Abstract
We have sequenced the complete genomes of 72 individuals affected with early-onset familial Alzheimer's disease caused by an autosomal dominant, highly penetrant mutation in the presenilin-1 (PSEN1) gene, and performed genome-wide association testing to identify variants that modify age at onset (AAO) of Alzheimer's disease. Our analysis identified a haplotype of single-nucleotide polymorphisms (SNPs) on chromosome 17 within a chemokine gene cluster associated with delayed onset of mild-cognitive impairment and dementia. Individuals carrying this haplotype had a mean AAO of mild-cognitive impairment at 51.0±5.2 years compared with 41.1±7.4 years for those without these SNPs. This haplotype thus appears to modify Alzheimer's AAO, conferring a large (~10 years) protective effect. The associated locus harbors several chemokines including eotaxin-1 encoded by CCL11, and the haplotype includes a missense polymorphism in this gene. Validating this association, we found plasma eotaxin-1 levels were correlated with disease AAO in an independent cohort from the University of California San Francisco Memory and Aging Center. In this second cohort, the associated haplotype disrupted the typical age-associated increase of eotaxin-1 levels, suggesting a complex regulatory role for this haplotype in the general population. Altogether, these results suggest eotaxin-1 as a novel modifier of Alzheimer's disease AAO and open potential avenues for therapy.
Introduction
Alzheimer's disease (AD) is an incurable and invariably fatal age-related neurodegenerative disease affecting millions of people worldwide with devastating consequences for those suffering, their families and society at large. The incidence of sporadic AD increases exponentially with age in people over 65 making AD one of the top 10 causes of death and the only one among these 10 lacking a prevention, disease-modifying treatment or cure. If available, treatments able to delay Alzheimer's age at onset (AAO) or to slow the progression of the disease would save billions of dollars in direct and indirect medical costs while increasing the quality of life for the millions of affected individuals.1 To inform such treatments, we sought to identify variants modifying the AAO of AD in one of the world's largest and best-characterized familial AD populations: the familial AD kindred in Antioquia, Colombia. In this population, a highly penetrant, autosomal dominant missense mutation encoding a glutamate-to-alanine substitution at codon 280 in the gene encoding presenilin-1 (PSEN1 E280A) causes early-onset familial AD at an average age of 49 years.2
AAO of AD is highly heritable in both familial and sporadic forms of the disease, with predicted genetic contributions of up to 78% in the sporadic late-onset form,3, 4 and over 90% in a monogenic early-onset familial form.5 The best known genetic modulator of AD AAO, the APOE ɛ4 allele, reduces AAO in both familial and sporadic AD,6, 7 suggesting pathologic commonalities underlying both forms of the disease. Although genome-wide association studies (GWAS) have identified several loci contributing to the risk of developing AD, the entire burden of genetic risk has not been accounted for and additional genetic modifiers of Alzheimer's risk and AAO are likely to exist.
Common confounders in GWAS hamper discoveries of genetic modifiers of disease and include the genetic and phenotypic heterogeneity of complex disease, population stratification, inaccurate phenotyping, misclassification of disease status, missing data and interactions between disease risk, onset and progression. Our present study mitigates most of these confounders by using an expertly phenotyped, geographically isolated founder population affected with a monogenic early-onset form of AD.8 Benefits conferred by our approach include a uniform genetic background enriched for founder haplotypes, and a reduction of environmental confounders such as differences in diet, education and socioeconomic status among study participants. Most importantly, our case-only study design includes individuals carrying a causal mutation in PSEN1 whose onset ages were accurately diagnosed. The accuracy of our phenotypic data was achieved by regular clinical monitoring of mutation carriers from 1995 through the present at minimum intervals of every 2 years.2 Identical diagnostic criteria were used on all participants, enabling accurate definition of distinct stages of the disease. Usage of whole-genome sequencing data rather than genotype microarray or exome sequencing provides a nearly complete assessment of genetic variation in this cohort, and obviates the need for statistical imputation of missing genotypes. The near-complete penetrance of disease risk among mutation carriers in our cohort allows us to disentangle risk from onset, and focus our study on the latter. Taken together, these benefits enhance our power to identify genetic variants modifying Alzheimer's AAO.
Neuroinflammation is a common feature of AD9 and GWAS have associated several immune-related genes with AD including ABCA7, CD33, CLU, CR1, MS4A10, 11 and recently TREM2.12, 13 Most of these associated genes are expressed in microglia,14 the resident immune cells of the brain and mediators of neuroinflammation.15 Growing evidence supports chronic inflammation as a major cause or contributor to Alzheimer's pathology.16 Chemokines, or chemotactic cytokines, are effector molecules of this inflammation.17 In particular, CCL2, a proinflammatory chemokine, is implicated in the chronic neuroinflammation accompanying AD,18 and its cerebrospinal fluid levels predict a faster rate of cognitive decline in AD.19 Identification of genetic variants related to neuroinflammation will inform targeted therapeutic approaches to treat or prevent AD.
