Summary
An early event in skeletal joint development is the specification of articular chondrocytes at the joint surface. Articular chondrocytes are distinct in producing lower levels of cartilage matrix and not being replaced by bone, yet how they acquire these properties remains poorly understood. Here, we show that two members of the Iroquois transcriptional repressor family, Irx7 and Irx5a, function to block chondrocyte maturation at the developing hyoid joint of zebrafish. These Irx factors suppress the production of cartilage matrix at the joint in part by preventing the activation of a col2a1a enhancer by Sox9a. Further, both zebrafish Irx7 and mouse IRX1 are able to repress cartilage matrix production in a murine chondrogenic cell line. Iroquois proteins may therefore have a conserved role in keeping chondrocytes in an immature state, with the lower levels of cartilage matrix produced by these immature cells contributing to joint flexibility.
Graphical Abstract

Introduction
Joints provide essential mobility to the vertebrate skeleton, with degeneration of joints in arthritis a leading cause of disability in humans (Hootman et al., 2012). Most joints follow a common developmental sequence. From an early pre-cartilage condensation, a subset of cells within the “interzone” are specified as articular chondrocytes while flanking cells mature into hypertrophic chondrocytes (Hartmann and Tabin, 2001 and Archer et al., 2003). Unlike growth plate chondrocytes, articular chondrocytes are not replaced by bone and rapidly shut off Col2a1 expression, which may aid in this tissue serving as a flexible cushion for joint surfaces. The larval zebrafish face has at least two sets of bilateral joints that appear to replicate the first stage of interzone formation: a “jaw joint” which articulates Meckel’s and palatoquadrate cartilages and a “hyoid joint” connecting hyomandibular and ceratohyal cartilages via a small interhyal cartilage (Figure 1A,B).
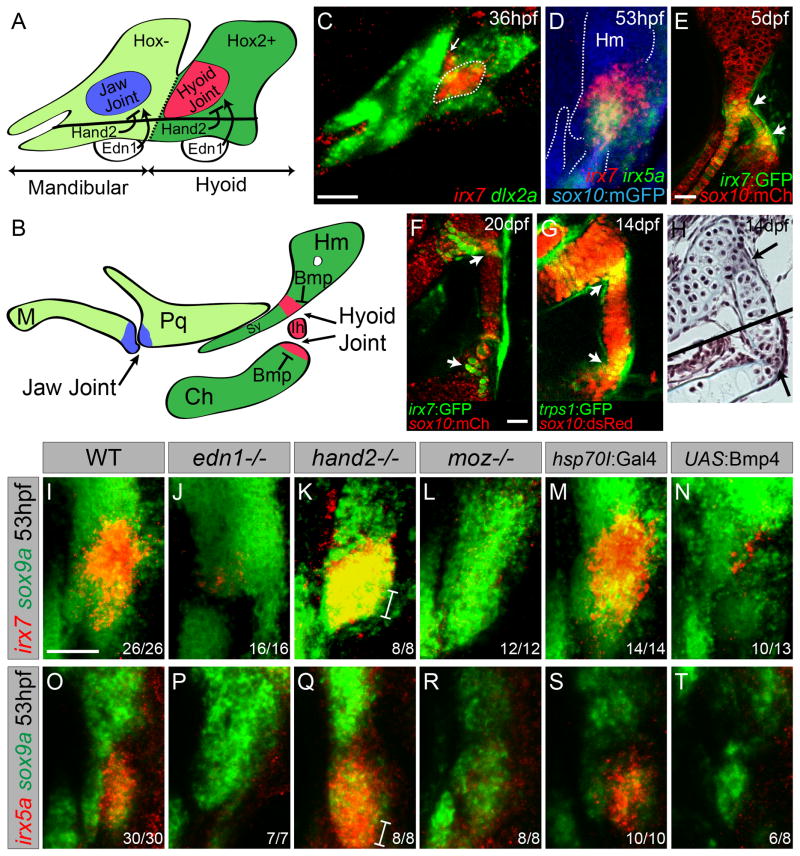
Figure 1. Regulation of irx7 and irx5a expression and character of the hyoid joint.
(A,B) Schematics of mandibular and hyoid arches at 36 hpf and cartilage derivatives at 5 dpf show early regulation of joints by Hand2 and Edn1 and late regulation by Bmp signaling.
(C) In situ hybridization at 36 hpf shows expression of irx7 (red) relative to dlx2a+ neural-crest-derived cells (green) in a domain of intermediate hyoid arch cells (dashed line), and in a portion of the first pharyngeal pouch (arrow).
(D) Relative to sox10:GFPCAAX+ cartilages (anti-GFP, blue), overlapping expression of irx7 (red) and irx5a (green) is seen at the developing hyoid joint.
(E,F) An irx7:GFP gene-trap line (green) shows increasingly restricted expression at the bipartite hyoid joint (arrows) relative to sox10:mCherryCAAX+ cartilages (red).
(G) trps1:GFP labels a similar population of cells as irx7 at each side of the hyoid joint (arrows) relative to sox10:dsRed+ cartilages.
(H) Trichrome staining shows that cells at the hyoid joints (arrows) are smaller and lack the large lacunae of neighboring hypertrophic chondrocytes. Black line denotes merging of two adjacent sections.
(I–T) Expression of irx7 and irx5a (red) relative to sox9a+ hyoid chondrocytes (green) in edn1, hand2, and moz mutants, as well as hsp70I:Gal4 control and hsp70I:Gal4; UAS:Bmp4 embryos subjected to 40–44 hpf heat-shock. Brackets show ventral expansion in hand2 mutants. M, Meckel’s; Pq, palatoquadrate; Sy, symplectic; Hm, hyomandibula; Ch, ceratohyal; Ih, interhyal. Numbers indicate proportion of animals showing the displayed phenotype. Scale bars = 30 μM. See also Figure S1.
