Abstract
Nanobodies are the recombinant antigen-recognizing domains of the minimalistic heavy chain-only antibodies produced by camels and llamas. Nanobodies can be easily generated, effectively optimized, and variously derivatized with standard molecular biology protocols. These properties have triggered the recent explosion in the nanobody use in basic and clinical research. This review focuses on the emerging use of nanobodies for understanding and monitoring protein dynamics on the scales ranging from isolated protein domains to live cells, from nanoseconds to hours. The small size and high solubility make nanobodies uniquely suited for studying protein dynamics by NMR. The ability to produce conformation-sensitive nanobodies in cells enables studies that link structural dynamics of a target protein to its cellular behavior. The link between in vitro and in-cell dynamics, afforded by nanobodies, brings the analysis of such important events as receptor signaling, membrane protein trafficking, and protein interactions to the next level of resolution.
Keywords: nuclear magnetic resonance (NMR); protein domain; protein dynamic; single-domain antibody (sdAb, nanobody); X-ray crystallography; nanobody; protein dynamics; protein structure; protein engineering
Introduction
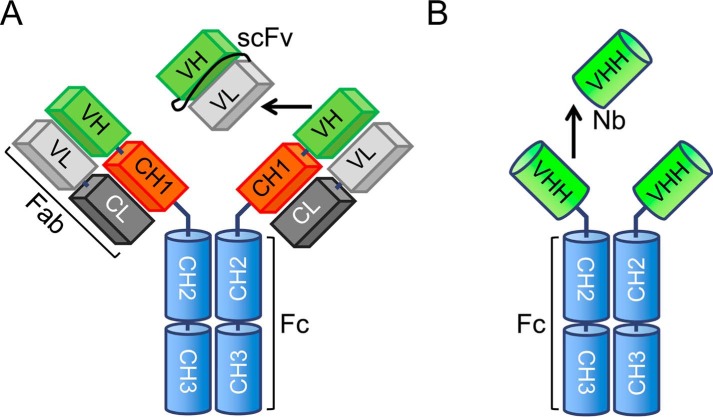
In addition to the conventional IgG antibodies, camelids and sharks produce unusual antibodies that lack light chains (Fig. 1, A and B) (1). These heavy chain-only antibodies retain full antigen specificity and binding affinity and can be further truncated without significant loss of their antigen-recognizing properties to produce isolated variable domains (VHH).2 The VHHs from camelids were initially trademarked as Nanobodies, but VHHs from llama are now more commonly employed for research. VHHs are also known as single domain antibodies.
FIGURE 1.
Domain structure of IgG antibodies (A) and heavy chain only camelid antibodies (B). The isolated variable domain of the latter is called nanobody.
Several unique properties of VHHs define their potential as research tools distinct from the conventional antibodies. First, nanobodies are much smaller. Although the IgG antibody consists of four chains with multiple domains and has a molecular mass of about 150 KDa, the nanobody is a single domain with a molecular mass of only about 15 KDa. Second, nanobodies can be easily screened for affinity and specificity using a wide spectrum of approaches ranging from phage display to NMR. Most importantly, nanobodies can be cloned, genetically or chemically modified, and produced in a recombinant form in various cells, and, potentially, in live organisms. Bacterial expression systems enable generation of purified nanobodies in milligram quantities per liter of culture, offering an unlimited supply of a selective reagent with consistent properties.
Importantly, nanobodies can be used to trace in real time and manipulate localization and activity of target proteins in eukaryotic cells (2–4). This allows correlating protein structural dynamics in vitro with the behavior of proteins in cells. The focus of this review is on the novel nanobody applications that span the full temporal and spatial scales of protein dynamics, from nanoseconds to hours, from isolated domains to whole cells. Such applications range from studies of fast protein dynamics by NMR, to detection and stabilization of functionally important transient protein conformations, to manipulating protein trafficking in the cell.
Generation of Nanobodies
A detailed review of nanobody production (Fig. 2) has been published recently (5). Briefly, llamas (or camels) are immunized with the antigen protein. When immune response develops, mRNA is isolated from lymphocytes, and a cDNA library of variable heavy chain domains is created by reverse transcription. The cDNA is used to express VHH as fusions with phage coat proteins (phage display), and the nanobodies are enriched by one or more rounds of panning against the immobilized antigen. Routinely, the selected nanobodies are expressed in Escherichia coli with a hexahistidine tag added to allow purification by nickel-nitrilotriacetic acid affinity chromatography, and a secretion signal sequence inserted to direct the expressed protein to the periplasm for easier purification and to enable disulfide bond formation.
FIGURE 2.
Schematic representation of the nanobody generation process, starting with the immunization of a camelid. PBMC, peripheral blood mononuclear cells; Ni-NTA, nickel-nitrilotriacetic acid; Nb, nanobody; IMAC, immobilized metal affinity chromatography; SEC, size exclusion chromatography.
Shark heavy chain antibodies IgNAR have also been used to derive single domain antibodies, which possess similar antigen binding mode to camelid VHHs (6), and have many of the same advantages as research tools and potential therapeutic agents (7).
Nanobodies Versus Fragments of the Conventional Antibodies as Tools for Structural Biology
The conventional antibodies, exemplified by the most common isotype IgG, consist of two heavy and two light chains (Fig. 1A) associated through the disulfide bonds and non-covalent interactions. The binding properties of the antibodies are defined by the six complementarity-determining regions (CDRs) located within the variable domains of the heavy (VH) and light (VL) chains. The large size, multichain composition, and requirement for essential disulfide bond formation complicate production of the recombinant IgGs. Two smaller antibody fragments have been developed. The Fab has a molecular mass of about 50 KDa and consists of the light chain and a truncated heavy chain, including the two variable domains. The smallest practical derivative of a conventional antibody, scFv (∼25 KDa), was produced by connecting VH and VL with an artificial peptide linker.
Because the association between the variable domains of the light and the heavy chains depends on the hydrophobic interactions, production and applications of the single isolated variable domains are hampered by poor protein solubility and aggregation. Although it is possible, in principle, to modify the amino acid sequence of VH or VL to eliminate the hydrophobic patches on the protein surface (8), such work is time-consuming and may be derailed by the unintended effects of such mutations. The heavy chain-only camelid antibodies, where variable domains (VHH) do not interact with any other domains, offer a much more practical starting point for producing the single-domain antibodies. Indeed, unlike the isolated human VH domains, nanobodies are highly soluble. In turn, solubility and folding properties of the human VH domains can be improved by introducing a few mutations at the contact surface with the VL domain based on the sequence comparison to the camelid VHH (8, 9).
