Abstract
Background
After myocardial infarction (MI), the left ventricle (LV) undergoes a wound healing response that includes the robust infiltration of neutrophils and macrophages to facilitate removal of dead myocytes as well as turnover of the extracellular matrix (ECM). Matrix metalloproteinase (MMP)-9 is a key enzyme that regulates post-MI LV remodeling.
Methods and Results
Infarct regions from wild type and MMP-9 null mice (n=8/group) analyzed by glycoproteomics showed that of 541 N-glycosylated proteins quantified, 45 proteins were at least two-fold up- or down-regulated with MMP-9 deletion (all p<0.05). Cartilage intermediate layer protein (CILP) and platelet glycoprotein 4 (CD36) were identified as having the highest fold increase in MMP-9 null mice. By immunoblotting, CD36 but not CILP decreased steadily over the time course post-MI, which identified CD36 as a candidate MMP-9 substrate. MMP-9 was confirmed in vitro and in vivo to proteolytically degrade CD36. In vitro stimulation of day 7 post-MI macrophages with MMP-9 or a CD36 blocking peptide reduced phagocytic capacity. Dual immunofluorescence revealed concomitant accumulation of apoptotic neutrophils in the MMP-9 null group compared to WT. In vitro stimulation of isolated neutrophils with MMP-9 decreased neutrophil apoptosis, indicated by reduced caspase-9 expression.
Conclusions
Our data reveals a new cell signaling role for MMP-9 through CD36 degradation to regulate macrophage phagocytosis and neutrophil apoptosis.
Keywords: extracellular matrix, proteomics, metalloproteinase, myocardial infarction, platelet glycoprotein 4
Introduction
Myocardial infarction (MI) is a significant contributor to the high morbidity and mortality rates associated with cardiovascular disease.1 Following MI, the left ventricle (LV) undergoes a wound healing response comprised of robust infiltration of inflammatory cells that regulate extracellular matrix (ECM) turnover to remove necrotic debris and form an infarct scar.2 In the mouse model of MI, the inflammatory response begins to subside by day 7, leading the way for an increase in ECM production and scar formation.
Matrix metalloproteinases (MMPs) are zinc-dependent enzymes that proteolytically process ECM proteins, cytokines, chemokines, growth factors, and adhesion molecules.3, 4 MMP cleavage can activate or inactivate a protein; for example, cleavage of interleukin (IL)-8 increases its activity while cleavage of Cxcl4 decreases its activity.5 MMP-9 is particularly important in the post-MI setting, as protein concentrations increase 3-fold in the infarct region during the first week post-MI.6, 7 MMP-9 deletion attenuates LV dilation post-MI, indicating a causal role for MMP-9.6, 8 Although a number of in vitro MMP-9 substrates have been identified, including collagens (IV, V, VII, X and XIV), gelatin, fibronectin, elastin, IL-8, Cxcl4, and IL-1β, the mechanisms whereby MMP-9 modulates post-MI LV remodeling has not been completely elucidated.9, 10
Methods
Detailed methods are described in the Supplemental Methods.
Mice
C57BL/6J wild type (WT) and MMP-9 null male and female mice of 4–7 months of age were used in this study (Supplemental Table S1). The MMP-9 null mice were generated by Zena Werb’s laboratory and backcrossed onto the C57BL/6J strain by Lynn Matrisian’s laboratory.11, 12 Mice were kept in a light-controlled environment with a 12:12 hour light-dark cycle and given free access to standard mice chow and water. All animal procedures were approved by the Institutional Animal Care and Use Committee at the University of Texas Health Science Center at San Antonio and the University of Mississippi Medical Center in accordance with the “Guide for the Care and Use of Laboratory Animals”. The mice underwent permanent coronary artery ligation surgery, to produce MI, as described previously.3, 11 Three sets of mice (n≥5/day/set) were used (Supplemental Table S1): one set was used for glycoproteomics, a second set was used for immunoblotting, immunohistochemistry, and immunofluorescence, and a third set was used for in vitro stimulation of macrophages and blood neutrophils. Supplemental Figure S1 illustrates the workflow of the study. For all procedures, samples were randomized and analyzed in a blinded manner.
Tissue samples and protein extraction
The LV infarct region was collected at 0, 1, 3, 5, and 7 days post-MI as described previously.3, 13 Because the insoluble fraction is enriched for ECM, we used that fraction for the glycoproteomic analysis. Immunoblotting also used the insoluble fraction for confirmation of glycoproteomic findings.
Mass spectrometry
The samples were digested with trypsin at a ratio of 1:50 (w/w, enzyme:protein) at 37°C overnight with gentle shaking. Peptide concentration was determined by BCA assay. N-linked glycopeptides were isolated from the tryptic peptides using the solid phase extraction of glycopeptides (SPEG) method previously reported.14, 15 The efficiency of glycopeptide capture was evaluated previously by Zhou et al.16 According to two standard proteins with isotope labeling, 85% of the glycoproteins were coupled to the hydrazide resin.
Peptides were analyzed by LC-MS/MS using a Q Exactive. MS/MS spectra were searched with SEQUEST using Proteome Discoverer (version 1.4) against the mouse RefSeq database (released in Nov 2014) containing 57,788 sequences. The False Discovery Rate was set at 0.01 to eliminate low-probability protein identifications. To eliminate false positive identifications of N-glycopeptides, we determined the extent of spontaneous deamidation on Asn residues satisfying the N-X-S/T sequence by profiling the unconjugated fraction of hydrazide beads without PNGase F treatment.17, 18 The rate of spontaneous deamidation in the consensus motif was 1.5% in our sample set. Glycosylation was confirmed for several proteins, including fibronectin, γ-sarcoglycan, and periostin (Supplemental Figure S2). Peptides were quantified by label-free relative quantification based on integrated peptide peak intensities using the SIEVE software, version 2.1. Unsupervised clustering was performed using normalized peak areas of the identified glycopeptides.
Proteomic data repository
The mass spectrometry proteomics data was deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (http://www.ebi.ac.uk/pride/) with the dataset identifier PXD001393.19
Bioinformatics analysis
For Gene Ontology (GO) analysis, proteins were classified to GO categories according to their cellular component and main biological processes using DAVID Bioinformatics tools (v6.7) with default settings.20, 21 For network interaction analysis, the list of proteins was submitted to several protein-protein interaction (PPI) databases, including Database of Interacting Proteins, Molecular INTeraction database, and the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database v9.1.22, 23 STRING identified the most proteins from our list, therefore it was used in this study to identify protein-protein interactions using database, literature, and experimental search parameters.23 The confidence score was set as high (0.7). The interaction network was visualized by Cytoscape.24
Immunoblot analysis
Immunoblotting was used to validate proteomic findings. The insoluble fraction from the infarct area of WT or MMP-9 null mice (n=8/group; 4 male and 4 female) were analyzed. Samples were pooled for time course analysis and analyzed individually for the day 7 post-MI analysis. The blots were examined using the IQ-TL image analysis software on luminescent image analyzer. The signal intensity of each sample was normalized to the densitometry value for the total protein of its corresponding lane.
RT-PCR analysis
RNA extraction was performed on the LVI tissue as previously described.25–27 CILP, CD36, Ltbp2, Col6a6, Col6a1, Lama5, and Fbln1 gene expression was assessed.
In vitro cleavage assay
Mouse CD36 recombinant protein was incubated with active MMP-2, MMP-9, or MMP-12 recombinant protein at a ratio of 2:1. Two negative controls (MMP alone or recombinant protein alone) were included. Samples were run on SDS-PAGE, followed by silver staining.
Immunofluorescence or immunohistochemistry
Five random images were captured at 40x magnification for analysis. Quantification was measured by Image-Pro Plus version 6.2. Representative images are shown at 40x or 60x magnification.
Phagocytosis Assay
Macrophages were isolated from day 7 post-MI LV of male mice, as previously described and stimulated with 1 ng/uL of active MMP-9, a CD36 blocking peptide (CD36i), or a combination of both (1:1 ratio).26, 28 Phagocytosis was measured using the Vybrant Phagocytosis Kit (Life Technologies), with quantification measured on the SpectraMax M3 Plate Reader (excitation: 480 nm; emission: 520 nm). Representative images are shown at 40X magnification.
