Summary
Protein concentrations evolve under greater evolutionary constraint than mRNA levels. Translation efficiency of mRNA represents the chief determinant of basal protein concentrations. This raises a fundamental question of how mRNA and protein levels are coordinated in dynamic systems responding to physiological stimuli. This report examines the contributions of mRNA abundance and translation efficiency to protein output in cells responding to oxygen stimulus. We show that changes in translation efficiencies, not mRNA levels, represent the major mechanism governing cellular responses to [O2] perturbations. Two distinct cap-dependent protein synthesis machineries select mRNAs for translation: the normoxic eIF4F and the hypoxic eIF4FH. O2-dependent remodeling of translation efficiencies enables cells to produce adaptive translatomes from preexisting mRNA pools. Differences in mRNA expression observed under different [O2] are likely neutral, as they are during evolution. We propose that mRNAs contain translation efficiency determinants for their triage by the translation apparatus on [O2] stimulus.
Keywords: Oxygen, hypoxia, cancer, translation, RNA sequencing, SILAC, eIF4E, eIF4E2, eIF4F, HIF
Graphical Abstract
Introduction
It is assumed that steady state mRNA levels represent an accurate proxy for protein expression. In most studies, the protein synthesis machinery is perceived as a passive participant in the regulation of gene expression that reflexively translates mRNA abundance into protein output. Recent studies have challenged this assumption by demonstrating a lack of correlation between protein and mRNA levels (Schwanhausser et al., 2011; Tian et al., 2004; Vogel et al., 2010; Wang et al., 2013). These studies provide strong evidence that translation efficiency is a superior predictor of steady state protein levels compared to mRNA levels, mRNA stability, and protein stability (Schwanhausser et al., 2011). Interestingly, a comparison of primates established that protein expression evolved under stronger constraints than mRNA levels, the latter being effectively neutral (Khan et al., 2013). These findings point to the evolution of complex regulatory processes of the translation apparatus to titrate protein output from highly divergent levels of cellular mRNAs. A biological role for alternative translation efficiency was recently reported for the transcriptionally silent system of Drosophila oocyte-to-embryo transition (Kronja et al., 2014), and in stem cell differentiation (Lu et al., 2009). How mRNA and protein abundance are coordinated in dynamic systems responding to a stimulus remains a fundamental question (Vogel, 2013).
Perturbations in environmental [O2] are observed in a wide array of physiological and pathological conditions including development, cardiovascular disease and cancer (Ratcliffe, 2013; Semenza, 2014). Cells exposed to hypoxia (i.e. low [O2]) activate a robust transcription program by the hypoxia-inducible factor (HIF) (Wang et al., 1995). HIF promotes the synthesis of key mRNAs that encode proteins involved in cellular O2 homeostasis. Hypoxia also elicits a fundamental reorganization of the cellular translation apparatus. In normoxia, the eIF4F complex typically initiates protein synthesis (Sonenberg and Hinnebusch, 2009). The cap-binding eIF4E, the RNA helicase eIF4A and the scaffold eIF4G comprise the three major components of eIF4F (Jackson et al., 2010). Hypoxia prevents binding of eIF4E to eIF4G thereby inhibiting eIF4F activity (Connolly et al., 2006; Koritzinsky et al., 2006; Liu et al., 2006). Hypoxic cells activate an alternative translation pathway that relies on the cap-binding eIF4E2 and the O2-regulated HIF-2α (Uniacke et al., 2012; Uniacke et al., 2014). Additional eIF4E-dependent and independent pathways, such as internal ribosome entry site (IRES), can be activated during hypoxia (Braunstein et al., 2007; Yi et al., 2013; Young et al., 2008). The profound reorganization of essential cellular pathways by [O2] provides an ideal system to examine the contributions of the transcription and translation machineries to protein output in response to a physiological stimulus. In this report, we present evidence that an O2-regulated global remodeling of translation efficiencies, rather than changes in transcript abundance, is the principal determinant of protein output to O2 deprivation.