A number of studies point to a critical balance between Gdf/Tgfβ and Bmp signaling in specifying articular versus hypertrophic chondrocyte fates at the interzone, yet their downstream targets are not well understood (Francis-West et al., 1999). Gdf5 and Gdf6 are expressed within the joint interzone and required for joint formation within the mouse limb (Settle et al., 2003). Conversely, the expression of Bmp antagonists, such as Chordin and Noggin, within the interzone suggests an important role of Bmp repression in promoting joint development (Brunet et al., 1998, Hartmann and Tabin, 2001 and Nichols et al., 2013). In support of this, Tgfβ and Bmps can bias the adoption of articular versus hypertrophic fates, respectively, upon chondrogenic differentiation of murine embryonic stem cells (Craft et al., 2013). Here, we show that transcriptional repression by Bmp signaling localizes expression of irx7 and irx5a to the developing hyoid joints.
Iroquois proteins are thought to function largely as transcriptional repressors (Cavodeassi et al., 2001). While there are three members of the Iroquois family in Drosophila (Gomez-Skarmeta et al., 1996 and McNeill et al., 1997), most terrestrial vertebrates have six members arranged into two genomic clusters: Irx1/2/4 and Irx3/5/6. These Irx genes are expressed within the developing avian and murine limbs, with the Irx1/2/4 cluster expressed in interzone regions of the digit joints and Irx3/5/6 in mesenchyme flanking each digit (McDonald et al., 2010). A role of Irx genes in limiting skeletal development is suggested by fusions of the distal phalanges in the spontaneous Fused toes mouse, which contains a large heterozygous deletion encompassing the Irx3/5/6 cluster and three other genes (Grotewold and Ruther, 2002 and Peters et al., 2002). Interestingly, an additional non-clustered Irx–gene irx7–exists only in fishes, and we find it to be expressed uniquely at the developing hyoid joint. Here, we show that Irx7, together with Irx5a, cell-autonomously repress high-level expression of cartilage matrix genes within the less mature chondrocytes of the joint interzone.
Results and Discussion
Expression of irx7 and irx5a within the developing hyoid joint
In an expression screen of the developing zebrafish face, we detected early and persistent expression of irx7 and irx5a within the joint-forming region of the second pharyngeal arch. At 36 hours post-fertilization (hpf), prior to cartilage differentiation, we observed restricted expression of irx7 within dlx2a+ mesenchymal cells of the intermediate hyoid arch, with additional weak expression in the first pharyngeal pouch (Figure 1C). Previous lineage tracing had shown this irx7+ intermediate domain to contribute to the hyoid joint and a joint-proximal extension of cartilage called the symplectic (Crump et al., 2006). At the onset of chondrogenesis (53 hpf), multi-color fluorescent in situs revealed overlapping expression of irx7 and irx5a at the developing hyoid joint, as well as in a zone connecting the nascent hyomandibular and symplectic cartilages (Figure 1D). By 72 hpf, irx7 and irx5a became further restricted to cells within and surrounding the hyoid joint, with additional irx5a expression along the posterior margin of the hyoid arch (Figure S1). Analysis of a SAGp11A gene-trap line, in which GFP was inserted in the irx7 locus (Kawakami et al., 2004), confirmed irx7 expression in the developing hyoid joint, as well as continued expression in the joint until at least 20 days-post-fertilization (dpf) (Figure 1E,F) (Figure S1). Comparison of irx7:GFP and trps1:GFP lines further shows that irx7-expressing cells maintain the immature chondrocyte marker trps1 (Figure 1F,G). Histology reveals that irx7 expression corresponds to two zones of compact chondrocytes that connect either side of the interhyal cartilage with hyomandibular and ceratohyal cartilages, which are themselves composed of larger chondrocytes embedded in lacunae (Figure 1H). Live imaging of sox10:dsRed+ chondrocytes shows that the larval hyoid joint functions as a hinge during respiration (Movie S1). Although the hyoid joint shows no cavitation, this motility and lack of chondrocyte hypertrophy at the joint is reminiscent to that seen at mammalian hinge joints, such as the interphalangeal joints of the digits.
In the mandibular arch, nkx3.2 (bapx1), which is required for jaw joint formation, is restricted to the intermediate domain by positive Edn1 and negative Hand2 function (Miller et al., 2003). Likewise, we find irx7 and irx5a expression to be lost in edn1 mutants and ventrally expanded in hand2 mutants (Figure 1I–K,O–Q). Whereas the jaw joint gene nkx3.2 is excluded from the hyoid arch by Moz-dependent Hox2 expression (Miller et al., 2004), hyoid joint expression of irx7 and irx5a instead requires Moz (Figure 1L,R). Similar dorsoventral signaling pathways therefore restrict irx7/5a and nkx3.2 to the joint-forming regions of the hyoid and mandibular arches, respectively, with Hox2 genes promoting the expression of irx7 at the expense of nkx3.2 in the hyoid arch.
In previous work, we had shown that Bmp signaling has a distinct role from Edn1 in specifying ventral at the expense of intermediate/joint fates (Zuniga et al., 2011). Here, by using a heat-shock-inducible Gal4/UAS system, we find that misexpression of Bmp4 ligand during later chondrogenic stages (40–44 hpf) disrupts hyoid joint formation without affecting overt dorsoventral patterning (Figure S1). These joint defects correspond with a loss of irx7 and irx5a expression in the hyoid arch, indicating that Bmp signaling negatively regulates Irx gene expression (Figure 1N,T). Moreover, this loss of Irx expression is not solely due to the reported upregulation of Hand2 by Bmp4 (Zuniga et al., 2011), as either early (20–24 hpf) or late (40–44 hpf) Bmp4 misexpression still inhibits irx7 expression in embryos lacking hand2 (Figure S1). Thus, distinct from its early role in dorsoventral patterning, Bmp signaling has a later role in restricting irx7/5a expression to the developing hyoid joint.
