Abstract
The magnesium transporter A (MgtA) is a specialized P-type ATPase, believed to import Mg2+ into the cytoplasm. In Salmonella typhimurium and Escherichia coli, the virulence determining two-component system PhoQ/PhoP regulates the transcription of mgtA gene by sensing Mg2+ concentrations in the periplasm. However, the factors that affect MgtA function are not known. This study demonstrates, for the first time, that MgtA is highly dependent on anionic phospholipids and in particular, cardiolipin. Colocalization studies confirm that MgtA is found in the cardiolipin lipid domains in the membrane. The head group of cardiolipin plays major role in activation of MgtA suggesting that cardiolipin may act as a Mg2+ chaperone for MgtA. We further show that MgtA is highly sensitive to free Mg2+ (Mg2+free) levels in the solution. MgtA is activated when the Mg2+free concentration is reduced below 10 μM and is strongly inhibited above 1 mM, indicating that Mg2+free acts as product inhibitor. Combined, our findings conclude that MgtA may act as a sensor as well as a transporter of Mg2+.
DOI: http://dx.doi.org/10.7554/eLife.11407.001
Research Organism: E. coli
eLife digest
Magnesium is an essential element for living cells, meaning that organisms from bacteria to humans need magnesium to survive. All cells are surrounded by a membrane made of fatty molecules called lipids, which is also embedded with proteins. Magnesium, like other metal ions, is transported inside cells across the cell’s membrane by specific membrane proteins.
A species of gut bacteria called E. coli has two separate magnesium transport systems: one that works at high concentrations of magnesium and one at lower concentrations. The latter system involves a membrane protein called magnesium transporter A (or MgtA for short), which works like a molecular pump. However, it was not known exactly how this transporter was affected by magnesium nor how sensitive it was to this divalent metal ion. It was also unclear whether MgtA worked alone in the bacterial membrane or if it worked in conjunction with other molecules.
Now Subramani et al. have managed to show that MgtA can sense magnesium ions down to micromolar concentrations, which is the equivalent to a pinch (1 gram) of magnesium salt in 10,000 liters of water. The experiments also showed that this detection system depended on a specific lipid molecule in the membrane called cardiolipin. MgtA and cardiolipin were found together in the membrane of living E. coli suggesting that the two do indeed work together.
The discovery that a membrane transporter that pumps ions needs cardiolipin to work suggests that cells could indirectly control the movement of ions by changing the levels of specific lipids in their membranes. Subramani et al. now hope to use techniques, such as X-ray crystallography, to visualize how magnesium and cardiolipin bind to MtgA and explore how the three molecules work together as a complete system. Information about these interactions could in the future help researchers understand how these bacteria try to protect themself in the hostile environment in the human gut or cells of the immune systems. Further studies of this system could be used to develop biological sensors for magnesium or to design antibiotics that interfere with the magnesium transporter to treat bacterial infections.
Introduction
Magnesium is the most abundant divalent cation in biological systems and is an essential requirement for all living cells (Reinhart, 1988). Mg2+ has diverse biological roles, ranging from being an essential cofactor in ATP-mediated enzymatic reactions to being a signaling molecule that activates important virulence systems in bacteria (Groisman et al., 2013). Mg2+ homeostasis is well studied in Gram-negative bacteria like S. typhimurium and E. coli (Papp-Wallace and Maguire, 2008). Three classes of Mg2+ transporters have been identified in bacteria: CorA, MgtE and MgtA (magnesium transporter A)/MgtB. Based on the Mg2+ transport studies, Snavely et al., proposed that CorA transports Mg2+ under normal Mg2+ levels, whereas MgtA and MgtB transport Mg2+ when bacteria faces low Mg2+ condition (Snavely et al., 1991). Later Garcia Vescovi et al., identified that the low Mg2+ levels in the periplasm activate the PhoQ/P system (Véscovi et al., 1996), which in turn induces the expression of genes essential for adapting the Mg2+ limiting environments (Monsieurs et al., 2005). One of the genes was found to be mgtA. In addition to PhoQ/P mediated activation, a Mg2+ sensing riboswitch at the 5’ leader region was found to activate the transcription of mgtA gene when the Mg2+ level in cytoplasm falls below a certain threshold (Cromie et al., 2006). Therefore, both the intracellular and extracellular Mg2+ concentrations regulate transcription of the mgtA gene. Upon translation, MgtA is believed to transport Mg2+ from the periplasm into the cytoplasm under Mg2+ depriving conditions (Snavely et al., 1989). It has been shown that deletion of the mgtA gene affects the survival of S. typhimurium at higher temperatures and also promotes lysis in Streptococcus pneumonia (O'Connor et al., 2009; Neef et al., 2011) .
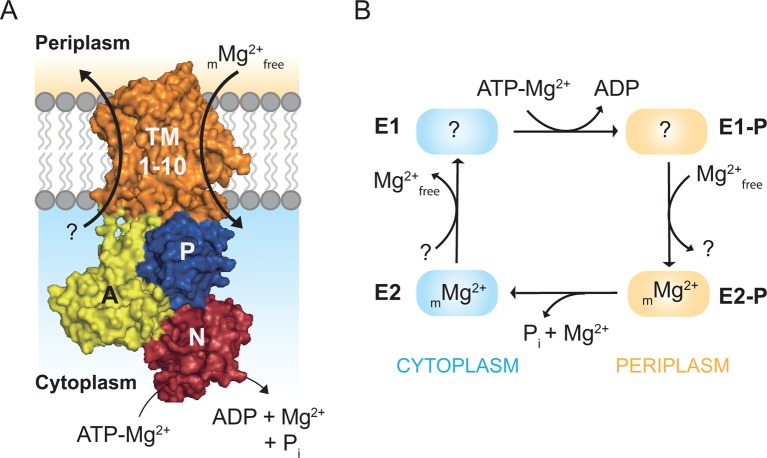
MgtA belongs to the P3 subfamily of P-type ATPases (Palmgren and Axelsen, 1998). The P3 family is subdivided into P3A and P3B. The P3A family is dominated by H+-ATPases found in plants (Palmgren, 2001), while P3B contains Mg2+ ATPases, found to be dominant amongst prokaryotes (Kühlbrandt, 2004). Recently, a close homolog of MgtA was reported in Petunia hybrida (PH1), which lacks the ability to induce a proton gradient. PH1 was proposed to support the proton pumping performed by the P3A proton pump PH5, but evidence for magnesium transport was not established (Faraco et al., 2014). In general, MgtA is found in bacteria, archaea, fungi and plants (See also Figure 1—figure supplement 1–2). MgtA has ten predicted transmembrane helices (TM), a nucleotide binding domain (N), a phosphorylation domain (P) and an actuator domain (A) (Figure 1A). The Post-Albers cycle describes the transport mechanism of a P-type ATPase (Albers, 1967; Post et al., 1969). During the transport cycle, P-type ATPases alternate between the E1 and E2 states with different intermediate conformational states (Palmgren and Nissen, 2011). The E1 state has high affinity and is open for ions binding from the cytoplasm. Ion binding induces autophosphorylation by transferring the γ-phosphate from an ATP molecule to a conserved aspartate residue in the DKTGT consensus motif of the P domain forming the E1P state (Lutsenko and Kaplan, 1995). In E. coli MgtA (ecMgtA), the phosphorylated aspartate corresponds to Asp 373. Phosphorylation induces domain rearrangements leading to the E2P state, which is now open to the periplasmic side. E2P has low affinity for the bound ions and high affinity for the counter ions (Mg2+ in case of MgtA). The exchange of counter ions dephosphorylates the enzyme and forms the E2 state. Further conformational changes return the enzyme back to the E1 state and the counterions are released into the cytoplasm, thus completing the transport cycle. A possible Post-Albers scheme with the four steps transport cycle can be visualized for MgtA and is given in Figure 1B. Since MgtA imports Mg2+ into the cytoplasm, Mg2+ is predicted to have high affinity to the E2P state. However, the stoichiometry and electrogenic nature of magnesium transport by MgtA remains unknown.
Figure 1. Magnesium uptake scheme for MgtA.
(A) Cartoon representation of MgtA showing transmembrane domain (TM), actuator domain (A), phosphorylation domain (P) and nucleotide binding domain (N). MgtA transports Mg2+ ions into cytoplasm as a function of ATP hydrolysis, (m) represent the unknown stoichiometry. (B) Proposed Post-Albers reaction scheme (Albers, 1967; Post et al., 1969) adapted for MgtA.
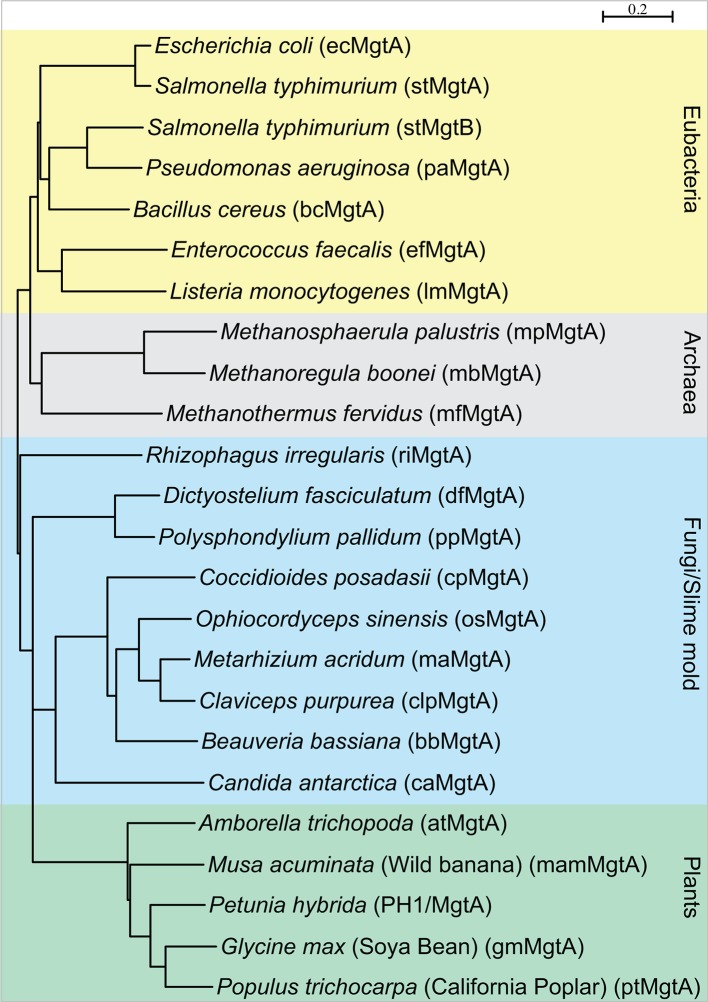
Figure 1—figure supplement 1. Phylogenetic tree showing distribution of MgtA homologs among four kingdom of life: eubacteria (yellow), archaea (grey), fungi and slime mold (blue) and the plant kingdom (green).
