Abstract
Conventional structural and chemical biology approaches are applied to macromolecules extrapolated from their native context. When this is done, important structural and functional features of macromolecules, which depend on their native network of interactions within the cell, may be lost. In-cell nuclear magnetic resonance is a branch of biomolecular NMR spectroscopy that allows macromolecules to be analyzed in living cells, at the atomic level. In-cell NMR can be applied to several cellular systems to obtain biologically relevant structural and functional information. Here we summarize the existing approaches and focus on the applications to protein folding, interactions, and post-translational modifications.
Keywords: cell compartmentalization, intracellular processing, molecular cell biology, nuclear magnetic resonance (NMR), post-translational modification (PTM), protein folding, structural biology
Overview of In-cell NMR Approaches
A cellular structural approach is needed if we want to characterize macromolecules, their interactions, and their functions while they retain their native, intracellular localization. In this respect, NMR spectroscopy is an ideal technique for studying macromolecules in living cells, as it is non-destructive and provides structural and biochemical details of macromolecules in solution, over a wide range of temperatures. The first in-cell NMR proof-of-principle was described by Serber et al. (1, 2), where the name “in-cell NMR” was coined to describe high-resolution NMR applied to obtain structural information of a macromolecule, e.g. a protein, inside living cells, as opposed to “in vivo NMR,” which often refers to the observation of small molecules in living organisms. In those works, the authors exploited recombinant protein expression in bacterial cells, an established sample preparation strategy for structural biology (Fig. 1a). With proper adaptations of the cell growth and induction protocol, globular soluble proteins (bacterial NmerA and human calmodulin) were overexpressed and isotopically labeled in Escherichia coli cells to a sufficient level to be detected by NMR above the other cellular components. Some important parameters were assessed, such as cell viability during the experiments, timing and type of isotopic labeling, and NMR line broadening. Uniform 15N-labeling was found to be the ideal choice in most cases, whereas uniform 13C labeling is often unsuitable for this type of experiment, due to the high natural occurrence of 13C (1.1%) and to the high amount of carbon atoms in biological molecules. [methyl-13C]Methionine labeling was shown to be a viable strategy to detect side chain carbon atoms with good selectivity against the cellular 13C background (3); other amino acid type-selective labeling strategies were also examined (2). Labeling of the protein of interest with non-natural amino acids containing 19F was also demonstrated to be a useful approach to investigate protein dynamics in the cellular environment (4, 5). Although 19F-containing amino acids tend to have large spectral overlap, the in-cell NMR spectra are virtually background-free. 19F allows for slower-tumbling molecules to be observed and is potentially useful for studying protein-drug interactions.
FIGURE 1.
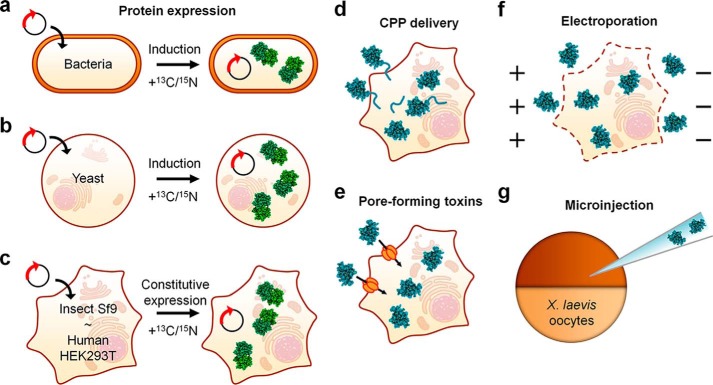
Overview of the existing sample preparation approaches for in-cell NMR. a–c, isotopically labeled proteins can be expressed (green) in bacterial cells (a) and yeast cells (b) by transforming the cells with expression vector(s) encoding the protein(s) of interest; proteins can be expressed in insect and human cells (c) by transfecting the cells with constitutive expression vectors. Isotopically enriched nutrients are added after induction/transfection. d–f, purified labeled proteins (blue) can be inserted in human cells by Cu,Zn-SOD1 CPP-mediated delivery (d), or by permeabilizing the cells either with pore-forming toxins (e) or via electroporation (f). g, proteins can be inserted in X. laevis oocytes by microinjection.
Further development of NMR in E. coli cells capitalized on the knowledge of bacterial protein expression systems. Burz et al. (6, 7) combined different systems for controlling expression of heterologous proteins, so that two or more proteins are overexpressed at different times in the same cells, and isotopic enrichment of a single protein can be performed. This approach, called STINT-NMR, allows protein-protein interactions to be investigated directly in bacterial cells, and was applied to characterize the interaction between ubiquitin and two ligands, a ubiquitin-binding peptide and the signal transducing adaptor protein STAM2 (6, 8). The same authors developed a similar approach, SMILI-NMR, which allows characterizing interactions of proteins with small molecules (9). The technique relies on STINT-NMR to produce a protein complex inside the cells, with only one partner isotopically labeled. The cells are then screened against a library of small molecules and monitored by in-cell NMR to detect changes in the complex due to protein-small molecule interactions occurring within the cells.
On the spectroscopy side, the application of advanced NMR methods increases the possibilities offered by in-cell NMR approaches. Structural investigation by NMR heavily relies on high-dimensionality (i.e. three-dimensional or more) heteronuclear experiments. Such experiments are necessary to perform a complete resonance assignment, which in turn serves as a starting point to solve the three-dimensional structure of a protein. However, they suffer from low sensitivity and require long experimental times, which are prohibitive considering the limited durability of living cell samples. Reduced sampling schemes decrease the time required for the acquisition of multidimensional NMR experiments (10), whereas fast-pulsing schemes decrease the interscan delay and increase the sensitivity by allowing the acquisition of more scans per unit of time (11). These approaches were shown to be beneficial when applied on bacterial cells (12) and were eventually used to determine a protein structure de novo, exclusively relying on in-cell NMR data (13). Direct 13C-detected NMR experiments were also proven to be useful to investigate intrinsically disordered proteins in cells (14, 15). Spectra of intrinsically disordered proteins have low dispersion of the 1H signals, whereas heteronuclear 13C-detected experiments benefit from improved resolution and relaxation properties over standard 1H-detected experiments (16).