Irx7 and Irx5a are Necessary for Hyoid Joint Formation
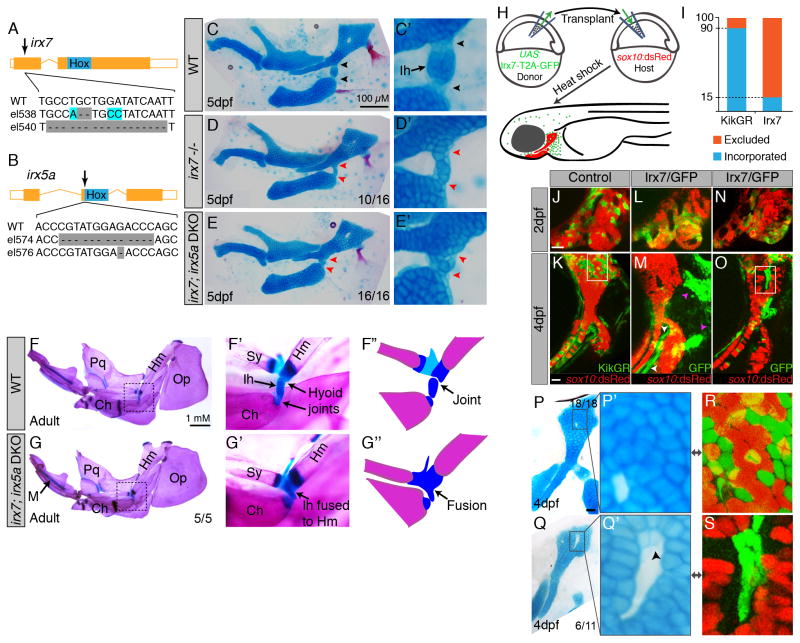
In order to assess requirements in hyoid joint development, we used TALE nucleases to generate early frame-shift alleles for irx7 and irx5a (Figure 2A,B). Whereas single irx5ael574 mutants had no apparent defects, 63% of irx7el538 mutants and 100% of irx7el538; irx5ael574 double mutants displayed specific losses of the hyoid joint. In the wild-type hyoid joint, cells on either side of the interhyal cartilage stain poorly with Alcian, a dye that detects sulfated proteoglycans characteristic of cartilage matrix (Figure 2C). In contrast, irx7el538 and irx7el538; irx5ael574 mutants had increased Alcian staining across the presumptive hyoid joint, with cells within the mutant hyoid interzone appearing larger than their wild-type counterparts (Figure 2D,E). Defects were also exclusive to the hyoid joint, consistent with the highly localized expression of irx7. Some irx7el538; irx5ael574 double mutants were adult viable despite the persistence of the fused hyoid joint (Figure 2G), with no apparent defects in other joints (including the jaw joint). In order to confirm that the observed phenotypes were due to loss of irx7 and irx5a, we also generated independent irx7el540 and irx5ae576 alleles and observed the same specific hyoid joint defects, either in irx7el540 alone or in all combinations of irx7 and irx5a alleles (Figure S2). We did not, however, observe the body axis defects reported using irx7 morpholinos, although we cannot exclude residual Irx7 protein in our mutants (Zhang et al., 2012). We also found no evidence in mutants for cross-regulation of irx7 and irx5a expression (Figure S2).
Figure 2. Requirements of Irx7 and Irx5a in hyoid joint formation.
(A,B) Schematics of irx7 and irx5a TALEN mutants show the position of out-of-frame deletions (arrows) relative to homeobox domains (blue boxes).
(C–E) Lateral views of mandibular and hyoid skeletons (left) and magnified views of the hyoid joints (right) show cartilages (Alcian Blue) and bones (Alizarin Red) at 5 dpf. In wild type, hyoid joint cells on either side of the Ih cartilage stain poorly with Alcian (black arrowheads). In irx7el538 and irx7el538; irx5ael574 mutants, the hyoid joint is dysmorphic with increased Alcian reactivity (red arrowheads).
(F,G) Dissected adult zebrafish facial skeletons (3 months-post-fertilization; size-matched). The hyoid joint (dashed box) is magnified in (F′, G′) and schematized in (F″, G″) to show joint fusions in irx7el538; irx5ael574 double mutants. Op, opercular bone.
(H) Scheme of neural crest transplants followed by 30–34 hpf heat-shocks to induce donor transgene expression.
(I) Whereas control kikGR-expressing cells contributed to hyoid cartilage in 18/20 cases, Irx7/GFP-expressing cells contributed to cartilage in only 11/72 cases (Fisher’s exact two-tailed test, p<0.0001).
(J–O) sox10:dsRed hosts received either control hsp70I:Gal4; UAS:kikGR or hsp70I:Gal4; UAS:Irx7-T2A-GFP donor neural crest precursors. Imaging at 2 dpf revealed contribution of donor cells to the mandibular and hyoid arches, and re-imaging at 4 dpf revealed contributions to sox10:dsRed+ hyoid cartilage. Arrowheads indicate contribution of Irx7/GFP-expressing cells to ligaments (white) and bones (magenta).
(P–S) Cell-specific loss of Alcian reactivity is seen in donor cells expressing Irx7/GFP (Jolma et al.) but not kikGR (magnified in P′, R is boxed region from K). Numbers indicate proportion of animals showing the displayed phenotype. Scale bars = 20 μM unless otherwise noted. See also Figure S2.
A related phenotype resulting from loss of irx7 was a shorter symplectic, which forms by the radial intercalation of cells adjacent to the nascent hyoid joint. In both irx7el538 and irx7el538; irx5ael574 mutants, the symplectic was composed of 30% fewer chondrocytes than in wild-type siblings (Figure S2). This loss of symplectic cells was reflected in a corresponding cell increase in the zone connecting the hyomandibula and symplectic, with total chondrocyte numbers unaffected. As previous time-lapse imaging had shown that symplectic cells originate from this connecting zone (Crump et al., 2004), Irx7/5a may thus be required for chondrocytes to rearrange into an extending symplectic. Interestingly, nkx3.2-deficient embryos also lack an element (the retroarticular) adjacent to the jaw joint (Miller et al., 2004), indicating common requirements for Irx7/5a and Nkx3.2 in coordinating joint formation with associated perichondral bones.
