Abstract
3JHNHα and 3JC′C′ couplings can be readily measured in isotopically enriched proteins and were shown to contain precise information on the backbone torsion angles, ϕ, sampled in disordered regions of proteins. However, quantitative interpretation of these couplings required the population of conformers with positive ϕ angles to be very small. Here, we demonstrate that this restriction can be removed by measurement of 3JC′Hα values. Even though the functional forms of the 3JC′Hα and 3JHNHα Karplus equations are the same, large differences in their coefficients enable accurate determination of the fraction of time that positive ϕ angles are sampled. A four-dimensional triple resonance HACANH[C′] E.COSY experiment is introduced to simultaneously measure 3JC′Hα and 3JHNC′ in the typically very congested spectra of disordered proteins. High resolution in these spectra is obtained by non-uniform sampling (in the 0.1-0.5% range). Application to the intrinsically disordered protein α-synuclein shows that while most residues have close-to-zero positive ϕ angle populations, up to 16% positive ϕ population is observed for Asn residues. Positive ϕ angle populations determined with the new approach agree closely with consensus values from protein coil libraries and prior analysis of a large set of other NMR parameters. The combination of 3JHNC′ and 3JC′C′ provides information about the amplitude of ϕ angle dynamics.
Keywords: backbone dynamics, backbone torsion angle, positive ϕ angle, intrinsically disordered protein, IDP, α-synuclein, Karplus equation, 3JC′Hα, 3JHNC′, backbone dynamics, protein NMR
1 Introduction
A survey of experimentally determined protein structures contained in the protein structure databank (PDB) shows that, with the exception of Gly, the vast majority of residues adopt negative values for the backbone torsion angle, ϕ. Well-ordered residues with positive ϕ angles are mostly restricted to type I′, type II, and type II′ tight β-turns and helix-terminating C-cap motifs. Transient occupation of conformations where one or more residues adopt positive ϕ angles may be of high functional relevance, and has been implicated in substrate recognition.1,2,3 Moreover, molecular dynamics simulations on folded proteins indicate that peptide plane flips associated with a change in the sign of ϕ are energetically feasible,4,5 suggesting that transient formation of conformations with a positive ϕ backbone torsion angle could be more common than implied by the analysis of static PDB X-ray structures. Adoption of positive ϕ values has been proposed to be particularly relevant in intrinsically disordered proteins (IDPs) and intrinsically disordered regions (IDRs) of amyloidogenic proteins, where they may impact the kinetics of fibril formation.6,7
Analysis of chemical shifts and other NMR parameters of IDPs and IDRs suggests low but non-vanishing populations of positive ϕ angle conformers,8,9,10,11,12 mostly following what is seen in statistical coil libraries extracted from the PDB.13,14 Traditionally, the primary markers for positive ϕ conformations are 1JCαHα, which is at a minimum in the αL region of Ramachandran space,15 and strong intraresidue HN-Hα NOEs.16 However, neither of these parameters provides reliable fractional populations of the positive ϕ conformations as the variations in their values associated with negative ϕ angles usually are large and not very well defined. The Karplus equation relating 3JC′Hα to ϕ indicates that positive ϕ angles, associated with a trans C′-N-Cα-Hα arrangement, have much larger 3JC′Hα values than those observed for negative ϕ angles.17 Here, we demonstrate that 3JC′Hα, in particular when used in conjunction with 3JHNHα and 3JC′C′, provides a precise quantitative measure for the fractional positive ϕ population of any given residue. We demonstrate that, even for the typically very poorly resolved spectral regions of disordered proteins, nearly complete sets of 3JC′Hα values can be obtained by recording the HACANH[C′] E.COSY spectrum in a four-dimensional (4D) fashion. This experiment then also yields 3JHNC′, providing yet another parameter to define the ϕ angle distribution. Relatively recently introduced non-uniform sampling (NUS) strategies permit the recording of 4D spectra at very high resolution and in relatively limited amounts of measurement time.18,19,20 This approach is used in our study, and the accuracy at which J couplings can be extracted from such NUS-reconstructed spectra is shown to scale approximately inversely with the square root of the sparsity of the acquired data. Thus, the accuracy depends inversely on the square root of measurement time, following the same rules as in fully sampled NMR spectra.
2 Experimental Section
2.1 NMR sample preparation
Uniformly 13C/15N-enriched, N-terminally acetylated21 wild-type α-synuclein (aS) protein was expressed and purified as described previously.22 The NMR sample contained 0.6 mM aS in 20 mM sodium phosphate, pH 6.0, 50 mM NaCl, and 5% D2O. The NMR sample of the third IgG-binding domain of protein G (GB3, 1.2 mM, uniformly 13C/15N-enriched) was prepared in 50 mM sodium phosphate, pH 6.5, 50 mM NaCl, and 5% D2O. The NMR sample of uniformly 13C/15N-enriched ubiquitin (2.8 mM) was prepared in 20 mM imidazole buffer, pH 6.0, containing 7% D2O.
2.2 NMR measurements
The 4D HACANH[C′] E.COSY spectrum of α-synuclein was recorded using the pulse sequence of Fig. 1, using a Bruker Avance III 600-MHz spectrometer equipped with a TCI cryogenic probe and a z-axis gradient coil. Non-uniform sampling with a sparsity of 0.55% was employed in order to achieve high resolution and thereby the most reliable peak positions and J coupling measurements, while keeping the total measuring time restricted to about three days. The time domain matrix consisted of 80* × 62* × 250* × 1024* complex data points, or acquisition times of 112 ms (t1, 1Hα), 28 ms (t2, 13Cα), 179 ms (t3, 15N), and 102 ms (t4, 1HN). The 1Hα, 13Cα and 15N carrier frequencies were set at 4.2, 56, and 118 ppm, respectively and 4 scans per FID were recorded. Additionally, 3D HA(CA)NH[C′] E.COSY spectra were recorded for ubiquitin and GB3, again using the pulse scheme of Fig. 1, but keeping the 13Cα evolution time t2 = 0. The 3D HN(COCO)NH pulse sequence23 was used at 900 MHz using a 2.5% NUS scheme, in order to obtain additional 3JC′C′ coupling values for α-synuclein residues that were previously inaccessible due to resonance overlap. The new 3D HN(COCO)NH spectrum was recorded with 200 ms acquisition times in both the t1 (15N) and t2 (15N) dimensions.
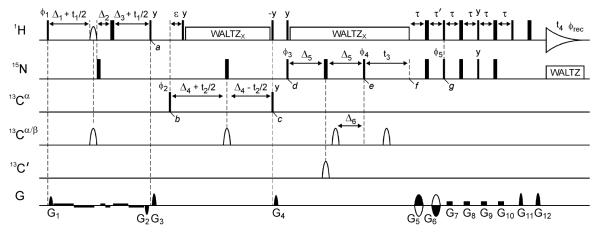
Fig. 1.
