Abstract
Nanodiscs are soluble nanoscale phospholipid bilayers which can self-assemble integral membrane proteins for biophysical, enzymatic or structural investigations. This means for rendering membrane proteins soluble at the single molecule level offers advantages over liposomes or detergent micelles in terms of size, stability, ability to add genetically modifiable features to the Nanodisc structure and ready access to both sides of the phospholipid bilayer domain. Thus the Nanodisc system provides a novel platform for understanding membrane protein function. We provide an overview of the Nanodisc approach and document through several examples many of the applications to the study of the structure and function of integral membrane proteins.
Keywords: Nanodiscs, Membrane Proteins, Self-Assembly
1. Introduction
Membrane proteins have been difficult to study from the mechanistic perspective as many of the biophysical and chemical techniques applicable to soluble enzymes fail to deal with insoluble aggregates. Ideally, one would prefer to have a membrane protein of interest in a solubilized state for ease in purification, functional biochemical assay, application of various biophysical methods and spectroscopies, crystallization for structure determination and biochemical manipulations that maintain the target protein in a stable state. Historically, membrane protein solubilization utilized detergents to form mixed detergent-protein-lipid micelles. However, detergent poses a hazard to membrane protein stability and the excess micellar phase can interfere with many assay techniques and often has non-ideal optical properties (absorbance and light scattering) as well as undesired partitioning of substrates and products into the excess detergent micelle. Detergent also presents technical obstacles during the manipulation of membrane proteins as they often co-concentrate with the protein target and can lead to inactive or denatured entities. Furthermore, many membrane protein systems require specific types of phospholipids to maintain active function, a requirement which is not mimicked by detergent micelles. Liposome preparations have been used to incorporate membrane proteins and this approach has been found to be useful when compartmentalization of each side of the bilayer is needed, as for example in the assay of ion channels. However liposomes are large, unstable and difficult to prepare with precisely controlled size and stoichiometry.
Nanodisc technology offers a solution to some of these challenges. In this approach, the membrane protein target is transiently solubilized with a detergent in the presence of phospholipids and an encircling amphipathic helical protein belt, termed a membrane scaffold protein (MSP) [1]. When the detergent is removed, by dialysis or adsorbtion to hydrophobic beads, the target membrane protein simultaneously assembles with phospholipids into a discoidal bilayer with the size controlled by the length of the MSP. The resultant Nanodiscs thus keep membrane proteins in solution, provide a native-like phospholipid bilayer environment that provides stability and functional requirements of the incorporated target and also allow control of the oligomeric state of the target membrane protein. Nanodiscs thus provide a cassette, rendering membrane proteins soluble at the single molecule level, and opening up structural-functional investigations that were heretofore limited to the class of soluble proteins and enzymes. Membrane proteins having many different topologies have been introduced into Nanodiscs (Table I). In addition, the provision of a soluble membrane surface with defined phospholipid composition has provided a means to investigate the mechanism of molecular recognition between protein and membranes. In the ensuing sections we highlight the utility of the Nanodisc platform through several specific examples and suggest future applications.
Table I.
Proteins, detergents and phospholipids used for Nanodisc formation.
| Target Protein Class | Phospholipids | Detergents |
|---|---|---|
| Single TM, Seven TM, Multi-TM, Cytochrome P450s, Multi-protein complex, Peripheral, Tethered | DPPC, DMPC, POPC, PC/PS, PC/PE, E. coli lipids, Sf9 membrane, PC/PG, PC/DOTAP, soy PC, egg PC, soy asolectin | CHAPS, cholate, Cymal, deoxycholate, digitonin, dihexanoyl PC, dodecylmaltoside, Emulgen 911, FOS-choline, octylglucoside, sodium dodecylsulfate, Triton X-100, Tween 20 |
