Summary
APOBEC family cytidine deaminases have been recently implicated as powerful mutators of cancer genomes. How APOBECs, which are ssDNA specific enzymes, gain access to chromosomal DNA is unclear. To ascertain the chromosomal ssDNA substrates of the APOBECs, we expressed APOBEC3A and APOBEC3B, the two most probable APOBECs mediating cancer mutagenesis, in a yeast model system. We demonstrate, using mutation reporters and whole genome sequencing, that APOBEC3A- and APOBEC3B-induced mutagenesis primarily results from the deamination of the lagging strand template during DNA replication. Moreover, our results indicate that both genetic deficiencies in replication fork-stabilizing proteins and chemical induction of replication stress greatly augment the mutagenesis of APOBEC3A and 3B. Taken together, these results strongly indicate that ssDNA formed during DNA lagging strand synthesis is a major substrate for APOBECs and may be the principal substrate in human cancers experiencing replication stress.
Introduction
Exposure to DNA damaging agents or deficiencies in DNA repair pathways commonly cause somatic mutations that underlie cellular transformation and carcinogenesis [reviewed in (Jackson and Bartek, 2009)]. We and others have recently identified the family of apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like (APOBEC) cytidine deaminases as an endogenous source of DNA damage that mutagenizes many human cancers (Burns et al., 2013a, Nik-Zainal et al., 2012, Roberts et al., 2013, Burns et al., 2013b, Roberts et al., 2012). These enzymes normally function within lipoprotein metabolism (Teng et al., 1993) and the immune system to restrict viral and retrotransposable elements [reviewed in (Refsland and Harris, 2013)]. However, if unrestrained, APOBEC enzymes can also act as potent mutators of chromosomal DNA, where they deaminate cytidines preferentially within the trinucleotide sequences, TCA and TCT (referred to collectively as TCW; the mutated base is underlined) (Refsland and Harris, 2013). Consequently, APOBEC-mutagenized tumors contain an over-abundance of C to T and C to G substitutions within TCW sequences (Roberts et al., 2013, Alexandrov et al., 2013, Burns et al., 2013b). This mutation signature is widespread among many human cancer types, occurring prominently in bladder, cervical, head and neck, breast, lung, and esophageal cancers, and can account for up to 70% of the total mutation load within a tumor (Alexandrov et al., 2013, Roberts et al., 2013, Saraconi et al., 2014, Burns et al., 2013b).
While it is accepted that APOBEC cytidine deaminases likely cause the extensive mutagenesis of TCW sequences in cancer, the identity of the responsible APOBECs and their chromosomal substrates are still under debate. Both APOBEC3A (A3A) and APOBEC3B (A3B) have access to the nucleus (Bogerd et al., 2006), can mutagenize human genes (Burns et al., 2013a, Thielen et al., 2010, Caval et al., 2014), and have elevated mRNA levels in human cancers, with A3B expression correlating more strongly than A3A expression with the total mutation load across multiple cancer types (Burns et al., 2013b, Roberts et al., 2013). Additionally, A3B has been shown to be the major source of deaminase activity and mutagenesis in a panel of human breast cancer cell lines (Burns et al., 2013a). However, a human polymorphism that involves the deletion of A3B predisposes individuals to breast cancers (Kidd et al., 2007, Komatsu et al., 2008, Xuan et al., 2013). This deletion has been shown to stabilize the A3A mRNA (Caval et al., 2014), suggesting that increased A3A expression may cause the cancer predisposition. Additional analysis of the sequence specificities of A3A and A3B in a yeast model system indicate that these enzymes prefer different nucleotides at the −2 position, targeting YTCW and RTCW, respectively (where Y = C or T; R = G or A). Both of these motifs are over-represented in APOBEC-mutagenized tumors, with specific tumors having either an A3A- or A3B-like mutation signature (Chan et al., 2015). Based on these observations, A3A and A3B are currently prime candidates as cancer mutators. However, an association of APOBEC1 expression with over-representation of TCW mutations in esophageal cancers (Saraconi et al., 2014) suggests that additional APOBECs may also be involved in mutagenesis in some cancers.
Biochemically, APOBEC family members prefer single stranded (ss) DNA as their substrate (Bransteitter et al., 2003, Suspene et al., 2004, Yu et al., 2004, Chen et al., 2006). Thus, double stranded chromosomal DNA should remain protected from these enzymes. However, normal cellular processes such as transcription, replication, and DNA repair frequently produce ssDNA intermediates that could serve as substrates for APOBEC-induced deamination, leading to the generation of mutations. Members of the human APOBEC family, including activation induced cytidine deaminase (AID) (Chaudhuri et al., 2003, Liu et al., 2008, Peters and Storb, 1996, Ramiro et al., 2003, Taylor et al., 2014) and APOBEC3G (A3G) (Taylor et al., 2014), as well as APOBEC homologs in lamprey (Lada et al., 2015), have all been shown to induce mutations with specificity to transcribed regions. Moreover, many APOBECs have been shown to deaminate ssDNA processively, resulting in the formation of closely spaced ‘mutation clusters’ (Pham et al., 2007). The induction of mutation clusters in yeast through expression of these enzymes is dependent on activity of the uracil DNA glycosylase Ung1, indicating that in addition to transcription intermediates, DNA repair intermediates can also serve as substrates (Taylor et al., 2013). In human cells, an A3A-induced increase in γ-H2AX foci requires cells to be in S-phase (Landry et al., 2011, Narvaiza et al., 2012), suggesting that this enzyme may deaminate ssDNA formed during DNA replication and ultimately induce DNA double strand breaks (DSBs) via subsequent enzymatic processing of the deoxyuridine by base excision repair. Nevertheless, the chromosomal substrates for A3A- and A3B-induced mutagenesis, remain undefined.
Here, we show that A3A and A3B mutagenize chromosomal DNA by deaminating the lagging strand template during DNA replication. Expression of either A3A or A3B in Saccharomyces cerevisiae caused strand-biased mutations in mutation reporters located to each side of a single, well-defined origin of replication. Similar strand-biased mutations were also present surrounding origins throughout the genome as evaluated by whole genome sequencing. A3A- and A3B-induced mutations 5′ of origins were primarily G to A substitutions, while 3′ of origins C to T substitutions predominated. This pattern of mutagenesis is consistent with deamination of ssDNA on the lagging strand template. Genetic deficiencies in proteins involved in replication fork stability (i.e. Rfa1 or Tof1) greatly augmented the mutagenicity of A3A and A3B, as did chemical inhibition of replication by hydroxyurea (HU). Thus, A3A and A3B enzymes may capitalize on the constant proliferation and/or replication stress occurring in cancers to mediate their mutagenic effects.