Pulse scheme of the HACANH[C′] E.COSY experiment. Narrow and wide lines indicate non-selective 90° and 180° pulses, respectively, except for the 90° pulses on 13Cα that have their duration adjusted to have no excitation of 13C'. 1H decoupling (γHB1 = 3.1 kHz; phase x) is applied using the WALTZ16 scheme.38 The carrier frequencies for 1H, 15N, 13Cα, 13Cα/β and 13C' are 4.7, 119, 56, 46 and 176 ppm, respectively. The 1Hα shaped pulse is of the ReBURP-type and has a duration of 3.5ms. The 13C' shaped pulse is of the hyperbolic-secant type and has a duration of 2 ms. The 13Cα/β shaped pulses are G3-shaped53 with a duration of 350μs, except for the pulse during 13Cα evolution (t2) which is of the ReBURP-type54 and has a duration of 420 μs. Note that the durations of all shaped pulses presented here are for measurements at 600 MHz 1H frequency; these values scale inversely with field strength and should be adjusted accordingly when experiments are recorded at a field strength that differs from 14.1 T. Delays: ε = 2.5 ms, τ = 2.6 ms, τ' = τ + t3(0), Δ1 = 1.7 ms - 0.92 × Prb/2 where Prb is the duration of the 1Hα ReBURP pulse and the scaling factor of 0.92 for 1JCαHα dephasing was measured experimentally for a pair of centered 1H ReBURP and 13C G3 shaped pulses with a duration ratio of 10:1, Δ2 = Δ1+Δ3, Δ3 = 0.17 ms, Δ4 = 14 ms, Δ5 = 15 ms and Δ6 = 14 ms. All pulse phases are x, unless specified. Phase cycling: ϕ1 = x; ϕ2 = x,-x; ϕ3 = x,x,-x,-x; ϕ4 = y,-y; ϕ5 = x; and ϕrec = x,-x,-x,x. The gradient pulses G1-G12 (z axis) have strengths of 7, −7, 30.1, 28.7, 25.9, 25.9, 3.5, 3.5, 4.9, 4.9, 25.9 and 25.9 G/cm, with durations of 0.05, 0.05, 2, 1.3, 1, 2, 2.4, 2.4, 2.4, 2.4, 0.2 and 0.503 ms, respectively. Weak gradients (0.35 G/cm) with a polarity indicated in the figure were applied during 1Hα evolution (t1) to minimize radiation damping of the solvent signal. Quadrature detection in the t1 and t2 dimensions was achieved by increasing ϕ1 and ϕ2, respectively, in the regular States-TPPI manner. Quadrature detection in the t3 dimension is obtained by the Rance-Kay method,37 inverting G5, G6 and ϕ5.
For NUS processing of the HACANH[C′] E.COSY and HN(COCO)NH spectra, we used the in-house written SMILE routine (Ying et al., unpublished), incorporated in the data processing software package NMRPipe.24 The digital resolution of the final 4D spectra was 1.4 Hz (1Hα, F1), 8.7 Hz (13Cα, F2), 1.4 Hz (15N, F3) and 2.4 Hz (1HN, F4) for aS, and 1.0 (15N, F1), 1.0 (15N, F2) and 2.4 Hz (1HN, F3) for the α-synuclein HN(COCO)NH spectrum. Peak picking and spectrum analysis was carried out with the programs NMRPipe and SPARKY.25
2.3 Coil Database
For all comparisons with backbone torsion angle distributions expected in disordered regions of proteins we used the recently introduced coil library of Mantsyzov et al. which consists of all longer (> 3 residue) segments that lack intramolecular H-bonding in the PDB.14 This library differs from the widely used Fitzkee coil library13 mostly by the absence of tight β-turns and was developed as a reference for the MERA program,14 which aims to provide a residue-specific ϕ/ψ distribution of IDPs and IDRs.
3 Measurement of 3JC'Hα from HACANH[C′] E.COSY
The exclusive-COSY (E.COSY) principle provides a convenient and accurate method for measurement of unresolved scalar coupling, JAB, between a pair of nuclear spins A and B. E.COSY experiments require the presence of a third nuclear spin, C, that has a large, resolvable coupling to spin A and whose chemical shift can be correlated in a multi-dimensional NMR experiment with spin B.26,27 Numerous E.COSY experiments have been introduced over the past few decades that permit the measurement of virtually every possible type of three-bond 1H-1H and heteronuclear J coupling in isotopically enriched proteins.17,28,29,30,31,32 A condition for accurate measurement of the JAB coupling from a B-C cross peak is that the spin-state of the passive spin, A, remains unperturbed. A reduction in the apparent coupling will result if the longitudinal relaxation time of spin A is insufficiently long relative to the delay durations used in the pulse scheme.33,17 So, on the one hand it is important to select from the two spins, A and B, the spin with the longest T1 relaxation time to function as the passive, unperturbed spin. On the other hand, as the JAB displacement is read out from the relative displacement in resonance frequency of spin B when monitoring the components of the B-C cross peak, it is important that the line width of spin B is narrow. For measurement of 3JC′Hα by an E.COSY experiment, the choice is whether the 13C′ or the 1Hα should act as the passive spin. The 13C′ T1 relaxation times for IDPs at 600 MHz 1H frequency and above are typically longer than 1 s, which is considerably longer than those of 1Hα in 13C-enriched proteins, thus favoring 13C′ to be the passive spin. On the other hand, the transverse relaxation time of 13C′ is longer than for 1Hα, favoring it to be the actively detected spin because it potentially offers higher frequency resolution. In our experiments, we have selected 13C′ to be the passive spin, but enhance the spectral resolution of the 1Hα spin by homonuclear decoupling it from 1HN and 1Hβ in a band-selective manner.34,35
3.1 Description of the pulse scheme
We here used the HACANH[C′] E.COSY pulse scheme (Fig. 1) for measurement of the 3JC′Hα and 3JHNC′ couplings. This triple resonance pulse scheme transfers magnetization in a unidirectional fashion from 1Hα via 13Cα and 15N to 1HN and is similar in design to the early H(CA)NNH experiment,36 but it lacks 13C′ decoupling and it uses gradient-enhanced transfer from 15N to 1H for sensitivity enhancement and improved solvent signal suppression.37 Other small but important differences will be discussed below. To minimize resonance overlap in crowded IDP spectra, we carry out the experiments in a 4D fashion, with evolution of 1Hα, 13Cα and 15N chemical shifts in the indirect dimensions, and direct detection of 1HN. The 13Cα evolution period can be omitted if the measurement is carried out as a 3D experiment, potentially useful for small proteins such as ubiquitin and GB3, where resonance overlap is less of a concern. However, this will remove the possibility to also measure 3JHNC′ from such a spectrum.
During the (t1, 1Hα) evolution period, a 1Hα band-selective pulse is paired with a non-selective 1H 180° pulse (separated by the short Δ2 delay, Fig. 1) to effectively invert all 1H spins other than 1Hα, thereby removing 1H-1H J-dephasing during t1 evolution and enhancing the attainable 1Hα spectral resolution. Note that 1Hα-1Hβ J-dephasing for Ser and Thr residues is not refocused during t1 because their 1Hβ resonate within the bandwidth that is inverted by the 1Hα band-selective pulse. Similarly, the geminal 1Hα-1Hα dephasing remains active for Gly residues. Evolution of 1JCαHα coupling during t1 is adjusted to be active for ~3.4 ms, with most of the 1JCαHα dephasing actually taking place during the pair of simultaneously applied band-selective 1Hα and 13Cα pulses. As indicated in Figure 1, very weak, antiphase gradients are also applied during the t1 evolution period to prevent radiation damping of the 1H2O magnetization, which otherwise would rotate 1Hα spins that resonate under the water resonance back to the z axis, making them effectively disappear from the spectrum. The antiphase G1/G2 gradient pulse pair serves to remove the effect of imperfections in the band-selective and non-selective 180° 1H pulses, applied during t1, while also eliminating any transverse 13C magnetization. At time point a, the relevant, selected magnetization then is described by 2Czα(i)Hzα(i) cos(δHαt1) (π2JHαC′t1) cos(π3JHαC′t1), where δHα is the 1Hα chemical shift frequency, and 2JHαC′ and 3JHαC′ are the two- and three-bond J couplings from 1Hα to the intraresidue and preceding 13C′, respectively.