Since in-cell NMR was established in bacterial cells, it was clear that it could potentially bridge the gap between structural and cellular biology. However, the technique would only unleash its full potential when applied to eukaryotic cells, and eventually mammalian cells. NMR in eukaryotic cells was first successfully performed on Xenopus laevis oocytes (17–19). Thanks to the large size of these cells, proteins, first produced by recombinant expression, can be then inserted into the oocytes by microinjection (Fig. 1g). This approach provides high labeling selectivity (almost no cellular background); however, it requires the protein to be highly concentrated before injection, to avoid excessive dilution in the oocyte cytoplasm. Notably, injection in X. laevis oocytes was also successfully applied to observe nucleic acids by in-cell NMR (20, 21). A further step forward was made by Inomata et al. (22), which reported the first example of in-cell NMR spectroscopy in cultured human cells. The authors devised a way — conceptually similar to microinjection, but on the molecular scale — to insert a purified protein inside the cells. Their approach relied on a fusion between the protein of interest and a cell-penetrating peptide (CPP)2 derived from the HIV-1 Tat protein (Fig. 1d). Alternatively, the CPP sequence could be covalently linked to the protein in vitro through a disulfide bond, which was then reduced in the cytoplasm, thus releasing the peptide-free protein. This approach was later applied to observe the human protein copper, zinc superoxide dismutase 1 (Cu,Zn-SOD1), in human cells. However, the protein had to be heavily modified to be efficiently imported (23). Ogino et al. (24) reported an alternative approach to translocate labeled proteins into cultured human cells, which relied on the use of pore-forming toxins (streptolysin O) to permeabilize the plasma membrane and allow the entrance of a sufficient amount of protein (Fig. 1e). By treating the cells with Ca2+, the plasma membrane could be resealed, thereby preventing cell death. More recently, protein electroporation was reported as an efficient method for inserting proteins into mammalian cells (25). Electroporation reversibly permeabilizes the plasma membrane, thereby letting a protein into the cells by passive diffusion (Fig. 1f). Conceptually different from protein insertion, intracellular protein expression was also shown to be achievable in cultured human cells by our research group (Fig. 1c) (26). The expression approach was made possible by exploiting recent advancements in mammalian protein production systems (27, 28). This approach has the advantage of investigating proteins directly in the cells where they are synthesized and does not require any import in other types of cells. It is therefore especially useful to study protein folding and maturation processes occurring immediately after protein synthesis in the cytoplasm (26, 29). In-cell NMR on proteins expressed directly in the cells can also be performed in yeast, as shown by Bertrand et al. (Fig. 1b), and in insect cells, as shown by Hamatsu et al. (31) (Fig. 1c). By inducing protein expression in yeast supplemented with different nutrients, the expressed protein is localized in different cellular compartments, and the effect of different subcellular environments on the protein can be investigated (30).
One intrinsic limitation of the technique is the limited cell lifetime inside the NMR tube, which often prevents the acquisition of experiments longer than few hours. Efforts to increase sample viability have been made, and resulted in the design of bioreactors that can be fitted inside an NMR spectrometer. Such bioreactors have been reported for both bacteria (32) and human cells (33). In both designs, a flow is applied that replaces the spent medium with fresh medium, providing nutrients and stabilizing the external pH. To reduce mechanical stress, the cells are encapsulated within a hydrogel, where they can still exchange nutrients and by-products, thus improving sample lifetime.
Effects of Crowding on Folding and Weak Interactions
In-cell NMR allows observation of structural and functional features of proteins within intact living cells at room temperature. Therefore, it is ideally applied to understand how the thermodynamics of protein folding are affected by the intracellular environment. Bacterial cells can be considered a suitable model system when studying proteins — either bacterial or eukaryotic — which are natively localized in the cytoplasm, as the physicochemical properties of the cytoplasm are comparable among different organisms, if functional interactions are not to be taken into account. A consequence of the high concentration of macromolecules (mainly proteins and nucleic acids) in the cytoplasm is macromolecular crowding. Its main effect on solute molecules is the excluded volume; due to steric repulsion between macromolecules, a large fraction of the total volume of the cytoplasm would not be accessible to other solute macromolecules, such as proteins. Therefore, the effective concentration of the latter, as well as their thermodynamic activity, would increase accordingly. As a consequence of this, the folding equilibrium of a protein is expected to shift toward the folded conformation, which minimizes the occupied volume. The crowding effect of the cytoplasm on protein folding was first observed through in-cell NMR by the Pielak group (34). In their work, Dedmon et al. (34) showed that FlgM, an intrinsically disordered protein from Salmonella enterica serovar Typhimurium, obtained a partially folded conformation in the cytoplasm of E. coli. As a consequence of folding, the amide crosspeaks of the C-terminal part of the intracellular protein were not detected in the NMR spectra, whereas those arising from the N terminus, which remained unfolded, could still be detected and remained unchanged when compared with the protein in solution. The resulting NMR spectrum was similar to that of FlgM interacting with the transcription factor σ28 in vitro, in which the C terminus of FlgM is known to be in exchange with a folded conformation bound to σ28. Recently, Smith et al. (35) measured the amide hydrogen exchange rates of intrinsically disordered proteins (α-synuclein and FlgM) in bacterial cells and showed that protein disorder still persisted in the crowded cellular environment.
When proteins act as crowding agents, additional phenomena can affect the protein folding landscape. Weak attractive interactions can occur between the crowding agents and a polypeptide, which stabilize more exposed conformations. This enthalpic contribution would therefore counterbalance the excluded volume, which is a purely entropic effect. The extent to which these two contributions affect the folding landscape of a protein depends on the intrinsic properties of the latter. Schlesinger et al. (36) proved this concept by showing a striking example of a protein that fails to reach the folded conformation in the bacterial cytoplasm. In the study, a variant of protein L from Peptostreptococcus magnus was investigated, which has a marginally stable folded conformation in vitro in the presence of K+ ions. In the bacterial cytoplasm, under similar concentration of K+ ions, the protein was mostly unfolded, indicating that interactions with other cytoplasmic components can overcome the excluded-volume effect, preventing folding of natively unstable globular proteins.