Results
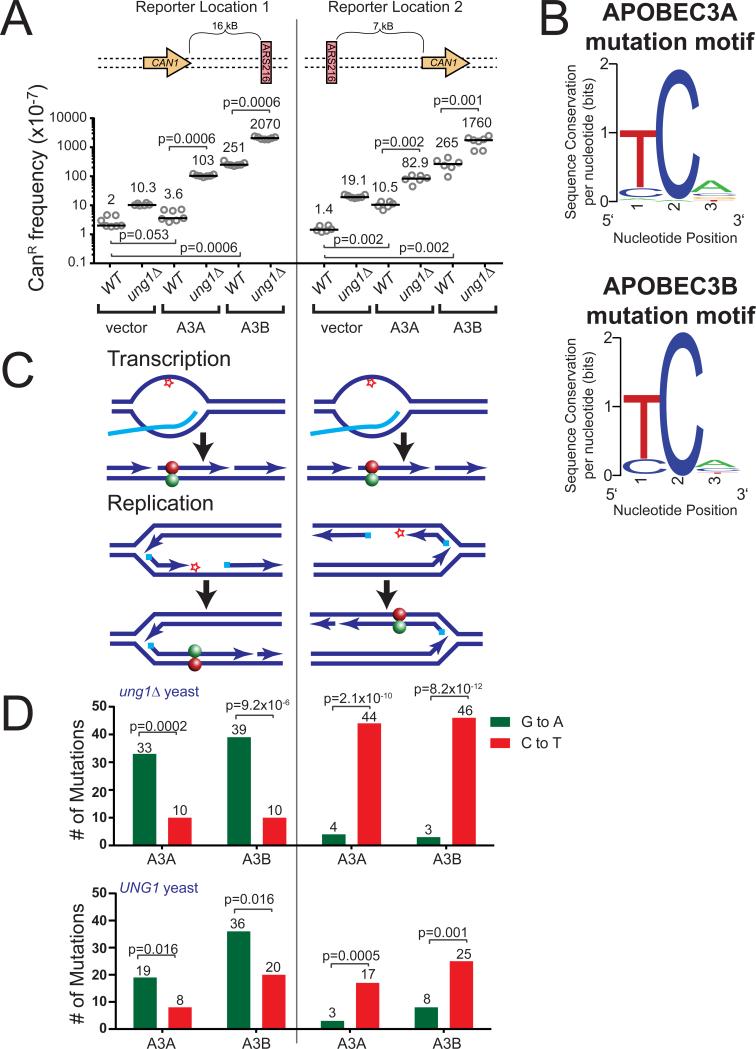
To determine the targets of A3A- and A3B-induced mutagenesis, we generated codon-optimized constructs and expressed these enzymes within strains of the yeast S. cerevisiae, which contained an array of forward mutation reporters (ADE2, URA3, and CAN1) inserted into the LYS2 gene on chromosome II. The A3A and A3B expression constructs produced similar amounts of their respective APOBEC mRNA (Figure S1) and were mutagenic, increasing the CAN1 mutation frequency approximately 2- to 189-fold compared to a vector only control (Figure 1A). As expected, deletion of the uracil-DNA glycosylase, Ung1, in A3A- and A3B-expressing strains further increased the CAN1 mutation frequency 7- to 29-fold, confirming that A3A and A3B expression was inducing mutations by deamination of cytidine to uridine within chromosomal DNA. We subsequently sequenced canavanine-resistant (CanR) clonal isolates from ung1Δ strains (to assess all APOBEC-induced deamination events) to identify the mutations that inactivated CAN1. Both A3A and A3B induced strand-biased mutations within their favored sequence motif (Figure 1B) (i.e. TCW or WGA on the opposite DNA strand). If A3A and A3B target transcriptional intermediates in CAN1, as has been shown for AID and A3G, the direction of CAN1 transcription would render the top DNA strand single stranded and A3A and A3B would produce C to T substitutions (Figure 1C). Surprisingly, both A3A and A3B produced predominantly G to A substitutions (Figure 1D and Table S1). This result indicates that these enzymes deaminated only the bottom DNA strand in the CAN1 gene. Thus, A3A and A3B appear to target a source of ssDNA other than transcription in this region of the yeast genome.
Figure 1. APOBEC3A and 3B induce strand-bias mutations around ARS216.
(A) Frequencies of canavanine-resistance (CanR) induced in WT and ung1Δ yeast following transformation with vector control plasmid, A3A expression plasmid, or A3B expression plasmid. CanR frequency was determined in isogenic strains with the CAN1 gene located either 16 kB 5′ of the origin of replication, ARS216, or 7 kB 3′ of ARS216. Horizontal bars and numeric values indicate the median frequency of six or seven independent replicates. Statistical significance was determined by two-tailed non-parametric Mann-Whitney Rank test. See also Figure S1. (B) Sequence logo describing the favored trinucleotide sequences mutated by A3A (top) and A3B (bottom) in ung1Δ yeast. See also Table S1. (C) Specific deamination of ssDNA formed in a transcription bubble would result in C to T transitions (red ball) within CAN1 regardless of whether the gene is positioned 5′ or 3′ of ARS216. In contrast, deamination of ssDNA formed during lagging strand synthesis would result in G to A substitutions (green ball) 5′ and C to T substitutions 3′ or the origin. (D) The number of G to A (green) and C to T (red) substitutions induced in the CAN1 gene (located 5′ and 3′ of ARS216) by A3A and A3B in ung1Δ and UNG1 yeast as determined by sequencing independent CanR isolates. P-values were determined using a one-tailed g-test goodness-of-fit comparing the ratio of C to T substitutions to G to A substitutions to an expected 1:1 ratio. See also Table S1.
The DNA template present during lagging strand synthesis is an additional source of ssDNA (reviewed in (Balakrishnan and Bambara, 2011)) that may serve as an alternative substrate for A3A and A3B within CAN1. Within this yeast strain, CAN1 is asymmetrically positioned between two neighboring origins of replication, being located 16 kB 5′ of a well-defined origin of replication, ARS216 and 57 kB 3′ of ARS215. Therefore, most replication forks traveling through CAN1 will originate from ARS216 and produce templating ssDNA during lagging strand synthesis on the bottom DNA stand. Consequently, deamination of this strand would result in G to A substitutions, which is consistent with our observed mutation spectrum. To test if A3A and A3B preferentially deaminate ssDNA formed in the lagging strand template, we expressed these enzymes in an isogenic yeast strain previously engineered to investigate the ability of the DNA alkylating agent, methyl methanesulfonate, to induce replication-associated mutation clusters (Roberts et al., 2012). The array of forward mutation reporters in this strain is positioned in an intergenic region 7 kB 3′ of ARS216 and 134 kB 5′ of the neighboring downstream origin, ARS220. Consequently, replication proceeds through CAN1 in this location in the opposite direction, causing lagging strand-associated ssDNA to be formed on the top DNA strand (Figure 1C). Thus, mutations in CAN1 stemming from DNA damage accumulating in replication-associated ssDNA are expected to be complementary base substitutions in each strain. Expression of A3A and A3B caused similar CanR frequencies with the CAN1 gene positioned 3′ of ARS216 (Figure 1A) as was observed with the reporter 5′ of the origin. However, sequencing of CanR mutants in the 3′ location produced a spectrum consisting predominantly of C to T substitutions (Figure 1D and Table S1), which is consistent with replication-associated ssDNA being the primary target of A3A and A3B within this locus. Similar strand-bias mutations consistent with the deamination of ssDNA in the lagging strand template also occurred in UNG proficient cells (Figure 1D and Table S1). This indicates that even with active repair of APOBEC-induced deoxyuridine, the formation of these lesions in replication-associated ssDNA is the major source of mutations in this system.
