Abstract
Electron transfer dissociation (ETD) is a valuable tool for protein sequence analysis, especially for the fragmentation of intact proteins. However, low product ion signal-to-noise often requires some degree of signal averaging to achieve high quality MS/MS spectra of intact proteins. Here we describe a new implementation of ETD on the newest generation of quadrupole-Orbitrap-linear ion trap Tribrid, the Orbitrap Fusion Lumos, for improved product ion signal-to-noise via ETD reactions on larger precursor populations. In this new high precursor capacity ETD implementation, precursor cations are accumulated in the center section of the high pressure cell in the dual pressure linear ion trap prior to charge-sign independent trapping, rather than precursor ion sequestration in only the back section as is done for standard ETD. This new scheme increases the charge capacity of the precursor accumulation event, enabling storage of approximately three fold more precursor charges. High capacity ETD boosts the number of matching fragments identified in a single MS/MS event, reducing the need for spectral averaging. These improvements in intra-scan dynamic range via reaction of larger precursor populations, which have been previously demonstrated through custom modified hardware, are now available on a commercial platform, offering considerable benefits for intact protein analysis and top down proteomics. In this work, we characterize the advantages of high precursor capacity ETD through studies with myoglobin and carbonic anhydrase.
Keywords: intact proteins, electron transfer dissociation, ion-ion reactions, instrumentation
INTRODUCTION
Interrogation of intact proteins via mass spectrometry (MS) has the potential to capture nearly all of the relevant information encoded in each protein, including primary sequence information, combinatorial patterns of post-translational modifications (PTMs), and protein gas-phase structure [1–5]. As molecular weight alone is largely insufficient for full protein characterization [6–9], tandem MS (MS/MS) is the key component of these top down sequencing methods, revealing both the primary sequence and protein modification state [10–13]. The emergence of new ion dissociation methods continues to drive top down proteomics [14–16] by offering valuable alternatives to traditional slow-heating methods (e.g., collision-activated dissociation, CAD). Electron transfer dissociation (ETD) leverages electron-driven radical rearrangements to promote cleavage of N-Cα bonds between amino acid residues, preserving labile post-translational modifications (PTMs) and providing extensive sequence-informative fragmentation of peptides and proteins [17–19]. Ideally suited for large, highly charged protein molecules, ETD has afforded important gains in top down proteomics, extending protein sequence coverage and enabling characterization of important PTMs and sequence variants [20–24].
Despite advancements in fragmentation methods and mass analyzers in the past decade, MS instrumentation remains a barrier to further progress in whole protein analysis [25–28]. Realizing the full potential of top down proteomics, especially when applied to large proteins, requires robust and comprehensive protein fragmentation, which continues to be a challenging endeavor. By providing high resolution/accurate mass (HR/AM) measurements for precursor and product ions with good sensitivity, Fourier-transform (FT) instruments are a well-suited MS platform for top down proteomics [29–33]. FT-MS instruments are easily coupled with other ion trapping devices (i.e., hybrid systems) to offer a considerable array of fragmentation methods, including CAD, higher-energy collisional dissociation (HCD), photo-activation, electron capture dissociation (ECD), and ETD [34–41]. A characteristic inherent to all ion trapping instruments, however, is that the number of ions that can be analyzed in a given scan is limited by a fixed number of charges that can be effectively contained and manipulated [42, 43]. The charge capacity of ion trapping devices becomes especially consequential for intact protein fragmentation, where product ion signal is often distributed amongst hundreds of potential fragment channels and increasingly complex isotopic distributions.
As protein mass increases, the efficacy of MS/MS on whole proteins notably diminishes; larger proteins carry more charge and have a greater number of dissociation channels. Not only does this increase spectral complexity, but it also limits precursor capacity (i.e., ion number) in ion trap reaction vessels [44–47]. For example, an ion trap with a charge capacity of approximately 300,000 charges can store ~ 30,000 precursor ions for the z = +10 charge state of ubiquitin (~8.6 kDa) but only roughly 9,400 precursor ions for the z = +32 charge state of carbonic anhydrase (~29 kDa). Compounding this, charge from those initial ion populations is potentially distributed across product ions from 75 backbone bonds for ubiquitin compared to carbonic anhydrase’s 258 backbone bonds. To improve the S/N of product ion measurements, spectral averaging (summing signal from several individual scans) is often required for MS/MS of even modest sized proteins. The tradeoff for the increase in S/N, however, is a significant increase in acquisition times required for generation of high quality spectra, accordingly limiting the sampling depth achievable in a given experiment. We conclude that increasing the number of precursor ion charges prior to initiation of the dissociation event is a direct way to improve S/N without spectral averaging [27, 48, 49]. Herein we describe modifications to ion processing and storage that permit increased precursor ion populations for ETD experiments – we call this method high capacity ETD (also called ETD High Dynamic range, or ETD HD).
For ETD on hybrid ion trap-Orbitrap systems, the size of the precursor population is limited by the precursor sequestration event in the dual cell quadrupole linear ion trap m/z analyzer (A-QLT) prior to the reaction [50, 51]. We have shown previously that a larger ETD reaction cell, called the multipurpose dissociation cell (MDC), can accommodate 6- to 10-fold larger initial populations of precursor ions, thereby alleviating the capacity restrictions imposed by using the A-QLT [52]. With the MDC, we achieved better ion statistics and increased the intra-scan dynamic range for protein fragmentation, leading to higher quality spectra (i.e., increased product ion S/N) with less spectral averaging required, which ultimately enabled better top down analyses of complex protein mixtures.
Hunt and co-workers described a different approach enabled by the development of a front-end ETD reagent source [53]. Here the A-QLT remained as the ion-ion reaction cell, but products from multiple rounds of ion-ion reactions were accumulated in the C-trap before a single mass analysis of all product ions in the Orbitrap, ultimately improving the S/N of MS/MS spectra. The promising results characterized in both the Hunt and Coon lab strategies have motivated us to develop an improved implementation of ETD on the newest generation of quadrupole-Orbitrap-linear ion trap Tribrid mass spectrometers [36].
Here we demonstrate that the ion capacity of the precursor accumulation event prior to the ETD reaction can be increased by changing where in the A-QLT precursor cations and reagent anions are stored. This new implementation of high capacity ETD on the newest generation of Orbitrap Fusion Lumos Tribrid platform allows use of larger precursor populations for ETD MS/MS scans, enabling higher product ion S/N over standard ETD, for a given spectral acquisition time. Ultimately this translates to more sequence-informative fragment ions and higher protein sequence coverage achieved with less spectral averaging in high capacity ETD.
METHODS
Materials, Reagents, and Sample Preparation
Myoglobin [P68082] and carbonic anhydrase [P00921] were purchased as mass spectrometry grade standards from Protea Biosciences (Morgantown, WV). Formic acid ampoules and acetonitrile were purchased from Thermo Scientific (Rockford, IL).
Mass Spectrometry Instrumentation
High precursor capacity ETD was implemented using the existing dual pressure linear ion trap (A-QLT) on the Orbitrap Fusion Lumos (Thermo Fisher Scientific, San Jose, CA). In standard ETD, the precursor sequestration event occurs by creating a DC potential well of approximately 2 volts in the back section of the high pressure cell (HPC). This holds the precursor cations in the back section of the HPC, while the center section and front section voltages of the HPC are set to allow for reagent anion accumulation. To enable high capacity ETD, instrument control code was modified to allow transfer of precursor ions directly from the ion routing multipole to the center section of the HPC for storage using a DC potential well of approximately 4 volts, omitting relocation of precursor ions to the back section prior to the ETD reaction (Figure 2a). Reagent accumulation is then achieved by holding the front section at a positive DC offset to establish the potential well for anions. Charge-sign independent trapping for the ion-ion reaction was then performed in the same fashion for both standard and high capacity ETD by setting all DC bias voltages to 0 V and applying axial confining RF voltages to the end lenses of the HPC.
ESI-MS/MS Analysis
Myoglobin (P68082) and carbonic anhydrase (P00921) were resuspended at approximately 10 picomole per microliter in 49.9:49.9:0.2 acetonitrile/water/formic acid, infused via syringe pump into the mass spectrometer at five microliters per minute through a 500 microliter syringe, and ionized with electrospray ionization (ESI) at +3.5 kV with respect to ground. For myoglobin, MS/MS scans were performed in the Orbitrap with unthresholded transient acquisition at a resolving power of 120,000 (full width at half maximum) at 200 m/z with a range of 200–2000 Th. Precursor ions were isolated with the mass selecting quadrupole with an isolation width of 10 m/z, and automatic gain control (AGC) targets values ranging from 100,000 to 1,000,000 charges as indicated. Transient averaging began after data acquisition was started so that scans with 1–100 transients averaged could be analyzed. An AGC target of 800,000 charges was used for fluoranthene reagent anions (m/z 202, isolated by the mass selecting quadrupole) for ETD and EThcD experiments, reaction times varied as indicated in Supplemental Table 1, and a normalized collision energy of 10 was used for EThcD. Analyses were performed in intact protein mode with a pressure of 3 mTorr in the ion-routing multipole. For carbonic anhydrase, MS/MS scans were performed in the Orbitrap at a both 120,000 and 240,000 resolving powers (at 200 m/z) with precursor AGC target values of 300,000 and 1,000,000 and a m/z range of 400–2000 Th. Transient averaging began after data acquisition was started so that scans with 1–200 transients averaged could be analyzed. The AGC target for fluoranthene reagent anions was set to 700,000 charges, reaction times varied as indicated in the text, and the pressure in the ion-routing multipole was set to 1 mTorr in intact protein mode.
