Abstract
Purpose
The purpose of this research was to study the association between the single nucleotide polymorphisms (SNPs) of the tektin-t gene and idiopathic asthenozoospermia.
Methods
We conducted sequence analyses of the tektin-t gene in 104 idiopathic asthenozoospermia and 102 fertile men with normospermic parameters in Sichuan, China.
Results
In this study, we found that allele 136 T (odds ratio [OR] 1.745, 95 % confidence interval [CI] 1.146–2.655, P = 0.009) was significantly increased in idiopathic asthenozoospermic patients compared with fertile men. This mutation substitutes a highly conserved arginine at position 46 to cysteine. Moreover, PolyPhen-2 analysis predicted that this variant was “probably damaging”. In addition, a novel heterozygous mutation, R207H (c.620G >A), was detected in five asthenozoospermic patients, while there was no detection of this genotype among the fertile candidates, indicating that the mutation was located within a conserved domain predicted by PolyPhen-2 analysis as “probably damaging” to the protein.
Conclusions
These results suggested that tektin-t variants (Arg/Cys + Cys/Cys) were probably one of the high risk genetic factors for idiopathic asthenozoospermia among males in Sichuan, China, while the R207H polymorphism may be associated with idiopathic asthenozoospermia risk.
Keywords: Genetic polymorphisms, Tektin-t, Asthenozoospermia, Sperm motility
Introduction
Infertility is a worldwide problem affecting about 15 % of couples trying to conceive, and male factor may partially or fully account for approximately 50 % of all infertility cases [1]. Genetic factors may account for a high percentage of male infertility. Four major semen anomalies (azoospermia, oligozoospermia, asthenozoospermia (AZS), and teratospermia) are present in almost 50 % of infertile couples and in approximately 90 % of infertile males [2]. In these cases, AZS is one of the major causes of male infertility second only to oligospermia [3]. AZS may exist as an isolated disorder, in association with other sperm anomalies or as parts of syndromic association. As an isolated disorder, AZS was found in as many as 18.71 % of patients presenting for male infertility and might be a significant factor in another 63.13 % of patients with combined oligo- and/or teratozoo-spermia [4]. Thus, AZS may be one of the main seminal pathologic infertilities among males.
Tektins have been proven to be indispensable for spermatozoa motility. Tektins were first isolated from sea urchin sperm flagellar microtubules [5] and named as tektin A, B, and C [6]. As the sequences of sea urchin tektin A, B, and C have been obtained [7–9], the facts were found that the tektin A and B were closely related, and the models of dimers and polymers molecules were devised [10]. Several functional studies have demonstrated tektins including tektin-2, tektin-3, tektin-4, and tektin-5 are critical for sperm motility [11–16]. These studies imply tektins play important roles in sperm function. Tanaka et al. [16] generated tektin-t-deficient mice and proved that females were fully fertile but males were infertile caused by debilitating sperm motility and impairing motility of both flagella and cilia. Moreover, Bhilawadikar R et al. [17] found that tektin-t levels were positively associated with sperm motility parameters, fertilization rate, embryo quality, and pregnancy rate. The human tektin-t (or h-tekB1 or Tektin-2) gene has been cloned [18–20], showing a specific expression in testis, and precisely localized in flagella of mature sperm. The localization of this protein and its association with sperm motility make it a good candidate gene for research into the causes of AZS. Therefore, we screened 104 idiopathic AZS patients as a case group and 102 fertile men as a control group to study the association of polymorphisms of the tektin-t gene with idiopathic AZS infertile patients in Sichuan, China.
Patients and methods
Patients
We recruited 104 male partners from successively enrolled couples who had their first infertility consultation in the Affiliate Reproductive Hospital Genitalia Hygiene Research Center (Sichuan, China) between May 2014 and March 2015. The asthenozoospermic infertility patients were aged between 23 and 44 years old and were not related. To screen the idiopathic AZS, patients with known diseases such as cryptorchidism, orchitis, epididymitis, varicocele, obstruction of the vas deferens, endocrine hypogonadism, karyotype anomalies, and Y chromosome microdeletions were excluded from the study. Patients with drug, alcohol, substance abuse, and tobacco use were also excluded. The control group consisted of 102 fertile and healthy males of a comparable age who had fathered at least one child. All subjects were given informed consent for molecular analysis of their semen samples and blood samples.