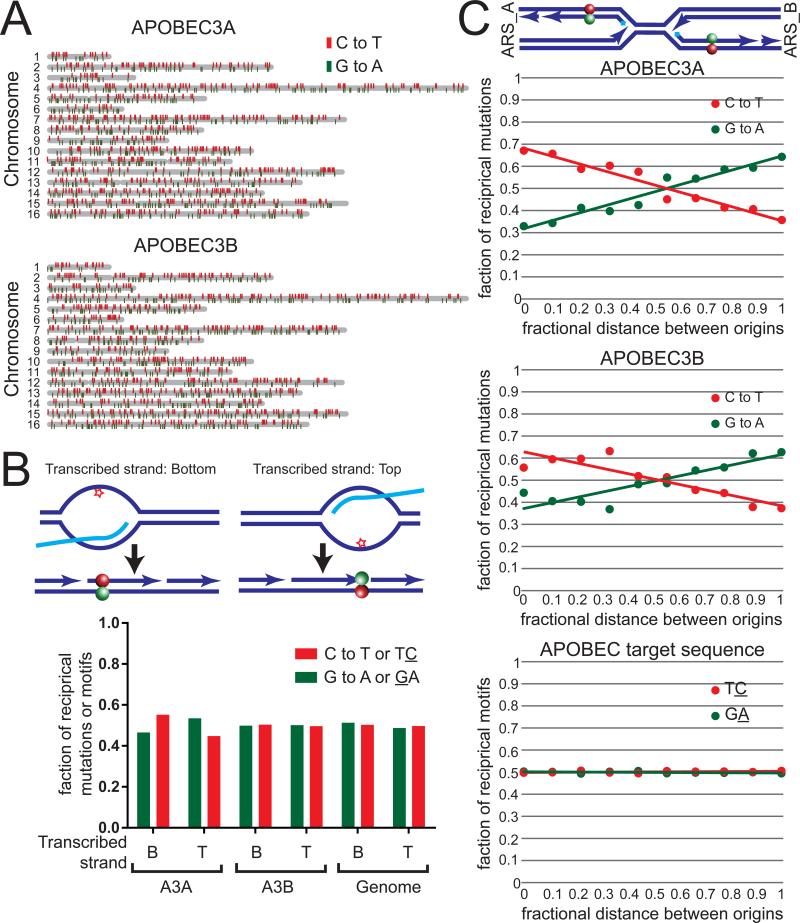
To ensure our results using the CAN1 reporter system were universal and not affected by specifics of the location of the reporter, we next assessed whether A3A and A3B mutagenized the lagging strand template elsewhere in the yeast genome. To do this, we constructed homozygous diploid ung1Δ yeast strains containing A3A and A3B expression cassettes integrated into the LEU2 gene on chromosome III. These stains enable maintenance of the expression constructs without selection, as well as accumulation of a significant number of APOBEC-induced mutations whose distribution is less likely to be altered by purifying selection (Lujan et al., 2014). We subsequently propagated outgrowth lines of these yeast for three months (~900 generations) and sequenced the genomes of independent clonal isolates to determine the location and identity of the APOBEC-induced mutations (Figure 2A and Table S2). Through this prolonged growth, A3A and A3B induced an estimated 0.09 and 0.16 mutations per genome per generation, respectively (Table S2). A3B-induced C to T and G to A mutations occurred in nearly equal abundance within genes transcribed on the top strand as well as in genes transcribed on the bottom strand (Figure 2B), indicating that deamination of the single stranded non-transcribed strand occurs infrequently. A3A expression induced a statistically significant (p<0.02 by two-tailed chi-square test), but slightly favors C to T mutations on the non-transcribed strand, indicating that this enzyme is capable of deaminating transcriptional intermediates at a low level. When we grouped mutations according to their relative distance between neighboring origins, both A3A and A3B predominantly induced C to T substitutions 3′ of origins and G to A substitutions to the 5′ side of origins (Figure 2C). This pattern occurred despite an equal representation of the APOBEC target motif between DNA strands across the genome (Figure 2C), and indicates that the lagging strand template, not the non-transcribed strand of genes, is the primary substrate for A3A and A3B in this system.
Figure 2. Genome-wide strand-bias of A3A and A3B mutations between neighboring origins of replication.
(A) Distribution of C to T (red) and G to A (green) mutations induced by A3A (top) and A3B (bottom). Yeast chromosomes are displayed in gray. (B) The relative abundance of A3A- and A3B-induced C to T mutations (red) and G to A mutations (green) in yeast genes +/− 500 nucleotides transcribed on the bottom strand (B) or the top strand (T). P-values were determined using a two-tailed chi-square test comparing the numbers of each mutation type to the number of A3A- and A3B-targeted TC (red) or GA (green) dinucleotides occurring in these regions. (C) The relative abundance of A3A- and A3B-induced C to T mutations (red) and G to A mutations (green) according to the fractional distance between neighboring replication origins. Lines represent linear trend lines fitting the fractional abundance of C to T (red) and G to A (green) mutations across the entire fractional distance between neighboring origins. A statistical significance of p<0.0001 was determined by comparing the number of each mutation type in each decile bin to the corresponding abundance of TC or GA dinucleotides, by chi-square test. See also Table S2.
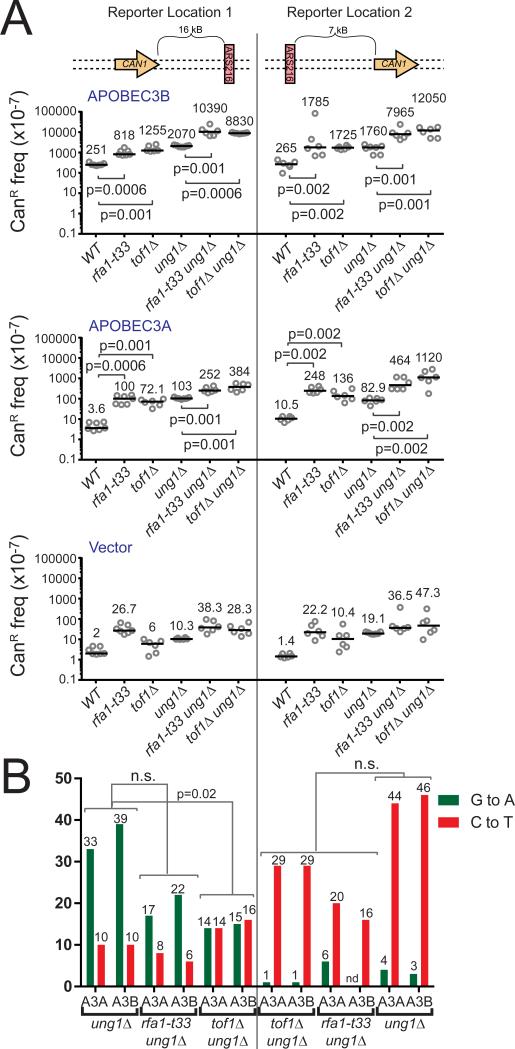
During cancer development, dysfunctional replication forks are common due to hyper-initiation of replication, which results in replication stress [reviewed in (Macheret and Halazonetis, 2015, Hills and Diffley, 2014)]. We therefore evaluated whether the loss of replication fork integrity provided additional opportunity for A3A and A3B to deaminate chromosomal DNA. We first assessed APOBEC-induced mutagenesis within yeast containing a hypomorphic RFA1 allele (t33; S373P). RFA1 encodes the large subunit of the ssDNA binding protein, Replication Protein A (RPA), which binds and protects the lagging strand template during DNA replication. Rfa1-t33 has been shown to have reduced ssDNA binding capacity compared to the wild type protein due to a point mutation in the DNA binding domain (Deng et al., 2014). Expression of either A3A or A3B in rfa1-t33 ung1Δ strains resulted in 2- to 6-fold higher CanR frequencies compared to ung1Δ strains (Figure 3A). The CAN1 mutation spectra on either side of ARS216 in rfa1-t33 ung1Δ strains retained an enrichment of substitutions at TCW sequences as well as strand-biased G to A changes 5′ and C to T changes 3′ of the origin (Figure 3B and Table S1). These results indicate that the increase in A3A- and A3B-induced mutagenesis in rfa1-t33 ung1Δ strains is the result of A3A and A3B acting on a more exposed lagging strand template. Thus, although A3A and A3B can access the lagging strand template, components of the replication fork complex provide some protection against these enzymes during normal replication. Surprisingly, we observed within rfa1-t33 ung1Δ strains a significant increase in the number of CanR mutants containing multiple APOBEC-induced mutations (p=0.01 by Fisher's Exact Test) (Table 1 and Table S1). Some CanR isolates contained up to 6 strand-coordinated mutations which were likely induced in a single event, suggesting that loss of full RPA functionality may facilitate the processivity of APOBEC enzymes (Lada et al., 2011, Pham et al., 2007). We tested an additional null strain to determine whether the increased APOBEC-induced mutagenesis observed in the rfa1-t33 strains is generalizable to any deficiency in replication fork stability. Tof1 functions with Csm3 at the replication fork to prevent uncoupling of the replicative polymerases from the MCM helicase, and deletions of these factors have been shown to result in the accumulation of replication-associated ssDNA in response to HU treatment (Katou et al., 2003). As in rfa1-t33 strains, A3A and A3B both induced 4- to 14-fold more CanR mutants in tof1Δ ung1Δ strains than in ung1Δ strains (Figure 3A) confirming that general loss of replication fork integrity increases the susceptibility of forks to APOBEC-induced deamination. Interestingly, sequencing of CAN1 in the tof1Δ ung1Δ strain revealed strand-biased mutations 3′ of ARS216, while C to T and G to A mutations occurred equally 5′ of the origin (Figure 3B and Table S1). This result suggests that A3A and A3B either gain access to ssDNA formed on both the leading and lagging strands in the absence of Tof1 (Katou et al., 2003) or that altered origin firing and replication fork progression in the tof1 null strains (Tourriere et al., 2005, Hayano et al., 2011) enables more forks originating from ARS215 to replicate the 5′ positioned CAN1 gene.