Data Analysis
MS/MS m/z spectra were deconvoluted with XTRACT (Thermo Fisher Scientific) using default parameters and a S/N threshold of 2. ProSight Lite [54] was used to generate matched fragments using a 10 ppm tolerance. ETD spectra were matched with c-, z-, and y-type ions, and EThcD spectra were also matched with those fragment types in addition to b-type ions. N-terminal methionines were removed from the protein sequences before matching with ProSight Lite, and carbonic anhydrase was matched with an additional sequence modification of N-terminal acetylation (+42.01 Da). Supplemental Figure 5 compares signal from fragments seen with ETD and high capacity ETD, and those unique only to high capacity ETD.
THEORY
Since precursor ion signal is distributed amongst product ions upon fragmentation, the size of the initial precursor population, and its subsequent effect on product ion S/N, is a critical consideration in tandem MS experiments. Ion trapping instruments use a fixed number of charges per scan, and the capacity of these devices is ultimately defined by the number of charges they can hold and manipulate. The maximum number of ions that can be effectively stored decreases linearly with the increasing ion charge, as shown by the expression:
| (1) |
where Ncharges is the number of stored charges, Zp is the charge of the precursor, and Nions is the number of ions that comprise a precursor population of charge Zp. This relationship becomes more consequential when analyzing intact proteins, as larger proteins tend to be more highly charged in standard electrospray ionization. This connection between charge state distribution and protein mass has been empirically modeled by Kelleher and co-workers [27]; according to their work, the theoretical charge state distribution of a protein as function of its molecular weight (MW) can be estimated as:
| (2) |
The charge state distributions shown in Figure 1a were obtained according to Equation 2, showing that the most intense predicted charge state for a smaller protein like ubiquitin is around z = +10, and the mode of the distribution increases as protein molecular weight increases (approximately z = +20 and z = +30 for myoglobin and carbonic anhydrase, respectively).
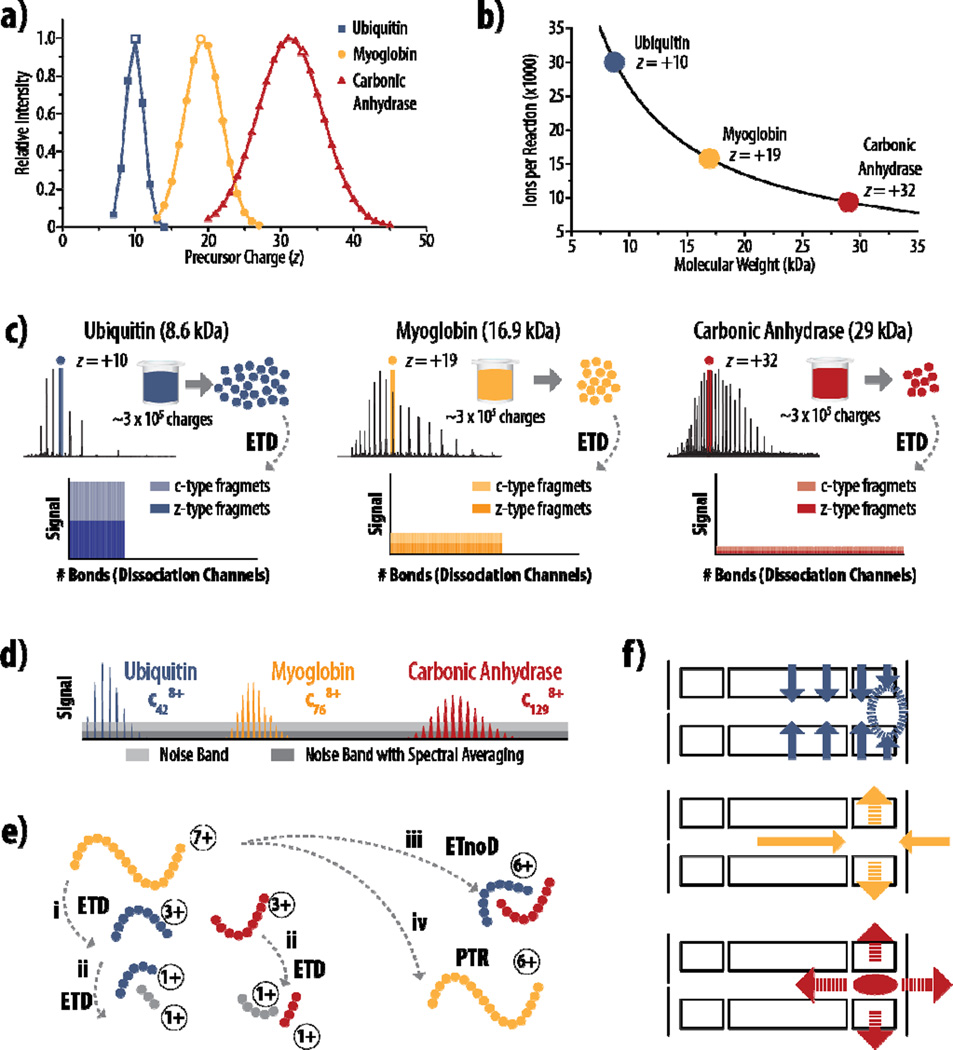
Figure 1. Challenges with product ion signal-to-noise following fragmentation of intact proteins.
a) Theoretical charge state distributions for ubiquitin, myoglobin, and carbonic anhydrase show that the absolute number of charges that precursors carry and the relative width of the charge state distribution both increase as protein mass increases. b) Considering precursors near the middle of the charge state distribution (panel a, open symbols), the number of ions stored for an MS/MS event is plotted vs. molecular weight, using a capacity of 300,000 charges. c) The S/N challenges inherent to fewer ions present in the precursor population is compounded by the increase in the number of dissociation channels as protein size increases, spreading the measureable signal across more fragment ions. d) Large proteins generate proportionally larger fragment ions, further exacerbating S/N problems, as larger fragments have broader isotope distributions and require more ions per fragment to lift peaks above the noise band (light grey). The relative S/N value can be improved by spectral averaging (dark grey), but this can substantially increase acquisition time and restrict throughput. e) ETD generates c-type (blue) and z-type (red) fragments (i), but competing pathways, like secondary dissociation events (ii), non-dissociative electron transfer (iii), and proton transfer reactions (iv), further dilute the signal seen for sequence informative product ions (c and z fragments). f) Three main factors contribute to the limitations on ion storage capacity of the back section of the high pressure cell: top, the confining rf field in the HPC is weakest near the lenses (dashed circle); middle, the axial dc confinement field that keeps ions in the back section of HPC is radially destabilizing (dashed arrows); bottom, space charge effects from the trapping of like-signed ions is both radially and axially destabilizing (dashed arrows), increasing with the amount of stored charge.
An increase in observed charge state for larger proteins equates to fewer ions that can be accumulated for MS/MS events in ion traps that have fixed charge capacities (Equation 1) – and fewer precursor ions mean less charge to be distributed across the resulting product reaction channels. Figure 1b shows the number of ions that can be accumulated for a standard ETD reaction (assuming a capacity of ~ 300,000 charges [50, 52]) for ubiquitin, myoglobin, and carbonic anhydrase. Here a precursor charge state with relatively high theoretical abundance (Figure 1a, open symbols) was selected for each protein so that all three precursor m/z values were within ~40 Th of each other. It is clear that the number of precursor ions stored decreases exponentially as protein size increases, meaning larger proteins already present challenges for product ion S/N. Moreover, the estimated number of dissociation channels increases linearly with increasing molecular weight, as approximated by the expression:
| (3) |
where Nchannels is the number of dissociation channels, NfragmentTypes describes the number of fragment types generated by the dissociation method (e.g., c- and z-type), Nbonds is the number of inter-residue bonds in the protein, MW is the molecular weight of the protein, and 111.1254 is the mass of averagine [55]. The last term in Equation 3 uses averagine as an approximation so that any protein of known molecular weight can be described without requiring knowledge of the number of amino acids comprising the primary sequence. If the residue count of a protein is known, Nbonds can be calculated simply by subtracting one from this total. Considering only the canonical ETD fragment series (c- and z-type) and assuming for simplicity that these fragments are the only channels across which signal can be distributed, Figure 1c illustrates the S/N challenges that arise from an already smaller number of precursor ions (Figure 1b) being spread across more fragments. To compound S/N challenges further, larger proteins generate larger fragments, which have broader isotope distributions. Depreciation in peak abundance due to the presence of naturally occurring isotopes can be expressed by the relationship:
| (4) |
which ultimately requires a greater number of ions for larger fragments to raise usable signal above the noise band (Figure 1d) [27, 56].