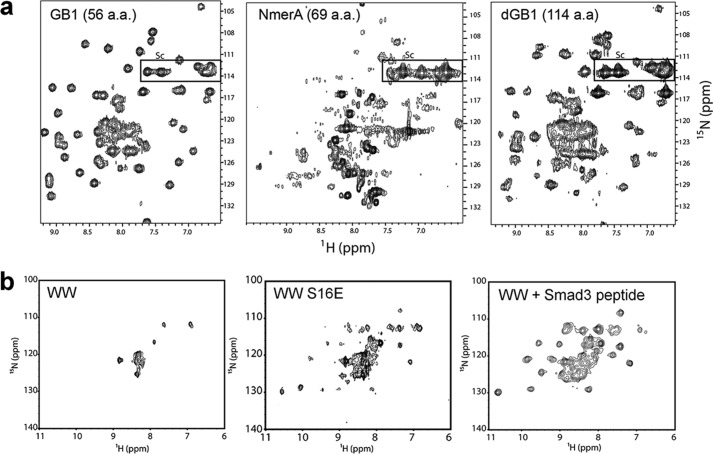
A common effect of the cellular environment on the in-cell NMR spectra of soluble proteins, which has been observed since the beginning, is the general broadening of the NMR resonances. The signal broadening is, in the simplest cases, due to a slower tumbling of the protein in the cytoplasm with respect to aqueous buffers. Intriguingly, globular proteins of similar size were shown to have very different NMR relaxation properties, and many of them tumble so slowly that they cannot be observed at all by in-cell NMR. Although line broadening is often observed in folded proteins, it is less likely to occur in disordered fragments. An example of such different behavior was shown in E. coli by Barnes et al. (37). A fusion construct was used consisting of ubiquitin (a folded protein) attached to α-synuclein (an intrinsically unfolded protein) with a flexible linker. In the bacterial cytoplasm, only the backbone resonances of α-synuclein were detected, whereas ubiquitin could only be detected upon lysing the cells. Wang et al. (38) characterized by in-cell NMR the rotational diffusion of three globular proteins in the bacterial cytoplasm: the B1 domain of staphylococcal protein G (GB1), a fusion of two GB1 domains, and the metal-binding domain of mercuric ion reductase (NmerA). By comparing the linewidth increase in cells, in lysates, and in a series of buffer solutions of increasing viscosity, they showed that the tumbling rate decreased differentially for each protein and did not correlate with the increased viscosity of the cytosol, nor with the molecular weight (Fig. 2a). Thus, the line broadening effect could not be attributed only to increased viscosity or excluded volume effects, and is mainly due to weak interactions between the soluble protein and the other cytoplasmic components.
FIGURE 2.
Effect of the molecular crowding on different proteins. a, in-cell 1H-15N correlation spectra of the B1 domain of staphylococcal protein G (GB1, left), mercuric ion reductase (NmerA, center), and a fusion of two GB1 domains (dGB1, right) in E. coli. Although dGB1 is larger than NmerA, it interacts less with the environment, giving rise to sharper NMR signals. a.a., amino acids. Adapted with permission from Ref. 38. Copyright (2011) American Chemical Society. b, in-cell 1H-15N correlation spectra of the Trp-Trp-binding module (WW) domain of the peptidyl-prolyl isomerase Pin1 in X. laevis oocytes. Unmodified WW (left) constantly interacts with the environment, resulting in signal loss; phosphorylation of Ser-16 (mimicked by S16E mutation, center) and interaction with a substrate (right) abrogate the nonspecific interactions, and the fast-tumbling WW domain can be detected by NMR. Adapted with permission from Ref. 45. Copyright (2013) American Chemical Society.
The extent of weak interactions in the presence of protein crowders was extensively investigated in vitro by the Pielak group. By measuring NMR relaxation properties of proteins in the presence of different crowding agents, they showed that proteins as crowding agents differ from synthetic polymers in that they exert weak, nonspecific interactions that impact protein translational and rotational diffusion (39) as well as folding stability (40, 41). More recently, the same research group devised a way to obtain quantitative, residue-level information on the folding thermodynamics of intracellular GB1. The authors measured by NMR the amide hydrogen-deuterium exchange on cell lysates; after quenching by acidification, the hydrogen-deuterium exchange occurred in the cells at different times (42). With this approach, they showed that the cytoplasm stabilizes the folded state of GB1, whereas protein crowding agents in vitro have an opposite effect, thus highlighting the intrinsic complexity of the intracellular milieu.
For some proteins, nonspecific interactions with the intracellular environment are strong enough to prevent NMR analysis even after diluting the cytoplasm upon cell lysis. Crowley et al. (43) probed such interactions by performing size-exclusion chromatography on cell lysates. They showed that in the bacterial lysate, cytochrome c (13 kDa) eluted with an apparent molecular mass of >150 MDa, due to interactions with cytosolic proteins. The effect was abolished in charge-inverted mutants, or with a high concentration of NaCl in the elution buffer, suggesting interactions of electrostatic nature. These charge interactions were also analyzed by observing a synthetic construct (ΔTat-GB1) in a bacterial lysate by size-exclusion chromatography and NMR (44).
In eukaryotes, post-translational modifications can alter the surface properties of a protein. In X. laevis oocytes and extracts, Luh et al. (45) showed that a peptidyl-prolyl isomerase (Pin1) interacts nonspecifically with the environment through the N-terminal Trp-Trp-binding module (WW) domain. Interestingly, upon substrate recognition, the nonspecific interactions between the WW domain and the environment are lost, and both specific and nonspecific interactions are abrogated when Pin1 contains a mutation that mimics phosphorylation of the WW domain (Fig. 2b).
Together, these works highlight the diversity of the possible protein-environment interactions and hint at the existence of a hidden layer of weak functional interactions between intracellular proteins, referred to as quinary interactions, which were predicted long ago but have since remained mostly uncharacterized (46).
Protein Interactions, Maturation, and Compartmentalization
The results discussed above demonstrate that the cellular environment can have an important role in fine-tuning the structural and dynamic properties of biological macromolecules. Therefore, in-cell NMR also stands as an ideal approach to determine how the functional properties of biomolecules, which in turn depend on their structure and dynamics, are affected by the environment. Of particular interest are the mechanisms that regulate the fate of proteins within the cells: from the initial folding, to co-factor binding or post-translational modifications, translocation in the relevant cellular compartment, and eventually, degradation. All these processes are strictly dependent on interactions with specific partners, which in turn are finely regulated inside the cell (e.g. they depend on partner and co-factor availability, properties of the environment, cellular localization).
