Abstract
RC/BTB2 is a binding partner of sperm associated antigen 16S (SPAG16S), which is regulator of spermiogenesis in mice, a process during which sperm flagella are formed. The expression of Rc/btb2 is also regulated by multicilin, a protein that controls ciliogenesis. Given that mouse Rc/btb2 mRNA is not only expressed in tissues bearing motile cilia, but also in tissues without motile cilia, we investigated whether RC/BTB2 plays a role in the general process of ciliogenesis by studying two somatic cells lines that have primary cilia, NIH3T3 and IMCD3. We discovered that the subcellular localization of RT/BTB2 in the NIH3T3 and IMCD3 cells encompasses the pathway for ciliogenesis. RC/BTB2 was found in the Golgi bodies and centrosomes, two key structures essential for normal ciliogenesis. Knockdown of Rc/btb2 gene expression in these cell lines disrupted ciliogenesis. The percentage of cells with primary cilia was significantly reduced in stable cell lines transduced with specific Rc/btb2 shRNA viruses compared to the control cells. When cilia were formed in the knockdown cells, they were significantly shorter than those in the control cells. Knockdown of Rc/btb2 expression did not affect cell proliferation and the cell cycle. Exogenous expression of RC/BTB2 in these stable knockdown cells restored ciliogenesis. These findings suggest that RC/BTB2 is a necessary component of the process of formation of primary cilia in somatic cells, perhaps through the transportation of cargos from Golgi bodies to centrosomes for cilia assembling.
Introduction
Cilia are microtubule-based hair-like organelles extending from the surface of most mammalian cells (Drummond 2012). Electron microscopic analysis of mammalian cells led to a model for the initial steps of primary cilium assembly (Pedersen and Rosenbaum 2008). These steps encompass the docking of a Golgi-derived vesicle to the distal end of the basal body. The basal body functions as a foundation for the construction of the cilia/flagella through intraflagellar transport (IFT) mechanism (Marshall 2008; Alieva and Vorobjev 2004; Oh and Katsanis 2012; Pazour and Rosenbaum 2002). Based on this model, both the Golgi bodies and basal bodies are important structures for normal ciliogenesis. The Golgi body is an organelle found in most eukaryotic cells. In mammals, a single Golgi apparatus complex is usually located near the cell nucleus. The Golgi apparatus has multiple functions; it is a site of general protein processing and sorting for proteins going through the secretory pathway (Nakamura et al. 2012). In addition, the Golgi apparatus is also involved in lipid transport and lysosome formation (D'Angelo et al. 2013; Raposo et al. 2007). The Golgi body also appears to function as a starting site, organizing cargo-containing vesicles destined for the cilia. Basal bodies are organelles formed from centrioles (Kobayashi and Dynlacht 2011). They are found at the base of eukaryotic cilia or flagella, and serve as a nucleation site for the growth of the axoneme microtubules. Thus, the basal body functions as the platform upon which the axoneme is built.
The mouse Rc/btb2 gene yields two major transcripts: 2.3 kb Rc/btb2-s, present in most somatic tissues; and 2.5 kb Rc/btb2-t, which contains a unique non-translated exon in its 5’-UTR that is only detected in the testis, where it is highly expressed in male germ cells (Wang et al. 2012). Recent studies demonstrated that during ciliogenesis, proteins passing the ciliary barrier region share a similar mechanism of translocation as nucleocytoplasmic transport (Dishinger et al. 2010; Kee and Verhey 2013). We previously reported that RC/BTB2 is expressed during acrosome formation in spermiogenesis (Wang et al. 2012). Because RC/BTB2 has a RCC1 domain that possibly functions in guanine nucleotide exchange on small GTP-binding proteins, we hypothesized that RC/BTB2 plays roles in transport processes involved in both acrosome formation and flagellogenesis in germ cells. Rc/btb2 is also expressed in somatic tissues (Wang et al. 2012). A recent study revealed that Rc/btb2 mRNA expression was regulated by multicilin during ciliogenesis (Stubbs et al. 2012), suggesting that this gene may have a function in normal ciliogenesis. To test the hypothesis that RC/BTB2 is critical to somatic cell ciliogenesis, we characterized RC/BTB2 protein localization and its role in cilia formation in mammalian IMCD3 and NIH3T3 cells by reducing Rc/btb2 mRNA expression through an shRNA strategy. Our findings demonstrate that RC/BTB2 is present in the subcellular structures that cover the pathway for ciliogenesis. Reducing expression of this gene results in a severe ciliogenesis defect with reduced cilia formation. These observations provide new insights into the role of RC/BTB2 in ciliogenesis.
Materials and Methods
Antibodies
A rabbit polyclonal anti-RC/BTB2 was generated previously in our laboratory (Wang et al. 2012). Mouse monoclonal anti-Golgin-97 (A-21270) was purchased from Life Technologies, anti-Golgi 58K Protein/Formiminotransferase Cyclodeaminase (FTCD) (G2404-.2 mL), anti-γ-tubulin (T6557-.2mL), and anti-acetylated tubulin (T7451-200 μL) antibodies were purchased from Sigma, and the concentrations used for immunofluorescence staining were 1.0 μg/ml, 1:100, 1:200, and 1:200, respectively. β-actin antibody was purchased from Cell Signaling (#4967S), and a 1:1000 dilution was used for Western blot analysis. The second antibodies used include Alexa Fluor 488-conjugated goat anti-mouse IgG (A-11001, 1:500; Invitrogen, NY, USA), Cy3-conjugated goat anti-rabbit IgG (A10520, 1:5000; Invitrogen), and HRP-linked anti-Rabbit IgG was obtained from GE Healthcare UK limited (NA934V), and a 1:2000 dilution was used for Western blot analysis.
Expression Vector Construct
cDNA containing the full length Rc/btb2 coding sequence was generated by RT-PCR using the following primers: forward: 5’-GGATCCATGGAAGAAGAAGTGCCTGGTTTC-3’, reverse: 5’-CTCGAGGATCAGTTCTTAAAGGCTCCAACTC-3’. After sequencing, the cDNA was cloned into the BamH1/XhoI site of the pcDNA3 vector, creating RC/BTB2/pcDNA3 plasmid.