Study populations and methods
Familial AD kindred in Antioquia, Colombia
The Colombian Alzheimer's Prevention Initiative registry comprises more than 4000 living members from an extended kindred affected with early-onset familial AD in Antioquia, Colombia. The familial AD subtype caused by the PSEN1 E280A mutation, except for its early-onset phenotype, clinically and pathologically resembles the more common sporadic form of AD, predicting that results obtained in this population may be generalizable to the sporadic AD population comprising 99% of all AD cases. Additional descriptions of the PSEN1 E280A Antioquian population, study participants, and relevant procedures have been previously and extensively described.2, 20 Enrollment in this study was pursuant to approval by the institutional review board at the Universidad de Antioquia and the Western Institutional Review Board.
In total, we obtained DNA from 117 individuals enrolled in the Colombian Alzheimer's Prevention Initiative registry. For these samples, available onset information was superimposed upon previously assembled pedigrees20 and we prioritized individuals with the most extreme AAO. To minimize spurious associations because of relatedness, we excluded one member of each pair of parent–offspring or sibships if concordant for extreme AAO. We did, however, retain three pairs of individuals who were discordant for their AAO. As a quality-control step to identify cryptic relatedness or sequencing duplicates, we estimated all pairwise relatedness by genome-wide identity in the 117 sequenced samples (6786 pairs). This analysis showed that the vast majority (98%) of all pairs of individuals in this cohort are estimated to be third-degree relatives or more distant (Supplementary Figure 1). Although removing related individuals is a common practice in GWAS, including relatives is recommended in certain cases,21 especially in GWAS of a quantitative trait such as AAO. After this selection process, 72 individuals met the study inclusion criteria of having (1) the causal PSEN1 E280A mutation and (2) accurate diagnosis for disease AAO.
UCSF Memory and Aging Center dementia cohort
The University of California San Francisco (UCSF) Memory and Aging Center Dementia cohort consisted of 152 carefully phenotyped older adults meeting the National Institute on Aging-Alzheimer's Association (NIA-AA) criteria for either mild-cognitive impairment22 or probable dementia because of AD.23 All patients underwent a medical history review and physical examination, a structured caregiver interview and neuropsychological tests (Table 1). To confirm the robustness of our findings, we elected to include a range of validated AD phenotypes (memory, executive, language and visuospatial predominant presentations), severity levels (Clinical Dementia Rating Scale of 0–2) and ages. Amyloid imaging biomarkers, obtained via positron emission tomography imaging with Pittsburgh compound B (11C-PiB)24 or Florbetapir F 18 (18F-AV-45)25 were available for 54 individuals, and all were positive. The study was approved by the UCSF institutional review board for human research. Consent for the study was provided by the participants or their assigned surrogate decision makers.
Table 1. Demographics and clinical characteristics of UCSF Alzheimer's Disease Cohort.
| UCSF Alzheimer's Disease Cohort: participant characteristics | Mean (or proportion) (s.d.) |
|---|---|
| Age | 67.8 (10.07) |
| Age at onset | 62.9 (9.70) |
| Age at onset to plasma draw (age gap)a | 4.89 (2.91) |
| Gender (% female) | 54% |
| Education (years) | 16.5 (2.35) |
| MMSE | 22.25 (6.11) |
| CDR | 0: 2% |
| 0.5: 52% | |
| 1.0: 36% | |
| 2.0: 10% | |
| APOE status | ⩾1 ɛ4 alleles: 49% |
| No ɛ4 allele: 51% | |
| Eotaxin levels (pg ml−1) | 173.2 (65.31) |
Abbreviations: APOE, apolipoprotein E; CDR, Clinical Dementia Rating Scale; MMSE, Mini Mental State Examination; UCSF, University of California San Francisco.
Number of years from age of onset until time of plasma draw.
Whole-genome sequencing and quality control
Whole-genome sequencing was performed by Complete Genomics (Mountain View, CA, USA).26 Reads were aligned to the human reference genome (National Center for Biotechnology Information Build 37, Genome Reference Consortium Human Build 37) and variants were called by Complete Genomics. As the average depth of sequencing was >50 × in our study, variant calling was reliable and accurate, with a presumed error rate on par with other studies using the Complete Genomics sequence data.27 Singleton variants are those found in only one individual in the study, are enriched for sequencing errors and cannot be associated with disease statistically; therefore, we excluded these variants from our data set. Note that this filter does not exclude rare variants per se, as variants rare in the general population may still be enriched in our geographically isolated founder population.
Association analyses
We used PLINK (v.1.07)28 to fit a linear regression model between genotypes and AAO of mild-cognitive impairment (MCI) as a quantitative trait. We also examined a model with covariate adjustment accounting for the number of APOE ɛ4 alleles (0, 1 or 2). The primary determinants of statistical power in GWAS include sample size and the frequency and effect size of the contributing variants. To address the small sample size in this study, we considered the significance threshold in a few ways. In the most conservative approach, we applied a Bonferroni-adjusted significance level of P<5E−08 to determine genome-wide significance. Another approach is to apply a threshold based on the effective number of independent tests. This threshold has been estimated at P<7.2E−08 for European populations.29 Because all carriers of the causal PSEN1 E280A mutation in the Colombian Alzheimer's cohort descended from a common ancestor of European descent,20 applying this empirical threshold is well justified in our cohort. Finally, we performed computationally intensive permutation procedures based on swapping phenotype labels to generate empirical P-values.