Results
Widespread remodeling of the translatome by O2
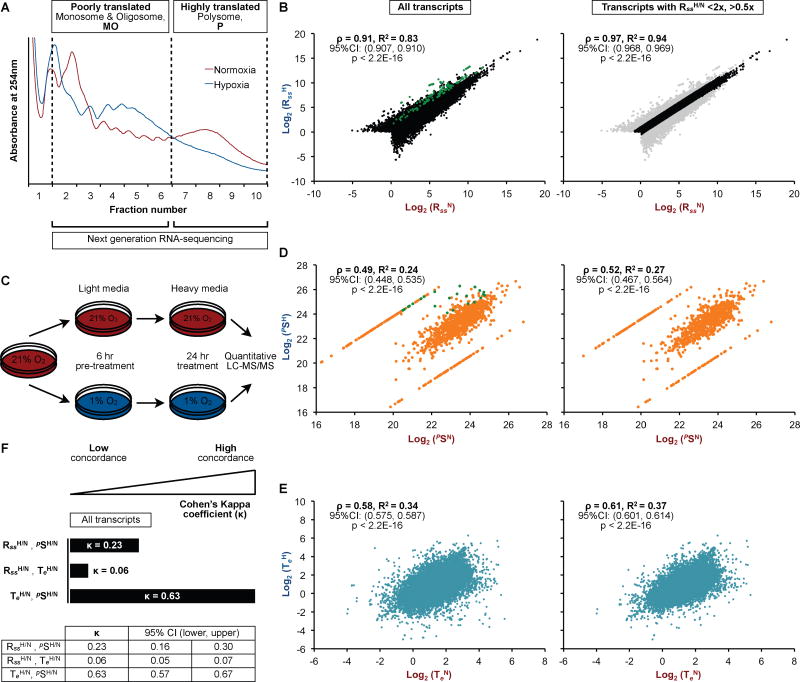
We investigated the role of mRNA expression and translation efficiency in a dynamic system associated with a robust transcription response to stimulus: oxygen tension. First, we isolated transcripts engaged by the protein synthesis machineries of cells maintained in normoxia (21% O2) or hypoxia (1% O2, 24h) (Figure 1A; S1A). Poorly translated mRNAs accumulate in the monosome and oligosome fractions (MO), while highly translated mRNAs are found in polysome (P) fractions (Figure 1A; S1A). Total RNA isolated from the MO and P fractions were subjected to high-throughput RNA-sequencing (RNA-Seq) (Figure 1A). Cellular RNA steady state level (Rss) was defined as the total read count (RC) from sequenced fractions: Rss=PRC+MORC. RNA-Seq analysis identified approximately 46,500 and 45,000 different transcripts in normoxic (RssN) and hypoxic (RssH) cells, respectively (Figure S1B, C). RssH and RssN displayed a high correlation (R2=0.83) (Figure 1B, S1D, left panels), with more than 77% of mRNAs within a range of 0.5-fold to 2-fold difference (Figure 1B, S1D, right panels). Targets of the HIF transcription program have high RssH/RssN ratios, as expected (Figure 1B, S1D, left panels, green). To determine protein output, we performed pulse-stable isotope labeling with amino acids in cell culture (pSILAC) analyses (Selbach et al., 2008) (Figure 1C). pSILAC identified more than 1,000 different newly synthesized proteins in normoxic (pSN) and hypoxic (pSH) cells. ~20% of proteins displayed pSH/pSN of ~1.0, whereas HIF targets exhibited high ratios (Figure S1E), confirming that this assay was capable of distinguishing between proteins with similar and different rates of synthesis. Interestingly, pSH/pSN displayed lower correlation (R2=0.24) (Figure 1D, S1F, left panels) than what would be predicted by RssH/RssN. The weak relationship between mRNA levels and protein output suggests that a switch in translation efficiency, rather than changes in transcript levels, may be the primary cellular response to O2 availability. To explore this possibility, we examined the translation efficiency (Te) of mRNAs identified by RNA-Seq of MO and P fractions (Te=PRC/MORC). TeH/TeN correlation (R2=0.34) (Figure 1E, S1G, left panels) was in good agreement with pSH/pSN. RssH/RssN had low concordance with TeH/TeN indicating that polysomal capture of transcripts cannot be simply implied by O2-regulated changes in steady state mRNA (Figure 1F, S1H). In contrast to RssH/RssN, TeH/TeN displayed higher concordance with pSH/pSN (Figure 1F). As the cellular response to O2 stimulus does encompass changes in mRNA steady state levels, we measured the relationship between RssH/RssN, pSH/pSN and TeH/TeN for transcripts displaying minimal variations in expression as a function of [O2]. Transcripts that display less than 2-fold difference between hypoxic and normoxic cells (Figure 1B, S1D, right panels) also produced highly variable protein outputs (Figure 1D, S1F, right panels) and TeH/TeN (Figure 1E, S1G, right panels) with similar concordances to those observed for the total mRNA population (Figure S1I). These results suggest that changes in [O2] causes a widespread remodeling of protein output that relies mostly on a systemic switch in translation efficiency and not on mRNA levels.
Figure 1. O2-dependent remodeling of the cellular translatome.
(A) RNA-Seq was performed on MO and P fractions of normoxic and hypoxic U87MG. MO/P demarcation was selected based on the induction pattern of hypoxic translation of several mRNAs (Uniacke et al., 2012; Uniacke et al., 2014). (B) Plots of Rss in hypoxic (RssH) versus normoxic (RssN) (left panel) U87MG (left panel) and transcripts with RssH/RssN (RssH/N) ratios between <2x and >0.5x (right panel, black). Transcripts with Rss <1 were excluded from analysis. (C) Schematic of pSILAC workflow. (D) Normalized heavy intensities of newly synthesized proteins determined by pSILAC in hypoxic (pSH) and normoxic (pSN) U87MG (left panel) and from transcripts with RssH/N ratios between <2x and >0.5x (right panel). Proteins that were only detected in normoxic (red) and hypoxic (blue) U87MG were given the maximum fold change observed. Canonical hypoxia-inducible genes are highlighted in green. (E) Plots of Te in hypoxic (TeH) versus normoxic (TeN) U87MG (left panel), and transcripts with RssH/N ratios between <2x and >0.5x (right panel). (F) Concordance analysis between RssH/N, pSH/N and TeH/N.
