Abstract
It has been appreciated for almost 20 years that members of the Chlamydiales possess a virulence-associated type III secretion mechanism. Given the obligate intracellular nature of these bacteria, defining exactly how type III secretion functions to promote pathogenesis has been challenging. We present a working model herein that is based on current evidence.
Keywords: Pathogenesis, effector, secretion
1. Introduction
Members of the order Chlamydiales represent highly successful parasites—or endosymbionts in the case of environmental chlamydiae—of eukaryotic hosts. Based on a host range that extends from single-celled amoeba to mammals, members of the Chlamydiales are often considered ubiquitous in nature. Medically important species include the respiratory pathogen Chlamydia pneumoniae and the zoonotic agent C. psittaci. However, C. trachomatis represents the major human health concern and is a leading cause of sexually transmitted disease (serovars D-K and LGV1-3) and infectious blindness (serovars A-C). Significant co-evolution with eukaryotic hosts represents one obvious reason for such success as a pathogen. Chlamydia and Chlamydia-like endosymbionts last shared a common ancestor an estimated 700 mya, and C. trachomatis has been speculated to have diverged with the appearance of human ancestors 6 mya [51]. Gene loss and evolution have resulted in a reductionist genome that is fine tuned to exploit an intracellular niche [46]. All Chlamydiaceae are obligate intracellular bacteria that exhibit a bi-phasic developmental cycle [2]. This unique cycle is initiated when infectious elementary bodies (EB) associate with and actively invade a host—typically an epithelial cell. EBs differentiate into metabolically active, yet non-infectious, reticulate bodies (RB) which actively divide until an unknown signal induces asynchronous conversion back to EBs. The cycle is concluded when host cells release EBs and RBs via lysis or extrusion [38]. Development occurs entirely within a parasitophorous vacuole termed an inclusion. Timing depends on the chlamydial species and host cell type, but is typically completed within 40–72 hrs.
The ability to subvert host cell biology via expression of a type III secretion system (T3SS) is a major factor in the success of Chlamydia as a parasite. The T3SS has received considerable attention as a paradigm in Gram-negative bacterial manipulation of single-celled, plant, insect, and vertebrate eukaryotic hosts. Often referred to as the injectisome, the T3S apparatus (T3SA) is a complex secretory nanomachine composed of >20 proteins that is stimulated upon contact with target host cells [31]. Chlamydial genomes contain genes encoding the ubiquitously conserved core components of the apparatus designated contact dependent secretion proteins CdsCJNQRSTUV. Based on homology to characterized systems, these components along with CdsDLP form the envelope-localized base needle complex [31]. CdsF is the extended needle subunit and CopBD represent apparent translocon proteins. It is now established that the T3SS represents an ancient exaptation of flagella that was manifested by the loss of essential flagellar motility genes and acquisition of components enabling protein secretion across the bacterial envelope and translocation through eukaryotic membranes [1]. The designation non-flagellar T3SS (NF-T3SS) is now commonly used to differentiate from flagellar systems. Based on phylogeny of the outer membrane (OM) secretin CdsC, the chlamydial NF-T3SS emerged and evolved separately from other systems [1]. However, basic secretion mechanisms are conserved since ectopically-expressed chlamydial T3S substrates can be recognized and secreted by other T3SSs [67, 24].
The NF-T3SS mediates vectorial injection of anti-host proteins termed effectors that exert changes in the host cell microenvironment that benefit the respective pathogen or symbiont [31]. A combination of approaches that include the use of heterologous T3SSs has been used to establish effectors deployed by chlamydiae. Although the full complement of proteins has not been established, there are several categories of effectors evident. For example, Inc proteins intercalate into the inclusion membrane and function to subvert processes such as vesicular trafficking [47]. Chlamydia also express effectors that target the host nucleus, endoplasmic reticulum and other host cell compartments. We will not discuss specific effector biology and refer readers to several recent reviews that cover this area [13, 69]. Details regarding T3SA content and assembly in Chlamydia spp can also be found elsewhere [10]. Our aim in this review is to explore recent data that address questions regarding how the T3S mechanism is manifested in Chlamydia. In some cases, we take the liberty of speculating on certain mechanisms with the goal of assimilating what is known into the context of chlamydial infection biology. We present a testable working model herein (Fig. 1) that addresses how the chlamydial apparatus functions at each stage of the infection process and how activity could integrate with the complex chlamydial developmental cycle. To stay consistent with much of the literature, we will use the “CTxxx” designation from C. trachomatis serovar D [62] to refer to genes/proteins that have not yet received formal names.
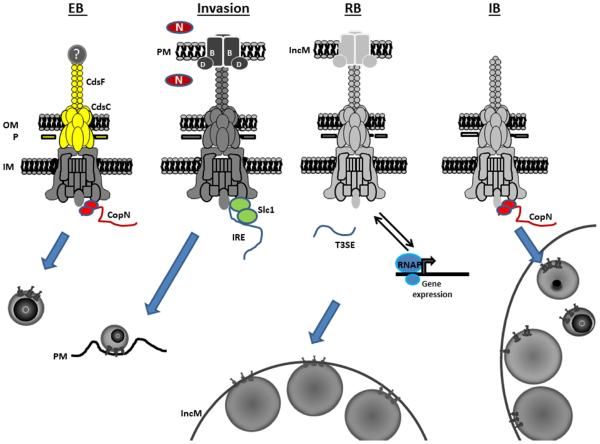
Figure 1.
Working model for T3S during chlamydial development. A completely assembled T3SA is present in the EB and spans the inner membrane (IM), periplasmic P-layer (P), and outer membrane (OM). Secretion activity is prevented by disulfide bonding within CdsF and CdsC (shown in yellow), and by positioning of the CopN plug on the cytoplasmic face of the T3SA. Secretion activity is activated upon contact of an EB with a host cell plasma membrane (PM). This results in displacement of a potential tip protein (?), secretion of CopN, and deployment of the translocon proteins CopB and CopD. The chaperone Slc1 can then mediate prioritized secretion of invasion related effectors (IRE) that orchestrate the invasion process. T3S by intracellular chlamydiae is maintained by association of the RB with the inclusion membrane (IncM) and subsequence de novo expression of T3SA genes (light gray). Chaperone-independent secretion becomes dominant and components of the T3SA affect chlamydial gene expression through interactions with RNAP. T3S activity ceases when CopN re-associates with the apparatus concomitant with RB conversion to IncM-dissociated intermediate bodies (IM).
