations (N138K and H270L) out of 7 resided in the pbp4 gene. In addition, an 11-bp deletion at nucleotide position -301 base pairs (bp) was present in the promoter sequence directly upstream of the pbp4 gene. A premature stop codon was also found in the gdpP gene of the SRT strain.
Similarly, mutations in pbp4 were noted in the CRT strain, which was selected in ceftaroline from a different parental strain (Coln). Two of 8 total coding single nucleotide variants (SNVs) were found to be associated with pbp4, including a coding change in the PBP4 protein also seen in SRT (N138K) and a single nucleotide insertion of a thymidine at nucleotide position -377 bp of the pbp4 promoter (S1 Table). The CRT strain also contained a 91 bp deletion in the pbp4 promoter beginning at nucleotide -274 bp and a G631S mutation in pbp2. Outside of the pbp genes, two deleted regions 60 bp and 12 bp in length were presented in CRT, resulting in truncations in two metabolic proteins (TdcB and IIABC).
The SgapT strain, also selected in ceftaroline, contained 24 total coding mutations relative to its parental SF8300ex strain. Of these, three of 24 mutations were found in pbp1 and pbp4 genes coding for PBP1 and PBP4, respectively. Two of the coding mutations were detected in the pbp4 gene (N138I and R200L) and a H499R mutation was found in pbp1. As in the CRT strain, a single nucleotide insertion of a thymidine at nucleotide position -377 bp was also present in the pbp4 promoter.
The CmTc strain, derived from parental strain Colnex, had a total of 9 coding mutations. Four of these mutations were mapped to previously identified mecA-independent beta-lactam resistance genes [13, 19], including two coding mutations in pbp4 (T201A, F241L), one mutation in gdpP (H443Y), and one mutation in pbp2 (D156N). In addition, the CmTc strain had the same -377 bp thymidine insertion in the pbp4 promoter as found in CRT and SgapT, as well as a T→G SNV that was not present in any of the other strains.
Comparison of the SpT strain to its grandparent SF8300ex strain demonstrated 24 coding SNVs in total. The SpT strain had four coding changes in the pbp4 gene; however two of these changes (E183V, F241R) had been deliberately introduced into the parental strain, and the remaining two (N138I, T201A) were acquired SNVs during selection in ceftaroline. In addition, one coding mutation (G581D) was present in pbp2 and a 3-bp deletion (N214del) was present in the gdpP gene. Importantly, as in SRT, CRT, SgapT, and CmTc, mutations in the pbp4 promoter were present, including a single nucleotide insertion of a thymidine at -377 bp and a single nucleotide deletion of a thymidine at -21 bp.
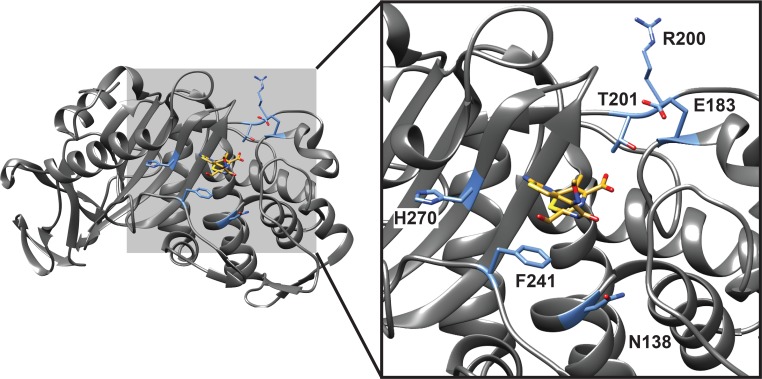
Across the 5 passaged daughter strains selected in ceftaroline or ceftobiprole, a total of 6 mutations in the pbp4 gene were identified (N138K, N138I, R200L, T201A, F241L, and H270L). Four of the 5 (80%) ceftaroline selections recovered a mutation to the Asn138 residue of pbp4 (N138K or N138I). Mapping of the 6 pbp4 mutations to the PBP4 crystal structure of Staphylococcus aureus Col strain in complex with cefotaxime (PDB 3HUM) revealed that all mutations were localized, as expected, to the cephalosporin-binding pocket (Fig 1). Importantly, all five of the ceftaroline selections also generated mutations to the pbp4 promoter, with 4 out of 5 (80%) selections demonstrating an identical single nucleotide thymidine insertion at -377 bp.
Fig 1. Mapping of pbp4 gene mutations to the crystal structure of the PBP4 protein of Staphylococcus aureus complexed with a cephalosporin antibiotic (cefaxotaxime).