During the subsequent (t2, 13Cα) constant-time evolution period between time points b and c, 1JCαN and 2JCαN are active for the full constant-time duration 2Δ4, and 1JCαHα rephasing is active for the duration ε, which is set to a compromise value (2.5 ms) that enables magnetization transfer to occur for both 13Cα methine and methylene (Gly) groups. The constant-time duration is set to 2Δ4 ≈ 1/1JCαCβ ≈ 28 ms such that the net effect of 1JCαCβ dephasing is eliminated. Broad-band 1H-decoupling with the WALTZ16 modulation scheme38 is used while care is taken that the water magnetization remains in a well-defined state, parallel to the decoupling radiofrequency field, thereby facilitating suppression of the intense water signal during the detection period. At time point c, following the 13Cα evolution period, the 2Czα(i)Hzα(i) terms from time point a are converted to 2Czα(i)Nz(i) sin(2π 1JCαNΔ4) cos(2π 2JCαNΔ4) + 2Czα(i)Nz(i+1) sin(2π 2JCαNΔ4) cos(2π 1JCαNΔ4). Undesired transverse magnetization terms are eliminated at this point by pulsed field gradient G4. Between time points d and e, prior to (t3, 15N) evolution, a 12C' filter is applied to ensure removal of any signals from isotopomers where 15N is bonded to a 12C' isotope (due to incomplete isotope labeling). In the absence of such a filter, we found that in the 15N dimension of the final spectrum, the 15N-{12C′} signal approximately overlaps with the downfield component of the 15N-{13C′} doublet,39 causing an artificial reduction in the apparent 3JC′Hα E.COSY splitting. A hyperbolic-secant-shaped 13C′ pulse is used during this 12C′ filter, such that complete 13C′ inversion is not adversely impacted by RF inhomogeneity.40 During the 12C′ filter, a 13Cα pulse is applied which rephases a significant fraction of the 2Czα(i)Ny(i) + 2Czα(i)Ny(i+1) terms present at time point d to yield 2Cz'(i-1)Ny(i) and 2Cz'(i)Ny(i+1) at point e. Therefore, at time point e, (t3, 15N) evolution starts from antiphase 15N-{13C′} magnetization, which is in-phase with respect to 1HN. Following t3 evolution (time point f) 15N magnetization is converted to the requisite antiphase 2NyHz terms during the duration τ + τ′, which also serves as the gradient-encoding delay (by oppositely signed gradient pulses G5 and G6), followed by standard gradient-enhanced transfer to 1HN,37 which is detected during the time t4. Note that 13C' decoupling is absent during all chemical shift evolution periods, leading to the desired E.COSY effect. However, since both 13C′(i) and 13C′(i-1) play a role in the evolution of the various terms, discussed above, the 4D E.COSY multiplet pattern is somewhat more complex than in typical E.COSY applications. Below, we will therefore briefly discuss the fine structure observed in these 4D multiplets.
3.2 J couplings contained in the HACANH[C′] E.COSY spectrum
The 4D HACANH[C′] E.COSY spectrum contains a rich array of J coupling information involving the backbone carbonyl nuclei. Next to the “active” 1JNC′ coupling, which yields the requisite well-resolved antiphase E.COSY splitting in the 15N dimension and which contains potentially useful information on H-bonding,41 the spectrum yields 1JC′Cα, 2JC′N, 3JHNC′, 2JC′Hα, and the main purpose of our study: 3JC′Hα. The values of 1JC′Cα, 2JC′N, and 2JC′Hα show relatively little variation with structure, and so far have not yet found much use in terms of protein structure analysis.42-44 Both 3JHNC′ and 3JC′Hα correlate with ϕ via well-defined Karplus equations and are of most immediate structural value. Below, we therefore briefly illustrate how these couplings are manifested in the 4D spectrum.
3.3 Measurement of 3JC′Hα and 2JC′Hα
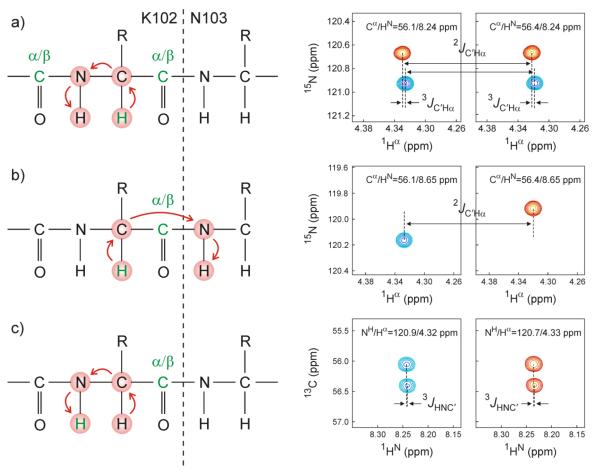
As discussed above, two types of magnetization transfer take place from 13Cα to 15N: intraresidue, mediated by 1JCαN and sequential, to 15N of residue i+1, mediated by 2JCαN. Figure 2a concerns the intraresidue correlation observed for K102, and illustrates the relevant magnetization transfer pathway. Note that in the 15N dimension, the cross peak is split by the presence of 1JC′N to the 13C′(i-1) of the preceding residue. The splitting is antiphase due to the presence of the 12C′ filter (between time points d and e in Fig. 1), and the displacement in the 1Hα dimension corresponds to the desired 3JC′Hα coupling. The spin state of the 13C′ is inverted between 1Hα and 15N evolution, giving rise to the negative slope of the doublets in Fig. 2a. However, after accounting for this sign inversion, and knowing that the sign of 1JC′N is negative (due to the negative magnetogyric ratio of 15N), this slope corresponds to a positive value for the 3JC′Hα(i-1,i) coupling. The Hα(i)➔HN(i) cross peak appears twice, as it is split in the 13Cα dimension by 13C′(i), therefore providing duplicate measurements for each 3JC′Hα coupling (Fig. 2a). Comparison of such pairs of measured 3JC′Hα couplings provides a measure for their reproducibility, and averaging their values is used to reduce their random uncertainty by √2.
Fig. 2.
Analysis of the 4D HACANH[C′] E.COSY spectrum, shown for couplings involving residue K102 in aS. The left hand side of each panel denotes the magnetization transfer pathway, with active and passive nuclei whose pairwise J coupling is measured shown in green. The right hand side of each panel shows small cross sectional regions through the 4D NMR spectrum, illustrating the measurement of various J couplings. (a) Measurement of 3JC'Hα and 2JC'Hα from 15N-1Hα cross sections. Two independent measurements are shown, separated in the orthogonal 13Cα dimension by 1JCαC'. Relative 1Hα displacement within each panel corresponds to 3JC'Hα; relative 1Hα displacement between the two panels corresponds to 2JC'Hα. (b) Alternative measurement of 2JC'Hα from the sequential 13Cα-1Hα (K102) to 15N-1HN (N103) cross peak. Relative displacement in the 1Hα dimension corresponds to 2JC'Hα. 15N displacement corresponds to 1JNC′, and 13Cα displacement to 1JCαC'. (c) Measurement of 3JHNC' from a 13Cα-1HN cross section. Relative 1HN displacement within each panel equals 3JHNC' and relative 13Cα displacement equals 1JCαC'. 15N displacement between the two panels equals 1JNC′; relative 1HN displacement between the two panels equals 2JHNC′.
Analogous to the 1JCαN transfer pathway discussed above, transfer via 2JCαN yields a Hα(i)➔HN(i+1) cross peak (Fig. 2b). Here, the 15N(i+1) and 13Cα(i) frequencies of the cross peak depend on the spin state of 13C′(i) only, yielding a single measurement of 2JC′Hα when extracting this value from the difference in 1Hα frequency of the two multiplet components. An upfield 1Hα displacement together with a downfield 13Cα displacement for the red versus the blue multiplet component in Fig. 2b points to opposite signs for 1JCαC′ and 2JC′Hα. Note that 2JC′Hα can also be obtained from Fig. 2a, from the relative 1Hα displacement of the duplicate doublets shown in its two panels.