All of these challenges are fundamental to top down proteomics, regardless of fragmentation type employed, but the characteristics of ETD itself also require consideration when examining product ion S/N in ETD tandem MS: 1) ETD reactions by their nature consume charge and 2) the signal in an ETD MS/MS spectrum exists in product ions from competing pathways, such as internal fragments from secondary ETD reactions, non-dissociative electron transfer, and proton transfer reactions (Figure 1e) [57–62]. Consumption of charge during the reaction limits the sensitivity of the MS/MS scan because S/N can be related to the number of charges present by:
| (5) |
assuming the presence of only thermal noise during detection, where vn is the thermal noise (estimated to be the equivalent of 20 charges, caused by the amplifier, in the Orbitrap) [48, 63]. Competing reaction pathways can reduce sequence-informative product ion yield, and even those products that do impart sequence information (i.e., c-type and z-type fragments) can exist in multiple charge states, consuming available signal for no additional gain in sequence coverage. We conclude that strategies for improving product ion S/N will be of considerable value for top down protein analysis.
Two of the most straight-forward practices to increase product ion S/N and effectively mitigate these challenges are 1) averaging signal from multiple spectra and 2) conducting reactions on larger precursor populations. Considering modern Fourier transform mass spectrometers, i.e., FT-ICR and Orbitrap systems, which are widely used for intact protein analysis and top down proteomics, spectral averaging consists of summing several individual time-domain signals. Note that transient is used here and throughout for simplicity to describe the time-domain signal from FT-MS measurements. Here, the signal amplitude increases proportionally with the number of transients averaged (Ntransients) while the noise increases as the square root of Ntransients:
| (6) |
making the relative gain in the S/N of a spectrum (GS/N) increase by the square root of Ntransients (Figure 1d) [64]. For example, the approximate 2-fold difference depicted in Figure 1d would require 4 averaged transients. While effective in generating high-quality MS/MS spectra, transient averaging requires significantly longer acquisition times, as even the fastest FT instruments can require several hundred milliseconds or longer per transient to achieve necessary HR/AM measurements [35, 37, 65]. To meet throughput demands of many experiments it is desirable to keep transient averaging to a minimum. Furthermore, extensive transient averaging may still fail to salvage low-level ions that are buried in the noise [66]; conversely, sufficiently large precursor populations can provide enough signal to boost these ions to a detectable level.
In commercially available linear ion trap-Orbitrap hybrid systems, ETD occurs in the high pressure cell (HPC) of the A-QLT. This cell is partitioned into three sections (front, center, and back) with linear dimensions of 12.5 mm, 35 mm, and 12.5 mm, respectively. With the introduction of a front-end reagent source on the Orbitrap Fusion Tribrid system, precursors are isolated by a mass-selecting quadrupole, accumulated in the HPC, and then sequestered in the back section of the cell with reagent trapping in the center and front sections (Figure 1f and Figure 2a). Three main factors influence the ion capacity of the back section of the HPC: (1) the RF radial confinement field on the HPC used to trap the ions; (2) the DC axial confinement field created by the DC potentials applied to the center section, back section, and end lens (Figure 2a); and (3) the space charge field exerted by the ions themselves. Increasing the number of ions sequestered into this back section causes each of these forces to contribute some form of ion destabilization. The RF field confining the ions radially weakens near the end lens due to fringe field affects, leading to a weaker confinement potential in a region of the back section that results in less efficient trapping (Figure 1f, top). Correspondingly, axial DC fields push ions into the back section from both axial directions, causing radial destabilization. As the amount of stored charge increases, ions will be pushed out radially from the stable regions of the trap into the RF field, resulting in micro-motion induced by the RF field that can cause collisional dissociation and/or ejection [67, 68]. This destabilization limits the number of precursors that can be effectively stored without unwanted fragmentation (Figure 1f, middle). Finally, the space charge forces exerted by the precursors of like charge is axially and radially destabilizing, increasing with the total amount of trapped charge and compounding the effects described for the other fields (Figure 1f, bottom).
Altering the precursor storage event so that precursor ions remain in the center section of the HPC, rather than sequester them to the back section, eliminates the challenges of both fringe field effects and smaller confinement fields. Additionally, this scheme provides a greater trapping volume, alleviating the destabilization effects of the DC axial confinement fields and space charge forces. Equation 7 estimates the relationship of the ion capacity/trapping volume of the sections of the linear ion trap and the section length:
| (7) |
where Ncenter/back and lcenter/back are the charge capacities and lengths of the center/back sections of the HPC, respectively [50, 69]. Because the center and back sections have the same general operating parameters, the ratio of charge capacities is approximately proportional to the ratio of the lengths of the two sections. Comparing the length of the center section (35 mm) and the back section (12.5 mm), we expect an approximate 3-fold increase in charge capacity when storing precursors in the center section instead of the back section. The current storage capacity of sequestration in the back section is estimated to be ~ 200,000 to ~500,000 charges [52], meaning precursor targets of 1,000,000 or more charges should be successfully stored using the new acquisition scheme. Note, using the center section for precursor ion storage affects the accumulation of reagent anions as well, which we discuss further in the next section. In this work we explore the benefits that can be achieved when altering the precursor accumulation event in the current A-QLT device to permit accumulation of more precursor charges in the center section.
RESULTS AND DISCUSSION
Implementing High Capacity ETD in a Dual Pressure Linear Ion Trap
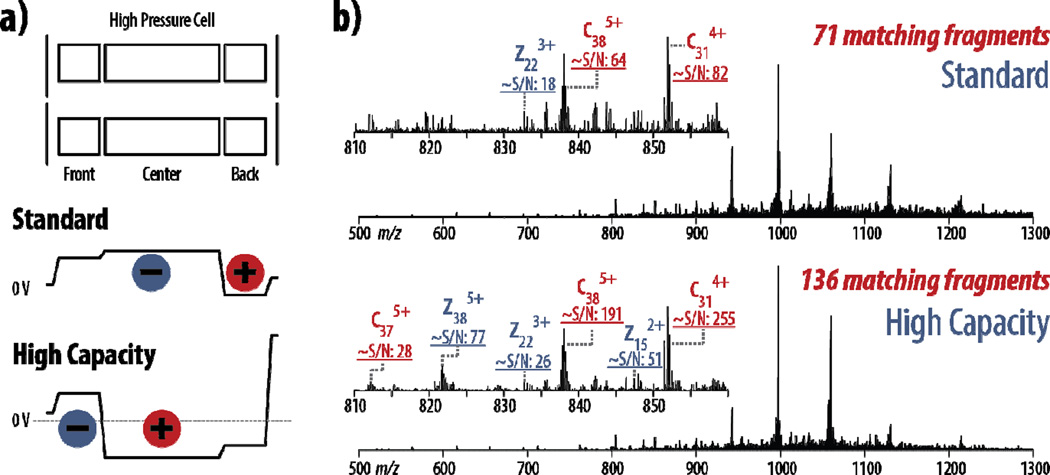
To address the limitations in ion capacity imposed by precursor sequestration in the back section of the HPC, we have employed a new implementation of ETD called high capacity ETD. Figure 2a illustrates how precursor storage differs between standard and high capacity ETD by showing the voltages employed relative to 0 volts during the reagent accumulation period. The practical implications of this change in precursor spatial confinement is highlighted in Figure 2b, showing ETD spectra from both the standard and high capacity implementations for the z = +18 precursor of myoglobin.
For both the standard and high capacity schemes, precursor AGC target values of 100,000, 200,000, 400,000, 600,000, 800,000, and 1,000,000 were investigated. In standard ETD, a target value of 400,000 produced the highest number of matched fragments (71), while higher target values did not translate to an increase in sequence-informative fragment ions. This is in accordance to the estimated capacity of ~200,000 to ~500,000 charges discussed above. In high capacity ETD, however, the largest AGC target value of 1,000,000 produced the most fragments (136), nearly doubling the number of fragments observed following standard ETD using the same amount of spectral averaging (two transients averaged for each). For product ions identified in both conditions, S/N values are approximately three-fold higher, sometimes more, in high capacity ETD. Additional fragment ion identifications were often due to the improved S/N, enabling confident charge state assignment and subsequent matching against theoretical values.
Note that the change in the precursor accumulation event necessitates changes in the reagent accumulation event. In standard ETD, reagent anions are accumulated in the center section of the HPC, while in high capacity ETD only the front section is used (Figure 1a). This now limits the capacity for reagent anion storage, although the ion capacity of the end section should be higher for the reagent anions than it is for the precursor cations. The higher relative capacity can be accounted for by both the significant difference in ion m/z between reagent anions (202 Th) and their precursor counterparts (especially in top down proteomics) and by the single charge of the reagent anions compared to highly charged protein precursors. Slightly smaller reagent anion populations may affect the pseudo-first order kinetics of the ETD reactions, as the reagent, which may no longer be in a large excess of the precursor population, could be depleted during the reaction. Indeed, loss of pseudo-first order kinetics due to reagent depletion was observed in these experiments, requiring longer reaction times to achieve sufficient fragmentation, even with the same reagent AGC target values (Supplemental Table 1). Using the spectra in part b of Figure 2 as an example, the standard ETD scheme using a precursor AGC target of 400,000 used a reaction time of 5 ms; the high capacity ETD with a precursor AGC target of 1,000,000 required a 25 ms reaction time to accrue a comparable level of progression of the ETD reaction.