Blood neutrophil isolation and stimulation
Neutrophils were isolated from the blood of male mice as previously described.27, 29 Cells were stimulated with 0.5 ng/uL of active MMP-9 or CD36i and harvested for RNA isolation and RT-PCR examination.
Statistical analysis
All analyses were performed blinded to groups, and data are presented as mean ± standard error of the mean (SEM). For two group comparisons, the nonparametric Wilcoxon rank sum test was used. For multiple group comparisons, the nonparametric Kruskal-Wallis test with Dunns post-test was used. A 2-way ANOVA was used for CD36 time course analysis. A value of r>0.30 was considered a strong correlation for the linear regression of CD36 and end diastolic dimension. A p value of <0.05 was considered statistically significant. Statistical analysis was performed using GraphPad InStat3 and Prism 5.
Results
Glycoproteomic analysis identified 45 MMP-9 dependent extracellular protein changes post-MI
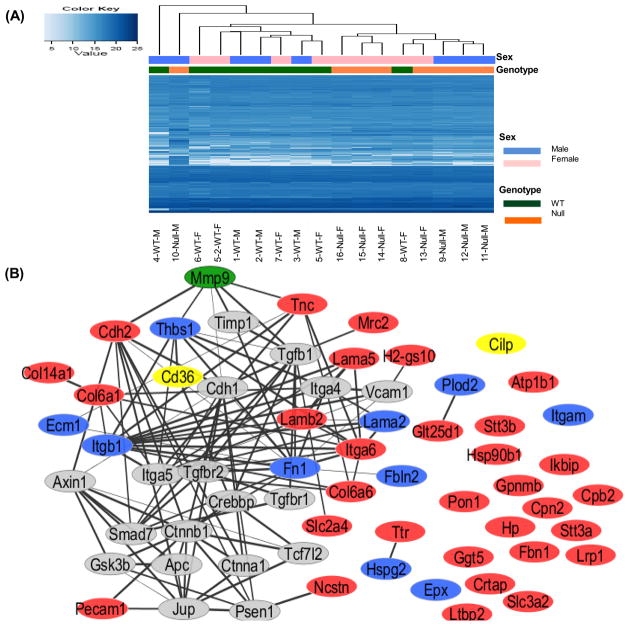
A total of 1161 unique N-linked glycopeptides were quantified by LC-MS with a 1% false discovery rate and these sites represented 541 unique glycoproteins (Supplemental Table S2). Unsupervised clustering was used to analyze the identified glycopeptides. Based on the peptide abundance, 7 of 8 WT samples and 7 of 8 MMP-9 null were clustered together, indicating good grouping based on genotype (Figure 1A). There was very poor clustering of males and females, indicating a lack of sex related differences.
Figure 1.
Glycoproteomic analysis identified MMP-9 dependent extracellular protein changes post-myocardial infarction (MI). (A) Unsupervised clustering of the identified glycopeptides was performed using normalized peak areas. WT samples and MMP-9 null clustered separately, with the exception of one WT and one null sample. Sex differences were not observed. (B) The changed proteins were analyzed by STRING (Search Tool for the Retrieval of Interacting Genes/Proteins v9.1) protein-protein interaction (PPI) software with high confidence (score >0.7) and visualized by Cytoscape. The majority of the changed proteins interacted with each other. Stronger associations are represented by thicker lines. Red nodes are the up-regulated proteins, blue nodes are the down-regulated proteins, and gray nodes present proteins associated with the changed proteins. MMP-9 is highlighted in green. CD36 and CILP, highlighted in yellow, were selected for immunoblot analysis.
A total of 122 proteins showed significant changes in MMP-9 null mice compared to WT mice (p<0.05; Supplemental Table S3). Combining a minimum two-fold change (fold change ≥2 or ≤0.5) and significance (p<0.05), there were 45 proteins (Table 1). Notably, 6 of the 45 proteins (fibronectin, collagen alpha-1, carboxypeptidase N subunit 2, beta-1 integrin, fibulin-2, and thrombospondin 1) had more than one glycopeptide identified with similar fold change, which increased the confidence for these proteins. Detailed information and quantification values can be found in Supplemental Table S2 and Supplemental Table S4. For the 20 single glycopeptides identified only once, the annotated MS/MS spectra are provided in Supplemental Figure S3. RT-PCR analysis was performed for 7 of the top 12 proteins (Supplemental Figure S4). Of note, none of the genes measured were significantly different between WT and MMP-9 null, indicating the glycoproteomic changes were not due to changes in transcription. This comparison between gene and protein levels highlights the importance of examining protein level changes in addition to gene level changes.
Table 1.
Proteins differentially expressed in MMP-9 null mice (all p < 0.05 and at least two-fold difference), ordered by normalized ratio (NR; null/WT; normalized to total ion current by SIEVE) to evaluate substrate accumulation in the absence of MMP-9.
| Accession number | Protein Description | Cellular component | Glycopeptides | NR | N-p Value |
|---|---|---|---|---|---|
| 27734196 | Cartilage intermediate layer protein (CILP) | Extracellular matrix | EQRPGQNCSNYTVR | 13.13 | 9.90E-07 |
| 568933000 | Platelet glycoprotein 4 (CD36) | Plasma membrane | QFWIFDVQNPDDVAK NSSK | 9.35 | 2.90E-02 |
| 158341636 | Latent-transforming growth factor beta-binding protein 2 | Extracellular region | DSSPQAAHVNHLSPPWGLNLTEK | 7.12 | 1.60E-02 |
| 6753138 | Sodium/potassium-transporting ATPase subunit beta-1 | Plasma membrane | LDWLGNCSGLNDDSYGYR | 6.74 | 8.60E-03 |
| 156616286 | Collagen alpha-6 (VI) | Extracellular matrix | LNANLLSSLWDTFQNK | 6.37 | 4.00E-02 |
| 6753484 | Collagen alpha-1 (VI) | Extracellular matrix | RNFTAADWGHSR | 5.25 | 3.70E-02 |
| 568920137 | Laminin subunit alpha-5 | Extracellular matrix | EQLQGINASSAAWAR | 5.09 | 4.80E-02 |
| 61651673 | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B | Endoplasmic reticulum membrane | TTLVDNNTWNNSHIALVGK | 4.81 | 2.70E-02 |
| 170784829 | Procollagen galactosyltransferase 1 | Unknown | TALWVATDHNTDNTSAILR | 4.34 | 2.30E-02 |
| 238637279 | 4F2 cell-surface antigen heavy chain | Plasma membrane | APLMPWNESSIFHIPRPVSLNMTVK | 4.33 | 2.00E-02 |
| 755498697 | Fibrillin-1 | Extracellular matrix | AWGTPCELCPSVNTSEYK | 4.29 | 3.20E-02 |
| CDSGFALDSEERNCTDIDECR | 0.45 | 4.50E-02 | |||
| 568911077 | Nicastrin | Plasma membrane | ANNSWFQSILK | 4.14 | 3.20E-02 |
| 755537649 | Platelet endothelial cell adhesion molecule | Plasma membrane | EKEDRPFHQAVVNDTQAFWHNK | 4.1 | 1.20E-02 |