The antibody-derived fragments, in particular Fab, have been widely used in structural biology to assist crystallization of difficult proteins, including several by now classic structures (10–12). Antibodies can promote crystallization by inducing order in the flexible regions of the target protein, improving crystal contacts, and increasing the hydrophilic surface area of the complex. Nanobodies can be used to assist protein crystallization essentially in the same way. Examples of the protein structures solved with the help of nanobodies include several membrane receptors and transporters, proteins of bacterial secretion systems, and others (13–20).
Nanobodies with known binding sites can be employed to map location of individual domains or proteins by cryo-electron microscopy (21). When the epitope is located at the interface of the interacting domains or subunits of a protein, the nanobody can be used to disrupt domain interactions (22). The main advantage of the nanobodies in such conventional applications is that a large panel of nanobodies can be easily screened to identify preferred epitopes or the best co-crystals between a nanobody and the target protein, or to reveal and stabilize unknown target conformers (23).
Nanobodies Versus Specific Binding Proteins Designed on Non-antibody Scaffolds
Natural antibodies inspired design of engineered proteins that bind to their targets with high affinity and specificity. In these constructs, a small protein domain with a high natural propensity for protein interactions is used as a scaffold for the target-specific binding sequences similar to the CDRs of the natural antibodies. Initially, a large library of potential binders with a partially randomized amino acid sequence in the binding site is created. The high-affinity binders are then selected by phage display or, recently, by ribosome display (24). Examples of such scaffold-protein affinity reagents (SPARs) (25) include DARPins (designed ankyrin repeat proteins). (26), monobodies, designed on the scaffold of human fibronectin III domain 3 (27), and also Affibody molecules (28) and anticalins (29).
Crystal structures of various SPARs in complex with their targets have been solved (30, 31), revealing the binding mode and illustrating their potential applications in structural biology. DARPins (32–34) and monobodies (35, 36) have been used to solve x-ray structures of such challenging targets as membrane transporters. Nanobodies and SPARs share many of the same advantages, such as small size, single domain composition, and the ease of producing recombinant proteins. Production of SPARs does not involve animal immunization, a cost-saving factor. On the other hand, nanobodies have inherently high affinity, whereas to achieve comparably high SPAR affinity, much larger synthetic libraries have to be generated and screened, a potentially daunting task. Some knowledge of the binding mode for the given target would allow using smaller biased libraries, but such knowledge is often unavailable for complex membrane proteins from mammalian cells. Another consideration in choosing between various SPARs and nanobodies is the preference for different epitope architectures, which is to a large extent determined by the shape of the scaffold protein (30). This aspect is discussed below in more detail for the nanobodies.
Structural Basis of the Distinctive Binding Properties of the Nanobodies
The VHH domain is composed of a folded β-sheet with three loops in the regions homologous to the CDRs of the IgG VH domains (Fig. 1). The length of CDR3 loop in the VHH can exceed 20 amino acid residues, as compared with the typical length of 9 (mouse) or 12 (human) in the conventional antibodies. The longer CDR3 loop can insert into the partially buried binding sites, as first seen in the structure of lysozyme in complex with a nanobody (37) (Fig. 3) (structure diagrams were generated using MOLMOL (38) and PyMOL (39)), and then in some other nanobody complexes (40–43). However, the binding mode of the nanobodies and the length of CDR3 loop can vary greatly (44, 45).
FIGURE 3.

Structure of a nanobody (orange) in complex with lysozyme (green). CDR loops 1–3 are shown in light blue, navy, and red respectively. H-bonds between the nanobody and lysozyme are shown as blue bars. Note that out of nine intermolecular H-bonds, eight involve CDR3.
A longer CDR3, the convex shape of the antigen-binding site, and the small size allow nanobodies to access epitopes that may be cryptic and non-antigenic for conventional antibodies. In fact, unlike the conventional antibodies, which more often detect linear or planar epitopes, many nanobodies bind to concave and discontinuous epitopes that only form in the folded protein (45). This property makes nanobodies valuable tools for probing conformational states of the target proteins, both in vitro and in cells. Nanobodies that bind discontinuous epitopes can be selected from the initial panel by competition with linear peptides or with an unfolded protein or by using the masked selection technique (46). Such nanobodies are particularly useful for stabilizing folded intermediates and specific protein conformations.
An interesting consequence of the preference for the discontinuous epitopes has been observed in the structure of the nanobody complex with a heterodimer of editosome proteins A3 and A6. These two proteins have overall low level of sequence identity, but a similar constellation of the essential residues in two different binding pockets, which caused nanobody binding to both A3 and A6 (19). This phenomenon can be exploited to select nanobodies against substrate- or ligand-binding sites, which usually have a high percentage of conserved residues across various species.
The stability of the nanobodies is on par with the more stable of the VH domains of the conventional antibodies, but a truly remarkable property of VHH is its ability to efficiently refold with full restoration of its antigen binding properties after thermal denaturation (47). This property opens interesting and largely unexplored possibilities for the nanobody use in protein folding studies. Because thermal denaturation of nanobodies is reversible, conceivably, nanobody binding could be turned on or off in the NMR tube by simply raising or lowering sample temperature.
Nanobodies Capture Transient Protein Conformations
In 2010, Kirchhofer et al. (44) published an exciting “proof-of-concept study” that demonstrated the utility of nanobodies for detecting and regulating conformational transitions of proteins in vitro and in cells. The authors identified two nanobodies, dubbed the Enhancer and the Minimizer, which had opposite effects on GFP fluorescence. Subsequent structural analysis revealed that each of the nanobodies induced subtle changes in the chromophore environment, thus modulating the absorption properties of GFP. This ability to manipulate GFP fluorescence enabled higher sensitivity and improved spatial resolution in studies of GFP-fused proteins in living cells. Recent development of nanobody applications for super-resolution microscopy combined with inventive methods of nanobody derivatization with fluorescent labels opens new and exciting possibilities for visualizing intracellular processes (48–52).