The eIF4F and eIF4FH protein synthesis machineries coordinate the O2-regulated translatomes
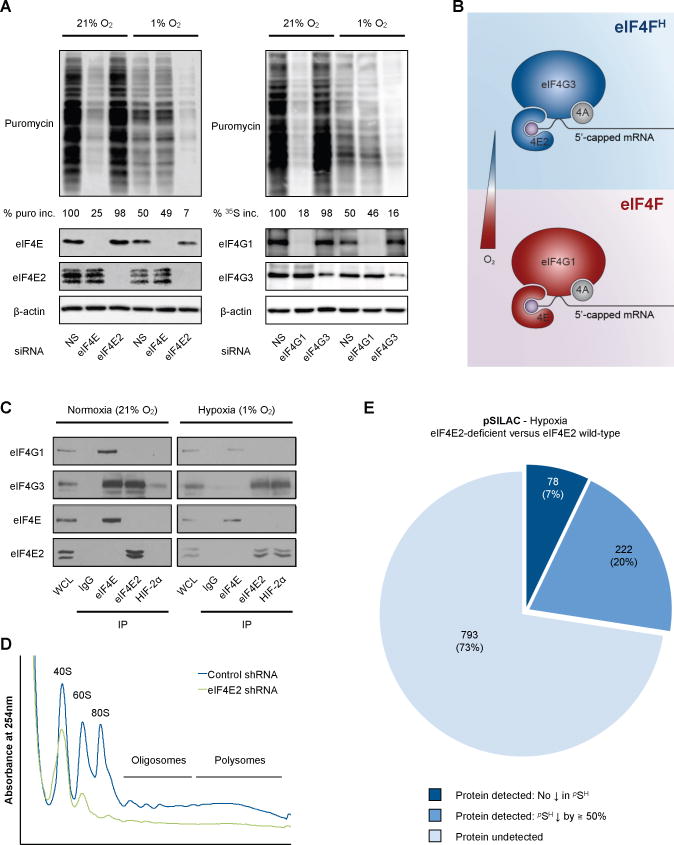
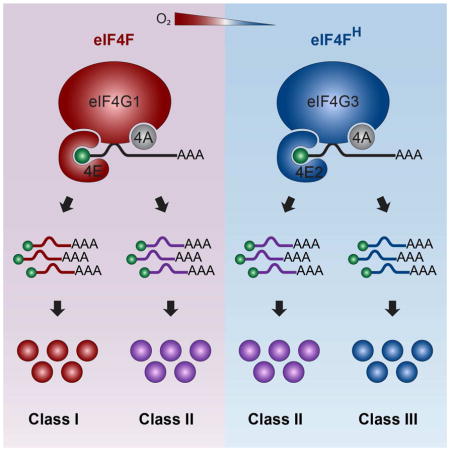
KEGG analysis revealed that hypoxic cells populate essentially the same functional pathways as their normoxic counterparts with different proteins while prioritizing certain processes over others (Figure S2A–C). Silencing elements of the canonical eIF4F translation initiation complex, namely eIF4E and eIF4G1, prevents the bulk of protein synthesis in normoxic cells (Figure 2A, Figure S2C), indicating that TeN (Figure S1H, left panel) and the normoxic translatome (Figure 1D, left panel) relies on this complex. On the other hand, eIF4E or eIF4G1 silencing had little effect on hypoxic global translation rates likely because of eIF4F inactivation by low [O2] (Connolly et al., 2006; Liu et al., 2006) (Figure 2A, Figure S2C). This implies the existence of a broad alternative hypoxic translation initiation complex that sustains TeH, which we term the hypoxic eIF4F (eIF4FH) (Figure 2B). Pull-down analysis revealed that eIF4FH consists of eIF4E2, eIF4A (Uniacke et al., 2012) and eIF4G3, a functional homolog of eIF4G1 (Figure 2C, Figure S2D). Silencing elements specific to eIF4FH essentially abolished global rate of translation in hypoxic cells with little effect on normoxic cells (Figure 2A, Figure S2C). Recruitment of mRNAs to polysomes of hypoxic cells was significantly impaired in eIF4E2-depleted (Figure 2D) or eIF4G3-depleted cells (Figure S2E). While this prevented a TeH analysis in eIF4FH-defective cells, pSILAC revealed that more than 90% of produced proteins observed in hypoxic cells were either not detectable or considerably reduced in eIF4E2-impaired cells (Figure 2E). These results demonstrate the existence of two major cap-dependent protein synthesis pathways (Figure 2B): the normoxic eIF4F (eIF4E-eIF4A-eIF4G1) and hypoxic eIF4FH (eIF4E2-eIF4A-eIF4G3) that remodel Te in response to O2 stimulus.
Figure 2. The eIF4F and eIF4FH translation machineries govern O2-dependent translatome remodeling.