| 568939471 | Serum paraoxonase/arylesterase 1 | Extracellular region | HANWTLTPLK | 4.05 | 3.80E-02 |
| 568926396 | Tenascin | Extracellular matrix | ASTEEVPSLENLTVTEAGWDGLR | 3.89 | 2.50E-02 |
| 31982236 | Integrin alpha 6 | Plasma membrane | LWNSTFLEEYSK | 3.76 | 5.00E-03 |
| 568991244 | Collagen alpha-1 (XIV) | Extracellular matrix | SFMVNWTQSPGK | 3.75 | 3.70E-02 |
| VVDKGNGSKPTSPEEVK | 2.18 | 3.60E-02 | |||
| 171846253 | Transmembrane glycoprotein NMB | Unknown | NLSDEIFLR | 3.66 | 3.20E-02 |
| 124494256 | Prolow-density lipoprotein receptor-related protein 1 | Plasma membrane | WTGHNVTVVQR | 3.62 | 2.40E-02 |
| 148747128 | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3A | Membrane | TILVDNNTWNNTHISR | 3.52 | 2.20E-02 |
| 6755863 | Endoplasmin | Plasma membrane | HNNDTQHIWESDSNEFSVIADPR | 3.36 | 3.60E-02 |
| 121674797 | Palmitoyl-protein thioesterase | Golgi apparatus | FFNDSIVDPVDSEWFGFYR | 3.29 | 4.40E-02 |
| 225543173 | Cartilage-associated protein | Extracellular matrix | DKWGLSDEHFQPRPEAVQFFNVTTLQK | 3.05 | 3.20E-02 |
| 569003077 | Cadherin-2 | Plasma membrane | RNWTINR | 3.03 | 1.00E-02 |
| 755537859 | Solute carrier family 2, facilitated glucose transporter member 4 | Plasma membrane | VIEQSYNATWLGR | 2.71 | 8.70E-03 |
| 31542891 | Gamma-glutamyltransferase 5 | Plasma membrane | LWDPSSHPGIQNISR | 2.65 | 2.50E-02 |
| 121583481 | Inhibitor of nuclear factor kappa-B kinase-interacting protein, isoform 1 | Endoplasmic reticulum membrane | FQNITDFWK | 2.52 | 4.70E-02 |
| 8850219 | Haptoglobin | Extracellular region | VVLHPNHSVVDIGLIK | 2.36 | 1.00E-02 |
| 568972029 | C-type mannose receptor 2 | Cell surface | ASNASKPGTLER | 2.18 | 3.80E-02 |
| 568962784 | Laminin subunit beta-2 | Extracellular matrix | AQAALDKANASR | 2.14 | 2.10E-02 |
| 7305599 | Transthyretin | Extracellular region | TLGISPFHEFADVVFTANDSGHR | 2.08 | 1.70E-02 |
| 147904569 | Carboxypeptidase N subunit 2 | Extracellular region | LQDLEITGSPVSNLSAHIFSNLSSLEK | 2.08 | 4.80E-02 |
| LSLDSNNLTALHPALFHNLSR | 2.02 | 2.50E-03 | |||
| 31982712 | Carboxypeptidase B2 | Extracellular region | IPFNVLMNNVEDLIEQQTFNDTVSPR | 2.07 | 2.50E-02 |
| 219521935 | MHC class I like protein GS10 | Plasma membrane | TLLSYYNQSAGGSHTIQVISGCEVGSDGR | 2.02 | 2.70E-03 |
| 45504394 | Integrin beta-1 | Plasma membrane | KDTCAQECSHFNLTK | 0.5 | 3.70E-02 |
| KENSSEICSNNGECVCGQCVCR | 0.37 | 2.90E-02 | |||
| 755522163 | Integrin alpha-M | Plasma membrane | TPVLNCSVAVCK | 0.49 | 5.20E-03 |
| 449083336 | Fibronectin | Extracellular matrix | WTPLNSSTIIGYR | 3.65 | 4.10E-02 |
| VFAVHQGRESNPLTAQQTTKLDAPTNLQFVNETDR | 0.5 | 1.60E-02 | |||
| HEEGHMLNCTCFGQGR | 0.43 | 2.50E-02 | |||
| RHEEGHMLNCTCFGQGR | 0.35 | 2.10E-02 | |||
| 269315863 | Collagen alpha-5 (VI) | Unknown | SNDSVLEPANR | 0.49 | 4.60E-02 |
| LEGVTMFAMGIEGANNTQLEDIVSYPSR | 0.44 | 4.20E-02 | |||
| SSANQSEFQQQIQK | 0.38 | 8.60E-03 | |||
| 166064058 | Slit homolog 2 protein | Unknown | DLEVLTLNNNNITR | 0.48 | 8.90E-03 |
| 218931165 | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 | Endoplasmic reticulum membrane | YNCSIESPR | 0.47 | 1.50E-02 |
| 568940646 | Fibulin-2 | Extracellular matrix | EGETCGAEDNDTCGVSLYK | 0.46 | 4.30E-03 |
| DLDECALGTHNCSEAETCHNIQGSFR | 0.37 | 1.40E-02 | |||
| 568930638 | Basement membrane-specific heparan sulfate proteoglycan core protein | Extracellular matrix | CDGDFDCEDRTDEANCSVK | 0.44 | 4.60E-02 |
| 47059073 | Thrombospondin 1 | Extracellular region | GCSSSATNVLLTLDNNVVNGSSPAIR | 0.39 | 2.30E-02 |
| VSCPIMPCSNATVPDGECCPR | 0.32 | 8.10E-03 | |||
| 145966840 | Eosinophil peroxidase | Cytoplasmic granule | ALLPFDNLHEDPCLLTNR | 0.28 | 7.50E-03 |
| 358030288 | Extracellular matrix protein 1 | Extracellular matrix | QIPGLIQNMTIR | 0 | 8.10E-06 |
glycosylation motif is indicated in bold
N-p value: Normalized p value
STRING analysis revealed that MMP-9 dependent changes work in concert as a network
To obtain an overview of the interconnections among the differentially expressed proteins, the protein list was submitted to the STRING protein-protein interaction (PPI) database and filtered for interactions of high confidence (score >0.7).23 In Figure 1B, the up-regulated proteins are shown in red, the down-regulated proteins are shown in blue, and proteins associated with the changed proteins but not identified in this study are shown in gray. Stronger associations are represented by thick black lines, whereas weaker associations are indicated by thin gray lines. MMP-9 is highlighted in green. The analysis revealed 20 of the 45 proteins (45%) had established interconnections. For example, platelet glycoprotein 4 (CD36) is connected with thrombospondin-1 (Thbs1), and the interaction between CD36 and Thbs1 stimulates pro-apoptotic signals.30, 31 In addition, several proteins co-regulated a common protein. For example, Tnc, Thbs1, Fn1, CD36 and Cdh2 directly or indirectly surround MMP-9 indicating these proteins act as a network.
CILP and CD36 had the highest fold increases in MMP-9 null mice
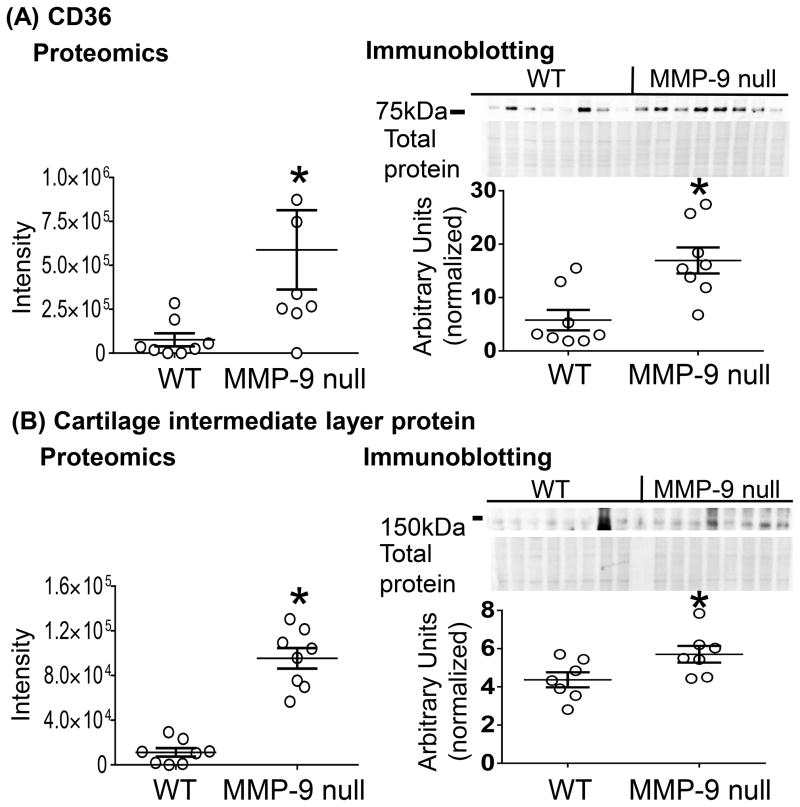
Glycoproteomics results identified cartilage intermediate layer protein (CILP) and CD36 as having the highest fold increase in MMP-9 null mice (Table 1). Since direct associations with MMP-9 had not been previously assigned, CD36 and CILP were selected for validation (highlighted in yellow in Figure 1B). Immunoblotting was performed on an independent set of mouse tissues and showed agreement with the proteomic results (Figure 2).