Figure 2. Increased product ion S/N with high capacity ETD.
a) In standard ETD, precursor cations are sequestered into the back section of the high pressure cell of the A-QLT prior to the reaction, while reagent anions are accumulated in the center and front sections. In high capacity ETD, precursor cations are accumulated in the center section, allowing larger precursor populations for increased product ion S/N. Black lines show DC potentials. b) Spectra from both standard and high capacity ETD scans (2 transients averaged for each) on the z = +18 precursor of myoglobin show that product ions have greater S/N in high capacity ETD, and this increase in product ion S/N allows more sequencing ions to be matched in high capacity ETD. Both spectra are on the same intensity scale.
Although this is a significant increase in reaction time, the resulting increase in total scan time is marginal. The standard ETD spectrum in Figure 2, with two averaged transients and an average precursor injection time of 4.3 ms, required 848 ms of total elapsed scan time for 71 fragments. The high capacity ETD spectrum, comparatively, with two averaged transients and a 12.4 ms average precursor injection time, required 904 ms of total elapsed scan time for 136 fragments. Thus, even though high capacity ETD required approximately 56 ms longer in total acquisition time, it nearly doubled the number of identifiable fragments generated. Even when averaging an additional transient (for a total acquisition time of 1,271 ms), the number of fragments identified in standard ETD increased to only 82 fragments. Furthermore, increasing the reaction time for standard ETD did not provide any beneficial information. Standard ETD at both an AGC target of 400,000 with an 11 ms reaction time (6 ms longer) and an AGC target of 1,000,000 with a 25 ms reaction time (20 ms longer) yielded fewer matching fragments, 64 and 55, respectively, indicating degrees of over-reaction.
High Capacity ETD for a Moderate Size Protein (~17 kDa)
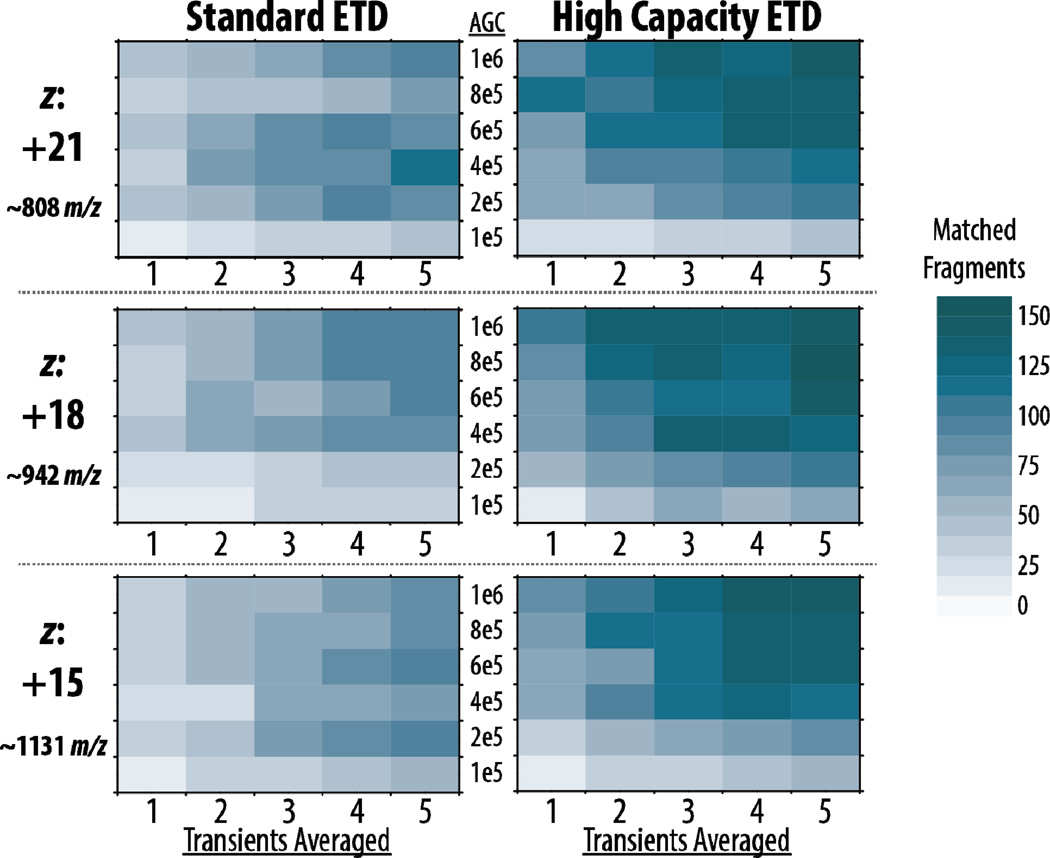
We extended our look at the benefits high capacity ETD can offer over standard ETD by investigating three charge states of myoglobin at six precursor AGC target values using varying amounts of spectral averaging. Figure 3 presents a heat map of the number of matched fragments generated from all three precursors selected. This includes data for each of the six AGC target values when averaging 1–5 transients. Several interesting trends arise – most notably the darker overall color of the high capacity heat maps that shows more fragments are being produced in high capacity ETD. For standard ETD, more fragments are generated from left to right as the number of transients averaged increases, but there is no distinct trend when moving from bottom to top in each heat map (i.e., increasing AGC target values), except for the increases seen when moving from 100,000 to higher targets. These results confirm that the storage capacity of the back section is approximately 200,000 to 400,000 charges, since these AGC targets have similar likelihoods of producing the same number of fragments as higher target values for a given number of averaged transients.
Figure 3. High capacity ETD generates more matched fragments than standard ETD.
Averaging only 1–2 transients with larger AGC targets in high capacity ETD provides as many or more matched fragment ions as averaging 4–5 transients in standard ETD.
High capacity ETD maps show a distinct pattern of darker colors (more matching fragments) for higher precursor targets (upper half), demonstrating that the new implementation of ETD permits reaction of larger precursor populations for improved product ion S/N and more sequence-informative fragments for a given acquisition time. Importantly, Figure 3 also demonstrates the improvements in fragment ion generation seen with high capacity ETD over standard ETD when considering a given number of transients averaged. Higher AGC target values — especially 800,000 and 1,000,000 — provide as many, if not more, matched fragments in two averaged transients as standard ETD can provide in five, and the benefits are striking when considering similar numbers of averaged transients for the two conditions. This may also indicate that approximately 2- to 5-fold more precursor ions can be stored successfully in high capacity ETD, if not more. Additionally, to eliminate differences in reaction time as a cause of the improvements observed with the high capacity scheme, we reacted precursors in the standard ETD scheme (HPC back section sequestration) for the duration used in high capacity ETD (Supplemental Figure 1). The larger precursor AGC targets required longer reactions times in high capacity ETD, so the number of fragments generated from standard ETD at high capacity reaction times is noticeably decremented due to over-reaction and generation of internal fragments, confirming that the benefits seen in high capacity ETD are attributed to the reaction of larger precursor populations.
