Abstract
Tuberculosis (TB) is a global health problem responsible for ~2 million deaths per year. Current antibiotic treatments are lengthy and fraught with compliance and resistance issues. There is a crucial need for additional approaches to provide a cost-effective means of exploring the design space for potential therapies. We discuss the use of mathematical and computational models in virtual experiments and virtual clinical trials both to develop new hypotheses regarding the disease and to provide a cost-effective means of discovering new treatment strategies.
INTRODUCTION
Despite years of scientific research and effort by world health organizations, tuberculosis (TB) remains a global health problem responsible for ~2 million deaths per year. TB is primarily a pulmonary disease, although there are some extrapulmonary manifestations as well. After infection with Mycobacterium tuberculosis (Mtb), humans develop primary (active) TB (10%) or latent infection (90%); reactivation of latent infection can occur leading to active TB [1]. Factors that result in these different infection outcomes in humans are not well-characterized. Of great concern is that TB persists as a latent infection in ~2 billion humans worldwide, providing a reservoir of potential disease and contagion. Immunosuppressive conditions increase susceptibility to primary and reactivation TB (reviewed in [2]). Drug susceptible TB can be effectively treated with a lengthy regimen (≥ 6 months) of multiple antibiotics. This complex treatment strategy is fraught with compliance issues, and some of these drugs, notably isoniazid (INH) and rifampin (RIF), have potential toxicity [3]. Drug resistant TB is a major problem worldwide and development of both new drugs and strategies is essential to prevent further spread of these strains. Single drug therapy is not permitted in treatment of active TB in humans as drug resistance can arise, and the standard of care must be adhered to. Thus, it is difficult to evaluate effects of new TB drugs or strategies in human clinical trials. The multi-scale nature of the disease also makes identification of new drug targets complex: interventions that show an effect at a molecular or single cell level or at a particular time scale may not have much impact on the overall course of infection. Thus there is a crucial need for novel approaches and platforms for developing, testing and optimizing new therapies for TB.
Systems Biology Approach
A systems biology approach allows us to integrate information from multiple model systems to understand, probe, and manipulate the immune response to Mtb infection. For drug discovery in TB, this includes, but is not limited to, data derived from humans, non-human primate (NHP) and other animal models of TB, and immunological and microbiological studies, together with mathematical and computational models (MCMs). In particular, MCMs provide a powerful method to integrate data generated in multiple experimental settings into a single framework for hypothesis generation and testing. Given the large numbers of possible combinations and timing of drugs to be tested, the design space of possible new strategies for TB treatment is not only enormous but also can be – due to the time and expense of working with animal models such as NHPs – inaccessible experimentally (Figure 1). Herein we briefly review MCMs of both bacterial and host dynamics during Mtb infection in lungs. We then suggest how MCMs can be used to augment existing experimental and clinical approaches by expansion and exploration of novel approaches for drug therapy for TB.
Figure 1.
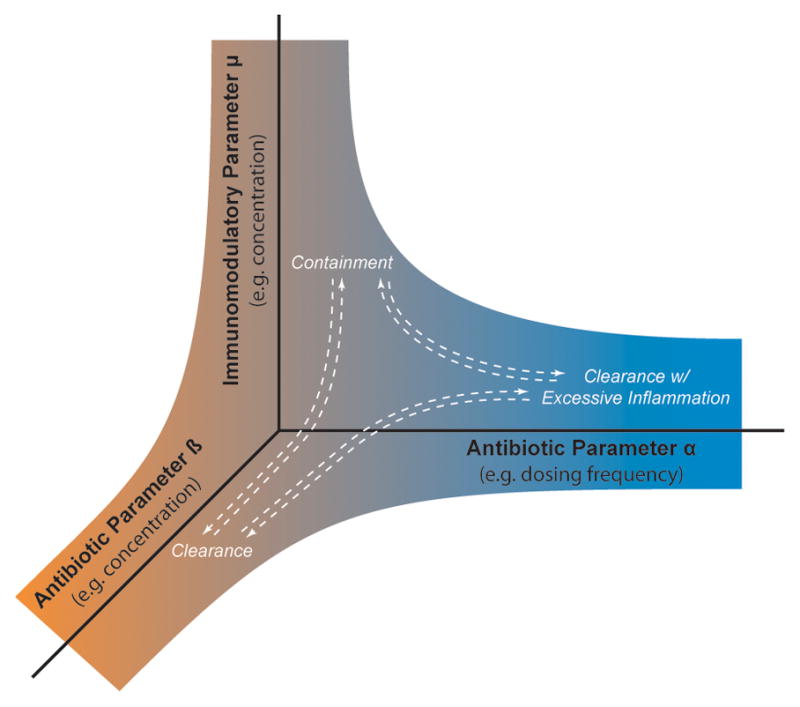
Design space for treatment development. The identities, dosing schedules, and concentrations of multiple antibiotics can be varied. Similarly, the drugs, dosing schedules, and concentrations of multiple immune-modulators can be varied. Only 3 such parameters are shown. White arrows indicate movement between treatment outcomes as parameters are varied.