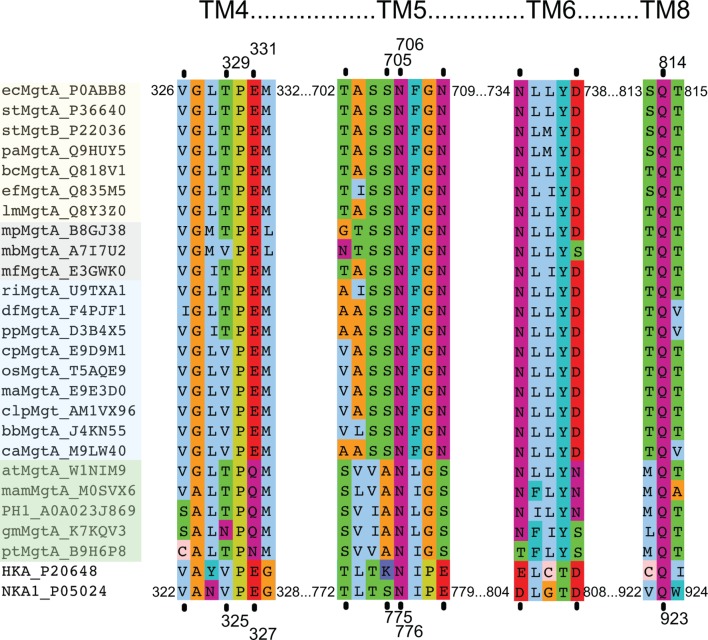
Figure 1—figure supplement 2. Multiple sequence alignment of selected MgtA homologs including the pig Na+/K+-ATPase (NKA1) and the human H+/K+-ATPase.
Over the past two decades, detailed studies of mgtA transcriptional regulation has provided a clear picture of the mechanisms through which S. typhimurium and E. coli tightly control mgtA gene expression (Groisman et al., 2013). Despite these detailed transcriptional regulatory studies, very little is known about the biochemical mechanism or factors influencing the Mg2+ transport by MgtA. Here, we present the first biochemical characterization of purified MgtA from E. coli. We show that anionic phospholipids, particularly cardiolipin (CL) are crucial for in vitro activation of MgtA. Further the overexpressed MgtA colocalizes with CL in vivo. We also show that MgtA ATPase activity is highly sensitive to Mg2+free concentration and that Mg2+ acts as an inhibitor at higher concentrations. Our findings reveal for the first time, the effect of anionic lipids and free Mg2+ on MgtA function.
Results
Expression and purification of MgtA
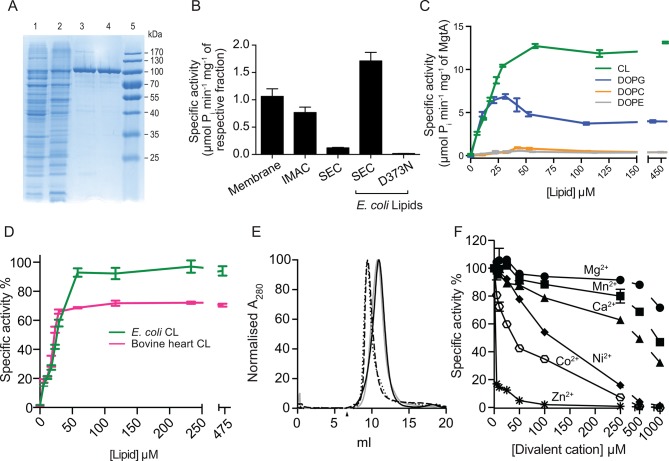
In this study, MgtA was successfully overexpressed in its native host E. coli with yields up to 4 mg per liter of LB medium. The overexpression of MgtA is clearly visible as a band both in the cell lysate and membrane fraction as shown in SDS-PAGE gel (Figure 2A). Initial affinity purification with His-tag yielded >85% pure protein, additional size exclusion chromatography (SEC) increased the purity up to >95%, resulting in a highly purified protein for in vitro biochemical studies.
Figure 2. Biochemical characterization and cardiolipin depencency.
(A) Samples taken throughout the purification process were subjected to SDS-PAGE. Lane 1- Cell lysate, Lane 2- Membrane fraction, Lane 3- Ni2+ column elute, Lane 4- SEC fractions. (B) An ATPase assay was performed with samples (5 μg) collected at each purification step. Total E. coli lipids (10 μg) were added to the SEC fraction. The inactive D373N mutant purified as wild-type MgtA and served as negative control. (C) Concentration-dependent activation of ATPase activity by the individual lipid component of E. coli inner membrane. Lipids prepared as described in the methods section were added to MgtA at indicated concentrations and phosphate release was measured. (D) Comparison of the ATPase activity induced by E. coli CL and bovine heart CL. 100% Specific activity represents 13 µM min-1 mg-1 of MgtA. (E). SEC profile of MgtA in the presence of CL and Mg2+. MgtA (—); MgtA with 5.0 mM MgCl2(—); MgtA with CL (−−); MgtA with CL and 5.0 mM MgCl2 (…). MgtA and CL were mixed at 1: 3500 molar ratio and incubated on ice for 30 min before SEC. The closed arrow indicates void volume. (F) Effect of divalent cations on ATP hydrolysis by MgtA. Mg2+(●), Mn2+(■), Ca2+(▲), Ni2+(♦), Zn2+( ), Co2+(
), Co2+( ). All the conditions with cations have a basal concentration of 3 mM Mg2+ and 3 mM ATP. B, C, E, F – Values plotted are mean ± SD (n = 3).
). All the conditions with cations have a basal concentration of 3 mM Mg2+ and 3 mM ATP. B, C, E, F – Values plotted are mean ± SD (n = 3).
DOI: http://dx.doi.org/10.7554/eLife.11407.006
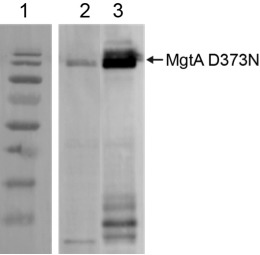
Figure 2—figure supplement 1. Purification of MgtA D373N: D373N mutant, which cannot be phosphorylated served as negative control.
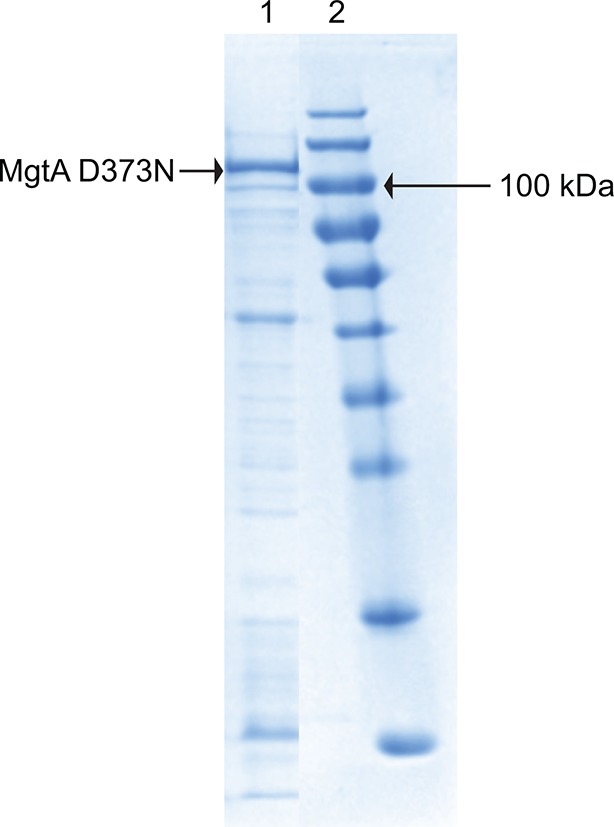
Figure 2—figure supplement 2. Effect of DOPE and DOPG on CL activated MgtA.
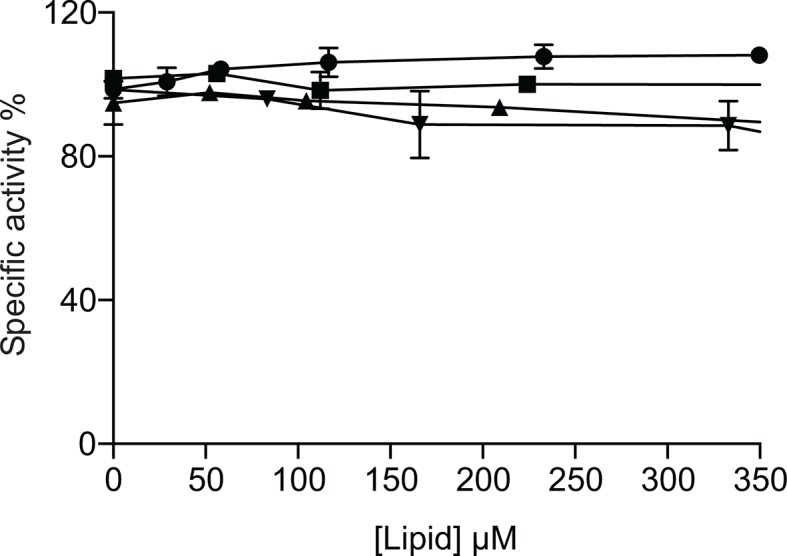
Figure 2—figure supplement 3. MgtA ATPase activity in the presence of inhibitors.
