Abstract
Background
There is increasing evidence that retinal microvascular diameters are associated with cardio- and cerebrovascular conditions. The shared genetic effects of these associations are currently unknown. The aim of this study was to increase our understanding of the genetic factors that mediate retinal vessel size.
Methods and Results
This study extends previous genome-wide association study results using 24,000+ multi-ethnic participants from 7 discovery and 5,000+ subjects of European ancestry from 2 replication cohorts. Using the Illumina HumanExome BeadChip, we investigate the association of single nucleotide polymorphisms (SNPs) and variants collectively across genes with summary measures of retinal vessel diameters, referred to as the central retinal venule equivalent (CRVE) and the central retinal arteriole equivalent (CRAE). We report 4 new loci associated with CRVE, one of which is also associated with CRAE. The 4 SNPs are rs7926971 in TEAD1 (p=3.1×10−11, minor allele frequency (MAF)=0.43), rs201259422 in TSPAN10 (p=4.4×10−9, MAF=0.27), rs5442 in GNB3 (p=7.0×10−10, MAF=0.05) and rs1800407 in OCA2 (p=3.4×10−8, MAF=0.05). The latter SNP, rs1800407, was also associated with CRAE (p=6.5×10−12). Results from the gene-based burden tests were null. In phenotype look-ups, SNP rs201255422 was associated with both systolic (p=0.001) and diastolic blood pressure (p=8.3×10−04).
Conclusions
Our study expands the understanding of genetic factors influencing the size of the retinal microvasculature. These findings may also provide insight into the relationship between retinal and systemic microvascular disease.
Keywords: genetics, microcirculation, meta-analysis, exome, retina
Microvascular disease, affecting blood vessels 100–300 µm in size, plays a significant role in many conditions such as diabetes, stroke, hypertension, coronary artery disease and cognitive decline. It is difficult to study the microvasculature in organs such as the heart and brain and it requires invasive methods. However, retinal venules and arterioles can be photographed non-invasively and their characteristics quantified using computer software.1 In addition, the physiology and embryology of the brain and retinal microvasculature is similar.2 Since evidence suggests changes in retinal vessels may provide an indirect indicator of similar changes in the brain,3 heart4 and kidneys,5 a better understanding of the genes associated the retinal blood vessels may provide insight into microvascular morbidity elsewhere in the body.
Current evidence suggests retinal vessel diameters are associated with a wide array of diseases, reflecting the role and contribution of microvascular disease processes in these conditions.6–8 For instance, narrower retinal arterioles is a sign of hypertension and remarkably, the association between blood pressure and smaller retinal arteriole diameter can be seen with current,9 past10 and incident hypertension.11
Based on previous studies, retinal venule diameter is associated with different pathophysiologic processes than retinal arterioles. Wider retinal venules are associated with hyperglycemia,12 dyslipidemia,13 measures of inflammation,13 obesity,14 stroke,15 subclinical cerebrovascular disease,16 coronary heart disease,15 cardiovascular mortality in younger participants,17 cognitive impairment18 and renal dysfunction in diabetics.19
Even as the evidence that retinal microvascular diameter is a marker of more generalized microvascular changes in other organs increases, our understanding of the genetic factors that control retinal vessel size is limited. Previous studies have shown retinal vessel diameters are more highly correlated among related than unrelated individuals. These values range from 0.23 (95% CI: 0.16, 0.31) in retinal venule diameter and 0.20 (95% CI: 0.12, 0.28) in retinal arteriole diameter of siblings compared with 0.03 and 0.04 in spouses.20 In a twin study, the heritability of retinal vessels diameters was estimated at 83% (95% CI: 73%–89%) for retinal venule diameter and 70% (95% CI: 54%–80%) for retinal arteriole diameter.21 A genome-wide linkage study estimated the heritability at 48–51% for retinal vessel diameter22 and showed evidence that linkage regions overlapped with regions previously associated with hypertension, the eNOS-related pathway, coronary heart disease and vasculogenesis. Results from our first genome-wide association studies found evidence for four novel loci associated with retinal venule diameter23 and one associated with retinal arteriole diameter.24 These studies only included Caucasians and were both smaller with 15,358 and 18,722 participants respectively.
Nevertheless, much of the heritability of retinal vessel diameters remains to be explained. It is theorized that rare single nucleotide polymorphisms (SNPs), those with a minor allele frequency (MAF) < 1%, account for some of this unexplained heritability. Recently, the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium cohorts were genotyped with the Illumina HumanExome BeadChip.25 In this study we combine 24,000+ participants from four ethnic groups (Caucasians, African-Americans, Asians and Hispanics) to investigate the association of single-variant SNPs and SNP variants collectively across genes with summary measures of retinal venule and arteriole diameters, referred to as the central retinal venule equivalent (CRVE) and the central retinal arteriole equivalent (CRAE).
Methods
Participants
The discovery phase included four members of the CHARGE Consortium;26 the Age, Gene, Environment, Susceptibility Study – Reykjavik (AGES),27 the Atherosclerosis Risk in Communities Study (ARIC),28 the Cardiovascular Health Study (CHS)29 and the Multi-Ethnic Study of Atherosclerosis (MESA).30 Three additional Asian cohorts also contributed data, the Singapore Chinese Eye Study (SCES),31, the Singapore Malay Eye Study (SiMES)32 and the Singapore Indian Eye Study (SINDI).31 The overall mix of ethnic backgrounds included European ancestry (60%), African Americans (14%), Asians (21%) and Hispanics (4%), Table 1.
Table 1.
Description of Cohorts
| Discovery | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ethnicity | Cohort | n | CRAE (µm) |
CRVE (µm) |
Age (years) |
SBP (mm Hg) |
BMI (kg/m2) |
FG (mmol/l) |
Sex (% male) |
HTN (%) |
DM (%) |
| White | AGES | 2,755 | 140 ± 14 | 202 ± 20 | 76 ± 5 | 143 ± 20 | 27 ± 4 | 5.8 ± 1.1 | 43 | 81 | 11 |
| N=14,740 | ARIC | 8,377 | 135 ± 13 | 199 ± 19 | 60 ± 6 | 122 ± 18 | 28 ± 5 | 6.0 ± 1.8 | 46 | 40 | 12 |
| CHS | 1,472 | 139 ± 14 | 197 ± 19 | 79 ± 4 | 134 ± 21 | 27 ± 4 | 5.6 ± 1.6 | 41 | 60 | 14 | |
| MESA | 2,136 | 143 ± 14 | 207 ± 21 | 64 ± 10 | 124 ± 21 | 28 ± 5 | 5.3 ± 1.1 | 49 | 39 | 8 | |
| AA | ARIC | 1,848 | 137 ± 12 | 209 ± 19 | 59 ± 6 | 130 ± 20 | 30 ± 7 | 6.8 ± 3.3 | 37 | 61 | 23 |
| N=3,345 | CHS | 287 | 140 ± 13 | 207 ± 21 | 76 ± 4 | 136 ± 22 | 29 ± 5 | 6.0 ± 2.3 | 37 | 69 | 21 |
| MESA | 1,300 | 145 ± 14 | 222 ± 22 | 63 ± 10 | 136 ± 23 | 30 ± 6 | 5.7 ± 1.8 | 47 | 61 | 19 | |
| Asian | MESA | 600 | 143 ± 14 | 215 ± 20 | 63 ± 10 | 124 ± 22 | 24 ± 3 | 5.6 ± 1.5 | 51 | 37 | 15 |
| N=5,113 | SCES | 1,771 | 141 ± 16 | 208 ± 21 | 58 ± 9 | 136 ± 19 | 24 ± 4 | * | 51 | 56 | 13 |
| SiMES | 2,101 | 140 ± 16 | 219 ± 22 | 59 ± 11 | 147 ± 23 | 26 ± 5 | * | 50 | 69 | 22 | |
| SINDI | 641 | 143 ± 13 | 209 ± 18 | 53 ± 8 | 128 ± 18 | 26 ± 5 | * | 47 | 32 | 0 | |
| Hispanic | MESA | 1,077 | 146 ± 14 | 217 ± 21 | 62 ± 10 | 129 ± 24 | 30 ± 5 | 6.0 ± 2.2 | 50 | 45 | 21 |
|
Replication | |||||||||||
| White | BMES | 448 | 187 ± 17 | 224 ± 19 | 65 ±7 | 148 ± 20 | 28 ± 4 | 5.5 ± 1.7 | 47 | 81 | 10 |
| White | RS* Genotyped | 2,550 | 147 ± 14 | 222 ± 21 | 69 ± 8 | 139 ± 22 | 26 ± 4 | 6.8 ± 2.4 | 48 | 59 | 7 |
| RS Imputed | 2,474 | 147 ± 15 | 222 ± 21 | 67 ± 8 | 138 ± 22 | 26 ± 4 | 6.9 ± 2.7 | 34 | 60 | 7 | |
Rotterdam did not genotype all of their subjects using the exome chip. For those that were not genotyped they imputed the SNP for the remaining subjects. For clarity we analyzed the two groups separately.