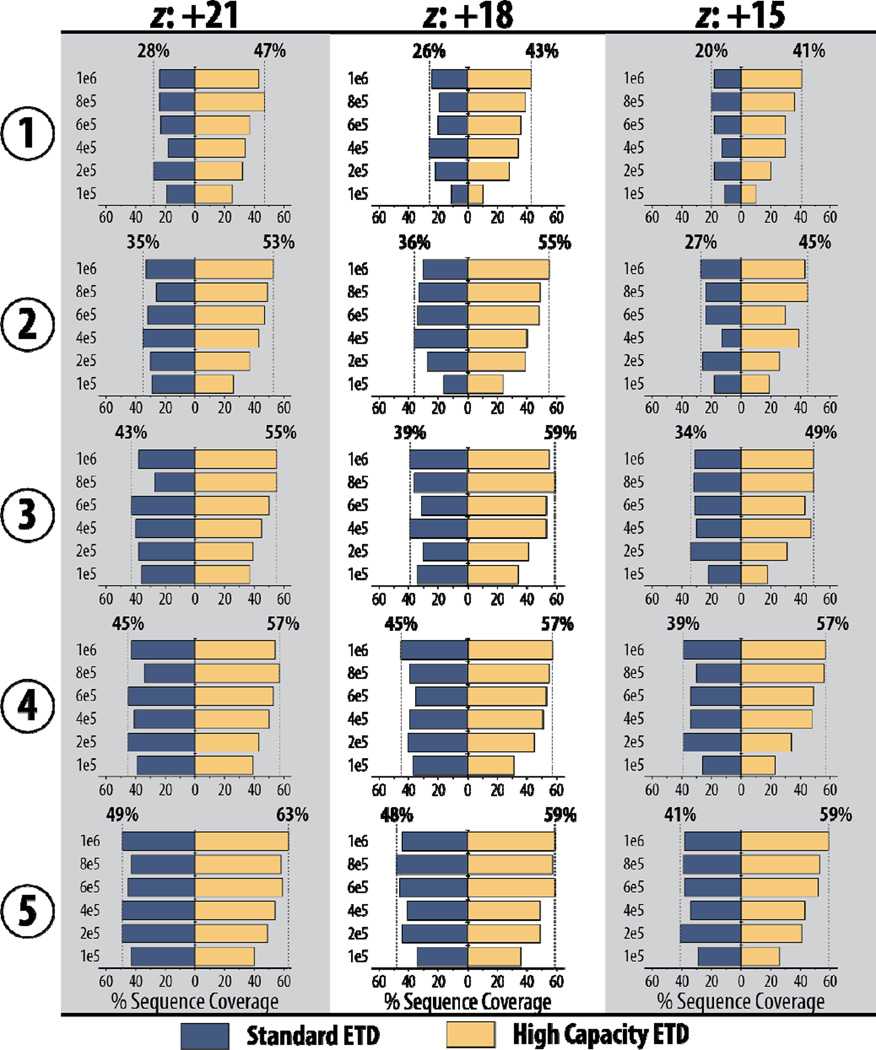
High capacity ETD afforded improved protein sequence coverage. These improvements for the three precursors of myoglobin are summarized in Figure 4. High capacity ETD provides more sequence coverage in two averaged transients than standard ETD can achieve with five averaged transients for all three precursor ion species. Even a single scan with high capacity ETD provides competitive sequence coverage values when compared to five averaged transients for standard ETD.
Figure 4. Higher quality spectra of high capacity ETD translate to greater protein sequence coverage with less spectral averaging required.
Numbers along the left indicate the number of averaged transients and the y-axes show precursor AGC target values.
As noted previously, the increased precursor injection times and longer reaction times in high capacity ETD can increase total spectral acquisition time slightly, even when using the same number of averaged transients, but Supplemental Figure 2 shows that these increases are minor relative to total scan time. The spread in the curves for different AGC target values in high capacity ETD (red) is greater than the curves in standard ETD (blue); this difference further demonstrates that larger precursor populations are indeed being retained when precursor target values are increased in high capacity ETD, whereas the size of the precursor population plateaus in standard ETD despite elevated AGC target values. Overall, the high capacity ETD scheme greatly improves the protein sequence coverage that can be obtained per second of data acquisition, which makes high capacity ETD highly advantageous when spectral quality must be balanced with acquisition time, as is needed in high-throughput top down (i.e., using online chromatography) proteomics experiments.
Even when increasing the degree of spectral averaging up to 100 averaged transients, high capacity ETD still provides increased sequence coverage (Figure 5). Here, as before, the AGC target values are set to the indicated values, although standard ETD cannot retain over ~400,000 precursors (see above). Sequence coverage for myoglobin with high capacity ETD and standard ETD remains similar, as expected, for precursor targets of 100,000 and 200,000 charges. Despite improvements seen in high capacity ETD with relatively small degrees of spectral averaging (1–10 averaged transients) for moderately large target values, i.e., 400,000 and 600,000, sequence coverage achieved for the two implementations does converge with high degrees of averaging (>50 transients). The largest AGC target values (800,000 and 1,000,000) provide consistent gains in sequence coverage with high capacity ETD even with significant spectral averaging, although the difference between the two still diminishes. This is likely because myoglobin is a moderately sized protein; for these proteins, high degrees of spectral averaging, while incurring significantly extended acquisition times, can mitigate S/N challenges and match the boosts seen from larger precursor populations.
Figure 5. High capacity ETD enables larger AGC target values that produce greater protein sequence coverage than standard ETD for myoglobin, even with many averaged transients.
Data here is shown for the z = +15 precursor of myoglobin.
Beyond traditional ETD fragmentation, we observed that high capacity ETD can aid hybrid fragmentation methods as well. EThcD, which uses beam-type collisional activation of ETD products after the ion-ion reaction [70, 71], can improve sequence coverage for precursor ions, especially those with low-charge density where precursor-to-product ion conversion efficiency is hindered by non-covalent interactions. We saw that the high capacity ETD scheme aided in fragment ion generation and protein sequence coverage with EThcD on the z = +15 precursor of myoglobin (Supplemental Figure 3). We surmised that high capacity ETD can be especially valuable for these hybrid fragmentation techniques, where secondary activation has to be carefully balanced since more fragmentation channels are being added to erode product ion signal. As expected, the best benefits to EThcD with the high capacity scheme were seen at the highest precursor targets.
High Capacity ETD for a Larger Protein (~29 kDa)
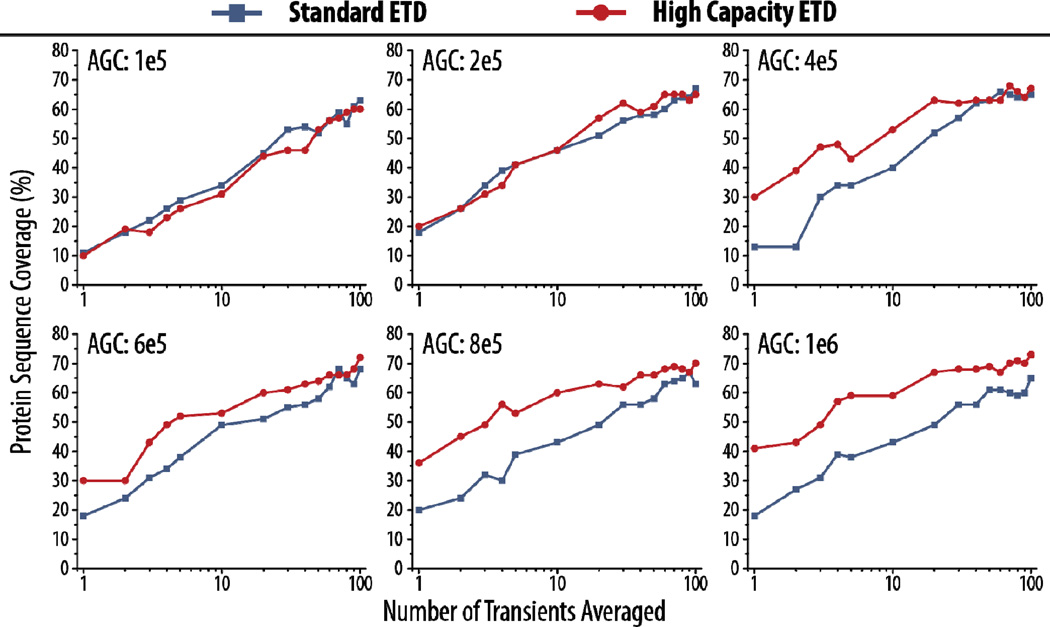
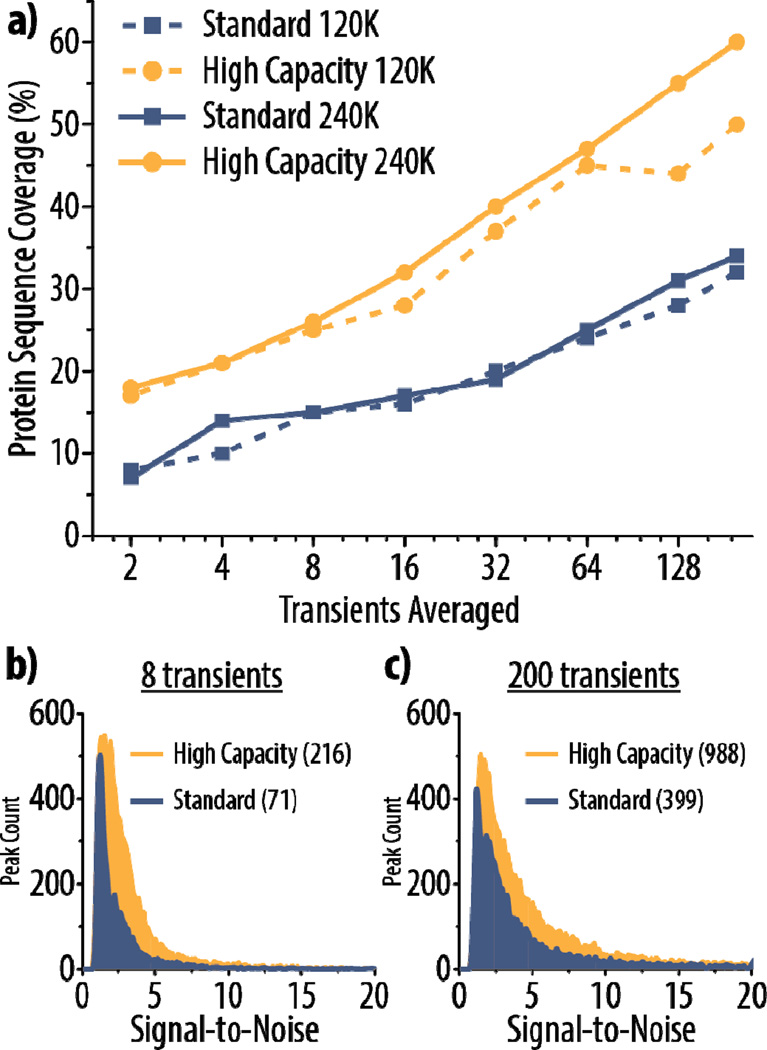
High capacity ETD is well-positioned to provide pronounced gains for larger proteins, where the number of dissociation channels is significantly greater and even considerable degrees of spectral averaging cannot approach the increases provided by reaction of large precursor populations [72, 73]. To investigate this, we reacted the z = +34 precursor of carbonic anhydrase (~29 kDa) using standard ETD (AGC target of 300,000) and high capacity ETD (AGC target of 1,000,000). To explore how the high capacity ETD and standard ETD schemes compare with higher resolution spectra, we also collected MS/MS spectra at two resolving powers (120K and 240K). First, the best reaction times to use for each condition were determined experimentally (Supplemental Figure 4), and reaction times of 4 ms and 7 ms were used for standard and high capacity ETD, respectively.
