Abstract
Background
Outcomes for children with high-risk neuroblastoma are poor, and improved understanding of the mechanisms underlying pathogenesis, recurrence and treatment resistance will lead to improved outcomes. Aberrant growth factor receptor expression and receptor tyrosine kinase signaling are associated with the pathogenesis of many malignancies. A germline polymorphism in the FGFR4 gene is associated with increased receptor expression and activity and with decreased survival, treatment resistance, and aggressive disease for many malignancies. We therefore investigated the role of this FGFR4 polymorphism in neuroblastoma pathogenesis.
Methods
Germline DNA from neuroblastoma patients and matched controls was assessed for the FGFR4 Gly/Arg388 polymorphism by RT-PCR. Allele frequencies were assessed for association with neuroblastoma patient outcomes and prognostic features. Degradation rates of the FGFR4 Arg388 and Gly388 receptors and rates of receptor internalization into the late endosomal compartment were measured.
Results
Frequency of the FGFR4 AA genotype and the prevalence of the A allele were significantly higher in patients with neuroblastoma than in matched controls. The Arg388 receptor demonstrated slower degradation than the Gly388 receptor in neuroblastoma cells and reduced internalization into multi-vesicular bodies.
Conclusions
The FGFR4 Arg388 polymorphism is associated with an increased prevalence of neuroblastoma in children, and this association may be linked to differences in FGFR4 degradation rates. Our study provides the first evidence of a role for FGFR4 in neuroblastoma, suggesting that FGFR4 genotype and the pathways regulating FGFR4 trafficking and degradation may be relevant for neuroblastoma pathogenesis.
Keywords: neuroblastoma, FGFR4, Arg388
Introduction
Neuroblastoma is the most common extra-cranial solid tumor in children, with an incidence of 10.5 cases per million children less than 15 years old [1]. Children with low and intermediate risk disease have an excellent prognosis, with long-term survival rates over 90%. However, less than 50% of children with high-risk disease achieve long-term survival despite aggressive treatment including multi-agent high-dose chemotherapy, surgery, radiation therapy, and biologic agents [1]. Those who do survive incur significant late effects associated with treatment. Improved understanding of the mechanisms underlying the pathogenesis, recurrence, and treatment resistance of neuroblastoma will lead to improved treatment and identify novel targets for future therapies.
Aberrant growth factor receptor (GFR) expression and receptor tyrosine kinase signaling are associated with the pathogenesis of many malignancies, and these kinases serve as targets for a number of novel therapies. Fibroblast growth factors (FGFs) and their cognate receptors (FGFR1-4) carry out key roles in multiple biologic processes including embryonic development, tissue repair, and angiogenesis [2]. FGFR activation and signaling have been shown to play important roles in tumorigenesis through stimulation of cell proliferation and migration and inhibition of cell death [2-7]. FGFR inhibition has also been shown to be effective in a variety of preclinical cancer models [8-12].
Among the FGFR family members, FGFR4 is the least understood. Unlike other FGFR family members, FGFR4 deletion does not result in embryonic lethality [13], and the kinase domain structure of FGFR4 is significantly different from FGFR1-3, suggesting the possibility of specific inhibition with reduced systemic toxicity [14]. FGFR4 expression and activity have been linked to the pathogenesis of a variety of cancers, including rhabdomyosarcoma [15,16] and adrenocortical carcinoma [17]. Neuroblastoma tumor cells have been shown to express the FGFR4 ligand basic FGF (bFGF) and FGFR family members [18,19], suggesting a potential role for FGFR4-mediated signaling in the pathogenesis of neuroblastoma.
A germline polymorphism in the FGFR4 gene (rs351855) results in the expression of FGFR4 containing arginine at codon 388 (Arg388) rather than the more common glycine (Gly388). This polymorphism has been shown to be associated with decreased survival rates, treatment resistance, and more aggressive disease in a variety of malignancies, including breast cancer, colon cancer, prostate cancer, soft tissue sarcomas, melanoma, lung adenocarcinoma, and head and neck squamous cell carcinoma [20-27]. Expression of the FGFR4 Arg388 variant results in increased cancer cell motility and invasiveness, and recent studies have shown that the FGFR4 Arg388 variant has markedly decreased degradation rates and increased phosphorylation after ligand binding when compared to the Gly388 variant [28]. In prostate cancer, malignant cells have also demonstrated that expression of the FGFR4 Arg388 variant leads to increased Ras/MAPK pathway activity and transcription of genes correlated with aggressive clinical behavior [29]. The roles of FGFR4 expression and function in the pathogenesis of neuroblastoma and the association of this FGFR4 polymorphism with neuroblastoma patient outcomes and prognostic features, however, have not been investigated.
Materials and Methods
Study Subjects
Banked peripheral blood samples from 126 patients with a confirmed diagnosis of neuroblastoma treated at Texas Children's Hospital between 1986 to 2011 were utilized for this analysis. Patients were consented for banking either at the time of an oncology clinic visit during active treatment (69.9%) or at the time of a long-term survivor clinic visit (30.1%). Saliva samples from 114 children recruited at a well-child visit or sports physical examination were frequency-matched to the neuroblastoma patients based on gender and race/ethnicity. All experiments utilizing patient samples and analyzing patient information were approved by the Baylor College of Medicine Institutional Review Board.
DNA Extraction and Genotyping
Germline DNA was extracted from peripheral blood samples (patients) and saliva (controls). Peripheral blood samples (3 ml) were collected in EDTA tubes and mixed with 9 mL of RBS lysis buffer (10 mM NH4Cl) in 15 mL conical polypropylene tubes. After a 10-minute incubation period at room temperature, the mixture was centrifuged, and the white blood cells (WBCs) were collected as a pellet. The WBCs were then re-suspended in 3 mL of nucleus lysis buffer (10 mM Tris-HCL, 400 mM NaCl and 2 mM EDTA, pH 8.2). The cell lysates were incubated at room temperature for 10 minutes with intermittent mixing. One mL of protein precipitation solution (6 M NaCl) was added to the tube, and the mixture was vortexed for 30 seconds. After centrifugation the supernatant was collected and transferred to a new tube. Isopropyl ethanol was added to the supernatant to precipitate the DNA. After centrifugation and removal of the supernatant, the DNA was washed twice with 70% alcohol and then air dried in the tube at room temperature for 10 minutes. Prior to usage, the DNA was stored at -20°C.