Figure 2.
MMP-9 deletion increased CD36 and CILP levels at day 7 post-myocardial infarction (MI). Immunoblotting analysis of the LV infarct (LVI) at day 7 post MI using individual mice. (A) CD36 and (B) CILP increased in MMP-9 null mice, which is consistent with the proteomic results (left panel). Data are represented as mean ± SEM. *p < 0.05 vs WT; n=8/group.
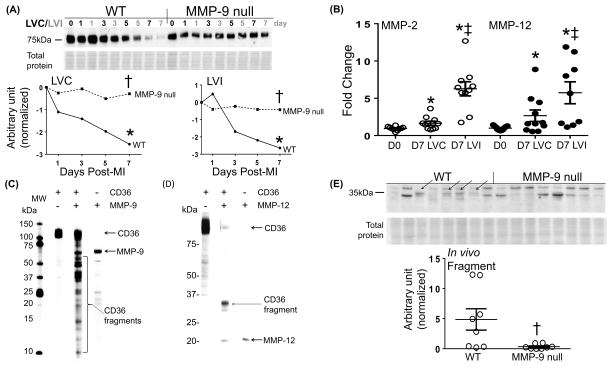
Since ECM remodeling post-MI is time and space dependent, we also investigated the protein expression of CILP and CD36 in the LV infarct (LVI) or remote control (LVC) regions over the post-MI time course.2 CILP continuously increased post-MI in both WT and MMP-9 null mice (Supplemental Figure S5). This pattern is not what we would expect if CILP was a substrate of MMP-9. For this reason, further analysis of CILP was not performed in this study. CD36 was robustly expressed at day 0, consistent with past reports of endothelial expression.32 CD36 showed a pattern consistent with being an MMP-9 substrate; namely, it decreased over time in the WT LVI, and this decrease was attenuated by MMP-9 deletion. Surprisingly, a similar pattern was found in the remote LVC (Figure 3A). The fact that CD36 decreased in the remote region indicates additional MMPs may also cleave CD36.
Figure 3.
MMP-9 regulates CD36 levels through proteolytic degradation. (A) Full length CD36 decreased over time in the LV infarct (LVI) of WT mice post-myocardial infarction (MI) and this decrease was attenuated by MMP-9 deletion. A similar pattern was found in the remote control (LVC). (B) MMP-2 and -12 increased in both the LVC and LVI at day 7 post-MI compared to day 0. In vitro cleavage assay showed recombinant CD36 was cleaved by active (C) MMP-9 and (D) MMP-12. Silver staining showed CD36 fragments as indicated by bracket. (E) Immunoblotting of the soluble fraction of homogenized LV tissue of WT and MMP-9 null mice using CD36 antibody showed a 35 kDa fragment detected largely in WT mice but not MMP-9 null, confirming CD36 is an in vivo substrate of MMP-9. Data are represented as mean ± SEM. *p < 0.05 vs respective D0; †p< 0.05 vs WT; ‡p< 0.05 vs D7 LVC; n=8/group.
MMP-9 regulates CD36 levels through proteolytic degradation
To assess in vitro proteolytic degradation, CD36 recombinant protein was incubated with active MMP-9. Because we found CD36 decreased over time in the remote region, we also incubated CD36 with active MMP-2 or MMP-12- two MMPs that increase in the remote region post-MI (Figure 3B). Fragments were visible with MMP-9 and MMP-12 incubation but not with MMP-2 incubation (Figure 3C and 3D). This demonstrates CD36 is cleaved by MMP-9 and MMP-12 in vitro, which may explain the decrease in CD36 levels observed in both the LVC and LVI samples. Interestingly, there were multiple bands generated with MMP-9 at molecular weights ranging from 50 to 10 kDa, illustrating MMP-9 not only generated a CD36 fragment but can further degrade it whereas MMP-12 only generated one major fragment. To confirm whether CD36 was cleaved by MMP-9 in vivo, immunoblotting was performed on the soluble fraction of homogenized LV tissue of WT and MMP-9 null mice. The results showed a 35 kDa fragment detected only in WT mice and not MMP-9 null, confirming CD36 is an in vivo substrate of MMP-9 (Figure 3E).
MMP-9 decreases macrophage phagocytosis
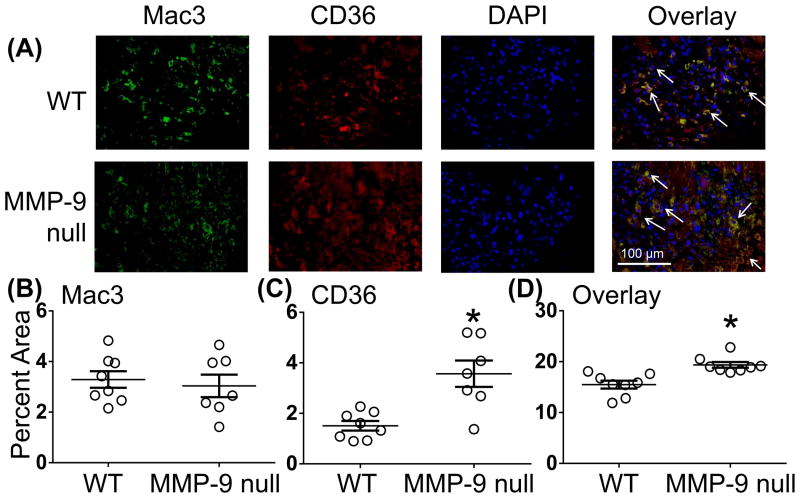
CD36 is known to be expressed by multiple cell sources, including macrophages and endothelial cells.32, 33 At day 7 post-MI, dual immunofluorescence revealed no significant difference in the number of macrophages present in the LVI in WT and MMP-9 null mice as previously shown.34 CD36 was significantly higher in MMP-9 null mice, consistent with glycoproteomic analysis. In addition, CD36 was co-localized with the macrophage marker Mac-3 (Figure 4). MMP-9 null mice showed higher levels of CD36+ macrophages compared to the WT mice at day 7 post-MI consistent with CD36 being reduced in WT.
Figure 4.
MMP-9 regulates macrophage expression of CD36. (A) Representative dual immunofluorescence images. (B) WT and MMP-9 null animals had similar numbers of Mac3 positive cells in the infarct at day 7 post-myocardial infarction (MI). (C) CD36 expression was increased in MMP-9 null animals similar to what was observed by proteomics and immunoblotting analysis. (D) Overlay analysis indicated CD36 was largely expressed by macrophages. Staining showed higher levels of CD36+ macrophages in the MMP-9 null mice at day 7 post-MI. Data are represented as mean ± SEM. *p< 0.05 vs WT; n=8/group.