Figure 3. Deletion of replication fork stability factors exacerbates A3A- and A3B-induced mutagenesis around ARS216.
(A) Frequencies of CanR induced in wild type, rfa1-t33, tof1Δ, ung1Δ, rfa1-t33 ung1Δ, and tof1Δ ung1Δ yeast following transformation with vector control plasmid, A3A expression plasmid, or A3B expression plasmid. Horizontal bars and numeric values indicate the median frequency of six or seven independent replicates. P-values were determined by two-tailed non-parametric Mann-Whitney Rank test. (B) The number of G to A (green) and C to T (red) substitutions induced in the CAN1 gene (located 5′ and 3′ of ARS216) by A3A and A3B in ung1Δ yeast in the genetic backgrounds mentioned in (A) as determined by sequencing independent CanR isolates. P-values were determined using a two-tailed Fisher's Exact test comparing the ratio of C to T substitutions to G to A substitutions between indicated genotypes. See also Table S1.
Table 1.
Summary of CAN1 sequencing results in strains expressing A3A and A3B
| Source of replication stress combined with A3A or A3B expression in ung1Δ strains | ||||
|---|---|---|---|---|
| None | 50mM HU | rfa1-t33 | tof1Δ | |
| Total CanR mutants sequenced | 190 | 122 | 108 | 120 |
| CanR mutants with APOBEC signature mutation(s) | 188 | 116 | 106 | 120 |
| Frequency of multiple strand-coordinated APOBEC-signature mutations | 0.011 | 0.025 | 0.056 | 0.008 |
| Number of mutations present in each CanR mutant with multiple strand-coordinated APOBEC-signature mutations | 2, 2 | 2, 2, 6 | 2, 2, 2, 2, 2, 5, 6 | 2 |
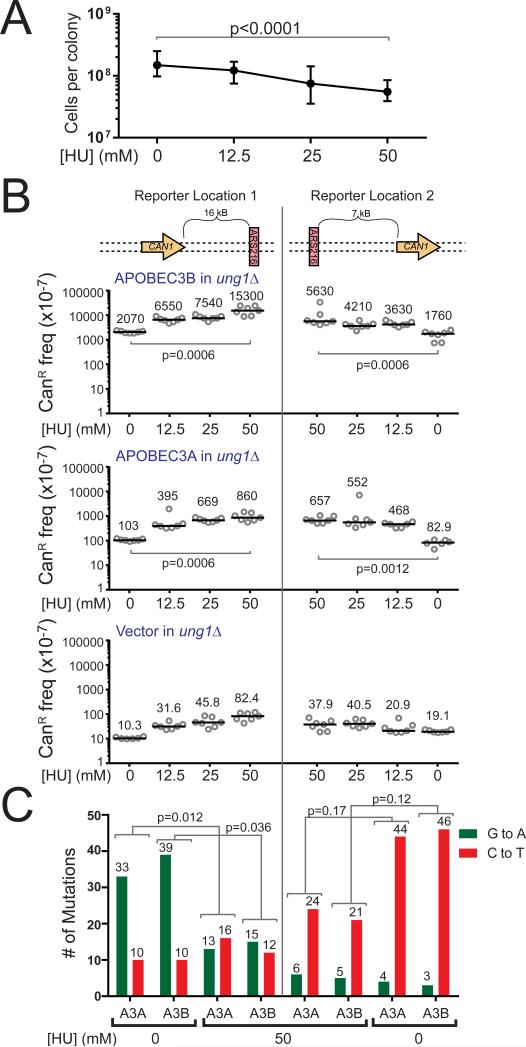
In addition to genetic replication fork defects, replication stress can be induced by exposure to drugs like HU (Zeman and Cimprich, 2014). HU inhibits the activity of ribonucleotide reductase, resulting in decreased deoxyribonucleotide pools and slowed DNA synthesis (Bianchi et al., 1986, Slater, 1973). We therefore decided to test whether replication stress with a fully competent replisome could augment APOBEC mutagenesis. We treated ung1Δ yeast strains containing either a vector control plasmid or A3A and A3B expression plasmids with HU for three days. Over this period, HU treated yeast displayed a dose-dependent decrease in cell growth indicating a slowing of replication and induction of replication stress (Figure 4A). In ung1Δ strains containing either A3A or A3B, HU treatment dramatically increased mutagenesis up to 8-fold over non-treated controls and 10- to 186-fold over corresponding vector transformed ung1Δ strains (Figure 4B). The mutation spectra in HU-treated strains maintained a strand-biased signature consistent with A3A and A3B mutagenizing the lagging strand template 3′ of ARS216. As seen in the tof1Δ ung1Δ strains, however, the mutational strand-bias 5′ of the origin was lost (Figure 4C and Table S1). This loss of strand-bias could result from HU treatment inducing ssDNA on both the leading and lagging strands through the functional uncoupling of the MCM helicase and replicative polymerases. However, for the HU treatment to differentially alter the mutation spectra in the two CAN1 locations, ssDNA would have to be formed in the lagging strand in a region specific manner. Alternatively, the loss of strand bias 5′ of ARS216 could be due to HU disrupting the timing of origin firing and the kinetics of replication (Poli et al., 2012), which could allow more forks originating from ARS215 to replicate CAN1. The synergistic elevation of CAN1 mutation frequency in HU treated yeast expressing A3A or A3B thus likely resulted from HU treatment increasing the amount of ssDNA substrate available to these enzymes. As in rfa1-t33 ung1Δ strains, several CanR isolates contained multiple strand-coordinated mutations within CAN1 (Table 1 and Table S1), reminiscent of previously reported DNA damage-induced mutation clusters and the APOBEC-induced kataegis events observed in human tumors (Nik-Zainal et al., 2012, Roberts et al., 2012). Therefore, chemically-induced replication stress greatly augments APOBEC mutagenesis, suggesting that other forms of replication stress that occur during carcinogenesis may likewise facilitate the extensive editing of tumor genomes by these enzymes.
Figure 4. Chemically-induced replication stress augments A3A- and A3B-induced mutagenesis.
(A) HU treatment causes a dose-dependent decrease in cell growth in ung1Δ yeast. Shown are medians and ranges for 18 independent replicates at each HU dose. P-values were determined by two-tailed non-parametric Mann-Whitney Rank test comparing untreated yeast to yeast treated with 50 mM HU. (B) Frequencies of CanR induced in ung1Δ yeast treated with 0, 12.5, 25, or 50 mM HU following transformation with vector control plasmid, A3A expression plasmid, or A3B expression plasmid. Horizontal bars and numeric values indicate the median frequency of six or seven independent replicates. P-values were determined by two-tailed non-parametric Mann-Whitney Rank test. (C) The number of G to A (green) and C to T (red) substitutions induced in the CAN1 gene by A3A and A3B in untreated or 50mM HU treated ung1Δ yeast as determined by sequencing independent CanR isolates. P-values were determined using a two-tailed Fisher's Exact test comparing the ratio of C to T substitutions to G to A substitutions between indicated treatments. See also Table S1.