Cell lines and cell culture
The murine renal epithelial cell line derived from inner medullary collecting duct (IMCD3) was maintained in a 1:1 mixture of DMEM and Ham's F12 medium supplemented with 10% fetal bovine serum (FBS, Gibco, Life Technologies, NY, USA). NIH3T3 cell was maintained in DMEM supplemented with 10% heat-inactivated fetal calf serum and 2mM glutamine.
To observe ciliogenesis, IMCD3 cells were cultured to confluency (Molla-Herman et al. 2010), and NIH3T3 cells were cultured in serum free medium (Kim et al. 2011). Two to three days later, the cells were either stained using an acetylated tubulin antibody or processed for scanning electronic microscopy.
Generation of stable Rc/btb2 knockdown mammalian cell lines
Short hairpin RNA (shRNA)-expressing lentivirus constructs were generated using pLV-RNAi vector (Biosettia, San Diego, CA). Three target sequences were designed based on mouse Rc/btb2 mRNA sequence: shRNA1: 5’-AAAAGGGACAACGAATCATGGTTTATTGGATCCAATAAACCATGATTCGTTGTCCC-3’, shRNA2: 5’-AAAAGGGCAAGTAGGATCTGGATCATTGGATCCAATGATCCAGATCCTACTTGCCC-3’, and shRNA3: 5’-AAAAGGCAGTAGGGTTGCTAGATCTTTGGATCCAAAGATCTAGCAACCCTACTGCC-3’. They were inserted to the pLV-RNAi plasmid following the manufacturer's protocol. The shRNA lentiviruses were propagated in 293FT cells using the same protocol as employed previously (Wu et al. 2013). The culture supernatants were used to infect IMCD3 or NIH3T3 cells. The GFP-positive cells were sorted out using flow cytometer 72 hours post virus infection to generate stable cell lines.
RT-PCR and quantitative PCR (qPCR)
Total RNA was isolated from adult male testes or indicated cell lines using TRIzol reagent (Invitrogen). The RNA concentration was measured using a NanoDrop 2000 Spectrophotometer (Thermo Scientific, Wilmington, Delaware), RNA integrity was not assessed. These total RNAs were reversed transcribed using the Transcritpor first strand cDNA synthesis kit from Roche. For regular PCR to examine Rc/btb2 mRNA expression, the primer set used in our previous study was applied, and the same Gapdh primer set was used for a control (Wang et al. 2012). To compare relative Rc/btb2 mRNA expression levels in the stable cell lines, qPCR was conducted on an ABI 7500 Thermocycler (Applied Biosystems) using the SYBR-green system from Bio-Rad with the primer set from OriGene (catalogue number: MP212564). For normalization of the Rc/btb2 expression, Gapdh was also amplified using the qPCR primer set from OriGene (MP205604).
Western blot analysis
Lysates collected from stable knockdown cells or cells transfected with RC/BTB2/pcDNA3 plasmid were loaded onto 10% sodium dodecyl sulfatepolyacrylamide gels, electrophoretically separated, and transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). Membranes were blocked with TBST (containing 5% nonfat dry milk and 0.05% Tween 20) and then incubated with the indicated antibodies at 4 °C overnight. After washing in TBST, the blots were incubated with second antibodies for 1 hr at room temperature. After washing, the proteins were detected with Super Signal chemiluminescent substrate (Pierce, Rockford, IL, USA).
Cell proliferation assay
Indicated stable cells were seeded into 24-well cell culture plates at a density of 5000/well. From the second day, cell number was counted for four continuous days. At each time point, triplicate experiments were conducted for each stable cell lines. A 5-day growth curve was graphed using time as the x-axis, and the cell number of the three replicates in each group was plotted as the y-axis.
Analysis of cell cycle by flow cytometry
For cell cycle profile analysis, cells were synchronized at G0 phase by serum starvation for 24 hr, and then allowed to reenter the cell cycle by supplying 10% serum for an additional 24 hr, and cell cycle was analyzed as described by Gray (Gray et al. 1986). Briefly, cells were detached with trypsin, washed with PBS, and fixed in cold 70% ethanol, resuspended in 1 mL PBS containing 30 μg/mL propidium iodine (Sigama) and 150 μg/mL RNase A (Sigma), incubated at 37 °C for 1 hr and stored at 4 °C until analyses. For each sample 10,000 cells were analyzed for their DNA-content using a Gallios A94291 cytometer by the Flow Cytometry Core at Virginia Commonwealth University. Data from NIH3T3 cells were analyzed using Diva software, and the cell populations were separated as G1, S, and G2. IMCD3 cells were analyzed using FCSExpress. The cell populations were categorized as M1, M2, and M3. G1=M1, S=M2, G2=M3.
Immunofluorescence staining of cultured mammalian cells
Cells were cultured in chambered slides. The prepared slides were fixed in 4% paraformaldehyde/PBS (containing 4% sucrose) at room temperature for 30 minutes, washed three times with PBS. The cells were permeabilized with 0.1% Triton X-100 (Sigma-Aldrich) at 37°C for 10 min and blocked with 10 % goat serum (in PBS) for 1 hr. The cells were washed three times with PBS and incubated with indicated antibodies at 4°C overnight. After extensive washing with PBS, the samples were incubated with either Alexa-488-conjugated anti-mouse IgG secondary antibody and/or Cy3-conjugated anti-rabbit IgG secondary antibody at room temperature for 1 hr. The slides were washed three times with PBS and mounted in VectaMount with DAPI (Vector Labs. Burlin-game, CA) and sealed with nail polish. Images were taken by confocal laser-scanning microscopy (Leica TCS-SP2 AOBS) and processed using Adobe Photoshop 5.0.
Scanning electron microscopy (SEM)
For SEM analysis, IMCD3 stable cells were cultured on glass cover slides. Two to three days after confluent, the cells were processed by standard procedure (Bray et al. 1993). Images were taken with a Zeiss EVO 50 XVP scanning electron microscope at Virginia Commonwealth University's Microscopy Facility. Cilia length was measured using the scale included in the images taken.
Ethics statement
Wild-type C57BL/6J black mice were purchased from Jackson Laboratory (Stock Number: 000664). All animal work was approved by Virginia Commonwealth University's Institutional Animal Care & Use Committee (Protocol AD10000167) in accordance with Federal and local regulations regarding the use of non-primate vertebrates in scientific research. Testes from adult males were collected for total RNA extraction using the methods described above.