Semen analyses were performed at least twice for all subjects according to the World Health Organization (WHO) recommendations [21]. At least one sperm sample per individual collected after 3–5 days of abstinence through masturbation was analyzed. These isolated asthenozoospermic infertility patients had concentration of spermatozoa >20 × 106/ml and rapid forward progressive motile sperm (grade A <25 %) and total progressive motile sperm (grades A + B <50 %) in fresh ejaculation according to the WHO (2010) criteria [21].
Extraction of genomic DNA
Total DNA of human spermatozoa was extracted using the EasyPure Blood Genomic DNA Kit (Transgen, Beijing, China). Briefly, the spermatozoa pellet was resuspended in sterile water and mixed with lysis solution containing 20-mg/ml proteinase K and Binding Buffer 3. Lysis was incubated at room temperature for 10 min. Lysates were added to centrifugal column to bind the DNA. Bound DNA was washed and then eluted from the centrifugal column. Quantification of the extracted genomic DNA was conducted by the spectrophotometry analysis. All of the DNA samples were stored at −20 °C until examination.
Polymerase chain reaction amplification and genotyping
PCR amplification of all exons of Tektin-t gene and the parts of flanking intronic sequence were performed using the primer sets as described in Table 1. PCR was performed in A200 Gradient Thermal cycler (Long Gene) in a total volume of 25-μL buffered solution containing 1.5 mM Mg2+ (Fermentas International Inc., Burlington, Ontario, Canada), 0.25 mM dNTPs (TransGen, Beijing, China), 0.2 μM of each primer (Sangon Biotech, Shanghai, China), 2 U of DNA polymerase (Fermentas International Inc., Burlington, Ontario, Canada), approximately 200 ng genomic DNA. The PCR conditions were as follows: initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation (94 °C, 1 min), annealing (56 °C, 1 min), extension (72 °C, 1 min), and a final extension at 72 °C for 5 min.
Table 1.
Primers used for the amplification of the 10 exons of the human Tektin-t gene
| Exon | Primer | Primer sequence | Size (bp) | PCR product size (bp) |
|---|---|---|---|---|
| 1 | tektin-t 1 F | 5′ AGTGAAAGCGCTGCCCTGAT 3′ | 20 | 384 |
| tektin-t 1R | 5′ GGAGAGCCCCGTGGGAAGAG 3′ | 20 | ||
| 2-3 | tektin-t 2 F | 5′ CAGAGCAAAATGGACAGGAA 3′ | 20 | 661 |
| tektin-t 3R | 5′ TAGCGCTTGTCCCCACTTTA 3′ | 20 | ||
| 4 | tektin-t 4 F | 5′ GCCATCACTCCCAGCTAA 3′ | 18 | 488 |
| tektin-t 4R | 5′ GCCAAAATGTAGCACGAGAG 3′ | 20 | ||
| 5-7 | tektin-t 5 F | 5′ CCTGTGGTGTCCCTTTAGTA 3′ | 20 | 894 |
| tektin-t 7 F | 5′ TCAGGCTCAGGAGCTTTGTG 3′ | 20 | ||
| 8-10 | tektin-t 8 F | 5′ ACTTCCTGCTGTGGGATGAG 3′ | 20 | 936 |
| tektin-t 10R | 5′ CCTCTGCATTTATTCCATTC 3′ | 20 |
F forward primer, R reverse primer
All PCR products were analyzed by electrophoresis using 2 % agarose gel prepared in 1 × TAE buffer containing DuRed nucleic acid gel stain (Abgent Biotechnology Co., Ltd., Suzhou, China) run at 120 V for 35 min at room temperature. Subsequently, the PCR products were sequenced with the ABI3730XL DNA Analyzer (Applied Biosystems, Foster City, CA, USA) using the BigDye fluorescence labeling Terminator Cycle Sequencing kit (Sangon Biotech, China). The sequence alignment of the Tektin-t gene was performed using DNAMAN. Sequences from the species were obtained from NCBI.