2. Attachment
Purified chlamydial EBs can be stimulated to release effectors via in vitro treatment with T3S-inducing buffers containing BSA and EDTA [39, 49, 61]. Although inefficient, these data confirm that EBs contain a complete T3SA capable of secretion. Given that activation of T3S activity is contact-dependent in other systems, it is probable that EBs are not actively secreting effectors prior to encountering a host cell. While there are sufficient ATP stores to potentially energize secretion, and some metabolic activity is possible, EBs are typically considered inert particles that are primed for infection [52]. This model is supported by quantitative proteomic analysis that indicates EBs are primed with high levels of invasion-related T3S chaperones and effectors [56]. One obvious question that arises is: what prevents premature deployment of this arsenal?
Chlamydia CopN, an ortholog of Yersinia YopN, likely represents part of the answer. Based on primary sequence analysis CopN represents a fusion corresponding to YopN and TyeA [59]. In the Yersinia T3SS, a complex of YopN and TyeA with the chaperones SycN and YscB associates with the T3SA to block secretion prior to activating signals [54]. This machinery appears to be conserved in Chlamydia [59]. The chlamydial T3S chaperones (T3SC) Scc1 and Scc4 form a complex and interact with C. pneumoniae CopN. Scc1/Scc4 interacts with the N-terminus of CopN and promotes CopN secretion in the heterologous Yersinia T3SS. By analogy to the Yersinia model, the Scc1/Scc4 heteroduplex would position CopN at the T3SS in EBs, yet secretion would be prevented by the TyeA-like domain of CopN. In agreement with this hypothesis, an additional chaperone—Scc3—was found to interact with the C-terminus of CopN [60]. Scc3 was shown to reduce the secretion of CopN by yersiniae [59]. This suggests Scc3 also plays a role in the negative regulation of the T3S. This has interesting implications since Scc3 also interacts with the translocator CopB (see chaperone discussion below).
In addition to regulatory factors that are common to T3SSs, Chlamydia-specific mechanisms likely also contribute to maintaining an inactive state prior to host cell contact. For example, we have hypothesized that Chlamydia-specific physiology could uniquely impact T3S activity. The EB envelope is highly cross-linked via inter-and intra-molecular disulfide bonds among cysteine-rich proteins [34]. The precise timing of alterations is unknown, yet these bonds must be reduced very early to enable EB conversion to the vegetative RB. Unlike orthologs in other T3SSs, the chlamydial needle subunit protein CdsF contains Cys residues [12]. Intermolecular disulfide bonds among CdsF subunits are apparent in EBs but not RBs [11], raising the possibility that disulfide bonding has evolved in Chlamydia to couple T3S activity with one of the hallmarks of the chlamydial developmental cycle. Based on modeling of other T3S needle proteins [25], the N-terminally located Cys residues of CdsF could be oriented in the lumen of the needle channel. In this scenario, CdsF disulfide bonds in EBs could preclude secretion. Whatever the role, it is clear that disulfide bonding in the chlamydial T3SS mirrors that of developmentally regulated events in the envelope.
3. Invasion
T3S is most likely activated upon intimate, irreversible attachment to host cells [20]. Once chlamydiae attach, unknown signals are transduced that lead to activation of T3S. Interestingly, host Protein Disulfide Isomerase (PDI) activity is required for invasion [3], raising the possibility that PDI functions to reduce surface-exposed disulfide bonds such as those found in CdsF. If disulfide bonds in CdsF are accessible to PDI, this could be one signal that triggers secretion. Indeed, it has been speculated that conformational changes in the N-terminus of needle subunits are linked to signal transduction [31]. The orientation of CdsF would certainly be consistent with this concept. CdsF-specific antibodies label surface filaments on C. trachomatis EBs [12]. These antibodies also indicate a polar distribution of CdsF and needles that orient toward the host cell filopodia [50] that are induced during chlamydial invasion [19]. In other T3SSs, a tip protein oriented at the terminus of the needle serves a role in activation. There is currently no direct evidence that the chlamydial T3SS contains a corresponding structure. The purification of chlamydial structures and high-resolution electron microscopy necessary to directly assess tip structures has not been achieved. C. trachomatis CT584 has been implicated as a potential tip protein based on predicted structure and biophysical evidence [44]. In support of this notion, CT584 is present in EBs [56], and like the corresponding C. pneumoniae protein Cpn0803 [66], interacts with CdsF [61]. CT584 [7] and Cpn0803 [66] can form hexamers, but solved structures do not resemble those of either class-1 or -2 tip proteins typical of either Salmonella SipD or Yersinia LcrV, respectively [31]. In addition, CT584 has recently been implicated as a potential chaperone [53]. Therefore, further investigation is warranted before designating CT584 as the tip protein.
Once activated, the T3S channel would first need to deploy the “gatekeeper” protein CopN followed by translocator proteins CopB and CopD. The fate of secreted CopN is unclear. C. trachomatis CopN appears to accumulate in the expanding inclusion membrane [29], yet CopN of C. pneumoniae, but not C. trachomatis, has been implicated as having effector function [5]. This function would be novel since corresponding gatekeepers in other T3SSs lack described effector activity. Now that an in vivo secretion assay is available for Chlamydia [49], it should be possible to directly assess whether CopN is translocated or remains within the inclusion lumen. Current evidence indicates that secreted CopB and CopD would form the invasion-related translocon enabling translocation of subsequently secreted effectors across the host membrane. Both CopB and CopD are present in EBs [56] and have structural characteristics consistent with translocator proteins [16, 17]. Consistent with translocator function, CopB accumulates in the inclusion membrane [30] and partitions as an integral membrane protein [21]. Interestingly, chlamydial genomes contain a potential duplication of translocator proteins. CopB2 and CopD2 possess many of the same structural characteristics of CopB and CopD. CopB2 can also localize to the inclusion membrane. Unlike CopB, CopB2 levels increase during mid-cycle development [21], raising the possibility that CopB mediates early and late translocation whereas CopB2 functions in the interim. However, questions regarding this possibility remain since CopB2 does not partition as a membrane protein and requires coiled-coil domains for inclusion membrane association [21].