Given the recent founder effect of this population and similar ancestry in this population, we did not adjust for population stratification in our model. Also, with the high endogamy and consanguinity in the cohort, we could not remove all related individuals from the analysis without markedly reducing our sample size. To address this, we performed association testing with the Mendel software package (v.14.3) using the Ped-GWAS option30 designed for use with a mixture of related and unrelated individuals. We performed association testing with the permutation option (—perm 1 000 000) to generate empirical P-values. We also conducted a separate linear regression analysis between genotypes and AAO of dementia, although only 60 individuals met inclusion criteria for this variable, limiting our sample size and hence detection power.
For the UCSF cohort, we implemented a general linear model, using AAO as the outcome variable, Clinical Dementia Rating (CDR) and APOE genotype as covariates, and eotaxin-1 levels as the primary predictor variable. We also controlled for patient age by including a variable representing the difference between disease onset and the age at which the samples were drawn (referred to hereafter as an 'age gap' variable). Statistical analyses were performed using the SAS software (SAS Institute, Cary, NC, USA).
CCL11 secretion assay
To determine the effect of the CCL11 A23T mutation on the secretion of this chemokine, wild-type and mutant coding sequences (CDS) of CCL11 (NM_002986) encoding the eotaxin precursor protein were cloned into a mammalian expression vector containing GFP (pEGFP-N1) and verified by Sanger sequencing. Four million HEK-293T cells were seeded onto a 10-cm dish the day before transfection with 8 μg of expression vector. Media were changed after 12 h, and the supernatant was collected 24 h after this for analysis. Transfections were performed in biological quadruplicate for each construct.
Genotyping
TaqMan single-nucleotide polymorphism (SNP) genotyping assays (Applied Biosystems, Foster City, CA, USA) for rs1129844 and rs9909184 and APOE were performed on genomic DNA extracted from the UCSF cohort.
ELISA measurements of eotaxin-1 levels in UCSF cohort
Following collection, each blood sample was centrifuged at 2000 g for 15 min at 4 °C with the resultant plasma divided into 500 μl aliquots and stored at −80 °C. All assays were conducted following the manufacturer's protocol for Human Chemokine Panel 1V-PLEX Plus Kit (Meso Scale Diagnostics, Rockville, MD, USA). Each multiplex array was scanned using an MESO QuickPlex SQ 120 (Meso Scale Diagnostics, Rockville, MD, USA). Manufacturer supplied software (Discover Workbench 4.0) was used to quantify the concentration of eotaxin-1 based on sample dilution and relative to the supplied in-assay standard curve. Nominal recovery for control eotaxin-1 levels remained between 111 and 120% with a coefficient of variation <10%. Eotaxin-1 coefficient of variation remained below 10% for 91%, and below 20% for 98% across all patient samples. Standard curve coefficients of variation for patient sample detection range remained below 10% with standard sample recovery at 100% (±5%) across all plates.
Functional variant prediction in the extremes for AAO
Individuals whose AAO manifests at the extremes of the trait distribution in the Antioquian familial AD kindred may harbor large-effect genetic variants modulating AAO that can be bioinformatically identified.31 Therefore, we categorized each individual as early- or late-onset for MCI if their AAO exceeded 1 s.d. of the population mean onset, in either direction. This threshold-based approach resulted in 12 and 16 individuals being classified as early and late onset, respectively, for MCI. We filtered the observed genetic variants by their frequency among early- and late-onset individuals, requiring the presence of a variant in at least four individuals of one category and its absence from the other group.
These filters yielded 23 726 variants, which we then annotated using ANNOVAR.32 Only 121 variants fell within protein-coding regions. We further filtered these variants based on their mappability scores and their false-positive rates in other whole-genome sequencing studies. We used Kaviar to assess the frequency of variants in the general population.33 None of the coding variants were predicted to cause protein truncation (nonsense variants) or frameshifts. Rather they encoded substitutions including 26 nonsynonymous variants. The nonsynonymous variant class of mutations comprises the majority of variants underlying human inherited disease,34 and may affect protein function. We predicted whether the identified nonsynonymous variants were likely to be deleterious to protein function using the SIFT35 and CADD36 algorithms, which are based on a variety of metrics including sequence conservation across species. An overview of these filtering steps is depicted in Supplementary Figure 2.