Saliva samples were collected using OrageneDNA collection kits (DNAGenotek, Kanata, Ontario, Canada). 500μL Oragene/saliva samples were incubated at 50°C for two hours, and 20μL of Oragene purifier was added to each sample. After incubation on ice for 10 minutes, samples were centrifuged and supernatants collected. 500μL of 100% ethanol was added to each sample, and the DNA precipitate was collected by centrifugation and resuspended in 100μL TE buffer.
To determine the distribution of the FGFR4 Arg388 and Gly388 alleles in patients and controls, germline DNA was subjected to PCR as previously described [21,25]. Genotyping was done with restriction-fragment length polymorphism (RFLP) analysis using BstN1 (New England Biolabs, Beverly, MA) digestion for 3 hours. The fragments were resolved on a 5% agarose gel and visualized by ethidium bromide staining. DNA isolated from prostate cancer cell lines DU145 and PC3 (provided by Dr. Michael Ittmann) was utilized as positive controls (Supplemental Figure 1).
Statistical Analysis of Polymorphism Effects
Statistical evaluations were performed using STATA statistical software (v12.1; StataCorp, College Station, TX). Differences in demographics between the patients and controls were calculated using the chi-squared test. The polymorphism was tested for deviation from Hardy-Weinberg equilibrium, and genotype and allele frequencies were compared between groups using the chi-squared test. Odds ratios and 95% confidence intervals were calculated using logistic regression. Survival rates of neuroblastoma patients were analyzed using the Kaplan-Meier method. Differences in median survival were compared using the log-rank test. P values less than 0.05 were considered significant.
Cell Culture
The characteristics of neuroblastoma cell lines used in this study have been previously described [30-34] and were purchased from the American Type Culture Collection (ATCC; Rockville, MD) or were generously provided by Susan Cohn (The University of Chicago Children's Hospital, Chicago, IL) or John Maris (Children's Hospital of Philadelphia, Philadelphia, PA). Cell lines were authenticated by STR DNA profiling prior to use. Cell lines were grown at 37°C in 5% CO2 in appropriate media (Invitrogen, Carlsbad, CA) supplemented with 10% heat inactivated fetal bovine serum (FBS, USB, Minneapolis, MN) and L-glutamine (Sigma Chemical Company, St. Louis, MO). Human Embryonic Kidney (HEK) 293T cells were cultured as a monolayer in 10-cm plastic dishes in appropriate media (Dulbecco's Modified Eagle Medium, Mediatech, Manassas, VA) containing 10% FBS under 5% CO2 at 37 °C.
Western blots for EGFR and FGFR4
Neuroblastoma cell lines were plated at approximately 80% confluence. Plates were washed twice with PBS and incubated in serum free media for 3 hours, followed by treatment with either 100 ng/mL recombinant human EGF (R&D Systems, Minneapolis) or 50 ng/mL recombinant human bFGF (R&D Systems) for 0, 15, 30, 60 or 90 minutes. Cells were washed with PBS and lysed with RIPA lysis buffer (50mM Tris pH 8.0, 150 mM NaCl, 1% Triton X-100, 0.5% Na deoxycholate, 0.1% SDS) with protease inhibitor (Sigma). 30 ug total denatured protein from each cell line was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes (Invitrogen) using standard techniques. Membranes were blocked in Odyssey blocking buffer (Li-Cor, Lincoln, NE) for 1 hour at room temperature and then incubated with primary antibodies to total EGFR (Thermo Scientific, Marietta, OH), FGFR-4 (Cell Signaling, Danvers, MA), and β-actin (A5316 or A5441; 1:5000; Sigma). Bound primary antibodies were incubated for 1 hour at room temperature with IRDye800 conjugated affinity purified anti-rabbit or anti-mouse secondary antibody (1:5000; Rockland, Gilbertsville, PA), and the signal was visualized on an Odyssey infrared imaging system (Li-Cor).
Cell Transfection
Cells were transfected with vectors encoding V5-tagged FGFR4Arg388 or FGFR4Gly388. Plasmid DNA was prepared (Qiagen, Valencia, CA), and transient transfections were performed (Lipofectamine 2000) following the manufacturer's protocol (Invitrogen). Briefly, cells were grown in wells of 6-well plates until approximately 90% confluent. 3μg of plasmid DNA were added to 250μL of Opti-MEM Reduced Serum Medium, mixed, and incubated for 5 minutes at room temperature. 3.5μL of lipofectamine 2000 reagent was added to 250μL of Opti-MEM Reduced Serum Medium, mixed, and incubated for 5 minutes at room temperature. The tubes were combined, mixed gently, and incubated for 20 minutes at room temperature. 500μL of this transfection mixture was then added to each well. After 48 hours, the cells were used for degradation experiments and in the cell-free sorting assays described below.
Degradation Assays
After 48 hours, transfected cells were washed with 1X PBS and starved in media without serum containing cycloheximide (30 μg/mL) for 2 hours at 37C°. Media was removed and cells were incubated in media supplemented with bFGF (50 ng/mL) and cycloheximide on ice for 1 hour. Subsequently cells were rinsed and returned to 4°C (time point 0), or incubated in media at 37°C with cycloheximide for 3, 6, 9, 12, and 24 hours. Cells were scraped, resuspended in PBS, and centrifuged (1500 × g for 10 min). Cell pellets were resuspended in RIPA buffer (1% Triton-X-100, 1% NP-400, 1% SDS, 150 mM NaCl, 50 mM Tris pH 8.0) and a protease inhibitor cocktail (112 μM PMSF, 3 μM aprotinin, 112 μM leupeptin, 17 μM pepstatin) and were incubated for 1 hour at 4°C with constant mixing. Lysates were centrifuged (1500 × g for 15 minutes) and supernatants were collected. 15μg protein from each sample was subjected to SDS-PAGE and Western blotting. Blots were stained with Ponceau S to ensure accuracy of protein loading. Proteins were visualized with enhanced chemiluminescence (ECL; Pierce, Thermo Scientific, Waltham, MA) and exposed to autoradiography film. Quantification of bands was performed using ImageJ (v 1.46r; NIH).