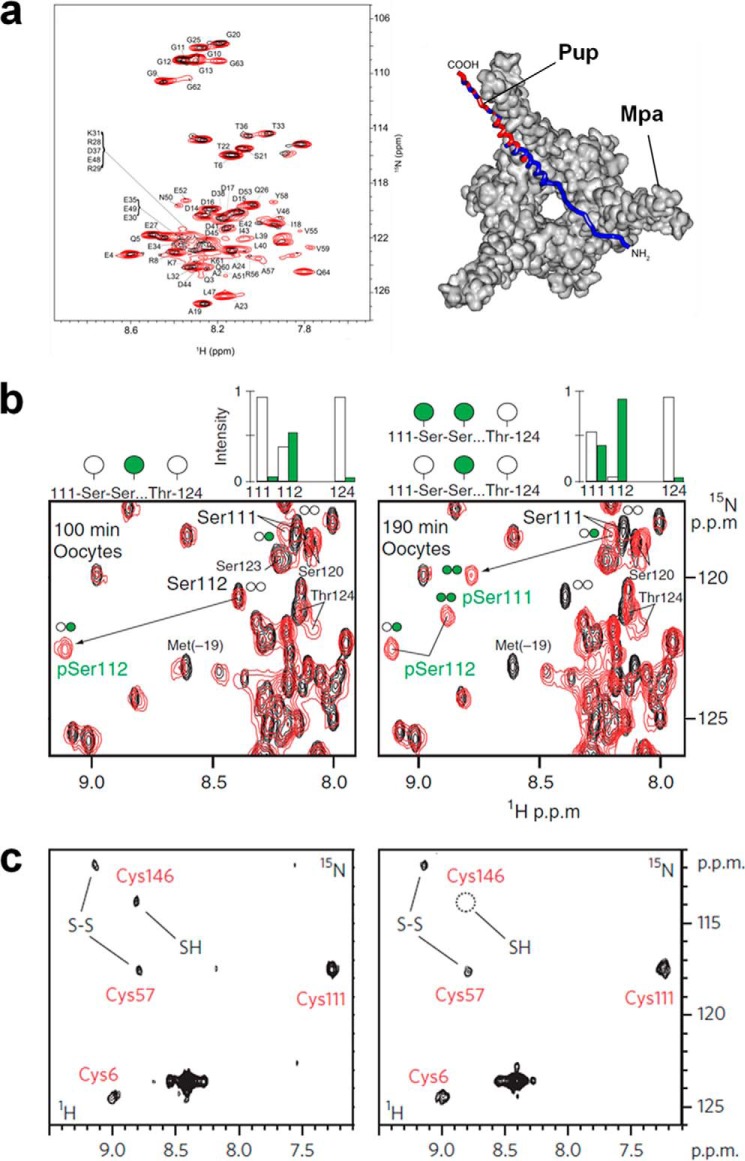
By applying the STINT-NMR method, Burz and Shekhtman (8) showed in E. coli cells how a ubiquitin-partner interaction changed upon phosphorylation of the partners. The authors monitored the interaction of ubiquitin (Ubq) with two components of the receptor tyrosine kinase endocytic sorting machinery (STAM2 and Hrs) separately and together (STAM2-Hrs heterodimer). In a separate set of experiments, tyrosine-kinase Fyn was also expressed, and the two interacting proteins were phosphorylated in-cell. Isotopically labeled Ubq was then expressed, and then monitored by in-cell NMR. Phosphorylation was found to weaken the interaction between Ubq and the interacting partners, whereas the mode of interaction between Ubq and either of its partners did not change between the binary and the ternary complexes. An advantage of this approach is that interactions involving proteins with a short lifetime, which are not easily purified, can be studied. The same approach was then applied to investigate how the mycobacterial proteasome regulates the degradation of the prokaryotic ubiquitin-like protein (Pup) (47). Pup-GGQ (precursor of the active Pup-GGE form) is a small and unstructured protein that exerts in prokaryotes a similar function to that of eukaryotic ubiquitin, by targeting proteins to the proteasomal machinery. In that work, Pup-GGQ was shown to interact weakly with the proteasomal ATPase (Mpa) through residues in its N and C terminus (Fig. 3a). Conversely, in the presence of the full proteasomal particle (Mpa-proteasome core particle complex), Pup-GGQ was extensively bound to Mpa through most of its residues, suggesting that it binds to both the mouth and the central cavity of Mpa hexamer.
FIGURE 3.
Applications of in-cell NMR to study protein function. a, in E. coli, the interaction between 15N-labeled Pup and unlabeled proteasomal ATPase (Mpa) causes the resonances from some residues of Pup to broaden (left, black spectrum). The broadened residues mapped on a structural model (right, in red) show the mode of interaction. Reprinted from Ref. 47 under CC BY license. b, time-resolved in-cell NMR of CK2-dependent phosphorylation of a 15N-labeled SV40 large T antigen peptide injected in X. laevis oocytes shows that Ser-111 and Ser-112 are sequentially phosphorylated with a two-step mechanism. pSer111, phospho-Ser-111; pSer112, phospho-Ser0112. Reprinted by permission from Macmillan Publishers Ltd., Nat. Struct. Mol. Biol. (49). Copyright (2008). c, the chaperone-dependent formation of an intramolecular disulfide bond (S–S) is observed by in-cell NMR on [15N]cysteine-labeled SOD1 expressed in human cells. In the absence of supplemented copper, partial disulfide formation occurs (left); when copper is supplemented, Cu-CCS-dependent disulfide formation is fully observed. Reprinted by permission from Macmillan Publishers Ltd., Nat. Chem. Biol. (26). Copyright (2013).
Protein-drug interactions can also be monitored by in-cell NMR, as shown both in E. coli (9) and in human cells (22), by observing binding of immunosuppressant drugs to the FK-506-binding protein. More recently, the interaction between cisplatin and the human copper chaperone Atox1 was characterized by NMR in bacterial cells by Arnesano et al. (48). Upon treatment of cells with cisplatin, the formation of a previously uncharacterized 1:1 {Pt(NH3)2}-Atox1 complex was observed first, followed by loss of the ammines and formation of a platinum-bridged Atox1 dimer. Thus, in-cell NMR shows great potential for drug design applications, as the effects of the cell environment on drug import, localization, and binding selectivity can be investigated.