3.4 Measurement of 3JHNC′
As mentioned above, the intraresidue Hα(i)➔HN(i) cross peak is split in the 13Cα dimension by 1JCαC′, with the upfield and downfield components corresponding to 13C′ in the ∣α> and ∣β> spin states, respectively. Therefore, when viewing 13Cα-1HN cross-sections of the 4D spectrum taken through the cross peak of Fig. 2a, an E.COSY tilt reflecting the intraresidue 3JHNC′ is seen, with the 1JCαC′ splitting in the 13Cα dimension, and 3JHNC′ in the 1HN dimension (Fig. 2c). Again this coupling is obtained in duplicate by viewing the two cross-sections obtained at δN±1JNC′/2 (Fig. 2c).
As with most 4D NMR experiments, the HACANH[C′] E.COSY spectrum can also be recorded in fewer dimensions, and here this can be done most conveniently by not incrementing the duration the 13Cα evolution period (i.e., keeping t2=0). However, in practice we find that overlap between the sequential and intraresidue Hα➔HN cross peaks is rather common, in particular in disordered proteins. In folded proteins, the 3D version of the experiment is a viable option, although reducing the dimensionality of the experiment in this manner removes the ability to measure 3JHNC′ from the spectrum.
3.5 Reproducibility of the measured 3JC′Hα and 3JHNC′ values
The random measurement uncertainty, σ, of a peak position in a frequency domain spectrum is directly proportional to the resonance line width, LW, and inversely related to the signal-to-noise ratio, SNR. To a fair approximation, it is given by:45
| (1) |
The uncertainty in the J splitting requires propagation of the uncertainty in both of its components and is therefore √2 larger. For time domain signals that have not fully decayed, as typically applies in the indirect dimensions of multi-dimensional NMR experiments, the attainable line width is approximately given by 1/AT, where AT is the length of the acquisition time in the dimension considered. This consideration indicates that it is important to sample the signals in the dimension where the coupling is measured for as long as practically feasible (up to ca 2×T2, where T2 is the decay constant of the signal), and that it is desirable to minimize the apparent decay rate through the removal of 1H-1H J splittings, as is done in the scheme of Fig.1 by band-selective homonuclear decoupling in the 1Hα dimension. Band-selective homonuclear decoupling in the detected 1HN dimension of the spectrum would be advantageous for enhancing 1HN resolution and thereby the accuracy of 3JHNC′. However, this mode of decoupling in the directly detected dimension is technically more challenging in terms of water signal suppression on cryogenic probes, which typically have only a single-axis pulsed field gradient accessory, and therefore was not used in our experiments. Note also that 1HN is only split by 1Hα, making homonuclear decoupling less advantageous than it is for 1Hα, where the couplings to both 1HN and 1Hβ are removed. Overall, the line width in the 1HN dimension remains somewhat larger than in the homonuclear decoupled 1Hα dimension, and with the applicable SNR (cf eq 1) being the same for defining the peak position precision, the random error in the 3JHNC′ will be correspondingly larger than for 3JC′Hα.
Each intraresidue Hα➔HN cross peak consists of four multiplet components, corresponding to the splitting by 13C′(i) and 13C′(i-1) (Fig. 2a,c) whereas each sequential Hα(i)➔HN(i+1) cross peak is split into doublets by 13C′(i) (Fig. 2b). Therefore, the number of resonances in the 4D spectrum approximately equals 6N, where N is the number of residues in the protein. This number typically is small relative to the total number of time domain data points sampled in the indirect frequency dimensions, meaning that the HACANH[C′] E.COSY spectrum is relatively sparse and well suited for the use of non-uniform sampling (NUS) techniques18,19,20,46,47,48. Here, spectral reconstruction was carried out by the in-house written SMILE routine of the NMRPipe program24 which integrates accurate spectral reconstruction with spectral resolution enhancement by using a method that effectively mimics multi-dimensional linear prediction.49
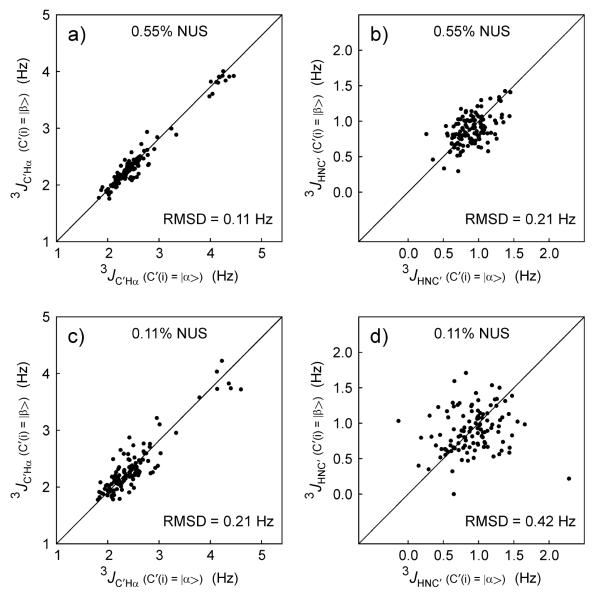
The availability of duplicate measurements for 3JHNC′ and 3JC′Hα makes evaluation of their random uncertainty straightforward, and permits evaluation of how closely this random error approaches the theoretical limit set by eq 1. As can be seen in Figure 3a, the pairwise root-mean-square deviation (RMSD) between the two sets of 3JC′Hα is 0.11 Hz, and averaging the two sets of values therefore reduces the random uncertainty to 0.06 Hz.
Fig. 3.
Reproducibility of the duplicate measurements of 3J couplings from the 4D NUS HACANH[C′] E.COSY spectrum of α-synuclein. Comparisons of the duplicate 3JC'Hα measurements (see right panels of Fig. 2a) obtained when (a) using 0.55% NUS sampling and (c) 0.11% NUS sampling. Analogous comparisons of the two 3JHNC' measurements (see right two panels of Fig. 2c) obtained when using (b) 0.55% and (d) 0.11% NUS sampling.
The average signal-to-noise ratio for intra-residue cross peaks is ca 120:1, and line widths are approximately 10 Hz (1Hα of non-T/S/G), ca 15-20 Hz for T/S/G; 25 Hz (13Cα), 5 Hz (15N) and 15-20 Hz (1HN). Eq 1 can then be used to predict precisions of ca 0.1 and 0.06 Hz for 3JHNC′ and 3JC′Hα, respectively. As can be seen from the pairwise RMSD values observed for the duplicate sets of 3JHαC′ and 3JHNC′ (Fig. 3), these predicted precisions fall close to their theoretical limit, indicating that the NUS reconstruction process used in our analysis faithfully reproduces peak positions.
The processed NMR spectrum containing the 3JHαC′ and 3JHNC′ splittings were generated from a 4D NUS time domain matrix that included measured data for only 0.55% of its elements, i.e., with 99.45% of them initially set to zero and subsequently adjusted during the iterative NUS processing procedure to values that approach those that would have been sampled if measurements for these time points had been made. As mentioned above, with only ca 800 resonances in the HACANH[C′] E.COSY spectrum of aS, the total number of time domain data points sampled in the indirect dimensions is far larger than this number of 800 resonances, making the NUS reconstruction feasible. It is therefore interesting to test whether a similar quality spectrum can be reconstructed from even fewer sampled data. For this purpose, we also reconstructed a 0.11% sampled data set, representing 20% of the same data used above for spectral reconstruction, and consequently yielding √5 lower SNR. As can be seen in Fig. 3c,d, the reproducibility of the measured J couplings then also decreases by a factor of ca √5, indicating that no additional uncertainty in extracted peak position is added by the NUS processing over what is expected on the basis of simple signal-to-noise considerations, even for a sparsity as low as 0.11%.