Results
Association of a novel locus on chromosome 17 modifying Alzheimer's AAO
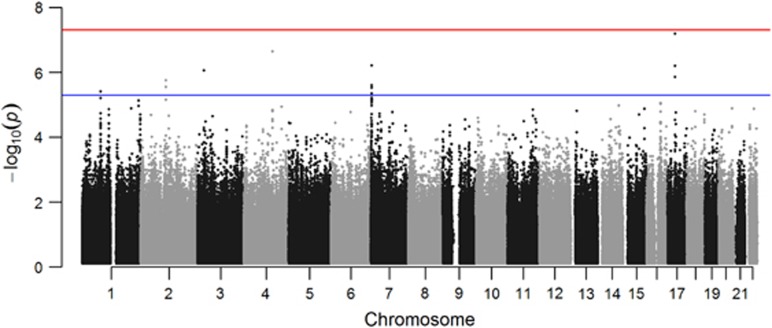
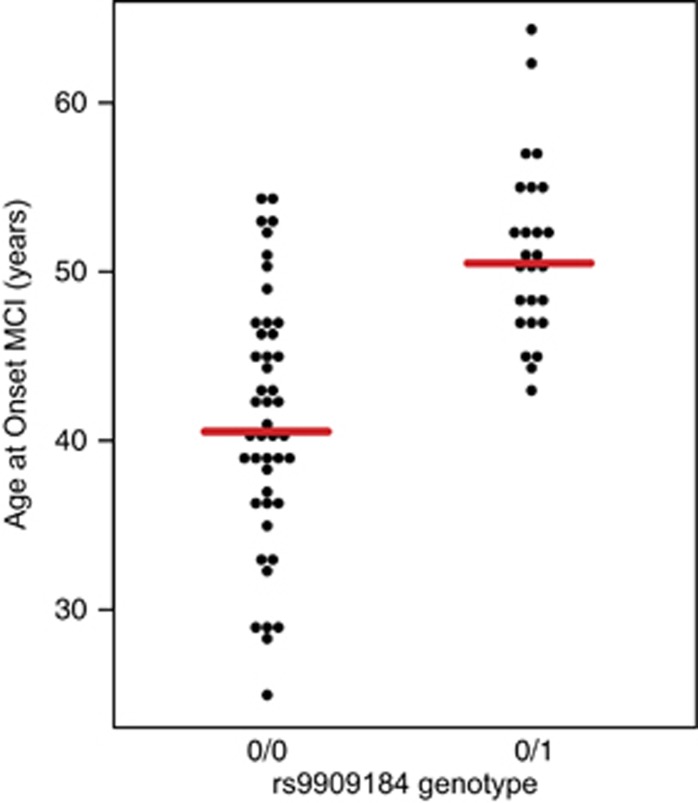
Association analysis revealed a cluster of single-nucleotide variants comprising a haplotype on chromosome 17 approaching genome-wide significance for association with AAO of MCI (best P-value 6.43E−08). This P-value is genome-wide significant at the empirical threshold for European populations29 and was well supported by association of multiple variants in the region (Figure 1 and Supplementary Table 1). Adjusted for APOE status, the regression model identified the same peak at chromosome 17 and improved the best P to a genome-wide significant 4.85E−08. APOE ɛ4 was not significant in this model. Permutation testing empirically supported the chromosome 17 peak as the top associated locus in our study (P=1E−06, the lowest achievable P-value; Supplementary Table 2). The regression model using AAO of dementia as the trait of interest also indicated this same region as the top hit, although the overall P-value was increased as a result of the diminished sample size for this variable (Supplementary Table 3). The Mendel software program, accounting for relatedness among the samples, identified the same locus as the top associated region in this study and achieved genome-wide significance (P=2.86E−08; Supplementary Table 4). Remarkably, the effect size of this haplotype was almost 10 years of delayed onset in carriers. Average AAO of MCI in carriers of the protective haplotype was 51.0±5.2 years compared with non-carriers age at MCI 41.1±7 years (mean±s.d.) (Figure 2).
Figure 1.
Manhattan plot of genome-wide association results for age of mild-cognitive impairment in our cohort. Red line indicates genome-wide significance (P<5E−08) and blue line indicates nominally associated loci (P<5E−06).
Figure 2.
Carriers of rs9909184, the top associated single-nucleotide polymorphism (SNP) in this study and marker of the protective haplotype, show a ~10-year delay in age at onset for mild-cognitive impairment (MCI).
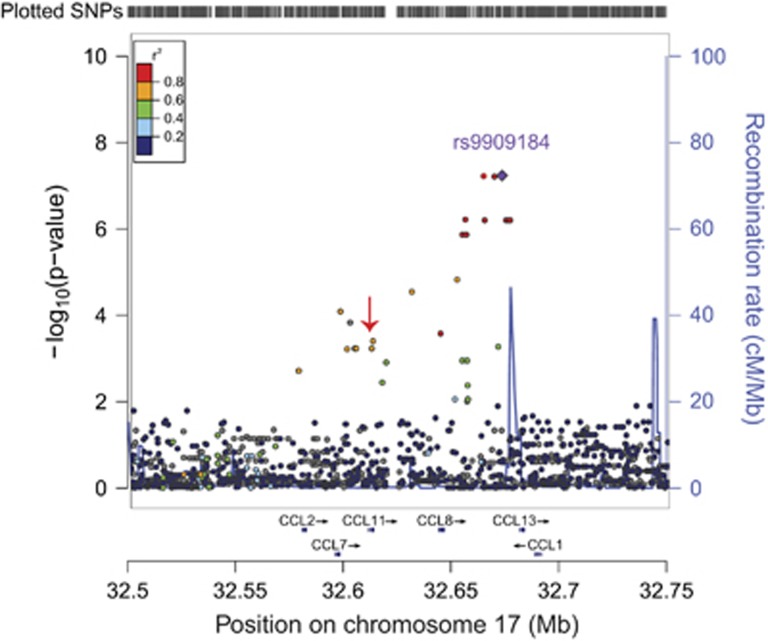
Examination of the regional association plot on chromosome 17 revealed that the associated haplotype spans several chemokines, including some proinflammatory factors reported to be elevated in AD (Figure 3).37 One chemokine subfamily, the monocyte chemoattractant proteins (MCPs), is implicated in neuronal death during sustained inflammation.38 Four major MCPs reside within the genomic neighborhood associated in this study—MCP1 encoded by the CCL2 gene, MCP2 encoded by CCL7 gene, MCP3 encoded by CCL8 gene and MCP4 encoded by CCL13 gene. Two additional genes in this locus include CCL1, encoding a well-characterized proinflammatory chemokine,39 and CCL11, which encodes eotaxin-1, a chemokine whose serum and cerebrospinal fluid levels increase with age and correlate with reduced neurogenesis,40 making it a promising candidate for modifying Alzheimer's AAO.