2. What is a Nanodisc?
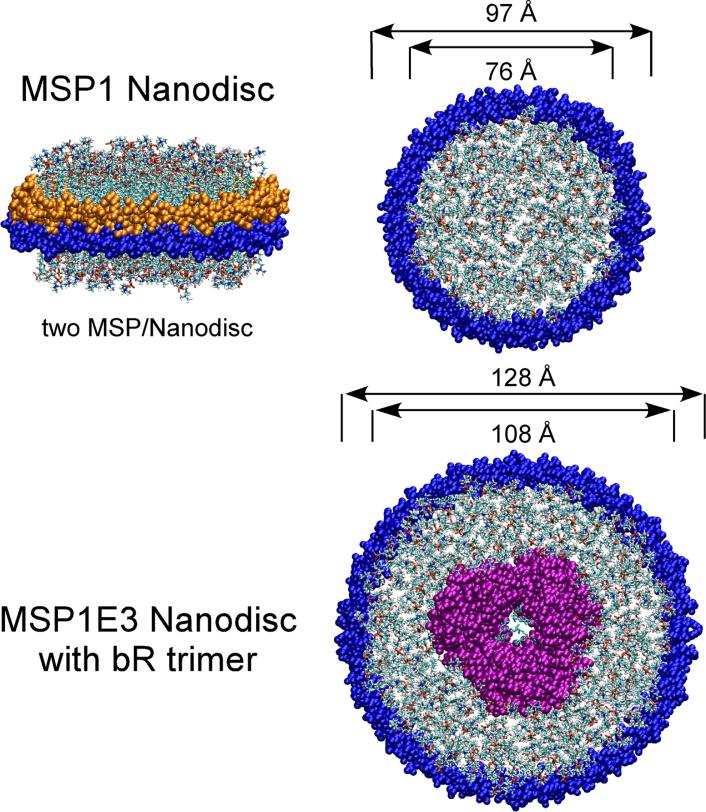
The Nanodisc is a non-covalent assembly of phospholipid and a genetically engineered “membrane scaffold protein” (MSP) which itself is based upon the sequence of human serum apolipoprotein A-I. The phospholipid associates as a bilayer domain while two molecules of MSP wrap around the edges of the discoidal structure in a belt-like configuration, one MSP covering the hydrophobic alkyl chains of each leaflet (Figure 1). A detailed picture of the Nanodisc self-assembly process has emerged from a combination of theoretical simulations using coarse grain and whole-atom molecular dynamics and solution x-ray scattering [2,3]. A critical component, the MSP, is related to the serum apolipoproteins that are the primary component of high density lipoproteins (rHDL). The latest MSP sequences were engineered into a synthetic gene optimized for expression in E. coli and include various affinity tags (6xHis, FLAG, Cys, etc.) and of varying lengths which control the overall Nanodisc size (see Table II). Although a relatively new technology, we have spent considerable effort over the past few years to characterize Nanodiscs and their assemblies with integral membrane protein targets. For instance, the phospholipid bilayer and structural organization of the Nanodisc has been probed by atomic force microscopy and analyzed using small angle x-ray scattering (SAXS), confirming that the Nanodisc contains a phospholipid bilayer with MSP associated at the edge [1,4,5]. The belt organization was directly proven by solid state magic angle spinning NMR of a uniformly labeled 13C,15N-labeled MSP [6].
Figure 1.
Illustrations of Nanodisc structures. Top panel: Nanodiscs composed of MSP1D1 and phospholipid shown in side view and top view. The two MSPs are colored orange and blue. Bottom panel: Nanodisc composed of MSP1E3D1, phospholipid and bR trimer (twenty-one transmembrane helices). The MSP1D1 and MSP1E3D1 structures are drawn to the same scale for comparison.
Table II.
| Phospholipid | |||||
|---|---|---|---|---|---|
| MSP type | POPC | DPPC | DMPC | Diameter, Å | bilayer area, Å2 |
| MSP1D1 | 61 | 82 | 77 | 98 | 4400 |
| MSP1E1D1 | 79 | 106 | 102 | 106 | 5700 |
| MSP1E2D1 | 103 | 134 | 122 | 119 | 7200 |
| MSP1E3D1 | 125 | 167 | 148 | 129 | 8900 |
Some simple rules and relationships arise from the belt-disk organization, providing a check for self-consistency. The diameter of the Nanodisc is dictated by the length of the MSP belt at the optimum lipid content. This relationship is supported by experimental SAXS and size exclusion chromatography (SEC) on data obtained using MSPs of different lengths [4,5]. MSP and apolipoprotein AI consist of 22-mer helical repeats punctuated by proline and glycine. A series of larger MSP lengths were constructed by adding additional 22-mer repeat units. Homogeneous populations of the resulting Nanodiscs are routinely characterized by SEC, analyzed for phospholipid content and structurally defined by solution x-ray scattering [4]. These biophysical efforts confirmed that Nanodisc diameter, MSP length, area per phospholipid and number of phospholipids per Nanodisc are all interrelated, as expected.
Another consequence of the MSP belt-length/Nanodisc diameter relationship is that a high yield of homogeneous Nanodiscs requires a defined ratio of phospholipid to MSP during the assembly process. If the lipid ratio during formation is too high, populations of large particles are formed along with the Nanodiscs because a higher area to perimeter is needed match the length of the hydrophobic MSP belt to the amount of phospholipid and total bilayer surface area. The apolipoprotein literature contains many descriptions of in vitro reconstituted high density lipoprotein (rHDL) particles containing integral numbers of apolipoprotein AI with two, three and four associated with rHDL of increasing diameter and phospholipid content [7,8]. MSP can also form lipid-poor particles if the ratio of lipid to MSP is too low or unfavorable conditions are used for disk formation. The phase diagram for assembling homogeneous Nanodisc preparations has been determined [1] and molecular dynamics simulations demonstrated the deformation of disks containing too few phospholipids because of the energetic requirement of matching the length of the MSP belt to the length of the bilayer hydrophobic edge [9,10]. Critical to obtaining homogeneous size assemblies is genetic engineering to delete from the Apo-AI sequence the amino terminal residues that have low affinity for the discoidal bilayer state [4].
Thus, Nanodiscs are an ideal model membrane system with defined size and phospholipid composition. The membrane can be composed of different mixtures of phospholipid types as well as other components such as cholesterol among others (Table I). Hence the bilayer can be tailored in composition to suit a membrane protein of interest and varied in composition to examine functional effects of the bilayer environment. The Nanodisc bilayer undergoes a phase transition similar to that of the pure phospholipid component, though shifted by a few degrees to higher temperatures and broadened due to the presence of the MSP [5,11] and are quite similar to proteoliposomes. The Nanodisc-membrane protein particle, due to its small size and robust nature, can be treated much as a soluble protein target would be, such as subjected to chromatography, rapid reaction methods, studied in solution phase at varying temperatures, frozen, lyophilized and attached to matrices or surfaces through engineered MSP.