(A) Global translation rates of U87MG transiently transfected with siRNA against the indicated proteins and non-silencing (NS) control siRNA were measured by puromycin incorporation. Loading was performed on an equal cell basis. %puro inc., percent puromycin incorporation. Immunoblots of silenced proteins are shown. β-actin was used as a loading control. (B) Schematic of eIF4FH and eIF4F. (C) Immunoblots of eIF4E, eIF4E2, and HIF-2α endogenous IPs in normoxic and hypoxic U87MG. WCL, 5% whole cell lysate. (D) Polysome profiles and (E) pSILAC analysis of hypoxic U87MG stably expressing eIF4E2-specific or non-silencing (NS) control shRNA.
Classification of three major mRNA classes based on O2-dependent translation efficiencies
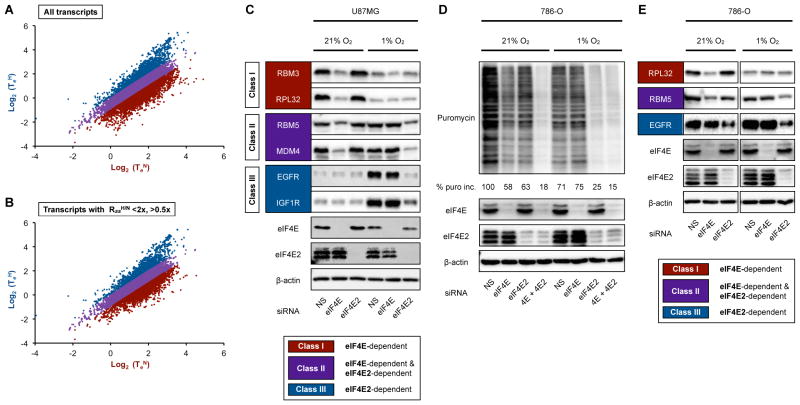
A closer examination of TeH/TeN ratios suggests that mRNAs can be divided into three O2-responsive classes. (Figure 3A, S3A, S3B, top panel). Class I mRNAs are efficiently translated in normoxia but less in hypoxia. Class II mRNAs are efficiently translated independently of [O2]. Class III mRNAs maintain or increase translation efficiency in hypoxia. Overall, Class I, II, and III represent ~25%, ~60%, and ~15% of the combined normoxic and hypoxic translatome, respectively. The presence and relative size of the three classes were maintained even for mRNAs that exhibited minimal RssH/RssN differences (Figure 3B, S3B, bottom panel, Figure 1B right panel). Five representative transcripts from each class were validated by qRT-PCR (Figure S3B). These results raise the intriguing possibility that cells express mRNA populations that are hard-wired for either normoxic eIF4F (Class I–II) or hypoxic eIF4FH (Class II–III) translation. Immunoblot analysis revealed that proteins derived from Class I mRNAs, e.g. RBM3 and RPL32, accumulate preferentially under normoxia and are predominantly dependent on eIF4F (Figure 3C). Class II proteins e.g. RBM5 and MDM4, can be synthesized by eIF4F and eIF4FH, respectively (Figure 3C). Proteins of Class III mRNAs, e.g. EGFR and IGF1R accumulate preferentially under hypoxia and are synthesized by eIF4FH (Figure 3C). To confirm the validity of Class I–III mRNAs, we tested our model in the renal carcinoma cell line 786-O. The eIF4E2/eIF4FH activator HIF-2α, which is normally degraded in normoxia, is constitutively active in 786-O as a consequence of VHL-deficiency (Maxwell et al., 1999). This provides the opportunity to examine eIF4F and eIF4FH operating in parallel within the same normoxic cellular context. Silencing of both eIF4E and eIF4E2 was required to reduce global translation to below 20% of control, confirming that both translation machineries are operative in normoxic 786-0 cells (Figure 3D). Protein accumulation of the Class I mRNA-derived RPL32 occurred in normoxic 786-O in an eIF4E- but not eIF4E2-dependent manner (Figure 3E). Class II protein RBM5 was dependent on both eIF4E and eIF4E2 under normoxic conditions (Figure 3E), as expected. Finally, the Class III protein EGFR was sensitive to eIF4E2, but not eIF4E depletion (Figure 3E). Only Class II and III proteins were produced in hypoxic 786-O due to the loss of eIF4F activity. These results suggest the existence of translation efficiency determinants that are hard-wired in mRNAs, which provide the basis for differential recruitment by the O2-regulated protein synthesis machineries eIF4F and eIF4FH.
Figure 3. Global reorganization of translation efficiency during O2 deprivation.
(A) Classification of all transcripts and (B) transcripts with RssH/N ratios between <2x and >0.5x into three major classes according to TeH/TeN ratios. Class I transcripts (TeH/TeN ≤0.5-fold, red); Class II transcripts (purple); Class III transcripts (TeH/TeN ≥1-fold, blue). Low abundance transcripts (Rss <10) were excluded from the analysis. (C) Immunoblots of representative proteins in normoxic and hypoxic U87MG from each class. (D) Global translation rates in normoxic and hypoxic 786-O transiently transfected with eIF4E-specific, eIF4E2-specific, or NS control siRNA were measured using puromycin incorporation. Immunoblots of silenced proteins are shown. (E) Immunoblots of representative proteins in normoxic and hypoxic 786-O as measured in (C).