Figure 3.
Regional association plot on chromosome 17 at the top associated single-nucleotide polymorphism (SNP) for age at onset of mild-cognitive impairment reveals the associated locus is a haplotype spanning a chemokine gene cluster. This haplotype includes a missense polymorphism (A23T) in the coding region of CCL11 (red arrow).
To identify the genetic variants potentially causal for the large-effect size of the association peak, we functionally annotated all single-nucleotide variants in linkage disequilibrium with our association peak. The haplotype associated with AAO of MCI comprises 22 single-nucleotide variants spanning almost 80 kb (Figure 3 and Supplementary Table 5). Of these, the only single-nucleotide variant located within the coding portion of a gene, rs1129844 (NG_012212.1:g.5208G>A, NP_002977.1:p.Ala23Thr), encodes an alanine-to-threonine missense polymorphism at codon 23 in eotaxin-1 (CCL11 A23T). This missense polymorphism lies directly at the signal peptide cleavage site of the eotaxin-1 precursor protein and is predicted to alter its cleavage in silico (Supplementary Figure 3).41 Differential secretion of mutant and wild-type eotaxin-1 in HEK-293T cells suggests that this mutation may enhance the secretion of eotaxin-1 (Supplementary Figure 3).
Association of eotaxin-1 levels with Alzheimer's AAO and effect of the associated haplotype on eotaxin-1 levels in sporadic AD
A prerequisite for generalizability of our results is the presence of this allele in the general population. Within the Colombian data set, rs1129844 (G>A) was observed with a minor allele frequency of 0.14. In the general population, the minor allele frequency for rs1129844 is identical (0.14), making this variant a fairly common polymorphism with a prevalence in the general population matching that of APOE ɛ4 (minor allele frequency=0.15). Given the presence of this haplotype in the general population, we tested whether plasma levels of eotaxin-1 correlate with AAO in an independent Alzheimer's cohort at the UCSF Memory and Aging Center (n=152). After controlling for CDR, APOE ɛ4 genotype and the age gap variable, we found that higher plasma eotaxin-1 levels were significantly correlated with higher AAO (F(6, 145)=2.81; P=0.012; β=0.0252, s.e.=0.0122; t-value=2.07, F-test for linear regression). APOE ɛ4 genotype was not significant in this model.
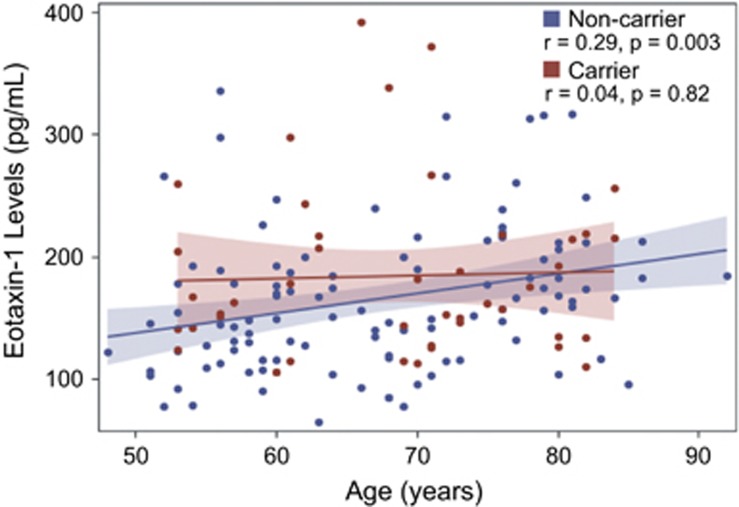
Finally, we asked whether the haplotype identified in the Antioquian early-onset Alzheimer's cohort affects eotaxin-1 levels in the UCSF Memory and Aging Center Alzheimer's cohort. In agreement with previous studies,40 eotaxin-1 levels increased with age in the total cohort. However, when we stratified these samples by the presence of the onset-associated haplotype, we found that this haplotype decoupled the relationship between age and plasma levels of eotaxin-1 (Figure 4). In other words, we observed a linear increase in eotaxin-1 levels for non-haplotype carriers but not for haplotype carriers. This decoupling suggests that this haplotype exerts a complex regulatory effect on eotaxin-1 levels. Although the age-associated increase in eotaxin-1 levels has been correlated with reduced neurogenesis and memory impairment,40 eotaxin-1 may elicit a hormetic response curve with normal or even neuroprotective effects within a certain interval, and deleterious effects at higher levels. By abrogating the linear increase between eotaxin-1 levels and age, the haplotype identified in this study may effectively constrain eotaxin-1 levels within this normal/protective realm. We observed a trend toward a protective effect of the associated haplotype on AAO in the UCSF cohort, although this effect was not significant (Supplementary Figure 5).