Irx7 inhibits cartilage differentiation cell-autonomously
In order to examine whether Irx7 functions within developing chondrocytes to inhibit their maturation, we performed early gastrula-stage transplants of hsp70I:Gal4; UAS:Irx7-T2A-GFP neural crest precursors, or control hsp70I:Gal4; UAS:kikGR precursors, into wild-type sox10:dsRed hosts. This sox10:dsRed transgene shows two waves of expression: first in all arch neural-crest-derived cells and then in chondrocytes. Subsequent heat shocks from 30–34 hpf induced clones of donor cells expressing Irx7/GFP or the control kikGR fluorescent protein within arches otherwise populated by sox10:dsRed+ host cells (Figure 2H). Whereas both kikGR- and Irx7/GFP-expressing donor cells efficiently populated the pharyngeal arches of hosts at 48 hpf, Irx7/GFP-expressing cells contributed to cartilage in only 15% of hosts at 4 dpf, compared to 90% of hosts for control cells (Figure 2I–O). The inefficient incorporation of Irx7/GFP-expressing cells into cartilage was not simply due to their loss as they extensively contributed to hyoid bones and ligaments. Moreover, in the 15% of hosts in which Irx7/GFP-expressing cells incorporated into cartilages, we observed decreased Alcian staining in a largely cell-autonomous manner (Figure 2Q,S). Irx7 thus acts very locally to block Alcian+ matrix production. At the normal hyoid joint, this paucity of Alcian+ cartilage matrix might contribute to the flexibility of attachment points.
Irx7 arrests chondrocyte maturation at the hyoid joint
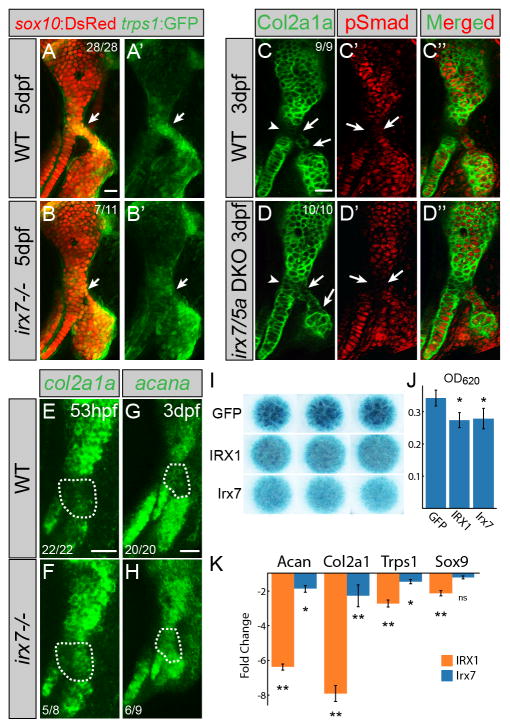
We next examined the mechanism by which Irx7 inhibits the deposition of Alcian+ matrix at the hyoid joint. As in mammals, zebrafish chondrocytes initially express SoxE factors (sox9a and sox10) and the trps1 gene (Nichols et al., 2013). Maturing chondrocytes then downregulate trps1 and upregulate a number of genes, in particular major constituents of cartilage matrix such as type II collagen (col2a1a) and aggrecan (acana). In irx7el538 mutants, we observed that trps1:GFP expression was reduced in the larval hyoid joint (Figure 3A,B). We also observed elevated col2a1a and acana expression and Col2a1 protein across the hyoid joint of irx7el538 mutants (Figure 3C–H). Further, we found that zebrafish Irx7 was sufficient to inhibit Alcian+ cartilage matrix production and Col2a1 and Acan expression in chondrogenic murine ATDC5 cells cultured as micromasses. Intriguingly, mouse IRX1 could similarly inhibit cartilage matrix production and gene expression, suggesting that inhibition of chondrocyte maturation may be a more general property of vertebrate Irx proteins (Figure 3I–K). Compared to inhibition of Col2a1 and Acan expression, Sox9 expression was unaffected by Irx7 misexpression and only modestly decreased by IRX1, consistent with Irx proteins functioning largely downstream of Sox9 to inhibit type II collagen and Aggrecan production. Irx7/IRX1 misexpression also modestly decreased Trps1 expression. That Irx7 is required for trps1:GFP expression at the zebrafish hyoid joint but not sufficient to induce Trps1 in vitro might reflect an indirect role of Irx7 in regulating Trps1, in line with previous data that Irx proteins function largely as transcriptional repressors (Cavodeassi et al., 2001).
Figure 3. Irx7 represses chondrocyte maturation.
(A,B) trps1:GFP (green) is reduced at the hyoid joint (arrows) of irx7el538 mutants compared to wild-type siblings. sox10:dsRed labels hyoid cartilages (red).
(C,D) Immunostaining shows increased Col2a1a protein (green) but no change in phospho-Smad1/5/8 (red) in the hyoid joints (arrows) and Hm-Sy connecting zone (arrowheads) of irx7el538; irx5ael574 mutants relative to wild-type siblings.
(E–H) In situ hybridization shows increased expression of col2a1a and acana in the hyoid joint region (dashed areas) of irx7el538 mutants relative to wild-type siblings. Numbers indicate proportion of animals showing the displayed phenotype. Scale bars = 20μM.
(I) Irx7 and murine IRX1 (but not GFP control) inhibit Alcian+ matrix accumulation in micromass cultures of ATDC5 cells grown in chondrogenic media for 7 days. Results were consistent across biological triplicates.
(J) Alcian blue content of the micromasses quantified by absorbance at 620 nm.
(K) Quantitative RT-PCR on RNA extracted from micromass cultures of ATDC5 cells grown in chondrogenic media for 7 days. Compared to GFP-expressing controls, expression of Col2a1, Acan, and Trps1 was reduced by IRX1 and Irx7 misexpression. Sox9 expression was reduced by IRX1 but not Irx7 misexpression. Error bars represent 95% confidence interval of the mean. *p<0.05 and **p<0.01 using Tukey HSD test. See also Figure S3.