Statistical analysis and the pathogenicity prediction
Hardy-Weinberg equilibrium and the comparison of genotype frequencies between patients and control groups were performed using the chi-squared (χ2) test. Using the unconditional logistic regression analysis to calculate odds ratio (OR) and 95 % confidence interval (95% CI) were to measure the risk associated with variant genotypes. P < 0.05 was considered to be statistically significant. All data were analyzed using Statistical Package for Social Sciences software version 20.0 (SPSS Inc., Chicago, IL, USA).
The prediction of the damaging effect of missense mutation to protein structure and function was performed using PolyPhen-2 bioinformatic program (http://genetics.bwh.harvard.edu/pph2/) [22]. For a false positive rate of 20 %, PolyPhen-2 achieved true positive prediction rates of 92 % and 73 % on HumDiv and HumVar datasets, respectively [22, 23]. We also used the ExPASy-ProtScale (http://web.expasy.org/protscale/) to analyze the changes in hydrophobicity of the tektin-t protein due to the mutation.
Results
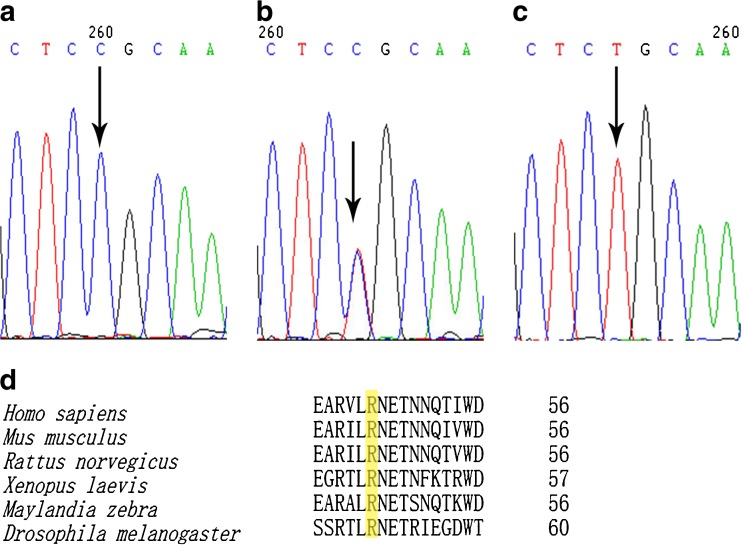
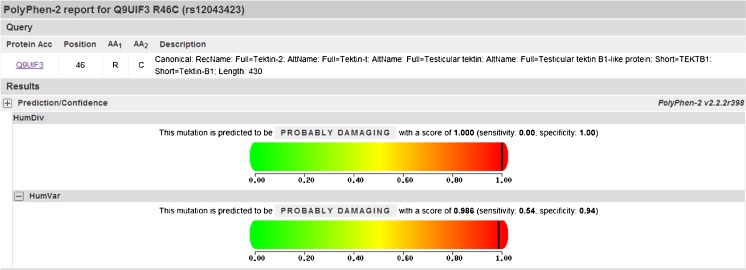
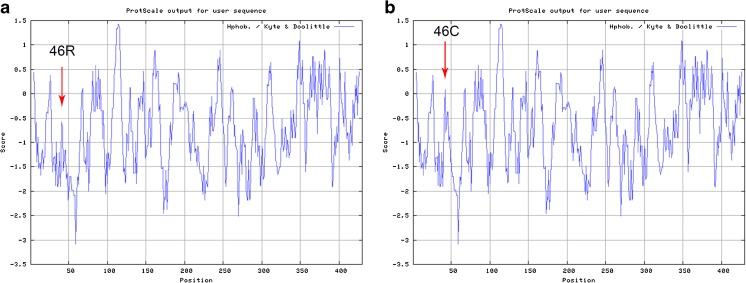
A descriptive comparison of our study population is presented in Table 2. After sequencing the 10 exons of tektin-t and the flanking intronic sequence of 104 AZS patients, we screened five nucleotide substitutions: R46C (c.136C >T) in exon3, S174S (c.522C >T) in exon 5, R207H (c.620G >A) in exon 5, K267N (c.801G >T) in exon 7, and T393T (c.1179A >G) in exon 10. The five nucleotide changes were all detected in the heterozygotes excepted for a part of c.136C >T. We confirmed that c.522C >T, c.801G >T, and c.1179A >C were common mutations after screening the 102 controls and analyzing online database; however the genotype and allele frequencies of the missense mutation c.136C >T changing the arginine at position 46 to Cysteine were different. The amino acid 46R (Fig. 1a–c) is located in a highly conserved domain from invertebrate to higher species (Fig. 1d). This mutation in both asthenozoospermic patients and controls were in Hardy-Weinberg equilibrium (χ2 = 1.5622, 0.9418; P = 0.4579, 0.6244, respectively). The frequencies of genotype CT (OR = 1.867, 95 % CI = 1.047–3.328, P = 0.034) and TT (OR = 3.503, 95 % CI = 1.138–10.778, P = 