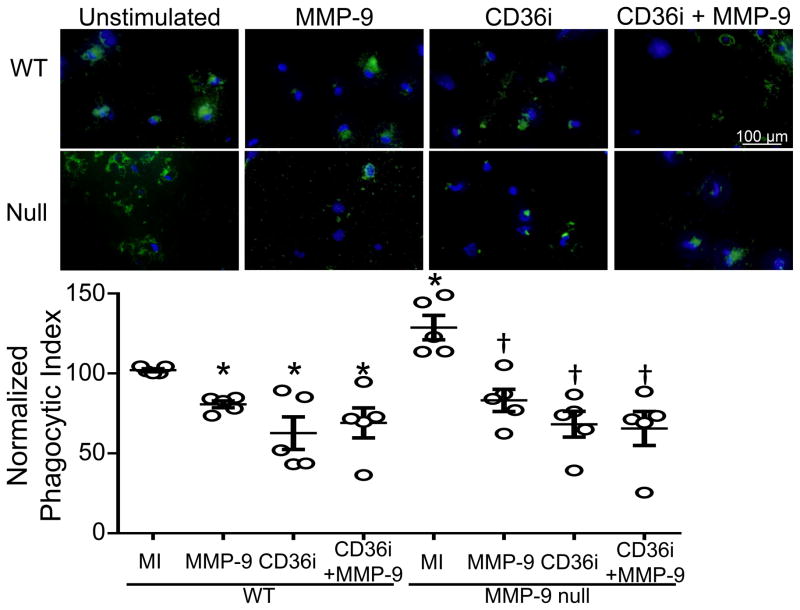
CD36 expression is required for macrophage mediated phagocytosis, thus we proposed MMP-9 cleavage of CD36 would decrease phagocytosis in WT mice.33 Macrophages isolated from the infarct of MMP-9 null mice exhibited a higher phagocytic index compared to WT macrophages (Figure 5). Stimulation with MMP-9 at concentrations similar to what is observed physiologically reduced phagocytosis by 30% in WT and 50% in MMP-9 null indicating MMP-9 regulates phagocytosis. MMP-9 null mice had higher levels of CD36+ macrophages which explains the greater reduction in phagocytic index. To test if this decrease was due to CD36 degradation, macrophages were incubated with CD36 blocking peptide (CD36i) or a mixture of MMP-9 and CD36i. Phagocytic potential decreased to a similar extent after incubation with CD36i or the mixture indicating MMP-9 regulates phagocytosis through CD36.
Figure 5.
MMP-9 regulates phagocytosis through CD36. As shown by the in vivo stimulated cells isolated from day 7 post-myocardial infarction left ventricle (MI), MMP-9 deletion increases macrophage phagocytic index compared to WT macrophages. MMP-9 incubation decreased phagocytosis in both WT and MMP-9 null. MMP-9, CD36 blocking peptide (CD36i) and the combination of MMP-9 and CD36i decreased phagocytosis to similar degrees, indicating MMP-9 regulates macrophage phagocytic potential through CD36. Quantification of phagocytosis was performed using the SpectraMax M3 Plate Reader. Data are represented as mean ± SEM. Representative images are at 40X magnification. Fluorescent Bioparticles are shown in green. DAPI staining is shown in blue. *p< 0.05 vs WT; †p< 0.05 vs vs null MI; n=5/group.
MMP-9 decreased apoptotic neutrophils in infarct region at day 7 post-MI
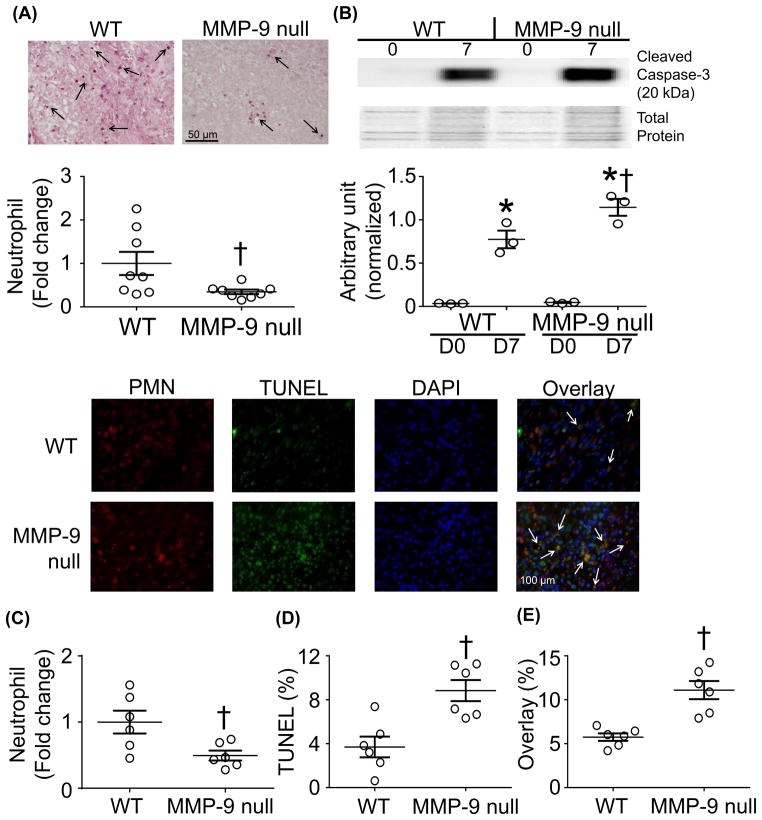
CD36 is required for the induction of macrophage-mediated phagocytosis of apoptotic neutrophils.35 If not removed in a timely manner, apoptotic neutrophils can continue to release granule components prolonging the inflammatory response.36 We showed MMP-9 can degrade CD36 post-MI leading to a decrease in phagocytosis but whether this leads to sustained neutrophil inflammation is unknown. To address this, the LVs of WT and MMP-9 null mice at day 7 post-MI were stained for neutrophils. MMP-9 null had lower neutrophils numbers in the infarct region compared to WT at day 7 post-MI (Figure 6A, C). In addition to facilitating the removal of apoptotic cells, CD36 also initiates the rapid activation of caspase-3 resulting in neutrophil apoptosis.33, 35 MMP-9 null mice had higher levels of caspase-3 and TUNEL staining compared to WT mice at day 7 post-MI (Figure 6B, D). Caspase-3–mediated spontaneous death in neutrophils is critical for modulating inflammatory responses.37, 38 To confirm the higher levels of cleaved caspase-3 were due to neutrophil apoptosis, dual immunofluorescence of PMNs and TUNEL was performed (Figure 6E). Overlay analysis showed higher numbers of apoptotic neutrophils with MMP-9 deletion. During active inflammation, neutrophil apoptosis is delayed leading to increased tissue damage.39
Figure 6.
MMP-9 decreases removal of apoptotic neutrophils post-myocardial infarction (MI). (A, C) MMP-9 null mice had lower numbers of neutrophils in the infarct at day 7 post-MI compared to WT. This decrease was coupled with an increase in the apoptotic markers (B) cleaved caspase-3 (pooled samples) and (D) terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) at day 7 post-MI. (E) Overlay analysis indicated lower numbers of apoptotic neutrophils in WT animals indicating prolonged neutrophil mediated inflammation at day 7 post-MI. Data are represented as mean ± SEM. *p< 0.05 vs respective D0; †p< 0.05 vs WT; n=8/group.
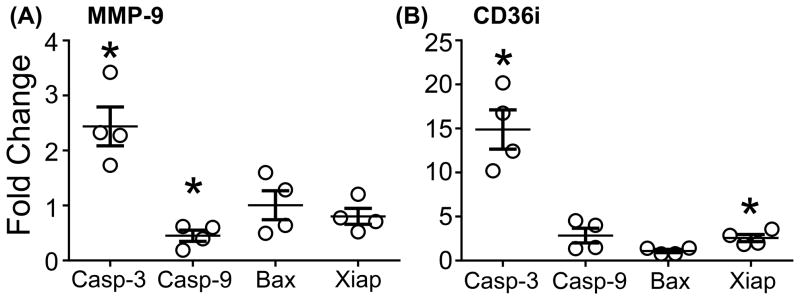
We hypothesized that a lack of CD36 degradation was the cause of increased apoptotic neutrophils observed in the MMP-9 null mice and tested this using an in vitro assay. Because neutrophils are known to undergo spontaneous apoptosis in the absence of extracellular stimuli,40–43 we used this natural process to induce apoptosis to limit exogenous influences. Incubating neutrophils with MMP-9 decreased the gene expression of the apoptotic marker caspase-9 by 2-fold and increased caspase-3, with no effect on Bax or Xiap (Figure 7). The increase in caspase-3 may be a compensatory response to the decrease in caspase-9. Caspase-9 is a critical upstream activator of the caspase pathway, and these results indicate that MMP-9 regulates apoptosis in vitro. CD36i increased caspase-3 and Xiap expression but did not affect caspase-9 or Bax. Xiap is an inhibitor of the caspase pathway. Our data indicates that while MMP-9 directly regulated neutrophil apoptosis, this effect was independent of CD36 degradation.
Figure 7.