We also find that Irx7 regulates cartilage maturation independently of Bmp signaling. Both wild types and irx7el538; irx5ael574 mutants had similarly lower levels of Bmp activity at the hyoid joint compared to neighboring chondrocytes as assessed by phosphorylation of Smad1/5/8 (Figure 3C′,D′). Combined with the ability of Bmp4 to inhibit irx7 and irx5a expression, these findings imply that Irx7/5a function downstream of Bmp signaling to inhibit chondrocyte maturation at the hyoid joint. Interestingly, we found the zone connecting the symplectic and hyomandibula also to be low in col2a1a and acana in wild types, with increased col2a1a and acana in irx7el538 mutants. The inability to maintain chondrocytes in an immature state may thus explain not only hyoid joint fusions but also the shortening of the joint-proximal symplectic cartilage in mutants, with accelerated cartilage matrix production entrapping cells before they can rearrange into stacks.
Irx7 directly inhibits a col2a1a enhancer
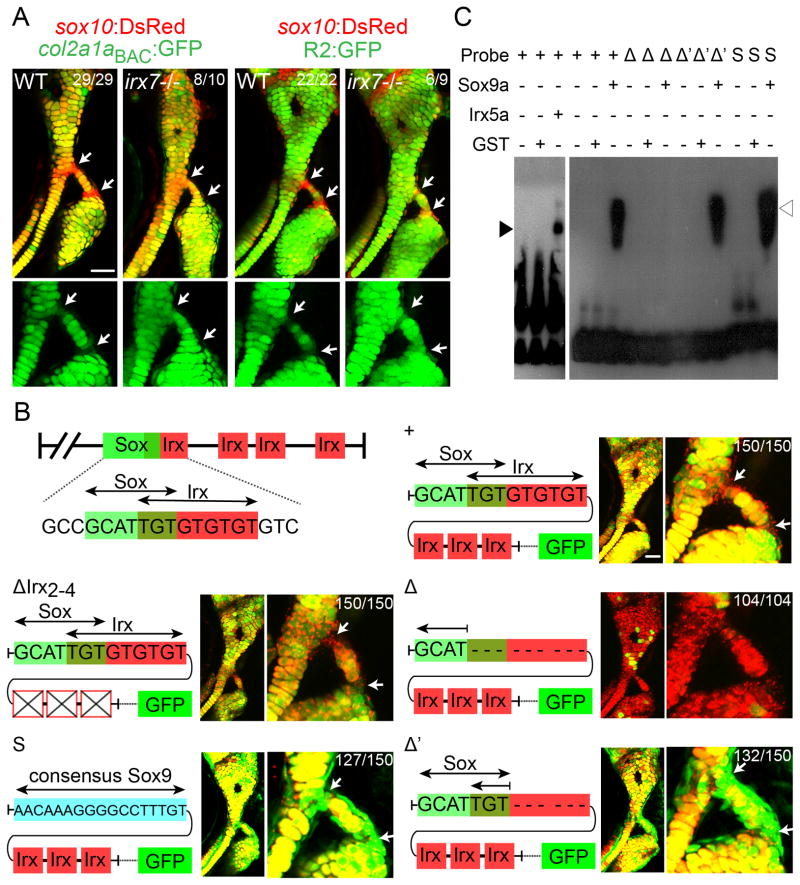
While our cell culture studies indicate that Irx proteins are sufficient to inhibit Col2a1 and Acan expression, this could reflect Irx binding to either protein co-factors or DNA regulatory regions. We therefore investigated whether Irx7 might inhibit col2a1a expression in zebrafish through binding to a well-characterized cartilage-specific “R2” enhancer (Dale and Topczewski, 2011). As with endogenous Col2a1a mRNA and protein and a newly constructed col2a1a BAC transgene, we find that an R2:GFP transgene is low at the wild-type hyoid joint and upregulated in irx7el538 mutant joints (Figure 4A). Based on previous analysis of human (Jolma et al., 2013 and Wingender et al., 2013) and Drosophila (Bilioni et al., 2005) consensus Iroquois binding motifs (ACA-nn/nnn-ACA and ACA-nn/nnn-TGT), we identified four putative Irx sites in the R2 enhancer. Whereas deletion of the second through fourth sites had no affect on R2:GFP expression, deletion of the first Irx site resulted in near complete loss of cartilage expression (Figure 4B). As this Irx site overlaps with a predicted Sox9 binding site (Mathelier et al., 2014), we reasoned that loss of Sox9 binding to this enhancer might account for the loss of cartilage transgene expression. Consistently, deleting only a portion of the Irx site without affecting the Sox9 site, or replacing it with a consensus Sox9-binding site lacking the Irx site, resulted in ectopic transgene expression within the hyoid joint (Figure 4B). An electrophoretic mobility shift assay further confirmed that both Sox9a and Irx5a proteins bind this overlapping site in vitro (Figure 4C). An attractive model then is that Irx proteins compete with or modify Sox9a activity at the R2 enhancer, thus dampening Sox9a-mediated activation of col2a1a. As Irx5 and Trps1 form a transcriptional complex with Irx3 in Xenopus (Bonnard et al., 2012), it will be interesting to test whether a similar complex involving Irx5a, Trps1, and Irx7 represses the expression of col2a1a, acana, and other chondrocyte matrix genes at the hyoid joint.
Figure 4. Irx genes directly repress a col2a1a enhancer.
(A) Relative to sox10:dsRed+ chondrocytes (red), col2a1aBAC:GFP and R2:GFP transgenes (green) show increased col2a1a-driven expression in hyoid joint cells (arrows) of irx7el538 mutants compared to wild-type siblings.
(B) Schematics show predicted Sox and Irx binding sites within the R2 enhancer of the col2a1a gene, as well as modified transgenic constructs. Confocal images at 6 dpf show the connection of the hyosymplectic and ceratohyal cartilages, with magnified images showing the bipartite hyoid joints (arrows). In all panels, the wild-type col2a1aBAC:mCherry-NTR transgene (red) displays lower levels in hyoid joints cells than neighboring chondrocytes. Transgenes with wild-type or modified R2 enhancers driving GFP are shown in green. Both the wild-type R2 enhancer and a version missing the second through fourth putative Irx sites are similar to wild-type col2a1aBAC:mCherry-NTR in being excluded from joints (yellow reflects overlap). Loss of the overlapping Sox/Irx site of the R2 enhancer abolishes most cartilage expression (red), and replacement with a Sox9 consensus site or deletion of just the Irx half-site results in ectopic joint expression (green). In the case of R2 enhancer missing the overlapping Sox/Irx site (Δ) 104 animals injected with the plasmid show similar expression and out of 30 founders screened, none showed more than just a sparse GFP expression in a few cartilage cells. For other panels experimental numbers are pooled for three independent stable transgenic lines. 50 embryos were examined for each line. Numbers indicate proportion of animals showing the displayed phenotype. Scale bars = 30 μM.