Secretion of translocators results in a complete T3SS that forms an infection synapse between the associated EB and host cell. At this point, invasion-related effectors (IRE) can be injected into the host cytosol where they can orchestrate chlamydial invasion. Thus far, three such effectors have been functionally described: TarP [23], TepP [22], and CT694 [36]. Given the complexity of chlamydial entry and early-cycle development, it is almost certain that many more will be identified with time.
4. Intracellular Development
EBs within the nascent inclusion rapidly differentiate into vegetative RBs, and cultures are considered essentially synchronous until RBs begin to asynchronously re-differentiate back to EBs later in development [48]. Throughout this time, chlamydiae need to secrete additional effectors that function to create and maintain the intracellular niche, subvert and evade host immunity (innate immune gene suppression via TepP, for example [22]), and protect host cell until completion of the developmental cycle (inhibition of apoptosis, for example [69]). Chlamydia Inc proteins represent perhaps the largest class of effector proteins. These integral membrane proteins intercalate into the inclusion membrane where they act as both scaffolds conferring inclusion integrity and as platforms for interactions with host cell proteins [47]. Based on the numbers of potential T3S substrates already identified, we have estimated that C. trachomatis may encode as many as 60–80 effectors [13]. Therefore, the chlamydial T3SA needs to orchestrate delivery of a large cohort of secretion substrates.
Chlamydial genes are expressed according to a temporal program that can be divided into at least three categories; early-, mid-, and late-cycle. Interestingly, it appears that once a gene is expressed, its expression is maintained. For example, the early-cycle effector IncG is expressed within an hour of infection and remains expressed throughout the remainder of the cycle [9]. This raises several interesting questions, one of which is: does the chlamydial apparatus discriminate among the many effectors in order to deploy them at specific times for specific functions? If so, how is this accomplished? De novo gene expression provides one obvious layer of selectivity. For example, homotypic fusion of inclusions in multiply infected cells does not occur until mid-cycle. IncA is essential for this event, and incA is not de novo expressed until mid-cycle development [32]. In addition, de novo expression of some invasion-related effectors such as CT694 does not occur until EBs begin to form. This is presumably to enable packaging of the effectors into EBs for subsequent infections. Hence, some degree of effector discrimination can be accomplished via temporal expression of genes. Although CT695 was recently shown to be secreted during late-cycle development [49], it is currently unclear whether this is true for other invasion-related effectors. Regardless, some mechanism exists whereby invasion-related effectors are retained during late-cycle development while other effectors clearly are not [56]. It is well established in other T3SSs that a hierarchy exists among T3S substrates [31]. Many of the mechanisms that function in those systems likely also apply to T3S in Chlamydia. We consider herein how elements of the T3SA and T3S-specific chaperones could contribute to substrate selectivity.
T3SS-expressing pathogens like Salmonella also encode many effectors. Some effectors function to enable entry into host cells while others are important during intracellular growth. Salmonellae employ differential expression and multiple secretion systems, designated SPI-1 and SPI-2, to separate these effector groups [31]. The chlamydial genome contains the coding capacity for a single complete system. How then does a single apparatus mediate deployment of numerous substrate classes? Intrinsic properties of the apparatus represent one mechanism. For example, we predict that the inner membrane component CdsU contributes to changes in substrate specificity. Cleavage of Yersinia YscU shifts secretion specificity from needle and translocator substrates to effectors [14], and this mechanism appears to be a conserved feature among T3SSs [31]. Cleavage of CdsU has not been tested. However, the flagellar homolog FhlB undergoes autocleavage at a conserved Asn-Pro sequence [28] to control the order of secretion for flagellar components. Blast searches of protein databases indicate that the Asn-Pro sequence is absolutely conserved in the Chlamydiales. While this might not be surprising since overall primary CdsU sequences are highly conserved among Chlamydia spp. (>83% identity), the Asn-Pro residues are also present in CdsU from the highly divergent Parachlamydia acanthamoebae. If CdsU plays a role similar to homologs, we anticipate the presence of full-length CdsU in EBs. Cleavage of CdsU would be apparent soon after attachment, and this would correlate with subsequent secretion of IREs.
Once the chlamydial T3SS is activated for effector secretion, secretion activity continues throughout development in metabolically active chlamydiae. For example, inclusion membrane-localized IncG can be detected within hours of infection. Secretion is obviously maintained since IncG can also be detected in membranes of large, mature inclusions [57]. Despite this apparent consistency, dynamic alterations likely manifest during intracellular development that influence T3SS selectivity. First, components of the core apparatus are de novo expressed during mid-cycle development [9]. Genes encoding components of the T3SA are dispersed in at least 10 operons that are predicted to respond to chlamydial sigma 66-mediated transcription [35]. Some subtle differences in precise timing are apparent [9]. Whether these differences have physiological relevance or are merely functions of technical aspects such as RT-PCR efficiency remains to be determined. Regardless, a complete apparatus would be established at times approximately corresponding to the first round of RB division [58].
This could provide an opportunity for a core T3SA differing in capacity from that mediating secretion since invasion. Although direct evidence is currently lacking, analysis of the apparent chlamydial T3S sorting platform provides some potential for changes in secretion selectivity. The T3S sorting platform functions in maintaining a hierarchy among secretion substrates and is composed of a cytoplasmic C-ring protein, an accessory protein, an ATPase and an apparent regulator of the ATPase. These proteins interact in a dynamic fashion to promote chaperone-dependent secretion (discussed below) and are designated in the unified nomenclature as SctQ, SctK, SctN, and SctL, respectively [31]. In the chlamydial system, CdsQ represents the C-ring protein whereas CdsN and CdsL represent the apparent ATPase and associated regulator. A protein corresponding to SctK has yet to be identified. However, C. trachomatis CT560 may represent SctK based purely on genomic positioning within a T3SS operon [35] and an apparent interaction with CdsQ [61]. In vitro studies have indicated ATPase activity for C. pneumoniae CdsN [65] that is regulated by CdsL [64]. Also consistent with other systems, interactions of CdsQ with itself, CdsN, CdsL and the multicargo T3SC Mcsc have been detected [61, 41, 65]. Additional protein-protein interaction findings further indicate that CdsQ represents a hub bridging the T3SA and secretion substrates [61]. Hence, it is presumed that the chlamydial sorting platform shares basic functions with those in other systems.