Translation efficiency controls protein production from HIF target mRNAs
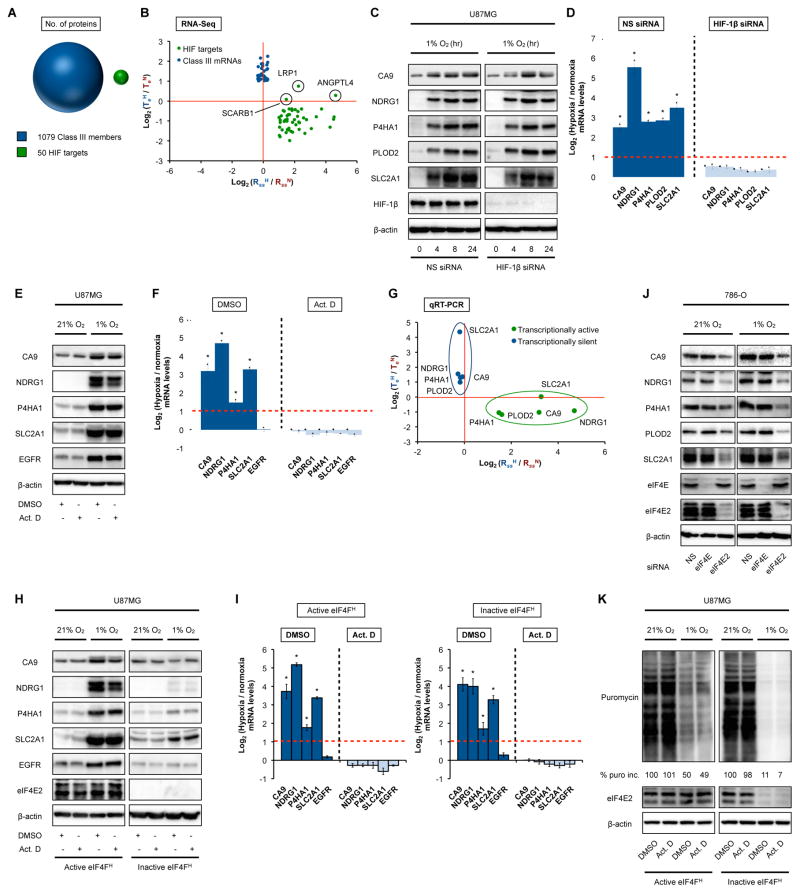
Hypoxia elicits a robust transcriptional response by HIF, which promotes the synthesis of genes involved in O2 homeostasis (Schodel et al., 2011; Wang et al., 1995). Exactly 50 canonical transcription targets of the HIF pathway were identified to exhibit an RssH/RssN ≥2 (Figure 1B, left panel), considerably less than proteins derived from Class III mRNAs with minimal change in RssH/NRss (Figure 4A). Interestingly, the majority of HIF targets showed decreased TeH/TeN (Figure 4B, S4A). Thus, we suspected that the substantial increase in HIF target proteins in hypoxia (Figure S4B) might be explained by the Class III property, rather than absolute increases in their respective mRNAs levels (Figure S4C). To test this, we examined the effect of silencing HIF-1β (ARNT), a mandatory subunit of the HIF transcription factor (Wang et al., 1995). HIF target proteins accumulated in HIF-1β-impaired hypoxic cells to similar levels as those observed in their control counterparts (Figure 4C), even though their corresponding mRNAs were not induced upon hypoxia (Figure 4D, Figure S4D). Likewise, cells treated with the general transcription inhibitor actinomycin D (Act. D) exhibited an accumulation of HIF target proteins to levels undistinguishable from those of untreated controls (Figure 4E) even in the absence of their respective mRNA induction (Figure 4F). Translation efficiency analysis revealed a substantial increase in TeH/TeN of HIF target mRNAs in transcriptionally silent cells, thereby revealing their identity as Class III mRNAs (Figure 4G). In agreement with Class III mRNA characteristics (Figure 3), HIF target mRNAs are selectively recruited for translation by the eIF4FH machinery in hypoxic cells regardless of cellular transcription competency (Figure 4H) and mRNA induction (Figure 4I) as well as in normoxic 786-0 cells where eIF4FH and eIF4F are simultaneously active (Figure 4J). These observations can be generalized on a global scale, as protein output is mostly unaffected in transcription-incompetent hypoxic cells while remaining dependent on eIF4E2 activity (Figure 4K). These results demonstrate that translation efficiency, rather than mRNA expression, is the primary determinant of protein levels in dynamic systems responding to a physiological stimulus, even in the presence of robust transcriptional activity.
Figure 4. Translation efficiency determines protein output in response to hypoxia.