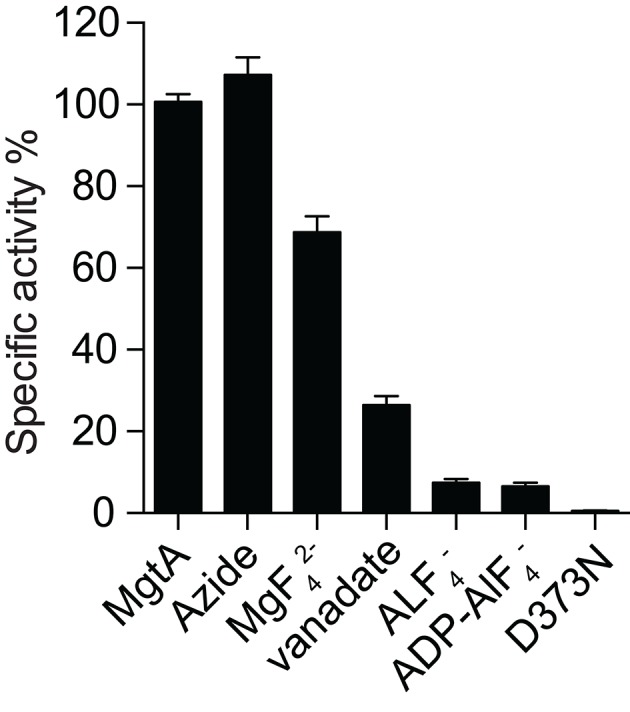
Figure 2—figure supplement 4. Influence of Ni2+ on Mg2+total induced ATPase activity of MgtA.
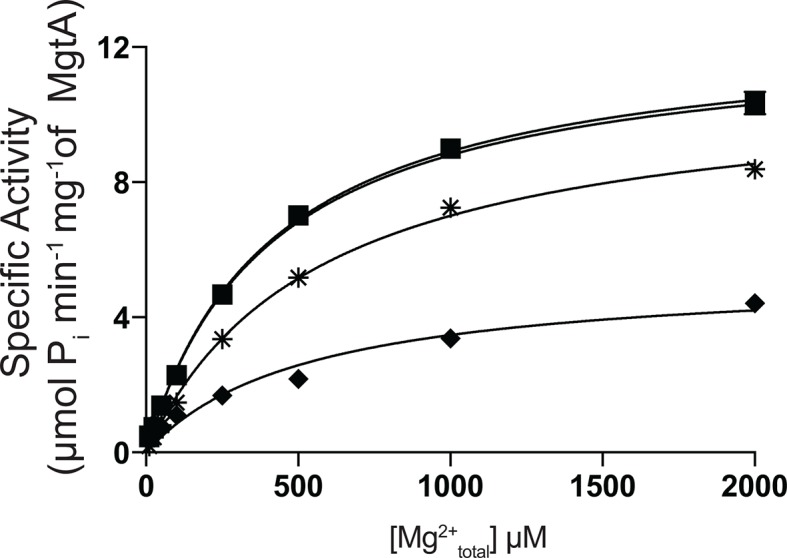
 ), 250 μM(♦) were added to indicated concentration of Mg2+total. 3mM ATP was used to all conditions. Values plotted are mean ± SD (n = 3).
), 250 μM(♦) were added to indicated concentration of Mg2+total. 3mM ATP was used to all conditions. Values plotted are mean ± SD (n = 3).ATPase activity of MgtA is restored by cardiolipin and phosphatidyl glycerol
To test whether detergent solubilization and further purification steps had any deleterious effects on MgtA function, we monitored the protein activity using an ATPase assay, which measures phosphate release. A decline in the ATPase activity was observed during purification of wild type (wt) MgtA, with more than 90% loss of the ATPase activity after the final SEC purification (Figure 2B). The gradual decrease in the ATPase activity between each purification step prompted us to test whether the ATPase activity of MgtA was lipid dependent. Addition of total lipid extract from E. coli recovered the ATPase activity (Figure 2B). We included an inactive mutant (D373N MgtA), which cannot be phosphorylated, as a negative control to rule out the ATPase activity from other endogenous ATPases. The mutant was purified equivalently to wtMgtA (Figure 2—figure supplement 1) and did not show any significant ATPase activity when supplemented with E. coli lipids.
The phospholipid composition of E. coli inner membrane has been reported to consist of ~75% phosphatidyl ethanolamine (PE), ~20% phosphatidyl glycerol (PG) and ~5% CL (Morein et al., 1996). To test whether the observed effect of lipids is attributed to a single lipid type, we performed the ATPase assay with increasing amounts of DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine), DOPG (1,2-dioleoyl-sn-glycero-3-phospho-(1'-rac-glycerol) and CL (isolated from E. coli) and found that only anionic lipids such as DOPG and E. coli CL restored the ATPase activity in a concentration-dependent manner. DOPG activated MgtA by thirty-fold, while CL by hundred-fold, relative to the lipid-free condition (Figure 2C). DOPE increased the ATPase activity only by three folds. Further, a non-native E. coli lipid, DOPC (1,2-Dioleoyl-sn-glycero-3-phosphocholine), did not show any measureable effect on the ATPase activity. Apparent Vmax and Km calculated for MgtA in the presence of CL are given in Table 1.
Table 1.
Kinetic property of purified MgtA. The apparent Vmax,Km, and the turnover number (Kcat) values were determined by least squares fit of the data from Figure 4C, as described in materials and methods.
| Parameters | 1 mM ATP | 3 mM ATP | 6 mM ATP |
|---|---|---|---|
| Vmax(μmol min-1 mg-1) | 14.0 ± 0.2 | 13.7 ± 0.2 | 14.5 ± 0.2 |
| Km(μM) | 15 ± 0.6 | 10 ± 0.6 | 10 ± 0.6 |
| Kcat (s-1) | 23 ± 0.3 | 22 ± 0.8 | 24 ± 0.2 |
The maximal ATPase activity observed with total E. coli lipid extract (1.7 μM Pi min-1 mg-1) was lower than the maximum ATPase activity observed with pure CL. The possible reasons could either be the low availability of CL in E. coli lipid extract or other lipid types like PE and PG competing for the lipid binding site in MgtA. To test the latter possibility, we added increasing amounts of DOPE and DOPG to pre-incubated MgtA-CL mixture and measured ATP hydrolysis. Only a minor decrease in the ATPase activity (<20%) (Figure 2—figure supplement 2) was observed, suggesting that other lipids are likely not competing with CL binding. Moreover, the extra DOPG did not show any additive effect over CL induced ATPase activity. The minor decrease in the ATPase activity is likely due to the presence of detergent C12E8 (Octaethylene glycol monododecyl ether) in which the lipids are dissolved (Figure 2—figure supplement 2). Therefore, the subsequent enzymatic studies were performed in the presence of E. coli CL extract unless stated otherwise.
The head group of CL plays a major role in MgtA activation
CL isolated from E. coli has a diverse fatty acid distribution, with 16:0–18:1 as the most dominant (~35%) and 16:0–17:0Δ as the second most dominant (~17%) species at positions 1 and 2 of sn-glycerol 3-phosphate (Yokota et al., 1980). To test whether the head group (diacylglycerol phosphate) of CL or the diversity of fatty acid chains plays a prominent role in activating MgtA, we performed ATPase assays in the presence of CL extracted from bovine heart (Figure 2D). All the four fatty acid chains of bovine heart CL are almost exclusively linoleate chains (18:2, 18:1, 18:3) (Shinzawa-Itoh et al., 2007; Schlame et al., 1993). The initial activation profile for both CL extracts are equivalent, which suggest that the head group plays the major role in the activation and not the chemical diversity found within the fatty acid chains in E. coli CL. However, the maximal ATPase activity in the presence of bovine heart CL was approximately 30% lower than what was observed with E. coli CL. This could mean that the longer linoleate chains from bovine heart CL are suboptimal for effective interaction between MgtA and CL.
MgtA functions as a monomer and is sensitive to P-type ATPase inhibitors
Whether MgtA acts as a monomer or dimer in the presence of CL will affect further interpretation of the enzymatic studies since CL has been shown to bind and facilitate oligomerization in other proteins like the nitrate reductase (Arias-Cartin et al., 2011) and SecYEG (Gold et al., 2010). The SEC elution profile shows a monodisperse peak corresponding to the size of a monomer (Figure 2E). The SEC profile of MgtA in the presence and absence of CL did not show signs of dimerization of MgtA. However, a significant shift in the elution peak was observed corresponding to a difference of ~50 kDa. This shift likely indicates an increase in lipid-protein-detergent micelle radius caused by binding of CL to MgtA. The presence of Mg2+ with CL does not show any further shift, confirming that MgtA function as a monomer in the described assay conditions. The general P-type ATPase inhibitor vanadate as well as specific inhibitors that affect different steps of Post-Albers cycle (Figure 2—figure supplement 3) inhibited the MgtA ATPase activity. AlF4- and ADP-AlF4- showed maximal inhibition, followed by vanadate, whereas MgF4- showed only a partial inhibition. Further, the typical F-type ATPase inhibitor azide had no effect on MgtA ATPase activity. Together these results confirmed that MgtA behaves like other thoroughly studied P-type ATPase like SERCA (Danko et al., 2004).