MMP-9 inhibits apoptosis in blood neutrophils. (A) Compared to unstimulated cells MMP-9 decreased neutrophil expression of caspase-9 and increased caspase-3 expression but had no effect on Bax or Xiap. (B) CD36 blocking peptide (CD36i) increased expression of capsase-3 in addition to the apoptotic inhibitor, Xiap, indicating that while MMP-9 does regulate apoptosis, it is not through CD36 degradation. Stimulated data was normalized to unstimulated data to illustrate fold change. Data are represented as mean ± SEM. *p< 0.05 vs unstimulated; n=4/group.
MMP-9 degradation of CD36 increases LV dilation post-MI
Excessive inflammation post-MI has been shown to impair infarct healing contributing to LV dysfunction.7, 8, 26 At day 7 post-MI, WT mice showed increased LV dilation (Table 2), indicated by elevated diastolic dimensions and volumes compared to baseline day 0 controls. This increase was attenuated in the absence of MMP-9, consistent with previous findings.11 Regression analysis showed decreased CD36 levels were linked to increases in end diastolic dimensions (R2=0.48; p < 0.05). MMP-9 deletion removed the link between CD36 and LV dilation.
Table 2.
While dilation of the left ventricle occurred in both WT and MMP-9 null mice at Day 7 post-myocardial infarction (MI), MMP-9 deletion attenuated this effect.
| WT | MMP-9 null | |||
|---|---|---|---|---|
|
| ||||
| Day 0 (n=8) | Day 7 (n=16) | Day 0 (n=8) | Day 7 (n=16) | |
| Infarct size (%) | NA | 53±3 | NA | 52±2 |
| Heart Rate (bpm) | 473±14 | 473±11 | 440±7 | 450±11 |
| LV Infarct Wall Thickness (mm) | 1.20±0.07 | 0.65±0.05* | 1.13±0.03 | 0.68±0.05* |
| End Diastolic Dimension (mm) | 3.47±0.10 | 5.79±0.18* | 3.52±0.05 | 5.14±0.24*† |
| End Systolic Dimension (mm) | 2.16±0.08 | 5.42±0.19* | 2.14±0.06 | 4.78±0.26* |
| Fractional Shortening % | 38±1 | 6±1* | 39±2 | 7±1* |
| End Diastolic Volume (uL) | 58±2 | 160±10* | 54±2 | 124±10*† |
| End Systolic Volume (uL) | 17±1 | 140±10* | 15±1 | 107±10* |
| Ejection Fraction (%) | 70±1 | 12±1* | 72±1 | 15±2* |
Day 0 are naive, no MI controls. Values are means ± SEM.
vs p<0.05 vs respective baseline
vs p<0.05 vs WT Day 7
Discussion
The goal of this study was to identify MMP-9 dependent signaling mechanisms in the post-MI LV. Our results showed that of 541 N-glycosylated proteins quantified, 45 proteins were at least two-fold up- or down-regulated with MMP-9 deletion; and CILP and CD36 were identified as having the highest fold increases in MMP-9 null mice. MMP-9 regulated CD36 levels through proteolytic degradation, decreasing macrophage phagocytosis and prolonging neutrophil inflammation. In addition, regression analysis showed decreased CD36 levels were linked to increases in end diastolic dimensions. Combined, our results reveal a role for MMP-9 in regulating CD36-mediated aspects of post-MI LV remodeling.
The proteins identified in this study are potential downstream mediators of MMP-9. Of the 45 differentially expressed proteins, at least five are known MMP-9 substrates, including fibronectin, collagen XIV, tenascin, laminin α5, and laminin β2.10 Future studies identifying the role of the other key proteins identified, such as CILP and latent TGF-beta binding protein (LTBP)-2, will be necessary to fully delineate MMP-9 post-MI mechanisms. This study validated CD36 as a novel in vivo MMP-9 substrate. CD36 degradation delayed inflammation resolution by decreasing macrophage mediated phagocytosis. Based on our in vitro results, MMP-9 also regulates neutrophil apoptosis through a pathway that is not dependent on CD36 degradation.
In addition to MMP-9, CD36 was also cleaved by MMP-12. Inhibition of MMP-12 post-MI is known to suppress neutrophil apoptosis leading to prolonged inflammation and worsened LV function.27 Interestingly, the in vitro cleavage assay showed multiple bands generated with MMP-9 whereas MMP-12 only generated one major fragment. This suggests that while MMP-12 cleaves CD36, MMP-9 degrades it further preventing CD36 biological activity. While MMP-9 increases only in the infarct area, MMP-12 increases in both the remote and infarct area. This is one explanation for why we see such a dramatic decrease in CD36 in the LVC. Lower levels of CD36 in the LVC could reduce fatty acid supply to the surviving myocytes, leading to a reduction in energy supply and contributing to infarct expansion.44
MMP-9 increases dramatically during the first week post-MI, and this lead to CD36 degradation. We focused our evaluation on day 7 post-MI, because this is a time when the early peak in MMP-9 concentrations is waning, as is the inflammatory process.45 This time point, therefore, allows us to monitor the end points of the inflammatory process and the beginning of the scar formation process. Our study identified macrophages as a source of CD36 in the infarct. CD36 is a multifunctional plasma membrane protein that plays a role in fatty acid transport, cell apoptosis, and inflammation.46 CD36 binds Thbs1 initiating apoptosis and the release of additional Thbs1 as a signal to recruit macrophages. The CD36/Thbs1 complex on the cell surface of the apoptotic cell acts as a ligand, which interacts with the CD36/αvβ3 complex on macrophages initiating phagocytosis of the apoptotic cell. Macrophage CD36 recognition and internalization of apoptotic cells inhibits the release of proinflammatory cytokines like tumor necrosis factor-α (TNF-α), interleukin (IL)-12, IL-1β, and IL-8 and initiates the anti-inflammatory response which is mediated by the release of IL-10 and transforming growth factor (TGF)β.33, 47 In our study, we showed CD36 levels decreased post-MI in WT only. This decrease was due to MMP-9 mediated degradation. Degradation of CD36 led to decreased macrophage phagocytosis at day 7 post-MI implicating MMP-9 as a key player in the persistence of the inflammatory response by mediating neutrophil removal.
Future directions
MMP-9 deletion has been proven to be beneficial post-MI in mice. However, the use of a non-specific inhibitor in clinical trials has shown to be inconclusive.48 Our study revealed that MMP-9 plays a vital role post-MI by delaying resolution of the inflammatory response after MI. MMPs are involved in multiple biological processes, including cell-surface-receptor cleavage and release, cytokine and chemokine activation and inactivation, and ECM turnover. Understanding the entire composite of MMP-9 roles will be important for translating these findings to the clinic. Identifying the effects of MMP-9 cleavage products may provide answers and serve as novel and more selective treatment strategies for post-MI patients. Future studies utilizing full-length CD36 as well as MMP-9 derived cleavage products to rescue the phenotype are warranted. In addition, future studies utilizing CD36 null mice to investigate CD36 degradation and cardiac remodeling would further evaluate the mechanism behind this connection.
In this study, we did not evaluate the effect of CD36 degradation on metabolism or energy components. CD36 also delivers fatty acids to the myocardium for physiological energy requirements. In response to MI, the LV utilizes more energy from glucose than fatty acids leaning towards a failing phenotype.46 MMP-9-mediated degradation of CD36 in the infarct area of the WT may reduce the fatty acid supply to the post-MI LV in WT, in contrast MMP-9 null LV showed higher levels of CD36 meeting the energy demands of the post-MI LV.46 Future studies with a major focus on post-MI metabolic measurements and different substrate inputs will advance the field.