The ability to identify and trap specific protein conformations using nanobodies has already found creative uses in elucidating the mechanisms of important cellular events, such as receptor-mediated signaling, trafficking, and protein complex assembly. In recent studies of epidermal growth factor receptor (EGFR) (53–55), the nanobodies enabled detection of a functionally silent EGFR heterodimer, which is distinct from the active ligand-bound conformer. EGF binding to the extracellular domain of EGFR triggers conformational changes and homodimerization, or heterodimerization with the other members of the EGFR family, such as ErbB2, initiating the signaling cascade. The existence of an inactive “tethered” dimer was predicted (53, 54), but its conformational status was difficult to define. Nevoltris et al. (55) isolated nanobodies selective either for the ligand-free or for the ligand-bound EGFR. Using these conformation-sensitive nanobodies and homogenous time-resolved fluorescence measurements, the authors demonstrated the presence at the plasma membrane of the EGFR/ErbB2 “pre-dimers,” which were structurally and functionally distinct from the activated dimers. Biological relevance of this finding was further demonstrated in cells using the wild-type ErbB receptors.
Inhibitory conformation-sensitive nanobodies were used to investigate the contribution of L-plastin to the formation of immune synapse (56). L-plastin is an actin-binding protein, which redistributes to the immune synapse following interaction between the T cells and the antigen-presenting cells. Nanobodies that trapped L-plastin in an inactive conformation affected several distinct steps in the formation of the synapse, including IL-2 secretion and T cell proliferation. Nanobodies against various domains of L-plastin further helped to dissect its interactions with the other proteins involved in the synapse formation. Thus, nanobodies enabled functional studies of L-plastin at the level of detail typically achieved by mutagenesis, but without potentially complicating the effects of mutations on protein folding or stability.
Of particular interest are nanobodies that trap the target protein in a defined conformational state and can be used to manipulate its activity in the cell. For example, mouse P-glycoprotein (Pgp) has been crystallized in complex with a nanobody that bound to one of the two nucleotide-binding domains in the Pgp dimer. This binding stabilized the inward-facing conformation of the Pgp dimer and inhibited Pgp catalytic cycle by interfering with the interaction of the two nucleotide-binding domains (57). The ability to modulate intracellular activity of Pgp, an ABC-type transporter involved in cancer cell resistance to chemotherapeutic drugs, suggests the potential for developing a conceptually new therapeutic strategy to fight drug resistance in cancer cells in the future.
A significant advantage of nanobodies lies in their amenability to further optimization for binding to the desired conformer by repeated rounds of selection from the initial cDNA libraries or by directed evolution of the previously selected nanobodies. This property was used extensively to assist crystallization of the active conformation of the β2-adrenergic receptor (βAR) by the Kobilka group. Crystallization of GPCRs in the active form had been a major challenge due to the conformational plasticity of these states. Multiple optimizing mutations, fusions with a helper protein, and agonists with very high binding affinity had been previously employed to stabilize GPCRs in the active conformation (58–60). A conformation-selective nanobody offered an elegant alternative to these methods.
A nanobody that showed preferential binding to βAR in the active state (61) was further optimized for binding affinity. A library of nanobody variants with partially randomized binding sites was subjected to multiple rounds of positive selection for stronger binding to βAR in the active conformation stabilized by a strong agonist. A round of negative selection against βAR bound to an inverse agonist removed variants with reduced conformational specificity. This selection process produced a nanobody with a 10-fold higher affinity, which was used to stabilize and successfully crystallize the active conformation of βAR in complex with adrenaline (62). A similar strategy was used to select the conformation-specific variants of a nanobody against the agonist-bound muscarinic acetylcholine receptor and led to a high-resolution structure of this GPCR (63).
One biologically important facet of the conformation-selective nanobodies is that by stabilizing the active conformation, they emulate the effect of the natural interacting partners of their target protein. Thus, the Nb80 nanobody used to stabilize the active state of βAR increased its affinity to the agonist isoproterenol by about 100-fold, perfectly matching the effect of the cognate G-protein, which shows strong binding cooperativity with the receptor agonists (61), and also stabilizes the active state of the receptor. In fact, crystal structures of the β2-adrenoreceptor in complex with either Nb80 or the Gs protein were found to be almost identical (61, 64).
The conformation-selective nanobodies help to investigate receptor-mediated signaling in live cells with an unprecedented insight into protein dynamics by bridging protein conformational transitions and complex intracellular functions (65). A recent elegant study employed a conformation-sensitive nanobody to demonstrate signaling by the activated and internalized β2−adrenergic receptor from the early endosomes (66). This study revealed that the internalized receptors contribute to the cellular cyclic AMP response within several minutes after agonist application, thus providing previously unavailable spatial and temporal information. Selectivity of nanobodies for the conformational, non-linear epitopes is also likely to reduce nonspecific “off-target” effects in intracellular applications.
Nanobodies and Intrinsically Disordered Proteins
Nanobodies have been used as crystallization chaperones for proteins with a high extent of intrinsic disorder, such as antitoxin MazE, a component of the programmed cell death system in bacteria (67). Remarkably, only 45% of amino acid residues were ordered in that structure. An x-ray structure of the human prion protein represents another interesting example of using nanobodies to reveal structural propensities of intrinsically disordered proteins (68). This structure shows expansion of a β-sheet in the usually disordered N-terminal region of PrPC, the normal form of the prion protein. Although formation of the additional β-strands was likely caused by the nanobody, this effect may mimic a step in the naturally occurring structural transition that leads to the increase in the β-strand content, and consequently to PrPSc aggregation, which culminates in amyloid fibril formation. Interestingly, the discontinuous nanobody epitope in PrPC includes the connecting loop leading to the newly formed β-strand, but not the strand itself, suggesting that the nanobody may have triggered a natural transition between the two energetically close conformations, rather than forced an artificial structure formation. This work suggests the possibility of using large nanobody panels to explore the dynamic conformational landscapes of the intrinsically disordered proteins to identify physiologically relevant transitions.
Nanobodies as Probes of Fast Protein Dynamics
Nanobodies open new opportunities for correlating fast protein dynamics with the function of protein in the cell. NMR spectroscopy is the most powerful method for studying protein dynamics on the nanosecond scale. For protein NMR, the small size of the nanobodies offers unique advantages and truly sets them apart from the conventional antibodies or their derivatives. Unlike x-ray crystallography or electron microscopy, solution NMR is increasingly difficult to use with larger proteins. This is mostly a consequence of the relationship between the rate of molecular tumbling of the protein molecule and NMR relaxation rates. The slower tumbling of larger proteins leads to fast relaxation, which produces broad NMR lines that result in significant peak overlap, deterioration of signal intensity, and eventually major loss of spectral information.