The influence of divalent cations other than Mg2+ on MgtA
Mg2+ transport by MgtA was studied in vivo and reported to show inhibition according to the following order of potency: Zn2+ ≥ Mg2+ > Ni2+ ≈ Co2+ > Ca2+ (Snavely et al., 1989). Utilizing our ATPase assay, we investigated the effect of divalent ions in vitro in the presence of CL and 3.0 mM Mg2+ at pH 7.0. We observed the order of inhibition to be Zn2+ > Co2+ > Ni2+ > Ca2+ > Mn2+ > Mg2+ (Figure 2F). Half maximal inhibition of ATPase activity was found to be at < 5 μM for Zn2+, 37 μM for Co2+, 100 μM for Ni2+, 500 μM for Ca2+ and 1 mM for Mn2+. These values agree reasonably well with the published values (Snavely et al., 1989). MgtA has been reported to transport Ni2+ as well as Mg2+ and Ni2+ acting as a competitive inhibitor (Snavely et al., 1991). The ATPase assay performed with increasing Mg2+ concentration at several fixed Ni2+ concentrations, reveal that Ni2+likely acts as a non-competitive inhibitor (Figure 2—figure supplement 4) rather than competitive.
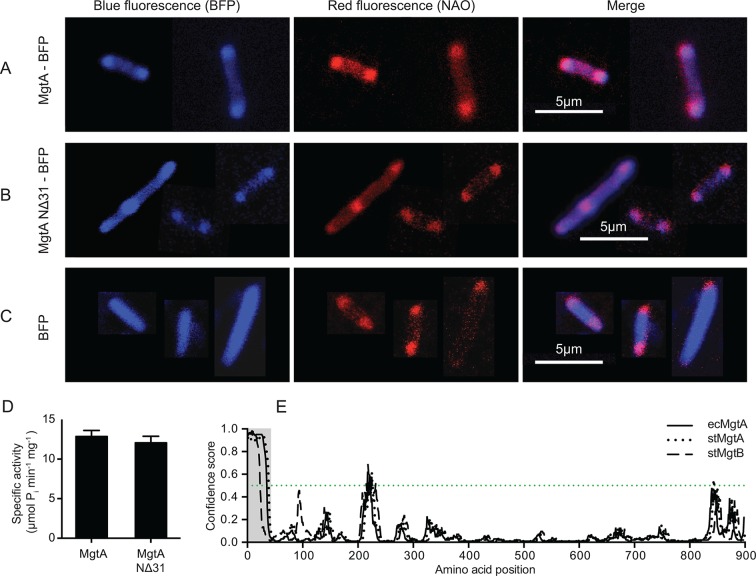
MgtA colocalizes with cardiolipin domains at the poles of E. coli
Studies with the fluorescent dye 10-N-Nonyl Acridine Orange (NAO) has shown that CL exists as enriched domains at the poles and division septa in E. coli (Fishov and Woldringh, 1999). NAO is widely used to image anionic phospholipids in bacteria and displays a specific red-shifted fluorescence emission when bound to CL (Mileykovskaya et al., 2001). The online web resource GenoBase: comprehensive resource database of Escherichia coli K-12 (Otsuka et al., 2015), reports protein localization images for most E. coli proteins. The genes were fused to GFP and overexpressed in the E. coli strain K-12. The confocal images of MgtA (gene locus: JW4201) fused to GFP from GenoBase showed clear fluorescence localized at the poles. Since our biochemical data showed that MgtA is activated in the presence of CL, we hypothesized that MgtA could colocalize with the CL rich domains in E. coli. To further expand the analysis, we decided to include an N-terminal deletion mutant as well (NΔ31-MgtA). The N-terminal of ecMgtA is rich in positively charged residues and is predicted to harbor intrinsically disordered regions. Similar disordered regions are observed in MgtA and the ortholog MgtB from S. typhimurium. The deletion mutant served two purposes; 1) to test whether the positively charged residues were important for the interaction with CL, since activation by lipids binding at the N-terminus had been reported for another P-type ATPase, ATP13A2 of the P5 subfamily (Faraco et al., 2014) and 2) to test whether this stretch was important for trafficking to the membrane. We generated constructs of MgtA and NΔ31-MgtA with blue fluorescent protein (BFP) fused at the C-terminus. The confocal images of E. coli C43(DE3) overexpressing MgtA-BFP confirmed that it was mainly concentrated at the poles and colocalized with the NAO-stained CL domains (Figure 3A). The NΔ31-MgtA-BFP was also found at the poles and colocalized with CL (Figure 3B). As a control, the overexpressed BFP alone did not colocalize with the NAO stained CL (Figure 3C). Furthermore, no significant difference in the ATPase activity between the purified wt and NΔ31 mutant (Figure 3D) was observed. This suggests that unlike ATP13A2, the intrinsic disorder predicted in the N-termini of both MgtA in E. coli and MgtA/B in S. typhimurium is not required for activation by lipids and do not play any role in membrane trafficking or targeting to the CL domains (Figure 3E).
Figure 3. Colocalization of MgtA and CL. in E. coli C43(DE3).
(A) Colocalization of MgtA–BFP with CL. (B) Colocalization of MgtA-NΔ31–BFP with CL. (C) BFP and NAO stained CL. (D) ATPase activity of MgtA and MgtA NΔ31 measured in the presence of 125 μM CL. No significant difference in the ATPase activity was observed. (E) Disordered regions predicted using DISOPRED3 (Jones and Cozzetto, 2015). The polypeptide chain is considered disordered when the prediction is above a confidence score of 0.5. Residues 1–35, marked in grey box shows the disordered region. D, Values plotted are mean ± SD (n = 3).
DOI: http://dx.doi.org/10.7554/eLife.11407.013
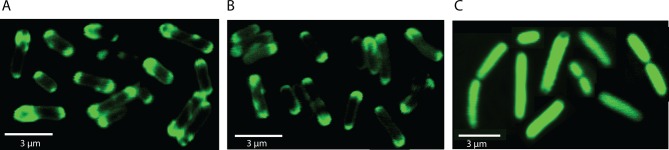
Figure 3—figure supplement 1. Localisation of wtMgtA and the non-phosphorylatable mutant D373N in E.coli C43(DE3) cells.
In addition to the colocalization experiments described above, we also tested localization of the nonphosphorylable mutant MgtA D373N to understand whether the membrane trafficking and co-localization of MgtA with CL at the poles were dependent on the catalytically active MgtA. We observed that the nonphosphorylable mutant D373N fused to green fluorescent protein (GFP) at the C-terminus showed similar localization at the poles (Figure 3—figure supplement 1). This leads us to conclude that the active transport of ions through MgtA does not affect trafficking to the membrane or its localization in the membrane.
Effect of Mg2+free on MgtA ATPase activity
Studying the Mg2+ dependency of purified MgtA with an ATPase assay is complicated as it is difficult to decouple the inherent Mg2+ dependency of ATP from the vectorial transport of Mg2+ by MgtA. To estimate the sensitivity of MgtA to Mg2+, we have to make the assumption that Mg2+ must be free in solution to be transported through MgtA. The Mg2+free indicates that the Mg2+ ions are coordinated only by water and not by ATP, which is also present in the assay mix. It is not possible to perform an ATPase assay with the concentrations of Mg2+free, ATPfree and Mg2+-ATP acting independent of each other. Thus, it becomes inevitable to obtain the desired information from an assay where one ligand is fixed (e.g. ATP) while varying the concentration of other ligand (e.g. Mg2+).
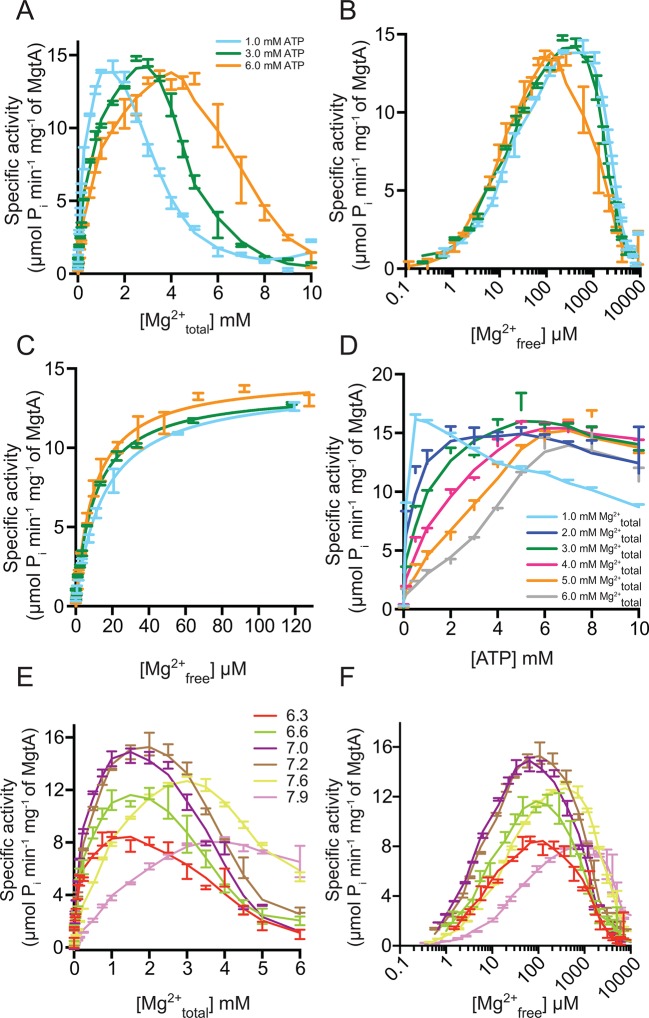
The ATPase activity was measured at three fixed concentrations (1.0, 3.0 and 6.0 mM) of ATP, with increasing concentrations of Mg2+ (Mg2+total) from 0 to 10.0 mM. This experiment showed steep activation of MgtA ATPase activity at low Mg2+total levels that peak when the molar ATP concentration is equivalent to the molar Mg2+total concentration (Figure 4A). A rapid decline in ATPase activity was observed at higher Mg2+total concentrations. For each of the ATP concentrations indicated, the ATPase activity reached equivalent maximum velocity (~14 μmol Pi min-1 mg-1). However, the activation profile was slightly shifted towards higher Mg2+total as the ATP concentration in assay buffer increased. This shift can be explained by the fact that higher ATP concentrations chelate more Mg2+ than lower ATP concentrations, thereby reducing the level of Mg2+free available for MgtA to transport, causing a delay in activation. This is verified by plotting the data from Figure 4A against the calculated Mg2+free (Calculated with MAXC, see materials and methods) (Figure 4B). We found that the activating concentration of Mg2+free was equivalent (~10.0 μM) for all three fixed ATP concentrations. Further, we observed that the ATPase activity peaked at 250 μM Mg2+free and was inhibited rapidly above 1.0 mM Mg2+free. Since the cytoplasmic Mg2+free concentration is ~1 mM (Silver and Clark, 1971; Froschauer et al., 2004), the inhibition that is observed could occur from the cytoplasmic side of MgtA. In that case, the inhibition can be explained as product inhibition, considering the Mg2+free transported into the cytoplasm to be the product. Together, these results indicate that Mg2+free regulates MgtA ATPase activity when the Mg2+free levels deviate from the 1 mM physiological threshold concentration. We also calculated the apparent Km for Mg2+free to be between 10–15 μM for the three ATP concentrations tested (Figure 4C, Table 1) which is only slightly lower than the reported ~29 μM from in vivo studies with S. typhimurium at 37°C (Snavely et al., 1989).