Granulomas and TB
The immune response to Mtb infection is determined by a myriad of host and bacterial factors operating at multiple length and time scales in lung and draining lymph nodes (LN). Briefly, infection occurs after inhalation of Mtb into the lungs. Resident antigen presenting cells such as macrophages and dendritic cells (DCs) take up Mtb. DCs traffic to lung draining LNs where T cells are primed. These T cells migrate to the lung and participate in the formation of a self-organizing spherical collection of cells termed a granuloma [4] (Figure 2A). Granulomas serve not only to functionally and spatially contain infection but also as a survival niche for the bacterium, highlighting the unique, complex and interdependent interaction between Mtb and host. Many data now support the idea that host-pathogen dynamics occurring within granuloma are the key to determining infection outcome [5]. In addition, new data suggest that each granuloma is distinct [6]. Thus, we focus on granulomas as the environments most likely to be impacted by new approaches to therapy.
Figure 2.
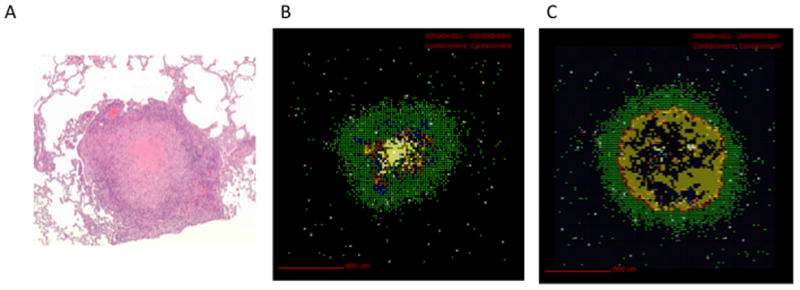
Experimental and virtual granulomas. (A) H&E stain of a NHP lung granuloma showing macrophages and lymphocytes surrounding a necrotic core (courtesy of Flynn Lab, University of Pittsburgh). (B) VE of a granuloma at day 200 post-infection, using a model similar to Ray et al. [8]. A well-circumscribed, stable granuloma that controls bacteria (but does not clear it) emerges. See also time courses at http://malthus.micro.med.umich.edu/lab/movies). Bacterial counts for this granuloma at day 200 are: 1309 intracellular Mtb, 825 extracellular Mtb, 302 nonreplicating Mtb in necrotic areas, and 2135 total Mtb. (C) VE representing a TNF knockout. Notice the much larger region of bacteria and significantly lower levels of activated macrophages. Bacterial counts for this granuloma at day 200 are: 3494 intracellular Mtb, 46601 Extracellular Mtb, 36 non-replicating Mtb in necrotic areas, and 50195 Total Mtb. For both (B) and (C) macrophages are shown as: resting (green), activated (blue), infected (orange), chronically infected (red). T cells are pink (CD4+) and purple (CD8+). Yellow shows bacterial concentrations and white hash marks represent necrotic regions.
Mathematical and Computational Models of Mtb infection
A tractable in silico model of the immune response during Mtb infection can provide a new tool for developing, understanding, predicting, and optimizing drug therapy for TB treatment. Because it is important to consider molecular, cellular and tissue-level events occurring over short and long times, as well as in different biological compartments (i.e. lung, lymph node, and blood), the model developed must ultimately be both multi-scale and multi-organ. Our groups have developed multiple generations of MCMs to explore the host-pathogen dynamics of Mtb infection in the lung and draining LNs [7–10]. We have had success building MCMs that capture stochastic events occurring at the cellular scale that manifest in an emergent fashion at the tissue scale as granulomas (Figure 2B). We use agent-based models that include macrophages (resting, infected, chronically infected, and activated), T cells (regulatory, cytotoxic, cytokine-producing), bacteria (extracellular, intracellular, non-replicating bacteria trapped in caseous/necrotic areas), and effector molecules (chemokines such as CXCR3 and CCR5-ligands: CCL2, CCL5, CXCL9/10/11; and well-studied pro- and anti-inflammatory cytokines IL-12, TNF, IFN-γ IL-10). Cells are discrete agents and effector molecules are tracked continuously and diffuse. In addition, we have developed multi-scale models that track the dynamics of molecular scale events relating to two key cytokines, TNF [11–13] and IL-10 [10]. Our current multi-scale model has been developed in both two and three dimensions. Models are validated against a variety of experimental data; NHP studies are particularly valuable as this animal model most closely replicates human infection (e.g. [14]). Thus we have the capability of studying molecular level, cellular, and tissue level events in our granuloma MCM that will provide a unique platform for studying drug therapy.