4 Karplus equation for 3JC′Hα
An empirically parameterized 3JC′Hα Karplus equations was introduced on the basis of values measured for ubiquitin using a 3D E.COSY method conceptually analogous to the 4D experiment of Fig. 1.32 Comparison of the previously measured and new ubiquitin data shows pairwise RMSD values of 0.29 Hz relative to the earlier measurements. Considering that this RMSD is more than √2 higher than the estimated uncertainty in the newly acquired data, we conclude that the newer data is of higher accuracy than the earlier data, and we will only use the new data for any fitting and analysis.
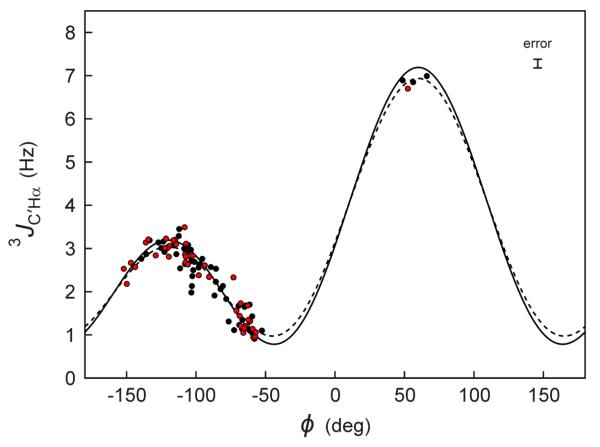
Best fits of the Karplus equation to the experimental 3JC′Hα values and the coordinates of the NMR-determined structure of ubiquitin (PDB entry 2MJB)50 and the RDC-refined structure of GB3 (PDB entry 2OED)23 are shown in Figure 4 (dashed lines). The RMSD between observed and Karplus-predicted 3JC′Hα values (dashed lines) equals 0.25 Hz, respectively. As expected on the basis of the higher measurement precision reached in the current study, this value is somewhat lower than obtained previously.17 However, it remains considerably higher than the random uncertainties in the measurement. This latter observation indicates that, similar to what was previously noted for the 3JC′C′ coupling,23 parameters other than ϕ can also influence these couplings. These other factors, which may include valence angle distortion of, for example, the τ angle (N-Cα-C′) and/or H-bonding of amide and carbonyl groups, are difficult to account for in experimental structures, and therefore limit the extent to which agreement between experimental 3JC′Hα and predicted values can be enforced during the calculation of structures of folded proteins. In IDPs and IDRs, the effects of factors other than ϕ on the 3J couplings are expected to average to roughly zero, and should permit interpretation of these couplings in a more quantitative fashion, which then can approach the experimental uncertainty in the measurement of these 3J couplings.14
Fig. 4.
Plot of experimental 3JC'Hα coupling values against the backbone torsion angle ϕ, for ubiquitin (black symbols) and GB3 (red). The backbone ϕ values are derived from RDC-refined structures of GB3 (PDB entry 2OED) and ubiquitin (PDB entry 2MJB), both obtained without 3JC'Hα data. The superimposed best-fit Karplus equation, shown as a dashed curve, is given by 3JC'Hα = 3.76 × cos2(ϕ−60°) + 1.95 × cos(ϕ−60°) + 1.23. After factoring out the effect of Gaussian ϕ angle fluctuations (σ = 0.226),52,51 the rigid limit Karplus equations (solid line) is given by: 3JC'Hα = 4.17 × cos2(ϕ−60°) + 2.00 × cos(ϕ−60°) + 1.02. Residues with elevated backbone dynamics (L12, D40, and G41 for GB3; T9-K11, D32-G35, G47, D52, and R72-G76 for ubiquitin) are excluded from the plot and the fitting process. The error bar (top right) corresponds to the random error (±1 standard deviation) based on the reproducibility of the 3JC'Hα measurements. The considerably larger RMSD of 0.25 Hz relative to the best-fitted Karplus curve (dashed line) reflects the impact of factors other than ϕ on the 3JC'Hα value.
The best-fit Karplus curve, described above, was derived using a single, static representation of the protein structure. However, it is well recognized that proteins are subject to modest amplitude ϕ-angle fluctuations, previously estimated to have an RMS amplitude of ca 13° in regions of well-ordered structure.51 When factoring out the effect of these backbone fluctuations,52 slightly different, “rigid-limit” Karplus parameters are obtained that should be applicable when refining a structure in terms of an ensemble of perfectly static conformers (solid lines in Fig. 4), and these latter curves are therefore used when describing an IDP or IDR in terms of an ensemble representation.14
Tables containing the experimental 3JC'Hα and 2JC'Hα values for α-synuclein, ubiquitin, and GB3, together with the α-synuclein 3JHNC′ values are included as Electronic Supplementary Information (ESI).
5 Fractional population of positive ϕ angles from 3J couplings
Under the assumption that all ϕ angles are negative, we previously were able to extract two parameters, the average <ϕ−> and its standard deviation σ, from two experimental observables: 3JHNHα and 3JC'C'.51 Although a small population, P+, of conformers with positive ϕ values in IDPs was shown to have minimal impact on the extracted <ϕ−> and σ values, the actual value of P+ could not be determined from that data. As we will show below, the large difference between 3JC'Hα coupling values for negative and positive values of ϕ now allows determination of P+. To this extent, we name the 3JC'Hα values that would correspond to the fractions of conformers with only negative and only positive ϕ angles 3JC'Hα(−) and 3JC'Hα(+), respectively. Considering that the positive ϕ angles sampled in the PDB, and in particular in the coil library,13,14 cluster relatively tightly around <ϕ+> values, this permits us to make an amino-acid-type-specific estimate for the average, <3JC'Hα(+)>, by using the positive-ϕ conformers of the coil library in concert with the rigid-limit Karplus equation. Note that the effect of dynamic averaging is included when calculating <3JC'Hα(+)> over the coil library distribution of positive ϕ conformers, and that it therefore is the rigid-limit parameterization of the Karplus curve that needs to be used for predicting <3JC'Hα(+)>.