3. How are Nanodiscs formed?
Nanodiscs assemble from a mixture of detergent/phospholipid micelles and MSP upon removal of the detergent. The phospholipid (PL) to MSP molar stoichiometry is critical in this process and is guided by considering the length of the MSP belt, which determines the energetic potential well for the optimal disc radius. If the exact PL/MSP ratio is used, there is complete self-assembly with a homogeneous size of Nanodiscs formed with little else in solution. If the ratio is slightly off, the excess PL or scaffold protein will appear as an aggregate in the void volume of a size exclusion column. If the ratio is far from optimum, however, a wide variety of lipoprotein particles are formed, certainly some Nanodiscs, but a large quantity of aggregates of varying size. Hence if one is trying a new lipid or lipid mixture it is imperative to determine the correct stoichiometry empirically. The importance of this parameter leads to a quandary when membrane protein targets are included. Here it is often not known how many lipids the target protein will displace when assembled into the bilayer and hence the optimal ratios of lipid to MSP are not known a priori. One experimental solution is to reconstitute with a large excess of empty Nanodiscs to minimize the ill-effects of the unknown contribution of target protein to the membrane surface area. If, however, one is constrained to assembling at a high ratio of target to Nanodiscs, then it is critical to carry out assembly at varying ratios of lipid with success monitored by homogeneous SEC results.
The detergent used for initial solubilization is also critical. Sodium cholate is ideal for the lipid fraction and is usually present at a 2:1 mole ratio to total PL, or higher. Other detergents can work equally well if the phospholipid goes fully into a clear solution of micelles. Some researchers are hesitant to expose their membrane protein to detergents like cholate, however the exposure time is brief and negative effects are attenuated by the presence of the phospholipid. Mixed detergents are another successful approach, with cholate solubilizing the lipid and the secondary detergent (alkyl maltoside or glucoside, polyoxyethylene glycols, phoscholines, CHAPS, etc.) dealing with the protein target (Table I). In all cases, the assembly process is initiated by removal of detergent by dialysis (for dialyzable detergents) or treatment with porous polystyrene beads (Biobeads SM2 or Amberlite XAD2). It should be noted that detergent removal by beads is both detergent and temperature-dependent. For information on bead-based detergent removal see [12,13]. Interestingly, we have shown that, contrary to intuition, the loss of MSP, phospholipid or other components by adsorption to the beads is minor. The temperature during assembly is also important, with Nanodiscs forming most efficiently near the phase transition temperature of the phospholipid. The reason for this may be construed as an effect of the phase behavior and possibly the size and organization of the phospholipid/detergent micelle at some point during, or perhaps throughout the process of detergent removal. For example, rHDL is known to form from mixtures of phospholipid vesicles and Apo AI at the phase transition temperature, presumably due to the presence of bilayer defects. Nanodisc formation depends upon the initial state of the mixture of phospholipid, cholate and MSP. Phospholipid in a mixed lamellar-micellar phase at the start of detergent removal correlates with poor Nanodisc formation and the presence of lipid-poor particles [1]. There is also the feeling, though with little experimental data, that the speed of detergent removal is also important. The desorption rate from target protein(s), lipid and scaffold are different and dependent on the choice of detergent(s) used. Hence the ratio of hydrophobic beads to protein/lipid is an additional parameter in the assembly process.
The addition of cholate to preformed Nanodiscs may be interpreted as the reverse of the process of Nanodisc formation as detergent is removed. The process of Nanodisc disassembly by addition of cholate was also studied by a combination of molecular dynamics simulation and SAXS [2]. Disassembly proceeds with cholate insinuating itself between the MSP and the edge of the phospholipid bilayer domain. Further additions of cholate result in a perturbation of the MSP, which starts to fluctuate in spatial structure. An even further increase in the cholate to phospholipid ratio results in the appearance of a more spherical particle shape with MSP still associated. The experimental SAXS results agreed qualitatively with X-ray scattering calculated from the output of the molecular dynamics simulations.
Although there have been several recent theoretical approaches to understanding the Nanodisc assembly process [2,3,9,14]clearly more effort is needed for a clear picture of the physics and kinetics.
4. How are proteins assembled into Nanodiscs?
In the majority of cases utilized in our laboratory, the membrane protein target of interest is completely pre-solubilized with a compatible detergent and mixed with the Nanodisc assembly components. Care is required in these steps since it is widely recognized that removal of a protein from the membrane can be most difficult: Even as the protein appears to be solubilized it can still be aggregated or undergo time-dependent aggregation [15]. If detergent solubilization is accomplished, however, it is very likely that the protein will self-assemble with PL and MSP into Nanodiscs when the detergent is removed. In a simple scenario one can think of the membrane protein as a solute in the phospholipid/detergent phase and Nanodisc formation proceeding as usual upon detergent removal. As detergent is removed, the relevant recognition events form target protein – lipid and lipid-lipid contacts as the Nanodisc bilayer formed, with the target protein ultimately incorporated into the bilayer in its native like configuration [9]. With more complex protein targets, the situation is more complicated due to multiple competing pathways such as occurs when the protein tends to self-aggregate [16]. The critical branch point is this non-productive self-aggregation and the formation of the correct protein-lipid contacts. The presence of large self-aggregates can usually be detected by size exclusion chromatography, although sometimes these elute near that of correctly formed Nanodiscs. If aggregates are suspected, a re-injection of a fraction of the peak can often be used to verify this scenario. A simple means to overcome the self-aggregation is to use a large excess of Nanodisc components in the reconstitution, i.e. excess lipid and MSP.