Although the size of protein envelope accessible by high-resolution NMR has been constantly expanding, proteins above 40 KDa remain challenging targets. Therefore complete IgG antibodies (150 KDa) and Fab fragments (50 KDa) are not well suited as tools for protein NMR. In contrast, nanobodies (12–15 KDa) can be successfully applied to studies of protein conformation and dynamics in solution by NMR.
Nanobodies can be used as NMR invisible probes, while the target proteins are isotopically labeled to record two-dimensional 1H,15N or 1H,13C chemical shift correlation spectra. Alternatively, isotopically 15N- and/or 13C-labeled nanobodies can be easily produced by standard isotope incorporation methods for proteins expressed in E. coli cells (69).
Nanobodies have been used to study the folding of the amyloid-forming variants of human lysozyme by NMR (70), as well as the interactions with the monomeric and fibril forms of α-synuclein (71, 72). These studies nicely illustrate the high quality and high information content of the NMR spectra that can be obtained for the protein-nanobody complexes. The binding sites of the nanobodies can be accurately mapped by NMR. Signals of the residues located at the contact surface with the nanobody show intensity loss, or chemical shift change, depending on the nanobody binding parameters. Crystal structures of several nanobody complexes with dihydrofolate reductase showed good agreement between NMR mapping and the x-ray structures (73).
The potential of nanobodies to analyze dynamics of multidomain proteins is illustrated in the recent studies of Wilson disease protein (ATP7B), a transmembrane copper transporter powered by ATP hydrolysis (Fig. 4A) (74). In cells, activity and localization of ATP7B are regulated by copper (75). The available data point to copper-dependent interactions between the six ferredoxin-like metal-binding domains (MBDs) located in the cytosolic N-terminal portion of ATP7B (Fig. 4A) as a basis of this regulation. However, analysis of the domain-domain interactions proved to be difficult due to their transient nature and the largely independent motions of MBDs revealed by NMR studies (76).
FIGURE 4.
Conformation and dynamics of metal-binding domains of ATP7B revealed by the nanobodies. A, a homology-based model of ATP7B structure (80) based on the x-ray structure of a copper-transporting ATPase CopA from Legionella pneumophila (81). A, N, and P denote the respective domains. Green ovals (MBDs) are the metal-binding domains of ATP7B absent in the CopA structure. B, a model of the chain of six MBD of ATP7B generated from the known MBD structures (red-yellow and cyan) (82–84). Nanobody bound to MBD3 is shown in orange. C, free MBD1–6 (left) and MBD1–6 with the 2R50 nanobody (orange) bound to MBD3 (right). Correlation times (τc), which characterize the rate of molecular tumbling, are shown next to each MBD. Changes in τc caused by the nanobody are color-coded as shown below. D, the difference between the transverse relaxation rates R2 in the absence and presence of the 2R50 or 1R1 nanobody is plotted as a function of the residue sequence number. The locations of individual MBDs in the primary protein sequence are shown. Negative difference (green→blue) indicates a faster relaxation (slower tumbling) with nanobody bound. Conversely, positive difference (green→red) corresponds to a slower relaxation in the presence of nanobody and thus faster tumbling.
NMR offers access to fast protein dynamics through the analysis of relaxation rates, R1 and R2, and heteronuclear NOEs. These parameters contain information on the degree of structural order (order parameter S2), the rate (rotational correlation time τc), and the extent of anisotropy (diffusion tensor component ratio D‖/D⊥) of the molecular motions. Transient interactions between protein domains will affect their mobility, and thus will be reflected in NMR relaxation parameters. However, to extract this information, reference data for non-interacting domains are required. By virtue of their uniquely small size, nanobodies can be used to selectively disrupt domain-domain interactions in the context of a full-length multidomain protein.
Two nanobodies were used to probe the dynamics of the 600-amino acid-long chain of six metal-binding domains of ATP7B (MBD1–6, Fig. 4B), one binding to MBD3, and the other to MBD4. The effects of the two nanobodies on the domain dynamics were strikingly different. Nanobody binding to MBD3 predictably increased its correlation time τc (slower tumbling), but decreased the correlation time of MBD1 and MBD2 (faster tumbling), to the values close to those observed for the isolated MBD2. Accelerated molecular motions of MBD1 and MBD2, in response to nanobody binding to MBD3, pointed to breaking interactions between the three domains through displacement (Fig. 4C). Thus, the anti-MBD3 nanobody revealed transient interactions of the domains, which were not detectable from the relaxation data without this differential approach (77). In contrast, the anti-MBD4 nanobody only caused deceleration of its target domain without any significant effect on the dynamics of the others (Fig. 4D), indicating that MBD4 does not significantly interact with the other domains.
The effect of nanobody binding to the MBDs was further studied in the cells where a stable expression of the nanobody facilitated trafficking of ATP7B from the trans-Golgi network to vesicles, mimicking the effect of copper binding to MBDs. To put domain dynamics into the context of protein function in the cell, nanobodies can be used to mimic interactions of multidomain proteins with their physiological partners, in the same key as described above for the GPCRs and their cognate G-proteins. In the case of ATP7B, the nanobody may mimic the effect of Atox1, a cytosolic copper chaperone protein that delivers copper to ATP7B in the cell (78). Atox1-Cu binding and subsequent copper transfer to MBD2 (79) may break interactions between MBD1, MBD2, and MBD3, in the same fashion as the nanobody binding to MBD3, producing a similar open conformation of MBD1–6 that triggers ATP7B trafficking.
In summary, nanobodies undoubtedly hold great potential for mechanistic studies of protein dynamics. A particularly exciting avenue is combining nanobody-assisted structural studies in vitro with the manipulation of the protein properties, activity, or localization in the cell, using the same nanobody expressed endogenously. Another promising set of applications is nanobody derivatization. Heavy metals for x-ray crystallography and small-angle x-ray scattering, paramagnetic labels for protein NMR, and fluorescent tags for in-cell work can be conveniently attached to the nanobodies, and this is in many cases preferable to the corresponding modifications of the protein of interest. A largely unexplored area is the use of nanobodies for protein folding studies, and their use in fast protein dynamics studies by NMR is just beginning. Many other nanobody applications that at this point escape the imagination of the authors will undoubtedly be developed in the near future.