In addition to these immune-centric models, detailed models of Mtb growth and metabolism have been published over the past decade and vary from phenomenological to metabolic. Magombedze and Mulder developed an ordinary differential equation model of Mtb including different Mtb phenotypes (dormant, latent and active), dosR gene regulation, nitric oxide and nutrient availability to evaluate the contribution of gene regulation to latent TB development [15]. Ankomah and Levin present a similar division of bacteria into subpopulations based on their antibiotic susceptibility and advocate development of more complex pharmacodynamic data for antibiotics that are used in combination [16]. Focusing more on metabolic pathways, Beste et al. built a metabolic network model for Mtb [17], and used flux balance and flux spectral analysis to deduce intracellular diets of Mtb [18] and to identify new metabolic pathways [19]. Shi et al. subsequently used this model in combination with gene expression data to show that Mtb redirects carbon flow from biosynthetic pathways to storage compound production under stressful conditions [20]. Palsson s group independently constructed a metabolic network for Mtb to predict gene essentiality [21]. That model was subsequently used to elucidate metabolic changes during Mtb adaptation to hypoxia [22], to predict effects of enzyme inhibition [23] and the effects of INH on Mtb metabolism [24]. What is presently not considered is the role of these bacterial dynamics (metabolism, gene regulation, growth, etc) in the context of the host and environmental influences within a granuloma. Our group is developing a next-generation granuloma model that includes these flux-balance bacterial models running within all of the bacteria agents. This allows for a more accurate representation of bacterial dynamics and mechanisms that are possible targets for drug therapies (unpublished).
Virtual experiments
MCMs can be used to apply two useful tools for drug development, testing and assessment: virtual experiments (VEs) and virtual clinical trials (VCTs). VEs allow us to generate predictions and test hypotheses as to, for example, aspects of the immune response to Mtb that most significantly contribute to granuloma formation and function. We can perform virtual knock-out or virtual depletion experiments to test effects of different system components, and we can simulate the effect of multiple interventions at once [8,13,25,26]. Finally, we can apply uncertainty and sensitivity analysis tools to predict specific “tipping points” in model system architecture that might be useful as therapeutic targets [27]. Granuloma outcomes in our models can be roughly classified into four categories - containment of bacteria, clearance of bacteria, non-containment of bacteria (dissemination), and containment of bacteria with excessive inflammation – these correspond well to experimentally observed situations, including the observed wide spectrum in between.
Virtual Clinical Trials
Currently, it costs 1 billion USD to test one new vaccine for TB in a clinical trial, and of course testing in humans is time-consuming and risky. VCTs can be used to simulate treatment in a collection of individuals with person-to-person variability. For example, VCTs suggested that the differential rates of reactivation TB in different populations were due to bioavailability of TNF following anti-TNF treatment [27]. In another study, variation in the number of memory T cells across a population was simulated to determine levels that offer effective protection against Mtb following vaccination [28,29]. Predictions from VCTs can be used to inform choices of drug scheduling, dosing etc in the next stage of clinical trial development for human populations.
Identification of drug targets
What host factors influence the ability of a granuloma to control infection? VEs can help us understand whether subtle changes in molecular, cellular or tissue-level host parameters might identify tipping points at which a granuloma transitions from being able to contain infection to not being able to do so (Figure 1). For example, Cilfone et al. [10], using a computational model that includes both the pro-inflammatory cytokine TNF and the anti-inflammatory cytokine IL-10, demonstrated that a balance of TNF and IL-10 concentrations is essential to infection control within a single granuloma. Too little TNF leads to poor infection control; too little IL-10 leads to infection control but excessive inflammation and thus tissue damage. The appropriate ratio of TNF to IL-10 concentrations leads to infection control with minimal inflammation. Granulomas that are out of balance may improve with the addition of antibodies or exogenous cytokines to shift from poorer outcomes towards containment. Our VEs with anti-TNF drugs, which are used as treatment for inflammatory diseases such as rheumatoid arthritis and psoriatic arthritis, also show the effect of disturbing this balance. Differences in TNF/drug binding kinetics and permeability explain the differential ability of particular drugs to cause reactivation TB, and suggest characteristics of anti-TNF drugs that may lessen the probability of doing so [12,26].