Figure 4.
The associated haplotype decouples plasma eotaxin-1 levels from age in haplotype carriers. Shaded regions show 95% confidence limits for the mean.
In silico identification of functional variants in extreme onset individuals
A complementary approach to association testing is bioinformatically screening the catalog of genetic variation we generated through whole-genome sequencing to identify putative functional variants. This bioinformatic screen for functional variants present in individuals at one extreme for AAO predicted eight variants to be deleterious to protein function (SIFT score ⩽0.1) (Supplementary Table 6). Among these was rs150955128 (NM_000418.3:c.554G>A, NP_000409.1:p.Arg185His), an arginine-to-histidine amino-acid substitution at codon 185 of the interleukin 4 receptor (IL4R). The variant is present in seven of the late-onset genomes and absent from the early-onset genomes. An additional 8 of the 72 PSEN1 mutation carriers also have this variant. Of the predicted deleterious variants, carriers of rs150955128 displayed the largest difference in AAO versus non-carriers (50.5 versus 43.2 years), suggesting a protective effect in carriers of this variant. This variant is present with a minor allele frequency around 1% (G>A) but the Exome Aggregation Consortium browser (http://exac.broadinstitute.org/) revealed that the variant is largely restricted to Latino populations and virtually non-existent in Asian, African or European populations. This finding also underscores the importance of including racially and ethnically diverse populations in GWAS.42
Discussion
Through whole-genome sequencing of 72 individuals affected with early-onset familial AD caused by an E280A mutation in PSEN1, we have identified a haplotype on chromosome 17 associated with delayed AAO of MCI and dementia. The identified haplotype spans several chemokines, including CCL2, a proinflammatory chemokine implicated in the chronic neuroinflammation accompanying AD.18 In our study, the associated haplotype confers ~10 years of protection against AD onset among carriers in the Antioquian early-onset AD cohort. In the general population, this haplotype is relatively common, with an expected prevalence of around one in four people. Within this haplotype, we identified a missense polymorphism in eotaxin-1 (CCL11 A23T) lying directly at the signal peptide cleavage site and therefore we predict this variant may alter the cleavage and secretion of this chemokine. Indeed, we found enhanced secretion of the variant protein in an in vitro expression study. Extensive pathological commonalities between the Antioquian familial AD and the general form of the disease, and the relatively common prevalence of the identified haplotype, lend hope that this haplotype functions similarly (i.e., protectively) in the general population. In the UCSF cohort, we observed a protective trend for this haplotype on AAO, which requires a larger sample to validate.
Eotaxin-1 levels increase throughout life and this increase is correlated with reduced neurogenesis.40 This raises the intriguing possibility that eotaxin-1 is a molecular effector of aging, the largest risk factor for developing AD. Here we show that plasma eotaxin-1 levels are correlated with AAO in an independent cohort of Alzheimer's patients, implicating this chemokine as a novel modulator of Alzheimer's AAO. Furthermore, carriers of the chromosome 17 haplotype in the UCSF cohort did not exhibit the typical increase of chemokine levels with age. As chemokines mediate both neuroprotection and injury,43 we postulate a hormesis response model for eotaxin-1 levels whereby low to moderate levels of this chemokine elicit a normal or protective response, but higher levels ultimately lead to neurodegeneration and memory impairment. By decoupling eotaxin-1 levels from age, the haplotype identified in this study may protect against the deleterious effects accompanying high levels of this chemokine.
The protein product of CCL11 resulting from differential signal peptide cleavage is predicted to retain additional amino-acid residues at its N terminus, a key region specifying binding and activity of chemokines.44 Recent structural studies have implicated a critical role for this region of eotaxin-1 in binding and activation of its receptor CCR3.45 Similar to other AD-associated immune-response genes, this receptor is expressed highly in microglia.14 Therefore, it is reasonable to hypothesize altered eotaxin-1 signaling modulates neuroinflammation among carriers of the CCL11 A23T mutation.
Individuals at the extremes of a trait distribution are likely to be enriched for functional variants underlying that trait. Through bioinformatic prediction of variants likely to alter gene function, we identified a Latino-specific variant in IL4R enriched in the latest-onset individuals in this study. Similar to the eotaxin-1 receptor, IL4R is also highly expressed in microglia14 and its ligand, the anti-inflammatory cytokine interleukin-4, attenuates AD symptoms in transgenic mice.46 Another ligand for IL4R is interleukin-13. Taken together, interleukin-4 and interleukin-13 induce clearance of β-amyloid and improve memory in transgenic mice.47 As these cytokines are known to cause the release of eotaxin-1 in certain human cell types upon binding IL4R,48 we suggest that eotaxin-1 release may mediate β-amyloid clearance. This finding thus dovetails with our implication of eotaxin-1 in modulating AAO, and highlights cytokine-mediated neuroinflammation as a promising pathway for therapeutic intervention in AD.