This work was supported by the Canadian Institutes of Health Research and Saskatchewan Health Research Foundation grants (to O. Y. D.) and by National Institutes of Health Grant GM067166 (to S. L.). This is the second article in the Thematic Minireview series “Modern Technologies for In-cell Biochemistry.” The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institutes of Health.
- VHH
- isolated variable heavy chain domain of the heavy chain-only camelid antibodies
- CDR
- complementarity-determining region
- SPAR
- scaffold protein affinity reagent
- GPCR
- G-protein-coupled receptor
- MBD
- metal-binding domain
- EGFR
- epidermal growth factor receptor
- βAR
- β2-adrenergic receptor
- Pgp
- P-glycoprotein
- PrP
- prion protein.
References
- 1. Hamers-Casterman C., Atarhouch T., Muyldermans S., Robinson G., Hamers C., Songa E. B., Bendahman N., and Hamers R. (1993) Naturally occurring antibodies devoid of light chains. Nature 363, 446–448 [DOI] [PubMed] [Google Scholar]
- 2. Broisat A., Hernot S., Toczek J., De Vos J., Riou L. M., Martin S., Ahmadi M., Thielens N., Wernery U., Caveliers V., Muyldermans S., Lahoutte T., Fagret D., Ghezzi C., and Devoogdt N. (2012) Nanobodies targeting mouse/human VCAM1 for the nuclear imaging of atherosclerotic lesions. Circ. Res. 110, 927–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Caussinus E., Kanca O., and Affolter M. (2012) Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat. Struct. Mol. Biol. 19, 117–121 [DOI] [PubMed] [Google Scholar]
- 4. Rothbauer U., Zolghadr K., Tillib S., Nowak D., Schermelleh L., Gahl A., Backmann N., Conrath K., Muyldermans S., Cardoso M. C., and Leonhardt H. (2006) Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods 3, 887–889 [DOI] [PubMed] [Google Scholar]
- 5. Pardon E., Laeremans T., Triest S., Rasmussen S. G., Wohlkönig A., Ruf A., Muyldermans S., Hol W. G., Kobilka B. K., and Steyaert J. (2014) A general protocol for the generation of Nanobodies for structural biology. Nat. Protoc. 9, 674–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stanfield R. L., Dooley H., Flajnik M. F., and Wilson I. A. (2004) Crystal structure of a shark single-domain antibody V region in complex with lysozyme. Science 305, 1770–1773 [DOI] [PubMed] [Google Scholar]
- 7. Zielonka S., Empting M., Grzeschik J., Könning D., Barelle C. J., and Kolmar H. (2015) Structural insights and biomedical potential of IgNAR scaffolds from sharks. MAbs 7, 15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barthelemy P. A., Raab H., Appleton B. A., Bond C. J., Wu P., Wiesmann C., and Sidhu S. S. (2008) Comprehensive analysis of the factors contributing to the stability and solubility of autonomous human VH domains. J. Biol. Chem. 283, 3639–3654 [DOI] [PubMed] [Google Scholar]
- 9. Tanha J., Nguyen T. D., Ng A., Ryan S., Ni F., and Mackenzie R. (2006) Improving solubility and refolding efficiency of human VHs by a novel mutational approach. Protein Eng. Des. Sel. 19, 503–509 [DOI] [PubMed] [Google Scholar]
- 10. Iwata S., Ostermeier C., Ludwig B., and Michel H. (1995) Structure at 2.8 Å resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature 376, 660–669 [DOI] [PubMed] [Google Scholar]
- 11. Jiang Y., Lee A., Chen J., Ruta V., Cadene M., Chait B. T., and MacKinnon R. (2003) X-ray structure of a voltage-dependent K+ channel. Nature 423, 33–41 [DOI] [PubMed] [Google Scholar]
- 12. Kane Dickson V., Pedi L., and Long S. B. (2014) Structure and insights into the function of a Ca2+-activated Cl− channel. Nature 516, 213–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Banner D. W., Gsell B., Benz J., Bertschinger J., Burger D., Brack S., Cuppuleri S., Debulpaep M., Gast A., Grabulovski D., Hennig M., Hilpert H., Huber W., Kuglstatter A., Kusznir E., Laeremans T., Matile H., Miscenic C., Rufer A. C., Schlatter D., Steyaert J., Stihle M., Thoma R., Weber M., and Ruf A. (2013) Mapping the conformational space accessible to BACE2 using surface mutants and cocrystals with Fab fragments, Fynomers and Xaperones. Acta Crystallogr. D Biol. Crystallogr. 69, 1124–1137 [DOI] [PubMed] [Google Scholar]
- 14. Baranova E., Fronzes R., Garcia-Pino A., Van Gerven N., Papapostolou D., Péhau-Arnaudet G., Pardon E., Steyaert J., Howorka S., and Remaut H. (2012) SbsB structure and lattice reconstruction unveil Ca2+ triggered S-layer assembly. Nature 487, 119–122 [DOI] [PubMed] [Google Scholar]
- 15. Ehrnstorfer I. A., Geertsma E. R., Pardon E., Steyaert J., and Dutzler R. (2014) Crystal structure of a SLC11 (NRAMP) transporter reveals the basis for transition-metal ion transport. Nat. Struct. Mol. Biol. 21, 990–996 [DOI] [PubMed] [Google Scholar]
- 16. Hassaine G., Deluz C., Grasso L., Wyss R., Tol M. B., Hovius R., Graff A., Stahlberg H., Tomizaki T., Desmyter A., Moreau C., Li X. D., Poitevin F., Vogel H., and Nury H. (2014) X-ray structure of the mouse serotonin 5-HT3 receptor. Nature 512, 276–281 [DOI] [PubMed] [Google Scholar]
- 17. Korotkov K. V., Pardon E., Steyaert J., and Hol W. G. (2009) Crystal structure of the N-terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody. Structure 17, 255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lam A. Y., Pardon E., Korotkov K. V., Hol W. G., and Steyaert J. (2009) Nanobody-aided structure determination of the EpsI:EpsJ pseudopilin heterodimer from Vibrio vulnificus. J. Struct. Biol. 166, 8–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Park Y. J., Pardon E., Wu M., Steyaert J., and Hol W. G. (2012) Crystal structure of a heterodimer of editosome interaction proteins in complex with two copies of a cross-reacting nanobody. Nucleic Acids Res. 40, 1828–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pathare G. R., Nagy I., Śledź P., Anderson D. J., Zhou H. J., Pardon E., Steyaert J., Förster F., Bracher A., and Baumeister W. (2014) Crystal structure of the proteasomal deubiquitylation module Rpn8-Rpn11. Proc. Natl. Acad. Sci. U.S.A. 111, 2984–2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schotte L., Thys B., Strauss M., Filman D. J., Rombaut B., and Hogle J. M. (2015) Characterization of poliovirus neutralization escape mutants of single-domain antibody fragments (VHHs). Antimicrob. Agents Chemother. 