0.023) were significantly increased compared with controls. The frequencies of allele 136 T (OR = 1.745, 95 % CI = 1.146–2.655, P = 0.009) showed a significant increase in asthenozoospermic patients. Overall, R46C showed a significant difference of distribution between the idiopathic asthenozoospermia and fertile controls (Table 3). Moreover, PolyPhen-2 analysis predicted that this mutation is “probably damaging” with the score of 0.986 on HumVar model (Fig. 2). In addition, a hydropathy plot of the R46C mutant polypeptide generated with the Kyte-Doolittle algorithm by using an online tool ExPASy-ProtScale demonstrated a further imbalance in its hydrophobicity caused by the a.136C >T mutation and may result in modification of the protein structure (Fig. 3).
Table 2.
Comparison of age and semen parameters between asthenozoospermic group and controls
| Clinical parameters | Asthenozoospermic group (n = 104)a | Control group (n = 102)a | P b |
|---|---|---|---|
| age (years) | 31.2 ± 6.2 | 34.9 ± 3.5 | 0.496 |
| Sperm concentration (×106/mL) | 38.0 ± 17.7 | 62.8 ± 16.3 | 0.018 |
| Rapid progressive motility (%) | 9.9 ± 5.9 | 37.9 ± 8.1 | <0.001 |
| Total progressive motility (%) | 21.2 ± 10.3 | 63.53 ± 12.8 | 0.006 |
aData are presented as mean ± SD
bThe comparison between groups was done with the Student’s t test. P < 0.05 was considered statistically significant
Fig. 1.
Sequencing analysis of the 136 C >T of exon 3 in the tektin-t gene with different allele expressions. a Wild-type homozygous (CC) genotype. b Heterozygous (CT) genotype. c Mutant homozygous (TT) genotype. d Sequence alignment of the tektin-t gene in six different species. The position of our reported mutated amino acid (R46C) is framed
Table 3.
Genotype and allele frequencies for tektin t C136T among idiopathic asthenozoospermia and controls
| Controls (N = 102) | Asthenozoospemia (N = 104) | Odds ratio (95 % CI) | P | |
|---|---|---|---|---|
| 136CC | (52.94 %)54 | (35.58 %)37 | ||
| 136CT | (42.16 %)43 | (52.88 %)55 | 1.867 (1.047–3.328) | 0.034* |
| 136TT | (4.90 %)5 | (11.54 %)12 | 3.503 (1.138–10.778) | 0.023* |
| 136 (CT + TT) | (47.06 %)48 | (64.42 %)67 | 2.037 (1.165–3.562) | 0.012* |
| 136C | (74.02 %)151 | (62.02 %)129 | ||
| 136 T | (25.98 %)53 | (37.98 %)79 | 1.745 (1.146–2.655) | 0.009** |
CI confidence interval
* P < 0.05
** P < 0.01
Fig. 2.