Functional interactions regulating eukaryotic proteins should ideally be studied in the correct cellular environment. For example, protein phosphorylation has a fundamental role in regulating protein activity and transducing information throughout the eukaryotic cell. Selenko et al. (49) investigated a sequence of phosphorylation events occurring to a substrate of CK2 by monitoring real-time phosphorylation events in vitro, in X. laevis egg extracts, and in intact live oocytes. They analyzed the regulatory region of the viral SV40 large T antigen, focusing on adjacent CK2 phosphorylation sites, and found that a stepwise sequence of phosphorylation events occurred, which required the substrate to detach from CK2 in the intermediate step. After in vitro characterization, the authors could observe the same mechanism in both X. laevis egg extracts andlive oocytes, by the action of endogenous CK2 (Fig. 3b). The same approach was used by Amata et al. (50) to study the phosphorylation of a disordered domain of c-Src in oocytes and egg extracts, where differential phosphorylation patterns pointed to indirect effects of the kinase-phosphatase networks.
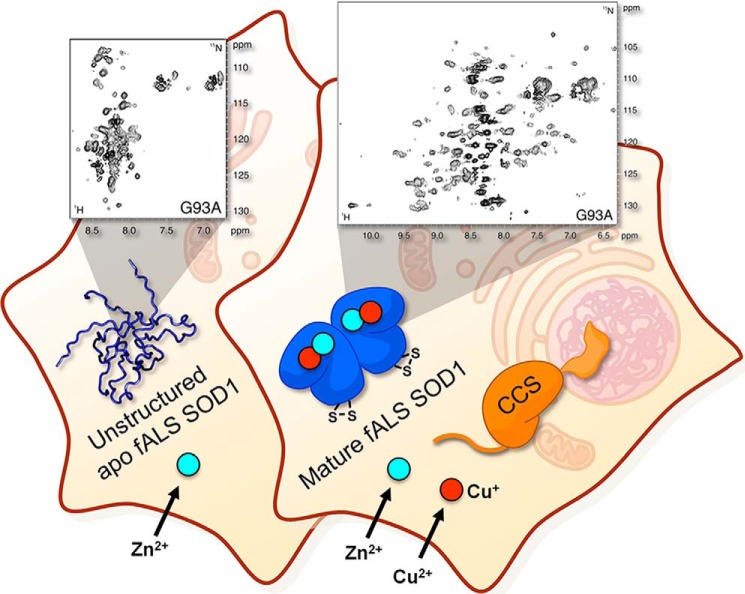
Many eukaryotic proteins require a series of maturation events to take place to reach their final, functional form. To investigate such processes by in-cell NMR, the protein should ideally be synthesized within the eukaryotic cell. Together with members of our research group, we developed and applied a method to observe, by NMR, maturation of proteins directly expressed in cultured human cells. We first applied this method to the maturation events of the human Cu,Zn-SOD1 metalloprotein, a conserved enzyme mainly localized in the cytoplasm, which protects the cell from oxidative damage (26, 51). We characterized by in-cell NMR the conformation of the intermediate maturation states of SOD1 by varying the abundance of the metal co-factors in the cell culture. In human cells, by enhancing the expression of the specific metallochaperone (CCS), we observed the CCS-dependent copper transfer and the formation of an internal disulfide bond, and revealed that CCS could promote disulfide formation in a previously unknown copper-independent mechanism (Fig. 3c). Mutations in the human SOD1 gene are related to the familial variant of amyotrophic lateral sclerosis (fALS), a fatal neurodegenerative disease. We recently analyzed the impaired folding mechanism of a set of fALS-linked SOD1 mutants in human cells by NMR (52). Although the mutations studied do not alter the structural properties of the metal-binding sites of SOD1, some of the mutants could not bind intracellular zinc, unlike the wild type protein, and were shown to irreversibly accumulate in an unstructured apo conformation, which was previously uncharacterized at residue level. Co-expression of CCS in the presence of copper rescued the maturation process of the mutant proteins, allowing them to reach the mature, folded form (Fig. 4).
FIGURE 4.
Disease-related mutations cause maturation defects in SOD1. fALS-linked mutations in the SOD1 gene impair zinc binding of the apo protein and cause the intracellular accumulation of an unstructured species detected by NMR in human cells (left), which is potentially a precursor of the cytotoxic aggregated SOD1 species that are a hallmark of the disease. Co-expression of CCS in the presence of excess copper rescues the mutant protein, preventing misfolding and allowing it to reach the mature, folded form (right). Reprinted by permission from Macmillan Publishers Ltd.,: Nat. Commun. (52). Copyright (2014).
Eukaryotic cells heavily rely on compartmentalization to separate different cellular processes. The vast majority of proteins are encoded by nuclear DNA and are synthesized in the cytoplasm. These proteins are then actively sorted and targeted toward the relevant organelles. In-cell NMR can be applied to understand how protein translocation mechanisms are regulated and how different subcellular environments affect protein function. Bertrand et al. (30) showed in yeast cells how the dynamic properties of ubiquitin change upon localization within different cellular compartments. By changing the growth medium composition, overexpressed ubiquitin could be either localized in the cytosol or targeted to protein storage bodies, where it had slower rotational diffusion. Mitochondrial protein import is another example of how the environment regulates protein function in each compartment. Our group showed by in-cell NMR that the import of Mia40, an oxidoreductase of the intermembrane space responsible for the oxidative folding of proteins in the intermembrane space, is regulated in the cytoplasm by a redox-controlled folding (29). Mia40 expressed in the cytoplasm of human cells adopts a folded conformation, due to the formation of two structural disulfide bonds, which cannot be imported into mitochondria. Co-expression of one of the two main thiol-disulfide-regulating proteins of the cytoplasm (glutaredoxin 1 or thioredoxin 1) caused Mia40 to remain unfolded, in an import-competent state. Finally, we recently showed that NMR can be performed on the intact mitochondria extracted from human cells (53). In-mitochondria NMR can provide precious structural information on mitochondrial proteins, and the approach can in principle be extended to other organelles.