59, 4695–4706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Koromyslova A. D., and Hansman G. S. (2015) Nanobody binding to a conserved epitope promotes norovirus particle disassembly. J. Virol. 89, 2718–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chaikuad A., Keates T., Vincke C., Kaufholz M., Zenn M., Zimmermann B., Gutiérrez C., Zhang R. G., Hatzos-Skintges C., Joachimiak A., Muyldermans S., Herberg F. W., Knapp S., and Müller S. (2014) Structure of cyclin G-associated kinase (GAK) trapped in different conformations using nanobodies. Biochem. J. 459, 59–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dreier B., and Plückthun A. (2012) Rapid selection of high-affinity binders using ribosome display. Methods Mol. Biol. 805, 261–286 [DOI] [PubMed] [Google Scholar]
- 25. Škrlec K., Štrukelj B., and Berlec A. (2015) Non-immunoglobulin scaffolds: a focus on their targets. Trends Biotechnol. 33, 408–418 [DOI] [PubMed] [Google Scholar]
- 26. Plückthun A. (2015) Designed ankyrin repeat proteins (DARPins): binding proteins for research, diagnostics, and therapy. Annu. Rev. Pharmacol. Toxicol. 55, 489–511 [DOI] [PubMed] [Google Scholar]
- 27. Koide S., Koide A., and Lipovšek D. (2012) Target-binding proteins based on the 10th human fibronectin type III domain (10Fn3). Methods Enzymol. 503, 135–156 [DOI] [PubMed] [Google Scholar]
- 28. Löfblom J., Feldwisch J., Tolmachev V., Carlsson J., Ståhl S., and Frejd F. Y. (2010) Affibody molecules: engineered proteins for therapeutic, diagnostic and biotechnological applications. FEBS Lett. 584, 2670–2680 [DOI] [PubMed] [Google Scholar]
- 29. Gebauer M., and Skerra A. (2012) Anticalins small engineered binding proteins based on the lipocalin scaffold. Methods Enzymol. 503, 157–188 [DOI] [PubMed] [Google Scholar]
- 30. Gilbreth R. N., and Koide S. (2012) Structural insights for engineering binding proteins based on non-antibody scaffolds. Curr. Opin. Struct. Biol. 22, 413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sennhauser G., and Grütter M. G. (2008) Chaperone-assisted crystallography with DARPins. Structure 16, 1443–1453 [DOI] [PubMed] [Google Scholar]
- 32. Du D., Wang Z., James N. R., Voss J. E., Klimont E., Ohene-Agyei T., Venter H., Chiu W., and Luisi B. F. (2014) Structure of the AcrAB-TolC multidrug efflux pump. Nature 509, 512–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eicher T., Cha H. J., Seeger M. A., Brandstätter L., El-Delik J., Bohnert J. A., Kern W. V., Verrey F., Grütter M. G., Diederichs K., and Pos K. M. (2012) Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc. Natl. Acad. Sci. U.S.A. 109, 5687–5692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Eicher T., Seeger M. A., Anselmi C., Zhou W., Brandstätter L., Verrey F., Diederichs K., Faraldo-Gómez J. D., and Pos K. M. (2014) Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB. Elife 3, e03145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lu M., Symersky J., Radchenko M., Koide A., Guo Y., Nie R., and Koide S. (2013) Structures of a Na+-coupled, substrate-bound MATE multidrug transporter. Proc. Natl. Acad. Sci. U.S.A. 110, 2099–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stockbridge R. B., Kolmakova-Partensky L., Shane T., Koide A., Koide S., Miller C., and Newstead S. (2015) Crystal structures of a double-barrelled fluoride ion channel. Nature 525, 548–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Desmyter A., Transue T. R., Ghahroudi M. A., Thi M. H., Poortmans F., Hamers R., Muyldermans S., and Wyns L. (1996) Crystal structure of a camel single-domain VH antibody fragment in complex with lysozyme. Nat. Struct. Biol. 3, 803–811 [DOI] [PubMed] [Google Scholar]
- 38. Koradi R., Billeter M., and Wüthrich K. (1996) MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55, 29–32 [DOI] [PubMed] [Google Scholar]
- 39. DeLano W. L. (2014) The PyMOL Molecular Graphics System, Version 1.7.4, Schrödinger, LLC, New York [Google Scholar]
- 40. De Genst E., Silence K., Decanniere K., Conrath K., Loris R., Kinne J., Muyldermans S., and Wyns L. (2006) Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc. Natl. Acad. Sci. U.S.A. 103, 4586–4591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nguyen V. K., Hamers R., Wyns L., and Muyldermans S. (2000) Camel heavy-chain antibodies: diverse germline VHH and specific mechanisms enlarge the antigen-binding repertoire. EMBO J. 19, 921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schmitz K. R., Bagchi A., Roovers R. C., van Bergen en Henegouwen P. M., and Ferguson K. M. (2013) Structural evaluation of EGFR inhibition mechanisms for nanobodies/VHH domains. Structure 21, 1214–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stijlemans B., Conrath K., Cortez-Retamozo V., Van Xong H., Wyns L., Senter P., Revets H., De Baetselier P., Muyldermans S., and Magez S. (2004) Efficient targeting of conserved cryptic epitopes of infectious agents by single domain antibodies: African trypanosomes as paradigm. J. Biol. Chem. 279, 1256–1261 [DOI] [PubMed] [Google Scholar]
- 44. Kirchhofer A., Helma J., Schmidthals K., Frauer C., Cui S., Karcher A., Pellis M., Muyldermans S., Casas-Delucchi C. S., Cardoso M. C., Leonhardt H., Hopfner K. P., and Rothbauer U. (2010) Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol. 17, 133–138 [DOI] [PubMed] [Google Scholar]