(C) An electrophoretic mobility shift assay shows binding of Irx5a (closed arrowhead) and Sox9a (open arrowhead) but not a control GST protein to a probe containing the wild-type Sox/Irx site (+). Sox9a binds a probe deleted for the Irx half-site (Δ′) but not the entire Sox/Irx site (Δ), and binding is restored by replacement of the Sox/Irx site with a consensus Sox9 binding site (S). See also Figure S4.
Remarkably, the existence of a hyoid joint only in fishes precisely correlates with presence of the irx7 gene in all sequenced fish genomes (including coelacanths) but not in any known tetrapod genome. During the transition to land, the hyoid skeleton that originally connected the jaw to the ear evolved to acquire a new function in sound transduction, with the columella and then stapes (both viewed as hyomandibula homologs) retreating into the middle ear and losing their connections to the hyoid bone (homologous to the fish ceratohyal). One possibility then is that loss of the hyoid joint during ear evolution resulted in the irx7 gene becoming dispensable for development, hence resulting in its loss from tetrapod genomes (see Figure S3 for a potential evolutionary history of irx7).
While the unique requirement of a single Irx gene at the zebrafish hyoid joint has allowed us to uncover roles for Irx7 in repressing chondrocyte maturation, this function likely extends to other members of the Irx family. First, loss of irx5a, a homolog of one of the members of the mammalian Irx3/5/6 cluster, enhances joint defects in irx7 mutants. Second, deletion of the Irx binding site from the col2a1a enhancer also results in ectopic expression at the mandibular and hyomandibula-otic joints (Figure S4A–C). Third, we observe expression of irx1b at the joint connecting the pectoral fin to the girdle, which corresponds to a zone of high Sox10 and low Col2a1 and Alcian+ matrix as seen in the hyoid joint (Figure S4D–F). Fourth, we find that Irx1 and Col2a1 expression similarly anti-correlate in the developing interphalangeal joints of the mouse paw (Figure S4G–I). Fifth, both zebrafish Irx7 and murine IRX1 are able to inhibit cartilage matrix production in a chondrogenic cell line. In the future, it will be interesting to examine the extent to which diverse members of the Irx family function to restrain the Sox9-mediated maturation of chondrocytes, including not only at joints but also within the periochondrium and other locations where cartilage differentiation needs to be tightly controlled.
Experimental Procedures
Zebrafish Lines
Zebrafish (Danio rerio) were staged as described (Kimmel et al., 1995). Reported lines include edn1tf216b (Miller et al., 2000), Df(chr01:hand2)s6 (Yelon et al., 2000), mozb719 (Miller et al., 2004), trps1j127aGt (Talbot et al., 2010), Tg(hsp70I:Gal4)kca4 (Scheer and Campos-Ortega, 1999), Tg(UAS:Bmp4; cmlc2:GFP)el49 (Zuniga et al., 2011), Tg(UAS:kikGR; α-crystallin:Cerulean)el377 and Tg(sox10:dsRED)el10 (Das and Crump, 2012). SAGp11A (irx7:GFP) was obtained from National Institute of Genetics (Kawakami et al., 2004 and Nagayoshi et al., 2008).
Tg(UAS:Irx7-T2A-GFP; α-crystallin:Cerulean)el613, Tg(sox10:GFPCAAX)el375, and Tg(sox10:mCherryCAAX)el361 were generated by one-cell-embryo injection of plasmids constructed with the Tol2kit (Kwan et al., 2007) and col2a1aBAC:GFP and col2a1aBAC:mCherry-NTR lines were generated according to (Shin et al., 2003) (see Supplementary Experimental Procedures). Modified versions of the R2 enhancer were made using IDT gblocks and cloned into the R2-E1b:EGFP plasmid (Dale and Topczewski, 2011). See Supplementary Experimental Procedures, Sequences of modified R2 enhancers and probes for EMSA. Null alleles for irx7 and irx5a were generated using the TALEN protocol of (Sanjana et al., 2012). See Supplementary Experimental Procedures for details on the design of TALENs and genotyping assays. For heat-shock induction, embryos were placed in a 40°C incubator for 4 hours and then transferred to 28.5°C. All phenotypes were scored before genotyping.
Histology and Skeletal Analysis
Cartilage and bone staining and fluorescent in situ hybridizations (including dlx2a and sox9a) were performed as described (Zuniga et al., 2010). For all other genes, templates were amplified and cloned into pCR-Blunt II-TOPO® vector for probe synthesis (see Supplementary Experimental Procedures, In situ probe templates).
Immunohistochemistry was performed as described (Crump et al., 2004 and Nichols et al., 2013) using mouse anti-Col2a1 (1:100; II-II6B3-DSHB), and anti-pSmad1/5/8 (1:200; gift of Ed Laufer), and secondary antibodies AlexaFluor488 goat anti-mouse (1:300; Life Technologies, A-11001) and AlexaFluor568 goat anti-rabbit (1:300; Life Technologies, A-11011). Staining with Trichrome/Gomori One-Step/Aniline Blue Kit was performed per manufacturer instructions (Newcomersupply).
Transplants
As described in (Crump et al., 2004), donor cells from UAS:kikGR; hsp70I:Gal4 or UAS:Irx7-T2A-GFP; hsp70I:Gal4 embryos were transplanted into the neural crest precursor domain of sox10:dsRed embryos at 6 hpf. Embryos were subjected to heat shock from 30–34 hpf to activate transgene expression, imaged by a confocal microscope at 48 hpf and 4 dpf, and fixed at 4 dpf for skeletal staining.