(A) RNA-Seq analysis identified 1079 different proteins derived from Class III mRNAs with RssH/N ratios between <2x and >0.5x (blue), and 50 proteins derived from HIF target mRNAs with RssH/N ratios ≥2 (green) (see Figure 1B). (B) Plot of change in Rss against change in Te for 50 HIF target mRNAs in hypoxic versus normoxic U87MG (see Figure 1B). Blue dots; representative Class III candidates with minimal change in Rss levels. (C) Immunoblots of HIF target proteins in U87MG transiently transfected with HIF-1β-specific or NS siRNA, and subjected to a hypoxic time course. (D) Corresponding 24 hr hypoxia/normoxia steady-state mRNA levels of proteins measured in (C). * denotes statistical significance (p<0.05) compared to 0 hr hypoxia. (E) Immunoblots of hypoxia-inducible proteins in U87MG treated with Act. D or DMSO for 20 min, followed by 6 hr of hypoxic or normoxic treatment. (F) Corresponding hypoxia/normoxia steady-state mRNA levels of proteins measured in (E). * denotes p<0.05 compared to the corresponding normoxia control. (G) Plot of change in mRNA levels against change in Te for 5 representative HIF target mRNAs in transcriptionally silent versus active U87MG under hypoxic versus normoxic conditions. (H) Immunoblots of HIF target proteins in U87MG stably expressing shRNA targeting eIF4E2 (inactive eIF4FH) or NS shRNA (active eIF4FH), and treated with Act. D as in (E). (I) Corresponding hypoxic induction of steady-state mRNA levels of proteins measured in (H). * denotes p≤0.05 compared to the corresponding normoxia control. (J) Immunoblots of HIF target proteins in normoxic and hypoxic 786-O transiently transfected with eIF4E-specific, eIF4E2-specific, or NS siRNA. (K) Global translation rates in normoxic and hypoxic U87MG transiently transfected with eIF4E2-specific or NS siRNA, and treated with Act. D as in (E), were measured using puromycin incorporation. Immunoblots of silenced proteins are shown.
Discussion
The demonstration that protein concentration is determined by translation efficiency rather than mRNA abundance (Schwanhausser et al., 2011), and that changes in mRNA levels are evolutionarily neutral (Khan et al., 2013) represent breakthroughs in our understanding of gene expression. These studies raise the question as to the role of mRNA level changes in response to physiological stimuli (Vogel, 2013). We show that cells reprogram protein output as a function of [O2] through a systemic switch in mRNA translation efficiencies. Two distinct cap-dependent protein synthesis machineries govern this phenomenon: the normoxic eIF4F and hypoxic eIF4FH. These two translational programs remodel the cellular translatome by triaging available mRNAs depending on [O2], with minimal reliance on changes in steady state transcript levels. Even hypoxia-inducible mRNAs, including HIF targets, are ultimately controlled at the level of translation efficiency and not changes in mRNA levels. We suggest that translation efficiency controls protein output on O2 stimulus, and that changes in mRNA levels may be effectively neutral, as they are during evolution.
We have defined three major classes of O2-responsive mRNAs. Class I and III mRNAs are exclusively recruited by eIF4F or eIF4FH, respectively. Class II mRNAs can be recruited by both, and undergo efficient translation regardless of [O2]. These findings suggest that genes have evolved O2-regulated Te determinants that enable their selective recruitment by eIF4F, or eIF4FH. In the case of Class III mRNAs, this may be explained, at least in part, by the presence of the RNA hypoxia response elements (rHRE). The rHRE recruits RBM4 that inhibits eIF4F-mediated translation (Lin et al., 2007) but facilitates eIF4FH-directed hypoxic protein synthesis (Uniacke et al., 2012). It is possible that Class I mRNA encode RNA element(s) that promote eIF4F activity while opposing eIF4FH under hypoxia. These O2-regulated Te determinants enable cells to triage the diverse mRNA populations in order to remodel protein output in response to changes in [O2]. From a broader perspective, it is tempting to speculate that mRNAs encode an array of Te determinants that help redefine the translatome on different stimuli. In support of this model, another cap-binding protein, eIF4E3, has been suggested to regulate translation under other settings (Landon et al., 2014). Thus, it is likely that cells have evolved multiple alternative translation initiation machineries that allow them to activate stimulus-specific translation efficiency programs (Andreev et al., 2015; Baudin-Baillieu et al., 2014; Kronja et al., 2014; Lu et al., 2009; McKinney et al., 2014; Ventoso et al., 2012; Young et al., 2008). These alternative machineries would reprogram global mRNA translation efficiencies in order to generate distinct, adaptive translatomes/proteomes. Finally, these findings imply that we should revisit the role of transcription-induced changes in mRNA levels in response to stimuli, as we have from the evolutionary perspective. The future challenge will be to decipher the roles of translation efficiency/machinery in fields of research that have been dominated by studies of transcriptional responses to cellular perturbations.
Experimental Procedures
Cell culture and reagents
U87MG human glioblastoma and 786-O human renal clear cell carcinoma cell lines were obtained from the American Type Culture Collection and propagated as suggested. Cells were maintained at 37°C in a 5% CO2, humidified incubator. Cells were subjected to hypoxia (1% O2, 24 hr unless otherwise stated) at 37°C in a 5% CO2, N2-balanced, humidified H35 HypOxystation (HypOxygen). Actinomycin D (Amresco) was added to cells at a final concentration of 1 μg/ml.