Initial Developments in Solid-state In-cell NMR
An intrinsic limitation of solution NMR is the need for fast rotational diffusion of the observed molecules. Although this already poses an upper limit to the molecular size, even small proteins, as discussed above, may interact with the intracellular environment up to a point where they cannot be detected by solution in-cell NMR. Additionally, proteins associated with membranes, DNA, or the cytoskeleton are not suitable targets for in-cell solution NMR. Magic angle spinning solid-state NMR (MAS NMR) can overcome such limitations. To circumvent the molecular tumbling rate limit, the sample needs to be spun at high speed within the magnetic field. Fast spinning can cause high mechanical stress to cellular samples and is therefore a major obstacle for proper in-(living) cell NMR experiments. Bacterial cells offer moderate resistance to mechanical forces and can tolerate the experimental conditions required by MAS NMR. Renault et al. (54, 55) applied MAS NMR to study the conformation of a small trans-membrane protein (the bacterial outer membrane protein OmpA) inside intact E. coli cells and isolated membranes. By freezing bacterial cells, Reckel et al. (56) showed that signals from soluble proteins, which form high molecular weight transient complexes with other cellular components, can be detected by MAS NMR. In both cases, complex isotopic labeling strategies and tailored SS-NMR experiments had to be devised to improve spectral quality and allow data interpretation. For the application of MAS NMR to mammalian cell samples, complex isotopic labeling and improved resistance to mechanical stress represent future challenges.
Future Directions
The ability to investigate macromolecules within living cells at an atomic level is an important asset for structural biology. In-cell NMR is a relatively young approach, and many of the methods reviewed here have not yet been applied to scenarios other than those described in the original articles. In other instances, the methods have already found applications, and novel biological findings have been reported. In fact, in-cell NMR has largely proven to be up to the task by providing information on protein structure, dynamics, interaction, and ultimately, function in multiple cellular environments. We believe that the applications described in this review really show the potential of the approach. E. coli cells provide a good model environment for the cytoplasm, and bacterial in-cell NMR is relatively cost-effective and easy to implement. However, future developments of the technique should — and will — be directed toward improved eukaryotic model systems. In particular, the availability of a general approach to allow in-cell NMR in many types of mammalian cell lines would push the boundaries of the technique, as it would allow unique atomic-level studies of disease-related protein alterations, such as aggregation phenomena, as well as other types of cellular stress, in cells with the meaningful phenotype. Development of solid-state NMR in eukaryotic cells would greatly increase the possible applications by overcoming the tumbling rate problem and by extending the method to proteins stably associated with cellular structures. As a closing remark, we believe that in-cell NMR should be combined with other cellular techniques (e.g. super-resolution microscopy and electron cryo-microscopy) to develop totally new, integrated approaches for structural cellular biology. As an example, in a recent work, we have combined in-cell NMR with synchrotron radiation x-ray fluorescence microscopy and optical microscopy on immobilized cell samples to correlate the metallation state(s) of human SOD1 (monitored by in-cell NMR) with its expression levels and metal distribution within the cells. We observed that the increase in total cellular zinc was correlated with the SOD1 metal binding event, thus showing that the approach can potentially be applied to investigate protein-metal interactions at the subcellular level (57).
This is the third article in the Thematic Minireview series “Modern Technologies for In-cell Biochemistry.” The authors declare that they have no conflicts of interest with the contents of this article.
- CPP
- cell-penetrating peptide
- Ubq
- ubiquitin
- Pup
- prokaryotic ubiquitin-like protein
- fALS
- familial variant of amyotrophic lateral sclerosis
- MAS
- magic angle spinning
- Cu,Zn-SOD1
- copper, zinc superoxide dismutase 1.
References
- 1. Serber Z., Keatinge-Clay A. T., Ledwidge R., Kelly A. E., Miller S. M., and Dötsch V. (2001) High-resolution macromolecular NMR spectroscopy inside living cells. J. Am. Chem. Soc. 123, 2446–2447 [DOI] [PubMed] [Google Scholar]
- 2. Serber Z., Ledwidge R., Miller S. M., and Dötsch V. (2001) Evaluation of parameters critical to observing proteins inside living Escherichia coli by in-cell NMR spectroscopy. J. Am. Chem. Soc. 123, 8895–8901 [DOI] [PubMed] [Google Scholar]
- 3. Serber Z., Straub W., Corsini L., Nomura A. M., Shimba N., Craik C. S., Ortiz de Montellano P., and Dötsch V. (2004) Methyl groups as probes for proteins and complexes in in-cell NMR experiments. J. Am. Chem. Soc. 126, 7119–7125 [DOI] [PubMed] [Google Scholar]
- 4. Li C., Wang G.-F., Wang Y., Creager-Allen R., Lutz E. A., Scronce H., Slade K. M., Ruf R. A. S., Mehl R. A., and Pielak G. J. (2010) Protein 19F NMR in Escherichia coli. J. Am. Chem. Soc. 132, 321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ye Y., Liu X., Zhang Z., Wu Q., Jiang B., Jiang L., Zhang X., Liu M., Pielak G. J., and Li C. (2013) 19F NMR spectroscopy as a probe of cytoplasmic viscosity and weak protein interactions in living cells. Chemistry 19, 12705–12710 [DOI] [PubMed] [Google Scholar]