Conclusion
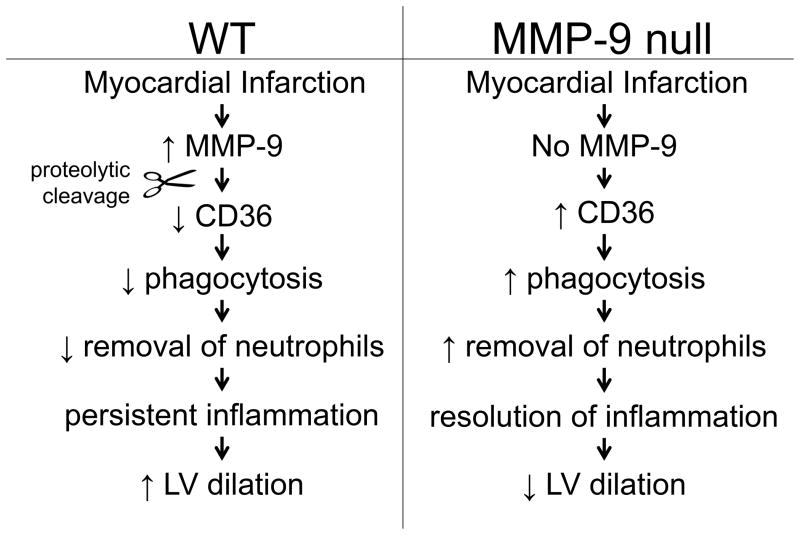
In summary, this study identified CD36 as an in vivo MMP-9 substrate. In the clinical setting, heart failure is marked by the persistent presence and activation of neutrophils due to their reduced clearance.49 Our study demonstrated that degradation of CD36 decreased macrophage phagocytosis and prolonged neutrophil inflammation leading to an enlarged LV post-MI (Figure 8). These results are the first to implicate MMP-generated decreased levels of CD36 as a possible marker for sustained inflammation and heart failure.
Figure 8.
Diagram depicting the roles of MMP-9 mediated CD36 degradation in LV remodeling post-myocardial infarction (MI). Following MI, MMP-9 degradation of CD36 resulted in decreased phagocytic index and prolonged neutrophil inflammation, leading to LV dysfunction.
Supplementary Material
Clinical Perspective.
In the United States, >40% of all deaths are attributed to cardiovascular disease (CVD), and more than half of these deaths are due to myocardial infarction (MI). Following MI, the cardiac extracellular matrix (ECM) is proteolytically cleaved by matrix metalloproteinases (MMPs). MMPs are involved in multiple biological processes, including cell-surface-receptor cleavage and release, cytokine and chemokine activation and inactivation, and ECM turnover. MMP-9 deletion has been shown to be beneficial post-MI in mice, however, the use of non-specific MMP inhibitors have been inconclusive in clinical trials. The current article used a proteomic strategy to identify novel MMP-9 substrates. Platelet glycoprotein 4, CD36, was validated as a novel in vivo MMP-9 substrate. Degradation of CD36 by MMP-9 decreased macrophage phagocytosis and prolonged neutrophil inflammation. This study revealed that MMP-9 deletion promotes resolution of the inflammatory response. Identifying the effects of MMP-9 cleavage products may provide novel therapeutic insight for post-MI patients.
Acknowledgments
We would like to thank the University of Mississippi Medical Center Center of Biostatics and Bioinformatics for their help with analysis of our datasets.
Funding Sources: We acknowledge support from American Heart Association for 13POST14350034 to KYD-P and 14POST18770012 to RPI, from NIH R00AT006704 to GVH, from Johns Hopkins Proteomics Center (N01-HV-00240) and Programs of Excellence in Glycosciences (PEG, P01HL107153) to HZ, from NIH/NHLBI HHSN 268201000036C (N01-HV-00244) for the San Antonio Cardiovascular Proteomics Center and R01 HL075360, HL051971, and GM104357 and from the Biomedical Laboratory Research and Development Service of the Veterans Affairs Office of Research and Development Award 5I01BX000505 to MLL.
Footnotes
Conflict of Interest Disclosures: None
References
- 1.Roger VL, Go AS, Lloyd-Jones DM, Benjamin EJ, Berry JD, Borden WB, et al. Heart disease and stroke statistics--2012 update: A report from the american heart association. Circulation. 2012;125:e2–e220. doi: 10.1161/CIR.0b013e31823ac046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jourdan-Lesaux C, Zhang J, Lindsey ML. Extracellular matrix roles during cardiac repair. Life Sci. 2010;87:391–400. doi: 10.1016/j.lfs.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zamilpa R, Zhang J, Chiao YA, de Castro Bras LE, Halade GV, Ma Y, et al. Cardiac wound healing post-myocardial infarction: A novel method to target extracellular matrix remodeling in the left ventricle. Methods Mol Biol. 2013;1037:313–324. doi: 10.1007/978-1-62703-505-7_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindsey ML. Mmp induction and inhibition in myocardial infarction. Heart Fail Rev. 2004;9:7–19. doi: 10.1023/B:HREV.0000011390.44039.b7. [DOI] [PubMed] [Google Scholar]
- 5.Van Lint P, Libert C. Chemokine and cytokine processing by matrix metalloproteinases and its effect on leukocyte migration and inflammation. J Leukoc Biol. 2007;82:1375–1381. doi: 10.1189/jlb.0607338. [DOI] [PubMed] [Google Scholar]
- 6.Ducharme A, Frantz S, Aikawa M, Rabkin E, Lindsey M, Rohde LE, et al. Targeted deletion of matrix metalloproteinase-9 attenuates left ventricular enlargement and collagen accumulation after experimental myocardial infarction. J Clin Invest. 2000;106:55–62. doi: 10.1172/JCI8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tao ZY, Cavasin MA, Yang F, Liu YH, Yang XP. Temporal changes in matrix metalloproteinase expression and inflammatory response associated with cardiac rupture after myocardial infarction in mice. Life Sci. 2004;74:1561–1572. doi: 10.1016/j.lfs.2003.09.042. [DOI] [PubMed] [Google Scholar]
- 8.Heymans S, Luttun A, Nuyens D, Theilmeier G, Creemers E, Moons L, et al. Inhibition of plasminogen activators or matrix metalloproteinases prevents cardiac rupture but impairs therapeutic angiogenesis and causes cardiac failure. Nat Med. 1999;5:1135–1142. doi: 10.1038/13459. [DOI] [PubMed] [Google Scholar]
- 9.Zamilpa R, Lopez EF, Chiao YA, Dai Q, Escobar GP, Hakala K, et al. Proteomic analysis identifies in vivo candidate matrix metalloproteinase-9 substrates in the left ventricle post-myocardial infarction. Proteomics. 2010;10:2214–2223. doi: 10.1002/pmic.200900587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zitka O, Kukacka J, Krizkova S, Huska D, Adam V, Masarik M, et al. Matrix metalloproteinases. Curr Med Chem. 2010;17:3751–3768. doi: 10.2174/092986710793213724. [DOI] [PubMed] [Google Scholar]
- 11.Lindsey ML, Escobar GP, Dobrucki LW, Goshorn DK, Bouges S, Mingoia JT, et al. Matrix metalloproteinase-9 gene deletion facilitates angiogenesis after myocardial infarction. Am J Physiol Heart Circ Physiol. 2006;290:H232–239. doi: 10.1152/ajpheart.00457.2005. [DOI] [PubMed] [Google Scholar]
- 12.Vu TH, Shipley JM, Bergers G, Berger JE, Helms JA, Hanahan D, et al. Mmp-9/gelatinase b is a key regulator of growth plate angiogenesis and apoptosis of hypertrophic chondrocytes. Cell. 1998;93:411–422. doi: 10.1016/s0092-8674(00)81169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Castro Bras LE, Ramirez TA, DeLeon-Pennell KY, Chiao YA, Ma Y, Dai Q, et al. Texas 3-step decellularization protocol: Looking at the cardiac extracellular matrix. J Proteomics. 2013;86:43–52. doi: 10.1016/j.jprot.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H, Li XJ, Martin DB, Aebersold R. Identification and quantification of n-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 15.Tian Y, Zhou Y, Elliott S, Aebersold R, Zhang H. Solid-phase extraction of n-linked glycopeptides. Nat Protoc. 2007;2:334–339. doi: 10.1038/nprot.2007.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Y, Aebersold R, Zhang H. Isolation of n-linked glycopeptides from plasma. Anal Chem. 2007;79:5826–5837. doi: 10.1021/ac0623181. [DOI] [PubMed] [Google Scholar]
- 17.Palmisano G, Melo-Braga MN, Engholm-Keller K, Parker BL, Larsen MR. Chemical deamidation: A common pitfall in large-scale n-linked glycoproteomic mass spectrometry-based analyses. J Proteome Res. 2012;11:1949–1957. doi: 10.1021/pr2011268. [DOI] [PubMed] [Google Scholar]
- 18.Wright HT. Nonenzymatic deamidation of asparaginyl and glutaminyl residues in proteins. Crit Rev Biochem Mol Biol. 1991;26:1–52. doi: 10.3109/10409239109081719. [DOI] [PubMed] [Google Scholar]