A precise dynamical and structural picture of membrane protein self-assembly into Nanodiscs is in its infancy. One interesting question is at what point in detergent removal is a membrane protein “trapped” within the forming Nanodisc structure? For example, the rate of detergent removal and Nanodisc formation could outpace the rate of productive membrane protein forming oligomers. There are examples of liposome reconstitutions where the speed of detergent removal and also type of detergent results in an enrichment of protein compared to phospholipid in a fraction of liposomes [16]. This outcome suggests a coexistence of different phases and preferential partitioning of membrane protein during removal of detergent. Experimental observations, as well as molecular dynamics, of what is occurring during the assembly of proteins into the Nanodisc and the properties of the mixed micelles of membrane protein, lipid, detergent and MSP along the path to Nanodisc formation would be very useful and an important inroad to understanding chemical and biological self-assembly.
5. Examples of proteins assembled into Nanodiscs
5.1 Cytochrome P450
A particularly powerful aspect of the Nanodisc system is that it can be used to isolate protein in a known monomeric or oligomeric state, a task difficult or impossible in liposomes or detergents. CYP 3A4, a human hepatic drug metabolizing cytochrome P450, is an example in which the state of the protein and thus function is affected by aggregation [17]. CYP3A4 shows higher apparent cooperativity of multiple testosterone binding and nearly full spin conversion of the heme iron upon ligand binding in Nanodiscs compared to detergent-solubilized preparations, with such differences attributed to detergent-induced effects and/or aggregation. In the absence of detergent, the aggregate displays multi-exponential kinetics of reduction by dithionite due to heterogeneity of the enzyme [18]. In contrast, monomeric CYP3A4 in Nanodiscs displays clean monophasic reduction kinetics. CYP3A4 in liposomes at high lipid stoichiometry also behaves as homogeneous monomer. At lower lipid stoichiometry heterogeneous behavior arises due to self-association of the CYP3A4, while Nanodiscs prohibit dynamic self-association as there is only one enzyme present within the particle. Similarly, redox potential measurements using Nanodiscs provide a homogeneous monomeric form of the enzyme for facile electrode interactions [19]. The Nanodisc thus represents a clean way to monitor molecular function of CYP in a bilayer as a monomeric species [20,21].
Assembly of multiple integral membrane proteins, starting with purified targets, was demonstrated in the case of CYP3A4 and its redox partner, cytochrome P450 reductase [21], as well as Arabidopsis CYP73A5, a cinnamate hydroxylase, with its P450 reductase using heterologously expressed crude membrane fractions [22]. With heterologously expressed proteins, isolating a Nanodisc with P450 and reductase is accomplished using differential affinity tags, in this case ADP-sepharose as affinity ligand for the reductase and metal chelate chromatography to bind the histidine tag on the CYP3A4. Nanodiscs can also be assembled directly from crude membrane preparations to afford the target protein and native lipids in the resulting Nanodisc bilayer [23]. Structural investigations of membrane proteins are also enabled by the Nanodisc technology. For example, Magic-angle solid state NMR using 13C,15N-labeled proteins was used to gain structural insight of the encircling MSP as well as incorporated human CYP3A4. [24].
5.2 Blood Coagulation and Human Tissue Factor
An interesting use of Nanodiscs to control the microenvironment around a protein was realized in investigations of blood clotting [25] where the activity is dependent on the phospholipid composition. The complex of the integral membrane protein tissue factor (TF) together with the soluble factor VIIa (FVIIa) initiates the blood coagulation cascade. The recruitment of FVIIa requires acidic phospholipid and calcium cation to react via the γ-carboxyglutamic acid (Gla) domains present in the factors. Acidic phospholipids such as phosphatidylserine (PS) tend to undergo cluster formation in large-scale bilayers due to multi-modal chelate interactions with divalent cations or charge interaction with positively charged proteins [26]. A Nanodisc is 600 fold smaller in bilayer surface area compared to a typical 100 nm diameter liposome, thus prohibiting the large-scale clustering of PS that occurs in liposomes and controlling the localized number of PS molecules [27]. This example also illustrates how the Nanodisc system can be used to provide coupling to a surface for precise measurement of macromolecular association. In this case the binding of factor X and factor VII were measured by surface plasmon resonance [25].