Another way to shift the balance of cytokines identified above is to target physiological processes that influence IL-10 or TNF concentrations or that make up the signaling pathways downstream of those cytokines. Our work gives several examples of this. First, VEs suggest that TNF-receptor internalization kinetics play a critical role in infection control within a granuloma, regulating a balance between paracrine and autocrine TNF-induced responses [9]. Internalization that is too slow leads to clearance with excessive inflammation, while internalization that is too rapid results in dissemination. Physiological values of internalization rates lead primarily to containment. Finally, VEs of pharmacological manipulation of the TNF-induced NFκB signaling pathway show improved infection control. Interestingly, we find that simultaneous targeting of several points along the pathway, rather than just one, produces (or leads to?) a better infection outcome [13], suggesting complex interactions are occurring between system factors that can be harnessed. Going forward, we envision that VEs will complement hands-on experiments help identify drug targets.
Optimization of antibiotic protocols
Short-course therapy for TB typically consists of a cocktail of 2–4 first-line antibiotics taken for 6–9 months. The first-line antibiotics for TB are isoniazid (INH), rifampin (RIF), pyrazinamide (PZA) and ethambutol (EMB), of which INH and RIF are arguably the most studied. Although antibiotic efficacy depends on many factors, certainly adequate exposure of the pathogen to antibiotics is key. Imaging technology has been used to demonstrate that spatial distributions of antibiotics in granulomas play a key role in treatment outcomes [30,31].
Even among approved antibiotics, the design space that includes various combinations, dosing, and timing is enormous. Recently Wallis et al. [32] counted 58 different antibiotic regimens being tested for TB treatment. Simulations of antibiotic regimens may prove useful in narrowing the treatment design space. Initial modeling studies have shed light on the success or failure of some regimens [33,34], and future work can additionally include consideration of the penetration of drugs into the granuloma [12]. For example, a computational model of the granuloma can be coupled to a pharmacokinetic model (e.g. [35]) describing blood antibiotic concentrations as a function of time.
Immunomodulation with antibiotic therapy
Combining immune modulation strategies (immunomodulation) with antibiotics is a potential strategy for enhancing treatment of TB [3]. Immunomodulation in this context refers to the addition/subtraction of cells and/or molecules (such as cytokines) in order to enhance the immune response. It stands to reason that boosting the immune system while reducing bacterial load could lead to more rapid and effective control of infection. However, the immune response to Mtb is complex and it remains unclear which are the appropriate immune responses to modulate to increase control of infection while simultaneously reducing pathology and pathogen load. Several strategies have been tried in murine models (e.g. addition of exogenous cytokines IFN-γ, GM-CSF, TNF, IL-12; reviewed in [36,37]) and a few in humans (addition of exogenous cytokines IL-2, GM-CSF, TNF, IFN-γ) [36,38], but results are inconclusive. Likely, successful immunomodulation therapy will involve combinations of immune mediators such as the molecules TNF, IL-10, IFN-γ, IL-12 and cells (effector and regulatory T cells) as well as chemokines (CXCR3 and CCR5-ligands). Again, appropriate delivery of both antibiotic and immunomodulatory agents to granulomas and proper timing, drug combinations and dosing are all likely to be key factors in successful therapy and ones that can be explored with MCMs. VEs and VCTs applied in this setting can help address the following questions: How can immunomodulation affect the establishment of latency, reactivation, and clearance? Which immunodulators are predicted to be the most effective at improving antibiotic outcome? Is there an optimal timing or combination of modulators and drugs that will minimize the development of active TB or reduce reactivation?
Conclusion
The development of new strategies for TB treatment is an urgent priority. We argue that virtual experiments and virtual clinical trials can complement experimental and clinical approaches toward this goal. The development of drug resistance adds additional complexity. While treatment of disease is an important goal, only vaccines will solve the TB problem. While not discussed in this article, new computational efforts also need to be extended in this direction. A systems biology approach combining multiple modalities is likely the only course to crossing the cure boundary for TB.