Statistical analysis
ANOVA test was used to showed statistical differences, the 2-tailed student's t-test was used for comparison of frequencies. Significance is defined as p < 0.05.
Results
Expression and localization of endogenous RC/BTB2 in NIH3T3 and IMCD3 cell lines
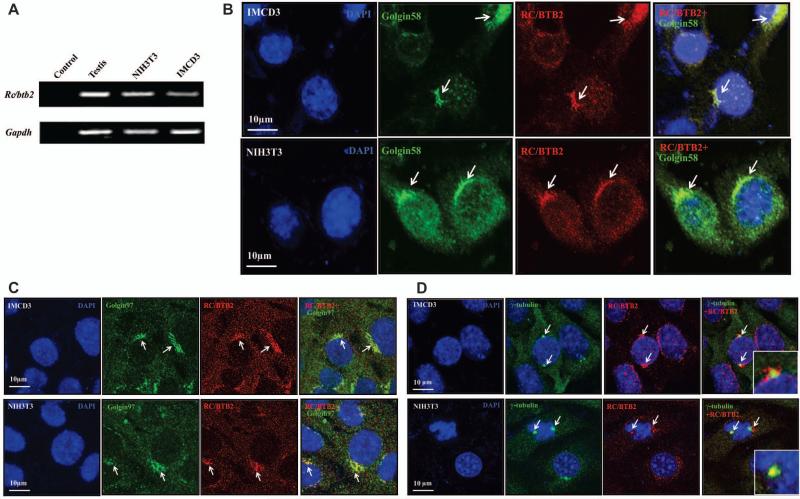
We localized RC/BTB2 in NIH3T3 and IMCD3 cells, two somatic cell lines widely used for ciliogenesis studies. RT-PCR analysis revealed that Rc/btb2 message RNA was expressed in both cell lines (Figure 1A). RC/BTB2 protein expression and localization were examined by immunofluorescence staining. The cells were double stained with an anti-RC/BTB2 antibody and an anti-Golgi 58K Protein antibody, or anti-Golgin 97 antibody or anti-γ-tubulin antibody. RC/BTB2 protein was not expressed throughout the whole cell body, instead, the signal was highly concentrated in specific regions, including nuclear membrane, and co-localized with Golgi 58K protein (Figure 1B), Golgin 97 (Figure 1C) and γ-tubulin (Figure 1D).
Figure 1. Rc/btb2 is expressed in somatic NIH3T3 and IMCD3 cells.
A. RT-PCR analysis of endogenous Rc/btb2 mRNA expression in NIH3T3 and IMCD3 cells. As in the mouse testicular sample, a specific Rc/btb2 mRNA signal is detected in both cell lines, indicating endogenous Rc/btb2 mRNA is expressed in these cells.
B, C, and D. Examination of endogenous RC/BTB2 protein localization by immunofluorescence staining. IMCD3 and NIH3T3 cells were cultured in chambered slides, and the cells were double stained by anti RC/BTB2 antibody and an antibody specific for Golgin58K protein (panel B), Golgin-97 (panel C) or centrosome (γ-tubulin, panel D). Notice that RC/BTB2 (red) is only concentrated in specific regions and co-localized with Golgin58K protein, Golgin-97 and γ-tubulin (arrows), indicating that RC/BTB2 is present in the Golgi body and centrosome.
Rc/btb2 knockdown does not affect cell proliferation and cell cycle in NIH3T3 and IMCD3 cells
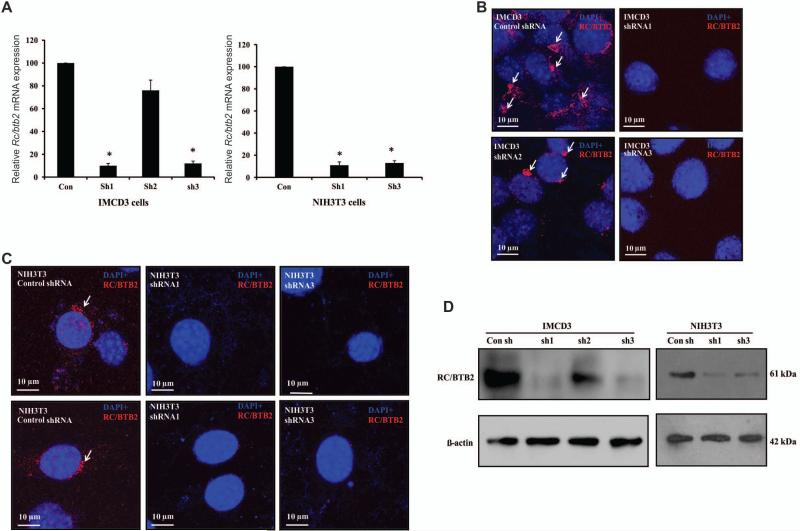
To explore the potential roles of the Rc/btb2 gene, we selected three regions based on the mouse Rc/btb2 cDNA sequence, and designed and constructed three cognate shRNA lentiviruses as well as an empty vector. The lentiviruses were used to generate stable IMCD3 cell lines. The knockdown efficiency of these shRNA constructs was evaluated by performing qPCR using total RNA extracted from the stable cells and immunofluorescence studies and Western blot analysis to assess RC/BTB2 protein expression. Compared to the control stable cell line generated from the empty vector, relative Rc/btb2 mRNA expression in shRNA1 and shRNA3 stable cells was reduced about 90%. In shRNA2 stable cells, Rc/btb2 was reduced about 30% (Figure 2A, left panel). shRNA1 and shRNA3 constructs were also used to generate stable NIH3T3 cells. Similar to the IMCD3 cells, Rc/btb2 mRNA expression was reduced by about 90% compared to the control NIH3T3 cells (Figure 2A, right panel).
Figure 2. Generation of stable IMCD3 and NIH3T3 cells with low Rc/btb2 expression.