Mammalian cytosol preparation
Cells (SK-N-AS or HEK 293T) were grown to approximately 75-80% confluence. Cells were washed with ice-cold PBS and scraped from the plate followed by centrifugation for 15 minutes at at 4°C at 2000 × g. The cell pellet was resuspended in 100μL of homogenization buffer (20mM HEPES pH 7.4, 0.25M sucrose, 2mM EGTA, 2mM EDTA, and 0.1mM DTT) containing protease inhibitor cocktail. Cells were then sonicated with a microprobe (Branson Sonifier 250, VWR Scientific). The lysate was centrifuged (2000 × g for 10 minutes at 4°C) to remove cell debris, and the resulting supernatant was centrifuged (100,000 × g for 1 hour at 4°C). The supernatant was collected and protein concentration was calculated using a Bradford assay.
Cell-free reconstitution of multivesicular body formation assay
The cell-free reconstitution of multivesicular body (MVB) formation and receptor sorting was performed as previously described [35,36]. Cells transfected with vectors encoding either V5-tagged FGFR4 Arg388 or FGFR4 Gly388 were serum-starved (1% Bovine Serum Albumin and 30 μg/mL cycloheximide in media) for 2 hours and stimulated with 50 ng/mL bFGF (with 30 μg/mL cycloheximide) in media for 6 hours. Cells were then placed on ice, scraped from the plate, and isolated by centrifugation at 1500 × g for 10 minutes at 4°C. The cell pellet was resuspended in 100μL of homogenization buffer and protease inhibitor cocktail. The cells were then drawn through a 30-gauge needle into a 1-ml syringe 30 times within approximately 6 minutes. The lysate was centrifuged (800 × g for 5 minutes) to remove debris, and the supernatant was further centrifuged (15 minutes at 1500 × g). Endosomal vesicle membranes were recovered from the resulting pellet and resuspended in homogenization buffer.
5μL endosomal membranes were placed on ice for 3 hours and incubated with trypsin (6μl of 0.27 μg/μL). For reactions with added cytosol, 15μL endosomal membranes were mixed with 6μL ATP regeneration system (2mM MgATP, 50μg/mL creatine kinase, 8mM phosphocreatine, 1mM DTT), 25μg mammalian cytosol (isolated as above) and homogenization buffer to a final volume of 50μL. All experimental reactions were incubated for 3 hours with gentle mixing at 37°C. After incubation, reactions were placed on ice and trypsin (6μL, 0.27μg/μL) was added for 30 minutes. The experimental reactions were centrifuged (15000 × g for 30 minutes at 4°C) while the control reactions remained on ice. Control reactions and experimental reaction pellets were resuspended in sample buffer. After boiling, proteins were separated by SDS-PAGE, followed by Western blotting with a murine antibody against the intracellular V5 tag of FGFR4 (Invitrogen, 1:5000) followed by goat anti-mouse polyclonal antibody conjugated to horseradish peroxidase (HRP) (Sigma-Aldrich Inc, 1:10,000). Proteins were detected with enhanced chemiluminescence (ECL; Pierce) and exposed to X-ray film. Quantification of bands was performed using ImageJ (v. 1.46r; NIH).
Statistical Analysis of degradation assays
Statistical significance was determined using a two-tailed Student's t test for independent samples. A p value of <0.05 was considered statistically significant.
Results
FGFR4 Polymorphism Distribution Among Cases and Controls
To evaluate the potential association between FGFR4 genotype and the risk of neuroblastoma, 126 neuroblastoma patient germline DNA samples and 114 control germline DNA samples were analyzed. The frequency distributions of select characteristics for the cases and controls are presented in Table I. The case group encompassed the spectrum of neuroblastoma stages, risk groups and biologic features. Approximately 18% of the cases had low risk disease, 24% had intermediate risk disease, and 48% had high-risk disease. Additionally, 28% of patients in our cohort had MYCN amplification. Comparison of the patient samples collected from long term survivor clinic and oncology clinic revealed no significant difference in age at diagnosis or ethnicity, but expected decreases in the percentage of patients with high risk disease and with tumors with MYCN amplification from the long term survivor cohort were seen (Supplemental Table I).
Table I. Characteristics of the Patient and Control Populations.
| Patients (n=126) | Controls (n=114) | P value | |
|---|---|---|---|
| Age | 0.0001 | ||
| <18 months | 58 | 0 | |
| >18 months | 68 | 114 | |
|
| |||
| Gender | 0.52 | ||
| Female | 61 (48%) | 50 (44%) | |
| Male | 65 (52%) | 64 (56%) | |
|
| |||
| Ethnicity | 0.3 | ||
| Hispanic | 30 (24%) | 31 (27%) | |
| NH White | 71 (56%) | 57 (50%) | |
| NH Black | 12 (10%) | 16 (14%) | |
| Other | 12 (10%) | 10 (9%) | |
|
| |||
| INSS Tumor Stage | |||
| Stage 1 | 10 (8%) | ||
| Stage 2 | 17 (13%) | ||
| Stage 3 | 30 (24%) | ||
| Stage 4 | 59 (47%) | ||
| Stage 4S | 8 (6%) | ||
| Unknown | 2 (2%) | ||
|
| |||
| Risk Group | |||
| Low | 22 (18%) | ||
| Intermediate | 30 (24%) | ||
| High | 61 (48%) | ||
| Unknown | 13 (10%) | ||
|
| |||
| MYCN status | |||
| Amplified | 35 (28%) | ||
| Non-amplified | 67 (53%) | ||
| Unknown | 24 (19%) | ||
We analyzed DNA from cases and controls by PCR and BstNI digestion for the presence of the FGFR4 Arg388 polymorphism. Frequency of the FGFR4 AA genotype was significantly higher in neuroblastoma patients than in controls, (OR 3.33, 95% confidence interval 1.0-11.1, p= 0.049; Table II). Additionally, the prevalence of the A allele was significantly increased in neuroblastoma patients compared to controls (OR 1.65, 95% CI 1.1-2.5, p=0.02; Table II). Comparison of long term survivor patients and oncology clinic patients revealed no significant difference in the allele frequencies between the groups (p=0.41, Supplemental Table II). These results suggest a potential association between FGFR4 genotype and the prevalence of neuroblastoma.