Discussion
Using both mutation reporter systems and whole genome sequencing, we have shown that A3A and A3B, the two most likely enzymes involved in the APOBEC-induced mutagenesis observed in cancer, strongly mutagenize the lagging strand template during DNA replication. In addition, mutations identified by whole genome sequencing lacked a strong transcriptional strand-bias. This observation indicates that A3A- and A3B-induced deamination of transcriptional intermediates is limited within this system and that ssDNA formed during lagging strand synthesis is the primary substrate for these enzymes. Other APOBEC enzymes have been shown to be capable of deaminating ssDNA formed during transcription. AID-induced deamination of immunoglobulin genes is dependent on transcription of the immunoglobulin locus (Peters and Storb, 1996) and AID-induced mutation frequencies can be greatly elevated by increasing the transcription level of mutation reporters in E. coli (Ramiro et al., 2003). In biochemical assays, AID, A3G, and A3A deamination can be made dependent on transcription of the target DNA (Pham et al., 2003, Pham et al., 2007, Pham et al., 2013, Love et al., 2012). Moreover, expression of AID or A3G in yeast yields an enrichment of mutations at active RNA polymerase pre-initiation sites (Taylor et al., 2014). These effects may be facilitated by extremely high levels of transcription, pausing of RNA polymerase, and formation of R-loops and secondary structures that would increase the size and time a transcribed region is single stranded. As such instances are likely limited to specific subsets of genes, the contribution of transcription-associated ssDNA to the overall distribution of APOBEC-induced mutations may be small. Supporting this, our global analysis of A3A- and A3B-induced mutations detected a small, but significant transcriptional strand-bias for A3A-induced mutations. Moreover, recent whole genome analyses of A3G-induced mutations in E. coli likewise observed a predominance of mutations originating from the deamination of cytidine in the lagging strand template over the non-transcribed strands of genes, regardless of uracil DNA glycosylase status (Bhagwat et al., 2016).
The mechanistic basis for how A3A and A3B choose their substrates is currently unclear. AID's specificity for transcription-associated ssDNA appears to be facilitated by interactions with multiple proteins present in the transcription apparatus (Chaudhuri et al., 2004, Pavri et al., 2010, Willmann et al., 2012) as well as long non-coding RNAs transcribed from the immunoglobulin switch regions (Zheng et al., 2015). In the absence of direct recruitment to a specific ssDNA substrate, the activity of the majority of APOBECs may be dictated by the abundance and persistence of the ssDNA intermediate. As transcription bubbles are relatively small (~10-20 nucleotides) (Pal et al., 2005, Choder and Aloni, 1988), APOBEC mutagenesis of transcription intermediates would appear to be limited. In comparison, both lagging strand synthesis and the homology-directed repair (HR) of DSBs produce significantly larger stretches of ssDNA (approximately 200 nucleotides [(Smith and Whitehouse, 2012) and references within] and 1-2 kilobases (Zhou et al., 2014, Chung et al., 2010), respectively) that A3A and A3B can mutagenize. Although HR resection tracks may produce more ssDNA per single DSB than the synthesis of one Okazaki fragment, the number of DSBs in a cell is likely very small compared to the number of Okazaki fragments formed during each round of DNA replication. Thus, the lagging strand template the favored substrate of multiple APOBEC enzymes based solely on the amount of ssDNA generated.
The susceptibility of ssDNA replication intermediates to A3A and A3B may be one of the key factors enabling the extensive APOBEC mutagenesis observed in many cancer types. The mutation spectra of other cancer mutagens, like UV light and tobacco smoke, display transcription-associated strand-biases presumably due to the favored repair of the transcribed stand by transcription-coupled nucleotide excision repair (Alexandrov et al., 2013). However, a small transcription related strand-bias among APOBEC mutations has only been observed specifically in highly expressed genes in two bladder cancer patients (Nordentoft et al., 2014). Recent analyses of the distribution of APOBEC signature mutations in breast and lung cancers have revealed that these mutations are enriched in gene-rich, early replicating regions of the genome (Kazanov et al., 2015). However, within these regions, APOBEC-induced mutations occurred equally in transcribed and non-transcribed DNA as well as on the transcribed and non-transcribed strands. More extensive computational analysis of 590 whole genome sequenced tumors from 14 tumor types revealed that overall APOBEC signature cancer mutations display strand-bias, but that these biases are more consistent with the enzymes deaminating cytidines within ssDNA on the lagging strand template than deamination of the non-transcribed strand during transcription (Haradhvala et al., 2016). This suggests that as in yeast and E. coli, replication and not transcription is the predominant APOBEC substrate within human tumors.
APOBEC-induced mutations could thus accumulate simply because of the large number of cell divisions and accompanying DNA replication that highly proliferative cancer cells undergo (Shibata and Lieber, 2010). Replication stress conditions that occur frequently in tumors likely further exacerbate this effect. Activation of oncogenes like Cyclin E or H-Ras during cancer development dysregulate the coordination of replication origin firing [reviewed in (Macheret and Halazonetis, 2015, Hills and Diffley, 2014)]. Consequently, many tumor cells over-replicate their genomes, which can result in the exhaustion of factors that maintain fork stability, and formation of even higher amounts of ssDNA (Toledo et al., 2013). Our results indicate that genetic disruption of replication fork integrity or chemical induction of replication stress facilitates A3A and A3B mutagenesis, indicating that oncogene-induced replication stress may do so as well. Supporting this, human tumors likely experiencing replication stress due to the mutation or silencing of the tumor-suppressor, FHIT, have reportedly higher numbers of APOBEC signature mutations in lung adenocarcinoma (Waters et al., 2015). Additionally, overexpression of the breast cancer-associated oncogene human epidermal growth factor-2 (Her2) in mice elevates DNA damage response signaling in tumors, which is an indicator of increased replication stress (Reddy et al., 2010). This observation coupled with our results indicating that replication stress increases A3A- and A3B-induced mutagenesis offer a possible explanation for the fact that breast cancers experiencing the overexpression of Her2 have been shown to have more APOBEC-signature mutations than other breast cancer subtypes (Roberts et al., 2013). Thus, the dysregulation of APOBEC expression and significant availability of their ideal substrate due to hyper-replication may produce a “perfect storm” enabling extreme mutagenesis in human cancer.
Experimental Procedures
Construction of Expression Plasmids
The expression plasmids used in this study were constructed from the centromeric yeast expression plasmid, pCM252 (Belli et al., 1998). The TRP1 selectable marker of pCM252 was exchanged for hygromycin resistance by co-transforming yeast with the plasmid and a HygroMX cassette, PCR amplified with the forward and reverse primers, 5′-AGGGCATTGGTGACTATTGAGCACGTGAGTATACGTGATTAAGCACACAAAGGCAGCTTGG AGTCGTACGCTGCAGGTCGAC and 5′-TAATCTAAGCGCATCACCAACATTTTCTGGCGTCAGTCCACCAGCTAACATAAAATGTAAGC TTCGATGAATTCGAGCTCG. Hygromycin-resistant, tryptophan-auxotrophic transformants where selected to identify isolates containing recombined plasmid (named pySR419) where the HygroMX cartridge had replaced TRP1 through homologous recombination. Pure plasmid was subsequently isolated and proper integration of HygroMX verified by restriction digest. A3A and A3B cDNAs were codon-optimized for expression in yeast and synthesized as gene blocks with appended 5′StuI and 3′ClaI restriction sites (DNA2.0, Menlo Park, CA). These gene blocks were digested, cloned into the StuI and ClaI sites of pySR419, and the resulting plasmids were sequenced to verify the integrity of A3A and A3B.
Yeast Strains and Culture
All yeast strain utilized in this study were constructed in the CG379 genetic background, a derivative of S288C (Morrison et al., 1991). Detailed descriptions of strain constructions can be found the supplemental experimental procedures. The genotypes of each strain used is listed in Table S3. Yeast were grown using standard techniques described in (Sherman et al., 1986) on standard rich media (YPDA) or synthetic complete (SC) media.
Determination of Mutation Frequencies
For all experiments measuring APOBEC-induced mutations, A3A and A3B were expressed at basal levels (i.e. without induction) from a tetracycline inducible promoter. To determine CanR frequencies for clonal strains transformed with either an empty vector control, A3A expression plasmid, or A3B expression plasmid, ~200 yeast cells were plated on YPDA media supplemented either with hygromycin (to select for maintenance of the plasmid) or hygromycin and HU and grown at 30°C for three days. Afterward, independent colonies were re-suspended in water and plated on synthetic complete media or SC-arginine supplemented with 0.006% canavanine (to select for CAN1 mutants) at appropriate dilutions to yield clearly independent colonies. Colonies were allowed to grow on selective and complete media for three days at 30°C and subsequently imaged and counted using Quantity One software (Bio-Rad, Hercules, CA). The frequency of CanR mutants was calculated using the4 following formula:
The medians of each the independent replicates were compared pairwise between different experimental conditions using the Mann-Whitney Rank Test.