Acknowledgments
This work was supported by NIH R01 HL106804 (awarded to DEK), R01 EB012579 and R01 HL 110811 (both awarded to DEK and JJL). We thank Elsje Pienaar and Nick Cilfone for assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jennifer J. Linderman, Email: linderma@umich.edu.
Denise E. Kirschner, Email: kirschne@umich.edu.
References
- 1.WHO. Global Tuberculosis Control. 2009. [Google Scholar]
- 2.Lin PL, Flynn JL. Understanding latent tuberculosis: a moving target. J Immunol. 2010;185 (1):15–22. doi: 10.4049/jimmunol.0903856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zumla A, et al. Advances in the development of new tuberculosis drugs and treatment regimens. Nature reviews Drug discovery. 2013;12 (5):388–404. doi: 10.1038/nrd4001. [DOI] [PubMed] [Google Scholar]
- 4.Flynn JL, et al. Macrophages and control of granulomatous inflammation in tuberculosis. Mucosal immunology. 2011;4 (3):271–278. doi: 10.1038/mi.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guirado E, Schlesinger LS. Modeling the Mycobacterium tuberculosis Granuloma - the Critical Battlefield in Host Immunity and Disease. Frontiers in Immunology. 2013;4:98. doi: 10.3389/fimmu.2013.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin PL, et al. Sterilization of granulomas is common in active and latent tuberculosis despite within-host variability in bacterial killing. Nature medicine. 2014;20 (1):75–79. doi: 10.1038/nm.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Segovia-Juarez JL, et al. Identifying control mechanisms of granuloma formation during M. tuberculosis infection using an agent-based model. J Theor Biol. 2004;231 (3):357–376. doi: 10.1016/j.jtbi.2004.06.031. [DOI] [PubMed] [Google Scholar]
- 8.Ray JC, et al. Synergy between individual TNF-dependent functions determines granuloma performance for controlling Mycobacterium tuberculosis infection. J Immunol. 2009;182 (6):3706–3717. doi: 10.4049/jimmunol.0802297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fallahi-Sichani M, et al. Multiscale computational modeling reveals a critical role for TNF-alpha receptor 1 dynamics in tuberculosis granuloma formation. J Immunol. 2011;186 (6):3472–3483. doi: 10.4049/jimmunol.1003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cilfone NA, et al. Multi-Scale Modeling Predicts a Balance of Tumor Necrosis Factor-alpha and Interleukin-10 Controls the Granuloma Environment during Mycobacterium tuberculosis Infection. PLoS One. 2013;8 (7):e68680. doi: 10.1371/journal.pone.0068680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fallahi-Sichani M, et al. Identification of key processes that control tumor necrosis factor availability in a tuberculosis granuloma. PLoS Comput Biol. 2010;6 (5):e1000778. doi: 10.1371/journal.pcbi.1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fallahi-Sichani M, et al. Differential risk of tuberculosis reactivation among anti-TNF therapies is due to drug binding kinetics and permeability. J Immunol. 2012;188 (7):3169–3178. doi: 10.4049/jimmunol.1103298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fallahi-Sichani M, et al. NF-kappaB Signaling Dynamics Play a Key Role in Infection Control in Tuberculosis. Front Physiol. 2012;3:170. doi: 10.3389/fphys.2012.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin PL, et al. Early events in Mycobacterium tuberculosis infection in cynomolgus macaques. Infect Immun. 2006;74 (7):3790–3803. doi: 10.1128/IAI.00064-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magombedze G, Mulder N. A mathematical representation of the development of Mycobacterium tuberculosis active, latent and dormant stages. Journal of Theoretical Biology. 2012;292:44–59. doi: 10.1016/j.jtbi.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 16.Ankomah P, Levin BR. Two-drug antimicrobial chemotherapy: a mathematical model and experiments with Mycobacterium marinum. PLoS pathogens. 2012;8 (1):e1002487. doi: 10.1371/journal.ppat.1002487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beste DJ, et al. GSMN-TB: a web-based genome-scale network model of Mycobacterium tuberculosis metabolism. Genome Biol. 2007;8 (5):R89. doi: 10.1186/gb-2007-8-5-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beste DJ, et al. (13)C-Flux Spectral Analysis of Host-Pathogen Metabolism Reveals a Mixed Diet for Intracellular Mycobacterium tuberculosis. Chemistry & biology. 2013;20 (8):1012–1021. doi: 10.1016/j.chembiol.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beste DJ, et al. (1)(3)C metabolic flux analysis identifies an unusual route for pyruvate dissimilation in mycobacteria which requires isocitrate lyase and carbon dioxide fixation. PLoS pathogens. 