Table II. Patient and Control FGFR4 Genotype and Allele Frequencies.
| Patients | Controls | OR (95% CI) | P value | |
|---|---|---|---|---|
| Genotype | ||||
| GG | 45 (36%) | 50 (44%) | 1 | |
| AG | 69 (55%) | 60 (53%) | 1.3 (1.8-2.2) | 0.36 |
| AA | 12 (9%) | 4 (3%) | 3.3 (1.0-11.1) | 0.049 |
| Allele | ||||
| G | 159 (63%) | 160 (70%) | 1 | |
| A | 93 (37%) | 68 (30%) | 1.7 (1.1-2.5) | 0.02 |
FGFR4 Genotype and Neuroblastoma Patient Outcomes and Prognostic Factors
FGFR4 gene polymorphisms have been linked to disease aggressiveness and patient outcomes in numerous adult tumors [20-27], but the association of FGFR4 genotype with neuroblastoma patient outcomes is unknown. We evaluated the association of FGFR4 genotype with outcomes and with clinical and biological features of tumors in 126 neuroblastoma patients. Of the neuroblastoma patients tested in our cohort, 31 (24%) experienced a relapse and 23 (18%) were deceased at the time of analysis. There was no significant difference in the event free (p=0.35) or overall (p=0.23) survival rates among patients with different FGFR4 genotypes (Supplemental Figure 2). Within the population of neuroblastoma patients, cases with the FGFR4 AA genotype were 2.5 times more likely to have tumors with MYCN amplification compared with those with genotypes AG and GG, although this association did not reach statistical significance (p=0.15; Table III). There was no significant difference in haplotype or genotype frequency in patients <18 months of age at diagnosis compared to frequencies in patients ≥18 months of age at diagnosis (Supplemental Table III). There was also no significant difference in the genotype frequencies between patients who were identified as high-risk patients and non-high risk patients (Table III), suggesting the link between FGFR4 genotype and the risk of neuroblastoma may be independent of disease risk categories and other prognostic variables.
Table III. FGFR4 polymorphism and MYCN amplification status or Risk group.
| Genotype | OR (95% CI) | P value | |||
|---|---|---|---|---|---|
| MYCN amplified | MYCN non-amplified | Unknown | |||
| GG | 13 (37%) | 21 (31%) | 11 (46%) | 1.0 | |
| AG | 18 (51%) | 42 (63%) | 9 (38%) | 0.5 (0.3-1.2) | 0.1 |
| AA | 4 (12%) | 4 (6%) | 4 (16%) | 1.8 (0.5-6.7) | 0.4 |
| High Risk | Non-High Risk | Unknown | |||
| GG | 24 (39%) | 16 (31%) | 5 (38%) | 1.0 | |
| AG | 33 (54%) | 30 (58%) | 6 (46%) | 0.7 (0.3-1.5) | 0.4 |
| AA | 4 (7%) | 6 (11%) | 2 (15%) | 0.6 (0.2-2.0) | 0.4 |
FGFR4 Receptor Trafficking
To evaluate the degradation rate of FGFR4 in neuroblastoma tumor cells, a panel of neuroblastoma tumor cell lines was analyzed for FGFR4 and EGFR protein levels at varying time points after exposure to ligand. The degradation of FGFR4 protein was delayed compared to EGFR, with no ligand-induced degradation observed at short time intervals (0-90 min) compared to 40-80% degradation observed with EGFR (Figure 1).
Figure 1. EGFR and FGFR4 degradation after ligand exposure.
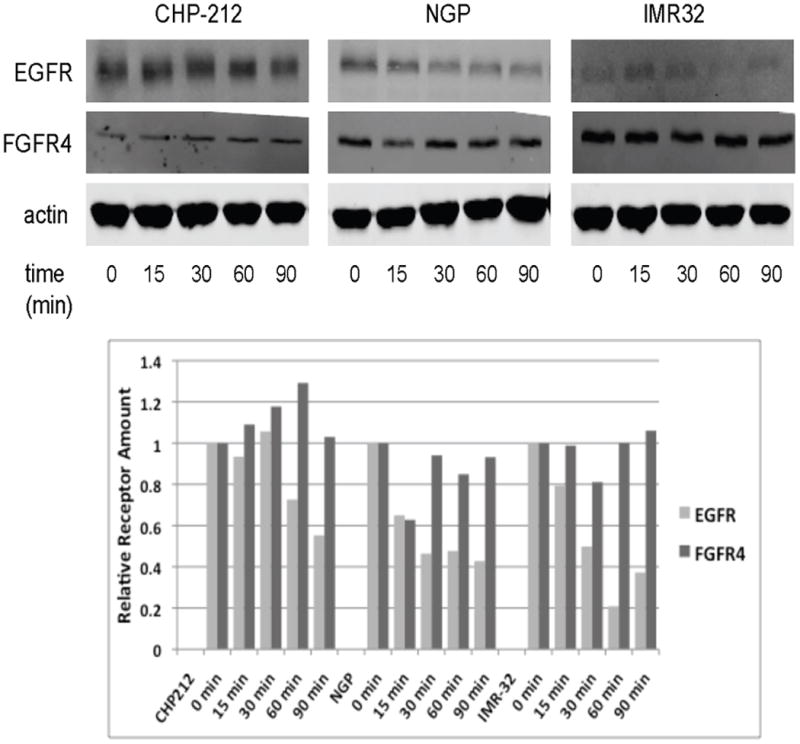
Top: Neuroblastoma cell lines were starved and then stimulated with either EGF (left) or basic FGF (right) for indicated times. Cell lysates were examined by immunoblotting for total EGFR (left) and FGFR4 (right) levels. β-actin was used as a protein loading control. Bottom: Band densities from the immunoblots were determined and calculated as a percentage of the total at time 0. The percentage of remaining EGFR and FGFR4 is plotted at each time point.
To evaluate the degradation rates of the FGFR4 Arg388 and Gly388 variants, SK-N-AS neuroblastoma cells and HEK293T cells were transfected with either V5-tagged FGFR4 Arg388 or FGFR4 Gly388 receptor constructs. Degradation of FGFR4 variants was examined after treating serum-starved cells with bFGF for 0-24hrs. Levels of the Arg388 receptor were higher at 9 and 12 hours compared to the Gly388 receptor in both neuroblastoma SK-N-AS cells and HEK293T cells, suggesting reduced receptor degradation (Figure 2) in both SK-N-AS neuroblastoma cells and HEK293T cells.