5.3 Bacteriorhodopsin
The light driven proton pump bacteriorhodopsin (bR) is a mainstay of membrane protein research. bR incorporates very efficiently into Nanodiscs as a monomer (70-90%) and the Nanodisc-bR monomer was used to address the structure and function of a multi-pass membrane protein Nanodisc assembly [28]. The resulting size and shape of the nanoparticle assembly was determined by size exclusion chromatography, atomic force microscopy and transmission electron microscopy. The stoichiometry and composition were measured spectroscopically and chemically, revealing that the assembled Nanodisc complex contained two MSP, one bR and 160 molecules of phospholipid (DMPC). Native function of bR was determined using spectroscopic identification of the M410 photocycle intermediate and retinal binding. Determination of phospholipid bilayer organization was inferred from measurement of fluorescent lipidic probe orientation in bR-Nanodiscs oriented on a glass surface and from electron microscopy with a resultant structural picture that is illustrated in Figure 1.
Membrane protein oligomers are a focus of interest and again bR proved to be a useful subject [29]. Bacteriorhodopsin forms 2-D crystals composed of trimers in its native state. bR in its trimeric form shows a positive and a negative peak in the circular dichroism spectrum of the chromophore due to exciton splitting arising from the geometry of the three chromophores thus providing an easy assay of oligomerization [30]. The reconstitution was optimized at three bR per Nanodisc by varying the phospholipid ratio and assessing the reconstitution by SEC. Four sizes of Nanodiscs were tested. Analysis of the main Nanodisc-bR peak revealed that the two smallest Nanodiscs did not exhibit the trimer circular dichroism signature presumably due to insufficient amounts of phospholipid. In larger Nanodiscs, more trimer formed than one would expect for bR incorporated with random topological orientation. Therefore, oligomerization must occur before Nanodiscs are completely assembled.
5.4 G-protein coupled receptors
Insight into the functions of the large family of 7-TM receptors has been aided by biochemical and biophysical studies of rhodopsin and the β2 adrenergic receptor. We have extensively used these systems to prove the utility of Nanodiscs for the broad class of GPCR drug targets. β2AR was one of the first receptors assembled into Nanodiscs [31], with efficient assembly (54% of starting activity recovered) and resulting agonist and antagonist association constants similar to literature values. β2AR-Nanodiscs also coupled to its G-protein (Gs), however the amount of coupling based on agonist binding to high-affinity receptor was low. The groups of Kobilka and Sunahara have subsequently used rHDL (formed using apolipoprotein A-I) to reconstitute β2AR and showed that full coupling could be achieved only at very high concentrations of G-protein added to the β2AR rHDL [32]. The solubility and therefore accessibility of G-protein is an issue when adding G-proteins highly modified with hydrophobic lipids to preformed receptor-Nanodiscs.
Rhodopsin is a light-activated GPCR present in photoreceptor cells of the retina and has been proposed to form dimers and higher order oligomers based on AFM and EM studies [33,34]. We reconstituted rhodopsin into our larger Nanodiscs with both one and two rhodopsin molecules per Nanodisc and compared their functionality [35]. The rhodopsin monomer Nanodiscs assemble at high yield in the presence of a five-fold excess of Nanodiscs. Two-rhodopsin Nanodiscs were formed at an assembly ratio of two rhodopsins per Nanodisc. One- and two-rhodopsin Nanodiscs could be separated and purified on sucrose density gradients. Both species were found to activate transducin with high efficiency, near the diffusional rate limit. The Nanodiscs containing two rhodopsins were half as efficient as Nanodiscs containing one rhodopsin on a per-rhodopsin basis. Binding of transducin was measured using an extra-metaII assay in which transducin binding converts MI absorbing at 460 nm to MII absorbing at 380 nm [36]. The result was that while the binding affinities were about the same, only half of the rhodopsin present in a two-rhodopsin Nanodisc could form MII at saturating amounts of transducin. One hypothesis is that dimers form and that only one subunit in a dimer can interact with transducin at a time due to steric reasons [35]. More recently, rHDL-rhodopsin monomers have also been used to address the possible requirement for dimerization [37]. The common conclusion of these investigations is that monomeric rhodopsin can efficiently conduct signal transduction.
5.5 Bacterial Chemoreceptor
More complex assemblies of integral membrane proteins are also possible to achieve in Nanodiscs. Tar is a bacterial chemoreceptor that can form trimers of dimers and extended arrays of trimers of dimers. To understand function of single dimers versus trimers of dimers, Tar was incorporated into Nanodiscs as a way of controlling the stoichiometry of interaction [38]. By using excess Nanodisc component, single dimers were found in Nanodiscs. Upon decreasing the Nanodisc component in the assembly mixture, multiple dimers were found per Nanodisc with the average number dependent on the assembly ratio. Functional assays for ligand binding, CheR catalyzed methylation, phospho-CheB-catalyzed deamination and kinase activity were performed on samples containing varying numbers of dimers. Single dimers were found to bind ligand, transmembrane signal, promote deamination and methylation, indicating that trimers are not necessary for these functions. However, kinase activation showed a peak value at an average of three dimers per Nanodisc.
5.6 The Peptide Translocon Complex
SecYEG is a protein translocon complex that requires oligomers for function. The protomer, a heterotrimer having 15 transmembrane helices was put into Nanodiscs as single protomers for functional studies [39,40]. A fundamental question was what functions can be attributed to protomer and what functions can be attributed to higher order oligomers. SecA, a soluble motor protein, was found to bind the SecYEG protomer. Additionally phosphatidylglycerol co-incorporated into the Nanodiscs was found to increase the affinity for SecA. Syd, a SecYEG interacting protein, was found to bind the SecYEG protomer and to displace SecA. This study demonstrates the use of Nanodiscs to control membrane protein association state in a membrane of defined composition to determine functional aspects of the membrane protein-soluble partner interactions.