A. Q-PCR analysis of relative Rc/btb2 mRNA expression in stable IMCD3 cells (left panel) and NIH3T3 cells (right panel). Notice that in IMCD3 cells, shRNA1 and shRNA3 reduced Rc/btb2 mRNA expression by about 90%, shRNA2 reduced it by 30%. Like in the IMCD3 cells, expression of Rc/btb2 mRNA in shRNA1 and shRNA3 stable NIH3T3 cells was also significantly lower than the control stable NIH3T3 cells. * p<0.05. B. Immunofluorescence staining of RC/BTB2 in stable IMCD3 cells. Notice that RC/BTB2 is expressed in the control and shRNA2 stable cells (arrows), but not in the shRNA1 and shRNA3 stable cells. C. Immunofluorescence staining of RC/BTB2 in stable NIH3T3 cells. Like in the IMCD3 cells, RC/BTB2 is expressed in the control stable cells, but not in the shRNA1 and shRNA3 stable cells. D. Analysis of endogenous RC/BTB2 protein expression in the stable IMCD3 and NIH3T3 cells. Notice that the 61 kDa RC/BTB2 protein was expressed in the control and shRNA2 cells. However, its expression was dramatically reduced in the shRNA1 and shRNA3 cells.
Knockdown efficiency was also examined by immunofluorescence staining by counting 500 cells for each line. When the stable cells were stained with RC/BTB2 antibody, the control and shRNA2 IMCD3 cells showed specific RC/BTB2 signals, however, no specific signals were detected in the stable cells generated with shRNA1 and shRNA3 constructs (Figure 2B). Similarly, RC/BTB2 protein was detected in the control NIH3T3 cells, but not in the shRNA1 and shRNA3 NIH3T3 cells (Figure 2C).
Expression of endogenous RC/BTB2 protein was also examined by Western blot analysis. In both IMCD3 and NIH3T3 cells, the expression of endogenous RC/BTB2 protein in shRNA1 and shRNA3 stable cells was dramatically reduced (Figure 2D).
Silencing Rc/btb2 gene expression in both IMCD3 and NIH3T3 cells had no obvious effect on cell morphology. Cells with different Rc/btb2 mRNA expression levels were indistinguishable (Figure 3A).
Figure 3. Knockdown of Rc/btb2 mRNA expression has no effect on cell proliferation and cell cycle.
A. Representative images of stable IMCD3 (a) and NIH3T3 (b) cells with different Rc/btb2 mRNA expression levels. There was no difference in morphology.
B. Cell proliferation in stable IMCD3 (left) and NIH3T3 (right) cells. The indicated stable cells were seeded into 24-well plates, and cell number was counted for five days. There were no differences in cell number among the cells with different Rc/btb2 expression levels. C. Cell cycle analysis by flow cytometry on stable NIH3T3 cells using Diva software. a, b, and c show representative results for control, shRNA1 and shRNA3 stable cells, d shows the statistical analysis of the three stable cell lines. Notice that cell populations at different cell cycle periods are similar between the stable cell lines. D. Cell cycle analysis by flow cytometry on stable IMCD3 cells using FCSExpress. a, b, and c show representative results for control, shRNA1 and shRNA2 stable cells, d shows the statistical analysis of the three stable cell lines.
To determine if silencing Rc/btb2 gene expression affects cell growth, equal numbers of IMCD3 or NIH3T3 cells with different Rc/btb2 expression levels were seeded into 24 well culture plates, and cell number was analyzed during five days. There was no significant difference in total cell numbers between groups (Figure 3B). The cell cycle was also analyzed by flow cytometry in both NIH3T3 (Figure 3C) and IMCD3 (Figure 3D) cells with different Rc/btb2 mRNA expression levels. Cell populations at different cell cycle periods were comparable to the control cells.
Ciliogenesis is disrupted in IMCD3 and NIH3T3 cells with low Rc/btb2 mRNA expression
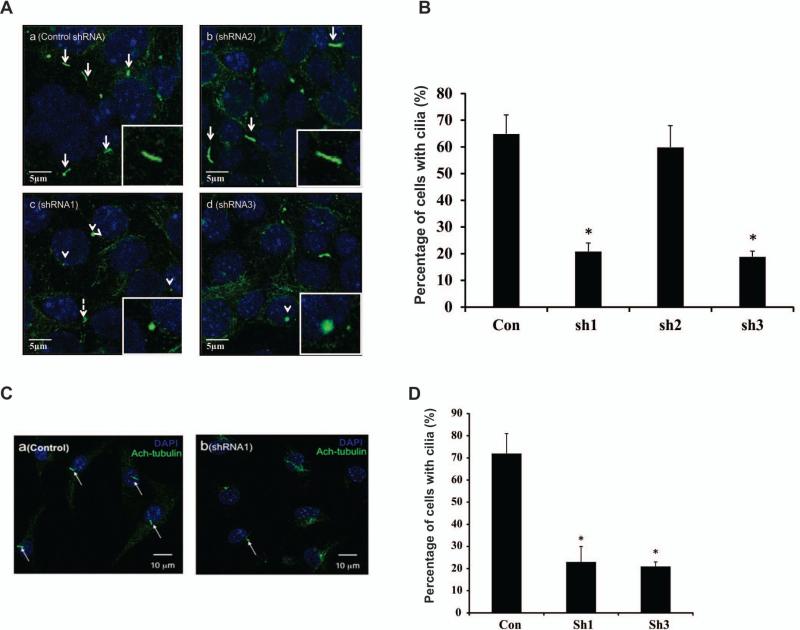
IMCD3 cells were cultured in chambered slides to examine ciliogenesis. Two to three days after the cells were confluent, they were stained with acetylated tubulin antibody. Most control and shRNA2 IMCD3 cells formed cilia. However, very few shRNA1 and shRNA3 cells formed cilia. When cilia were formed, the ciliary axonemes appeared truncated (Figure 4A). The total cell number and cells with cilia were counted and percentage of ciliated cells was calculated. More than 60% of control and shRNA2 stable cells formed cilia, only about 20% shRNA1 and shRNA3 stable cells formed cilia (Figure 4B). Ciliogenesis was also examined in stable NIH3T3 cells. The stable cells were seeded in chamber slides, and after serum starvation for 24-48 hr, the cells were stained with acetylated tubulin antibody. Similar to the IMCD3 stable cells, most control NIH3T3 cells formed cilia, but few shRNA1 and shRNA3 cells did (Figure 4C). The percentage of ciliated cells was also similar to the stable IMCD3 cells (Figure 4D).
Figure 4. Analysis of ciliogenesis by immunofluorescence staining of stable cell lines with different Rc/btb2 mRNA expression levels.