Figure 4. Mg2+and pH dependency of MgtA.
(A) ATP hydrolysis was measured with increasing concentration Mg2+ at fixed ATP concentrations. Irrespective of the ATP concentration used, maximum ATPase activity was observed at around 13 μM Pi min-1 mg-1 and increasing Mg2+ concentration decreases the ATPase activity. (B) Data from [A] was plotted against Mg2+free in assay condition determined using MAXC as mentioned in the methods section. (C) Data from [B] was plotted in the range from 0 to 120 μM Mg2+free and used for kinetic calculations summarized in Table 1. (D) ATP hydrolysis was measured with increasing ATP concentration at various fixed Mg2+ concentrations. Since ATP acts as Mg2+ chelator, the Mg2+free concentration decreases with increasing ATP concentration. (E) ATP hydrolysis measured at various fixed pH conditions while increasing the Mg2+ concentration. Bis-tris propane at indicated pH values was used as buffer. (F) Data from [E] was plotted against calculated Mg2+free. A-F, Values plotted are mean ± SD (n = 3).
DOI: http://dx.doi.org/10.7554/eLife.11407.016
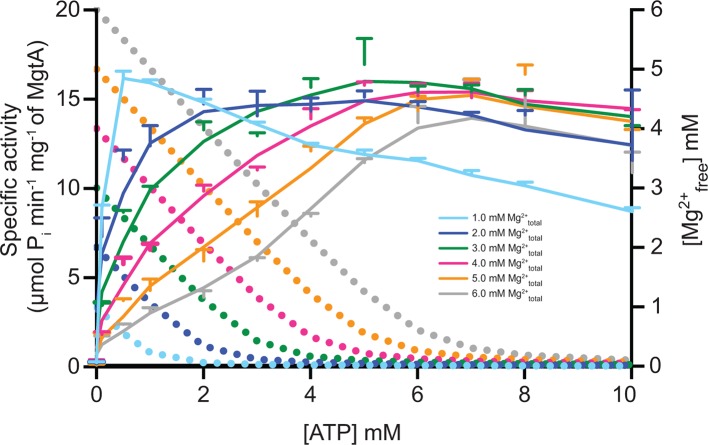
Figure 4—figure supplement 1. ATP dependency of MgtA at fixed Mg2+ concentration.
Effect of increasing ATPconcentration on the ATPase activity of MgtA
Correspondingly in a complementary experiment, the ATPase activity was measured at six fixed Mg2+total concentrations with increasing ATP concentrations from 0–10.0 mM (Figure 4D). As observed in Figure 4A, the ATPase activity peaked when the molar concentration of ATP was equivalent to the indicated levels of molar Mg2+total. However, a slight decrease in the ATPase activity was observed at higher concentrations of ATP for all fixed Mg2+total. This could indicate that the ATPfree is competing for the Mg2+-ATP binding site, thereby reducing the MgtA-Mg2+-ATP complex formation. However, interestingly, the activation curves were shifted to higher concentrations of ATP as the Mg2+total in the reaction mix increased. This delay in activation at higher concentrations of fixed Mg2+total once again indicates that high levels of Mg2+free acts as inhibitor as observed in Figure 4B, and visualized in Figure 4—figure supplement 1. When excess Mg2+free is present at lower ATP concentrations, the ATPase activity is inhibited. As the ATP concentration increases, it chelates more Mg2+free thereby activating MgtA. These results further confirm that MgtA is more sensitive to Mg2+free levels in the environment.
Effect of pH on MgtA
Protons often act as counter ions for P-type ATPases and as MgtA is classified as a P3–type ATPase it is expected that MgtA may export H+ or another undetermined substance in exchange for Mg2+ import (Faraco et al., 2014; Kehres and Maguire, 2002). To check the influence of pH on MgtA, we tested the MgtA Mg2+total dependency at different pH values (Figure 4E). Maximum ATPase activity was observed at pH 7.2. However, at all pH values, a decrease in ATPase activity at higher Mg2+total concentration was observed corresponding to product inhibition as seen in Figure 4A. When the ATPase activity was plotted against Mg2+free (Figure 4F), it became clear that in the pH range from 6.3 to 7.2, the activation and inhibition profiles were equivalent with only decrease in the maximum ATPase activity when the pH is below 7.0. At pH 7.6 and pH 7.9, the affinity for Mg2+free was reduced, but the inhibition by Mg2+free was equivalent to the other pH ranges. Our data do not show any clear proton dependent Mg2+ affinity for MgtA. Considering that the MgtA homolog PH1 from P. hybrida did not show proton transport activity (Faraco et al., 2014), we have to question whether proton counter transport occurs in MgtA. Both MgtA and PH lack the membrane imbedded arginine, which is important for proton translocation as described for the P3A-type (H+) ATPases, Pma1 (Arg 695) in yeast (Dutra et al., 1998) and AHA2 (Arg 655) in Arabidopsis (Buch-Pedersen and Palmgren, 2003; Pedersen et al., 2007). These observations suggest that MgtA may be importing magnesium without the need for proton as counter ion. Since enzymatic activity in general is affected by changes in pH, the pH dependent ATPase activity changes we observe may not be linked to proton translocation. The role of protons as counterions for MgtA therefore awaits further verification.
Discussion
Over the past two decades, identification of different Mg2+ transporters in bacteria and their link to the virulence system PhoP/PhoQ (Groisman et al., 2013) has prompted new studies into these transporters. A key Mg2+ transporter is MgtA, which is expressed under low Mg2+ conditions sensed by the PhoP/PhoQ system. A detailed picture of the transcriptional regulation of mgtA is available. However, the in vitro enzymatic characterization of MgtA has been missing and the factors that affect the enzymatic functions are not known. Towards the aim of enzymatic characterization of MgtA, we purified MgtA and demonstrated the tight association of MgtA with CL, and sensitivity of MgtA towards Mg2+, providing the first step in understanding the P3B-type ATPases.
In this study, MgtA was overexpressed and purified from E. coli. The purification process decreased the ATPase activity of MgtA, indicating the removal of an activating compound or an interacting partner. Addition of total E. coli lipids restored the ATPase activity, leading to successful reactivation of purified MgtA. Addition of DOPE, DOPG and CL independently to an ATPase assay revealed that MgtA was only activated by anionic phospholipids. Furthermore, CL showed the strongest activation as compared to DOPG, while DOPE had no effect on ATPase activity. This indicated the importance of specific lipid-protein interactions in the membrane. Our results are the first to link activation by CL to any P-type ATPase. CL has often been linked to membrane proteins connected with the oxidative phosphorylation in mitochondria and membrane proteins involved with lipid metabolism (Planas-Iglesias et al., 2015). Lipids exert their effect on membrane proteins either by binding to specific sites or by changing the environment immediate to the protein, thus facilitating oligomerization (Ernst et al., 2010). MgtA does not exhibit oligomerization in presence of CL or CL-Mg2+ in vitro Thus, CL likely causes a small structural change directly on the protein surface either by binding to a specific site or by facilitating Mg2+free recognition. Our data suggests that the CL binding does not occur at the N-terminus but at an yet unidentified site.
Addition of other lipid types (e.g. DOPE and DOPG) to MgtA pre-incubated with CL did not affect the ATPase activity. This suggests that other lipid types do not interfere with CL binding to MgtA and hints that the head group of the lipids plays an important role in the activation. The preferential selection of CL in the presence of other lipid types hint that MgtA function may be regulated in vivo by changing the local concentration of CL. Since CL have been shown to form domains or rafts at the poles and division septa in E. coli (Renner and Weibel, 2011), we hypothesized that MgtA could be localized in these CL domains. Our confocal studies indeed showed that MgtA is confined to the poles and colocalized with NAO stained CL. Several publications have validated the use of NAO to stain CL-rich domains (Mileykovskaya et al., 2001; Mileykovskaya and Dowhan, 2000; Mileykovskaya and Dowhan, 2009). However, a recent study questions the specificity of NAO to CL, and shows that NAO stains both anionic phospholipids, PG and CL in the membrane and at the poles of E. coli (Oliver et al., 2014). Considering the discrepancies in NAO staining, it is not possible to distinguish which anionic lipid among CL and PG would have the most pronounced effect on MgtA in vivoin the E. coli inner membrane. It has earlier been reported that expression of MgtA and consequent removal of Mg2+ from the periplasm by MgtA further activated the sensor kinase PhoQ of the two-component system PhoQ/PhoP. This positive feedback regulation increased the level phosphorylated PhoP, which is necessary for the expression of subset of genes expressed under low Mg2+ condition Park and Groisman, 2014It is interesting to note that PhoP of the two-component system PhoQ/P, which activates MgtA expression, also goes to one pole in S. typhimurium when activated by phosphorylation (Sciara et al., 2008).Our observation that MgtA is localized in CL rich domains in the poles, thus connect the localization observed for activated PhoP and MgtA. However the exact molecular mechanism behind the similar localization pattern of MgtA-CL and PhoP remains to be determined.