Quantitative comparison of protein levels in EBs and RBs suggest an interesting wrinkle in this presumption. While components of the core apparatus such as CdsD did not change in abundance, levels of CdsQ and CdsN were greatly reduced in RBs compared to EBs [56]. It is important to emphasize that these proteins are not totally absent in RBs. For example, transcription of cdsQ and cdsN occurs during this time [9], and CdsQ is certainly detectible in RB lysates via immunoblot [61]. One potential explanation for decreased levels of CdsQ has arisen from recent evidence indicating alternative translation of the C-ring protein. An internal translation start is apparent in Salmonella SsaQ [73] and Yersinia YscQ [18]. The alternatively translated C-terminus of YscQ is essential for secretion and exists in an apparent 2:1 ratio with full-length YscQ [18]. By analogy, the proportion of full-length CdsQ could be reduced in the active T3SA. Chlamydial CdsQ does contain a potential Met codon at position 207. The alternative translation product could have eluded detection in proteomic analyses [56] due to lack of tryptic peptides and in immunoblots since CdsQ antibodies were generated against a recombinant protein containing only the N-terminus of CdsQ [61].
This explanation, however, seems intuitively inconsistent with the fact that as many as 22 full-length copies of SctQ form the C-ring [26]. A more intriguing possibility is that, as suggested [56], RBs may have reduced capacity (compared to EBs) for chaperone-dependent secretion of effectors. The sorting platform represents a peripherally associated, mobile portion of the T3SA that fluctuates between an apparatus-associated and cytoplasmic localization [26]. In the flagellar T3SS, lack of secretion in a SctQ deficient strain can be overcome by overexpression of the ATPase [42]. However, overall CdsN levels are also comparatively reduced in RBs. Interestingly, chlamydial genomes contain genes encoding redundant components of the T3SS that more closely resemble flagellar components [10]. These include CT717 (CdsN paralog) and CT718 (CdsL paralog), raising the possibility that these components could substitute for deficiencies in CdsN. Indeed, the C. pneumoniae paralog of FliI does have ATPase activity [63]. Unfortunately, CT717 was not detectible under any conditions in proteomic studies [56] so the question remains open. Importantly the T3S ATPase has been implicated in the recognition of secretion substrates [31]. It is therefore possible that the mid-cycle T3SA assembles in a subtly different configuration. Other replacement components are possible since paralogs of inner membrane components CdsV and CdsJ are apparent in Chlamydia. Under one potential scenario, chaperones would be employed during invasion and early-cycle infection to prioritize secretion of cognate effectors while chaperone-independent secretion would predominate during mid-cycle. Although speculative, this would be consistent with observations that i) flagellar homologs are transcribed earlier than NF-T3S components [9, 45] and ii) levels of some T3S chaperones are also reduced in mid-cycle RBs [56].
Chaperones are obvious candidates for factors contributing to a secretion hierarchy. Depending on their respective role, T3SCs directly interact with T3S substrates to prevent aggregation among interacting substrates, maintain the substrates in a secretion competent state, and pilot the substrate to the T3SA [31]. T3SCs typically have limited sequence similarity, but tend to be small (ca. 15–20 kDa), have an acidic isoelectric point, and bind as homo- or heterodimers to a discrete N-terminally localized domain on the cognate substrate. Genomic sequencing provided the first evidence that Chlamydia express proteins typical of T3SCs [62]. These included Class I (CT043, CT088, and CT663) and Class II (CT576, CT862, and CT274). Since then, experimental evidence has identified additional T3SCs including CT260, CT584, CT665, CT667 and CT670.
Most of the currently identified chaperones function in assembly and regulation of the T3SA. As indicated already, Scc1 (CT088) and Scc4 (CT663) promote CopN secretion as a heterodimeric chaperone [59]. According to our model, they would function in secretion of EB-localized CopN. The chaperones must also be functional later since accumulation of secreted CopN is detectible at the inclusion membrane during mid-cycle development [29]. Scc3 (CT862) also interacts with CopN [61], yet interferes with CopN secretion [59]. Interestingly, Scc3 also interacts with the translocon protein CopB [30]. This is consistent with the occurrence of a tetratricopeptide repeat (TPR) domain typical of translocon-specific chaperones [31]. Although it is unknown how this chaperone functions, Scc3 links the gatekeeper CopN with the subsequently secreted translocon proteins. Therefore, Scc3 could function in the secretion hierarchy exhibited by these T3S substrates. It is unclear how Scc3 activity might mesh with Scc2 which is also an apparent translocon chaperone [30, 61, 16, 17]. However, Scc2 is much more abundant in EBs than Scc3 [22]. In addition, chlamydial CT274 is also a TPR-containing chaperone. However, interactions of CT274 with both apparent T3SA (CT668) and effector (CT161) proteins [61] indicate a potentially divergent role for this chaperone. Clearly, much more work is required to untangle what is likely a very complex and dynamic aspect of chlamydial T3S. The situation with CdsE (CT665) and CdsG (CT667) is more straightforward. These proteins represent Class III chaperones that stabilize needle subunits and prevent premature needle polymerization. CdsE and CdsG form a heterodimer capable of interacting with CdsF [61, 12]. CdsF-containing projections are apparent on EBs [12], and nearly all of EB-localized CdsF contains intermolecular disulfide bonds [11]. This suggests that secretion of CdsF is not required during the invasion process. Hence CdsE and CdsG would function as typical needle chaperones to secreted CdsF during and after mid-cycle de novo production of T3SAs. Finally, CT670 represents an apparent homolog of Yersinia YscO that binds to the apparent YscP homolog CT671 [43]. By analogy, CdsO would serve as a section chaperone for CdsP. If true, this would have important implications for regulation of the Chlamydia T3SS since Yersinia YscP and its homologs represent a molecular ruler that contributes to substrate selectivity [31].
Given the number of indicated chlamydial T3S effectors, the apparent dearth of corresponding secretion chaperones is a bit surprising. Currently, Slc1 (CT043) and Mcsc (CT260) represent the only identified chaperones dedicated to effectors. Both of these proteins function as Class IB multi-cargo chaperones. Slc1 is abundant in EBs [56] and binds the IREs to promote their secretion [22, 15, 53]. Therefore, this chaperone is clearly involved in prioritizing secretion of effectors required for invasion. Mcsc interacts with Cap1 and two Incs, CT225 and CT618 [61], all of which are de novo expressed during early development in C. trachomatis [9]. This represents only a very few of secreted effectors. It remains to be seen whether these chaperones function for other effectors or whether more chaperones exist in the chlamydial genome. Regardless, Slc1 and Mcsc indicate a prominent role of chaperones in earl-cycle development and would be consistent with a model where chaperone-independent secretion predominates later in development. It is also consistent with the fact that even chaperones effectors have chaperone-independent, N-terminal secretion signals [31]. This apparently holds true in Chlamydia since Subtil et al [67] were able to show secretion of numerous Incs by the surrogate Shigella T3SS using only the N-terminal signal fused to Cya. One could argue that this secretion is a product of ectopic overexpression in a heterologous system. However, the recently acquired ability to transform Chlamydia was recently applied to show that IncD lacking its N-terminal sequence was not secreted by chlamydiae [8]. Overall, these data would suggest that chaperones have primary roles in assembly/regulation of the T3SA and prioritizing effectors needed to establish a protected replication niche.