CRAE-t: central retinal arteriole equivalent-trunk; CRVE: central retinal venule equivalent; SBP: systolic blood pressure; BMI: body mass index; FG: fasting glucose; HTN: hypertension; DM: diabetes mellitus; AA: African-American; AGES: Age, Gene, Environment, Susceptibility Study – Reykjavik; ARIC: Atherosclerosis Risk in Communities Study; CHS: Cardiovascular Health Study; MESA: Multi-Ethnic Study of Atherosclerosis; SCES: Singapore Chinese Eye Study; SiMES: Singapore Malay Eye Study; SINDI: Singapore Indian Eye Study; BMES: Blue Mountain Eye Study; RS: Rotterdam Study
The replication cohorts included the Rotterdam Study (RS)33 and a subset of the Blue Mountain Eye Study (BMES)34 that had exome chip data. Additional details of each participating study are provided in the Data Supplement. All participating cohorts secured approval from their respective institutional review boards, and all participants provided written informed consent in accordance with the Declaration of Helsinki.
Retinal Phenotypes
The central retinal venule equivalent (CRVE) and central retinal arteriole equivalent (CRAE) are summary measures of the six largest retinal microvascular diameters. Standardized protocols and software were developed at the University of Wisconsin.35 Retinal photographs were centered on the optic nerve. Photographs were obtained through pharmacologically dilated (AGES, BMES, RS) or un-dilated pupils of one (ARIC, CHS) or both eyes. Photographs were digitized using a high-resolution scanner (ARIC, CHS, RS) or digitally captured. All digital retinal images were analyzed with a semi-automated measurement system and the calibers of all retinal venules and arterioles were measured in the area located between half and one disc-diameter from the optic disc margin. Trunk vessels were measured using the Parr-Hubbard-Knudtson formulas36 and measures are expressed in micrometers (µm).
Exome Chip
Cohort participants (AGES, ARIC, CHS, MESA, RS, SCES, SINDI) were genotyped with the Illumina HumanExome BeadChip v1.0 (Illumina, Inc., San Diego, CA) and v1.1 for SiMES. It covers putative functional exonic variants selected from over 12,000 individual exome and whole-genome sequences. It consists of 247,870 markers comprised primarily of 219,621 non-synonymous single nucleotide polymorphisms (SNPs) representing European, African, Chinese and Hispanic populations. Additional markers include splice-site variation, stop codons, promoter region SNPs, GWAS tag markers, ancestry informative markers, X / Y / mitochondrial variants, SNPs in the extended major histocompatibility complex region, human leukocyte antigen tag variants, identical by descent markers and insertion-deletions. (Exome-array design: http://genome.sph.umich.edu/wiki/Exome_Chip_Design)
Genotyping and Quality Control
Approximately 62,000 ethnically diverse samples from the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium were genotyped with the Illumina HumanExome BeadChip. The raw data files for the samples were assembled into a single project for joint calling and are described in detail elsewhere.25 Briefly, Illumina’s GenTrain version 2.0 clustering algorithm in GenomeStudio or zCall37 was used for genotype calling. A total of 8,997 variants failed quality control measures and were eliminated from all analyses. Additional details regarding genotyping and QC performed centrally and by each study are summarized in the Data Supplement. Annotation of the v1.0 exome chip variants are expressed with dbNSFP v2.0.
Statistical Analysis
The discovery phase included single variant tests and gene-based association analyses in which CRVE and CRAE were analyzed separately. Where applicable, each of the seven cohorts, (AGES, ARIC, CHS, MESA, SCES, SiMES and SINDI) analyzed their participants by ethnicity, Caucasian, African-American, Asian or Hispanic. This resulted in 12 ethnic specific groups that were combined for meta-analysis to elucidate any ethnic specific differences in the results. For the single variant tests, genome wide significance was set at p=2.0×10−7 (0.05/~250,000 variants). Significance for the gene-based tests was set at 2.5×10−6 (0.05/20,000 genes). To validate findings from the discovery process, a replication phase was performed using two independent cohorts (BMES and RS). Replication was considered significant at p<0.05 and the direction of the effect was consistent with discovery.
Diabetes was defined as fasting blood glucose≥126 mg/dL [7.0 mmol/L], current use of insulin or other hypoglycemic agent. If fasting blood glucose was not available then a random glucose>198 mg/dL [11.0 mmol/L] was used. Hypertension was defined as current use of blood pressure medication or systolic/diastolic blood pressure (SBP/DBP)≥140/90 mm Hg.
Residuals were analyzed because results using untransformed CRVE & CRAE led to inflation of our findings due to extreme phenotypes in the adjusted analysis and small minor-allele frequencies of many of the SNPs, Figures 1SA & 2SA. Residuals were derived by linear regression for each outcome adjusting for age (years), sex (male–yes/no), BMI (kg/m2), SBP (Hg mm), diabetes (yes/no), hypertension (yes/no), the first 5 ethnic and study-specific principle components and study site (where applicable). An inverse normal transformation of the residuals was performed. Each cohort then analyzed the transformed residuals using the R package seqMeta or SkatMeta (http://www.r-project.org) with linear regression adjusting for the SNP using an additive genetic model, the first 5 ethnic and study-specific principle components and study site. Adjusting for population stratification in the calculation of the residuals alone would not be enough to correct for any similar stratification which may occur in our primary analysis induced by the inclusion of SNP data. Model based standard errors were used in the analysis.
Inverse variance weighted fixed-effects meta-analysis was performed by ethnic group for single variant tests and gene-based association analyses including the Sequence Kernel Association Test (SKAT)38 and the T1 Count Test39 using the R package seqMeta. Gene-based burden tests were included to aggregate evidence from multiple rare/low-frequency variants. We used a minor allele frequency threshold of 0.01 in the T1 Test and 0.05 in the SKAT Test.
The Data Supplement contains additional information on the choice of p-value for replication, choice of minor allele frequency threshold for the burden tests, information regarding forest and regional association plots, and details of the the in-silico algorithms used to predict whether SNPs would be damaging.
Rotterdam did not genotype all of their subjects using the exome chip. For those that were not genotyped they imputed the SNPs for the remaining subjects. For clarity we analyzed the two groups separately. Details of the imputation are supplied in the Data Supplement. Replication of gene based results was carried out using Rotterdam data only.
The CHARGE Exome Chip Blood Pressure Consortium provided phenotype look-ups for 4 newly and 4 previously identified SNPs associated with CRVE or CRAE. Blood pressure phenotypes included SBP, DBP and hypertension.
Results
Table 1 displays the demographic and covariate characteristics stratified by ethnic group within participating cohorts where available. A total of 24,342 participants had measures of CRVE and 24,345 had measures of CRAE. After marker-level and sample-level quality control 223,566 variants were available for analyses of CRVE and 240,931 for CRAE.
Table 2 contains the results from the discovery phase of our meta-analysis for the single variant tests. Three CRVE SNPs reached exome-wide significance, rs7926971 (p=7.0×10−9) in TEAD1, rs201259422 (p=8.0×10−9) in TSPAN10 and rs5442 (p=5.8×10−8) in GNB3. Two other single SNPs of interest were carried forward for replication, rs1800407 (p=2.1x 10−7) in OCA2 and rs2306765 (p=6.9×10−6) in GSG1. One CRAE SNP reached exome-wide significance, rs1800407 (p=4.5×10−10) in OCA2. During replication, 3 CRVE SNPs (rs7926971 in TEAD1, rs5442 in GNB3, rs1800407 in OCA2) and 1 CRAE SNP (rs1800407 in OCA2) all reached nominal significance (p<0.05) or better. SNPs rs201259422 in TSPAN10 and rs2306765 in GSG1 did not replicate, but the direction and effect size for rs201259422 in TSPAN10 was consistent in the replication sample, and overall increased the statistical significance in the combined sample. Figures 1 – 6, show the regional association plots for each of the SNPs carried forward for replication. No additional ancestry specific SNPs reached exome-wide significance for CRVE or CRAE.