The left panel shows the entire complex, whereas the right panel shows a zoomed image of the cephalosporin binding pocket. The Staphylococcus aureus strain depicted in the crystal structure (PDB 3HUM) [30], Col, is the parental strain of the Colnex strain used in the current study (Table 1). Mutant residues in PBP4 identified by selection with ceftaroline or ceftobiprole are highlighted in blue. The ligand marked in yellow is cefotaxime.
Recurrent mutations across multiple selections identify additional candidate genes for mecA-independent beta-lactam resistance
After characterizing mutations in the 6 genes known to be associated with beta-lactam resistance genes (Macheboeuf et al., 2006, Zapun et al., 2008), we next examined remaining coding mutations in other genes that were represented in at least 3 of the 6 ceftaroline or ceftobiprole daughter strains (S1 Table). Interestingly, only 3 additional genes met this criterion (Table 2). Strains CmTc, SgapT, and SpT contained coding mutations in transcription termination factor Rho. The S14 family endopeptidase ClpX and PP2C protein phosphatase genes were found mutated in the ceftobiprole-resistant SRB strain as well as the ceftaroline-resistant SRT and SgapT strains. Both SgapT and SRB strains included mutation of the same residue (glycine 169) in the PP2C protein phosphatase, a highly conserved MRSA gene on the basis of NCBI conserved domain searches (family cd00143) [20].
Table 2. Additional coding mutations present in at least three selections in this study.
Mutated genes from Table 1 are not included.
| CmTc | SRT | SRB | SgapT | SpT | Locus tag | Gene | Description |
|---|---|---|---|---|---|---|---|
| Q31X | G169S | G169D | USA300HOU_1156 | pp2C | possible PP2C protein phosphatase | ||
| P323L | E354fsX358 | V381E | USA300HOU_1666 | clpX | S14 family endopeptidase ClpX | ||
| F241L | E356V | F83delinsX | USA300HOU_2109 | rho | transcription termination factor Rho |
Discussion
In this study, we performed whole-genome sequencing of 9 total parent-daughter strains of S. aureus using an Illumina NGS platform to identify novel mutations associated with high-level, mecA-independent resistance to fifth-generation cephalosporins. Notably, all 5 ceftaroline selections generated mutations in the pbp4 promoter, while 4 of 5 (80%) selections were associated with pbp4 SNVs coding for a mutation in the ASN138 residue of PBP4. Previously undescribed mutations in additional candidate genes such as ClpX endopeptidase, PP2C protein phosphatase and termination transcription factor Rho were also detected in multiple screens. Our data suggest that mutations in the pbp4 gene or its promoter are strongly associated with resistance to fifth-generation cephalosporins, although further biochemical characterization will be needed to confirm these findings. These results also illustrate the power of whole-genome sequencing of bacterial isolates to rapidly identify key genetic loci involved in resistance.
Previous work on ceftaroline resistance in S. aureus has suggested that PBP4 has poor affinity for ceftaroline [19, 21–23]. However, loss of the pbp4 gene has been shown to confer a significant reduction in methicillin resistance, and it has been suggested this may be due to epistatic interactions of PBP4 with PBP2/2a that facilitate resistance to beta-lactam antibiotics [24]. Interestingly, a previously reported MRSA strain PVI [25], which is known to overexpress pbp4, is similar to the ceftaroline-resistant strains reported here in that the PVI strain also contains a deletion in the pbp4 promoter (of size 90 bp) and an adenosine, rather than thymidine, insertion at position -377 bp. This suggests that the pbp4 promoter mutations isolated in this study may also lead to PBP4 overexpression and promote resistance. Docking analysis of 19 beta-lactams with all available PBP structures identified Asn138 as part of the PBP4 active site (Fig 1) and, importantly, ceftaroline has higher affinity for PBP4 than the PBP1 and PBP2 proteins [26].
Our genomic sequencing also reveals mutations in new candidate genes such as ClpX endopeptidase, Rho transcription termination factor and PP2C protein phosphatase, and it is plausible that these mutations may contribute to high-level resistance to fifth-generation cephalosporins. Deletion of the ClpX gene in MRSA strain USA300 was previously shown to increase the level of resistance to beta-lactam antibiotics [27]. However, this is the first reported identification of mutations in ClpX occurring from selection with a beta-lactam antibiotic (ceftaroline / ceftobiprole). The Rho transcription termination factor has been shown to be associated with reduced susceptibility to beta-lactam antibiotics, with Rho-null mutants demonstrating reduced sensitivity to cephalosporins [28]. Finally, a PP2C-type protein phosphatase (IreP) has been implicated in a signal transduction system that controls cephalosporin resistance in Enterococcus faecalis [29].
Supporting Information
Tabs indicate the changes in a given strain relative to the comparison strain (“SRT-SF8300” refers to changes present in strain SRT when compared to strain SF8300).