Activation by both anionic lipids has been observed for other membrane proteins, including the anaerobic respiratory complex in E. coli (Arias-Cartin et al., 2011), the mechanosensitive channel MscL (Powl et al., 2003), the protein translocon complex SecYEG (Gold et al., 2010) and the cell division proteins MinD and MinE (Renner and Weibel, 2012). Thus, it is reasonable to conclude that MgtA is active in an anionic phospholipid environment. This diversity of lipid binding could arise from the structural similarity between PG and CL. In bacteria, CL is synthesized by the fusion of two PG molecules (Tropp, 1997). While CL carries two negative charges in the head group, PG carries only one. This difference in the charge could contribute either to efficient binding of lipid or efficient presentation of substrate (Mg2+) to MgtA. It has been shown that CL can bind divalent cations like Mg2+ and undergo lipid phase changes as a function of Mg2+ concentration(Vail and Stollery, 1979). It is therefore tempting to speculate that CL could act as a Mg2+ reservoir or as a chaperone for easy substrate presentation to MgtA. The Cu+-ATPases from the P1B-type ATPase family are well known to require protein chaperones that are either part of the Cu+-ATPases or expressed as separate proteins in cytoplasm to bind and deliver Cu+ to the Cu+-ATPases (Arguello et al., 2007). Our observation with bovine heart CL indicates that the head group of cardiolipin plays the main role in activation of MgtA relative to the composition of fatty acid tails. This, it is intriguing to speculate that the head group of CL could function as the Mg2+ chaperone for MgtA and its function could differ depending on the side it associates with MgtA. If the headgroup of CL faces the periplasmic side, then it may act as a chaperon for Mg2+, assisting the uptake of Mg2+. If the headgroup faces the cytoplasmic side, it may play a role in sensing the Mg2+ levels in cytoplasm, thereby regulating MgtA activity. However to further address these questions, a crystal structure of CL bound to MgtA will be necessary as it is not possible to determine the sidedness of the CL headgroup binding site in MgtA in the present study.
Surprisingly, the effect of Mg2+ on the ATPase activity of MgtA was more pronounced than expected. MgtA is activated already below 10 μM Mg2+free. The activation is followed by a rapid decrease in activity as the Mg2+free concentration rises above 1 mM (Figure 4B). All P-type ATPases require Mg2+ for their activity. Even though our data does not conclusively establish the transport of Mg2+ by MgtA, it shows that MgtA is highly sensitive to the Mg2+free and the level of sensitivity is different from other P-type ATPases. The related H+ ATPase from Neurospora crassa also shows decrease in ATPase activity at higher Mg2+ concentration (Brooker and Slayman, 1983). However, it is not completely inhibited even above 20 mM of Mg2+total, whereas MgtA is completely inhibited above 5 mM. MgtA presents a strikingly narrow activity range induced by Mg2+ as compared to the H+ ATPase. Overall, we conclude that MgtA is more sensitive to Mg2+ concentrations in vitro than any other studied P-type ATPase. This sensitivity towards Mg2+ points out that Mg2+ is the ion transported through MgtA in vivo. The narrow activity range suggests that MgtA could have more Mg2+ binding sites other than the transport sites to sense the varying Mg2+free concentrations.
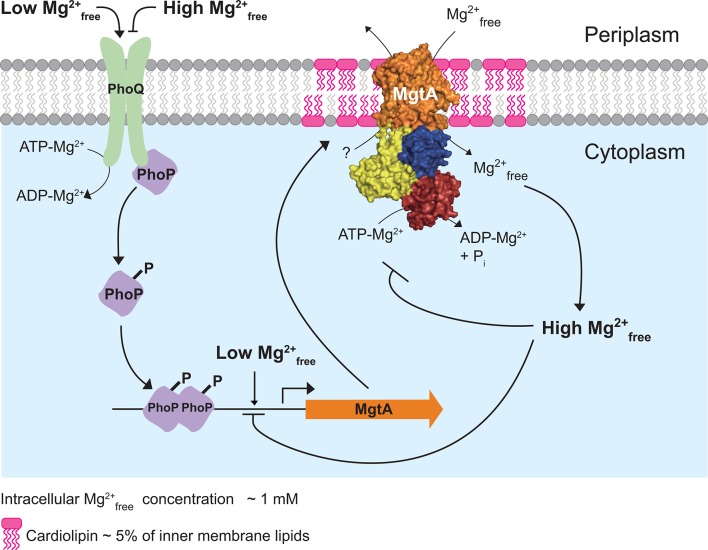
The Mg2+free concentration of E. coli and S. enterica cytoplasms is found to be ~1 mM (Silver and Clark, 1971; Froschauer et al., 2004). The observation that MgtA transports Mg2+ into the cytoplasm and its ATPase activity decreases above the physiological 1 mM Mg2+free, strengthens the notion that the cytoplasmic Mg2+free concentration also regulates MgtA internally. Additionally, it has been reported that increasing cytoplasmic Mg2+ concentrations negatively regulates mgtA-coding region (Cromie et al., 2006). Based on these results we extend the model, initially proposed by Groisman and coworkers (Cromie et al., 2006; Park et al., 2010) in which PhoP/PhoQ senses low Mg2+ in the periplasm and activates mgtA transcription. When the cytoplasmic Mg2+ falls below a threshold, the riboswitch allows transcription of mgtA. Our data adds further evidence to the post translated system, in which MgtA is targeted to the PG-CL domains at the poles and transport Mg2+ into the cytoplasm, thereby satisfying the cellular needs. When the Mg2+ concentration in the cytoplasm exceeds the threshold, Mg2+free negatively regulates both MgtA in PG-CL domain and mgtA coding region (Figure 5). In conclusion, our data supports that MgtA acts both as a transporter and as asensor for Mg2+.
Figure 5. Model illustrating the regulation of Mg2+ uptake by MgtA.
When PhoQ senses low Mg2+free (<50 μM) in periplasm, it phosphorylates PhoP. This activates PhoP and it promotes transcription of the mgtA gene. MgtA protein is targeted to CL-rich region in bacterial inner membrane. Association of MgtA with CL is essential for its activity. MgtA imports Mg2+free available in periplasm to the cytoplasm of bacteria, thereby increasing cytoplasmic Mg2+free concentration. When the cytoplasmic Mg2+free concentration reaches a threshold (~1 mM), MgtA is inhibited both at the transcriptional and the post-translational level.
Materials and methods
Lipids were purchased from Avanti Polar Lipids, Alabaster, Alabama. Dodecyl- β-D- Maltopyranoside (DDM) was purchased from Chemical Point UG, Deisenhofen, Germany. Octaethylene glycol monododecyl ether (C12E8) was purchased from Nikko Chemicals, Tokyo, Japan. Adenoside 5’-triphosphate disodium salt hydrate (ATP) was purchased from Sigma-Aldrich, Norway. All other chemicals, including primers were purchased as grade (BioUltra) available from Sigma-Aldrich.
Cloning and heterologous expression of MgtA
The ORF of mgtA was amplified from E. coli genomic DNA using primers, 5´ GGCCATGGCTTTTAAAGAAATTTTTACCCGGCTC 3´ and 5´ GGCTCGAGTCATTG CCAGCCGTAACGACGG 3´ purchased from Sigma-Aldrich. The PCR product was inserted into pETM11 vector (EMBL) between NcoI and XhoI restriction sites. The D373N mutant was created using the Quickchange Lightning mutagenesis kit (Agilent Technologies, Matrix AS, Norway). The sequence of the resulting constructs was verified. Then the MgtA-pETM11 plasmid was transformed into E. coli C43 (DE3) cells and plated on Luria Bertani (LB) media with 1.5% Agar and 50 µg/ml kanamycin. Colonies of transformed C43 (DE3) cells were inoculated in LB media containing 50 μg/ml kanamycin and incubated at 37°C for 16 hr. The following day, 1% of the overnight culture was added to LB media with 50 μg/ml kanamycin and incubated with shaking at 37°C until OD600nm reached ~0.6. Protein expression was induced by adding 1 mM Isopropyl-β-D-1-thiogalactopyranoside and grown at 18°C with shaking (200 rpm). Cells were harvested after 16 hr by centrifuging at 7000 ×g for 10 min. Cell pellets were stored at -20°C until use.
Purification
Cell pellet was suspended at 1:10 ratio in buffer A (50 mM HEPES pH 7, 100 mM K2S04, 10% glycerol, 1 mM Phenylmethylsulfonyl fluoride (PMSF), 5 mM β-mercaptoethanol) and lysed using High Pressure Homogenizer (C5 model, Avestin, Germany) at 15,000 psi. Unlysed cells and inclusion bodies were removed by centrifugation at 20,000 × g for 20 min. Resulting supernatant was centrifuged at 100,000 × g for 2 hr. Mixed membrane pellets were weighed and suspended at 1:10 ratio in buffer B (25 mM HEPES pH7, 100 mM K2S04, 5% glycerol, 1 mM PMSF, 5 mM β-mercaptoethanol) using a Dounce homogenizer (Sigma-Aldrich). Membranes were solubilized with 1% DDM for 4 hr. Solubilized membrane suspension was passed through His trap FF (GE Healthcare, Norway). The column was washed with 10 column volumes (CV) of buffer B supplemented with 20 mM Imidazole and ~3 critical micelle concentration (CMC) DDM (0.6 mM). Protein was eluted with buffer B supplemented with 250 mM Imidazole and 3 CMC DDM. Eluted fractions were pooled, concentrated and subjected to size exclusion chromatography (SEC) using a S200 sepharose 16/600 column with Buffer C (25 mM HEPES pH 7, 100 mM K2S04, 5% glycerol, 1 mM DL-Dithiothreitol (DTT), 3 CMC DDM). Fractions from SEC were pooled, concentrated, flash frozen and stored at -80°C until use. Protein concentration was measured using Bradford method. A description of how the critical steps in the purification was overcome, to yield monodisperse MgtA, was described in detail and published at Bio-protocol (Subramani and Morth, 2016).