- 45. Rudolph M. J., Vance D. J., Cheung J., Franklin M. C., Burshteyn F., Cassidy M. S., Gary E. N., Herrera C., Shoemaker C. B., and Mantis N. J. (2014) Crystal structures of ricin toxin's enzymatic subunit (RTA) in complex with neutralizing and non-neutralizing single-chain antibodies. J. Mol. Biol. 426, 3057–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Even-Desrumeaux K., Nevoltris D., Lavaut M. N., Alim K., Borg J. P., Audebert S., Kerfelec B., Baty D., and Chames P. (2014) Masked selection: a straightforward and flexible approach for the selection of binders against specific epitopes and differentially expressed proteins by phage display. Mol. Cell. Proteomics 13, 653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. van der Linden R. H., Frenken L. G., de Geus B., Harmsen M. M., Ruuls R. C., Stok W., de Ron L., Wilson S., Davis P., and Verrips C. T. (1999) Comparison of physical chemical properties of llama VHH antibody fragments and mouse monoclonal antibodies. Biochim. Biophys. Acta 1431, 37–46 [DOI] [PubMed] [Google Scholar]
- 48. Ashour J., Schmidt F. I., Hanke L., Cragnolini J., Cavallari M., Altenburg A., Brewer R., Ingram J., Shoemaker C., and Ploegh H. L. (2015) Intracellular expression of camelid single-domain antibodies specific for influenza virus nucleoprotein uncovers distinct features of its nuclear localization. J. Virol. 89, 2792–2800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bleck M., Itano M. S., Johnson D. S., Thomas V. K., North A. J., Bieniasz P. D., and Simon S. M. (2014) Temporal and spatial organization of ESCRT protein recruitment during HIV-1 budding. Proc. Natl. Acad. Sci. U.S.A. 111, 12211–12216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li Z., Theile C. S., Chen G. Y., Bilate A. M., Duarte J. N., Avalos A. M., Fang T., Barberena R., Sato S., and Ploegh H. L. (2015) Fluorophore-conjugated Holliday junctions for generating super-bright antibodies and antibody fragments. Angew. Chem. Int. Ed Engl. 54, 11706–11710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Platonova E., Winterflood C. M., Junemann A., Albrecht D., Faix J., and Ewers H. (2015) Single-molecule microscopy of molecules tagged with GFP or RFP derivatives in mammalian cells using nanobody binders. Methods 88, 89–97 [DOI] [PubMed] [Google Scholar]
- 52. Ries J., Kaplan C., Platonova E., Eghlidi H., and Ewers H. (2012) A simple, versatile method for GFP-based super-resolution microscopy via nanobodies. Nat. Methods 9, 582–584 [DOI] [PubMed] [Google Scholar]
- 53. Gadella T. W. Jr., and Jovin T. M. (1995) Oligomerization of epidermal growth factor receptors on A431 cells studied by time-resolved fluorescence imaging microscopy: a stereochemical model for tyrosine kinase receptor activation. J. Cell Biol. 129, 1543–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Moriki T., Maruyama H., and Maruyama I. N. (2001) Activation of preformed EGF receptor dimers by ligand-induced rotation of the transmembrane domain. J. Mol. Biol. 311, 1011–1026 [DOI] [PubMed] [Google Scholar]
- 55. Nevoltris D., Lombard B., Dupuis E., Mathis G., Chames P., and Baty D. (2015) Conformational nanobodies reveal tethered epidermal growth factor receptor involved in EGFR/ErbB2 predimers. ACS Nano. 9, 1388–1399 [DOI] [PubMed] [Google Scholar]
- 56. De Clercq S., Zwaenepoel O., Martens E., Vandekerckhove J., Guillabert A., and Gettemans J. (2013) Nanobody-induced perturbation of LFA-1/L-plastin phosphorylation impairs MTOC docking, immune synapse formation and T cell activation. Cell. Mol. Life Sci. 70, 909–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ward A. B., Szewczyk P., Grimard V., Lee C. W., Martinez L., Doshi R., Caya A., Villaluz M., Pardon E., Cregger C., Swartz D. J., Falson P. G., Urbatsch I. L., Govaerts C., Steyaert J., and Chang G. (2013) Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain. Proc. Natl. Acad. Sci. U.S.A. 110, 13386–13391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cherezov V., Rosenbaum D. M., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Kuhn P., Weis W. I., Kobilka B. K., and Stevens R. C. (2007) High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science 318, 1258–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rosenbaum D. M., Cherezov V., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Yao X. J., Weis W. I., Stevens R. C., and Kobilka B. K. (2007) GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function. Science 318, 1266–1273 [DOI] [PubMed] [Google Scholar]
- 60. Weis W. I., and Kobilka B. K. (2008) Structural insights into G-protein-coupled receptor activation. Curr. Opin. Struct. Biol. 18, 734–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rasmussen S. G., Choi H. J., Fung J. J., Pardon E., Casarosa P., Chae P. S., Devree B. T., Rosenbaum D. M., Thian F. S., Kobilka T. S., Schnapp A., Konetzki I., Sunahara R. K., Gellman S. H., Pautsch A., Steyaert J., Weis W. I., and Kobilka B. K. (2011) Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature 469, 175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ring A. M., Manglik A., Kruse A. C., Enos M. D., Weis W. I., Garcia K. C., and Kobilka B. K. (2013) Adrenaline-activated structure of β2-adrenoceptor stabilized by an engineered nanobody. Nature 502, 575–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kruse A. C., Ring A. M., Manglik A., Hu J., Hu K., Eitel K., Hübner H., Pardon E., Valant C., Sexton P. M., Christopoulos A., Felder C. C., Gmeiner P., Steyaert J., Weis W. I., Garcia K. C., Wess J., and Kobilka B. K. (2013) Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature 504, 101–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rasmussen S. G., DeVree B. T., Zou Y., Kruse A. C., Chung K. Y., Kobilka T. S., Thian F. S., Chae P. S., Pardon E., Calinski D., Mathiesen J. M., Shah S. T., Lyons J. A., Caffrey M., Gellman S. H., Steyaert J., Skiniotis G., Weis W. I., Sunahara R. K., and Kobilka B. K. (2011) Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 477, 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Staus D. P., Wingler L. M., Strachan R. T., Rasmussen S. G., Pardon E., Ahn S., Steyaert J., Kobilka B. K., and Lefkowitz R. J. (2014) Regulation of β2-adrenergic receptor function by conformationally selective single-domain intrabodies. Mol. Pharmacol. 