Figure 6a demonstrates that high capacity ETD affords greater sequence coverage than standard ETD for up to 200 transients averaged at both 120K and 240K resolving powers. In fact, high capacity ETD at 120K outperforms standard ETD at 240K. To show that the gains seen with high capacity ETD can be attributed to increases in product ion S/N, we plotted histograms of S/N values for product ions from ETD spectra (at 240K) with 8 (Figure 6b) and 200 (Figure 6c) transients averaged for both high capacity and standard ETD. The distributions are only shown up to S/N 20 to emphasize the region where the majority of the peaks lie, but the maximum S/N values for each condition are given in parentheses in the figure legends. Expected patterns arise: the distributions are shifted toward higher S/N values in high capacity ETD, and the distributions are broader with higher S/N values when 200 versus 8 transients are averaged. Note that 8 averaged transients with high capacity ETD provide similar sequence coverage to that seen with 200 averaged transients in standard ETD. While a scan using 8 averaged transients still requires approximately 2–4 seconds to acquire at 120K (Supplemental Figure 2), high capacity ETD makes more thorough characterization of larger proteins on chromatographic timescales a realistic goal.
Figure 6. High capacity ETD provides superior results for characterization of carbonic anhydrase (~29 kDa).
Protein sequence coverage for the z = +34 precursor of carbonic anhydrase using either high capacity or standard ETD is shown in (a) with varying degrees of spectral averaging and at two different resolving powers (120K and 240K). Histograms display the distribution of signal-to-noise (S/N) of peaks in high capacity and standard ETD spectra using 8 or 200 averaged transients (b and c, respectively) at a resolving power of 240K. The y-axes show the peak count for a given S/N value (bin size = 0.1). The maximum S/N for a peak in each spectrum is given in parentheses in the figure, although the histograms only display the distributions up to 20 S/N to highlight the region where the majority of the peaks lie.
CONCLUSION
We have enabled the accumulation and retention of 2- to 5-fold more precursors for ETD reactions by altering the region in the ion trap where precursor ions are stored during reagent ion injection. When holding precursor cations in the center section of the high pressure cell of a dual cell quadrupole linear ion trap, as many as 1,000,000 charges or more can be stored for subsequent ion-ion reactions. This increase in precursor ion capacity boosts the signal-to-noise of product ions, producing higher quality MS/MS spectra with only minor increases in acquisition time. High capacity ETD facilitates a more robust characterization of intact protein cations – a single scan can achieve fragment ion production and protein sequence coverage equivalent to approximately five averaged scans of standard ETD. Overall, high capacity ETD improves the compromise between S/N improvements and spectral acquisition speed while still enabling enhanced MS/MS data quality for intact proteins, regardless the degree of spectral averaging. Moreover, high capacity ETD has been implemented using commercially accessible hardware and is available on the newest generation of quadrupole-Orbitrap-linear ion trap Tribrid mass spectrometer (Orbitrap Fusion Lumos), giving it a distinct advantage over earlier approaches that required custom modified devices.
The improvements in MS/MS characterization of intact proteins with high capacity ETD will advance top down proteomics by providing more robust fragmentation on a chromatographic time-scale. This new implementation of ETD also benefits hybrid dissociation methods like EThcD, which are demonstrating promise as new approaches to intact protein fragmentation approaches. Future work will focus on how high capacity ETD can benefit other hybrid dissociation techniques, e.g., ultraviolet photo-dissociation (UVPD)-ETD methods [74] and activated ion ETD (AI-ETD) [75, 76], with an emphasis on how this improved approach to ETD can be employed in large-scale proteome characterizations. With the implementation of high capacity ETD, we present a straightforward strategy to improve tandem mass spectra of intact proteins; accordingly, this approach is implemented on the Orbitrap Fusion Lumos and maintains all of the benefits of conducting ion-ion reactions in the dual-cell quadrupole linear ion trap.
Supplementary Material
ACKNOWLEDGMENT
The authors gratefully acknowledge support from Thermo Fisher Scientific and NIH grant R01 GM080148. N.M.R. was funded through an NSF Graduate Research Fellowship (DGE-1256259). We also thank Graeme McAlister for helpful discussions.
REFERENCES
- 1.Kelleher NL. Peer Reviewed: Top-Down Proteomics. Anal. Chem. 2004;76:196A–203A. [PubMed] [Google Scholar]
- 2.Chait BT. Chemistry. Mass spectrometry: bottom-up or top-down? Science. 2006;314:65–66. doi: 10.1126/science.1133987. [DOI] [PubMed] [Google Scholar]
- 3.Zhang H, Cui W, Wen J, Blankenship RE, Gross ML. Native electrospray and electron-capture dissociation FTICR mass spectrometry for top-down studies of protein assemblies. Anal. Chem. 2011;83:5598–5606. doi: 10.1021/ac200695d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Konermann L, Vahidi S, Sowole M. a: Mass Spectrometry Methods for Studying Structure and Dynamics of Biological Macromolecules. Anal. Chem. 2013 doi: 10.1021/ac4039306. [DOI] [PubMed] [Google Scholar]
- 5.McLafferty FW, Breuker K, Jin M, Han X, Infusini G, Jiang H, Kong X, Begley TP. Top-down MS, a powerful complement to the high capabilities of proteolysis proteomics. FEBS J. 2007;274:6256–6268. doi: 10.1111/j.1742-4658.2007.06147.x. [DOI] [PubMed] [Google Scholar]
- 6.Carr SA, Hemling ME, Bean MF, Roberts GD. Integration of mass spectrometry in analytical biotechnology. Anal. Chem. 1991;63:2802–2824. doi: 10.1021/ac00024a003. [DOI] [PubMed] [Google Scholar]
- 7.Mann M, Højrup P, Roepstorff P. Use of mass spectrometric molecular weight information to identify proteins in sequence databases. Biol. Mass Spectrom. 1993;22:338–345. doi: 10.1002/bms.1200220605. [DOI] [PubMed] [Google Scholar]
- 8.Mann M, Wilm M. Error-Tolerant Identification of Peptides in Sequence Databases by Peptide Sequence Tags. Anal. Chem. 1994;66:4390–4399. doi: 10.1021/ac00096a002. [DOI] [PubMed] [Google Scholar]
- 9.Yu L, Xiong Y-M, Polfer NC. Periodicity of monoisotopic mass isomers and isobars in proteomics. Anal. Chem. 2011;83:8019–8023. doi: 10.1021/ac201624t. [DOI] [PubMed] [Google Scholar]
- 10.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 11.Coon J, Syka JEP, Shabanowitz J, Hunt DF. Tandem Mass Spectrometry for Peptide and Protein Sequence Analysis. Biotechniques. 2005;38:519–523. doi: 10.2144/05384TE01. [DOI] [PubMed] [Google Scholar]
- 12.Smith LM, Kelleher NL. Proteoform: a single term describing protein complexity. Nat. Methods. 2013;10:186–187. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Breuker K, Jin M, Han X, Jiang H, McLafferty FW. Top-down identification and characterization of biomolecules by mass spectrometry. J. Am. Soc. Mass Spectrom. 2008;19:1045–1053. doi: 10.1016/j.jasms.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zubarev R, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein. A nonergodic process. J. Am. Chem. Soc. 1998;120:3265–3266. [Google Scholar]
- 15.Shaw JB, Li W, Holden DD, Zhang Y, Griep-Raming J, Fellers RT, Early BP, Thomas PM, Kelleher NL, Brodbelt JS. Complete protein characterization using top-down mass spectrometry and ultraviolet photodissociation. J. Am. Chem. Soc. 2013;135:12646–12651. doi: 10.1021/ja4029654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou M, Wysocki VH. Surface induced dissociation: dissecting noncovalent protein complexes in the gas phase. Acc. Chem. Res. 2014;47:1010–1018. doi: 10.1021/ar400223t. [DOI] [PubMed] [Google Scholar]