PolyPhen-2 analysis predicting the pathogenicity, the p.R46C substitution on the tektin-t protein
Fig. 3.
Hydropathy plot for the tektin-t protein prepared in the Expasy ProtScale Website according to the Kyte and Doolittle algorithm. The hydrophobicity of the wild-type tektin-t protein a. is compared to the mutant form, including the novel p.R46C mutation b. The hydrophobicity scores of p.R46C of the tektin-t protein are higher than the wild type. The mutated site is indicated with an arrow
A novel heterozygous missense mutation (c.620G > A) was dectected in five asthenospermic patients (4.8 %), but this mutation was absent in controls, suggesting that it may be associated with asthenospermia. PolyPhen-2 analysis predicted that this mutation is “probably damaging” with the score of 0.999 on HumVar model.
Discussion
Sperm with low motility has been identified as one of the important factors of male infertility [24]. AZS is a phenotype often present in primary ciliary dyskinesia (PCD) that is a rare autosomal recessive genetic disorder which could cause a defect in the action of the cilia lining of the respiratory tract and the flagella of sperm cells [25–27]. About 50 % of all affected PCD patients have laterality defects such as situs inversus (heterotaxy), this syndrome is referred as Kartagener Syndrome (KS) [28]. Approximately, 90 % of PCD/KS male patients are affected by AZS and most of them showing dynein genes mutation [16]. It has been demonstrated that tektins participated in the assembly of axonemal microtubules and offered stability to axonemal microtubules [29]. Tektins are a highly conserved family of flagellar and ciliary proteins in many species from invertebrate to higher species, such as Chlamydomonas, sea urchins, zebrafish, hedgehog, rodents, and humans [18, 19, 28, 30, 31]. In mammals, tektins exist in the testis, brain, retina, and other tissues containing ciliated cells [32, 33]. Tektins are insoluble α-helical proteins and are related to intermediate filament (IF) proteins and nuclear lamins. Thus, tektins may play a fundamental role in ciliary movement [10]. The human tektin-t gene has been first cloned in 2002, and tektin-t protein is localized to the tail of mature sperm [19]. Its highly conserved sequence suggested that tektin-t has an important role in the formation of ciliary and flagellar [17, 34].
In our study, we found a significant difference in frequency of the missense mutation (c.136C >T) of the tektin-t gene between idiopathic asthenozoospermic patients and the control groups. The incidences of the missense tektin-t gene mutation (R46C) (heterozygote [CT] and homozygote [TT]) were 64.42 % in asthenozoospermic patients and 47.06 % in the controls, showing a significant increase between the patients and the controls (OR = 2.037, 95 % CI = 1.165–3.562, P = 0.012) which indicated that this mutation could be a possible risk (OR = 1.745, 95 % CI = 1.146–2.655, P = 0.009) for AZS. These results were consistent with the facts that the mutated amino acid is located in a highly conserved position in the tektin-t protein (Fig. 1d), and this variant was analyzed using PolyPhen-2 with a result of “probably damaging” (Fig. 2). Additionally, the hydrophobicity of the arginine at the position 46 of the tektin-t protein was significantly different from the wild type, and the substitutions may result in modification of the protein structure (Fig. 3). Moreover, a novel mutation R207H was found in our study in five patients and predicted that this change is “probably damaging” with the score of 0.999 on HumVar model by PolyPhen-2 analysis. In addition, the frequency of R207H is more in Asian population (0.005) than non-Asian population (0.00003) which supports our finding.