5.1 Iterative procedure for determination of P+
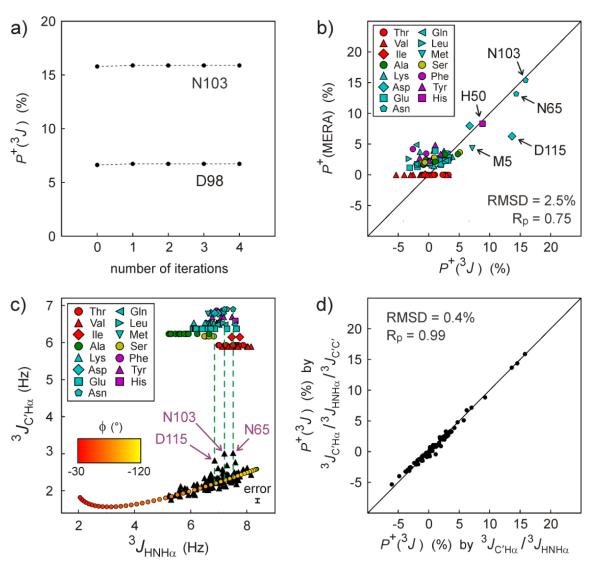
Since 3JHNHα and 3JC'C' coupling values for averaged negative ϕ angles in coil regions (<ϕ−>, typically between −80 and −100°) are similar to the coupling values averaged for positive ϕ angles, experimental 3JHNHα and 3JC'C' couplings are not much impacted by a small fraction of conformers with positive ϕ values. Therefore, to a first approximation, they may be ignored when calculating residue-specific <ϕ−> and σ values.51 These <ϕ−> and σ values then can be used to calculate 3JC'Hα(−) from the dynamics-corrected52 form of the 3JC'Hα Karplus equation (see also Section 6, eqn (4)). Next, P+ for each residue can be derived from
| (2) |
where 3JC'Hα is the experimentally observed value. In a subsequent iteration, this P+ value can be used to recalculate the 3JHNHα(−) and 3JC'C'(−) values, yielding an improved estimate for <ϕ−> and σ, and an improved 3JC'Hα(−) value. This then allows a better estimate of P+ in this second iteration. More iterations could be carried out, but in practice the procedure converges rapidly (Fig. 5a) and a single iteration suffices. With an RMSD of only 2.5%, excellent agreement is seen (Fig. 5b) between the P+ values derived in this manner and those previously obtained by MERA analysis14 of a large set of NMR input parameters (incl. 13C and 15N chemical shifts, 1H-1H NOEs, and five different types of 1J, 2J and 3J couplings).
Fig. 5.
Deriving the fractional population of positive ϕ angles, P+, in IDPs and IDRs. (a) P+ for α-synuclein residues N103 and D98 during successive iterations of 3JHNHα, 3JC'C', and 3JC'Hα analysis (see main text). (b) Comparison of the P+ derived from iterative 3J analysis (3JC'Hα, 3JHNHα, and 3JC'C') with P+ obtained with the MERA webserver14 without using 3JC'Hα as input parameter. Different residue types are presented by the symbols shown in the inset. Full results are listed in ESI Table S5. (c) Extracting P+ from only 3JC'Hα and 3JHNHα by interpolation. Red-to-yellow color shaded points correspond to the calculated correlation between 3JC'Hα and 3JHNHα dynamic Karplus equations (eqn 4), assuming σ = 29.5° (average IDP σ value previously derived for α-synuclein51). Shaped symbols mark the residue-type specific <3JC'Hα> values calculated from the Mantsyzov coil library for residues with ϕ >0.14 Pairs of 3JHNHα and 3JC'Hα values are shown as black filled triangles, all in the 3JC'Hα = 2-3 Hz range. Residues with 3JC'Hα that are significantly higher than expected from 3JHNHα when assuming P+ = 0 are marked by arrows and residue number. The error bar (bottom right) corresponds to the random error (±1 standard deviation) in the 3JC'Hα measurements. The random error in 3JHNHα (±0.05 Hz) is too small for display purposes. (d) Comparison of P+ calculated by iterative analysis of 3JC'Hα, 3JHNHα and 3JC'C' with those of simplified analysis (panel c method) of 3JC'Hα and 3JHNHα.
5.2 Direct determination of P+ from 3JHNHα and 3JC'Hα
As an alternative to the above iterative method, a good approximation for P+ can be obtained directly from the experimental 3JHNHα and 3JC'Hα values without requiring knowledge of 3JC'C': The fact that to a good approximation 3JHNHα and 3JC'Hα are linearly correlated over the range from −60° < ϕ < −120° (Fig. 5c) permits a direct prediction of 3JC'Hα (3JC'Hαpred,neg) under the assumption that only this range of ϕ is populated. In practice, higher values may be observed, and P+ can be calculated from
| (3) |
where 3JC'Hαpred,pos is the residue-type-specific predicted value for positive coil angles, spanning a narrow range from 6 to 7 Hz for the different residue types (Fig. 5c; ESI Table S4). In practice, the iterative analysis of 3JC'Hα 3JC'C' and 3JHNHα and the simple interpolation method of Fig. 5c yield very similar P+ values (Fig. 5d).
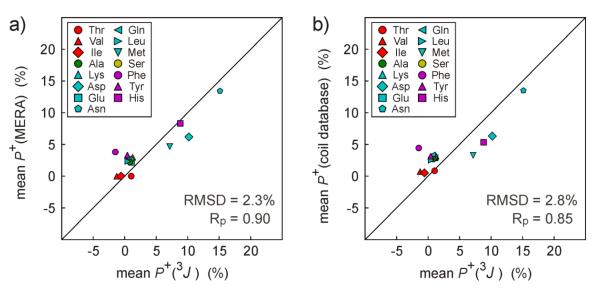
The newly acquired 3JC'Hα data allows us to revisit the propensities to adopt positive ϕ angles for different residue types in the IDP α-synuclein. We find that P+ values, averaged by residue type, agree very closely with those obtained from MERA analysis (Fig. 6a), which did not use 3JC'Hα as an input parameter. The only slight outlier in the correlation concerns Phe residues, which show a weak propensity (~4%) for positive ϕ angles, both in the MERA results and in the coil database (Fig. 6b), whereas 3JC'Hα points to a vanishingly low population. However, with 3JC'Hα data available for only two Phe residues in α-synuclein, the statistical uncertainty in the Phe <P+> values obtained for α-synuclein is rather large.
Fig. 6.
Positive ϕ angle propensity in α-synuclein for different residue types. (a) Comparison of P+ obtained from MERA analysis (lacking 3JC'Hα as input) with results of iterative analysis of 3JC'Hα, 3JHNHα and 3JC'C'. (b) Comparison of P+ obtained from iterative analysis of 3JC'Hα, 3JHNHα and 3JC'C' with residue-type averaged P+ values of the Mantsyzov coil library.14
6 Amplitudes of ϕ angle fluctuations from 3J couplings
Assuming that 3JHNHα and 3JC'C' couplings only depend on the intervening backbone torsion angle, ϕ, the amplitude of ϕ angle fluctuations (assumed to follow a Gaussian distribution) can be extracted from such a pair of measurements.51 This approach relies on the motion-corrected Karplus equation,52
| (4) |
where A'=A exp(−2σ2), B' =B exp(−σ2/2), and C'=C+A[1-exp(−2σ2)]/2 , with A, B, and C being the rigid limit Karplus coefficients, θ is the averaged intervening dihedral angle, and σ is its standard deviation.
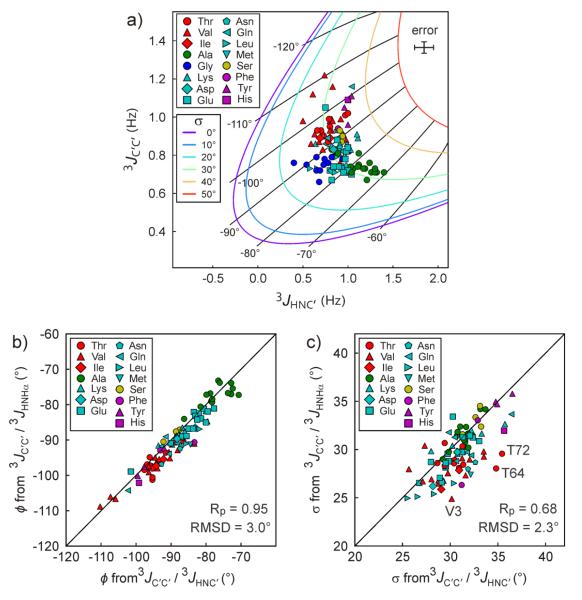
Our current study measures 3JC′Hα and 3JHNC′ and this pair of couplings, or any other pairwise combination of the four sets of 3JC′Hα, 3JHNC′, 3JC'C', or 3JHNHα can be used for analysis of the amplitude of σ, provided that their respective Karplus equations are not linearly related. Previously, we used the combination of 3JHNHα and 3JC'C' to extract residue-specific σ values for α-synuclein.51 Here, we demonstrate that σ values can be extracted from pairs of 3JC′C′ and 3JHNC′ values (Fig. 7). Colored lines in Figure 7a correlate the 3JHNHα and 3JC'C values predicted by their respective Karplus equations for different values of <ϕ> (marked by black, radial spokes), with different colors corresponding to different amplitudes of σ. As can be seen from the figure, the plot of 3JHNC′ versus 3JC′C′ values for α-synuclein places the σ values of all residues in the 25-35° range. With an RMSD of only 3°, the residue-specific <ϕ> values extracted from this figure agree closely with the corresponding <ϕ> values previously obtained from 3JHNHα and 3JC′C′ analysis (Fig. 7b), and similarly, with an RMSD of only 2.3°, the σ values also agree very well with our prior study (Fig. 7c).51
Fig. 7.