Polysome fractionation and RNA sequencing
Polysome fractionations were performed essentially as previously described (Franovic et al., 2007). Total RNA were isolated from individual fractions by standard phenol/chloroform extraction and ethanol precipitation following proteinase K treatment. Equal volumes of individual fractions from four independent experiments were pooled to yield the MO (fractions 2–6) and P (fractions 7–10) samples. cDNA library construction (Ovation RNA-Seq V2, NuGEN), sequencing runs (NextSeq 500, Illumina), and raw data processing were performed by Cofactor Genomics (St. Louis, MO, USA). RNA-Seq data are available via the NCBI SRA (accessions SRP065114, SRP065127).
pSILAC and mass spectrometry (MS)
Cells grown in “light” (R0K0) media were subjected to 1% O2 or 21% O2 pre-treatment for 6 hr. Light media was then replaced with “heavy” (R10K8) media, and cells were left to grow at 1% O2 or 21% O2 for 24 hr. Total cellular protein were then harvested using a 9M urea lysis buffer, and subjected to LC-MS/MS analysis. MS data are available via ProteomeXchange (identifier PXD003037).
Global protein synthesis measurements
Global protein synthesis was measured by puromycin (Gibco, Life Technologies) incorporation (1 μg/ml) for 30 min followed by immunoblot analysis with an anti-puromycin antibody (Kerafast).
RNA interference (RNAi)
siRNA (GE Dharmacon) were transfected at a final concentration of 100 nM using Effectene (Qiagen). shRNA (GE Dharmacon) were stably introduced as previously described (Uniacke et al., 2012).
Statistical analysis
All experiments were performed at least three independent times, unless otherwise stated. Student’s t-tests were performed on immunoblot and qRT-PCR measurements (mean ± SEM). Pearson correlation coefficients (ρ) were calculated for RNA-Seq and pSILAC analyses. Cohen’s Kappa coefficients (κ) were calculated to assess concordance between changes in Te, Rss, and pS.
Additional details are provided in Supplemental Experimental Procedures.
Supplementary Material
Highlights.
O2 stimulus reprograms protein output by altering mRNA translation efficiency
eIF4FH mediates hypoxic cap-dependent protein synthesis
eIF4F and eIF4FH triage mRNAs to generate O2-responsive translatomes
Hypoxia-inducible proteins are controlled by translation efficiency, not mRNA levels
Acknowledgments
S.L. is funded by the Sylvester Comprehensive Cancer Center and grants from the National Institutes of General Medical Sciences (1R01GM115342) and the National Cancer Institute (1R01CA200676). S.L. was funded by a grant from the Canadian Institutes of Health (CIHR). J.U. is funded by a grant from the Cancer Research Society. J.J.D.H is a recipient of a CIHR Postdoctoral Fellowship.
Abbreviations
- pSILAC (pS)
pulse-stable isotope labeling with amino acids in cell culture
- pSN
normoxic pSILAC output
- pSH
hypoxic pSILAC output
- Rss
RNA steady-state level
- RssN
normoxic Rss
- RssH
hypoxic Rss
- Te
Translation efficiency
- TeN
normoxic Te
- TeH
hypoxic Te
Footnotes
Author contributions
J.J.D.H., M.W., S.K.C., S.T., S.L.E., J.R.K., and J.U. performed the experiments. J.J.D.H., T.E.A., J.U., and S.L. conceived the experiments. J.J.D.H., M.W., S.B., M.L.G., J.U., and S.L. analyzed the data. D.K. and S.C. performed statistical analysis. J.J.D.H. and S.L. wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andreev DE, O’Connor PB, Zhdanov AV, Dmitriev RI, Shatsky IN, Papkovsky DB, Baranov PV. Oxygen and glucose deprivation induces widespread alterations in mRNA translation within 20 minutes. Genome biology. 2015;16:90. doi: 10.1186/s13059-015-0651-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin-Baillieu A, Legendre R, Kuchly C, Hatin I, Demais S, Mestdagh C, Gautheret D, Namy O. Genome-wide translational changes induced by the prion [PSI+] Cell reports. 2014;8:439–448. doi: 10.1016/j.celrep.2014.06.036. [DOI] [PubMed] [Google Scholar]
- Braunstein S, Karpisheva K, Pola C, Goldberg J, Hochman T, Yee H, Cangiarella J, Arju R, Formenti SC, Schneider RJ. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Molecular cell. 2007;28:501–512. doi: 10.1016/j.molcel.2007.10.019. [DOI] [PubMed] [Google Scholar]
- Connolly E, Braunstein S, Formenti S, Schneider RJ. Hypoxia inhibits protein synthesis through a 4E-BP1 and elongation factor 2 kinase pathway controlled by mTOR and uncoupled in breast cancer cells. Molecular and cellular biology. 2006;26:3955–3965. doi: 10.1128/MCB.26.10.3955-3965.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franovic A, Gunaratnam L, Smith K, Robert I, Patten D, Lee S. Translational up-regulation of the EGFR by tumor hypoxia provides a nonmutational explanation for its overexpression in human cancer. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13092–13097. doi: 10.1073/pnas.0702387104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nature reviews Molecular cell biology. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan Z, Ford MJ, Cusanovich DA, Mitrano A, Pritchard JK, Gilad Y. Primate transcript and protein expression levels evolve under compensatory selection pressures. Science. 