- 17.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coon JJ, Ueberheide B, Syka JEP, Dryhurst DD, Ausio J, Shabanowitz J, Hunt DF. Protein identification using sequential ion/ion reactions and tandem mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9463–9468. doi: 10.1073/pnas.0503189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Udeshi ND, Compton PD, Shabanowitz J, Hunt DF, Rose KL. Methods for analyzing peptides and proteins on a chromatographic timescale by electron-transfer dissociation mass spectrometry. Nat. Protoc. 2008;3:1709–1717. doi: 10.1038/nprot.2008.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui W, Rohrs HW, Gross ML. Top-down mass spectrometry: recent developments, applications and perspectives. Analyst. 2011;136:3854–3864. doi: 10.1039/c1an15286f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tran JC, Zamdborg L, Ahlf DR, Lee JE, Catherman AD, Durbin KR, Tipton JD, Vellaichamy A, Kellie JF, Li M, Wu C, Sweet SMM, Early BP, Siuti N, LeDuc RD, Compton PD, Thomas PM, Kelleher NL. Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature. 2011;480:254–258. doi: 10.1038/nature10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Russell JD, Scalf M, Book AJ, Ladror DT, Vierstra RD, Smith LM, Coon JJ. Characterization and quantification of intact 26S proteasome proteins by real-time measurement of intrinsic fluorescence prior to top-down mass spectrometry. PLoS One. 2013;8:e58157. doi: 10.1371/journal.pone.0058157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durbin KR, Fellers RT, Ntai I, Kelleher NL, Compton PD. Autopilot: an online data acquisition control system for the enhanced high-throughput characterization of intact proteins. Anal. Chem. 2014;86:1485–1492. doi: 10.1021/ac402904h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhoads TW, Rose CM, Bailey DJ, Riley NM, Molden RC, Nestler AJ, Merrill AE, Smith LM, Hebert AS, Westphall MS, Pagliarini DJ, Garcia BA, Coon JJ. Neutron-encoded mass signatures for quantitative top-down proteomics. Anal. Chem. 2014;86:2314–2319. doi: 10.1021/ac403579s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Catherman AD, Skinner OS, Kelleher NL. Top Down proteomics: facts perspectives. Res. Commun. Biochem. Biophys. 2014;445:683–693. doi: 10.1016/j.bbrc.2014.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia BA. What does the future hold for Top Down mass spectrometry? J. Am. Soc. Mass Spectrom. 2010;21:193–202. doi: 10.1016/j.jasms.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 27.Compton PD, Zamdborg L, Thomas PM, Kelleher NL. On the scalability and requirements of whole protein mass spectrometry. Anal. Chem. 2011;83:6868–6874. doi: 10.1021/ac2010795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siuti N, Kelleher NL. Decoding protein modifications using top-down mass spectrometry. Nat. Methods. 2007;4:817–821. doi: 10.1038/nmeth1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kellie JF, Tran JC, Lee JE, Ahlf DR, Thomas HM, Ntai I, Catherman AD, Durbin KR, Zamdborg L, Vellaichamy A, Thomas PM, Kelleher NL. The emerging process of Top Down mass spectrometry for protein analysis: biomarkers, protein-therapeutics, and achieving high throughput. Mol. Biosyst. 2010;6:1532–1539. doi: 10.1039/c000896f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang T-Y, McLuckey SA. Top-down protein characterization facilitated by ion/ion reactions on a quadrupole/time of flight platform. Proteomics. 2010;10:3577–3588. doi: 10.1002/pmic.201000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makarov A, Denisov E. Dynamics of ions of intact proteins in the Orbitrap mass analyzer. J. Am. Soc. Mass Spectrom. 2009;20:1486–1495. doi: 10.1016/j.jasms.2009.03.024. [DOI] [PubMed] [Google Scholar]
- 32.Bogdanov B, Smith RD. Proteomics by FTICR mass spectrometry: Top down and bottom up. Mass Spectrom. Rev. 2005;24:168–200. doi: 10.1002/mas.20015. [DOI] [PubMed] [Google Scholar]
- 33.Mann M, Kelleher NL. Precision proteomics: the case for high resolution and high mass accuracy. Proc. Natl. Acad. Sci. U. S. A. 2008;105:18132–18138. doi: 10.1073/pnas.0800788105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McAlister GC, Berggren WT, Griep-Raming J, Horning S, Makarov A, Phanstiel D, Stafford G, Swaney DL, Syka JEP, Zabrouskov V, Coon JJ. A proteomics grade electron transfer dissociation-enabled hybrid linear ion trap-orbitrap mass spectrometer. J. Proteome Res. 2008;7:3127–3136. doi: 10.1021/pr800264t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michalski A, Damoc E, Lange O, Denisov E, Nolting D, Müller M, Viner R, Schwartz J, Remes P, Belford M, Dunyach J-J, Cox J, Horning S, Mann M, Makarov A. Ultra high resolution linear ion trap Orbitrap mass spectrometer (Orbitrap Elite) facilitates top down LC MS/MS and versatile peptide fragmentation modes. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.O111.013698. O111.013698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Senko MW, Remes PM, Canterbury JD, Mathur R, Song Q, Eliuk SM, Mullen C, Earley L, Hardman M, Blethrow JD, Bui H, Specht A, Lange O, Denisov E, Makarov A, Horning S, Zabrouskov V. Novel parallelized quadrupole/linear ion trap/Orbitrap tribrid mass spectrometer improving proteome coverage and peptide identification rates. Anal. Chem. 2013;85:11710–11714. doi: 10.1021/ac403115c. [DOI] [PubMed] [Google Scholar]
- 37.Ahlf DR, Compton PD, Tran JC, Early BP, Thomas PM, Kelleher NL. Evaluation of the compact high-field orbitrap for top-down proteomics of human cells. J. Proteome Res. 2012;11:4308–4314. doi: 10.1021/pr3004216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Håkansson K, Chalmers MJ, Quinn JP, McFarland MA, Hendrickson CL, Marshall AG. Combined Electron Capture and Infrared Multiphoton Dissociation for Multistage MS/MS in a Fourier Transform Ion Cyclotron Resonance Mass Spectrometer. Anal. Chem. 2003;75:3256–3262. doi: 10.1021/ac030015q. [DOI] [PubMed] [Google Scholar]
- 39.Haselmann KF, Budnik BA, Olsen JV, Nielsen ML, Reis CA, Clausen H, Johnsen AH, Zubarev RA. Advantages of external accumulation for electron capture dissociation in Fourier transform mass spectrometry. Anal. Chem. 2001;73:2998–3005. doi: 10.1021/ac0015523. [DOI] [PubMed] [Google Scholar]
- 40.Vasicek La, Ledvina AR, Shaw J, Griep-Raming J, Westphall MS, Coon JJ, Brodbelt JS. Implementing photodissociation in an Orbitrap mass spectrometer. J. Am. Soc. Mass Spectrom. 2011;22:1105–1108. doi: 10.1007/s13361-011-0119-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Syka JEP, Marto JA, Bai DL, Horning S, Senko MW, Schwartz JC, Ueberheide B, Garcia B, Busby S, Muratore T, Shabanowitz J, Hunt DF. Novel Linear Quadrupole Ion Trap/FT Mass Spectrometer: Performance Characterization and Use in the Comparative Analysis of Histone H3 Post-translational Modifications. J. Proteome Res. 2004;3:621–626. doi: 10.1021/pr0499794. [DOI] [PubMed] [Google Scholar]
- 42.March RE. An Introduction to Quadrupole Ion Trap Mass Spectrometry. J. Mass Spectrom. 1997;32:351–369. [Google Scholar]
- 43.Louris JN, Cooks RG, Syka JEP, Kelley PE, Stafford GC, Todd JFJ. Instrumentation, applications, and energy deposition in quadrupole ion-trap tandem mass spectrometry. Anal. Chem. 1987;59:1677–1685. [Google Scholar]
- 44.Stephenson JL, McLuckey SA, Reid GE, Wells JM, Bundy JL. Ion/ion chemistry as a top-down approach for protein analysis. Curr. Opin. Biotechnol. 2002;13:57–64. doi: 10.1016/s0958-1669(02)00285-9. [DOI] [PubMed] [Google Scholar]
- 45.Reid GE, McLuckey SA. “Top down” protein characterization via tandem mass spectrometry. J. Mass Spectrom. 2002;37:663–675. doi: 10.1002/jms.346. [DOI] [PubMed] [Google Scholar]
- 46.Scherperel G, Reid GE. Emerging methods in proteomics: top-down protein characterization by multistage tandem mass spectrometry. Analyst. 2007;132:500–506. doi: 10.1039/b618499p. [DOI] [PubMed] [Google Scholar]