Extracting the amplitude of ϕ angle fluctuations from 3JC′C′ and 3JHNC'. (a) Plot of 3JC′C′ versus 3JHNC′ for α-synuclein. The rigid limit (after factoring out σ=13° motions) 3JHNC′ Karplus parameterization17 was used: 3JHNC′ = 4.78 × cos2(ϕ) + 0.86 × cos(ϕ) − 0.23 Hz. Solid lines show correlations between 3JC′C′ and 3JHNC′ predicted by the dynamics-corrected Karplus equations for ϕ angles (ϕ values marked by black “radial spokes”), with the different colors representing the amplitude of σ. The error bars (top right) corresponds to the random error (±1 standard deviation) of the 3JC'Hα and 3JC' C' measurements. (b) ϕ and (c) σ values that are extracted from the 3JC′C′ and 3JHNC' analysis are compared to those from 3JC′C′ and 3JHNHα analysis, reported previously.51
7 Concluding Remarks
We have shown that 3JC'Hα and 3JHNC' can be measured simultaneously and at high precision by using a 4D NMR pulse sequence. Only when such experiments are carried out in conjunction with very sparse non-uniform sampling does it become experimentally feasible to use long acquisition times in all four dimensions. These long acquisition times are required for obtaining the very high spectral resolution that is needed for precise measurement of very small J couplings. Although many of the widely used spectral analysis software packages are not yet fully optimized for convenient analysis of the extremely large 4D matrices, there is no conceptual or technical problem in obtaining resonance positions at the same accuracy as in spectra of lower dimensionality. Our analysis shows that the uncertainty in the extracted 4D peak positions is primarily determined by the intrinsic signal to noise ratio in the acquired time domain data. Indeed, the uncertainty in peak positions when using only a fraction of this data for NUS reconstruction is found to scale approximately with the inverse square root of the size of this fraction.
The large difference in 3JC'Hα values for residues with negative and positive ϕ angles makes this coupling particularly useful for identifying fractional populations of positive ϕ angles in disordered regions of a protein. In particular, when used in conjunction with 3JHNHα, the uncertainty in populations of positive ϕ conformers can be detected with uncertainties of only a few percent. Positive ϕ populations determined in this manner for α-synuclein agree well with those from the recent MERA analysis14 which used far more experimental data, but not 3JC'Hα. Results obtained for α-synuclein are also in very close agreement with positive ϕ propensities seen in a recently introduced large coil library, extracted from non-H-bonded residues in the PDB, suggesting that the new method is not only precise but also accurate.
As previously shown for the combination of 3JHNHα and 3JC'C', the newly measured 3JC'Hα and 3JHNC' values can be combined with either 3JHNHα or 3JC'C' to extract the average of the negative ϕangles sampled by a given IDP or IDR residue, and the standard deviation relative to this average value. Comparison of <ϕ−> and σ values derived from 3JC'C' and 3JHNC' agree very well with the earlier analysis and confirm that such an analysis is quite robust.
It has been hypothesized, mostly based on molecular dynamics simulations, that formation of α-pleated sheet structures may be a common conformational transition intermediate on the pathway to amyloid formation.6 The requisite Ramachandran map flips from αRαL to ββ for pairs of adjacent residues is commonly observed when comparing X-ray crystal structures of closely related proteins,7 providing some experimental support that such transitions are energetically feasible. α-Synuclein clearly falls in the category of amyloidogenic proteins, but its intrinsic disorder previously made it difficult to assess the presence of elevated transient population of α-pleated sheet backbone conformations. Analysis of our 3JC'Hα data finds no evidence for elevated population of the positive ϕ angles associated with α-pleated sheet, and propensities of its residues to adopt positive ϕ angles closely correlate with the residue-type specific propensities seen in the coil library. We note, however, that this observation does not preclude the formation of α-pleated sheet structures as an intermediate, but simply that if present the population of such structures must be very low.
For folded proteins, factors other than the intervening torsion angle ϕ are found to have a non-negligible impact on 3JC'Hα. Although this limits the accuracy at which ϕ angles can be extracted from 3JC'Hα, it also offers new opportunities to gain access to these additional factors, which include deviations from idealized geometry, H-bonding, and χ1 angles. We are currently exploring the feasibility of developing quantitative relations that extend the Karplus equations to take these additional factors into account.
Supplementary Material
Acknowledgements
We thank F. Li for providing GB3 and ubiquitin samples, and J. L. Baber, Y. Shen, and A. Grishaev for technical support. NUS reconstruction utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, Md. (http://biowulf.nih.gov). This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases and by the Intramural Antiviral Target Program of the Office of the Director, NIH. J.H.L. is the recipient of a KVSTA Fellowship.