5.7 Receptor Tyrosine Kinase
Epidermal growth factor receptor (EGFR) has been assembled into Nanodiscs to confer stability of its kinase activity [41]. The EGF-bound dimer in detergent was used as starting material for Nanodisc assembly. Single EGFR dimers were placed into Nanodiscs and kinase activity was demonstrably stabilized in Nanodiscs, as well as liposomes, with 80% activity remaining at 24 hours compared to 28% remaining activity in detergent.
6. Nanodiscs on surfaces
A powerful feature of Nanodiscs is the ability to attach the membrane protein stabilized in Nanodiscs to surfaces via tags on the membrane scaffold protein (MSP). Various affinity tags can be attached to the MSP. For instance, the MSP 6xHis tag has been used to bind PS-containing Nanodiscs to a Ni-NTA biosensor chip to measure binding of factor X and of arrestin using SPR [25,42]. Further, SPR was used to measure binding of cholera toxin to ganglioside-containing Nanodiscs which were immobilized using similar methodology [43]. Nanodiscs containing rhodopsin were patterned via binding of the 6xHis tags on Nanodiscs to a nickel charged triaza-terminated self-assembled monolayer of alkanethiols. Interaction of light-activated rhodopsin with its G-protein, transducin, was observed using SAMDI-TOF (self-assembled monolayers for matrix assisted laser desorption ionization) mass spectrometry [44]. The detection is label-free and enables identification of the G-protein specificity of a receptor based on mass.
LSPR (localized surface plasmon resonance) is observed when light interacts with metal nanoparticles. LSPR is highly dependent on the refractive index of the surrounding media and upon analyte binding the extinction maximum wavelength of the nanoparticle shifts, providing a mechanism for chemical and biological sensing. The localized nature of the plasmon makes it much more sensitive to changes in the local environment compared to gold films typically used for SPR. CYP3A4 in Nanodiscs was covalently coupled using carbodiimide chemistry to silver nanoparticle surfaces generated using nanosphere lithography. Drug binding to CYP3A4-Nanodisc on the nanoparticle surface was detected using LSPR wavelength shift [45]. The device was also able to detect the mode of interaction of drugs with the enzyme because of the strong coupling between the molecular resonances of heme in cytochrome P450 and the nanoparticles LSPR.
Nanodiscs also simply adsorb to glass or mica surfaces reproducibly with the bilayer plane parallel to the surface [46]. Such adsorption has been used to orient Nanodiscs and protein containing Nanodiscs for interrogation by atomic force microscopy [47-50]. Functional lipidic groups may also be used such as biotinylated lipid that will interact with streptavidin treated surfaces [42,51]. Nanodiscs containing biotinylated lipid have been patterned using microfluidic channels and challenged with streptavidin-coated quantum dots flowed through the same channels. (Goluch 2008).
7. Membrane Protein Structures via Nanodiscs?
Structure determination of membrane proteins is a widely-sought goal. Solid state magic angle spinning NMR of a membrane protein in Nanodiscs is an example of the use of Nanodiscs in this field [24]. Lyukmanova et al. have used rHDL particles formed with apolipoprotein A-I to incorporate the membrane active peptide antiamoebin-I and the potassium channel KcsA for solution NMR to obtain topological information on antiamoebin-I and to demonstrate the promising uses Nanodisc-like particles for high resolution solution NMR of membrane proteins [52,53]. High throughput screening has also been enabled using solution NMR [54]. The case for cryoEM of membrane proteins and 3-D particle reconstructions using Nanodiscs has also seen recent success.
8. What new applications of Nanodiscs can be envisioned?
Relevant technological applications should take advantage of the properties of the Nanodisc such as its small size compared to liposomes, less light scattering, faster diffusion, stability in shear flow, access to both sides of the protein in solution, the ability to add probes to the Nanodiscs and a means for surface attachment Thus far the only device-type technologies reported using Nanodiscs are the SAMDI-TOF [44], LSPR sensors [45] and microfluidic pattering [42] and recent results using single wall carbon nanotubes and cantilever detection modalities. Reports of apolipoprotein-based nanoparticles in cell-free expression of membrane proteins might be an emerging use of Nanodisc-like particles [55-57]. Bacteriorhodopsin co-expressed with apolipoprotein AI in the presence of liposomes and retinal cofactor appears to form active bR in nanoparticles. Preformed rHDL were also used with similar results where the soluble fraction of several membrane proteins put into the cell free expression increased to various extents [56].
Acknowledgements
This work was supported by Grants GM 33775 and GM 31756 to S.G.S.