Imaging
Confocal microscopy was performed on a Zeiss LSM5 microscope using ZEN software. Skeletons were imaged on a Leica DM2500 microscope. Levels were adjusted in Adobe Photoshop CS5, with identical adjustments applied to images from the same data set.
In vitro cartilage differentiation assay
Retroviral expression vectors, pMXs_GFP-P2A-IRX1, pMXs_Irx7-T2A-GFP, and pMXs_GFP, were assembled by Gateway cloning (Invitrogen) and transduction was performed as described (Takahashi et al., 2007). ATDC5 cells were infected with two pools of viral supernatant during a 48-hour period. GFP-positive cells were FACS purified and cultured as micromasses at a density of 105 cells per well. Samples were collected at day 7 of culture. Continued transgene expression was confirmed by RT-qPCR for GFP, irx7, Irx1, and Gapdh (See Supplementary Experimental Procedures). In vitro cartilage induction and RT-qPCR was performed as described (Brown et al., 2008 and Schmittgen and Livak, 2008). qPCR data was analyzed using ΔΔCT method. See Supplementary Experimental Procedures for extended details and sequence of qPCR primers.
Electrophoretic Mobility Shift Assay
Zebrafish Irx5a and Sox9a and negative control GST proteins were translated using the PURExpress in vitro protein synthesis kit (NEB). Biotin-labeled probes were obtained from IDT (See Supplementary Experimental Procedures, Sequences of modified R2 enhancers and probes for EMSA). Proteins and probes in binding buffer were incubated at room temperature for 30 minutes and then run on a 5% TBE polyacrylamide gel. Chemiluminescent nucleic acid detection was performed per manufacturer instructions (Pierce).
Statistical Analysis
Fisher’s exact two-tailed test was used to test the significance of Irx7 overexpression on contribution of donor cells to cartilage (Figure 2I). Percentage of transplanted animals in which donor cells contributed to the cartilage is shown in each case. One-way ANOVA followed by Tukey-Kramer HSD test was performed for multiple pairwise comparisons (Figures 3J–K, and S2H). Error bars show 95% confidence interval of the mean.
Supplementary Material
Acknowledgments
We thank Megan Matsutani and Jennifer DeKoeyer Crump for fish care, Ed Laufer for the pSmad1/5/8 antibody, and Koichi Kawakami and the National BioResource Project for the SAGp11A line. Funding was by NIH R01 DE018405 and March of Dimes (J.G.C.), NIH T32 (A.A. and L.M.) and Giannini (L.M.) fellowships, and the Loyola University Chicago’s Provost office (R.M.D. and S.D.).
Footnotes
Author Contributions
A.A., L.M., S.P, S.D., and R.D. performed the experiments. X.H., A.K.I., S.G., J.K.I., A.P.M, and F.V.M. assisted with mammalian experiments. A.A. and J.G.C. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Archer CW, Dowthwaite GP, Francis-West P. Development of synovial joints. Birth defects research Part C, Embryo today : reviews. 2003;69:144–155. doi: 10.1002/bdrc.10015. [DOI] [PubMed] [Google Scholar]
- Bilioni A, Craig G, Hill C, McNeill H. Iroquois transcription factors recognize a unique motif to mediate transcriptional repression in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:14671–14676. doi: 10.1073/pnas.0502480102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnard C, Strobl AC, Shboul M, Lee H, Merriman B, Nelson SF, Ababneh OH, Uz E, Guran T, Kayserili H, et al. Mutations in IRX5 impair craniofacial development and germ cell migration via SDF1. Nature genetics. 2012;44:709–713. doi: 10.1038/ng.2259. [DOI] [PubMed] [Google Scholar]
- Brown AJ, Alicknavitch M, D’Souza SS, Daikoku T, Kirn-Safran CB, Marchetti D, Carson DD, Farach-Carson MC. Heparanase expression and activity influences chondrogenic and osteogenic processes during endochondral bone formation. Bone. 2008;43:689–699. doi: 10.1016/j.bone.2008.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet LJ, McMahon JA, McMahon AP, Harland RM. Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Science. 1998;280:1455–1457. doi: 10.1126/science.280.5368.1455. [DOI] [PubMed] [Google Scholar]
- Cavodeassi F, Modolell J, Gomez-Skarmeta JL. The Iroquois family of genes: from body building to neural patterning. Development. 2001;128:2847–2855. doi: 10.1242/dev.128.15.2847. [DOI] [PubMed] [Google Scholar]
- Craft AM, Ahmed N, Rockel JS, Baht GS, Alman BA, Kandel RA, Grigoriadis AE, Keller GM. Specification of chondrocytes and cartilage tissues from embryonic stem cells. Development. 2013;140:2597–2610. doi: 10.1242/dev.087890. [DOI] [PubMed] [Google Scholar]
- Crump JG, Swartz ME, Eberhart JK, Kimmel CB. Moz-dependent Hox expression controls segment-specific fate maps of skeletal precursors in the face. Development. 2006;133:2661–2669. doi: 10.1242/dev.02435. [DOI] [PubMed] [Google Scholar]
- Crump JG, Swartz ME, Kimmel CB. An Integrin-Dependent Role of Pouch Endoderm in Hyoid Cartilage Development. PLoS Biol. 2004;2:E244. doi: 10.1371/journal.pbio.0020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale RM, Topczewski J. Identification of an evolutionarily conserved regulatory element of the zebrafish col2a1a gene. Developmental biology. 2011;357:518–531. doi: 10.1016/j.ydbio.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Crump JG. Bmps and id2a act upstream of Twist1 to restrict ectomesenchyme potential of the cranial neural crest. PLoS genetics. 2012;8:e1002710. doi: 10.1371/journal.pgen.1002710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis-West PH, Parish J, Lee K, Archer CW. BMP/GDF-signalling interactions during synovial joint development. Cell and tissue research. 1999;296:111–119. doi: 10.1007/s004410051272. [DOI] [PubMed] [Google Scholar]
- Gomez-Skarmeta JL, Diez del Corral R, de la Calle-Mustienes E, Ferre-Marco D, Modolell J. Araucan and caupolican, two members of the novel iroquois complex, encode homeoproteins that control proneural and vein-forming genes. Cell. 1996;85:95–105. doi: 10.1016/s0092-8674(00)81085-5. [DOI] [PubMed] [Google Scholar]