Preparation of lipid stocks
Lipid dissolved in chloroform was dried under a nitrogen stream. Dried lipid film was resuspended in MilliQ water at a final concentration of 10 mg/ml with vigorous shaking until no visible aggregates could be detected. 20.0 mM C12E8 was added to the lipid suspension and solubilized with shaking for 1 hr at RT. Fresh lipid stocks were prepared prior to enzymatic assays.
Enzymatic assay
ATP hydrolysis was measured by detecting the released inorganic phosphate following the protocol described by Cariani et, al., (Cariani et al., 2004). Reaction buffer contain final concentration of 25.0 mM HEPES pH 7.0, 200 mM KCl, in the presence or absence of 0.25 mM Na2MoO4, 5.0 mM NaN3 and 20.0 mM KNO3. Last three substances were added to inhibit the F-Type ATPase, phosphatase and pyrophosphatase. Unless otherwise indicated, 0.25 µg of MgtA and 116 µM CL were used in the assay. The assay components except ATP-Mg2+ were mixed to final volume of 60 μl and preincubated at 37°C for 10 min. Reaction was initiated by addition of 3 mM ATP-Mg2+ and further incubated for 10 min at 37°C. The reaction was terminated and phosphate content was detected by adding 75 μl of Solution I (prepared fresh every time by dissolving 0.3 g of ascorbic acid in 3.5 ml water, then adding 5 ml of 1 M HCL, 0.5 ml of 10% ammonium molybdate and 1.5 ml of 20% Sodium Dodecyl Sulphate) and incubated for 10 min on ice. Then, 125 μl of solution II (3.5% sodium citrate and 3.5% bismuth citrate in 1.0 M HCl) was added and incubated at RT for 10 min. Absorbance was measured at 690 nm. The online web service MAXC (maxchelator.stanford.edu) was used to calculate Mg2+free levels in the presence of any given concentrations of ATP, pH at 37°C. Apparent Vmax, and Km, were calculated by fitting the curves (specific activity (SA) as a function of Mg2+free) using Michaelis Menten equation (Y = Vmax*X/Km+X) provided by GraphPad Prism6 (www.graphpad.com). Kcat was estimated using the formula Kcat = Vmax/[MgtA].
Sequence analysis
The sequences of the MgtA homologs were identified using the HMMER web server (Finn et al., 2011) using default settings with ecMgtA (uniprot code P0ABB8) as search sequence. Selected subsets of MgtA homologs were chosen from each kingdom and aligned using MAFFT (Katoh et al., 2002) and edited manually in Jalview version 2.8.2 (Troshin et al., 2011), the phylogenetic three was built in Seaview, using the BioNJ algorithm (Gascuel, 1997).
Confocal microscopy
MgtA and NΔ31MgtA were cloned into pRSET-BFP vector between BamHI and NcoI. E. coli C43(DE3) cells were transformed with MgtA-pRSET-BFP NΔ31MgtA-pRSET-BFP, MgtAD373N-GFP, empty pRSET-BFP or empty pCFGP vector. Cells were grown overnight in LB medium with 100 µg/ml ampicillin at 37°C, 180 rpm. Fresh overnight cultures were diluted 1:100 in LB medium with 100 µg/ml ampicillin and grown to OD600 of 0.6. IPTG at 1 mM was added to induce protein expression.
When indicated, 200 nM nonyl acridine orange (NAO) was simultaneously added to the culture to visualize CL membrane domains. After 3 hr at 37°C, 500 µl of each culture was centrifuged for 5 min at 4000 rpm and 450 µl of the supernatant was removed. The cells were resuspended by pipetting and 2 µl was immobilized on a poly-D-lysine coated glass dish (P35GC-1.5-14-C, MatTek) covered with a 1% agarose-LB pad containing 1 mM IPTG.
Fluorescent images were viewed with a LSM510 (Zeiss) microscope and a 63X oil immersion objective. To minimize the toxicity of high-energy excitation light, the focus was set under phase-contrast conditions and then fluorescence images were captured shortly after the shift to high-energy excitation light. Blue fluorescence from BFP tags (excitation 405 nm, emission 420–480 nm) and red fluorescence from enriched and dimerized NAO in CL domains (excitation 488 nm, emission 600–650 nm) were detected. Fluorescence from GFP was acquired at excitation 488 nm and emission 510–560 nm. Images were obtained and processed using ImageJ software (National Institutes of Health).
Acknowledgement
The authors would like to thank Robert Brooker, Klaus Fendler and William Stanley for critical reading of the manuscript. Hari Shroff, Prahathees Eswara Moorthy and Damian Dalle Nogare for useful suggestions on how to perform the bacterial imaging. The Norwegian Research Council Funding (FRIMEDBIO) # ES486454 and NCMM core Funding supported this study.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Funding Information
This paper was supported by the following grant:
Norges Forskningsråd ES486454 to Saranya Subramani, Harmonie Perdreau-Dahl, Jens Preben Morth.
Additional information
Competing interests
The authors declare that no competing interests exist.
Author contributions
SS, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article, Contributed unpublished essential data or reagents.
HP-D, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article.
JPM, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article, Contributed unpublished essential data or reagents.
References
- Albers RW. Biochemical aspects of active transport. Annual Review of Biochemistry. 1967;36:727–756. doi: 10.1146/annurev.bi.36.070167.003455. [DOI] [PubMed] [Google Scholar]
- Argüello JM, Eren E, González-Guerrero M. The structure and function of heavy metal transport P1B-ATPases. BioMetals. 2007;20:233–248. doi: 10.1007/s10534-006-9055-6. [DOI] [PubMed] [Google Scholar]
- Arias-Cartin R, Grimaldi S, Pommier J, Lanciano P, Schaefer C, Arnoux P, Giordano G, Guigliarelli B, Magalon A. Cardiolipin-based respiratory complex activation in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7781–7786. doi: 10.1073/pnas.1010427108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooker RJ, Slayman CW. Effects of Mg2+ ions on the plasma membrane [H+]-ATPase of Neurospora crassa. II. Kinetic studies. The Journal of Biological Chemistry. 1983;258:8833–8838. [PubMed] [Google Scholar]
- Buch-Pedersen MJ, Palmgren MG. Conserved Asp684 in transmembrane segment M6 of the plant plasma membrane p-type proton pump AHA2 is a molecular determinant of proton translocation. Journal of Biological Chemistry. 2003;278:17845–17851. doi: 10.1074/jbc.M212729200. [DOI] [PubMed] [Google Scholar]
- Cariani L, Thomas L, Brito J, del Castillo JR. Bismuth citrate in the quantification of inorganic phosphate and its utility in the determination of membrane-bound phosphatases. Analytical Biochemistry. 2004;324:79–83. doi: 10.1016/j.ab.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Cromie MJ, Shi Y, Latifi T, Groisman EA. An RNA sensor for intracellular Mg2+ Cell. 2006;125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- Danko S, Yamasaki K, Daiho T, Suzuki H. Distinct natures of beryllium fluoride-bound, aluminum fluoride-bound, and magnesium fluoride-bound stable analogues of an ADP-insensitive phosphoenzyme intermediate of sarcoplasmic reticulum Ca2+-ATPase: changes in catalytic and transport sites during phosphoenzyme hydrolysis. The Journal of Biological Chemistry. 2004;279:14991–14998. doi: 10.1074/jbc.M313363200. [DOI] [PubMed] [Google Scholar]
- Dutra MB, Ambesi A, Slayman CW. Structure-function relationships in membrane segment 5 of the yeast Pma1 h+-ATPase. Journal of Biological Chemistry. 1998;273:17411–17417. doi: 10.1074/jbc.273.28.17411. [DOI] [PubMed] [Google Scholar]
- Ernst AM, Contreras FX, Brügger B, Wieland F. Determinants of specificity at the protein-lipid interface in membranes. FEBS Letters. 2010;584:1713–1720. doi: 10.1016/j.febslet.2009.12.060. [DOI] [PubMed] [Google Scholar]
- Faraco M, Spelt C, Bliek M, Verweij W, Hoshino A, Espen L, Prinsi B, Jaarsma R, Tarhan E, de Boer AH, Di Sansebastiano GP, Koes R, Quattrocchio FM. Hyperacidification of vacuoles by the combined action of two different p-ATPases in the tonoplast determines flower color. Cell Reports. 2014;6:32–43. doi: 10.1016/j.celrep.2013.12.009. [DOI] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Research. 2011;39:W29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishov I, Woldringh CL. Visualization of membrane domains in escherichia coli. Molecular Microbiology. 1999;32:1166–1172. doi: 10.1046/j.1365-2958.1999.01425.x. [DOI] [PubMed] [Google Scholar]
- Froschauer EM, Kolisek M, Dieterich F, Schweigel M, Schweyen RJ. Fluorescence measurements of free [Mg2+] by use of mag-fura 2 in salmonella enterica. FEMS Microbiology Letters. 2004;237:49–55. doi: 10.1016/j.femsle.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Gascuel O. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Molecular Biology and Evolution. 1997;14:685–695. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- Gold VA, Robson A, Bao H, Romantsov T, Duong F, Collinson I. The action of cardiolipin on the bacterial translocon. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10044–10049. doi: 10.1073/pnas.0914680107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman EA, Hollands K, Kriner MA, Lee EJ, Park SY, Pontes MH. Bacterial Mg2+ homeostasis, transport, and virulence. Annual Review of Genetics. 2013;47:625–646. doi: 10.1146/annurev-genet-051313-051025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Cozzetto D. DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics. 2015;31:857–863. doi: 10.1093/bioinformatics/btu744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Research. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehres DG, Maguire ME. Structure, properties and regulation of magnesium transport proteins. BioMetals. 2002;15:261–270. doi: 10.1023/A:1016078832697. [DOI] [PubMed] [Google Scholar]