Abbreviations
- DDM
dodecylmaltoside
- DMPC
dimyristoylphosphatidylcholine
- DPPC
dipalmitoylphosphatidylcholine
- MSP
membrane scaffold protein
- OG
octylglucoside
- PC
phosphatidylcholine
- PL
phospholipid
- POPC
1-palmitoyl-2-oleoyl phosphatidylcholine
- PS
phosphatidylserine
- rHDL
reconstituted high density lipoprotein
- SAXS
small angle x-ray scattering
- SEC
size exclusion chromatography
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bayburt TH, Grinkova YV, Sligar SG. Nano Lett. 2002;2:853–856. [Google Scholar]
- 2.Shih AY, Freddolino PL, Sligar SG, Schulten K. Nano Lett. 2007;7:1692–1696. doi: 10.1021/nl0706906. [DOI] [PubMed] [Google Scholar]
- 3.Shih AY, Freddolino PL, Arkhipov A, Schulten K. J. Struct. Biol. 2007;157:579–592. doi: 10.1016/j.jsb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. J. Am. Chem. Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 5.Denisov IG, McLean MA, Shaw AW, Grinkova YV, Sligar SG. J. Phys. Chem. B. 2005;109:15580–15588. doi: 10.1021/jp051385g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Kijac AZ, Sligar SG, Rienstra CM. Biophys. J. 2006;91:3819–3828. doi: 10.1529/biophysj.106.087072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swaney JB. J. Biol. Chem. 1980;255:877–881. [PubMed] [Google Scholar]
- 8.Brouillette CG, Jones JL, Ng TC, Kercret H, Chung BH, Segrest JP. Biochemistry. 1984;23:359–367. doi: 10.1021/bi00297a027. [DOI] [PubMed] [Google Scholar]
- 9.Shih AY, Denisov IG, Phillips JC, Sligar SG, Schulten K. Biophys. J. 2005;88:548–556. doi: 10.1529/biophysj.104.046896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Catte A, et al. Biophys. J. 2006;90:4345–4360. doi: 10.1529/biophysj.105.071456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaw AW, McLean MA, Sligar SG. FEBS Lett. 2003;556:260–264. doi: 10.1016/s0014-5793(03)01400-5. [DOI] [PubMed] [Google Scholar]
- 12.Rigaud J-L, Mosser G, Lacapere J-J, Olofsson A, Levy D, Ranck J-L. J. Struct. Biol. 1997;118:226–235. doi: 10.1006/jsbi.1997.3848. [DOI] [PubMed] [Google Scholar]
- 13.Rigaud JL, Levy D, Mosser G, Lambert O. Eur. Biophys. J. 1998;27:305–319. [Google Scholar]
- 14.Shih AY, Arkhipov A, Freddolino PL, Sligar SG, Schulten K. J. Phys. Chem. B. 2007;111:11095–11104. doi: 10.1021/jp072320b. [DOI] [PubMed] [Google Scholar]
- 15.Møller JV, Andersen JP, le Maire M. In: Progress in Protein Lipid Interactions. Watts A, Pont J.J.H.H.M.d., editors. Vol. 2. Elsevier; Amsterdam/New York: 1986. pp. 147–196. [Google Scholar]
- 16.Levy D, Gulik A, Bluzat A, Rigaud JL. Biochim. Biophys. Acta. 1992;1107:283–298. doi: 10.1016/0005-2736(92)90415-i. [DOI] [PubMed] [Google Scholar]
- 17.Baas BJ, Denisov IG, Sligar SG. Arch. Biochem. Biophys. 2004;430:218–228. doi: 10.1016/j.abb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 18.Davydov DR, Fernando H, Baas BJ, Sligar SG, Halpert JR. Biochemistry. 2005;44:13902–13913. doi: 10.1021/bi0509346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das A, Grinkova YV, Sligar SG. J. Am. Chem. Soc. 2007;129:13778–13779. doi: 10.1021/ja074864x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denisov IG, Grinkova YV, Baas BJ, Sligar SG. J. Biol. Chem. 2006;281:23313–23318. doi: 10.1074/jbc.M605511200. [DOI] [PubMed] [Google Scholar]
- 21.Denisov IG, Baas BJ, Grinkova YV, Sligar SG. J. Biol. Chem. 2007;282:7066–7076. doi: 10.1074/jbc.M609589200. [DOI] [PubMed] [Google Scholar]
- 22.Duan H, Civjan NR, Sligar SG, Schuler MA. Arch. Biochem. Biophys. 2004;424:141–153. doi: 10.1016/j.abb.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 23.Civjan NR, Bayburt TH, Schuler MA, Sligar SG. BioTechniques. 2003;35:556–563. doi: 10.2144/03353rr02. [DOI] [PubMed] [Google Scholar]
- 24.Kijac AZ, Li Y, Sligar SG, Rienstra CM. Biochemistry. 2007;46:13696–13703. doi: 10.1021/bi701411g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaw AW, Pureza VS, Sligar SG, Morrissey JH. J. Biol. Chem. 2007;282:6556–6563. doi: 10.1074/jbc.M607973200. [DOI] [PubMed] [Google Scholar]
- 26.Yang L, Glaser M. Biochemistry. 1996;35:13966–13974. doi: 10.1021/bi9610008. [DOI] [PubMed] [Google Scholar]