(XLSX)
Acknowledgments
This work was funded in part by NIH grant R01-AI100291 (to HFC), NIH grant R01-HL105704 (to CYC), and an Abbott Pathogen Discovery Award (to CYC). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data Availability
All genomic data from this study have been deposited publicly in NCBI under BioProject PRJNA293093.
Funding Statement
This work was funded in part by NIH (National Institutes of Health) grant R01-AI100291 (to HFC), NIH grant R01-HL105704 (to CYC), and an Abbott Pathogen Discovery Award (to CYC). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Boucher HW, Corey GR. Epidemiology of methicillin-resistant Staphylococcus aureus. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2008;46 Suppl 5:S344–9. 10.1086/533590 . [DOI] [PubMed] [Google Scholar]
- 2.Chambers HF. The changing epidemiology of Staphylococcus aureus? Emerging infectious diseases. 2001;7(2):178–82. 10.3201/eid0702.700178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dantes R, Mu Y, Belflower R, Aragon D, Dumyati G, Harrison LH, et al. National burden of invasive methicillin-resistant Staphylococcus aureus infections, United States, 2011. JAMA internal medicine. 2013;173(21):1970–8. 10.1001/jamainternmed.2013.10423 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beresford E, Biek D, Jandourek A, Mawal Y, Riccobene T, Friedland HD. Ceftaroline fosamil for the treatment of acute bacterial skin and skin structure infections. Expert review of clinical pharmacology. 2014;7(2):123–35. 10.1586/17512433.2014.884457 . [DOI] [PubMed] [Google Scholar]
- 5.Corey GR, Wilcox M, Talbot GH, Friedland HD, Baculik T, Witherell GW, et al. Integrated analysis of CANVAS 1 and 2: phase 3, multicenter, randomized, double-blind studies to evaluate the safety and efficacy of ceftaroline versus vancomycin plus aztreonam in complicated skin and skin-structure infection. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2010;51(6):641–50. 10.1086/655827 . [DOI] [PubMed] [Google Scholar]
- 6.File TM Jr, Low DE, Eckburg PB, Talbot GH, Friedland HD, Lee J, et al. Integrated analysis of FOCUS 1 and FOCUS 2: randomized, doubled-blinded, multicenter phase 3 trials of the efficacy and safety of ceftaroline fosamil versus ceftriaxone in patients with community-acquired pneumonia. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2010;51(12):1395–405. 10.1086/657313 . [DOI] [PubMed] [Google Scholar]
- 7.Alm RA, McLaughlin RE, Kos VN, Sader HS, Iaconis JP, Lahiri SD. Analysis of Staphylococcus aureus clinical isolates with reduced susceptibility to ceftaroline: an epidemiological and structural perspective. The Journal of antimicrobial chemotherapy. 2014;69(8):2065–75. 10.1093/jac/dku114 . [DOI] [PubMed] [Google Scholar]
- 8.Biedenbach DJ, Alm RA, Lahiri SD, Reiszner E, Hoban DJ, Sahm DF, et al. In Vitro Activity of Ceftaroline against Staphylococcus aureus Isolated in 2012 from Asia-Pacific Countries: AWARE Surveillance Program. Antimicrobial agents and chemotherapy. 2015. 10.1128/AAC.01867-15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones RN, Mendes RE, Sader HS. Ceftaroline activity against pathogens associated with complicated skin and skin structure infections: results from an international surveillance study. The Journal of antimicrobial chemotherapy. 2010;65 Suppl 4:iv17–31. 10.1093/jac/dkq252 . [DOI] [PubMed] [Google Scholar]
- 10.Chan LC, Basuino L, Diep B, Hamilton S, Chatterjee SS, Chambers HF. Ceftobiprole- and ceftaroline-resistant methicillin-resistant Staphylococcus aureus. Antimicrobial agents and chemotherapy. 2015;59(5):2960–3. 10.1128/AAC.05004-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Long SW, Olsen RJ, Mehta SC, Palzkill T, Cernoch PL, Perez KK, et al. PBP2a mutations causing high-level Ceftaroline resistance in clinical methicillin-resistant Staphylococcus aureus isolates. Antimicrobial agents and chemotherapy. 2014;58(11):6668–74. 10.1128/AAC.03622-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banerjee R, Gretes M, Basuino L, Strynadka N, Chambers HF. In vitro selection and characterization of ceftobiprole-resistant methicillin-resistant Staphylococcus aureus. Antimicrobial agents and chemotherapy. 2008;52(6):2089–96. 10.1128/AAC.01403-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banerjee R, Gretes M, Harlem C, Basuino L, Chambers HF. A mecA-negative strain of methicillin-resistant Staphylococcus aureus with high-level beta-lactam resistance contains mutations in three genes. Antimicrobial agents and chemotherapy. 2010;54(11):4900–2. 10.1128/AAC.00594-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17(1). [Google Scholar]
- 15.Li H. Pairfq—Research Computing Center Wiki [updated 5 March 2015]. Available: https://wiki.gacrc.uga.edu/wiki/Pairfq. Accessed 17 August 2015.