- Kühlbrandt W. Biology, structure and mechanism of p-type ATPases. Nature Reviews Molecular Cell Biology. 2004;5:282–295. doi: 10.1038/nrm1354. [DOI] [PubMed] [Google Scholar]
- Lutsenko S, Kaplan JH. Organization of p-type ATPases: significance of structural diversity. Biochemistry. 1995;34:15607–15613. doi: 10.1021/bi00048a001. [DOI] [PubMed] [Google Scholar]
- Mileykovskaya E, Dowhan W, Birke RL, Zheng D, Lutterodt L, Haines TH. Cardiolipin binds nonyl acridine orange by aggregating the dye at exposed hydrophobic domains on bilayer surfaces. FEBS Letters. 2001;507:187–190. doi: 10.1016/S0014-5793(01)02948-9. [DOI] [PubMed] [Google Scholar]
- Mileykovskaya E, Dowhan W. Visualization of phospholipid domains in escherichia coli by using the cardiolipin-specific fluorescent dye 10-n-nonyl acridine orange. Journal of Bacteriology. 2000;182:1172–1175. doi: 10.1128/JB.182.4.1172-1175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileykovskaya E, Dowhan W. Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochimica Et Biophysica Acta. 2009;1788:2084–2091. doi: 10.1016/j.bbamem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsieurs P, De Keersmaecker S, Navarre WW, Bader MW, De Smet F, McClelland M, Fang FC, De Moor B, Vanderleyden J, Marchal K. Comparison of the PhoPQ regulon in escherichia coli and salmonella typhimurium. Journal of Molecular Evolution. 2005;60:462–474. doi: 10.1007/s00239-004-0212-7. [DOI] [PubMed] [Google Scholar]
- Morein S, Andersson A, Rilfors L, Lindblom G. Wild-type escherichia coli cells regulate the membrane lipid composition in a "window" between gel and non-lamellar structures. The Journal of Biological Chemistry. 1996;271:6801–6809. doi: 10.1074/jbc.271.12.6801. [DOI] [PubMed] [Google Scholar]
- Morth JP, Pedersen BP, Toustrup-Jensen MS, Sørensen TL, Petersen J, Andersen JP, Vilsen B, Nissen P. Crystal structure of the sodium-potassium pump. Nature. 2007;450:1043–1049. doi: 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- Neef J, Andisi VF, Kim KS, Kuipers OP, Bijlsma JJ. Deletion of a cation transporter promotes lysis in streptococcus pneumoniae. Infection and Immunity. 2011;79:2314–2323. doi: 10.1128/IAI.00677-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor K, Fletcher SA, Csonka LN. Increased expression of Mg(2+) transport proteins enhances the survival of salmonella enterica at high temperature. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:17522–17527. doi: 10.1073/pnas.0906160106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver PM, Crooks JA, Leidl M, Yoon EJ, Saghatelian A, Weibel DB. Localization of anionic phospholipids in escherichia coli cells. Journal of Bacteriology. 2014;196:3386–3398. doi: 10.1128/JB.01877-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka Y, Muto A, Takeuchi R, Okada C, Ishikawa M, Nakamura K, Yamamoto N, Dose H, Nakahigashi K, Tanishima S, Suharnan S, Nomura W, Nakayashiki T, Aref WG, Bochner BR, Conway T, Gribskov M, Kihara D, Rudd KE, Tohsato Y, Wanner BL, Mori H. GenoBase: comprehensive resource database of escherichia coli k-12. Nucleic Acids Research. 2015;43:D606–617. doi: 10.1093/nar/gku1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren MG, Axelsen KB. Evolution of p-type ATPases. Biochimica Et Biophysica Acta. 1998;1365:37–45. doi: 10.1016/S0005-2728(98)00041-3. [DOI] [PubMed] [Google Scholar]
- Palmgren MG, Nissen P. P-type ATPases. Annual Review of Biophysics. 2011;40:243–266. doi: 10.1146/annurev.biophys.093008.131331. [DOI] [PubMed] [Google Scholar]
- Palmgren MG. Plant plasma membrane H+-ATPases: Powerhouses for Nutrient Uptake. Annual Review of Plant Physiology and Plant Molecular Biology. 2001;52:817–845. doi: 10.1146/annurev.arplant.52.1.817. [DOI] [PubMed] [Google Scholar]
- Papp-Wallace KM, Maguire ME. Magnesium transport and magnesium homeostasis. EcoSal Plus. 2008;3 doi: 10.1128/ecosalplus.5.4.4.2. [DOI] [PubMed] [Google Scholar]
- Park S-Y, Groisman EA. Signal-specific temporal response by the salmonella PhoP/PhoQ regulatory system. Molecular Microbiology. 2014;91:135–144. doi: 10.1111/mmi.12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Cromie MJ, Lee EJ, Groisman EA. A bacterial mRNA leader that employs different mechanisms to sense disparate intracellular signals. Cell. 2010;142:737–748. doi: 10.1016/j.cell.2010.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen BP, Buch-Pedersen MJ, Morth JP, Palmgren MG, Nissen P. Crystal structure of the plasma membrane proton pump. Nature. 2007;450:1111–1114. doi: 10.1038/nature06417. [DOI] [PubMed] [Google Scholar]
- Planas-Iglesias J, Dwarakanath H, Mohammadyani D, Yanamala N, Kagan VE, Klein-Seetharaman J. Cardiolipin interactions with proteins. Biophysical Journal. 2015;109:1282–1294. doi: 10.1016/j.bpj.2015.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post RL, Kume S, Tobin T, Orcutt B, Sen AK. Flexibility of an active center in sodium-plus-potassium adenosine triphosphatase. The Journal of General Physiology. 1969;54:306–326. doi: 10.1085/jgp.54.1.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powl AM, East JM, Lee AG. Lipid−protein interactions studied by introduction of a tryptophan residue: the mechanosensitive channel MscL. Biochemistry. 2003;42:14306–14317. doi: 10.1021/bi034995k. [DOI] [PubMed] [Google Scholar]
- Reinhart RA. Magnesium metabolism. Archives of Internal Medicine. 1988;148:2415–2420. doi: 10.1001/archinte.1988.00380110065013. [DOI] [PubMed] [Google Scholar]
- Renner LD, Weibel DB. Cardiolipin microdomains localize to negatively curved regions of escherichia coli membranes. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6264–6269. doi: 10.1073/pnas.1015757108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner LD, Weibel DB. MinD and MinE interact with anionic phospholipids and regulate division plane formation in escherichia coli. The Journal of Biological Chemistry. 2012;287:38835–38844. doi: 10.1074/jbc.M112.407817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlame M, Brody S, Hostetler KY. Mitochondrial cardiolipin in diverse eukaryotes. comparison of biosynthetic reactions and molecular acyl species. European Journal of Biochemistry. 1993;212:727–733. doi: 10.1111/j.1432-1033.1993.tb17711.x. [DOI] [PubMed] [Google Scholar]
- Sciara MI, Spagnuolo C, Jares-Erijman E, Véscovi EG. Cytolocalization of the PhoP response regulator in salmonella enterica: modulation by extracellular Mg2+ and by the SCV environment. Molecular Microbiology. 2008;70:479–493. doi: 10.1111/j.1365-2958.2008.06427.x. [DOI] [PubMed] [Google Scholar]
- Shinzawa-Itoh K, Aoyama H, Muramoto K, Terada H, Kurauchi T, Tadehara Y, Yamasaki A, Sugimura T, Kurono S, Tsujimoto K, Mizushima T, Yamashita E, Tsukihara T, Yoshikawa S. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. The EMBO Journal. 2007;26:1713–1725. doi: 10.1038/sj.emboj.7601618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver S, Clark D. Magnesium transport in escherichia coli. The Journal of Biological Chemistry. 1971;246:569–576. [PubMed] [Google Scholar]
- Snavely MD, Florer JB, Miller CG, Maguire ME. Magnesium transport in salmonella typhimurium: 28Mg2+ transport by the CorA, MgtA, and MgtB systems. Journal of Bacteriology. 1989;171:4761–4766. doi: 10.1128/jb.171.9.4761-4766.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snavely MD, Gravina SA, Cheung TT, Miller CG, Maguire ME. Magnesium transport in salmonella typhimurium. regulation of mgtA and mgtB expression. The Journal of Biological Chemistry. 1991;266:824–829. [PubMed] [Google Scholar]
- Subramani S, Morth J. Heterologous expression and purification of the magnesium transporter A (MgtA) in Escherichia coli. Bio-Protocol. 2016;6:e2001. doi: 10.21769/BioProtoc.2001. [DOI] [Google Scholar]
- Tropp BE. Cardiolipin synthase from escherichia coli. Biochimica Et Biophysica Acta. 1997;1348:192–200. doi: 10.1016/S0005-2760(97)00100-8. [DOI] [PubMed] [Google Scholar]
- Troshin PV, Procter JB, Barton GJ. Java bioinformatics analysis web services for multiple sequence alignment--JABAWS:MSA. Bioinformatics. 2011;27:2001–2002. doi: 10.1093/bioinformatics/btr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vail WJ, Stollery JG. Phase changes of cardiolipin vesicles mediated by divalent cations. Biochimica Et Biophysica Acta. 1979;551:74–84. doi: 10.1016/0005-2736(79)90354-7. [DOI] [PubMed] [Google Scholar]
- Véscovi EG, Soncini FC, Groisman EA. Mg2+ as an extracellular signal: environmental regulation of salmonella virulence. Cell. 1996;84:165–174. doi: 10.1016/S0092-8674(00)81003-X. [DOI] [PubMed] [Google Scholar]
- Yokota K, Kanamoto R, Kito M. Composition of cardiolipin molecular species in escherichia coli. Journal of Bacteriology. 1980;141:1047–1051. doi: 10.1128/jb.141.3.1047-1051.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]