Clarifying the precise relationship between the identified haplotype and eotaxin-1 levels, and identifying the primary cellular sources and regulators of this chemokine in both healthy and diseased states, warrant further study. As the incidence of AD doubles approximately every 5 years,49 delaying the onset by this amount would consequently halve the disease incidence. In this study, we identified a protective haplotype conferring a ~10-year protective effect in one monogenic early-onset Alzheimer's population. Therapies based on this protective haplotype offer the potential to reduce markedly the incidence of AD while enhancing the quality of life of millions of individuals.
Acknowledgments
We gratefully acknowledge all the patients and their family members who enabled this study. This study was supported by the University of Luxembourg–Institute for Systems Biology Program, Colciencias (proyecto 115-408-20543 and contracto 277–2007). Dr. Bettcher was supported by NIH-NIA grant K23 AG042492-01 and Alzheimer‘s Association grant NIRP-12-259223. We also thank a generous donor for enabling the Meso Scale Diagnostics equipment purchase.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
Supplementary Material
References
- 1Lo AW, Ho C, Cummings J, Kosik KS. Parallel discovery of Alzheimer's therapeutics. Sci Transl Med 2014; 6: 3008228. [DOI] [PubMed] [Google Scholar]
- 2Acosta-Baena N, Sepulveda-Falla D, Lopera-Gomez CM, Jaramillo-Elorza MC, Moreno S, Aguirre-Acevedo DC et al. Pre-dementia clinical stages in presenilin 1 E280A familial early-onset Alzheimer's disease: a retrospective cohort study. Lancet Neurol 2011; 10: 213–220. [DOI] [PubMed] [Google Scholar]
- 3Li YJ, Scott WK, Hedges DJ, Zhang FY, Gaskell PC, Nance MA et al. Age at onset in two common neurodegenerative diseases is genetically controlled. Am J Hum Genet 2002; 70: 985–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Pedersen NL, Posner SF, Gatz M. Multiple-threshold models for genetic influences on age of onset for Alzheimer disease: findings in Swedish twins. Am J Med Genet 2001; 105: 724–728. [DOI] [PubMed] [Google Scholar]
- 5Marchani EE, Bird TD, Steinbart EJ, Rosenthal E, Yu CE, Schellenberg GD et al. Evidence for three loci modifying age-at-onset of Alzheimer's disease in early-onset PSEN2 families. Am J Med Genet B 2010; 153B: 1031–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 1993; 261: 921–923. [DOI] [PubMed] [Google Scholar]
- 7Pastor P, Roe CM, Villegas A, Bedoya G, Chakraverty S, Garcia G et al. Apolipoprotein Eepsilon4 modifies Alzheimer's disease onset in an E280A PS1 kindred. Ann Neurol 2003; 54: 163–169. [DOI] [PubMed] [Google Scholar]
- 8Lopera F, Ardilla A, Martinez A, Madrigal L, Arango-Viana JC, Lemere CA et al. Clinical features of early-onset Alzheimer disease in a large kindred with an E280A presenilin-1 mutation. JAMA 1997; 277: 793–799. [PubMed] [Google Scholar]
- 9Heneka MT, Carson MJ, Khoury JE, Landreth GE, Brosseron F, Feinstein DL et al. Neuroinflammation in Alzheimer's disease. Lancet Neurol 2015; 1: 388–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet 2011; 43: 429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet 2009; 41: 1094–1099. [DOI] [PubMed] [Google Scholar]
- 12Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E et al. TREM2 variants in Alzheimer's disease. N Engl J Med 2013; 368: 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J et al. Variant of TREM2 associated with the risk of Alzheimer's disease. N Engl J Med 2013; 368: 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 2014; 34: 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Gonzalez-Scarano F, Baltuch G. Microglia as mediators of inflammatory and degenerative diseases. Annu Rev Neurosci 1999; 22: 219–240. [DOI] [PubMed] [Google Scholar]
- 16Krstic D, Knuesel I. Deciphering the mechanism underlying late-onset Alzheimer disease. Nat Rev Neurol 2013; 9: 25–34. [DOI] [PubMed] [Google Scholar]
- 17Baggiolini M, Dewald B, Moser B. Human chemokines: an update. Annu Rev Immunol 1997; 15: 675–705. [DOI] [PubMed] [Google Scholar]
- 18Sokolova A, Hill MD, Rahimi F, Warden LA, Halliday GM, Shepherd CE. Monocyte chemoattractant protein-1 plays a dominant role in the chronic inflammation observed in Alzheimer's disease. Brain Pathol 2009; 19: 392–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Westin K, Buchhave P, Nielsen H, Minthon L, Janciauskiene S, Hansson O. CCL2 is associated with a faster rate of cognitive decline during early stages of Alzheimer's disease. PLoS One 2012; 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Lalli MA, Cox HC, Arcila ML, Cadavid L, Moreno S, Garcia G et al. Origin of the PSEN1 E280A mutation causing early-onset Alzheimer's disease. Alzheimers Dement 2013; 13: 02840–02849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Visscher PM, Andrew T, Nyholt DR. Genome-wide association studies of quantitative traits with related individuals: little (power) lost but much to be gained. Eur J Hum Genet 2008; 16: 387–390. [DOI] [PubMed] [Google Scholar]