- 19.Vizcaino JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Rios D, et al. Proteomexchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 21.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, et al. String 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deleon-Pennell KY, Bras LE, Lindsey ML. Circulating lipopolysaccharide resets cardiac homeostasis in mice through a matrix metalloproteinase-9 dependent mechanism. Physiol Rep. 2013;1:e00079. doi: 10.1002/phy2.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeLeon-Pennell KY, de Castro Bras LE, Iyer RP, Bratton DR, Jin YF, Ripplinger CM, et al. P. Gingivalis lipopolysaccharide intensifies inflammation post-myocardial infarction through matrix metalloproteinase-9. J Mol Cell Cardiol. 2014;76C:218–226. doi: 10.1016/j.yjmcc.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iyer RP, Patterson NL, Zouein FA, Ma Y, Dive V, de Castro Bras LE, et al. Early matrix metalloproteinase-12 inhibition worsens post-myocardial infarction cardiac dysfunction by delaying inflammation resolution. Int J Cardiol. 2015;185:198–208. doi: 10.1016/j.ijcard.2015.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zamilpa R, Kanakia R, Cigarroa Jt, Dai Q, Escobar GP, Martinez H, et al. Cc chemokine receptor 5 deletion impairs macrophage activation and induces adverse remodeling following myocardial infarction. Am J Physiol Heart Circ Physiol. 2011;300:H1418–H1426. doi: 10.1152/ajpheart.01002.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oh H, Siano B, Diamond S. Neutrophil isolation protocol. J Vis Exp. 2008;17:745. doi: 10.3791/745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin L, Kim E, Ratan R, Lee FS, Cho S. Genetic variant of bdnf (val66met) polymorphism attenuates stroke-induced angiogenic responses by enhancing anti-angiogenic mediator cd36 expression. J Neurosci. 2011;31:775–783. doi: 10.1523/JNEUROSCI.4547-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seftor RE, Seftor EA, Stetler-Stevenson WG, Hendrix MJ. The 72 kda type iv collagenase is modulated via differential expression of alpha v beta 3 and alpha 5 beta 1 integrins during human melanoma cell invasion. Cancer Res. 1993;53:3411–3415. [PubMed] [Google Scholar]
- 32.Swerlick RA, Lee KH, Wick TM, Lawley TJ. Human dermal microvascular endothelial but not human umbilical vein endothelial cells express cd36 in vivo and in vitro. J Immunol. 1992;148:78–83. [PubMed] [Google Scholar]
- 33.Fadok VA, Warner ML, Bratton DL, Henson PM. Cd36 is required for phagocytosis of apoptotic cells by human macrophages that use either a phosphatidylserine receptor or the vitronectin receptor (alpha v beta 3) J Immunol. 1998;161:6250–6257. [PubMed] [Google Scholar]
- 34.Lindsey ML, Escobar GP, Dobrucki LW, Goshorn DK, Bouges S, Mingoia JT, et al. Matrix metalloproteinase-9 gene deletion facilitates angiogenesis after myocardial infarction. Am J Physiol Heart Circ Physiol. 2006;290:H232–H239. doi: 10.1152/ajpheart.00457.2005. [DOI] [PubMed] [Google Scholar]
- 35.Ma Y, Yabluchanskiy A, Lindsey ML. Neutrophil roles in left ventricular remodeling following myocardial infarction. Fibrogenesis Tissue Repair. 2013;6:11. doi: 10.1186/1755-1536-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bratton DL, Henson PM. Neutrophil clearance: When the party is over, clean-up begins. Trends Immunol. 2011;32:350–357. doi: 10.1016/j.it.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loison F, Zhu H, Karatepe K, Kasorn A, Liu P, Ye K, et al. Proteinase 3-dependent caspase-3 cleavage modulates neutrophil death and inflammation. J Clin Invest. 2014;124:4445–4458. doi: 10.1172/JCI76246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Savill J, Hogg N, Ren Y, Haslett C. Thrombospondin cooperates with cd36 and the vitronectin receptor in macrophage recognition of neutrophils undergoing apoptosis. J Clin Invest. 1992;90:1513–1522. doi: 10.1172/JCI116019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheah FC, Hampton MB, Darlow BA, Winterbourn CC, Vissers MC. Detection of apoptosis by caspase-3 activation in tracheal aspirate neutrophils from premature infants: Relationship with nf-kappab activation. J Leukoc Biol. 2005;77:432–437. doi: 10.1189/jlb.0904520. [DOI] [PubMed] [Google Scholar]
- 40.Kirschnek S, Vier J, Gautam S, Frankenberg T, Rangelova S, Eitz-Ferrer P, et al. Molecular analysis of neutrophil spontaneous apoptosis reveals a strong role for the pro-apoptotic bh3-only protein noxa. Cell Death Differ. 2011;18:1805–1814. doi: 10.1038/cdd.2011.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frumento G, Ottonello L, Bertolotto M, Franchello S, Melioli G, Dallegri F. Spontaneous apoptosis in neutrophils is associated with downregulation of hla class i and is prevented by ligation of class i. J Leukoc Biol. 2000;68:873–880. [PubMed] [Google Scholar]
- 42.Savill JS, Wyllie AH, Henson JE, Walport MJ, Henson PM, Haslett C. Macrophage phagocytosis of aging neutrophils in inflammation. Programmed cell death in the neutrophil leads to its recognition by macrophages. J Clin Invest. 1989;83:865–875. doi: 10.1172/JCI113970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akgul C, Moulding DA, Edwards SW. Molecular control of neutrophil apoptosis. FEBS Lett. 2001;487:318–322. doi: 10.1016/s0014-5793(00)02324-3. [DOI] [PubMed] [Google Scholar]
- 44.Irie H, Krukenkamp IB, Brinkmann JF, Gaudette GR, Saltman AE, Jou W, et al. Myocardial recovery from ischemia is impaired in cd36-null mice and restored by myocyte cd36 expression or medium-chain fatty acids. Proc Natl Acad Sci U S A. 2003;100:6819–6824. doi: 10.1073/pnas.1132094100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patterson NL, Iyer RP, de Castro Bras LE, Li Y, Andrews TG, Aune GJ, et al. Using proteomics to uncover extracellular matrix interactions during cardiac remodeling. Proteomics Clin Appl. 2013;7:516–527. doi: 10.1002/prca.201200100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bonen A, Tandon NN, Glatz JF, Luiken JJ, Heigenhauser GJ. The fatty acid transporter fat/cd36 is upregulated in subcutaneous and visceral adipose tissues in human obesity and type 2 diabetes. Int J Obes (Lond) 2006;30:877–883. doi: 10.1038/sj.ijo.0803212. [DOI] [PubMed] [Google Scholar]
- 47.Fadok VA, Bratton DL, Konowal A, Freed PW, Westcott JY, Henson PM. Macrophages that have ingested apoptotic cells in vitro inhibit proinflammatory cytokine production through autocrine/paracrine mechanisms involving tgf-beta, pge2, and paf. J Clin Invest. 1998;101:890–898. doi: 10.1172/JCI1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hudson MP, Armstrong PW, Ruzyllo W, Brum J, Cusmano L, Krzeski P, et al. Effects of selective matrix metalloproteinase inhibitor (pg-116800) to prevent ventricular remodeling after myocardial infarction: Results of the premier (prevention of myocardial infarction early remodeling) trial. J Am Coll Cardiol. 2006;48:15–20. doi: 10.1016/j.jacc.2006.02.055. [DOI] [PubMed] [Google Scholar]
- 49.Benites-Zapata VA, Hernandez AV, Nagarajan V, Cauthen CA, Starling RC, Tang WH. Usefulness of neutrophil-to-lymphocyte ratio in risk stratification of patients with advanced heart failure. Am J Cardiol. 2015;115:57–61. doi: 10.1016/j.amjcard.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.