Table 2.
Summary of findings
| CRVE | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Discovery | effect | Replication | Combined | ||||||||||||
| SNP | gene | function | maf | n | allele | beta* | se | p | n | beta | se | p | beta | se | p |
| rs7926971 | TEAD1 | Intron | 0.43 | 24,342 | G | 0.05 | 0.01 | 7.0x10−9 | 5,472 | 0.06 | 0.02 | 0.001 | 0.05 | 0.01 | 3.1x10−11 |
| rs201259422 | TSPAN10 | missense | 0.27 | 22,566 | A | −0.06 | 0.01 | 8.0x10−9 | 2,998 | −0.04 | 0.03 | 0.16 | −0.06 | 0.01 | 4.4x10−9 |
| rs5442 | GNB3 | missense | 0.05 | 22,566 | A | −0.12 | 0.02 | 5.8x10−8 | 5,472 | −0.10 | 0.04 | 0.003 | −0.11 | 0.02 | 7.0x10−10 |
| rs1800407 | OCA2 | missense | 0.05 | 22,566 | T | −0.12 | 0.02 | 2.1x10−7 | 5,472 | −0.09 | 0.04 | 0.05 | −0.11 | 0.02 | 3.4x10−8 |
| rs2306765 | GSG1 | missense | 0.05 | 24,342 | G | −0.10 | 0.02 | 6.9x10−6 | 5,024 | 0.14 | 0.07 | 0.03 | −0.07 | 0.02 | 3.5x10−4 |
|
CRAE | |||||||||||||||
| rs1800407 | OCA2 | missense | 0.05 | 22,569 | T | −0.14 | 0.02 | 4.5x10−10 | 5,472 | −0.13 | 0.04 | 0.004 | −0.14 | 0.02 | 6.5x10−12 |
Beta and standard errors represent inverse-quantile normalized traits not direct measures of CRVE and CRAE-t.
CRVE: central retinal venule equivalent; CRAE: central retinal arteriole equivalent; SNP: single nucleotide polymorphisms; maf: minor allele frequency; se: standard error; p: p-value
Figure 1.
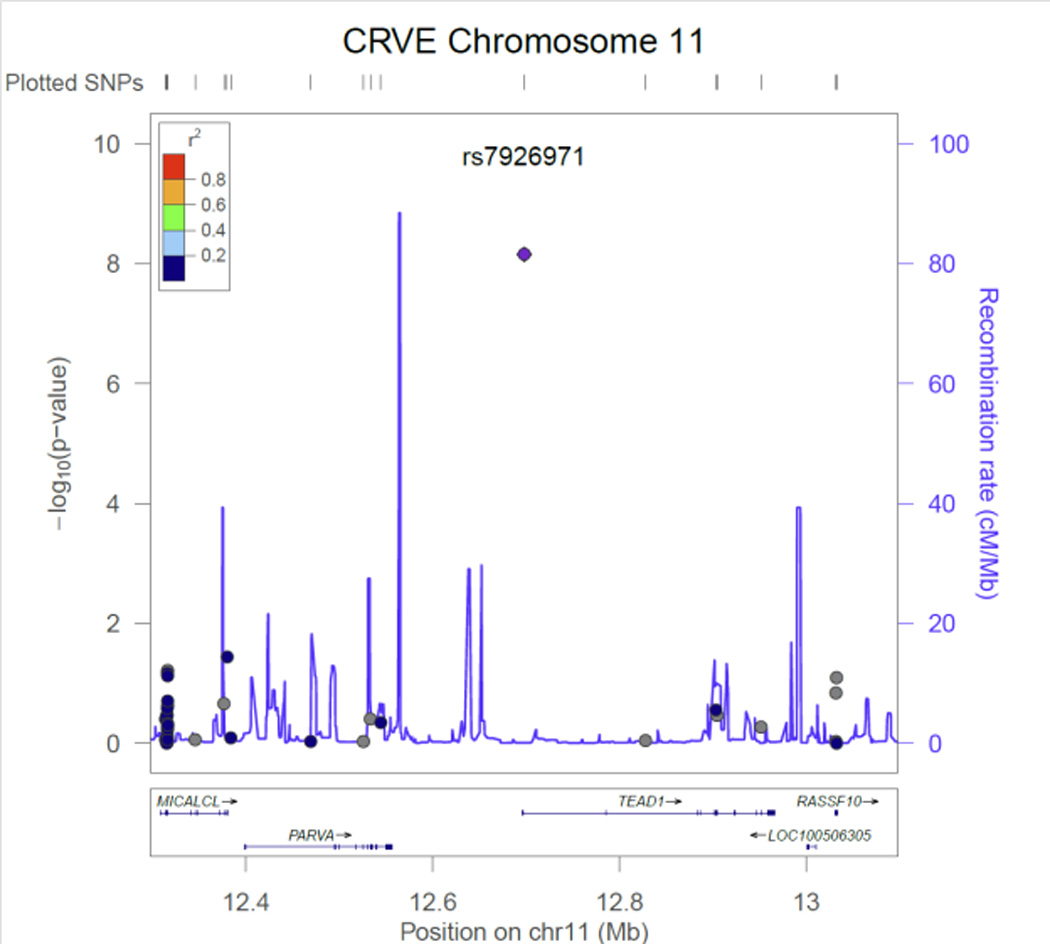
CRVE regional association plot for SNP rs7926971 in TEAD1.
Figure 6.
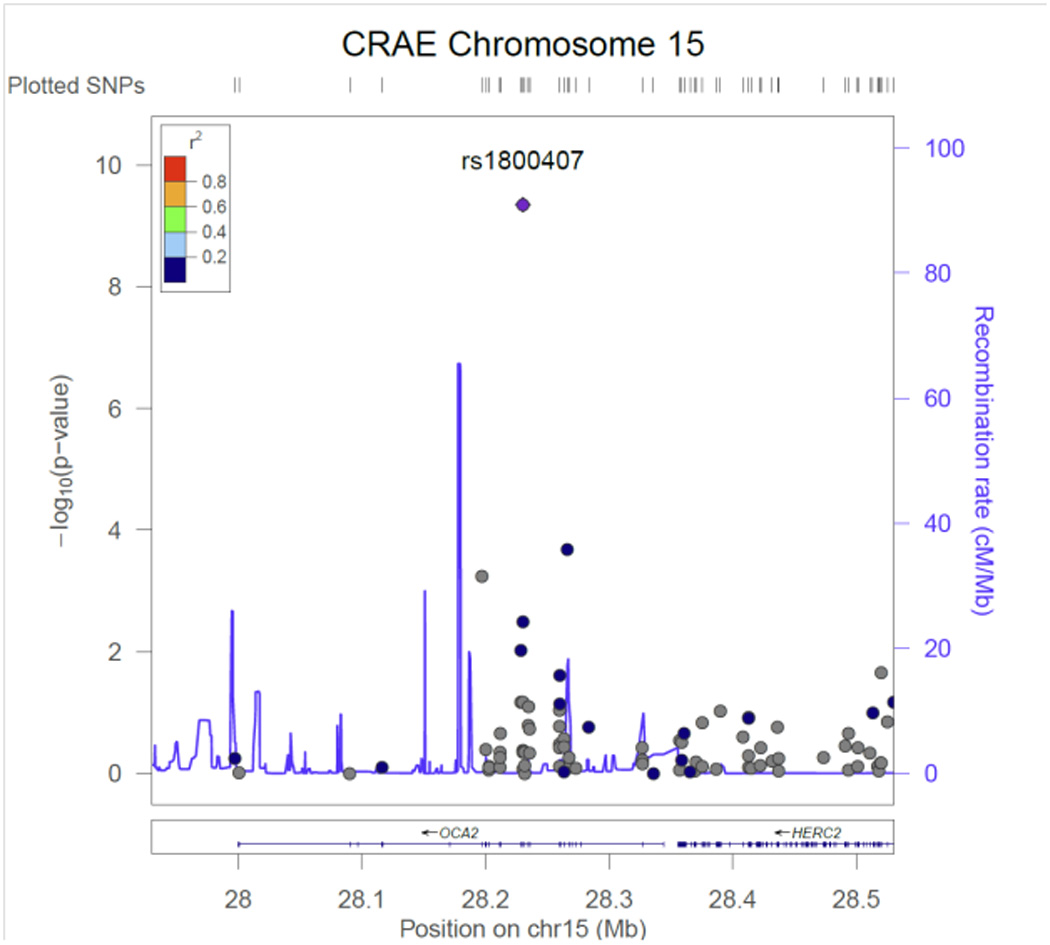
CRAE regional association plot for SNP rs1800407in OCA2.