5. Interactions with development and shutting off secretion activity
It is clear that the T3SS is intimately linked with biphasic developmental cycle manifested in Chlamydia. Secretion activity commences with contact of an EB with a eukaryotic cell and must be subsequently silenced when RBs differentiate back into EBs. A central question has been whether the T3SS activity is merely responding to developmental ques or does the T3SS play an active role in orchestrating those changes. As noted above, disulfide bonding within the chlamydial apparatus correlates with developmentally responsive bonds in envelope proteins. RB-localized T3SSs are oriented toward the inclusion membrane during intracellular development [27], and it has been suggested that the ability of the T3SA apparatus to interact with the inclusion membrane represents a developmental switch [72]. Although preliminary, details are beginning to emerge that are consistent with T3S activity in Chlamydia being directly linked to the developmental genetic program.
Recent evidence has implicated a component of the chlamydial T3SS interacting with RNA polymerase (RNAP). C. trachomatis CT398 has structural similarity to the Helicobacter flagellar protein FlgZ [6]. This study further revealed interactions of CT398 with the alternative sigma factor RpoN, raising the possibility that CT398 could modulate activity of RNAP. Although potential activities were not investigated, CT398 was also found to interact with the T3S ATPase regulators CdsL and CT718. CT398 was designated CdsZ, and the authors speculated, based on analogy with FlgZ, that CdsZ may facilitate co-translational secretion of effectors. This intriguing possibility would fit into our working model of T3S in Chlamydia. Perhaps CdsZ is one factor that functions during intracellular development when chaperone-independent mechanisms are most prevalent.
In addition to their role as secretion chaperones, T3SCs also appear to function in linking T3S activity with gene regulation in Chlamydia. As indicated above Scc1 and Scc4 serve as secretion chaperones for CopN. Scc4 has also been shown to modulate RNAP through interactions with the σ-subunit and σ66 [55, 33]. The interaction was shown to inhibit RNAP activity in vitro in a σ66 dependent fashion. This observation has important implications since σ66 apparently controls not only gene expression for the T3SA [35], but also genes required for vegetative chlamydial growth [68]. Importantly, both Scc1 and CopN were found to antagonize the inhibitory activity of Scc4 [33]. If the in vitro data reflect a physiologically relevant situation in Chlamydia, then secretion of CopN would release Scc4 at a time when σ66 is becoming important. However, Scc1 would still be available to interact with Scc4 and would theoretically prevent inhibition of σ66-RNAP-mediated transcription. The question then becomes, when would Scc4 be free to exert effects on RNAP? Rao, et al [55] suggested that high levels of Scc4 during late cycle development could result in a pool of Scc4 capable of interacting with, and inhibiting, σ66-RNAP. Therefore, Scc4 may directly link secretion activity with developmental gene regulation in Chlamydia in such a way as to promote a shift toward EB formation. This would be consistent with data indicating that a subset of late-cycle genes associated with RB differentiation to EBs are σ28-dependent [68], and Scc4 does not interfere with the σ28-RNAP [55].
These data highlight open questions regarding how T3S activity is turned off during RB to EB differentiation. The most logical scenario is one where detachment of the RB correlates with loss of secretion activity. Hence T3S would be inactive in intermediate bodies. According to our working model, CopN secretion would cease. Whether this occurs due to substrate specificity switching where full-length CdsU predominates or alterations in chaperone function remains untested. Perhaps reconstitution of disulfide bonds via re-oxidation of cys residues in the T3SA and chlamydial envelope also contributes. Chlamydial redox state appears to be independent of the host cell environment and DsbJ has been proposed to function in late-cycle disulfide bond formation [70]. This is an attractive hypothesis, but more factors are likely involved. As a T3S substrate, CdsF would be secreted without a periplasmic intermediate and likely does not have direct access to DsbJ. Regardless, some environmental que results in positioning of the CopN/Scc1/Scc4/Scc3 complex at the cytoplasmic face of the T3SA to prevent further effector secretion. Pools of IREs could then accumulate for use in subsequent rounds of infection, and the process would be primed to start all over again.
6. Concluding remarks
Our understanding of how T3S contributes to chlamydial pathogenesis has come a long way since it initial discovery in C. psittaci [37]. Cumulatively, the data have revealed a complex picture where an arsenal of diverse, host-interactive effectors contribute to the stealthy subversion and evasion of eukaryotic cellular biology. As indicated in our discussion, many gaps still remain to be filled. The recently acquired ability to genetically manipulate Chlamydia will certainly facilitate further investigation. Engineered mutation of effector genes is already enabling reverse genetic approaches for elucidation of effector function [40, 22]. In addition, the ability to epitope tag secretion substrates to assay for effector secretion during chlamydial infection will eliminate dependence on surrogate T3SSs [8, 49, 4]. Given the apparent essential nature of T3S, it is doubtful that direct inactivation of T3SA genes will be possible. As an obligate intracellular parasite, any loss of function mutations would result in the inability to propagate the chlamydial strains. However, the emergence of inducible gene expression should allow alternative approaches [71, 8]. It should be possible to use anti-sense RNA or dominant negative approaches to conditionally disrupt T3S activity. While significant work will be required, we make the safe prediction that these novel approaches will answer many questions and provide definitive evidence for just how important T3S is for Chlamydia pathogenesis.