- 27.Morrissey JH, Pureza V, Davis-Harrison RL, Sligar SG, Ohkubo YZ, Tajkhorshid E. Thromb. Res. 2008;122:S23–S26. doi: 10.1016/S0049-3848(08)70014-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bayburt TH, Sligar SG. Protein Sci. 2003;12:2476–2481. doi: 10.1110/ps.03267503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bayburt TH, Grinkova YV, Sligar SG. Arch. Biochem. Biophys. 2006;450:215–222. doi: 10.1016/j.abb.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 30.Heyn MP, Cherry RJ, Dencher NA. Biochemistry. 1981;20:840–849. doi: 10.1021/bi00507a029. [DOI] [PubMed] [Google Scholar]
- 31.Leitz AJ, Bayburt TH, Barnakov AN, Springer BA, Sligar SG. BioTechniques. 2006;40:601–612. doi: 10.2144/000112169. [DOI] [PubMed] [Google Scholar]
- 32.Whorton Matthew R, Bokoch Michael P, Rasmussen Soren GF, Huang B, Zare Richard N, Kobilka B, Sunahara Roger K. Proc. Natl Acad. Sci. U S A. 2007;104:7682–7687. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. Nature. 2003;421:127–128. doi: 10.1038/421127a. [DOI] [PubMed] [Google Scholar]
- 34.Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. FEBS Lett. 2004;564:281–288. doi: 10.1016/S0014-5793(04)00194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bayburt TH, Leitz AJ, Xie G, Oprian DD, Sligar SG. J. Biol. Chem. 2007;282:14875–14881. doi: 10.1074/jbc.M701433200. [DOI] [PubMed] [Google Scholar]
- 36.Emeis D, Kühn H, Reichert J, Hofmann KP. FEBS Lett. 1982;143:29–34. doi: 10.1016/0014-5793(82)80266-4. [DOI] [PubMed] [Google Scholar]
- 37.Whorton MR, Jastrzebska B, Park PSH, Fotiadis D, Engel A, Palczewski K, Sunahara RK. J. Biol. Chem. 2008;283:4387–4394. doi: 10.1074/jbc.M703346200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boldog T, Grimme S, Li M, Sligar SG, Hazelbauer GL. Proc. Natl. Acad. Sci. U. S. A. 2006;103:11509–11514. doi: 10.1073/pnas.0604988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alami M, Dalal K, Lelj-Garolla B, Sligar SG, Duong F. EMBO J. 2007;26:1995–2004. doi: 10.1038/sj.emboj.7601661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dalal K, et al. J. Biol. Chem. 2009;284:7897–7902. doi: 10.1074/jbc.M808305200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mi L-Z, Grey MJ, Nishida N, Walz T, Lu C, Springer TA. Biochemistry. 2008;47:10314–10323. doi: 10.1021/bi801006s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goluch ED, Shaw AW, Sligar SG, Liu C. Lab on a Chip. 2008;8:1723–1728. doi: 10.1039/b806733c. [DOI] [PubMed] [Google Scholar]
- 43.Borch J, Torta F, Sligar SG, Roepstorff P. Anal. Chem. 2008;80:6245–6252. doi: 10.1021/ac8000644. [DOI] [PubMed] [Google Scholar]
- 44.Marin VL, Bayburt TH, Sligar SG, Mrksich M. Angew. Chem. Int. Ed. 2007;46:8796–8798. doi: 10.1002/anie.200702694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Das A, Zhao J, Schatz GC, Sligar SG, Van Duyne RP. Anal. Chem. 2009;81:3754–3759. doi: 10.1021/ac802612z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carlson JW, Jonas A, Sligar SG. Biophys. J. 1997;73:1184–1189. doi: 10.1016/S0006-3495(97)78150-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bayburt TH, Carlson JW, Sligar SG. J. Struct. Biol. 1998;123:37–44. doi: 10.1006/jsbi.1998.4007. [DOI] [PubMed] [Google Scholar]
- 48.Bayburt TH, Carlson JW, Sligar SG. Langmuir. 2000;16:5993–5997. [Google Scholar]
- 49.Bayburt TH, Sligar SG. Proc. Natl. Acad. Sci. U. S. A. 2002;99:6725–6730. doi: 10.1073/pnas.062565599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blanchette CD, et al. Biochim. Biophys. Acta. 2009;1788:724–731. doi: 10.1016/j.bbamem.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 51.Nath A, Koo PK, Rhoades E, Atkins WM. J. Am. Chem. Soc. 2008;130:15746–15747. doi: 10.1021/ja805772r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shenkarev ZO, et al. Biochemistry (Moscow) 2009;74:756–765. doi: 10.1134/s0006297909070086. [DOI] [PubMed] [Google Scholar]
- 53.Lyukmanova EN, et al. J. Am. Chem. Soc. 2008;130:2140–2141. doi: 10.1021/ja0777988. [DOI] [PubMed] [Google Scholar]
- 54.Glueck JM, Wittlich M, Feuerstein S, Hoffmann S, Willbold D, Koenig BW. J. Am. Chem. Soc. 2009;131:12060–12061. doi: 10.1021/ja904897p. [DOI] [PubMed] [Google Scholar]
- 55.Cappuccio JA, et al. Mol. Cell. Proteomics. 2008;7:2246–2253. doi: 10.1074/mcp.M800191-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Katzen F, et al. J. Proteome Res. 2008;7:3535–3542. doi: 10.1021/pr800265f. [DOI] [PubMed] [Google Scholar]
- 57.Katzen F, Peterson TC, Kudlicki W. Trends Biotechnol. 2009;27:455–460. doi: 10.1016/j.tibtech.2009.05.005. [DOI] [PubMed] [Google Scholar]