Zuccarello et al. [35], analyzed 90 isolated non-syndromic AZS patients and found a heterozygous mutation A229V in one patient and predicted that a.a 229 holds some important roles in the tektins’ functions. However, there was no detection of this mutation in our study. Furthermore, the R46C mutation was found to be associated with asthenozoospermia by statistical analysis and predicted to have a possible damaging effect to the protein structure and function, while this change was also found in previous study [35], but was regarded as a common SNP. In addition, the frequency of R46C is higher in Asian population (0.24) than non-Asian population (0.006), which is inconsistent with our result, which could be explained by the difference in geographical location. Moreover, a novel mutation R207H was found in our study in five patients, this mutation was never found in the previous study. The differences between our study and the previous study could be explained by many factors as follows: the difference in geographical location and genetic background of the study populations, different screening standards of samples, and study sample size.
Conclusion
In conclusion, this is the first Chinese study on the association between SNPs of the tektin-t gene and idiopathic asthenozoospermia. Our results suggest that the tektin-t variant (R46C) is probably associated with asthenozoospermia. We also found a novel missense (R207H) variant in five asthenozoospermic patients but absent in fertile men, suggesting that this mutation might be associated with asthenozoospermia. Further studies are needed to confirm our results with larger groups of participants. Analyzing the structure of the tektin-t protein and functional studies should be taken to demonstrate these variants affect tektin-t protein activity, protein synthesis, and flagellar impairment of mature sperm. Furthermore, future studies including subjects of different ethnic and geographic origins should be conducted for tektin-t gene combined with other genes associated with AZS.
Acknowledgments
We thank the Affiliate Reproductive Hospital Genitalia Hygiene Research Center (Sichuan, China) for providing us with a platform for the analysis of the semen parameters, and we are grateful for the contributions of the patients and controls who participated in this study.
Footnotes
Capsule These results suggested that tektin-t variants (Arg/Cys + Cys/Cys) were probably one of the high risk genetic factors for idiopathic asthenozoospermia among males in Sichuan, China, while the R207H polymorphism may be associated with idiopathic asthenozoospermia risk.
Contributor Information
Shao-hong Zhang, Email: hnlyzhangsh@163.com.
Xian-ping Ding, Phone: +86-028-85413096, Email: brainding@scu.edu.cn.
References
- 1.Huynh T, Mollard R, Trounson A. Selected genetic factors associated with male infertility. Hum Reprod Update. 2002;8(2):183–98. doi: 10.1093/humupd/8.2.183. [DOI] [PubMed] [Google Scholar]
- 2.De Kretser D, Baker H. Infertility in men: recent advances and continuing controversies. J Clin Endocrinol Metab. 1999;84(10):3443–50. doi: 10.1210/jcem.84.10.6101. [DOI] [PubMed] [Google Scholar]
- 3.Asero P, Calogero AE, Condorelli RA, Mongioi L, Vicari E, Lanzafame F, et al. Relevance of genetic investigation in male infertility. J Endocrinol Investig. 2014;37(5):415–27. doi: 10.1007/s40618-014-0053-1. [DOI] [PubMed] [Google Scholar]
- 4.Curi S, Ariagno J, Chenlo P, Mendeluk G, Pugliese M, Sardi Segovia L, et al. Asthenozoospermia: analysis of a large population. Syst Biol Reprod Med. 2003;49(5):343–9. doi: 10.1080/01485010390219656. [DOI] [PubMed] [Google Scholar]