2011;7 (7):e1002091. doi: 10.1371/journal.ppat.1002091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi L, et al. Carbon flux rerouting during Mycobacterium tuberculosis growth arrest. Molecular microbiology. 2010;78 (5):1199–1215. doi: 10.1111/j.1365-2958.2010.07399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jamshidi N, Palsson BO. Investigating the metabolic capabilities of Mycobacterium tuberculosis H37Rv using the in silico strain iNJ661 and proposing alternative drug targets. BMC systems biology. 2007;1:26. doi: 10.1186/1752-0509-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang X, et al. Modeling phenotypic metabolic adaptations of Mycobacterium tuberculosis H37Rv under hypoxia. PLoS computational biology. 2012;8 (9):e1002688. doi: 10.1371/journal.pcbi.1002688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang X, et al. A systems biology framework for modeling metabolic enzyme inhibition of Mycobacterium tuberculosis. BMC Syst Biol. 2009;3:92. doi: 10.1186/1752-0509-3-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhat AG, et al. Modeling metabolic adjustment in Mycobacterium tuberculosis upon treatment with isoniazid. Systems and synthetic biology. 2010;4 (4):299–309. doi: 10.1007/s11693-011-9075-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medina-Franco JL, et al. Shifting from the single to the multitarget paradigm in drug discovery. Drug discovery today. 2013;18 (9–10):495–501. doi: 10.1016/j.drudis.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fallahi-Sichani M, et al. A systems biology approach for understanding granuloma formation and function in tuberculosis. In: McFadden J, DBaAK, editors. Systems biology of tuberculosis. Springer; 2013. [Google Scholar]
- 27.Marino S, et al. A methodology for performing global uncertainty and sensitivity analysis in systems biology. J Theor Biol. 2008;254 (1):178–196. doi: 10.1016/j.jtbi.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marino S, et al. Differences in reactivation of tuberculosis induced from anti-TNF treatments are based on bioavailability in granulomatous tissue. PLoS Comput Biol. 2007;3 (10):1909–1924. doi: 10.1371/journal.pcbi.0030194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sud D, et al. Contribution of CD8+ T cells to control of Mycobacterium tuberculosis infection. J Immunol. 2006;176 (7):4296–4314. doi: 10.4049/jimmunol.176.7.4296. [DOI] [PubMed] [Google Scholar]
- 30.Kjellsson MC, et al. Pharmacokinetic evaluation of the penetration of antituberculosis agents in rabbit pulmonary lesions. Antimicrobial agents and chemotherapy. 2012;56 (1):446–457. doi: 10.1128/AAC.05208-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prideaux B, et al. High-sensitivity MALDI-MRM-MS imaging of moxifloxacin distribution in tuberculosis-infected rabbit lungs and granulomatous lesions. Analytical chemistry. 2011;83 (6):2112–2118. doi: 10.1021/ac1029049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wallis RS, et al. Month 2 culture status and treatment duration as predictors of tuberculosis relapse risk in a meta-regression model. PLoS One. 2013;8 (8):e71116. doi: 10.1371/journal.pone.0071116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goutelle S, et al. Population modeling and Monte Carlo simulation study of the pharmacokinetics and antituberculosis pharmacodynamics of rifampin in lungs. Antimicrobial agents and chemotherapy. 2009;53 (7):2974–2981. doi: 10.1128/AAC.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magombedze G, et al. Mathematical Modeling of Chemotherapy of Human TB Infection. Journal of Biological Systems. 2006;14 (04):509–553. [Google Scholar]
- 35.Wilkins JJ, et al. Population pharmacokinetics of rifampin in pulmonary tuberculosis patients, including a semimechanistic model to describe variable absorption. Antimicrobial agents and chemotherapy. 2008;52 (6):2138–2148. doi: 10.1128/AAC.00461-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Churchyard GJ, et al. Advances in immunotherapy for tuberculosis treatment. Clin Chest Med. 2009;30 (4):769–782. ix. doi: 10.1016/j.ccm.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 37.Rook GA, et al. Immunotherapeutics for tuberculosis in experimental animals: is there a common pathway activated by effective protocols? The Journal of infectious diseases. 2007;196 (2):191–198. doi: 10.1086/518937. [DOI] [PubMed] [Google Scholar]
- 38.Wallis RS. Reconsidering adjuvant immunotherapy for tuberculosis. Clin Infect Dis. 2005;41 (2):201–208. doi: 10.1086/430914. [DOI] [PubMed] [Google Scholar]