Data Supplement Table 1S contains a more detailed summary of the top SNPs associated with CRVE including the four SNPs we have previously reported23 and the other SNPs in high linkage disequilibrium with those four SNPs. Similarly, Data Supplement Table 2S contains a more detailed summary of the top SNPs associated with CRAE including a SNP in high linkage disequilibrium with the SNP we have previously reported.24 All of the previously reported GWAS findings reached exome-wide significance in the present study. These tables include the annotation of the v1.0 exome chip variants.
Results from the gene-based T1 burden tests were null for both CRVE and CRAE. In the SKAT test, GSG1 was associated with CRVE (p=1.3×10−6) and OCA2 was associated with CRAE (p=2.1×10−8). In GSG1, there were 9 SNPs with cumulative minor allele frequency (MAF) of 0.06. We repeated gene-based testing by excluding rs2306765 after which the gene was not associated with CRVE (cumulative MAF of 0.01 and p=0.01). For CRAE in OCA2, there were 38 SNPs included with cumulative MAF of 0.09. Similarly, the gene was not associated with CRAE after excluding rs1800407 (cumulative MAF of 0.04 and p=0.03). Neither gene, GSG1 for CRVE or OCA2 in CRAE reached nominal significance in replication.
Data Supplement Figures 3S – 8S show the direction of effects across all studies. These Figures showed no overall heterogeneity in the meta-analysis results any of the SNPs There was evidence the effect size in MESA Hispanics for rs7926971 (TEAD1) differed from other ethnic groups (p=0.01), Figure 3S, and that the effect size in MESA Asians for rs201259422 (TSPAN10) differed from other Asian cohorts (p=0.02), Figure 4S.
A look-up of the 4 newly and 4 previously identified SNPs associated with CRVE or CRAE was performed by the CHARGE Exome Chip Blood Pressure Consortium, Table 3S. Of the 8 SNPs, 5 SNPs were either associated with SBP, DBP, hypertension or a combination of the three phenotypes. However, after correction for multiple testing, (p=0.05/24=0.002), only 2 SNPs remained associated with blood pressure phenotypes, rs201259422 in TSPAN10 and rs10774625 in ATXN2.
Discussion
Our collaboration has identified four new SNPs associated with retinal venule diameter and one new SNP associated with retinal arteriole diameter bringing the total to eight SNPs now associated with venule diameter and two with arteriole diameter. After adjusting for age, sex, BMI, diabetes (Y/N), hypertension (Y/N) and SBP, the new variants explained 1% of the total variability in CRVE (4 SNPs) and <½% for CRAE (1 SNP). This increased to 2% for CRVE (8 SNPs) and ½% for CRAE (2 SNPs) with all known variants.
Four SNPs identified in our discovery cohort showed evidence of replication in our study; three SNPs for CRVE in TEAD1, GNB3 and OCA2 and one SNP for CRAE in OCA2. The SNP in TSPAN10 for CRVE did not replicate, but the direction of the association and the combined discovery and replication results provide some support for its association with CRVE. Our inability to replicate outright may have been due in part to the small size of the replication cohort and smaller effect sizes compared to those seen in some of the other SNPs. The SNP in GSG1 did show a significant p-value in replication but the association was in the opposite direction from what was observed in discovery. It is of note; none of the new findings were low frequency SNPs. The SNPs in TSPAN10 and OCA2 were not genotyped or imputed in our previous GWAS. The SNP in TEAD1 was included as an imputed SNP. The SNP in GNB3 was genotyped but results did not pass quality control. It is unclear if genotyping, covariate adjustment, smaller effect sizes or a combination of these factors resulted in the conflicting results in our GWAS and the present study.
SNP rs5442 in GNB3 is a predicted damaging non-synonymous SNP on chromosome 12. Each copy of the minor allele decreased CRVE 2.3 ± 0.4 µm. (This decrease and the changes cited below are the observed effects on CRVE and CRAE in the untransformed analyses, not the beta and standard errors from the inverse-quantile normalized traits.) Heterotrimeric guanine nucleotide-binding proteins (G proteins), integrate signals between receptors and effector proteins, are composed of an alpha, a beta, and a gamma subunit and are expressed in all tissues.40 A separate SNP (C825T) in this gene is associated with essential hypertension, obesity and dyspepsia.40, 41
SNP rs1800407 in OCA2 is a predicted damaging non-synonymous SNP on chromosome 15. Each copy of the minor allele decreased CRAE and CRVE, 1.7 ± 0.3 and 2.2 ± 0.4 µm respectively. OCA2 encodes the human homolog of the mouse p (pink-eyed dilution) gene.42 The encoded protein is believed to be an integral membrane protein involved in small molecule transport, specifically tyrosine, which is a precursor to melanin synthesis. It is involved in mammalian pigmentation, where it may control skin color variation and act as a determinant of brown or blue eye color. Mutations in this gene result in type 2 oculocutaneous albinism.43
SNP rs7926971 in TEAD1 is an intronic SNP on chromosome 11. Each copy of the minor allele increased CRVE 0.5 ± 0.1 µm. TEAD1 encodes a ubiquitous transcriptional enhancer factor that is a member of the TEA/ATTS domain family. This protein directs the transactivation of a wide variety of genes and, in placental cells, also acts as a transcriptional repressor. Mutations in this gene cause Sveinsson's chorioretinal atrophy.44 The expression of TEAD1 is significantly induced during smooth muscle cell phenotypic modulation and negatively correlates with smooth muscle-specific gene expression. Mechanistically, TEAD1 competes with myocardin for binding to serum response factor (SRF), resulting in disruption of myocardin and SRF interactions and thereby attenuating expression of smooth muscle-specific genes.45
SNP rs201259422 in TSPAN10 (tetraspanin 10) is a predicted damaging SNP on chromosome 17. Each copy of the minor allele decreased CRVE 1.0 ± 0.2 µm. TSPAN10 encodes a transcript that is expressed in human RPE & choroid.46 Tetraspanins are widespread, numerous and largely mysterious in function but are involved in diverse processes such as cell activation and proliferation, adhesion and motility, differentiation, and cancer.47 Two related tetraspan proteins of the retina, ROM1 and RDS, are both involved in retinal degenerations.48
The SNP in OCA2 and the previously reported SNP rs17421627 in LINC00461 adjacent to MEF2C23, 24 represent two SNPs now that are associated with both CRAE & CRVE. None of the other previously reported SNPs associated with CRVE were associated with CRAE in our data, nor was the SNP in TEAD1, Table 1S. The associations between CRAE and the SNPs in TSPAN10 and GNB3 are weaker than for CRVE but not trivial. Despite the strong correlation between CRVE and CRAE it appears that not only is retinal venule diameter associated with different pathophysiologic processes than retinal arterioles but that the genetic control of retinal venules and retinal arterioles is not identical either.
It is unknown why more genetic loci have been identified for CRVE than CRAE. CRVE has slightly less measurement error than CRAE35 but is not likely the major reason. Second, hypertension and medications to treat hypertension influence retinal arterioles (CRAE) more than venules (CRVE). Consequently, we adjusted for both systolic blood pressure and medication use in our analyses. Finally, there are anatomical differences in the two types of vessels. Unlike retinal venules, retinal arterioles auto-regulate blood flow in response to changes in perfusion pressure and like the cerebral vasculature are largely independent of extrinsic neurogenic factors.49 This may make genetic effects on CRAE smaller than the corresponding effects on CRVE and thus may take larger sample sizes to detect, for instance see rs20125942 (TSPAN10) in Table 3S.