- 22Albert MS, DeKosky ST, Dickson D, Dubois B, Feldman HH, Fox NC et al. The diagnosis of mild cognitive impairment due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement 2011; 7: 270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR Jr, Kawas CH et al. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement 2011; 7: 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24Klunk WE, Engler H, Nordberg A, Wang Y, Blomqvist G, Holt DP et al. Imaging brain amyloid in Alzheimer's disease with Pittsburgh Compound-B. Ann Neurol 2004; 55: 306–319. [DOI] [PubMed] [Google Scholar]
- 25Wong DF, Rosenberg PB, Zhou Y, Kumar A, Raymont V, Ravert HT et al. In vivo imaging of amyloid deposition in Alzheimer disease using the radioligand 18 F-AV-45 (florbetapir [corrected] F 18). J Nucl Med 2010; 51: 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 2010; 327: 78–81. [DOI] [PubMed] [Google Scholar]
- 27Roach JC, Glusman G, Smit AF, Huff CD, Hubley R, Shannon PT et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science 2010; 328: 636–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet Epidemiol 2008; 32: 227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30Lange K, Papp JC, Sinsheimer JS, Sripracha R, Zhou H, Sobel EM. Mendel: the Swiss army knife of genetic analysis programs. Bioinformatics 2013; 29: 1568–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31Lalli MA, Garcia G, Madrigal L, Arcos-Burgos M, Arcila ML, Kosik KS et al. Exploratory data from complete genomes of familial Alzheimer disease age-at-onset outliers. Hum Mutat 2012; 33: 1630–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010; 38: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33Glusman G, Caballero J, Mauldin DE, Hood L, Roach JC. Kaviar: an accessible system for testing SNV novelty. Bioinformatics 2011; 27: 3216–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34Krawczak M, Ball EV, Fenton I, Stenson PD, Abeysinghe S, Thomas N et al. Human gene mutation database-a biomedical information and research resource. Hum Mutat 2000; 15: 45–51. [DOI] [PubMed] [Google Scholar]
- 35Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res 2003; 31: 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014; 46: 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37Ishizuka K, Kimura T, Igata-yi R, Katsuragi S, Takamatsu J, Miyakawa T. Identification of monocyte chemoattractant protein-1 in senile plaques and reactive microglia of Alzheimer's disease. Psychiatry Clin Neurosci 1997; 51: 135–138. [DOI] [PubMed] [Google Scholar]
- 38Conductier G, Blondeau N, Guyon A, Nahon JL, Rovere C. The role of monocyte chemoattractant protein MCP1/CCL2 in neuroinflammatory diseases. J Neuroimmunol 2010; 224: 93–100. [DOI] [PubMed] [Google Scholar]
- 39Luo Y, Laning J, Devi S, Mak J, Schall TJ, Dorf ME. Biologic activities of the murine beta-chemokine TCA3. J Immunol 1994; 153: 4616–4624. [PubMed] [Google Scholar]
- 40Villeda SA, Luo J, Mosher KI, Zou B, Britschgi M, Bieri G et al. The ageing systemic milieu negatively regulates neurogenesis and cognitive function. Nature 2011; 477: 90–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 2011; 8: 785–786. [DOI] [PubMed] [Google Scholar]
- 42Haga SB. Impact of limited population diversity of genome-wide association studies. Genet Med 2010; 12: 81–84. [DOI] [PubMed] [Google Scholar]
- 43Wang J, Asensio VC, Campbell IL. Cytokines and chemokines as mediators of protection and injury in the central nervous system assessed in transgenic mice. Curr Top Microbiol Immunol 2002; 265: 23–48. [DOI] [PubMed] [Google Scholar]
- 44Crump MP, Gong JH, Loetscher P, Rajarathnam K, Amara A, Arenzana-Seisdedos F et al. Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1. EMBO J 1997; 16: 6996–7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45Millard CJ, Ludeman JP, Canals M, Bridgford JL, Hinds MG, Clayton DJ et al. Structural basis of receptor sulfotyrosine recognition by a CC chemokine: the N-terminal region of CCR3 bound to CCL11/eotaxin-1. Structure 2014; 22: 1571–1581. [DOI] [PubMed] [Google Scholar]
- 46Kiyota T, Okuyama S, Swan RJ, Jacobsen MT, Gendelman HE, Ikezu T. expression of anti-inflammatory cytokine interleukin-4 attenuates Alzheimer's disease-like pathogenesis in APP+PS1 bigenic mice. FASEB J 2010; 24: 3093–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47Kawahara K, Suenobu M, Yoshida A, Koga K, Hyodo A, Ohtsuka H et al. Intracerebral microinjection of interleukin-4/interleukin-13 reduces beta-amyloid accumulation in the ipsilateral side and improves cognitive deficits in young amyloid precursor protein 23 mice. Neuroscience 2012; 207: 243–260. [DOI] [PubMed] [Google Scholar]
- 48Moore PE, Church TL, Chism DD, Panettieri RA Jr, Shore SA. IL-13 and IL-4 cause eotaxin release in human airway smooth muscle cells: a role for ERK. Am J Physiol Lung Cell Mol Physiol 2002; 282: L847–L853. [DOI] [PubMed] [Google Scholar]
- 49Kukull WA, Higdon R, Bowen JD, McCormick WC, Teri L, Schellenberg GD et al. Dementia and Alzheimer disease incidence: a prospective cohort study. Arch Neurol 2002; 59: 1737–1746. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.