Figure 2. Effects of Glycine-to-Arginine substitution at position 388 of FGFR4 on receptor degradation.
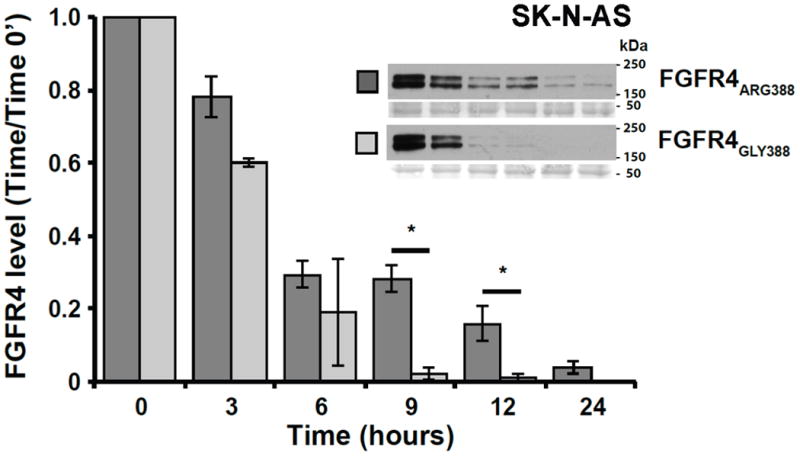
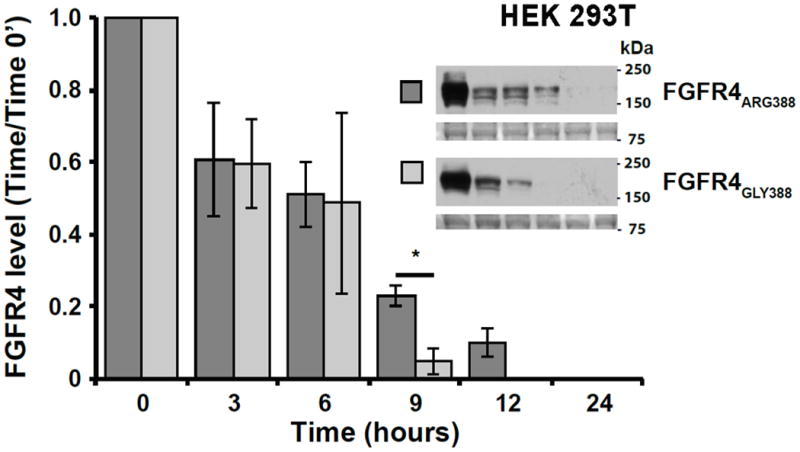
Degradation of V5-tagged Gly388 and Arg388 FGFR4 was examined in SK-N-AS (A) and HEK293T (B) transfected cells. FGFR4 levels were assessed by Western blot at 0, 3, 6, 9, 12, and 24 hours after ligand exposure for each cell line. Images of Ponceau S staining of unrelated areas are shown to indicate equal levels of protein loading. Data represent the mean ± S.E. for 4 (SK-N-AS) and 3 (HEK293T) experiments, *denotes p < 0.05.
To examine a possible mechanism underlying the differences in degradation between FGFR4 receptor isoforms, we next examined whether movement of FGFR4 from the limiting endosomal membrane into internal vesicles of the late endosome/multi-vesicular body (MVB) differed between the two FGFR4 isoforms. In cell-free assays performed as previously described [35,36], internalization of the FGFR4 Arg388 variant into internal MVB vesicles was reduced compared to the FGFR4 Gly388 variant in endosomes isolated from HEK 293T cells and from SK-N-AS cells, although the reduction observed in SK-N-AS cells did not reach statistical significance (Figure 3). This decreased internalization of the FGFR4 Arg388 variant into internal MVB vesicles may explain, at least in part, the observed reduction in degradation and may provide a mechanistic link to the increased prevalence of neuroblastoma seen in children with the FGFR4 Arg388 polymorphism.
Figure 3. Effects of Glycine-to-Arginine substitution at position 388 of FGFR4 on sorting of FGFR4 into internal MVB vesicles.
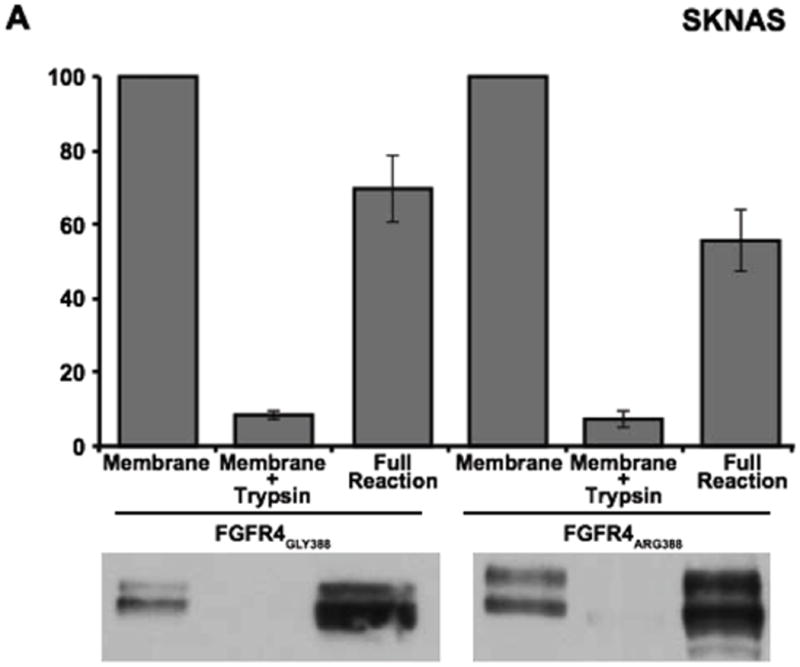
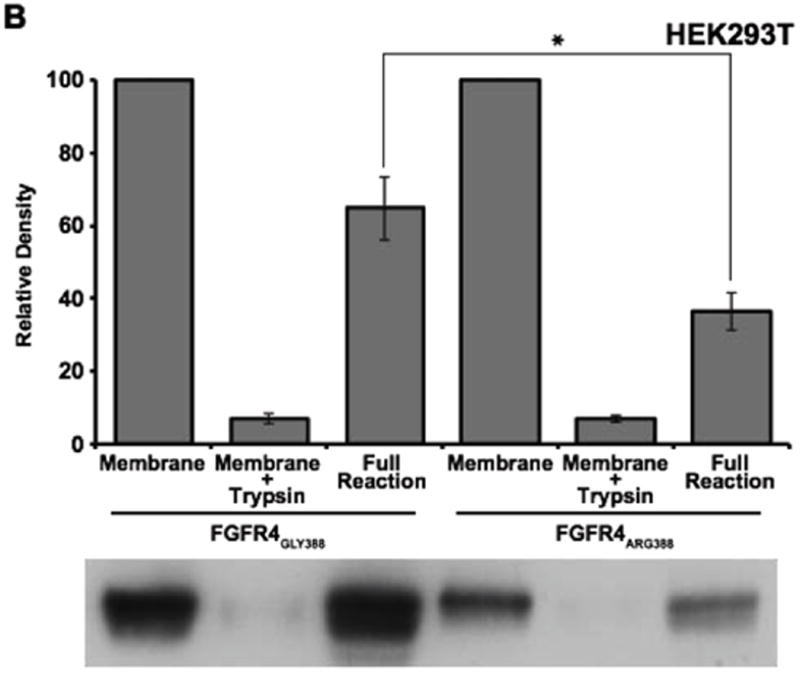
SK-N-AS (A) and HEK293T (B) cells were starved and pulsed (6 hours) with bFGF (50 ng/mL) to induce internalization of FGFR4 from the plasma membrane. Partially purified membranes containing FGFR4 were incubated with ATP and cytosol for 3 hours at 37°C. The amount of FGFR4 protected from trypsin digestion was analyzed as previously described (35). Data represents the mean +/- S.E (n=4) normalized to the control. *denotes P<0.05.
Discussion
Children with high-risk neuroblastoma have extremely poor outcomes, and additional understanding of the pathways involved in neuroblastoma pathogenesis will likely provide strategies to improve outcomes. FGFR4 expression and activity have been linked to the pathogenesis of a variety of cancers, and a germline polymorphism in the FGFR4 gene, resulting in the expression of FGFR4 Arg388 rather than the more common Gly388, is associated with decreased survival rates, treatment resistance, and more aggressive disease in patients with a variety of malignancies [20-27]. We speculate that our results demonstrate a potential association between the FGFR4 genotype and the prevalence of neuroblastoma in children and also suggest that this association may be linked to differences in FGFR4 degradation rates.
We have identified an increase in the prevalence of neuroblastoma among individuals who have the FGFR4 Arg388 allele. Our data did not, however, show a significant difference in 5-year survival rates or a trend toward worse outcomes for patients with the variant allele as studies in adult cancers have shown. This difference may be related to our relatively small sample size or that many of our patient samples were collected at the time of follow up in long-term survivor clinic, potentially biasing our sample toward patients with better outcomes. Our sample included only 12 patients with the AA genotype, and all of these patients were alive at the time of analysis. We also were unable to identify a