Two of the eight SNPs associated with CRVE or CRAE were associated with blood pressure phenotypes, rs201259422 on TSPAN10 and rs10774325 on ATXN2. SNP rs10774625 (ATXN2) was previously reported to be associated with blood pressure.50 However, to our knowledge this is the first reported association of rs201259422 (TSPAN10) with blood pressure phenotypes. Current thinking is that wider retinal venules and narrower retinal arterioles are associated with increased risk of hypertension,51 but that relationship does not hold for rs201259422 (TSPAN10) & rs10774625 (ATXN2) in our analyses, Table 3S. Other SNPs, like rs1800407 (OCA2) and rs17421627 (MEF2C) had relatively large effects on retinal vessel caliber. However, there was no obvious relationship with blood pressure phenotypes. These findings may reflect developmental differences independent of cumulative or secondary effects occurring over an individual’s lifetime. SNPs rs7926971 (TEAD1) and rs2287921 (RASIP1) show an association with CRVE but not CRAE or blood pressure phenotypes. The significance of these finding remains to be determined.
Strengths of this study include a large sample size, an ethnically diverse population-based sample, quantitative phenotypes assessed in a similar manner, and genotyping performed on the exome array enriched for coding variants with many cohorts involved in joint calling. Weaknesses include a small replication cohort, cross-sectional measures and a phenotype that is a summary measure of the six largest retinal arteriole and venule diameters.
In summary, this study increases our understanding about genes associated with retinal venule and arteriole diameter. However, considerable work remains to be done to determine how these genes act, if their effects vary over time, whether these genes interact or are subject to epigenetic factors and ultimately whether these genes contribute to the risk of microvascular disease in other organs or are associated with major conditions such as diabetes, stroke, hypertension, coronary disease and cognitive decline.
Supplementary Material
Figure 2.
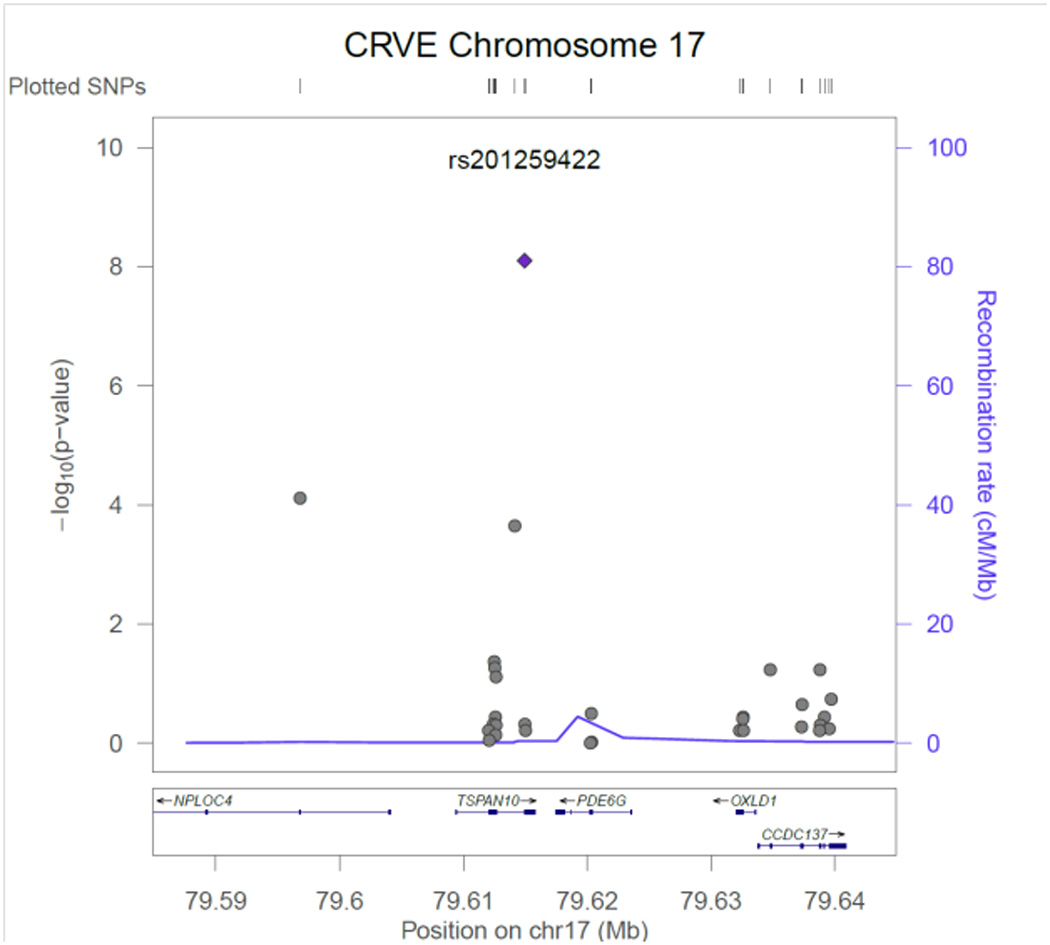
CRVE regional association plot for SNP rs201259422in TSPAN10.
Figure 3.
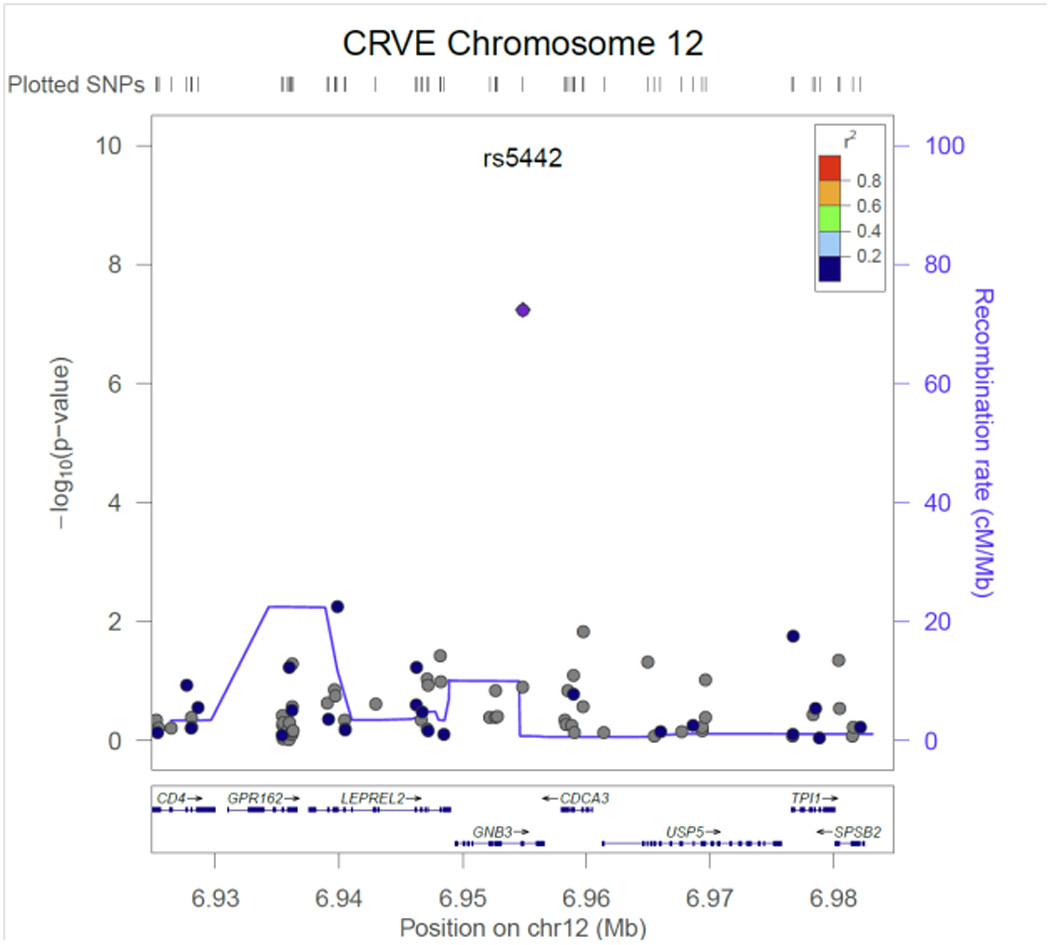
CRVE regional association plot for SNP rs5442in GNB3.
Figure 4.