- 6. Burz D. S., Dutta K., Cowburn D., and Shekhtman A. (2006) Mapping structural interactions using in-cell NMR spectroscopy (STINT-NMR). Nat. Methods. 3, 91–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burz D. S., Dutta K., Cowburn D., and Shekhtman A. (2006) In-cell NMR for protein-protein interactions (STINT-NMR). Nat. Protoc. 1, 146–152 [DOI] [PubMed] [Google Scholar]
- 8. Burz D. S., and Shekhtman A. (2008) In-cell biochemistry using NMR spectroscopy. PLoS One 3, e2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xie J., Thapa R., Reverdatto S., Burz D. S., and Shekhtman A. (2009) Screening of small molecule interactor library by using in-cell NMR spectroscopy (SMILI-NMR). J. Med. Chem. 52, 3516–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hoch J. C., Maciejewski M. W., Mobli M., Schuyler A. D., and Stern A. S. (2014) Nonuniform sampling and maximum entropy reconstruction in multidimensional NMR. Acc. Chem. Res. 47, 708–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schanda P., and Brutscher B. (2005) Very fast two-dimensional NMR spectroscopy for real-time investigation of dynamic events in proteins on the time scale of seconds. J. Am. Chem. Soc. 127, 8014–8015 [DOI] [PubMed] [Google Scholar]
- 12. Reardon P. N., and Spicer L. D. (2005) Multidimensional NMR spectroscopy for protein characterization and assignment inside cells. J. Am. Chem. Soc. 127, 10848–10849 [DOI] [PubMed] [Google Scholar]
- 13. Sakakibara D., Sasaki A., Ikeya T., Hamatsu J., Hanashima T., Mishima M., Yoshimasu M., Hayashi N., Mikawa T., Wälchli M., Smith B. O., Shirakawa M., Güntert P., and Ito Y. (2009) Protein structure determination in living cells by in-cell NMR spectroscopy. Nature. 458, 102–105 [DOI] [PubMed] [Google Scholar]
- 14. Bertini I., Felli I. C., Gonnelli L., Kumar M. V., V., and Pierattelli R. (2011) 13C direct-detection biomolecular NMR spectroscopy in living cells. Angew. Chem. Int. Ed. Engl. 50, 2339–2341 [DOI] [PubMed] [Google Scholar]
- 15. Felli I. C., Gonnelli L., and Pierattelli R. (2014) In-cell 13C NMR spectroscopy for the study of intrinsically disordered proteins. Nat. Protoc. 9, 2005–2016 [DOI] [PubMed] [Google Scholar]
- 16. Felli I. C., and Pierattelli R. (2014) Novel methods based on 13C detection to study intrinsically disordered proteins. J. Magn. Reson. 241, 115–125 [DOI] [PubMed] [Google Scholar]
- 17. Sakai T., Tochio H., Tenno T., Ito Y., Kokubo T., Hiroaki H., and Shirakawa M. (2006) In-cell NMR spectroscopy of proteins inside Xenopus laevis oocytes. J. Biomol. NMR. 36, 179–188 [DOI] [PubMed] [Google Scholar]
- 18. Selenko P., Serber Z., Gadea B., Ruderman J., and Wagner G. (2006) Quantitative NMR analysis of the protein G B1 domain in Xenopus laevis egg extracts and intact oocytes. Proc. Natl. Acad. Sci. U.S.A. 103, 11904–11909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Serber Z., Selenko P., Hänsel R., Reckel S., Löhr F., Ferrell J. E. Jr., Wagner G., and Dötsch V. (2006) Investigating macromolecules inside cultured and injected cells by in-cell NMR spectroscopy. Nat. Protoc. 1, 2701–2709 [DOI] [PubMed] [Google Scholar]
- 20. Hänsel R., Foldynová-Trantírková S., Löhr F., Buck J., Bongartz E., Bamberg E., Schwalbe H., Dötsch V., and Trantírek L. (2009) Evaluation of parameters critical for observing nucleic acids inside living Xenopus laevis oocytes by in-cell NMR spectroscopy. J. Am. Chem. Soc. 131, 15761–15768 [DOI] [PubMed] [Google Scholar]
- 21. Hänsel R., Foldynová-Trantírková S., Dötsch V., and Trantírek L. (2013) Investigation of quadruplex structure under physiological conditions using in-cell NMR. Top. Curr. Chem. 330, 47–65 [DOI] [PubMed] [Google Scholar]
- 22. Inomata K., Ohno A., Tochio H., Isogai S., Tenno T., Nakase I., Takeuchi T., Futaki S., Ito Y., Hiroaki H., and Shirakawa M. (2009) High-resolution multi-dimensional NMR spectroscopy of proteins in human cells. Nature. 458, 106–109 [DOI] [PubMed] [Google Scholar]
- 23. Danielsson J., Inomata K., Murayama S., Tochio H., Lang L., Shirakawa M., and Oliveberg M. (2013) Pruning the ALS-associated protein SOD1 for in-cell NMR. J. Am. Chem. Soc. 135, 10266–10269 [DOI] [PubMed] [Google Scholar]
- 24. Ogino S., Kubo S., Umemoto R., Huang S., Nishida N., and Shimada I. (2009) Observation of NMR signals from proteins introduced into living mammalian cells by reversible membrane permeabilization using a pore-forming toxin, streptolysin O. J. Am. Chem. Soc. 131, 10834–10835 [DOI] [PubMed] [Google Scholar]
- 25. Bekei B. (2013) In-cell NMR Spectroscopy in Mammalian Cells. Ph.D. thesis, Freie Universität Berlin, Germany [Google Scholar]
- 26. Banci L., Barbieri L., Bertini I., Luchinat E., Secci E., Zhao Y., and Aricescu A. R. (2013) Atomic-resolution monitoring of protein maturation in live human cells by NMR. Nat. Chem. Biol. 9, 297–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aricescu A. R., Lu W., and Jones E. Y. (2006) A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D. Biol. Crystallogr. 62, 1243–1250 [DOI] [PubMed] [Google Scholar]
- 28. Zhao Y., Bishop B., Clay J. E., Lu W., Jones M., Daenke S., Siebold C., Stuart D. I., Jones E. Y., and Aricescu A. R. (2011) Automation of large scale transient protein expression in mammalian cells. J. Struct. Biol. 175, 209–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Banci L., Barbieri L., Luchinat E., and Secci E. (2013) Visualization of redox-controlled protein fold in living cells. Chem. Biol. 20, 747–752 [DOI] [PubMed] [Google Scholar]
- 30. Bertrand K., Reverdatto S., Burz D. S., Zitomer R., and Shekhtman A. (2012) Structure of proteins in eukaryotic compartments. J. Am. Chem. Soc. 134, 12798–12806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hamatsu J., O'Donovan D., Tanaka T., Shirai T., Hourai Y., Mikawa T., Ikeya T., Mishima M., Boucher W., Smith B. O., Laue E. D., Shirakawa M., and Ito Y. (2013) High-resolution heteronuclear multidimensional NMR of proteins in living insect cells using a baculovirus protein expression system. J. Am. Chem. Soc. 135, 1688–1691 [DOI] [PubMed] [Google Scholar]