2013;342:1100–1104. doi: 10.1126/science.1242379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koritzinsky M, Magagnin MG, van den Beucken T, Seigneuric R, Savelkouls K, Dostie J, Pyronnet S, Kaufman RJ, Weppler SA, Voncken JW, et al. Gene expression during acute and prolonged hypoxia is regulated by distinct mechanisms of translational control. The EMBO journal. 2006;25:1114–1125. doi: 10.1038/sj.emboj.7600998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronja I, Yuan B, Eichhorn SW, Dzeyk K, Krijgsveld J, Bartel DP, Orr-Weaver TL. Widespread changes in the posttranscriptional landscape at the Drosophila oocyte-to-embryo transition. Cell reports. 2014;7:1495–1508. doi: 10.1016/j.celrep.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landon AL, Muniandy PA, Shetty AC, Lehrmann E, Volpon L, Houng S, Zhang Y, Dai B, Peroutka R, Mazan-Mamczarz K, et al. MNKs act as a regulatory switch for eIF4E1 and eIF4E3 driven mRNA translation in DLBCL. Nature communications. 2014;5:5413. doi: 10.1038/ncomms6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JC, Hsu M, Tarn WY. Cell stress modulates the function of splicing regulatory protein RBM4 in translation control. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:2235–2240. doi: 10.1073/pnas.0611015104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Cash TP, Jones RG, Keith B, Thompson CB, Simon MC. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Molecular cell. 2006;21:521–531. doi: 10.1016/j.molcel.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Markowetz F, Unwin RD, Leek JT, Airoldi EM, MacArthur BD, Lachmann A, Rozov R, Ma’ayan A, Boyer LA, et al. Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature. 2009;462:358–362. doi: 10.1038/nature08575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- McKinney C, Zavadil J, Bianco C, Shiflett L, Brown S, Mohr I. Global reprogramming of the cellular translational landscape facilitates cytomegalovirus replication. Cell reports. 2014;6:9–17. doi: 10.1016/j.celrep.2013.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe PJ. Oxygen sensing and hypoxia signalling pathways in animals: the implications of physiology for cancer. The Journal of physiology. 2013;591:2027–2042. doi: 10.1113/jphysiol.2013.251470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schodel J, Oikonomopoulos S, Ragoussis J, Pugh CW, Ratcliffe PJ, Mole DR. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood. 2011;117:e207–217. doi: 10.1182/blood-2010-10-314427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annual review of pathology. 2014;9:47–71. doi: 10.1146/annurev-pathol-012513-104720. [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Stepaniants SB, Mao M, Weng L, Feetham MC, Doyle MJ, Yi EC, Dai H, Thorsson V, Eng J, et al. Integrated genomic and proteomic analyses of gene expression in Mammalian cells. Molecular & cellular proteomics: MCP. 2004;3:960–969. doi: 10.1074/mcp.M400055-MCP200. [DOI] [PubMed] [Google Scholar]
- Uniacke J, Holterman CE, Lachance G, Franovic A, Jacob MD, Fabian MR, Payette J, Holcik M, Pause A, Lee S. An oxygen-regulated switch in the protein synthesis machinery. Nature. 2012;486:126–129. doi: 10.1038/nature11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uniacke J, Perera JK, Lachance G, Francisco CB, Lee S. Cancer cells exploit eIF4E2-directed synthesis of hypoxia response proteins to drive tumor progression. Cancer research. 2014;74:1379–1389. doi: 10.1158/0008-5472.CAN-13-2278. [DOI] [PubMed] [Google Scholar]
- Ventoso I, Kochetov A, Montaner D, Dopazo J, Santoyo J. Extensive translatome remodeling during ER stress response in mammalian cells. PLoS One. 2012;7:e35915. doi: 10.1371/journal.pone.0035915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C. Evolution. Protein expression under pressure. Science. 2013;342:1052–1053. doi: 10.1126/science.1247833. [DOI] [PubMed] [Google Scholar]
- Vogel C, de Abreu RS, Ko D, Le SY, Shapiro BA, Burns SC, Sandhu D, Boutz DR, Marcotte EM, Penalva LO. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Molecular systems biology. 2010;6:400. doi: 10.1038/msb.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Cui Y, Jin J, Guo J, Wang G, Yin X, He QY, Zhang G. Translating mRNAs strongly correlate to proteins in a multivariate manner and their translation ratios are phenotype specific. Nucleic acids research. 2013;41:4743–4754. doi: 10.1093/nar/gkt178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi T, Papadopoulos E, Hagner PR, Wagner G. Hypoxia-inducible factor-1alpha (HIF-1alpha) promotes cap-dependent translation of selective mRNAs through up-regulating initiation factor eIF4E1 in breast cancer cells under hypoxia conditions. The Journal of biological chemistry. 2013;288:18732–18742. doi: 10.1074/jbc.M113.471466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RM, Wang SJ, Gordan JD, Ji X, Liebhaber SA, Simon MC. Hypoxia-mediated selective mRNA translation by an internal ribosome entry site-independent mechanism. The Journal of biological chemistry. 2008;283:16309–16319. doi: 10.1074/jbc.M710079200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.