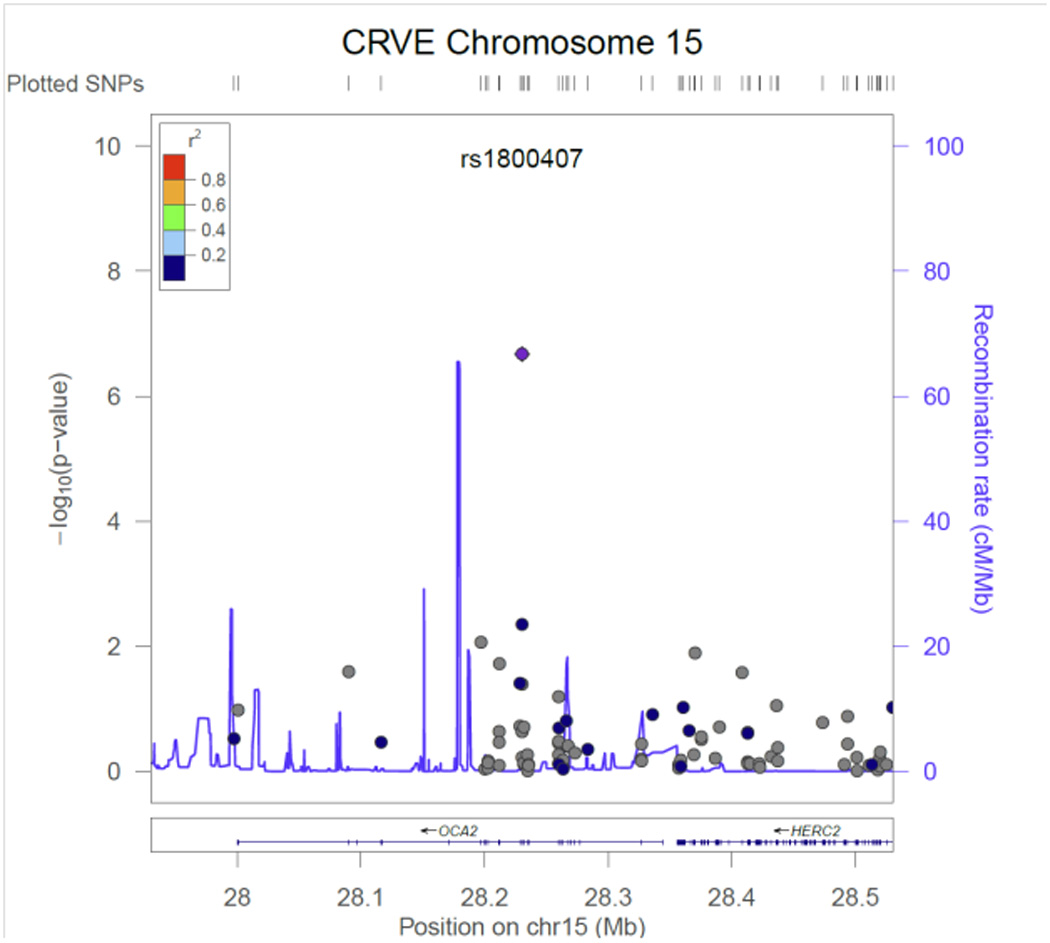
CRVE regional association plot for SNP rs1800407in OCA2.
Figure 5.
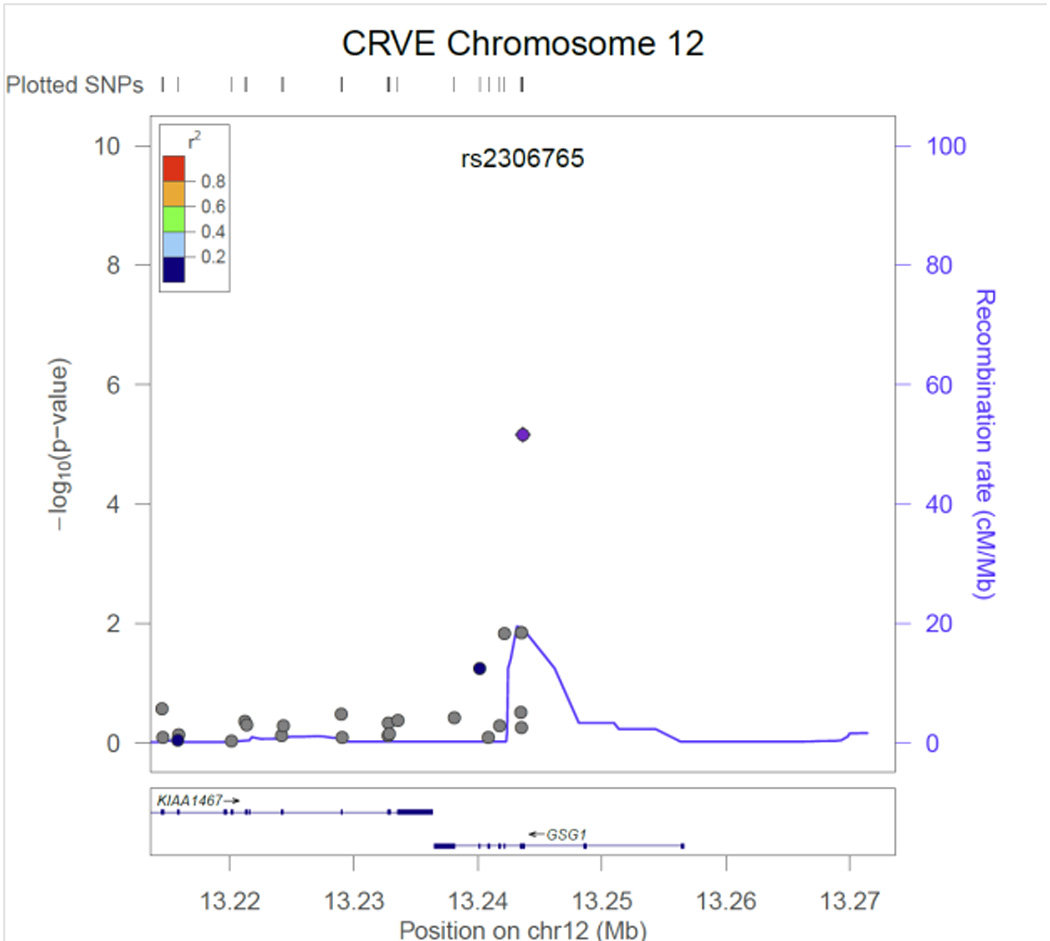
CRVE regional association plot for SNP rs2306765 in GSG1.
Clinical Perspective.
Microvascular disease, affecting blood vessels 100–300 µm in size, plays a significant role in many conditions such as diabetes, stroke, hypertension, coronary artery disease and cognitive decline. It is difficult to study the microvasculature in organs such as the heart and brain and it requires invasive methods. However, retinal venules and arterioles can be photographed non-invasively and their characteristics quantified using computer software. In addition, the physiology and embryology of the brain and retinal microvasculature is similar. Since evidence suggests changes in retinal vessels may provide an indirect indicator of similar changes in the brain, heart and kidneys, a better understanding of the genes associated the retinal blood vessels may provide insight into microvascular morbidity elsewhere in the body.