CAN1 Mutation Spectra
Following three days growth at 30°C on YPDA media containing either hygromycin or hygromycin and HU, independent colonies of ung1Δ strains transformed with either A3A or A3B expression constructs were replica plated to SC-arginine media supplemented with 0.006% canavanine and allowed to grow at 30°C for an additional three days. A single CanR papillae was selected per independent colony and was additionally clonally isolated by single cell streaking. Genomic DNA was isolated from these clones and the CAN1 gene was PCR amplified (primers listed in Table S4). CAN1 PCR products were Sanger sequenced (Eton Biosciences, San Diego, CA and GenScript, Piscataway, NJ) and mutations inactivating CAN1 (Table S1) identified using the Geneious software package (Biomatters Limited). The preferred trinucleotide sequences mutated in CAN1 following A3A or A3B expression were generated using webLogo (http://weblogo.berkeley.edu/logo.cgi) (Crooks et al., 2004).
Whole Genome Sequencing and Mutation Calling
The accumulation of APOBEC-induced mutations was conducted similar to (Lujan et al., 2014). Briefly, ~200 diploid yeast cells deficient in ung1 and containing A3A or A3B integrated into the LEU2 locus of chromosome III were plated on rich media and grown at 30°C for three days. 10-20 independent colonies for each yeast strain were selected and passaged through bottlenecks for three months (~900 generations) on rich media at 30°C to establish outgrowth lines. After the three month growth period, a single clonal isolate from each line was obtained. Total genomic DNA was isolated from these clones as well as from a sample of the bulk yeast prior to passaging (time zero). These genomic DNAs were subjected to high throughput paired-end sequencing similar to (Roberts et al., 2012) on an Illumina HiSeq2500 (San Diego, CA). Raw sequencing reads can be accessed from the NCBI short read archive database under accession number SRP067952. Paired-end reads were mapped with CLC genomics Workbench 7.5 (Qiagen) using default settings at greater than 50x coverage to a reference of ySR128 based on the publically available S288C reference (obtained from the Saccharomyces Genome Database, http://www.yeastgenome.org) and constructed as in (Roberts et al., 2012). Reads mapping to multiple locations in the genome were discarded. Mutations were called similarly to (Sakofsky et al., 2014). Briefly, homozygous and heterozygous mutations were identified as base differences relative to the reference sequence that occurred in greater than 45% of reads. All mutations were covered by at least 9 reads. Due to being likely mapping artifacts, mutations occurring in regions annotated as “Repeat Regions” and “LTR” in the S288C S. cerevisiae reference were removed from analysis as were mutations that occur at positions where mapping of reads from a non-mutagenized reference strain resulted in greater than 20% of reads containing non-reference sequence. Mutations that occurred in strains prior to outgrowth (identified as mutations occurring in the time zero sample) were also removed.
Analyses of Whole Genome Mutation Distribution
The position of genes and origins of replication were obtained from the annotations in the S288C reference UCSC sacCer2 (downloaded from NCBI on June 16, 2008) and converted into the coordinates of the ySR128 reference. APOBEC-induced mutations were placed into groups based upon whether they occurred in genes transcribed on the top or bottom DNA strand, and their location in relationship to the percent distance between neighboring origins. The number of C to T substitutions was compared to the number of G to A substitutions and the frequency of the favored APOBEC target sequence.
Statistical analyses
All statistical analysis were conducted as described in the text using the Graphpad Prism software.
Supplementary Material
Acknowledgements
The authors would like to thank Drs. Dmitry Gordenin and Dale Ramsden for advice and helpful comments, Dr. Levi O'Loughlin for aid in passaging APOBEC expressing yeast, Dr. Yiyong Liu for preliminary Illumina sequencing of propagated yeast strains, and Dr. John Wyrick for providing the constructs to edit yeast with the CRISPR/Cas9 system. This work is supported by an NIH grant R00ES022633-03 from NIEHS awarded to S.A.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors contributions:
L.C., J.H., T.M., and S.A.R. designed the experiments, executed them, and wrote the manuscript. E.P.M and P.A.M. performed whole-genome sequencing. S.A.R. oversaw the project.
References
- ALEXANDROV LB, NIK-ZAINAL S, WEDGE DC, APARICIO SA, BEHJATI S, BIANKIN AV, BIGNELL GR, BOLLI N, BORG A, BORRESEN-DALE AL, BOYAULT S, BURKHARDT B, BUTLER AP, CALDAS C, DAVIES HR, DESMEDT C, EILS R, EYFJORD JE, FOEKENS JA, GREAVES M, HOSODA F, HUTTER B, ILICIC T, IMBEAUD S, IMIELINSK M, JAGER N, JONES DT, JONES D, KNAPPSKOG S, KOOL M, LAKHANI SR, LOPEZ-OTIN C, MARTIN S, MUNSHI NC, NAKAMURA H, NORTHCOTT PA, PAJIC M, PAPAEMMANUIL E, PARADISO A, PEARSON JV, PUENTE XS, RAINE K, RAMAKRISHNA M, RICHARDSON AL, RICHTER J, ROSENSTIEL P, SCHLESNER M, SCHUMACHER TN, SPAN PN, TEAGUE JW, TOTOKI Y, TUTT AN, VALDES-MAS R, VAN BUUREN MM, VAN 'T VEER L, VINCENT-SALOMON A, WADDELL N, YATES LR, ZUCMAN-ROSSI J, FUTREAL PA, MCDERMOTT U, LICHTER P, MEYERSON M, GRIMMOND SM, SIEBERT R, CAMPO E, SHIBATA T, PFISTER SM, CAMPBELL PJ, STRATTON MR. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BALAKRISHNAN L, BAMBARA RA. Eukaryotic lagging strand DNA replication employs a multi-pathway mechanism that protects genome integrity. J Biol Chem. 2011;286:6865–70. doi: 10.1074/jbc.R110.209502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BELLI G, GARI E, PIEDRAFITA L, ALDEA M, HERRERO E. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 1998;26:942–7. doi: 10.1093/nar/26.4.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BHAGWAT AS, HAO W, TOWNES JP, LEE H, TANG H, FOSTER PL. Strand-biased Cytosine deamination at the Replication Fork causes Cytosine to Thymine Mutations in Escherichia coli. PNAS. 2016 doi: 10.1073/pnas.1522325113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BIANCHI V, PONTIS E, REICHARD P. Changes of deoxyribonucleoside triphosphate pools induced by hydroxyurea and their relation to DNA synthesis. J Biol Chem. 1986;261:16037–42. [PubMed] [Google Scholar]
- BOGERD HP, WIEGAND HL, HULME AE, GARCIA-PEREZ JL, O'SHEA KS, MORAN JV, CULLEN BR. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci U S A. 2006;103:8780–5. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRANSTEITTER R, PHAM P, SCHARFF MD, GOODMAN MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci U S A. 2003;100:4102–7. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BURNS MB, LACKEY L, CARPENTER MA, RATHORE A, LAND AM, LEONARD B, REFSLAND EW, KOTANDENIYA D, TRETYAKOVA N, NIKAS JB, YEE D, TEMIZ NA, DONOHUE DE, MCDOUGLE RM, BROWN WL, LAW EK, HARRIS RS. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013a;494:366–70. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BURNS MB, TEMIZ NA, HARRIS RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nature genetics. 2013b;45:977–983. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAVAL V, SUSPENE R, SHAPIRA M, VARTANIAN JP, WAIN-HOBSON S. A prevalent cancer susceptibility APOBEC3A hybrid allele bearing APOBEC3B 3'UTR enhances chromosomal DNA damage. Nat Commun. 2014;5:5129. doi: 10.1038/ncomms6129. [DOI] [PubMed] [Google Scholar]