- 16.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–9. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macheboeuf P, Contreras-Martel C, Job V, Dideberg O, Dessen A. Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol Rev. 2006;30(5):673–91. 10.1111/j.1574-6976.2006.00024.x . [DOI] [PubMed] [Google Scholar]
- 18.Zapun A, Contreras-Martel C, Vernet T. Penicillin-binding proteins and beta-lactam resistance. FEMS Microbiol Rev. 2008;32(2):361–85. 10.1111/j.1574-6976.2007.00095.x . [DOI] [PubMed] [Google Scholar]
- 19.Leski TA, Tomasz A. Role of penicillin-binding protein 2 (PBP2) in the antibiotic susceptibility and cell wall cross-linking of Staphylococcus aureus: evidence for the cooperative functioning of PBP2, PBP4, and PBP2A. Journal of bacteriology. 2005;187(5):1815–24. 10.1128/JB.187.5.1815-1824.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Das AK, Helps NR, Cohen PT, Barford D. Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 A resolution. The EMBO journal. 1996;15(24):6798–809. [PMC free article] [PubMed] [Google Scholar]
- 21.Kosowska-Shick K, McGhee PL, Appelbaum PC. Affinity of ceftaroline and other beta-lactams for penicillin-binding proteins from Staphylococcus aureus and Streptococcus pneumoniae. Antimicrobial agents and chemotherapy. 2010;54(5):1670–7. 10.1128/AAC.00019-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendes RE, Deshpande LM, Smyth DS, Shopsin B, Farrell DJ, Jones RN. Characterization of methicillin-resistant Staphylococcus aureus strains recovered from a phase IV clinical trial for linezolid versus vancomycin for treatment of nosocomial pneumonia. Journal of clinical microbiology. 2012;50(11):3694–702. 10.1128/JCM.02024-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moisan H, Pruneau M, Malouin F. Binding of ceftaroline to penicillin-binding proteins of Staphylococcus aureus and Streptococcus pneumoniae. The Journal of antimicrobial chemotherapy. 2010;65(4):713–6. 10.1093/jac/dkp503 . [DOI] [PubMed] [Google Scholar]
- 24.Memmi G, Filipe SR, Pinho MG, Fu Z, Cheung A. Staphylococcus aureus PBP4 is essential for beta-lactam resistance in community-acquired methicillin-resistant strains. Antimicrobial agents and chemotherapy. 2008;52(11):3955–66. 10.1128/AAC.00049-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henze UU, Berger-Bachi B. Penicillin-binding protein 4 overproduction increases beta-lactam resistance in Staphylococcus aureus. Antimicrobial agents and chemotherapy. 1996;40(9):2121–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar K, Anitha P, Sivasakthi V, Bag S, P l, A A, et al. In silico study of penicillin derivatives and cephalosporins for upper respiratory tract bacterial pathogens. 3 Biotech. 2014;4:241–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baek KT, Grundling A, Mogensen RG, Thogersen L, Petersen A, Paulander W, et al. beta-Lactam resistance in methicillin-resistant Staphylococcus aureus USA300 is increased by inactivation of the ClpXP protease. Antimicrobial agents and chemotherapy. 2014;58(8):4593–603. 10.1128/AAC.02802-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee YH, Helmann JD. Mutations in the primary sigma factor sigmaA and termination factor rho that reduce susceptibility to cell wall antibiotics. Journal of bacteriology. 2014;196(21):3700–11. 10.1128/JB.02022-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kristich CJ, Little JL, Hall CL, Hoff JS. Reciprocal regulation of cephalosporin resistance in Enterococcus faecalis. mBio. 2011;2(6):e00199–11. 10.1128/mBio.00199-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Navratna V, Nadig S, Sood V, Prasad K, Arakere G, Gopal B. Molecular basis for the role of Staphylococcus aureus penicillin binding protein 4 in antimicrobial resistance. Journal of bacteriology. 2010;192(1):134–44. 10.1128/JB.00822-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tabs indicate the changes in a given strain relative to the comparison strain (“SRT-SF8300” refers to changes present in strain SRT when compared to strain SF8300).
(XLSX)
Data Availability Statement
All genomic data from this study have been deposited publicly in NCBI under BioProject PRJNA293093.