- 5.Linck RW, Albertini DF, Kenney DM, Langevin GL. Tektin filaments: chemically unique filaments of sperm flagellar microtubules. Prog Clin Biol Res. 1982;80:127–32. doi: 10.1002/cm.970020724. [DOI] [PubMed] [Google Scholar]
- 6.Linck RW, Amos LA, Amos WB. Localization of tektin filaments in microtubules of sea-urchin sperm flagella by immunoelectron microscopy. J Cell Biol. 1985;100(1):126–35. doi: 10.1083/jcb.100.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Norrander JM, Amos LA, Linck RW. Primary structure of tektin A1: comparison with intermediate filament proteins and a model for its association with tubulin. Proc Natl Acad Sci U S A. 1992;89:8567–71. doi: 10.1073/pnas.89.18.8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen R, Perrone CA, Amos LA, Linck RW. Tektin B1 from ciliary microtubules - primary structure as deduced from the cDNA sequence and comparison with tektin A1. J Cell Sci. 1993;106:909–18. doi: 10.1242/jcs.106.3.909. [DOI] [PubMed] [Google Scholar]
- 9.Norrander JM, Perrone CA, Amos LA, Linck RW. Structural comparison of tektins and evidence for their determination of complex spacings in flagellar microtubules. J Mol Biol. 1996;257(2):385–97. doi: 10.1006/jmbi.1996.0170. [DOI] [PubMed] [Google Scholar]
- 10.Setter PW, Malvey-Dorn E, Steffen W, Stephens RE, Linck RW. Tektin interactions and a model for molecular functions. Exp Cell Res. 2006;312(15):2880–96. doi: 10.1016/j.yexcr.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 11.Roy A, Yan W, Burns KH, Matzuk MM. Tektin3 encodes an evolutionarily conserved putative testicular microtubules-related protein expressed preferentially in male germ cells. Mol Reprod Dev. 2004;67(3):295–302. doi: 10.1002/mrd.20025. [DOI] [PubMed] [Google Scholar]
- 12.Roy A, Lin Y-N, Agno JE, DeMayo FJ, Matzuk MM. Tektin 3 is required for progressive sperm motility in mice. Mol Reprod Dev. 2009;76(5):453–9. doi: 10.1002/mrd.20957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takiguchi H, Murayama E, Kaneko T, Kurio H, Toshimori K, Iida H. Characterization and subcellular localization of Tektin 3 in rat spermatozoa. Mol Reprod Dev. 2011;78(8):611–20. doi: 10.1002/mrd.21352. [DOI] [PubMed] [Google Scholar]
- 14.Roy A, Lin YN, Agno JE, DeMayo FJ, Matzuk MM. Absence of tektin 4 causes asthenozoospermia and subfertility in male mice. FASEB J. 2007;21(4):1013–25. doi: 10.1096/fj.06-7035com. [DOI] [PubMed] [Google Scholar]
- 15.Cao W, Ijiri TW, Huang AP, Gerton GL. Characterization of a novel tektin member, TEKT5, in mouse sperm. J Androl. 2011;32(1):55–69. doi: 10.2164/jandrol.109.009456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka H, Iguchi N, Toyama Y, Kitamura K, Takahashi T, Kaseda K, et al. Mice deficient in the axonemal protein Tektin-t exhibit male infertility and immotile-cilium syndrome due to impaired inner arm dynein function. Mol Cell Biol. 2004;24(18):7958–64. doi: 10.1128/MCB.24.18.7958-7964.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhilawadikar R, Zaveri K, Mukadam L, Naik S, Kamble K, Modi D, et al. Levels of Tektin 2 and CatSper 2 in normozoospermic and oligoasthenozoospermic men and its association with motility, fertilization rate, embryo quality and pregnancy rate. J Assist Reprod Genet. 2013;30(4):513–23. doi: 10.1007/s10815-013-9972-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iguchi N, Tanaka H, Fujii T, Tamura K, Kaneko Y, Nojima H, et al. Molecular cloning of haploid germ cell-specific tektin cDNA and analysis of the protein in mouse testis. FEBS Lett. 1999;456:315–21. doi: 10.1016/S0014-5793(99)00967-9. [DOI] [PubMed] [Google Scholar]