Acknowledgments
Funding Sources: AGES: This research was supported by the National Institute of Health through the Intramural Research Program of the National Institute of Aging (ZIAAG007380) and the National Eye Institute (ZIAEY00401), National Institute of Health contract number N01-AG-1-2100, Hjartavernd (the Icelandic Heart Association), the Althingi (Icelandic Parliament), and the University of Iceland Research Fund. We are indebted to IHA clinic staff and to the AGES participants whose decision to volunteer made this study possible. The funders had no role in data collection, management, analysis and interpretation of the data, preparation, writing and approval of the manuscript, or decision to submit the manuscript for publication. ARIC: The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute (NHLBI) contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C and HHSN268201100012C), R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. Support for the exome chip genotyping and joint calling was provided by Building on GWAS for NHLBI-diseases: the U.S. CHARGE consortium through the National Institutes of Health American Recovery and Reinvestment Act of 2009 (5RC2HL102419) (PI: E. Boerwinkle). We thank the staff and participants of the ARIC study for their important contributions. CHS: This research was supported by NHLBI contracts HHSN268201200036C, HHSN268200800007C, N01HC55222, N01HC75150, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086; and NHLBI grants U01HL080295, HL080295, HL087652, HL103612, HL120393, HL105756, HL068986 with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through AG023629 from the National Institute on Aging (NIA). A full list of CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi.htm. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. MESA – MESA Family – MESA SHARe – CTSI – DRC: This research was supported by the Multi-Ethnic Study of Atherosclerosis (MESA) contracts N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, by grant HL071205 and by UL1-DR-001079 from NCRR. Funding for MESA SHARe genotyping was provided by NHLBI Contract N02-HL-6-4278. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The authors thank the participants of the MESA study, the Coordinating Center, MESA investigators, and study staff for their valuable contributions. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org. SCES, SINDI & SiMES: The Singapore Chinese Eye Study (SCES), Singapore Malay Eye Study (SiMES) and Singapore Indian Eye Study (SINDI) were supported by the National Medical Research Council (NMRC), Singapore (grants 0796/2003, IRG07nov013, IRG09nov014, NMRC 1176/2008, STaR/0003/2008, CG/SERI/2010), and Biomedical Research Council (BMRC), Singapore (08/1/35/19/550 and 09/1/35/19/616). Ching-Yu Cheng is supported by an award from NMRC (CSA/033/2012). The Singapore Tissue Network and the Genome Institute of Singapore, Agency for Science, Technology and Research, Singapore provided services for tissue archival and genotyping, respectively. BMES: The Blue Mountains Eye Study has been supported by the Australian National Health & Medical Research Council, Canberra Australia (Grant Numbers 974159, 211069, 457349, 512423, 475604, 529912, and the funding for Centre for Clinical Research Excellence in Translational Clinical Research in Eye Diseases, CCRE in TCR-Eye). The cost of genotyping was funded by the NEI/NIH as part of the International AMD Genomics Consortium (IAMDGC), an international collaborative effort to investigate genetics of age-related macular degeneration. Genotyping was performed at the Center for Inherited Diseases Research (CIDC) at the Johns Hopkins University, USA. We also acknowledge the funding body National Institutes of Health Research (NIHR) Biomedical Research Centre for Ophthalmology, Moorfields Eye Hospital and UCL Institute of Ophthalmology, London, UK. RS: The generation and management of the Illumina exome chip v1.0 array data for the Rotterdam Study (RS-I) was executed by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, Rotterdam, The Netherlands. The Exome chip array data set was funded by the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, from the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO)-sponsored Netherlands Consortium for Healthy Aging (NCHA; project nr. 050-060-810); the Netherlands Organization for Scientific Research (NWO; project number 184021007) and by the Rainbow Project (RP10; Netherlands Exome Chip Project) of the Biobanking and Biomolecular Research Infrastructure Netherlands (BBMRI-NL; www.bbmri.nl). We thank Ms. Mila Jhamai, Ms. Sarah Higgins, and Mr. Marijn Verkerk for their help in creating the exome chip database, and Carolina Medina-Gomez, MSc, Lennard Karsten, MSc, and Linda Broer PhD for QC and variant calling. Variants were called using the best practice protocol developed by Grove et al. as part of the CHARGE consortium exome chip central calling effort.
Footnotes
Conflict of Interest Disclosures: None
References
- 1.Liew G, Wang JJ, Mitchell P, Wong TY. Retinal vascular imaging: a new tool in microvascular disease research. Circ Cardiovasc Imaging. 2008;1:156–161. doi: 10.1161/CIRCIMAGING.108.784876. [DOI] [PubMed] [Google Scholar]
- 2.Wong TY, Klein R, Klein BE, Tielsch JM, Hubbard L, Nieto FJ. Retinal microvascular abnormalities and their relationship with hypertension, cardiovascular disease, and mortality. Surv Ophthalmol. 2001;46:59–80. doi: 10.1016/s0039-6257(01)00234-x. [DOI] [PubMed] [Google Scholar]
- 3.Cheung CY, Ong YT, Ikram MK, Chen C, Wong TY. Retinal microvasculature in Alzheimer's disease. J Alzheimers Dis. 2014;42(Suppl 4):S339–S352. doi: 10.3233/JAD-141596. [DOI] [PubMed] [Google Scholar]
- 4.Sasongko MB, Wong TY, Wang JJ. Retinal arteriolar changes: intermediate pathways linking early life exposures to cardiovascular disease? Microcirculation. 2010;17:21–31. doi: 10.1111/j.1549-8719.2009.00007.x. [DOI] [PubMed] [Google Scholar]
- 5.Skyler JS. Microvascular complications. Retinopathy and nephropathy. Endocrinol Metab Clin North Am. 2001;30:833–856. doi: 10.1016/s0889-8529(05)70218-8. [DOI] [PubMed] [Google Scholar]
- 6.Cheung CY, Ikram MK, Klein R, Wong TY. The clinical implications of recent studies on the structure and function of the retinal microvasculature in diabetes. Diabetologia. 2015;58:871–885. doi: 10.1007/s00125-015-3511-1. [DOI] [PubMed] [Google Scholar]
- 7.Cheung CY, Ikram MK, Sabanayagam C, Wong TY. Retinal microvasculature as a model to study the manifestations of hypertension. Hypertension. 2012;60:1094–1103. doi: 10.1161/HYPERTENSIONAHA.111.189142. [DOI] [PubMed] [Google Scholar]
- 8.Al-Fiadh AH, Wong TY, Kawasaki R, Clark DJ, Patel SK, Freeman M, et al. Usefulness of retinal microvascular endothelial dysfunction as a predictor of coronary artery disease. Am J Cardiol. 2015;115:609–613. doi: 10.1016/j.amjcard.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 9.Wong TY, Klein R, Klein BE, Meuer SM, Hubbard LD. Retinal vessel diameters and their associations with age and blood pressure. Invest Ophthalmol Vis Sci. 2003;44:4644–4650. doi: 10.1167/iovs.03-0079. [DOI] [PubMed] [Google Scholar]
- 10.Leung H, Wang JJ, Rochtchina E, Wong TY, Klein R, Mitchell P. Impact of current and past blood pressure on retinal arteriolar diameter in an older population. J Hypertens. 2004;22:1543–1549. doi: 10.1097/01.hjh.0000125455.28861.3f. [DOI] [PubMed] [Google Scholar]
- 11.Wong TY, Shankar A, Klein R, Klein BE, Hubbard LD. Prospective cohort study of retinal vessel diameters and risk of hypertension. BMJ. 2004;329:79. doi: 10.1136/bmj.38124.682523.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikram MK, Janssen JA, Roos AM, Rietveld I, Witteman JC, Breteler MM, et al. Retinal vessel diameters and risk of impaired fasting glucose or diabetes: the Rotterdam study. Diabetes. 2006;55:506–510. doi: 10.2337/diabetes.55.02.06.db05-0546. [DOI] [PubMed] [Google Scholar]
- 13.Wong TY, Islam FM, Klein R, Klein BE, Cotch MF, Castro C, et al. Retinal vascular caliber, cardiovascular risk factors, and inflammation: the multi-ethnic study of atherosclerosis (MESA) Invest Ophthalmol Vis Sci. 2006;47:2341–2350. doi: 10.1167/iovs.05-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang JJ, Taylor B, Wong TY, Chua B, Rochtchina E, Klein R, et al. Retinal vessel diameters and obesity: a population-based study in older persons. Obesity (Silver Spring) 2006;14:206–214. doi: 10.1038/oby.2006.27. [DOI] [PubMed] [Google Scholar]