A. Representative images of stable IMCD3 cells. The cells were stained with acetylated tubulin antibody. Most control (a) and shRNA2 (b) stable cells formed normal appearing cilia (white arrows and inserts). Most shRNA1 (c) and shRNA3 (d) stable IMCD3 cells had truncated or shortened cilia on the cell surface (white arrow heads and inserts).
B. Percentage of cells with cilia in the stable IMCD3 cells. The percentage of cells with cilia was significantly lower in shRNA1 and shRNA3 stable cells. Three independent experiments were conducted, and in each experiment about 500 cells were analyzed for each cell line. * p<0.05.
C. Representative images of stable NIH3T3 cells. Most control cells formed normal looking cilia (a), only a few shRNA1 and shRNA3 cells formed cilia (b). The white arrows point to cilia.
D. Percentage of cells with cilia in the stable NIH3T3 cells. The percentage of cells with cilia was significantly lower in shRNA1 and shRNA3 stable cells. Three independent experiments were conducted, and in each experiment about 500 cells were analyzed for each cell line. * p<0.05.
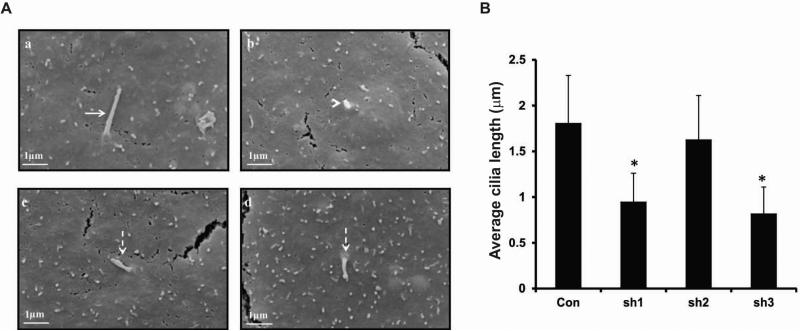
To further examine ciliogenesis in these stable cells, SEM was carried out on IMCD3 cells with different Rc/btb2 mRNA expression levels. Consistent with the immunofluorescence results, most control IMCD3 cells formed cilia, and all the cilia were normal looking, extending from the cell surface (Supplemental Figure 1 and Figure 5Aa). However, very few shRNA1 and shRNA3 cells formed cilia. Some cells just had small protrusions on the cell surface, and some cells formed cilia, but the cilia were much shorter than those on the control cells (Supplemental Figure 2 and Figure 5Ab-d). Like the control cells, most shRNA2 stable cells formed cilia (Supplemental Figure 3). Quantitative analysis revealed that the length of primary cilia on shRNA1 and shRNA3 stable IMCD3 cells was significantly shorter than the control IMCD3 cells and the shRNA2 IMCD3 cells. There was no significant difference in cilia length between the control and shRNA2 stable IMCD3 cells (Figure 5B).
Figure 5. Analysis of ciliogenesis in stable cell lines with different Rc/btb2 mRNA expression levels by scanning electronic microscopy.
A. Representative images of stable control shRNA IMCD3 cells (a) and shRNA1 IMCD3 cells (b-d). High magnification images showed that each control cell had one normal appearing cilium (arrow in a). However, some cells with low Rc/btb2 mRNA (shRNA1 stable cells) expression levels had only small protrusions on the cell surface (arrow head in b). When cilia were formed, most appeared to be shorter than cilia on the control cells (dashed arrows in c, d).
B. Average length of cilia formed in the stable IMCD3 cells. Three independent experiments were conducted. For each stable cell line, twenty cilia were randomly selected and cilia length was measured, and statistical differences were calculated. * p<0.05.
Exogenous expression of RC/BTB2 protein in the stable knockdown cells restores normal ciliogenesis
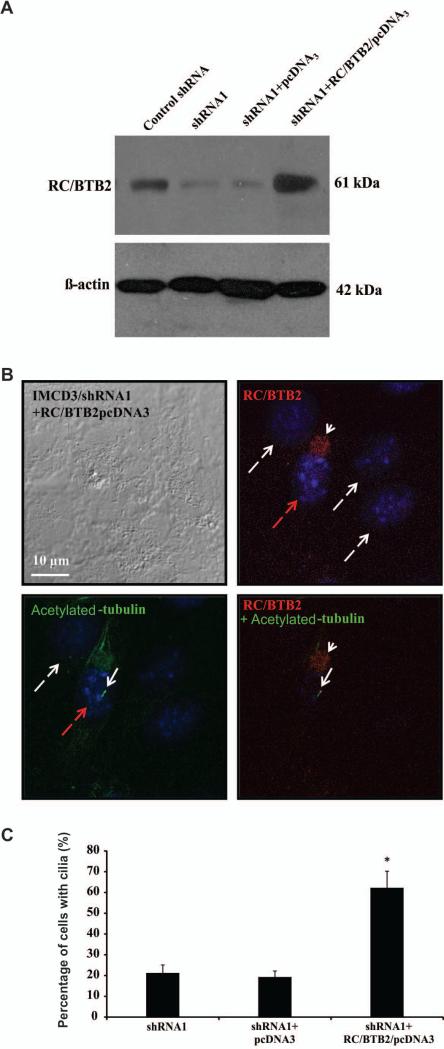
To determine if ciliogenesis defects in the stable knockdown cells were due to reduced RC/BTB2 expression, shRNA1 stable IMCD3 cells were transfected with RC/BTB2/pcDNA3 plasmid or empty pcDNA3 plasmid. Western blot analysis demonstrated that RC/BTB2 protein expression level was increased in the shRNA1 stable IMCD3 cells once transfected with the plasmid expressing RC/BTB2 (Figure 6A). The cells were double stained with an anti-RC/BTB2 antibody and an anti-acetylated tubulin antibody. Overexpressed RC/BTB2 appeared to accumulate as granules closely attached to the nuclei, and the cells expressing RC/BTB2 showed normal cilia (Figure 6B), indicating normal ciliogenesis had been restored in these cells. The percentage of cells with normal cilia was determined from the analysis of 300 cells showing RC/BTB2 expression and 300 cells without RC/BTB2 expression in the RC/BTB2/pcDNA3 plasmid transfected cells, and 300 cells transfected with empty pcDNA3 plasmid. The percentage of cells with cilia was significantly increased cells expressing RC/BTB2 (Figure 6C).