- 19.Iguchi N, Tanaka H, Nakamura Y, Nozaki M, Fujiwara T, Nishimune Y. Cloning and characterization of the human tektin-t gene. Mol Hum Reprod. 2002;8(6):525–30. doi: 10.1093/molehr/8.6.525. [DOI] [PubMed] [Google Scholar]
- 20.Wolkowicz MJ, Naaby-Hansen S, Gamble AR, Reddi PP, Flickinger CJ, Herr JC. Tektin B1 demonstrates flagellar localization in human sperm. Biol Reprod. 2002;66(1):241–50. doi: 10.1095/biolreprod66.1.241. [DOI] [PubMed] [Google Scholar]
- 21.WHO . WHO laboratory manual for the examination and processing of human semen. Geneva: World Health Organization; 2010. [Google Scholar]
- 22.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;Chapter 7:Unit7 20. [DOI] [PMC free article] [PubMed]
- 24.Hull MGR, Clazener CMA, Kelly NJ, Conway DI, Foster PA, Hinton RA, et al. Population study of cases, treatment, and outcome of infertility. Br Med J. 1985;291:1693–7. doi: 10.1136/bmj.291.6510.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Narayan D, Krishnan SN, Upender M, Ravikumar TS, Mahoney MJ, Dolan TF, Jr, et al. Unusual inheritance of primary ciliary dyskinesia (Kartagener’s syndrome) J Med Genet. 1994;31(6):493–6. doi: 10.1136/jmg.31.6.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang SM, Li HB, Wang JX, Shi YC, Cheng HB, Wang W, et al. Morphological characteristics and initial genetic study of multiple morphological anomalies of the flagella in China. Asian J Androl. 2015;17(3):513–5. doi: 10.4103/1008-682X.146100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luconi M. Pathophysiology of sperm motility. Front Biosci. 2006;11(1):1433. doi: 10.2741/1894. [DOI] [PubMed] [Google Scholar]
- 28.Chodhari R, Mitchison HM, Meeks M. Cilia, primary ciliary dyskinesia and molecular genetics. Paediatr Respir Rev. 2004;5(1):69–76. doi: 10.1016/j.prrv.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 29.Linck R, Fu X, Lin J, Ouch C, Schefter A, Steffen W, et al. Insights into the structure and function of ciliary and flagellar doublet microtubules: tektins, Ca2 + −binding proteins, and stable protofilaments. J Biol Chem. 2014;289(25):17427–44. doi: 10.1074/jbc.M114.568949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka Y, Okada Y, Hirokawa N. FGF-induced vesicular release of Sonic hedgehog and retinoic acid in leftward nodal flow is critical for left-right determination. Nature. 2005;435(7039):172–7. doi: 10.1038/nature03494. [DOI] [PubMed] [Google Scholar]
- 31.Ha Y. A tektin homologue is decreased in chlamydomonas mutants lacking an axonemal inner-arm dynein. Mol Biol Cell. 2004;15(5):2105–15. doi: 10.1091/mbc.E03-11-0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Norrander J, Larsson M, Stahl S, Hoog C, Linck R. Expression of ciliary tektins in brain and sensory development. J Neurosci. 1998;18(21):8912–8. doi: 10.1523/JNEUROSCI.18-21-08912.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amos LA. The tektin family of microtubule-stabilizing proteins. Genome Biol. 2008;9(7):229. doi: 10.1186/gb-2008-9-7-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mariappa D, Aladakatti RH, Dasari SK, Sreekumar A, Wolkowicz M, van der Hoorn F, et al. Inhibition of tyrosine phosphorylation of sperm flagellar proteins, outer dense fiber protein-2 and tektin-2, is associated with impaired motility during capacitation of hamster spermatozoa. Mol Reprod Dev. 2010;77(2):182–93. doi: 10.1002/mrd.21131. [DOI] [PubMed] [Google Scholar]
- 35.Zuccarello D, Ferlin A, Garolla A, Pati MA, Moretti A, Cazzadore C, et al. A possible association of a human tektin-t gene mutation (A229V) with isolated non-syndromic asthenozoospermia: case report. Hum Reprod. 2008;23(4):996–1001. doi: 10.1093/humrep/dem400. [DOI] [PubMed] [Google Scholar]