significant association between FGFR4 genotype and patient treatment risk group or tumor MYCN status. One possible explanation is that the FGFR4 genotype is important in early stages of tumorigenesis but not critical in disease aggressiveness, but additional studies would be requried to establish this association. Further analysis of a larger cohort of neuroblastoma patient samples collected uniformly at the time of diagnosis will also be needed to determine if indeed the FGFR4 genotype is associated with other neuroblastoma prognostic variables or has an impact on outcomes in children with neuroblastoma.
The association between the FGFR4 Arg388 polymorphism and prevalence of particular cancers and more aggressive disease has been well established. However, the mechanism by which this polymorphism is associated with cancer incidence and risk is not known. Prior studies have demonstrated that the FGFR4 Arg388 variant is associated with increased receptor stability and sustained signaling [28,29]. In neuroblastoma tumor cells, the FGFR4 Arg388 variant demonstrated delayed degradation, consistent with the prior findings in prostate cancer cells [28]. Our results also suggest a potential link between FGFR4 trafficking and degradation and the pathogenesis of cancers such as neuroblastoma that requires further investigation.
We have previously shown that UBE4B plays a crucial role in GFR trafficking and degradation in neuroblastoma tumor cells, suggesting a previously uncharacterized link between GFR trafficking and degradation and tumorigenesis [36,37]. Dysregulation of GFR trafficking is clearly emerging as an important mechanism for oncogenesis in a variety of human cancers. A large and growing body of evidence clearly demonstrates that disruption of the GFR trafficking pathways can alter cell expression and activity of GFRs, leading to the development and progression of cancer [38,39]. Therefore, developing a better understanding of the GFR trafficking pathways is critical in the understanding of the process of tumorigenesis.
The results of these studies provide important and novel information about the previously undescribed roles and mechanisms of FGFR4 genotype in children with neuroblastoma. Our results identified a significant increase in the frequency of the A allele in the patients with neuroblastoma and an increased frequency of the AA genotype in those with neuroblastoma compared to controls. We have also demonstrated a difference in the degradation of the FGFR4 Arg388 and Gly388 isoforms. Our study provides the first suggestion of a role for FGFR4 in the pathogenesis of neuroblastoma, and additional studies will be required to confirm this association and further investigate the underlying molecular mechanisms.
Supplementary Material
Germline DNA was isolated from patient samples and RT-PCR for the FGFR4 polymorphism was performed. DNA was isolated, subjected to degradation with BstNI, separated on agarose gels, and patient genotypes were determined. A single 109bp band is expected for the Gly388 allele, while fragments of 82 and 27bp characterize the Arg388 allele. Control samples from prostate cancer cell lines DU145 (GG) and PC3 (AA) were used for comparison.
FGFR4 genotypes were determined in 126 neuroblastoma patients, and overall and event-free Kaplan Meier curves were created using FGFR4 genotype and patient outcome information and STATA statistical software (v12.1).
Acknowledgments
The authors wish to acknowledge Natalie Sirisaengtaksin's assistance with preparation of the manuscript and Fatih Okcu for assistance in obtaining patient and control samples for analysis. We also wish to thank Hyundai Hope on Wheels Foundation for their support.