Acknowledgements
We thank Drs. Kate Wolf and Konrad Mueller for critical reading of the manuscript. KAF is supported by a Public Health Service grant from the National Institutes of Health, NIAID (AI065530).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Abby SS, Rocha EPC. The non-flagellar type III secretion system evolved from the bacterial flagellum and diversified into host-cell adapted systems. PLoS Genet. 2012;8:e1002983. doi: 10.1371/journal.pgen.1002983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].AbdelRahman Y, Belland R. The chlamydial developmental cycle. FEMS Microbiol Rev. 2005;29:949–59. doi: 10.1016/j.femsre.2005.03.002. [DOI] [PubMed] [Google Scholar]
- [3].Abromaitis S, Stephens RS. Attachment and entry of Chlamydia have distinct requirments for host protein disulfide isomerase. PLoS Pathog. 2009;5:e1000357. doi: 10.1371/journal.ppat.1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Agaisse H, Derré I. Expression of the effector protein IncD in Chlamydia trachomatis mediates mecruitment of the lipid transfer protein CERT and the endoplasmic reticulum-resident Protein VAPB to the inclusion membrane. Infect Immun. 2014;82:2037–47. doi: 10.1128/IAI.01530-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Archuleta TL, Du Y, English CA, Lory S, Lesser C, Ohi MD, et al. The Chlamydia effector chlamydial outer protein N (CopN) sequesters tubulin and prevents microtubule assembly. J Biol Chem. 2011;286:33992–8. doi: 10.1074/jbc.M111.258426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Barta ML, Battaile KP, Lovell S, Hefty PS. Hypothetical protein cT398 (CdsZ) interacts with σ54 (RpoN)-holoenzyme and the type iii secretion export apparatus in Chlamydia trachomatis. Protein Sci. 2015;10:2746. doi: 10.1002/pro.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Barta ML, Hickey J, Kemege KE, Lovell S, Battaile KP, Hefty PS. Structure of CT584 from Chlamydia trachomatis refined to 3.05 A resolution. Acta Crystallographica Section F. 2013;69:1196–201. doi: 10.1107/S1744309113027371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bauler LD, Hackstadt T. Expression and targeting of secreted proteins from Chlamydia trachomatis. J Bacteriol. 2014;196:1325–34. doi: 10.1128/JB.01290-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Belland RJ, Zhong G, Crane DD, Hogan D, Sturdevant D, Sharma J, et al. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. PNAS. 2003;100:8478–83. doi: 10.1073/pnas.1331135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Betts-Hampikian HJ, Fields KA. The chlamydial type III secretion mechanism: revealing cracks in a tough nut. Frontiers in Microbiol. 2010;1:114. doi: 10.3389/fmicb.2010.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Betts-Hampikian HJ, Fields KA. Disulfide bonding within components of the Chlamydia type III secretion apparatus correlates with development. J Bacteriol. 2011;193:6950–9. doi: 10.1128/JB.05163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Betts HJ, Twiggs LE, Sal MS, Wyrick PB, Fields KA. Bioinformatic and biochemical evidence for the identification of the type III secretion system needle protein of Chlamydia trachomatis. J Bacteriol. 2008;190:1680–90. doi: 10.1128/JB.01671-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Betts HJ, Wolf K, Fields KA. Effector protein modulation of host cells: examples in the Chlamydia spp. arsenal. Curr Opin Microbiol. 2009;12:81–7. doi: 10.1016/j.mib.2008.11.009. [DOI] [PubMed] [Google Scholar]
- [14].Björnfot A-C, Lavander M, Forsberg Å , Wolf-Watz H. Autoproteolysis of YscU of Yersinia pseudotuberculosis is important for regulation of expression and secretion of yop proteins. J Bacteriol. 2009;191:4259–67. doi: 10.1128/JB.01730-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Brinkworth AJ, Malcolm DS, Pedrosa AT, Roguska K, Shahbazian S, Graham JE, et al. Chlamydia trachomatis Slc1 is a type III secretion chaperone that enhances the translocation of its invasion effector substrate TARP. Mol Microbiol. 2011;82:131–44. doi: 10.1111/j.1365-2958.2011.07802.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bulir D, Waltho D, Stone C, Liang S, Chiang C, Mwawasi K, et al. Chlamydia Outer Protein (Cop) B from Chlamydia pneumoniae possesses characteristic features of a type III secretion (T3S) translocator protein. BMC Microbiol. 2015;15:163. doi: 10.1186/s12866-015-0498-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bulir DC, Waltho DA, Stone CB, Mwawasi KA, Nelson JC, Mahony JB. Chlamydia pneumoniae CopD translocator protein plays a critical role in type III secretion (T3S) and infection. PLoS ONE. 2014;9:e99315. doi: 10.1371/journal.pone.0099315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bzymek KP, Hamaoka BY, Ghosh P. Two translation products of Yersinia yscQ assemble to form a complex essential to type III secretion. Biochemistry. 2012;51:1669–77. doi: 10.1021/bi201792p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Carabeo R, Grieshaber S, Fischer E, Hackstadt T. Chlamydia trachomatis induces remodeling of the actin cytoskeleton during attachment and entry into HeLa cells. Infect Immun. 2002;70:3793–803. doi: 10.1128/IAI.70.7.3793-3803.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Carabeo RA, Hackstadt T. Isolation and characterization of a mutant chinese hamster ovary cell line that is resistant to Chlamydia trachomatis Infection at a Novel Step in the Attachment Process. Infect Immun. 2001;69:5899–904. doi: 10.1128/IAI.69.9.5899-5904.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chellas-Gery B, Wolf K, Tisoncik J, Hackstadt T, Fields KA. Biochemical and immunolocalization analyses of putative type III secretion translocator proteins CopB and CopB2 of Chlamydia trachomatis reveal significant distinctions. Infect Immun. 2011;79:3036–45. doi: 10.1128/IAI.00159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen Y-S, Bastidas RJ, Saka HA, Carpenter VK, Richards KL, Plano GV, et al. The Chlamydia trachomatis type III secretion chaperone Slc1 engages multiple early effectors, including TepP, a tyrosine-phosphorylated protein required for the recruitment of CrkI-II to nascent inclusions and innate immune signaling. PLoS Pathog. 2014;10:e1003954. doi: 10.1371/journal.ppat.1003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Clifton DR, Fields KA, Grieshaber SS, Dooley CA, Fischer ER, Mead DJ, et al. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. PNAS. 2004;101:10166–71. doi: 10.1073/pnas.0402829101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].da Cunha M, Milho C, Almeida F, Pais S, Borges V, Mauricio R, et al. Identification of type III secretion substrates of Chlamydia trachomatis using Yersinia enterocolitica as a heterologous system. BMC Microbiology. 2014;14:40. doi: 10.1186/1471-2180-14-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Deane JE, Roversi P, Cordes FS, Johnson S, Kenjale R, Daniall S, et al. Molecular model of a type three secretion system needle: implications for host cell sensing. PNAS. 2006;103:12529–33. doi: 10.1073/pnas.0602689103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Diepold A, Kudryashev M, Delalez NJ, Berry RM, Armitage JP. Composition, formation, and regulation of the cytosolic c-ring, a dynamic component of the type III secretion injectisome. PLoS Biol. 2015;13:e1002039. doi: 10.1371/journal.pbio.1002039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dumoux M, Clare DK, Saibil HR, Hayward RD. Chlamydiae assemble a pathogen synapse to hijack the host endoplasmic reticulum. Traffic. 2012;13:1612–27. doi: 10.1111/tra.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ferris HU, Furukawa Y, Minamino T, Kroetz MB, Kihara M, Namba K, et al. FlhB regulates ordered export of flagellar components via autocleavage mechanism. J Biol Chem. 2005;280:41236–42. doi: 10.1074/jbc.M509438200. [DOI] [PubMed] [Google Scholar]
- [29].Fields K, Hackstadt T. Evidence for the secretion of Chlamydia trachomatis CopN by a type III secretion mechanism. Mol Microbiol. 2000;38:1048–60. doi: 10.1046/j.1365-2958.2000.02212.x. [DOI] [PubMed] [Google Scholar]
- [30].Fields KA, Fischer ER, Mead DJ, Hackstadt T. Analysis of putative Chlamydia trachomatis chaperones Scc2 and Scc3 and their use in the identification of type III secretion substrates. J Bacteriol. 2005;187:6466–78. doi: 10.1128/JB.187.18.6466-6478.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Galán JE, Lara-Tejero M, Marlovits TC, Wagner S. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Ann Rev Microbiol. 2014;68:415–38. doi: 10.1146/annurev-micro-092412-155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hackstadt T, Scidmore-Carlson M, Shaw E, Fischer E. The Chlamydia trachomatis IncA protein is required for homotypic vesicle fusion. Cell Microbiol. 1999;1:119–30. doi: 10.1046/j.1462-5822.1999.00012.x. [DOI] [PubMed] [Google Scholar]
- [33].Hanson BR, Slepenkin A, Peterson EM, Tan M. Chlamydia trachomatis type III secretion proteins regulate transcription. J. Bacteriol. 2015;197:3238–44. doi: 10.1128/JB.00379-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hatch TP. Disulfide cross-linked envelope proteins: the functional equivalent of peptidoglycan in chlamydiae? J. Bacteriol. 1996;178:1–5. doi: 10.1128/jb.178.1.1-5.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hefty PS, Stephens RS. Chlamydial type III secretion system is encoded on ten operons preceded by sigma 70-like promoter elements. J Bacteriol. 2007;189:198–206. doi: 10.1128/JB.01034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hower S, Wolf K, Fields KA. Evidence that CT694 is a novel Chlamydia trachomatis T3S substrate capable of functioning during invasion or early cycle development. Mol Microbiol. 2009;72:1423–37. doi: 10.1111/j.1365-2958.2009.06732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hsia RC, Pannekoek Y, Ingerowski E, Bavoil PM. Type III secretion genes identify a putative virulence locus of Chlamydia. Mol Microbiol. 1997;25:351–9. doi: 10.1046/j.1365-2958.1997.4701834.x. [DOI] [PubMed] [Google Scholar]
- [38].Hybiske K, Stephens RS. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. PNAS. 2007;104:11430–5. doi: 10.1073/pnas.0703218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Jamison WP, Hackstadt T. Induction of type III secretion by cell-free Chlamydia trachomatis elementary bodies. Microb Pathog. 2008;45:435–40. doi: 10.1016/j.micpath.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Johnson CM, Fisher DJ. Site-specific, insertional inactivation of incA in Chlamydia trachomatis using a Group II Intron. PLoS ONE. 2013;8:e83989. doi: 10.1371/journal.pone.0083989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Johnson DL, Stone CB, Mahony JB. Interactions between CdsD, CdsQ, and CdsL, three putative Chlamydophila pneumoniae type III secretion proteins. J Bacteriol. 2008;190:2972–80. doi: 10.1128/JB.01997-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Konishi M, Kanbe M, McMurry JL, Aizawa S-I. Flagellar formation in c-ring-defective mutants by overproduction of FliI, the ATPase specific for flagellar type III secretion. J Bacteriol. 2009;191:6186–91. doi: 10.1128/JB.00601-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lorenzini E, Singer A, Singh B, Lam R, Skarina T, Chirgadze NY, et al. Structure and protein-protein interaction studies on Chlamydia trachomatis protein CT670 (YscO Homolog) J Bacteriol. 2010;192:2746–56. doi: 10.1128/JB.01479-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Markham AP, Jaafar ZA, Kemege KE, Middaugh CR, Hefty PS. Biophysical characterization of Chlamydia trachomatis CT584 supports its potential role as a type III secretion needle tip protein. Biochemistry. 2009;48:10353–61. doi: 10.1021/bi901200y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Maurer AP, Mehlitz A, Mollenkopf HJ, Meyer TF. Gene expression profiles of Chlamydophila pneumoniae during the developmental cycle and iron depletion-mediated persistence. PLoS Pathog. 2007;3:752–69. doi: 10.1371/journal.ppat.0030083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Merhej V, Royer-Carenzi M, Pontarotti P, Raoult D. Massive comparative genomic analysis reveals convergent evolution of specialized bacteria. Biol Direct. 2009;4:4–13. doi: 10.1186/1745-6150-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Moore ER, Ouellette SP. Reconceptualizing the chlamydial inclusion as a pathogen-specified parasitic organelle: an expanded role for Inc Proteins. Front Cell Infect Microbiol. 2014;4:157. doi: 10.3389/fcimb.2014.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Moulder JW. Interaction of chlamydiae and host cells in vitro. Microbiol Rev. 1991;55:143–90. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Mueller KE, Fields KA. Application of β-lactamase reporter fusions as an indicator of protein secretion during infections with the obligate intracellular pathogen Chlamydia trachomatis. PLoS One. 2015;10:e135295. doi: 10.1371/journal.pone.0135295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Nans A, Saibil HR, Hayward RD. Pathogen-host reorganization during Chlamydia invasion revealed by cryo-electron tomography. Cell Microbiol. 2014;16:1457–72. doi: 10.1111/cmi.12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Nunes A, Gomes JP. Evolution, phylogeny, and molecular epidemiology of Chlamydia. Infection, Genetics and Evolution. 2014;23:49–64. doi: 10.1016/j.meegid.2014.01.029. [DOI] [PubMed] [Google Scholar]