- 47.Louris JN, Brodbelt-Lustig JS, Graham Cooks R, Glish GL, van Berkel GJ, McLuckey SA. Ion isolation and sequential stages of mass spectrometry in a quadrupole ion trap mass spectrometer. Int. J. Mass Spectrom. Ion Process. 1990;96:117–137. [Google Scholar]
- 48.Limbach PA, Grosshans PB, Marshall AG. Experimental determination of the number of trapped ions, detection limit, and dynamic range in Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 1993;65:135–140. [Google Scholar]
- 49.Tsybin YO, Witt M, Baykut G, Håkansson P. Electron capture dissociation Fourier transform ion cyclotron resonance mass spectrometry in the electron energy range 0–50 eV. Rapid Commun. Mass Spectrom. 2004;18:1607–1613. doi: 10.1002/rcm.1525. [DOI] [PubMed] [Google Scholar]
- 50.Schwartz JC, Senko MW, Syka JEP. A two-dimensional quadrupole ion trap mass spectrometer. J. Am. Soc. Mass Spectrom. 2002;13:659–669. doi: 10.1016/S1044-0305(02)00384-7. [DOI] [PubMed] [Google Scholar]
- 51.Second TP, Blethrow JD, Schwartz JC, Merrihew GE, MacCoss MJ, Swaney DL, Russell JD, Coon JJ, Zabrouskov V. Dual-pressure linear ion trap mass spectrometer improving the analysis of complex protein mixtures. Anal. Chem. 2009;81:7757–7765. doi: 10.1021/ac901278y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rose CM, Russell JD, Ledvina AR, McAlister GC, Westphall MS, Griep-Raming J, Schwartz JC, Coon JJ, Syka JEP. Multipurpose dissociation cell for enhanced ETD of intact protein species. J. Am. Soc. Mass Spectrom. 2013;24:816–827. doi: 10.1007/s13361-013-0622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Earley L, Anderson LC, Bai DL, Mullen C, Syka JEP, English AM, Dunyach J-J, Stafford GC, Shabanowitz J, Hunt DF, Compton PD. Front-end electron transfer dissociation: a new ionization source. Anal. Chem. 2013;85:8385–8390. doi: 10.1021/ac401783f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fellers RT, Greer JB, Early BP, Yu X, LeDuc RD, Kelleher NL, Thomas PM. ProSight Lite: Graphical software to analyze top-down mass spectrometry data. Proteomics. 2014 doi: 10.1002/pmic.201570050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Senko MW, Beu SC, McLaffertycor FW. Determination of monoisotopic masses and ion populations for large biomolecules from resolved isotopic distributions. J. Am. Soc. Mass Spectrom. 1995;6:229–233. doi: 10.1016/1044-0305(95)00017-8. [DOI] [PubMed] [Google Scholar]
- 56.Rockwood AL. Relationship of Fourier transforms to isotope distribution calculations. Rapid Commun. Mass Spectrom. 1995;9:103–105. [Google Scholar]
- 57.Coon JJ, Syka JEP, Schwartz JC, Shabanowitz J, Hunt DF. Anion dependence in the partitioning between proton and electron transfer in ion/ion reactions. Int. J. Mass Spectrom. 2004;236:33–42. [Google Scholar]
- 58.Compton PD, Strukl JV, Bai DL, Shabanowitz J, Hunt DF. Optimization of electron transfer dissociation via informed selection of reagents and operating parameters. Anal. Chem. 2012;84:1781–1785. doi: 10.1021/ac202807h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McLuckey Sa, Stephenson JL. Ion/ion chemistry of high-mass multiply chargedions. Mass Spectrom. Rev. 1999;17:369–407. doi: 10.1002/(SICI)1098-2787(1998)17:6<369::AID-MAS1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 60.Rose CM, Rush MJP, Riley NM, Merrill AE, Kwiecien NW, Holden DD, Mullen C, Westphall MS, Coon JJ. A Calibration Routine for Efficient ETD in Large-Scale Proteomics. J. Am. Soc. Mass Spectrom. 2015 doi: 10.1007/s13361-015-1183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McLuckey SA, Stephenson JL, Asano KG. Ion/ion proton-transfer kinetics: implications for analysis of ions derived from electrospray of protein mixtures. Anal. Chem. 1998;70:1198–1202. doi: 10.1021/ac9710137. [DOI] [PubMed] [Google Scholar]
- 62.Good DM, Wirtala M, McAlister GC, Coon JJ. Performance characteristics of electron transfer dissociation mass spectrometry. Mol. Cell. Proteomics. 2007;6:1942–1951. doi: 10.1074/mcp.M700073-MCP200. [DOI] [PubMed] [Google Scholar]
- 63.Makarov A, Denisov E, Kholomeev A, Balschun W, Lange O, Strupat K, Horning S. Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal. Chem. 2006;78:2113–2120. doi: 10.1021/ac0518811. [DOI] [PubMed] [Google Scholar]
- 64.Marshall AG, Comisarow MB. Fourier transform methods in spectroscopy. J. Chem. Educ. 1975;52:638. [Google Scholar]
- 65.Makarov A, Denisov E, Lange O. Performance evaluation of a high-field Orbitrap mass analyzer. J. Am. Soc. Mass Spectrom. 2009;20:1391–1396. doi: 10.1016/j.jasms.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 66.Fornelli L, Damoc E, Thomas PM, Kelleher NL, Aizikov K, Denisov E, Makarov A, Tsybin YO. Analysis of intact monoclonal antibody IgG1 by electron transfer dissociation Orbitrap FTMS. Mol. Cell. Proteomics. 2012;11:1758–1767. doi: 10.1074/mcp.M112.019620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sannes-Lowery K, Griffey RH, Kruppa GH, Speir JP, Hofstadler SA. Multipole storage assisted dissociation, a novel in-source dissociation technique for electrospray ionization generated ions. Rapid Commun. Mass Spectrom. 1998;12:1957–1961. doi: 10.1002/(SICI)1097-0231(19981215)12:23<1957::AID-RCM418>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 68.Sannes-Lowery KA, Hofstadler SA. Characterization of multipole storage assisted dissociation: implications for electrospray ionization mass spectrometry characterization of biomolecules. J. Am. Soc. Mass Spectrom. 2000;11:1–9. doi: 10.1016/S1044-0305(99)00110-5. [DOI] [PubMed] [Google Scholar]
- 69.Campbell JM, Collings BA, Douglas DJ. A new linear ion trap time-of-flight system with tandem mass spectrometry capabilities. Rapid Commun. Mass Spectrom. 1998;12:1463–1474. doi: 10.1002/rcm.440. [DOI] [PubMed] [Google Scholar]
- 70.Frese CK, Altelaar AFM, van den Toorn H, Nolting D, Griep-Raming J, Heck AJR, Mohammed S. Toward full peptide sequence coverage by dual fragmentation combining electron-transfer and higher-energy collision dissociation tandem mass spectrometry. Anal. Chem. 2012;84:9668–9673. doi: 10.1021/ac3025366. [DOI] [PubMed] [Google Scholar]
- 71.Brunner AM, Lossl P, Liu F, Huguet R, Mullen C, Yamashita M, Zabrouskov V, Makarov A, Altelaar AFM, Heck AJR. Benchmarking multiple fragmentation methods on an Orbitrap Fusion for top-down phospho-proteoform characterization. Anal. Chem. 2015;87:4152–4158. doi: 10.1021/acs.analchem.5b00162. [DOI] [PubMed] [Google Scholar]
- 72.Fornelli L, Parra J, Hartmer R, Stoermer C, Lubeck M, Tsybin YO. Top-down analysis of 30–80 kDa proteins by electron transfer dissociation time-of-flight mass spectrometry. Anal. Bioanal. Chem. 2013;405:8505–8514. doi: 10.1007/s00216-013-7267-5. [DOI] [PubMed] [Google Scholar]
- 73.Han X, Jin M, Breuker K, McLafferty FW. Extending top-down mass spectrometry to proteins with masses greater than 200 kilodaltons. Science. 2006;314:109–112. doi: 10.1126/science.1128868. [DOI] [PubMed] [Google Scholar]
- 74.Cannon JR, Holden DD, Brodbelt JS. Hybridizing ultraviolet photodissociation with electron transfer dissociation for intact protein characterization. Anal. Chem. 2014;86:10970–10977. doi: 10.1021/ac5036082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Riley NM, Westphall MS, Coon JJ. Activated Ion Electron Transfer Dissociation for Improved Fragmentation of Intact Proteins. Anal. Chem. 2015;87:7109–7116. doi: 10.1021/acs.analchem.5b00881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao Y, Riley NM, Sun L, Hebert AS, Yan X, Westphall MS, Rush MJP, Zhu G, Champion MM, Medie FM, Champion PAD, Coon JJ, Dovichi NJ. Coupling Capillary Zone Electrophoresis with Electron Transfer Dissociation and Activated Ion Electron Transfer Dissociation for Top-Down Proteomics. Anal. Chem. 2015;87:5422–5429. doi: 10.1021/acs.analchem.5b00883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.