Funding Sources: Hyundai Hope on Wheels foundation Hope Grant (to PEZ)
Glossary
- FGFR
Fibroblast growth factor receptor
- GFR
Growth factor receptor
- MVB
Multi-vesicular body
Footnotes
Conflict of Interest Statement: The authors have no conflicts of interest to disclose.
References
- 1.Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362:2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nat Rev Drug Discov. 2009;8(3):235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwabi-Addo B, Ozen M, Ittmann M. The role of fibroblast growth factors and their receptors in prostate cancer. Endocr Relat Cancer. 2004;11(4):709–724. doi: 10.1677/erc.1.00535. [DOI] [PubMed] [Google Scholar]
- 4.Powers CJ, McLeskey SW, Wellstein A. Fibroblast growth factors, their receptors and signaling. Endocr Relat Cancer. 2000;7(3):165–197. doi: 10.1677/erc.0.0070165. [DOI] [PubMed] [Google Scholar]
- 5.Ozen M, Giri D, Ropiquet F, et al. Role of fibroblast growth factor receptor signaling in prostate cancer cell survival. J Natl Cancer Inst. 2001;93(23):1783–1790. doi: 10.1093/jnci/93.23.1783. [DOI] [PubMed] [Google Scholar]
- 6.Reiland J, Kempf D, Roy M, et al. FGF2 binding, signaling, and angiogenesis are modulated by heparanase in metastatic melanoma cells. Neoplasia. 2006;8(7):596–606. doi: 10.1593/neo.06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wesche J, Haglund K, Haugsten EM. Fibroblast growth factors and their receptors in cancer. Biochem J. 2011;437(2):199–213. doi: 10.1042/BJ20101603. [DOI] [PubMed] [Google Scholar]
- 8.Gozgit JM, Wong MJ, Moran L, et al. Ponatinib (AP24534), a multitargeted pan-FGFR inhibitor with activity in multiple FGFR-amplified or mutated cancer models. Mol Cancer Ther. 2012;11(3):690–699. doi: 10.1158/1535-7163.MCT-11-0450. [DOI] [PubMed] [Google Scholar]
- 9.Gowardhan B, Douglas DA, Mathers ME, et al. Evaluation of the fibroblast growth factor system as a potential target for therapy in human prostate cancer. Br J Cancer. 2005;92(2):320–327. doi: 10.1038/sj.bjc.6602274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pardo OE, Latigo J, Jeffery RE, et al. The fibroblast growth factor receptor inhibitor PD173074 blocks small cell lung cancer growth in vitro and in vivo. Cancer Res. 2009;69(22):8645–8651. doi: 10.1158/0008-5472.CAN-09-1576. [DOI] [PubMed] [Google Scholar]
- 11.Zhao GLWY, Chen D, et al. A Novel, Selective Inhibitor of Fibroblast Growth Factor Receptors That Shows a Potent Broad Spectrum of Antitumor Activity in Several Tumor Xenograft Models. Mol Cancer Ther. 2011;10:2200–2210. doi: 10.1158/1535-7163.MCT-11-0306. [DOI] [PubMed] [Google Scholar]
- 12.Gavine PR, Mooney L, Kilgour E, Thomas AP, Al-Kadhimi K, Beck S, Rooney C, Coleman T, Baker D, Mellor MJ, Brooks AN, Klinowska T. AZD4547: An Orally Bioavailable, Potent, and Selective Inhibitor of the Fibroblast Growth Factor Receptor Tyrosine Kinase Family. Cancer Res. 2012;72:2045–56. doi: 10.1158/0008-5472.CAN-11-3034. [DOI] [PubMed] [Google Scholar]
- 13.Yu C, Wang F, Kan M, Jin C, Jones RB, Weinstein M, Deng CX, McKeehan WL. Elevated Cholesterol Metabolism and Bile Acid Synthesis in Mice Lacking Membrane Tyrosine Kinase Receptor FGFR4. J Biol Chem. 2000;275:15482–9. doi: 10.1074/jbc.275.20.15482. [DOI] [PubMed] [Google Scholar]
- 14.Heinzle CEZ, Paur J, Grasl-Kraupp B, et al. Is Fibroblast Growth Factor Receptor 4 a Suitable Target of Cancer Therapy? Curr Pharm Des. 2014;20(17):2881–2898. doi: 10.2174/13816128113199990594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paulson V, Chandler G, Rakheja D, Galindo RL, Wilson K, Amatruda JF, Cameron S. High-Resolution Array CGH Identifies Common Mechanisms that Drive Embryonal Rhabdomyosarcoma Pathogenesis. Genes Chomosomes Cancer. 2011;50:397–408. doi: 10.1002/gcc.20864. [DOI] [PubMed] [Google Scholar]
- 16.Davicioni E, Finckenstein FG, Shahbazian V, Buckley JD, Triche TJ, Anderson MJ. Identification of a PAX-FKHR Gene Expression Signature That Defines Molecular Classes and Determines the Prognosis of Alveolar Rhabdomyosarcomas. Cancer Res. 2006;66:6936–46. doi: 10.1158/0008-5472.CAN-05-4578. [DOI] [PubMed] [Google Scholar]
- 17.West SNNGA, Pounds S, Figueredo BC, et al. Gene Expression Profiling of Childhood Adrenocortical Tumors. Cancer Res. 2007;67:600–608. doi: 10.1158/0008-5472.CAN-06-3767. [DOI] [PubMed] [Google Scholar]
- 18.Schweigerer L, Ledoux D, Fleischmann G, et al. Enhanced MYCN oncogene expression in human neuroblastoma cells is associated with altered FGF receptor expression and cellular growth response to basic FGF. Biochem Biophys Res Commun. 1991;179(3):1449–1454. doi: 10.1016/0006-291x(91)91735-u. [DOI] [PubMed] [Google Scholar]
- 19.Janet T, Ludecke G, Otten U, et al. Heterogeneity of human neuroblastoma cell lines in their proliferative responses to basic FGF, NGF, and EGF: correlation with expression of growth factors and growth factor receptors. J Neurosci Res. 1995;40(6):707–715. doi: 10.1002/jnr.490400602. [DOI] [PubMed] [Google Scholar]
- 20.Spinola M, Leoni V, Pignatiello C, et al. Functional FGFR4 Gly388Arg polymorphism predicts prognosis in lung adenocarcinoma patients. J Clin Oncol. 2005;23(29):7307–7311. doi: 10.1200/JCO.2005.17.350. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Stockton DW, Ittmann M. The fibroblast growth factor receptor-4 Arg388 allele is associated with prostate cancer initiation and progression. Clin Cancer Res. 2004;10:6169–6178. doi: 10.1158/1078-0432.CCR-04-0408. [DOI] [PubMed] [Google Scholar]