Figure 6. Re-expression of RC/BTB2 in the stable IMCD3 knockdown cells rescue ciliogenesis defect phenotype observed in the stable knockdown cells.
A. Western blot analysis of RC/BTB2 expression in the transfected stable shRNA knockdown IMCD3 cells. Notice that endogenous RC/BTB2 expression was dramatically reduced in the stable shRNA1 cells. However, its expression was increased when these cells were transfected with RC/BTB2/pcDNA3 plasmid, but not the empty pcDNA3 plasmid.
B. Representative images of the RC/BTB2 stable knockdown cells transfected with RC/BTB2/pcDNA3 plasmid stained by an anti-RC/BTB2 antibody and an anti-acetylated tubulin antibody. RC/BTB2 was labeled in red (white arrow head) and acetylated tubulin in green (white arrow). Notice that the cell expressing RC/BTB2 (with red dashed arrow) forms a normal like cilium. No cilia were observed in the cells without RC/BTB2 signal (white dashed arrows).
C. Percentage of cells with cilia with or without RC/BTB2 expression in the stable knockdown IMCD3 cells (shRNA1) transfected with RC/BTB2/pcDNA3 plasmid or empty pcDNA3 plasmid.
In each experiment, the cell number with cilia was counted from 100 cells with RC/BTB2 expression and 100 without RC/BTB2 expression in the RC/BTB2/pcDNA3 transfected cells, and 100 cells transfected with empty pcDNA3 plasmid, and percentages were calculated. The data were from three independent experiments. *p<0.05.
Discussion
The Golgi body packages proteins before they are sent to their destinations. It is considered to be the starting point of ciliogenesis (Emmer et al. 2010). In our initial studies, we reported that RC/BTB2 is localized in the acrosome of spermatids (Wang et al. 2012). The acrosome is a cap-like structure derived partly from the Golgi body(Martinez-Menarguez et al. 1996; Tang et al. 1982; Thorne-Tjomsland et al. 1988). Acrosome biogenesis is a multi-step process including vesicular trafficking and organelle migration (Berruti and Paiardi 2011). Golgi-derived vesicles have also been proposed to contain proteins essential for sperm tail assembling (Kierszenbaum et al. 2011; Lerer-Goldshtein et al. 2010; Moreno et al. 2000; Zhou et al. 2011).
RC/BTB2 is not only expressed in germ cells. Given that the gene is up-regulated during ciliogenesis induced by multicilin (Stubbs et al. 2012), and that the Rc/btb2 gene has a somatic isoform, we hypothesized that Rc/btb2 is important for ciliogenesis. To test this hypothesis, we chose to examine two mammalian cell lines that are widely used for the study of ciliogenesis, IMCD3 and NIH3T3 cells (Marley et al. 2013; Schneider et al. 2009; Schroder et al. 2007; Tammachote et al. 2009). Rc/btb2 mRNA is expressed in both of these cell lines. Moreover, in both cell lines RC/BTB2 protein is co-localized with Golgin-58K protein, Golgin-97 and γ-tubulin, markers for Golgi body and centrosome, respectively, two key structures involved in ciliogenesis. This subcellular distribution supports our hypothesis that RC/BTB2 plays a role in ciliogenesis.
To further test this hypothesis, we silenced gene expression using the shRNA technique. Three shRNA constructs were made that targeted different regions of the Rc/btb2 mRNA sequence. Ciliogenesis defects in the shRNA1 and shRNA3 stable cell lines appeared to be due to the knockdown effect on Rc/btb2 gene expression, as the control stable cells and shRNA2 stable cells which had only a 30% reduction in Rc/btb2 mRNA expression, had no obvious ciliogenesis defects, and exogenous expression of RC/BTB2 in the stable knockdown cells restored ciliogenesis.
In normal cells, cilia are dynamically regulated during cell cycle progression: they are present in G0 and G1 cells, and usually in S/G2 cells, but almost invariably are resorbed before entering into mitosis, re-appearing post-cytokinesis (Avidor-Reiss and Gopalakrishnan 2013; Paridaen et al. 2013). It is unlikely the ciliogenesis defect in the knockdown cells was due to impaired cell cycle, since flow cytometry analysis of the cell cycle for these stable cell lines indicated that cell populations at different cycle periods were similar, and there was no difference in cell proliferation between the control and knockdown stable cells.
Immunofluorescence staining and SEM experiments not only revealed a reduction in the percentage of ciliated cells, but also demonstrated that even if some shRNA1 or shRNA3 cells formed cilia, the length was significantly shorter than the primary cilia of the control cells. Many shRNA1 or shRNA3 cells just had small protrusion extending from the cell surface, suggesting a failed attempt to form a primary cilium.
The detailed mechanisms by which RC/BTB2 functions in the formation of primary cilia remains to be determined. We previously demonstrated that when RC/BTB2 fused to GFP was expressed in CHO cells, the protein was first present in small vesicles, and these vesicles gradually fused to form larger ones and collected close to the nuclear membrane. This movement pattern was highly suggestive of a role for RC/BTB2 in vesicle trafficking. Thus, it is possible that RC/BTB2 functions as a carrier to transport cargo along the ciliogenesis pathway required for the assembly of primary cilia. If this notion is correct, targeting Rc/btb2 should result in failure of the transport of cilia components from the Golgi body to the centrioles and consequently inability to construct a primary cilium or incomplete ciliogenesis.
The major function of primary cilia is to sense a variety of external stimuli. It has been shown that several signaling pathways are present in primary cilia, including platelet-derived growth factor receptor αα, Wnt, Hedgehog and calcium signaling (Seeger-Nukpezah and Golemis 2012). In addition, defects in primary cilia structure/function give rise to developmental disorders and genetic diseases (Oh EC and Katsanis 2012). Motile cilia are capable of clearing mucus over tracheal epithelial cells in the airway, moving cerebrospinal fluid over ependymal cells in brain ventricles, transporting eggs over the epithelial cells of the female reproductive tract and driving sperm to fertilize eggs. Thus, understanding the regulation of ciliogenesis elucidates mechanisms of cilia-related diseases. The complex architecture of the cilium allows for multiple independent regulatory mechanisms to control its composition, and allow its function to emerge from its form. These mechanisms include intraflagellar transport (Kozminski et al. 1993; Rosenbaum and Witman 2002), the role of transcription factors, such as Foxj1and Rfx (Look et al. 2001; Choksi et al. 2014), the cytoskeletal system (Walczak et al. 2013), small G proteins (Qin 2012), and autophagy (Orhon et al. 2014).