- 15.Wong TY, Kamineni A, Klein R, Sharrett AR, Klein BE, Siscovick DS, et al. Quantitative retinal venular caliber and risk of cardiovascular disease in older persons: the cardiovascular health study. Arch Intern Med. 2006;166:2388–2394. doi: 10.1001/archinte.166.21.2388. [DOI] [PubMed] [Google Scholar]
- 16.Ikram MK, De Jong FJ, Van Dijk EJ, Prins ND, Hofman A, Breteler MM, et al. Retinal vessel diameters and cerebral small vessel disease: the Rotterdam Scan Study. Brain. 2006;129:182–188. doi: 10.1093/brain/awh688. [DOI] [PubMed] [Google Scholar]
- 17.Wang JJ, Liew G, Wong TY, Smith W, Klein R, Leeder SR, et al. Retinal vascular calibre and the risk of coronary heart disease-related death. Heart. 2006;92:1583–1587. doi: 10.1136/hrt.2006.090522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Jong FJ, Schrijvers EM, Ikram MK, Koudstaal PJ, de Jong PT, Hofman A, et al. Retinal vascular caliber and risk of dementia: the Rotterdam study. Neurology. 2011;76:816–821. doi: 10.1212/WNL.0b013e31820e7baa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein R, Klein BE, Moss SE, Wong TY. Retinal vessel caliber and microvascular and macrovascular disease in type 2 diabetes: XXI: the Wisconsin Epidemiologic Study of Diabetic Retinopathy. Ophthalmology. 2007;114:1884–1892. doi: 10.1016/j.ophtha.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 20.Lee KE, Klein BE, Klein R, Knudtson MD. Familial aggregation of retinal vessel caliber in the beaver dam eye study. Invest Ophthalmol Vis Sci. 2004;45:3929–3933. doi: 10.1167/iovs.04-0462. [DOI] [PubMed] [Google Scholar]
- 21.Taarnhoj NC, Larsen M, Sander B, Kyvik KO, Kessel L, Hougaard JL, et al. Heritability of retinal vessel diameters and blood pressure: a twin study. Invest Ophthalmol Vis Sci. 2006;47:3539–3544. doi: 10.1167/iovs.05-1372. [DOI] [PubMed] [Google Scholar]
- 22.Xing C, Klein BE, Klein R, Jun G, Lee KE, Iyengar SK. Genome-wide linkage study of retinal vessel diameters in the Beaver Dam Eye Study. Hypertension. 2006;47:797–802. doi: 10.1161/01.HYP.0000208330.68355.72. [DOI] [PubMed] [Google Scholar]
- 23.Ikram MK, Sim X, Jensen RA, Cotch MF, Hewitt AW, Ikram MA, et al. Four novel Loci (19q13, 6q24, 12q24, and 5q14) influence the microcirculation in vivo. PLoS Genet. 2010;6:e1001184. doi: 10.1371/journal.pgen.1001184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sim X, Jensen RA, Ikram MK, Cotch MF, Li X, MacGregor S, et al. Genetic loci for retinal arteriolar microcirculation. PLoS One. 2013;8:e65804. doi: 10.1371/journal.pone.0065804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grove ML, Yu B, Cochran BJ, Haritunians T, Bis JC, Taylor KD, et al. Best practices and joint calling of the HumanExome BeadChip: the CHARGE Consortium. PLoS One. 2013;8:e68095. doi: 10.1371/journal.pone.0068095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Psaty BM, O'Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JI, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2:73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris TB, Launer LJ, Eiriksdottir G, Kjartansson O, Jonsson PV, Sigurdsson G, et al. Age, Gene/Environment Susceptibility-Reykjavik Study: multidisciplinary applied phenomics. Am J Epidemiol. 2007;165:1076–1087. doi: 10.1093/aje/kwk115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. Am J Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 29.Fried LP, Borhani NO, Enright P, Furberg CD, Gardin JM, Kronmal RA, et al. The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991;1:263–276. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- 30.Bild DE, Bluemke DA, Burke GL, Detrano R, Diez Roux AV, Folsom AR, et al. Multi-ethnic study of atherosclerosis: objectives and design. Am J Epidemiol. 2002;156:871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 31.Lavanya R, Jeganathan VS, Zheng Y, Raju P, Cheung N, Tai ES, et al. Methodology of the Singapore Indian Chinese Cohort (SICC) eye study: quantifying ethnic variations in the epidemiology of eye diseases in Asians. Ophthalmic Epidemiol. 2009;16:325–336. doi: 10.3109/09286580903144738. [DOI] [PubMed] [Google Scholar]
- 32.Foong AW, Saw SM, Loo JL, Shen S, Loon SC, Rosman M, et al. Rationale and methodology for a population-based study of eye diseases in Malay people: The Singapore Malay eye study (SiMES) Ophthalmic Epidemiol. 2007;14:25–35. doi: 10.1080/09286580600878844. [DOI] [PubMed] [Google Scholar]
- 33.Hofman A, Brusselle GG, Darwish Murad S, van Duijn CM, Franco OH, Goedegebure A, et al. The Rotterdam Study: 2016 objectives and design update. Eur J Epidemiol. 2015;30:661–708. doi: 10.1007/s10654-015-0082-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitchell P, Smith W, Attebo K, Wang JJ. Prevalence of age-related maculopathy in Australia. The Blue Mountains Eye Study. Ophthalmology. 1995;102:1450–1460. doi: 10.1016/s0161-6420(95)30846-9. [DOI] [PubMed] [Google Scholar]
- 35.Hubbard LD, Brothers RJ, King WN, Clegg LX, Klein R, Cooper LS, et al. Methods for evaluation of retinal microvascular abnormalities associated with hypertension/sclerosis in the Atherosclerosis Risk in Communities Study. Ophthalmology. 1999;106:2269–2280. doi: 10.1016/s0161-6420(99)90525-0. [DOI] [PubMed] [Google Scholar]
- 36.Knudtson MD, Lee KE, Hubbard LD, Wong TY, Klein R, Klein BE. Revised formulas for summarizing retinal vessel diameters. Curr Eye Res. 2003;27:143–149. doi: 10.1076/ceyr.27.3.143.16049. [DOI] [PubMed] [Google Scholar]
- 37.Goldstein JI, Crenshaw A, Carey J, Grant GB, Maguire J, Fromer M, et al. zCall: a rare variant caller for array-based genotyping: genetics and population analysis. Bioinformatics. 2012;28:2543–2545. doi: 10.1093/bioinformatics/bts479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morris AP, Zeggini E. An evaluation of statistical approaches to rare variant analysis in genetic association studies. Genet Epidemiol. 2010;34:188–193. doi: 10.1002/gepi.20450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klenke S, Kussmann M, Siffert W. The GNB3 C825T polymorphism as a pharmacogenetic marker in the treatment of hypertension, obesity, and depression. Pharmacogenet Genomics. 2011;21:594–606. doi: 10.1097/FPC.0b013e3283491153. [DOI] [PubMed] [Google Scholar]
- 41.Dai F, Liu Y, Shi H, Ge S, Song J, Dong L, et al. Association of genetic variants in GNbeta3 with functional dyspepsia: a meta-analysis. Dig Dis Sci. 2014;59:1823–1830. doi: 10.1007/s10620-014-3057-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rinchik EM, Bultman SJ, Horsthemke B, Lee ST, Strunk KM, Spritz RA, et al. A gene for the mouse pink-eyed dilution locus and for human type II oculocutaneous albinism. Nature. 1993;361:72–76. doi: 10.1038/361072a0. [DOI] [PubMed] [Google Scholar]
- 43.Visser M, Kayser M, Grosveld F, Palstra RJ. Genetic variation in regulatory DNA elements: the case of OCA2 transcriptional regulation. Pigment Cell Melanoma Res. 2014;27:169–177. doi: 10.1111/pcmr.12210. [DOI] [PubMed] [Google Scholar]
- 44.Fossdal R, Jonasson F, Kristjansdottir GT, Kong A, Stefansson H, Gosh S, et al. A novel TEAD1 mutation is the causative allele in Sveinsson's chorioretinal atrophy (helicoid peripapillary chorioretinal degeneration) Hum Mol Genet. 2004;13:975–981. doi: 10.1093/hmg/ddh106. [DOI] [PubMed] [Google Scholar]
- 45.Liu F, Wang X, Hu G, Wang Y, Zhou J. The transcription factor TEAD1 represses smooth muscle-specific gene expression by abolishing myocardin function. J Biol Chem. 2014;289:3308–3316. doi: 10.1074/jbc.M113.515817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wistow G, Bernstein SL, Wyatt MK, Fariss RN, Behal A, Touchman JW, et al. Expressed sequence tag analysis of human RPE/choroid for the NEIBank Project: over 6000 non-redundant transcripts, novel genes and splice variants. Mol Vis. 2002;8:205–220. [PubMed] [Google Scholar]
- 47.Maecker HT, Todd SC, Levy S. The tetraspanin superfamily: molecular facilitators. FASEB J. 1997;11:428–442. [PubMed] [Google Scholar]
- 48.Bascom RA, Schappert K, McInnes RR. Cloning of the human and murine ROM1 genes: genomic organization and sequence conservation. Hum Mol Genet. 1993;2:385–391. doi: 10.1093/hmg/2.4.385. [DOI] [PubMed] [Google Scholar]
- 49.Busija DW, Heistad DD. Factors involved in the physiological regulation of the cerebral circulation. Rev Physiol Biochem Pharmacol. 1984;101:161–211. doi: 10.1007/BFb0027696. [DOI] [PubMed] [Google Scholar]
- 50.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ding J, Wai KL, McGeechan K, Ikram MK, Kawasaki R, Xie J, et al. Retinal vascular caliber and the development of hypertension: a meta-analysis of individual participant data. J Hypertens. 2014;32:207–215. doi: 10.1097/HJH.0b013e32836586f4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