- 22.Morimoto Y, Ozaki T, Ouchida M, et al. Single nucleotide polymorphism in fibroblast growth factor receptor 4 at codon 388 is associated with prognosis in high-grade soft tissue sarcoma. Cancer. 2003;98(10):2245–2250. doi: 10.1002/cncr.11778. [DOI] [PubMed] [Google Scholar]
- 23.Streit S, Mestel DS, Schmidt M, et al. FGFR4 Arg388 allele correlates with tumour thickness and FGFR4 protein expression with survival of melanoma patients. Br J Cancer. 2006;94(12):1879–1886. doi: 10.1038/sj.bjc.6603181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.da Costa Andrade VC, Parise OJ, Hors CP, et al. The fibroblast growth factor receptor 4 (FGFR4) Arg388 allele correlates with survival in head and neck squamous cell carcinoma. Exp Mol Pathol. 2007;82(1):53–57. doi: 10.1016/j.yexmp.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Bange J, Prechtl D, Cheburkin Y, Specht K, Harbeck N, Schmitt M, Knyazeva T, Muller S, Gartner S, Sures I, Wang H, Imyanitov E, Haring HU, Knayzev P, Iacobelli S, Hofler H, Ullrich A. Cancer progression and tumor cell motility are associated with the FGFR4 Arg(388) allele. Cancer Res. 2002;62:840–847. [PubMed] [Google Scholar]
- 26.Thussbas C, Nahrig J, Streit S, Bange J, Kriner M, Kates R, Ulm K, Kiechle M, Hoefler H, Ullrich A, Harbeck N. FGFR4 Arg388 allele is associated with resistance to adjuvant therapy in primary breast cancer. J Clin Oncol. 2006;24:3747–3755. doi: 10.1200/JCO.2005.04.8587. [DOI] [PubMed] [Google Scholar]
- 27.Xu W, Li Y, Wang X, Chen B, Wang Y, Liu S, Xu J, Zhao W, Wu J. FGFR4 Transmembrane Domain Polymorphism and Cancer Risk: A Meta-Analysis Including 8555 Subjects. Eur J Cancer. 2010;46:3332–8. doi: 10.1016/j.ejca.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Yu W, Cai Y, et al. Altered fibroblast growth factor receptor 4 stability promotes prostate cancer progression. Neoplasia. 2008;10(8):847–856. doi: 10.1593/neo.08450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu WFS, Dakhova O, Creighton CJ, et al. FGFR-4 Arg38 Enhances Prostate Cancer Progression Via Extracellular Signal-Related Kinase and Serum Response Factor Signaling. Clin Cancer Res. 2011;17:4355–4366. doi: 10.1158/1078-0432.CCR-10-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlesinger HR, Gerson JM, Moorhead PS, Maguire H, Hummeler K. Establishment and Characterization of human Neuroblastoma Cell Line. Cancer Research. 1976;36:3094–3100. [PubMed] [Google Scholar]
- 31.Brodeur GM, Green AA, Hayes FA, Williams KJ, Williams DL, Tsiatis AA. Cytogenetic Features of Human Neuroblastomas and Cell Lines. Cancer Res. 1981;41:4678–86. [PubMed] [Google Scholar]
- 32.Tumilowicz JJ, Nichols WW, Cholon JJ, Greene AE. Definition of a Continuous Human Cell Line Derived from. Neuroblastoma Cancer Research. 1970;30:2110–2118. [PubMed] [Google Scholar]
- 33.Helson L, Helson C. Human neuroblastoma cells and 13-cis-retinoic acid. J Neurooncol. 1985;3:39–41. doi: 10.1007/BF00165170. [DOI] [PubMed] [Google Scholar]
- 34.Scorsone K, Zhang L, Woodfield SE, Hicks J, Zage PE. The Novel Kinase Inhibitor EMD1214063 Is Effective Against Neuroblastoma. Invest New Drugs. 2014;32:815–24. doi: 10.1007/s10637-014-0107-4. [DOI] [PubMed] [Google Scholar]
- 35.Sun W, Vida TA, Sirisaengtaksin N, et al. Cell-free reconstitution of multivesicular body formation and receptor sorting. Traffic. 2010;11(6):867–876. doi: 10.1111/j.1600-0854.2010.01053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sirisaengtaksin N, Gireud M, Yan Q, et al. UBE4B protein couples ubiquitination and sorting machineries to enable epidermal growth factor receptor (EGFR) degradation. J Biol Chem. 2014;289(5):3026–3039. doi: 10.1074/jbc.M113.495671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zage PE, Sirisaengtaksin N, Liu Y, et al. UBE4B levels are correlated with clinical outcomes in neuroblastoma patients and with altered neuroblastoma cell proliferation and sensitivity to epidermal growth factor receptor inhibitors. Cancer. 2013;119(4):915–923. doi: 10.1002/cncr.27785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zage PE, Bean AJ. Growth Factor Receptor Trafficking as a Potential Therapeutic Target in Pediatric Cancer. Front Biol. 2012;7:1–13. [Google Scholar]
- 39.Mellman I, Yarden Y. Endocytosis and Cancer. Cold Spring Harb Perspect Biol. 2013;5:a016949. doi: 10.1101/cshperspect.a016949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Germline DNA was isolated from patient samples and RT-PCR for the FGFR4 polymorphism was performed. DNA was isolated, subjected to degradation with BstNI, separated on agarose gels, and patient genotypes were determined. A single 109bp band is expected for the Gly388 allele, while fragments of 82 and 27bp characterize the Arg388 allele. Control samples from prostate cancer cell lines DU145 (GG) and PC3 (AA) were used for comparison.
FGFR4 genotypes were determined in 126 neuroblastoma patients, and overall and event-free Kaplan Meier curves were created using FGFR4 genotype and patient outcome information and STATA statistical software (v12.1).