85, 472–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Irannejad R., Tomshine J. C., Tomshine J. R., Chevalier M., Mahoney J. P., Steyaert J., Rasmussen S. G., Sunahara R. K., El-Samad H., Huang B., and von Zastrow M. (2013) Conformational biosensors reveal GPCR signalling from endosomes. Nature 495, 534–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Loris R., Marianovsky I., Lah J., Laeremans T., Engelberg-Kulka H., Glaser G., Muyldermans S., and Wyns L. (2003) Crystal structure of the intrinsically flexible addiction antidote MazE. J. Biol. Chem. 278, 28252–28257 [DOI] [PubMed] [Google Scholar]
- 68. Abskharon R. N., Giachin G., Wohlkonig A., Soror S. H., Pardon E., Legname G., and Steyaert J. (2014) Probing the N-terminal β-sheet conversion in the crystal structure of the human prion protein bound to a nanobody. J. Am. Chem. Soc. 136, 937–944 [DOI] [PubMed] [Google Scholar]
- 69. Marley J., Lu M., and Bracken C. (2001) A method for efficient isotopic labeling of recombinant proteins. J. Biomol. NMR 20, 71–75 [DOI] [PubMed] [Google Scholar]
- 70. De Genst E., Chan P. H., Pardon E., Hsu S. T., Kumita J. R., Christodoulou J., Menzer L., Chirgadze D. Y., Robinson C. V., Muyldermans S., Matagne A., Wyns L., Dobson C. M., and Dumoulin M. (2013) A nanobody binding to non-amyloidogenic regions of the protein human lysozyme enhances partial unfolding but inhibits amyloid fibril formation. J. Phys. Chem. B 117, 13245–13258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. De Genst E. J., Guilliams T., Wellens J., O'Day E. M., Waudby C. A., Meehan S., Dumoulin M., Hsu S. T., Cremades N., Verschueren K. H., Pardon E., Wyns L., Steyaert J., Christodoulou J., and Dobson C. M. (2010) Structure and properties of a complex of α-synuclein and a single-domain camelid antibody. J. Mol. Biol. 402, 326–343 [DOI] [PubMed] [Google Scholar]
- 72. Guilliams T., El-Turk F., Buell A. K., O'Day E. M., Aprile F. A., Esbjörner E. K., Vendruscolo M., Cremades N., Pardon E., Wyns L., Welland M. E., Steyaert J., Christodoulou J., Dobson C. M., and De Genst E. (2013) Nanobodies raised against monomeric α-synuclein distinguish between fibrils at different maturation stages. J. Mol. Biol. 425, 2397–2411 [DOI] [PubMed] [Google Scholar]
- 73. Oyen D., Wechselberger R., Srinivasan V., Steyaert J., and Barlow J. N. (2013) Mechanistic analysis of allosteric and non-allosteric effects arising from nanobody binding to two epitopes of the dihydrofolate reductase of Escherichia coli. Biochim. Biophys. Acta 1834, 2147–2157 [DOI] [PubMed] [Google Scholar]
- 74. Lutsenko S., Barnes N. L., Bartee M. Y., and Dmitriev O. Y. (2007) Function and regulation of human copper-transporting ATPases. Physiol. Rev. 87, 1011–1046 [DOI] [PubMed] [Google Scholar]
- 75. Hasan N. M., and Lutsenko S. (2012) Regulation of copper transporters in human cells. Curr. Top. Membr. 69, 137–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Banci L., Bertini I., Cantini F., Massagni C., Migliardi M., and Rosato A. (2009) An NMR study of the interaction of the N-terminal cytoplasmic tail of the Wilson disease protein with copper(I)-HAH1. J. Biol. Chem. 284, 9354–9360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Huang Y., Nokhrin S., Hassanzadeh-Ghassabeh G., Yu C. H., Yang H., Barry A. N., Tonelli M., Markley J. L., Muyldermans S., Dmitriev O. Y., and Lutsenko S. (2014) Interactions between metal-binding domains modulate intracellular targeting of Cu(I)-ATPase ATP7B, as revealed by nanobody binding. J. Biol. Chem. 289, 32682–32693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Pufahl R. A., Singer C. P., Peariso K. L., Lin S. J., Schmidt P. J., Fahrni C. J., Culotta V. C., Penner-Hahn J. E., and O'Halloran T. V. (1997) Metal ion chaperone function of the soluble Cu(I) receptor Atx1. Science 278, 853–856 [DOI] [PubMed] [Google Scholar]
- 79. Walker J. M., Huster D., Ralle M., Morgan C. T., Blackburn N. J., and Lutsenko S. (2004) The N-terminal metal-binding site 2 of the Wilson's Disease Protein plays a key role in the transfer of copper from Atox1. J. Biol. Chem. 279, 15376–15384 [DOI] [PubMed] [Google Scholar]
- 80. Schushan M., Bhattacharjee A., Ben-Tal N., and Lutsenko S. (2012) A structural model of the copper ATPase ATP7B to facilitate analysis of Wilson disease-causing mutations and studies of the transport mechanism. Metallomics. 4, 669–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gourdon P., Liu X. Y., Skjørringe T., Morth J. P., Møller L. B., Pedersen B. P., and Nissen P. (2011) Crystal structure of a copper-transporting PIB-type ATPase. Nature 475, 59–64 [DOI] [PubMed] [Google Scholar]
- 82. Achila D., Banci L., Bertini I., Bunce J., Ciofi-Baffoni S., and Huffman D. L. (2006) Structure of human Wilson protein domains 5 and 6 and their interplay with domain 4 and the copper chaperone HAH1 in copper uptake. Proc. Natl. Acad. Sci. U.S.A. 103, 5729–5734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Banci L., Bertini I., Cantini F., Rosenzweig A. C., and Yatsunyk L. A. (2008) Metal binding domains 3 and 4 of the Wilson disease protein: solution structure and interaction with the copper(I) chaperone HAH1. Biochemistry 47, 7423–7429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Dolgova N. V., Nokhrin S., Yu C. H., George G. N., and Dmitriev O. Y. (2013) Copper chaperone Atox1 interacts with the metal-binding domain of Wilson's disease protein in cisplatin detoxification. Biochem. J. 454, 147–156 [DOI] [PubMed] [Google Scholar]