- CHAN K, ROBERTS SA, KLIMCZAK LJ, STERLING JF, SAINI N, MALC EP, KIM J, KWIATKOWSKI DJ, FARGO DC, MIECZKOWSKI PA, GETZ G, GORDENIN DA. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat Genet. 2015;47:1067–72. doi: 10.1038/ng.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHAUDHURI J, KHUONG C, ALT FW. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 2004;430:992–8. doi: 10.1038/nature02821. [DOI] [PubMed] [Google Scholar]
- CHAUDHURI J, TIAN M, KHUONG C, CHUA K, PINAUD E, ALT FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–30. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- CHEN H, LILLEY CE, YU Q, LEE DV, CHOU J, NARVAIZA I, LANDAU NR, WEITZMAN MD. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol. 2006;16:480–5. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- CHODER M, ALONI Y. RNA polymerase II allows unwinding and rewinding of the DNA and thus maintains a constant length of the transcription bubble. J Biol Chem. 1988;263:12994–3002. [PubMed] [Google Scholar]
- CHUNG WH, ZHU Z, PAPUSHA A, MALKOVA A, IRA G. Defective resection at DNA double-strand breaks leads to de novo telomere formation and enhances gene targeting. PLoS Genet. 2010;6:e1000948. doi: 10.1371/journal.pgen.1000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CROOKS GE, HON G, CHANDONIA JM, BRENNER SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–90. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DENG SK, GIBB B, DE ALMEIDA MJ, GREENE EC, SYMINGTON LS. RPA antagonizes microhomology-mediated repair of DNA double-strand breaks. Nat Struct Mol Biol. 2014;21:405–12. doi: 10.1038/nsmb.2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARADHVALA N, POLAK P, STOJANOV P, COVINGTON KR, SHINBROT E, HESS J, RHEINBAY E, KIM J, MARUVKA Y, BRAUNSTEIN LZ, KAMBUROV A, HANAWALT PC, WHEELER DA, KOREN A, LAWRENCE MS, GETZ G. Mutational strand asymmetries across cancer reveal mechanisms of DNA damage and repair. Cell. 2016 doi: 10.1016/j.cell.2015.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAYANO M, KANOH Y, MATSUMOTO S, MASAI H. Mrc1 marks early-firing origins and coordinates timing and efficiency of initiation in fission yeast. Mol Cell Biol. 2011;31:2380–91. doi: 10.1128/MCB.01239-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HILLS SA, DIFFLEY JF. DNA replication and oncogene-induced replicative stress. Curr Biol. 2014;24:R435–44. doi: 10.1016/j.cub.2014.04.012. [DOI] [PubMed] [Google Scholar]
- JACKSON SP, BARTEK J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–8. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KATOU Y, KANOH Y, BANDO M, NOGUCHI H, TANAKA H, ASHIKARI T, SUGIMOTO K, SHIRAHIGE K. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424:1078–83. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- KAZANOV MD, ROBERTS SA, POLAK P, STAMATOYANNOPOULOS J, KLIMCZAK LJ, GORDENIN DA, SUNYAEV SR. APOBEC-Induced Cancer Mutations Are Uniquely Enriched in Early-Replicating, Gene-Dense, and Active Chromatin Regions. Cell Rep. 2015;13:1103–9. doi: 10.1016/j.celrep.2015.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIDD JM, NEWMAN TL, TUZUN E, KAUL R, EICHLER EE. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 2007;3:e63. doi: 10.1371/journal.pgen.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOMATSU A, NAGASAKI K, FUJIMORI M, AMANO J, MIKI Y. Identification of novel deletion polymorphisms in breast cancer. Int J Oncol. 2008;33:261–70. [PubMed] [Google Scholar]
- LADA AG, KLIVER SF, DHAR A, POLEV DE, MASHARSKY AE, ROGOZIN IB, PAVLOV YI. Disruption of Transcriptional Coactivator Sub1 Leads to Genome-Wide Re-distribution of Clustered Mutations Induced by APOBEC in Active Yeast Genes. PLoS Genet. 2015;11:e1005217. doi: 10.1371/journal.pgen.1005217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LADA AG, WAISERTREIGER IS, GRABOW CE, PRAKASH A, BORGSTAHL GE, ROGOZIN IB, PAVLOV YI. Replication protein A (RPA) hampers the processive action of APOBEC3G cytosine deaminase on single-stranded DNA. PLoS One. 2011;6:e24848. doi: 10.1371/journal.pone.0024848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LANDRY S, NARVAIZA I, LINFESTY DC, WEITZMAN MD. APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep. 2011;12:444–50. doi: 10.1038/embor.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU M, DUKE JL, RICHTER DJ, VINUESA CG, GOODNOW CC, KLEINSTEIN SH, SCHATZ DG. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–5. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- LOVE RP, XU H, CHELICO L. Biochemical Analysis of Hypermutation by the Deoxycytidine Deaminase APOBEC3A. Journal of Biological Chemistry. 2012;287:30812–30822. doi: 10.1074/jbc.M112.393181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUJAN SA, CLAUSEN AR, CLARK AB, MACALPINE HK, MACALPINE DM, MALC EP, MIECZKOWSKI PA, BURKHOLDER AB, FARGO DC, GORDENIN DA, KUNKEL TA. Heterogeneous polymerase fidelity and mismatch repair bias genome variation and composition. Genome Res. 2014;24:1751–64. doi: 10.1101/gr.178335.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MACHERET M, HALAZONETIS TD. DNA replication stress as a hallmark of cancer. Annu Rev Pathol. 2015;10:425–48. doi: 10.1146/annurev-pathol-012414-040424. [DOI] [PubMed] [Google Scholar]
- MORRISON A, BELL JB, KUNKEL TA, SUGINO A. Eukaryotic DNA polymerase amino acid sequence required for 3'----5' exonuclease activity. Proc Natl Acad Sci U S A. 1991;88:9473–7. doi: 10.1073/pnas.88.21.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NARVAIZA I, LANDRY S, WEITZMAN MD. APOBEC3 proteins and genomic stability: the high cost of a good defense. Cell Cycle. 2012;11:33–8. doi: 10.4161/cc.11.1.18706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIK-ZAINAL S, ALEXANDROV LB, WEDGE DC, VAN LOO P, GREENMAN CD, RAINE K, JONES D, HINTON J, MARSHALL J, STEBBINGS LA, MENZIES A, MARTIN S, LEUNG K, CHEN L, LEROY C, RAMAKRISHNA M, RANCE R, LAU KW, MUDIE LJ, VARELA I, MCBRIDE DJ, BIGNELL GR, COOKE SL, SHLIEN A, GAMBLE J, WHITMORE I, MADDISON M, TARPEY PS, DAVIES HR, PAPAEMMANUIL E, STEPHENS PJ, MCLAREN S, BUTLER AP, TEAGUE JW, JONSSON G, GARBER JE, SILVER D, MIRON P, FATIMA A, BOYAULT S, LANGEROD A, TUTT A, MARTENS JW, APARICIO SA, BORG A, SALOMON AV, THOMAS G, BORRESEN-DALE AL, RICHARDSON AL, NEUBERGER MS, FUTREAL PA, CAMPBELL PJ, STRATTON MR. Mutational Processes Molding the Genomes of 21 Breast Cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NORDENTOFT I, LAMY P, BIRKENKAMP-DEMTRODER K, SHUMANSKY K, VANG S, HORNSHOJ H, JUUL M, VILLESEN P, HEDEGAARD J, ROTH A, THORSEN K, HOYER S, BORRE M, REINERT T, FRISTRUP N, DYRSKJOT L, SHAH S, PEDERSEN JS, ORNTOFT TF. Mutational context and diverse clonal development in early and late bladder cancer. Cell Rep. 2014;7:1649–63. doi: 10.1016/j.celrep.2014.04.038. [DOI] [PubMed] [Google Scholar]
- PAL M, PONTICELLI AS, LUSE DS. The role of the transcription bubble and TFIIB in promoter clearance by RNA polymerase II. Mol Cell. 2005;19:101–10. doi: 10.1016/j.molcel.2005.05.024. [DOI] [PubMed] [Google Scholar]