In the present study, we only investigated the role of RC/BTB2 in primary ciliogenesis. Our previous studies suggested that RC/BTB2 is highly expressed in the testis, an organ that generates sperm with motile flagella. RC/BTB2 is also regulated by multicilin, which controls motile ciliogenesis. Collectively, the latter observations indicate that RC/BTB2 also has a role in motile ciliogenesis. Interestingly, Latil suggested that RC/BTB2 might be a tumor suppressor in human (Latil et al. 2003). Recent studies revealed that tumorigenesis is closely associated with ciliogenesis (Kuehn et al. 2007), raising the possibility that RC/BTB2 has dual functions in both ciliogenesis and tumorigenesis.
In summary, we characterized RC/BTB2 protein localization in somatic IMCD3 and NIH3T3 cells. Its subcellular localization strongly suggests that it is has an important role in ciliogenesis, perhaps through the mechanism of vesicle trafficking. The in vitro knockdown experiments documented that RC/BTB2 is essential for the normal formation of primary cilia.
Supplementary Material
Acknowledgements
This research was supported by NIH grant HD076257, Virginia Commonwealth University Presidential Research Incentive Program (PRIP) and Massey Cancer Award (to ZZ), Natural Science Foundation of China (81300536 to LZ), Natural Science Funds of Hubei Province of China (2013CFB331 to LZ, 2012FFB04904 to ZZ), and the Youth Talents of Science and Technology Projects of Health Department of Hubei Province of China (QJX2012-22 to YS).
Confocal microscopy and scanning EM was performed the Imaging Core of Virginia Commonwealth University (5P30NS047463).
References
- Alieva IB, Vorobjev IA. Vertebrate primary cilia: a sensory part of centrosomal complex in tissue cells, but a “sleeping beauty” in cultured cells? Cell Biol Int. 2004;28:139–50. doi: 10.1016/j.cellbi.2003.11.013. [DOI] [PubMed] [Google Scholar]
- Avidor-Reiss T, Gopalakrishnan J. Cell Cycle Regulation of the Centrosome and Cilium. Drug Discov Today Dis Mech. 2013;10:e119–e124. doi: 10.1016/j.ddmec.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berruti G, Paiardi C. Acrosome biogenesis: Revisiting old questions to yield new insights. Spermatogenesis. 2011;1:95–98. doi: 10.4161/spmg.1.2.16820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray DF, Bagu J, Koegler P. Comparison of hexamethyldisilazane (HMDS), Peldri II, and critical-point drying methods for scanning electron microscopy of biological specimens. Microsc Res Tech. 1993;26:489–95. doi: 10.1002/jemt.1070260603. [DOI] [PubMed] [Google Scholar]
- Choksi SP, Lauter G, Swoboda P, Roy S. Switching on cilia: transcriptional networks regulating ciliogenesis. Development. 2014;141:1427–41. doi: 10.1242/dev.074666. [DOI] [PubMed] [Google Scholar]
- D'Angelo G, Uemura T, Chuang CC, Polishchuk E, Santoro M, Ohvo-Rekila H, Sato T, Di Tullio G, Varriale A, D'Auria S. Vesicular and non-vesicular transport feed distinct glycosylation pathways in the Golgi. Nature. 2013;501:116–20. doi: 10.1038/nature12423. [DOI] [PubMed] [Google Scholar]
- Dishinger JF, Kee HL, Jenkins PM, Fan S, Hurd TW, Hammond JW, Truong YN, Margolis B, Martens JR, Verhey KJ. Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-beta2 and RanGTP. Nat Cell Biol. 2010;12:703–10. doi: 10.1038/ncb2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond IA. Cilia functions in development. Curr Opin Cell Biol. 2012;24:24–30. doi: 10.1016/j.ceb.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmer BT, Maric D, Engman DM. Molecular mechanisms of protein and lipid targeting to ciliary membranes. J Cell Sci. 2010;123:529–36. doi: 10.1242/jcs.062968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JW, Dolbeare F, Pallavicini MG, Beisker W, Waldman F. Cell cycle analysis using flow cytometry. Int J Radiat Biol Relat Stud Phys Chem Med. 1986;49:237–55. doi: 10.1080/09553008514552531. [DOI] [PubMed] [Google Scholar]
- Kee HL, Verhey KJ. Molecular connections between nuclear and ciliary import processes. Cilia. 2013;2:11. doi: 10.1186/2046-2530-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierszenbaum AL, Rivkin E, Tres LL, Yoder BK, Haycraft CJ, Bornens M, Rios RM. GMAP210 and IFT88 are present in the spermatid golgi apparatus and participate in the development of the acrosome-acroplaxome complex, head-tail coupling apparatus and tail. Dev Dyn. 2011;240:723–36. doi: 10.1002/dvdy.22563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Zaghloul NA, Bubenshchikova E, Oh EC, Rankin S, Katsanis N, Obara T, Tsiokas L. Nde1-mediated inhibition of ciliogenesis affects cell cycle re-entry. Nat Cell Biol. 2011;13:351–60. doi: 10.1038/ncb2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Dynlacht BD. Regulating the transition from centriole to basal body. J Cell Biol. 2011;193:435–44. doi: 10.1083/jcb.201101005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci U S A. 1993;90:5519–23. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn EW, Walz G, Benzing T. Von hippel-lindau: a tumor suppressor links microtubules to ciliogenesis and cancer development. Cancer Res. 2007;67:4537–40. doi: 10.1158/0008-5472.CAN-07-0391. [DOI] [PubMed] [Google Scholar]
- Latil A, Chene L, Mangin P, Fournier G, Berthon P, Cussenot O. Extensive analysis of the 13q14 region in human prostate tumors: DNA analysis and quantitative expression of genes lying in the interval of deletion. Prostate. 2003;57:39–50. doi: 10.1002/pros.10272. [DOI] [PubMed] [Google Scholar]
- Lerer-Goldshtein T, Bel S, Shpungin S, Pery E, Motro B, Goldstein RS, Bar-Sheshet SI, Breitbart H, Nir U. TMF/ARA160: A key regulator of sperm development. Dev Biol. 2010;348:12–21. doi: 10.1016/j.ydbio.2010.07.033. [DOI] [PubMed] [Google Scholar]