- Grotewold L, Ruther U. The Fused toes (Ft) mouse mutation causes anteroposterior and dorsoventral polydactyly. Developmental biology. 2002;251:129–141. doi: 10.1006/dbio.2002.0817. [DOI] [PubMed] [Google Scholar]
- Hartmann C, Tabin CJ. Wnt-14 plays a pivotal role in inducing synovial joint formation in the developing appendicular skeleton. Cell. 2001;104:341–351. doi: 10.1016/s0092-8674(01)00222-7. [DOI] [PubMed] [Google Scholar]
- Hootman JM, Helmick CG, Brady TJ. A public health approach to addressing arthritis in older adults: the most common cause of disability. American journal of public health. 2012;102:426–433. doi: 10.2105/AJPH.2011.300423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, et al. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Developmental cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, Buchman S, Chen CY, Chou A, Ienasescu H, et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic acids research. 2014;42:D142–147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald LA, Gerrelli D, Fok Y, Hurst LD, Tickle C. Comparison of Iroquois gene expression in limbs/fins of vertebrate embryos. Journal of anatomy. 2010;216:683–691. doi: 10.1111/j.1469-7580.2010.01233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill H, Yang CH, Brodsky M, Ungos J, Simon MA. mirror encodes a novel PBX-class homeoprotein that functions in the definition of the dorsal-ventral border in the Drosophila eye. Genes & development. 1997;11:1073–1082. doi: 10.1101/gad.11.8.1073. [DOI] [PubMed] [Google Scholar]
- Miller CT, Maves L, Kimmel CB. moz regulates Hox expression and pharyngeal segmental identity in zebrafish. Development. 2004;131:2443–2461. doi: 10.1242/dev.01134. [DOI] [PubMed] [Google Scholar]
- Miller CT, Schilling TF, Lee K, Parker J, Kimmel CB. sucker encodes a zebrafish Endothelin-1 required for ventral pharyngeal arch development. Development. 2000;127:3815–3828. doi: 10.1242/dev.127.17.3815. [DOI] [PubMed] [Google Scholar]
- Miller CT, Yelon D, Stainier DY, Kimmel CB. Two endothelin 1 effectors, hand2 and bapx1, pattern ventral pharyngeal cartilage and the jaw joint. Development. 2003;130:1353–1365. doi: 10.1242/dev.00339. [DOI] [PubMed] [Google Scholar]
- Nagayoshi S, Hayashi E, Abe G, Osato N, Asakawa K, Urasaki A, Horikawa K, Ikeo K, Takeda H, Kawakami K. Insertional mutagenesis by the Tol2 transposon-mediated enhancer trap approach generated mutations in two developmental genes: tcf7 and synembryn-like. Development. 2008;135:159–169. doi: 10.1242/dev.009050. [DOI] [PubMed] [Google Scholar]
- Nichols JT, Pan L, Moens CB, Kimmel CB. barx1 represses joints and promotes cartilage in the craniofacial skeleton. Development. 2013;140:2765–2775. doi: 10.1242/dev.090639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters T, Ausmeier K, Dildrop R, Ruther U. The mouse Fused toes (Ft) mutation is the result of a 1.6-Mb deletion including the entire Iroquois B gene cluster. Mammalian genome : official journal of the International Mammalian Genome Society. 2002;13:186–188. doi: 10.1007/s00335-001-2142-7. [DOI] [PubMed] [Google Scholar]
- Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nature protocols. 2012;7:171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer N, Campos-Ortega JA. Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mech Dev. 1999;80:153–158. doi: 10.1016/s0925-4773(98)00209-3. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nature protocols. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Settle SH, Jr, Rountree RB, Sinha A, Thacker A, Higgins K, Kingsley DM. Multiple joint and skeletal patterning defects caused by single and double mutations in the mouse Gdf6 and Gdf5 genes. Developmental biology. 2003;254:116–130. doi: 10.1016/s0012-1606(02)00022-2. [DOI] [PubMed] [Google Scholar]
- Shin J, Park HC, Topczewska JM, Mawdsley DJ, Appel B. Neural cell fate analysis in zebrafish using olig2 BAC transgenics. Methods in cell science : an official journal of the Society for In Vitro Biology. 2003;25:7–14. doi: 10.1023/B:MICS.0000006847.09037.3a. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Okita K, Nakagawa M, Yamanaka S. Induction of pluripotent stem cells from fibroblast cultures. Nature protocols. 2007;2:3081–3089. doi: 10.1038/nprot.2007.418. [DOI] [PubMed] [Google Scholar]
- Talbot JC, Johnson SL, Kimmel CB. hand2 and Dlx genes specify dorsal, intermediate and ventral domains within zebrafish pharyngeal arches. Development. 2010;137:2507–2517. doi: 10.1242/dev.049700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E, Schoeps T, Donitz J. TFClass: an expandable hierarchical classification of human transcription factors. Nucleic acids research. 2013;41:D165–170. doi: 10.1093/nar/gks1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelon D, Ticho B, Halpern ME, Ruvinsky I, Ho RK, Silver LM, Stainier DY. The bHLH transcription factor hand2 plays parallel roles in zebrafish heart and pectoral fin development. Development. 2000;127:2573–2582. doi: 10.1242/dev.127.12.2573. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yang Y, Trujillo C, Zhong W, Leung YF. The expression of irx7 in the inner nuclear layer of zebrafish retina is essential for a proper retinal development and lamination. PloS one. 2012;7:e36145. doi: 10.1371/journal.pone.0036145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga E, Rippen M, Alexander C, Schilling TF, Crump JG. Gremlin 2 regulates distinct roles of BMP and Endothelin 1 signaling in dorsoventral patterning of the facial skeleton. Development. 2011;138:5147–5156. doi: 10.1242/dev.067785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga E, Stellabotte F, Crump JG. Jagged-Notch signaling ensures dorsal skeletal identity in the vertebrate face. Development. 2010;137:1843–1852. doi: 10.1242/dev.049056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.