- PAVRI R, GAZUMYAN A, JANKOVIC M, DI VIRGILIO M, KLEIN I, ANSARAHSOBRINHO C, RESCH W, YAMANE A, REINA SAN-MARTIN B, BARRETO V, NIELAND TJ, ROOT DE, CASELLAS R, NUSSENZWEIG MC. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143:122–33. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PETERS A, STORB U. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 1996;4:57–65. doi: 10.1016/s1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- PHAM P, BRANSTEITTER R, PETRUSKA J, GOODMAN MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–7. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- PHAM P, CHELICO L, GOODMAN MF. DNA deaminases AID and APOBEC3G act processively on single-stranded DNA. DNA Repair (Amst) 2007;6:689–92. doi: 10.1016/j.dnarep.2007.01.001. author reply 693-4. [DOI] [PubMed] [Google Scholar]
- PHAM P, LANDOLPH A, MENDEZ C, LI N, GOODMAN MF. A biochemical analysis linking APOBEC3A to disparate HIV-1 restriction and skin cancer. J Biol Chem. 2013;288:29294–304. doi: 10.1074/jbc.M113.504175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POLI J, TSAPONINA O, CRABBE L, KESZTHELYI A, PANTESCO V, CHABES A, LENGRONNE A, PASERO P. dNTP pools determine fork progression and origin usage under replication stress. EMBO J. 2012;31:883–94. doi: 10.1038/emboj.2011.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAMIRO AR, STAVROPOULOS P, JANKOVIC M, NUSSENZWEIG MC. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nature immunology. 2003;4:452–456. doi: 10.1038/ni920. [DOI] [PubMed] [Google Scholar]
- REDDY JP, PEDDIBHOTLA S, BU W, ZHAO J, HARICHARAN S, DU YC, PODSYPANINA K, ROSEN JM, DONEHOWER LA, LI Y. Defining the ATM-mediated barrier to tumorigenesis in somatic mammary cells following ErbB2 activation. Proc Natl Acad Sci U S A. 2010;107:3728–33. doi: 10.1073/pnas.0910665107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REFSLAND EW, HARRIS RS. The APOBEC3 family of retroelement restriction factors. Curr Top Microbiol Immunol. 2013;371:1–27. doi: 10.1007/978-3-642-37765-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROBERTS SA, LAWRENCE MS, KLIMCZAK LJ, GRIMM SA, FARGO D, STOJANOV P, KIEZUN A, KRYUKOV GV, CARTER SL, SAKSENA G, HARRIS S, SHAH RR, RESNICK MA, GETZ G, GORDENIN DA. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–6. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROBERTS SA, STERLING J, THOMPSON C, HARRIS S, MAV D, SHAH R, KLIMCZAK LJ, KRYUKOV GV, MALC E, MIECZKOWSKI PA, RESNICK MA, GORDENIN DA. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Molecular cell. 2012;46:424–35. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAKOFSKY CJ, ROBERTS SA, MALC E, MIECZKOWSKI PA, RESNICK MA, GORDENIN DA, MALKOVA A. Break-induced replication is a source of mutation clusters underlying kataegis. Cell Rep. 2014;7:1640–8. doi: 10.1016/j.celrep.2014.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SARACONI G, SEVERI F, SALA C, MATTIUZ G, CONTICELLO SG. The RNA editing enzyme APOBEC1 induces somatic mutations and a compatible mutational signature is present in esophageal adenocarcinomas. Genome Biol. 2014;15:417. doi: 10.1186/s13059-014-0417-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHERMAN F, FINK GF, HICKS JB. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor; New York: 1986. [Google Scholar]
- SHIBATA D, LIEBER MR. Is there any genetic instability in human cancer? DNA Repair (Amst) 2010;9:858. doi: 10.1016/j.dnarep.2010.04.011. discussion 859-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SLATER ML. Effect of reversible inhibition of deoxyribonucleic acid synthesis on the yeast cell cycle. J Bacteriol. 1973;113:263–70. doi: 10.1128/jb.113.1.263-270.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SMITH DJ, WHITEHOUSE I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 2012;483:434–8. doi: 10.1038/nature10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUSPENE R, SOMMER P, HENRY M, FERRIS S, GUETARD D, POCHET S, CHESTER A, NAVARATNAM N, WAIN-HOBSON S, VARTANIAN JP. APOBEC3G is a single-stranded DNA cytidine deaminase and functions independently of HIV reverse transcriptase. Nucleic acids research. 2004;32:2421–9. doi: 10.1093/nar/gkh554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAYLOR BJ, NIK-ZAINAL S, WU YL, STEBBINGS LA, RAINE K, CAMPBELL PJ, RADA C, STRATTON MR, NEUBERGER MS. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. Elife. 2013;2:e00534. doi: 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAYLOR BJ, WU YL, RADA C. Active RNAP pre-initiation sites are highly mutated by cytidine deaminases in yeast, with AID targeting small RNA genes. Elife. 2014;3:e03553. doi: 10.7554/eLife.03553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TENG B, BURANT CF, DAVIDSON NO. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science. 1993;260:1816–9. doi: 10.1126/science.8511591. [DOI] [PubMed] [Google Scholar]
- THIELEN BK, MCNEVIN JP, MCELRATH MJ, HUNT BV, KLEIN KC, LINGAPPA JR. Innate immune signaling induces high levels of TC-specific deaminase activity in primary monocyte-derived cells through expression of APOBEC3A isoforms. The Journal of biological chemistry. 2010;285:27753–66. doi: 10.1074/jbc.M110.102822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TOLEDO LI, ALTMEYER M, RASK MB, LUKAS C, LARSEN DH, POVLSEN LK, BEKKER-JENSEN S, MAILAND N, BARTEK J, LUKAS J. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013;155:1088–103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- TOURRIERE H, VERSINI G, CORDON-PRECIADO V, ALABERT C, PASERO P. Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol Cell. 2005;19:699–706. doi: 10.1016/j.molcel.2005.07.028. [DOI] [PubMed] [Google Scholar]
- WATERS CE, SALDIVAR JC, AMIN ZA, SCHROCK MS, HUEBNER K. FHIT loss-induced DNA damage creates optimal APOBEC substrates: Insights into APOBEC-mediated mutagenesis. Oncotarget. 2015;6:3409–19. doi: 10.18632/oncotarget.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILLMANN KL, MILOSEVIC S, PAUKLIN S, SCHMITZ KM, RANGAM G, SIMON MT, MASLEN S, SKEHEL M, ROBERT I, HEYER V, SCHIAVO E, REINA-SAN-MARTIN B, PETERSEN-MAHRT SK. A role for the RNA pol II-associated PAF complex in AID-induced immune diversification. J Exp Med. 2012;209:2099–111. doi: 10.1084/jem.20112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XUAN D, LI G, CAI Q, DEMING-HALVERSON S, SHRUBSOLE MJ, SHU XO, KELLEY MC, ZHENG W, LONG J. APOBEC3 deletion polymorphism is associated with breast cancer risk among women of European ancestry. Carcinogenesis. 2013;34:2240–3. doi: 10.1093/carcin/bgt185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YU Q, KONIG R, PILLAI S, CHILES K, KEARNEY M, PALMER S, RICHMAN D, COFFIN JM, LANDAU NR. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol. 2004;11:435–42. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- ZEMAN MK, CIMPRICH KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHENG S, VUONG BQ, VAIDYANATHAN B, LIN JY, HUANG FT, CHAUDHURI J. Non-coding RNA Generated following Lariat Debranching Mediates Targeting of AID to DNA. Cell. 2015;161:762–73. doi: 10.1016/j.cell.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHOU Y, CARON P, LEGUBE G, PAULL TT. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res. 2014;42:e19. doi: 10.1093/nar/gkt1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.