- [52].Omsland A, Sager J, Nair V, Sturdevant DE, Hackstadt T. Developmental stage-specific metabolic and transcriptional activity of Chlamydia trachomatis in an axenic medium. PNAS. 2012;109:19781–5. doi: 10.1073/pnas.1212831109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Pais SV, Milho C, Almeida F, Mota LJ. Identification of novel type III secretion chaperone-substrate complexes of Chlamydia trachomatis. PLoS ONE. 2013;8:e56292. doi: 10.1371/journal.pone.0056292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Plano G, Schesser K. The Yersinia pestis type III secretion system: expression, assembly and role in the evasion of host defenses. Immunol Res. 2013;57:237–45. doi: 10.1007/s12026-013-8454-3. [DOI] [PubMed] [Google Scholar]
- [55].Rao X, Deighan P, Hua Z, Wang J, Luo M, Wang J, et al. A regulator from Chlamydia trachomatis modulates the activity of RNA polymerase through direct interaction with the beta subunit and the primary sigma subunit. Genes Dev. 2009;23:1818–29. doi: 10.1101/gad.1784009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Saka HA, Thompson JW, Chen Y-S, Kumar Y, Dubois LG, Moseley MA, et al. Quantitative proteomics reveals metabolic and pathogenic properties of Chlamydia trachomatis developmental forms. Mol Microbiol. 2011;82:1185–203. doi: 10.1111/j.1365-2958.2011.07877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Scidmore-Carlson M, Shaw E, Dooley C, Fischer E, Hackstadt T. Identification and characterization of a Chlamydia trachomatis early operon encoding four novel inclusion membrane proteins. Mol Microbiol. 1999;33:753–65. doi: 10.1046/j.1365-2958.1999.01523.x. [DOI] [PubMed] [Google Scholar]
- [58].Shaw EI, Dooley CA, Fischer ER, Scidmore MA, Fields KA, Hackstadt T. Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol Microbiol. 2000;37:913–25. doi: 10.1046/j.1365-2958.2000.02057.x. [DOI] [PubMed] [Google Scholar]
- [59].Silvia-Herzog E, Joseph SS, Avery A, Wolf K, Fields KA, Plano GV. Scc1 (CP0432) and Scc4 (CP0033) function as a type III secretion chaperone for CopN of Chlamdyia pneumoniae. J Bacteriol. 2011;193:3490–6. doi: 10.1128/JB.00203-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Slepenkin A, de la Maza LM, Peterson EM. Interaction between components of the type III secretion system of Chlamydiaceae. J Bacteriol. 2005;187:473–9. doi: 10.1128/JB.187.2.473-479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Spaeth KE, Chen YS, Valdivia RH. The Chlamydia type III secretion system C-ring engages a chaperone-effector protein complex. PLoS Pathog. 2009;5:e1000579. doi: 10.1371/journal.ppat.1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–9. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- [63].Stone CB, Bulir DC, Gilchrist JD, Toor RK, Mahony JB. Interactions between flagellar and type III secretion proteins in Chlamydia pneumoniae. BMC Microbiol. 2010;10:1471–2180. doi: 10.1186/1471-2180-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Stone CB, D.C. B, Emdin CA, Pirie RM, Porfilio EA, Slootstra JW, et al. Chlamydia pneumoniae CdsL regulates CdsN ATPase activity and disruption with a peptide mimetic prevents bacterial invasion. Frontiers in Microbiol. 2011;2:21. doi: 10.3389/fmicb.2011.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Stone CB, Johnson DL, Bulir DC, Gilchrist JD, Mahony JB. Characterization of the putative type III secretion ATPase CdsN (Cpn0707) of Chlamydophila pneumoniae. J Bacteriol. 2008;190:6580–8. doi: 10.1128/JB.00761-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Stone CB, Sugiman-Marangos S, Bulir DC, Clayden RC, Leighton TL, Slootstra JW, et al. Structural characterization of a novel Chlamydia pneumoniae type III secretion-associated protein, Cpn0803. PLoS One. 2012;7:e30220. doi: 10.1371/journal.pone.0030220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Subtil A, Delevoye C, Balana M, Tastevin L, Perrinet S, Dautry-Varsat A. A directed screen for chlamydial proteins secreted by a type III mechanism identifies a translocated protein and numerous other new candidates. Mol Microbiol. 2005;56:1636–47. doi: 10.1111/j.1365-2958.2005.04647.x. [DOI] [PubMed] [Google Scholar]
- [68].Tan M. Temporal gene regulation during the chlamydial developmental cycle. In: Tan M, Bavoil P, editors. Intracellular Pathogens I: Chlamydiales. ASM Press; Washington, DC: 2012. pp. 149–69. [Google Scholar]
- [69].Valdivia R. Chlamydia effector proteins and new insights into chlamydial cellular microbiology. Curr Opin Microbiol. 2008;11:53–9. doi: 10.1016/j.mib.2008.01.003. [DOI] [PubMed] [Google Scholar]
- [70].Wang X, Schwarzer C, Hybiske K, Machen TE, Stephens RS. Developmental stage oxidoreductive states of Chlamydia and infected host cells. mBio. 2014;5:e01924. doi: 10.1128/mBio.01924-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Wickstrum J, Sammons LR, Restivo KN, Hefty PS. Conditional gene expression in Chlamydia trachomatis using the Tet System. PLoS ONE. 2013;8:e76743. doi: 10.1371/journal.pone.0076743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Wilson DP, Timms P, McElwain DL, Bavoil P. Type III secretion, contact-dependent model for the intracellular development of Chlamydia. Bull Math Biol. 2006;68:161–78. doi: 10.1007/s11538-005-9024-1. [DOI] [PubMed] [Google Scholar]
- [73].Yu X-J, Liu M, Matthews S, Holden DW. Tandem Translation generates a chaperone for the Salmonella type III secretion system protein SsaQ. J Biol Chem. 2011;286:36098–107. doi: 10.1074/jbc.M111.278663. [DOI] [PMC free article] [PubMed] [Google Scholar]