- 32. Sharaf N. G., Barnes C. O., Charlton L. M., Young G. B., and Pielak G. J. (2010) A bioreactor for in-cell protein NMR. J. Magn. Reson. 202, 140–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kubo S., Nishida N., Udagawa Y., Takarada O., Ogino S., and Shimada I. (2013) A gel-encapsulated bioreactor system for NMR studies of protein-protein interactions in living mammalian cells. Angew. Chem. Int. Ed. Engl. 52, 1208–1211 [DOI] [PubMed] [Google Scholar]
- 34. Dedmon M. M., Patel C. N., Young G. B., and Pielak G. J. (2002) FlgM gains structure in living cells. Proc. Natl. Acad. Sci. U.S.A. 99, 12681–12684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith A. E., Zhou L. Z., and Pielak G. J. (2015) Hydrogen exchange of disordered proteins in Escherichia coli. Protein Sci. 24, 706–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schlesinger A. P., Wang Y., Tadeo X., Millet O., and Pielak G. J. (2011) Macromolecular crowding fails to fold a globular protein in cells. J. Am. Chem. Soc. 133, 8082–8085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barnes C. O., Monteith W. B., and Pielak G. J. (2011) Internal and global protein motion assessed with a fusion construct and in-cell NMR spectroscopy. Chembiochem 12, 390–391 [DOI] [PubMed] [Google Scholar]
- 38. Wang Q., Zhuravleva A., and Gierasch L. M. (2011) Exploring weak, transient protein-protein interactions in crowded in vivo environments by in-cell nuclear magnetic resonance spectroscopy. Biochemistry 50, 9225–9236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Y., Li C., and Pielak G. J. (2010) Effects of proteins on protein diffusion. J. Am. Chem. Soc. 132, 9392–9397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miklos A. C., Sarkar M., Wang Y., and Pielak G. J. (2011) Protein crowding tunes protein stability. J. Am. Chem. Soc. 133, 7116–7120 [DOI] [PubMed] [Google Scholar]
- 41. Wang Y., Sarkar M., Smith A. E., Krois A. S., and Pielak G. J. (2012) Macromolecular crowding and protein stability. J. Am. Chem. Soc. 134, 16614–16618 [DOI] [PubMed] [Google Scholar]
- 42. Monteith W. B., and Pielak G. J. (2014) Residue level quantification of protein stability in living cells. Proc. Natl. Acad. Sci. U.S.A. 111, 11335–11340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Crowley P. B., Chow E., and Papkovskaia T. (2011) Protein interactions in the Escherichia coli cytosol: an impediment to in-cell NMR spectroscopy. Chembiochem 12, 1043–1048 [DOI] [PubMed] [Google Scholar]
- 44. Kyne C., Ruhle B., Gautier V. W., and Crowley P. B. (2015) Specific ion effects on macromolecular interactions in Escherichia coli extracts. Protein Sci. 24, 310–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Luh L. M., Hänsel R., Löhr F., Kirchner D. K., Krauskopf K., Pitzius S., Schäfer B., Tufar P., Corbeski I., Güntert P., and Dötsch V. (2013) Molecular crowding drives active Pin1 into nonspecific complexes with endogenous proteins prior to substrate recognition. J. Am. Chem. Soc. 135, 13796–13803 [DOI] [PubMed] [Google Scholar]
- 46. McConkey E. H. (1982) Molecular evolution, intracellular organization, and the quinary structure of proteins. Proc. Natl. Acad. Sci. U.S.A. 79, 3236–3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maldonado A. Y., Burz D. S., Reverdatto S., and Shekhtman A. (2013) Fate of pup inside the Mycobacterium proteasome studied by in-cell NMR. PLoS One 8, e74576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Arnesano F., Banci L., Bertini I., Felli I. C., Losacco M., and Natile G. (2011) Probing the interaction of cisplatin with the human copper chaperone Atox1 by solution and in-cell NMR spectroscopy. J. Am. Chem. Soc. 133, 18361–18369 [DOI] [PubMed] [Google Scholar]
- 49. Selenko P., Frueh D. P., Elsaesser S. J., Haas W., Gygi S. P., and Wagner G. (2008) In situ observation of protein phosphorylation by high-resolution NMR spectroscopy. Nat. Struct. Mol. Biol. 15, 321–329 [DOI] [PubMed] [Google Scholar]
- 50. Amata I., Maffei M., Igea A., Gay M., Vilaseca M., Nebreda A. R., and Pons M. (2013) Multi-phosphorylation of the intrinsically disordered unique domain of c-Src studied by in-cell and real-time NMR spectroscopy. Chembiochem 14, 1820–1827 [DOI] [PubMed] [Google Scholar]
- 51. Banci L., Barbieri L., Bertini I., Cantini F., and Luchinat E. (2011) In-cell NMR in E. coli to monitor maturation steps of hSOD1. PLoS One 6, e23561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Luchinat E., Barbieri L., Rubino J. T., Kozyreva T., Cantini F., and Banci L. (2014) In-cell NMR reveals potential precursor of toxic species from SOD1 fALS mutants. Nat. Commun. 5, 5502. [DOI] [PubMed] [Google Scholar]
- 53. Barbieri L., Luchinat E., and Banci L. (2014) Structural insights of proteins in sub-cellular compartments: in-mitochondria NMR. Biochim. Biophys. Acta. 1843, 2492–2496 [DOI] [PubMed] [Google Scholar]
- 54. Renault M., Tommassen-van Boxtel R., Bos M. P., Post J. A., Tommassen J., and Baldus M. (2012) Cellular solid-state nuclear magnetic resonance spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 109, 4863–4868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Renault M., Pawsey S., Bos M. P., Koers E. J., Nand D., Tommassen-van Boxtel R., Rosay M., Tommassen J., Maas W. E., and Baldus M. (2012) Solid-state NMR spectroscopy on cellular preparations enhanced by dynamic nuclear polarization. Angew. Chem. Int. Ed Engl. 51, 2998–3001 [DOI] [PubMed] [Google Scholar]
- 56. Reckel S., Lopez J. J., Löhr F., Glaubitz C., and Dötsch V. (2012) In-cell solid-state NMR as a tool to study proteins in large complexes. Chembiochem 13, 534–537 [DOI] [PubMed] [Google Scholar]
- 57. Luchinat E., Gianoncelli A., Mello T., Galli A., and Banci L. (2015) Combining in-cell NMR and x-ray fluorescence microscopy to reveal the intracellular maturation states of human superoxide dismutase 1. Chem. Commun. (Camb.) 51, 584–587 [DOI] [PubMed] [Google Scholar]