Footnotes
Electronic supplementary information (ESI) available. See DOI: xxxxxxxxxx
References
- 1.Ramakrishnan C, Dani VS, Ramasarma T. Protein Eng. 2002;15:783–798. doi: 10.1093/protein/15.10.783. [DOI] [PubMed] [Google Scholar]
- 2.Fitzgerald CE, Patel SB, Becker JW, Cameron PM, Zaller D, Pikounis VB, O'Keefe SJ, Scapin G. Nature Struct. Biol. 2003;10:764–769. doi: 10.1038/nsb949. [DOI] [PubMed] [Google Scholar]
- 3.Honndorf VS, Coudevylle N, Laufer S, Becker S, Griesinger C. Angew. Chemie Int. Ed. 2008;47:3548–3551. doi: 10.1002/anie.200705614. [DOI] [PubMed] [Google Scholar]
- 4.Fadel AR, Jin DQ, Montelione GT, Levy RM. J. Biomol. NMR. 1995;6:221–226. doi: 10.1007/BF00211787. [DOI] [PubMed] [Google Scholar]
- 5.Piana S, Lindorff-Larsen K, Shaw DE. Proc. Natl. Acad. Sci. U. S. A. 2013;110:5915–5920. doi: 10.1073/pnas.1218321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armen RS, DeMarco ML, Alonso DOV, Daggett V. Proc. Natl. Acad. Sci. U. S. A. 2004;101:11622–11627. doi: 10.1073/pnas.0401781101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milner-White EJ, Watson JD, Qi G, Hayward S. Structure. 2006;14:1369–1376. doi: 10.1016/j.str.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 8.Marsh JA, Neale C, Jack FE, Choy W-Y, Lee AY, Crowhurst KA, Forman-Kay JD. J. Mol. Biol. 2007;367:1494–1510. doi: 10.1016/j.jmb.2007.01.038. [DOI] [PubMed] [Google Scholar]
- 9.Kragelj J, Ozenne V, Blackledge M, Jensen MR. ChemPhysChem. 2013;14:3034–3045. doi: 10.1002/cphc.201300387. [DOI] [PubMed] [Google Scholar]
- 10.Jensen MR, Salmon L, Nodet G, Blackledge M. J. Am. Chem. Soc. 2010;132:1270–1272. doi: 10.1021/ja909973n. [DOI] [PubMed] [Google Scholar]
- 11.Salmon L, Bouvignies G, Markwick P, Lakomek N, Showalter S, Li DW, Walter K, Griesinger C, Bruschweiler R, Blackledge M. Angew. Chem. Int. Ed. 2009;48:4154–4157. doi: 10.1002/anie.200900476. [DOI] [PubMed] [Google Scholar]
- 12.Krzeminski M, Marsh JA, Neale C, Choy W-Y, Forman-Kay JD. Bioinformatics. 2013;29:398–399. doi: 10.1093/bioinformatics/bts701. [DOI] [PubMed] [Google Scholar]
- 13.Fitzkee NC, Fleming PJ, Rose GD. Proteins. 2005;58:852–854. doi: 10.1002/prot.20394. [DOI] [PubMed] [Google Scholar]
- 14.Mantsyzov AB, Shen Y, Lee JH, Hummer G, Bax A. J. Biomol. NMR. 2015 doi: 10.1007/s10858-015-9971-2. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vuister GW, Delaglio F, Bax A. J. Am. Chem. Soc. 1992;114:9674–9675. [Google Scholar]
- 16.Mantsyzov AB, Maltsev AS, Ying J, Shen Y, Hummer G, Bax A. Protein Sci. 2014;23:1275–1290. doi: 10.1002/pro.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang AC, Bax A. J. Am. Chem. Soc. 1996;118:2483–2494. [Google Scholar]
- 18.Rovnyak D, Frueh DP, Sastry M, Sun ZYJ, Stern AS, Hoch JC, Wagner G. J. Magn. Reson. 2004;170:15–21. doi: 10.1016/j.jmr.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 19.Orekhov VY, Dubovskii PV, Yamada H, Akasaka K, Arseniev AS. J. Biomol. NMR. 2000;17:257–263. doi: 10.1023/a:1008346414720. [DOI] [PubMed] [Google Scholar]
- 20.Bermel W, Bertini I, Felli IC, Gonnelli L, Kozminski W, Piai A, Pierattelli R, Stanek J. J. Biomol. NMR. 2012;53:293–301. doi: 10.1007/s10858-012-9639-0. [DOI] [PubMed] [Google Scholar]
- 21.Johnson M, Coulton AT, Geeves MA, Mulvihill DP. PLoS One. 2010;5 doi: 10.1371/journal.pone.0015801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maltsev AS, Ying JF, Bax A. Biochemistry. 2012;51:5004–5013. doi: 10.1021/bi300642h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li F, Lee JH, Grishaev A, Ying J, Bax A. ChemPhysChem. 2015;16:572–578. doi: 10.1002/cphc.201402704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 25.Goddard TD, Kneller DG. Editon edn University of California; San Francisco: 2008. [Google Scholar]
- 26.Griesinger C, Sorensen OW, Ernst RR. J. Am. Chem. Soc. 1985;107:6394–6396. [Google Scholar]
- 27.Griesinger C, Sørensen OW, Ernst RR. J. Magn. Reson. 1987;75:474–492. [Google Scholar]
- 28.Montelione GT, Wagner G. J. Am. Chem. Soc. 1989;111:5474–5475. [Google Scholar]
- 29.Olsen HB, Ludvigsen S, Sørensen OW. Journal of Magnetic Resonance, Series A. 1993;105:321–322. [Google Scholar]
- 30.Weisemann R, Rüterjans H, Schwalbe H, Schleucher J, Bermel W, Griesinger C. J. Biomol. NMR. 1994;4:231–240. doi: 10.1007/BF00175250. [DOI] [PubMed] [Google Scholar]
- 31.Meissner A, Schulte-Herbruggen T, Sorensen OW. J. Am. Chem. Soc. 1998;120:3803–3804. [Google Scholar]
- 32.Wang AC, Bax A. J. Am. Chem. Soc. 1995;117:1810–1813. [Google Scholar]
- 33.Norwood TJ, Jones K. J. of Magn. Reson. Ser. A. 1993;104:106–110. [Google Scholar]
- 34.Hammarström A, Otting G. J. Am. Chem. Soc. 1994;116 [Google Scholar]
- 35.Ying J, Roche J, Bax A. J. Magn. Reson. 2014;241:97–102. doi: 10.1016/j.jmr.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kay LE, Ikura M, Bax A. J. Magn. Reson. 1991;91:84–92. [Google Scholar]
- 37.Kay LE, Keifer P, Saarinen T. J. Am. Chem. Soc. 1992;114:10663–10665. [Google Scholar]
- 38.Shaka AJ, Keler J, Freeman R. J. Mag. Res. 1983;53:313–340. [Google Scholar]
- 39.Ikura M, Krinks M, Torchia DA, Bax A. FEBS Lett. 1990;266:155–158. doi: 10.1016/0014-5793(90)81528-v. [DOI] [PubMed] [Google Scholar]
- 40.Silver MS, Joseph RI, Hoult DI. Nature. 1984;310:681–683. doi: 10.1038/310681a0. [DOI] [PubMed] [Google Scholar]
- 41.Juranic N, Ilich PK, Macura S. J. Am. Chem. Soc. 1995;117:405–410. [Google Scholar]
- 42.Schmidt JM, Zhou S, Rowe ML, Howard MJ, Williamson RA, Loehr F. Proteins-Structure Function and Bioinformatics. 2011;79:428–443. doi: 10.1002/prot.22893. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt JM, Hua Y, Loehr F. Proteins-Structure Function and Bioinformatics. 2010;78:1544–1562. doi: 10.1002/prot.22672. [DOI] [PubMed] [Google Scholar]
- 44.Schmidt JM, Howard MJ, Maestre-Martinez M, Perez CS, Loehr F. Magn. Reson. Chem. 2009;47:16–30. doi: 10.1002/mrc.2337. [DOI] [PubMed] [Google Scholar]
- 45.Kontaxis G, Clore GM, Bax A. J. Magn. Reson. 2000;143:184–196. doi: 10.1006/jmre.1999.1979. [DOI] [PubMed] [Google Scholar]
- 46.Kazimierczuk K, Stanek J, Zawadzka-Kazimierczuk A, Kozminski W. Prog. Nucl. Magn. Reson. Spectrosc. 2010;57:420–434. doi: 10.1016/j.pnmrs.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 47.Hyberts SG, Milbradt AG, Wagner AB, Arthanari H, Wagner G. J. Biomol. NMR. 2012;52:315–327. doi: 10.1007/s10858-012-9611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coggins BE, Werner-Allen JW, Yan A, Zhou P. J. Am. Chem. Soc. 2012;134:18619–18630. doi: 10.1021/ja307445y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu GA, Bax A. J. Magn. Reson. 1992;98:192–199. [Google Scholar]
- 50.Maltsev AS, Grishaev A, Roche J, Zasloff M, Bax A. J. Am. Chem. Soc. 2014;136:3752–3755. doi: 10.1021/ja4132642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee JH, Li F, Grishaev A, Bax A. J. Am. Chem. Soc. 2015;137:1432–1435. doi: 10.1021/ja512593s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bruschweiler R, Case DA. J. Am. Chem. Soc. 1994;116:11199–11200. [Google Scholar]
- 53.Emsley L, Bodenhausen G. Chem. Phys. Lett. 1990;165:469–476. [Google Scholar]
- 54.Geen H, Freeman R. J. Magn. Reson. 1991;93:93–141. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.