- Look DC, Walter MJ, Williamson MR, Pang L, You Y, Sreshta JN, Johnson JE, Zander DS, Brody SL. Effects of paramyxoviral infection on airway epithelial cell Foxj1 expression, ciliogenesis, and mucociliary function. Am J Pathol. 2001;159:2055–69. doi: 10.1016/S0002-9440(10)63057-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marley A, Choy RW, von Zastrow M. GPR88 reveals a discrete function of primary cilia as selective insulators of GPCR cross-talk. PLoS One. 2013;8:e70857. doi: 10.1371/journal.pone.0070857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Menarguez JA, Geuze HJ, Ballesta J. Evidence for a nonlysosomal origin of the acrosome. J Histochem Cytochem. 1996;44:313–20. doi: 10.1177/44.4.8601690. [DOI] [PubMed] [Google Scholar]
- Molla-Herman A, Ghossoub R, Blisnick T, Meunier A, Serres C, Silbermann F, Emmerson C, Romeo K, Bourdoncle P, Schmitt A. The ciliary pocket: an endocytic membrane domain at the base of primary and motile cilia. J Cell Sci. 2010;123:1785–95. doi: 10.1242/jcs.059519. [DOI] [PubMed] [Google Scholar]
- Moreno RD, Ramalho-Santos J, Chan EK, Wessel GM, Schatten G. The Golgi apparatus segregates from the lysosomal/acrosomal vesicle during rhesus spermiogenesis: structural alterations. Dev Biol. 2000;219:334–49. doi: 10.1006/dbio.2000.9606. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Wei JH, Seemann J. Modular organization of the mammalian Golgi apparatus. Curr Opin Cell Biol. 2012;24:467–74. doi: 10.1016/j.ceb.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh EC, Katsanis N. Cilia in vertebrate development and disease. Development. 2012;139:443–8. doi: 10.1242/dev.050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orhon I, Dupont N, Pampliega O, Cuervo AM, Codogno P. Autophagy and regulation of cilia function and assembly. Cell Death Differ. 2014 doi: 10.1038/cdd.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paridaen JT, Wilsch-Brauninger M, Huttner WB. Asymmetric inheritance of centrosome-associated primary cilium membrane directs ciliogenesis after cell division. Cell. 2013;155:333–44. doi: 10.1016/j.cell.2013.08.060. [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Rosenbaum JL. Intraflagellar transport and cilia-dependent diseases. Trends Cell Biol. 2002;12:551–5. doi: 10.1016/s0962-8924(02)02410-8. [DOI] [PubMed] [Google Scholar]
- Pedersen LB, Rosenbaum JL. Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr Top Dev Biol. 2008;85:23–61. doi: 10.1016/S0070-2153(08)00802-8. [DOI] [PubMed] [Google Scholar]
- Qin H. Regulation of intraflagellar transport and ciliogenesis by small G proteins. Int Rev Cell Mol Biol. 2012;293:149–68. doi: 10.1016/B978-0-12-394304-0.00010-5. [DOI] [PubMed] [Google Scholar]
- Raposo G, Marks MS, Cutler DF. Lysosome-related organelles: driving post-Golgi compartments into specialisation. Curr Opin Cell Biol. 2007;19:394–401. doi: 10.1016/j.ceb.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–25. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Schneider L, Stock CM, Dieterich P, Jensen BH, Pedersen LB, Satir P, Schwab A, Christensen ST, Pedersen SF. The Na+/H+ exchanger NHE1 is required for directional migration stimulated via PDGFR-alpha in the primary cilium. J Cell Biol. 2009;185:163–76. doi: 10.1083/jcb.200806019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder JM, Schneider L, Christensen ST, Pedersen LB. EB1 is required for primary cilia assembly in fibroblasts. Curr Biol. 2007;17:1134–9. doi: 10.1016/j.cub.2007.05.055. [DOI] [PubMed] [Google Scholar]
- Seeger-Nukpezah T, Golemis EA. The extracellular matrix and ciliary signaling. Curr Opin Cell Biol. 2012;24:652–61. doi: 10.1016/j.ceb.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs JL, Vladar EK, Axelrod JD, Kintner C. Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nat Cell Biol. 2012;14:140–7. doi: 10.1038/ncb2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammachote R, Hommerding CJ, Sinders RM, Miller CA, Czarnecki PG, Leightner AC, Salisbury JL, Ward CJ, Torres VE, Gattone VH., 2nd Ciliary and centrosomal defects associated with mutation and depletion of the Meckel syndrome genes MKS1 and MKS3. Hum Mol Genet. 2009;18:3311–23. doi: 10.1093/hmg/ddp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang XM, Lalli MF, Clermont Y. A cytochemical study of the Golgi apparatus of the spermatid during spermiogenesis in the rat. Am J Anat. 1982;163:283–94. doi: 10.1002/aja.1001630402. [DOI] [PubMed] [Google Scholar]
- Thorne-Tjomsland G, Clermont Y, Hermo L. Contribution of the Golgi apparatus components to the formation of the acrosomic system and chromatoid body in rat spermatids. Anat Rec. 1988;221:591–8. doi: 10.1002/ar.1092210205. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Gayek S, Ohi R. Microtubule-depolymerizing kinesins. Annu Rev Cell Dev Biol. 2013;29:417–41. doi: 10.1146/annurev-cellbio-101512-122345. [DOI] [PubMed] [Google Scholar]
- Wang J, Teves ME, Shen X, Nagarkatti-Gude DR, Hess RA, Henderson SC, Strauss JF, 3rd, Zhang Z. Mouse RC/BTB2, a member of the RCC1 superfamily, localizes to spermatid acrosomal vesicles. PLoS One. 2012;7:e39846. doi: 10.1371/journal.pone.0039846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Mukherjee A, Lebman DA, Fang X. Gene expression of the lysophosphatidic acid receptor 1 is a target of transforming growth factor beta. Oncogene. 2013;32:3198–206. doi: 10.1038/onc.2012.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Fabian L, Bayraktar JL, Ding HM, Brill JA, Chang HC. Auxilin is required for formation of Golgi-derived clathrin-coated vesicles during Drosophila spermatogenesis. Development. 2011;138:1111–20. doi: 10.1242/dev.057422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.