Abstract
Members of the receptor tyrosine kinase family (RTK) have been shown to be present in the nucleus of cells; however, the mechanisms underlying their trafficking to the nucleus, and their relevance once there are poorly understood. In the present study, we focus on the RTK ErbB3 and elucidate the mechanisms regulating its trafficking. We show that heregulin-stimulation induces trafficking of phosphorylated ErbB3 from the plasma membrane to the nucleus via a clathrin-independent mechanism. Nuclear import of ErbB3 occurs via importin β1, which drives the receptor through the nuclear pore complex. In the nucleus, ErbB3 interacts with transcription complexes, and thereby has a role in transcriptional regulation. Our results also demonstrate that ErbB3 nuclear localization is transient as it is exported out of the nucleus by the nuclear receptor protein crm-1. Analysis of normal, regenerating tissues, and tumors showed that ErbB3 nuclear translocation is a common event in proliferating tissues.
Keywords: nuclear translocation, proliferation, receptor endocytosis, receptor tyrosine kinase, signal transduction
Introduction
The epidermal growth factor receptor family (EGFR,3 ErbB2, ErbB3, and ErbB4) comprises some of the best characterized members of the receptor tyrosine kinases (RTKs). Their activation influences cellular signaling via the mitogen-activated protein kinase (MAPK)/ERK and the phosphatidylinositol 3-kinase (PI3K)-AKT pathways, both of which regulate essential cellular mechanisms including cell proliferation, survival, and differentiation. Dysregulation of these pathways is regularly found in carcinogenesis, tumor progression, and metastasis (1, 2). Furthermore, overexpression of EGFR, ErbB2, and ErbB3 in tumor tissue is often associated with poor patient outcome (3). ErbB3 is the least studied member of the EGFR family, However, recent evidence supports a key role for ErbB3 in cell transformation and malignancy of tumors (4). In addition, therapeutic interventions directed toward EGFR family members lead to the activation of ErbB3, which in turn is associated with the development of chemoresistance (5–8). However, the consequences of signaling activation and trafficking of ErbB3, as well as its functional relevance are poorly understood.
Previous reports have shown that EGFR family members are present in the nucleus, either as full-length molecules, as in the case of EGFR (9) and ErbB2 (10), or truncated as for γ-secretase-cleaved ErbB4 (11). In the present study, we demonstrate that nuclear translocation of ErbB3 is not limited to cancer, but appears to be a general phenomenon in proliferating tissues. To clarify ErbB3 trafficking from the cell surface to the nucleus, and its function once there, we applied a comprehensive approach using subcellular fractionation, cell surface protein labeling, immunostaining, and live cell imaging to investigate the transfer of ErbB3 to the nucleus. In addition, we studied the activation state, the translocation mechanisms, and the nuclear entry of ErbB3. To understand its function in the nucleus, we investigated the prevalence of nuclear ErbB3 in both controlled and diseased tissues. Our findings support an alternative pathway to the canonical signaling, which leads to a direct interaction of the surface receptor with the transcriptional machinery in the nucleus.
Experimental Procedures
Cell Culture
BT474 and T47D cells (from Cell Lines Service, Germany) were cultivated in DMEM/F12 (1:1) medium (PAN Biotech) supplemented with 10% heat inactivated FCS (PAN Biotech), 2 mm glutamine (PAN Biotech), and antibiotics. MCF-7 cells (DSMZ, Germany) were cultivated in DMEM (PAN Biotech) supplemented with 10% heat-inactivated FCS, 1 mm sodium pyruvate (Sigma), 10 μg/ml insulin (Sigma), MEM non-essential amino acids (Invitrogen), and antibiotics. HEK 293T cells were cultivated in DMEM medium supplemented with 10% heat inactivated FCS and antibiotics. Cells were maintained in humidified incubators with 5% CO2 at 37 °C. DSMZ confirmed cell line identity and purity by DNA fingerprinting.
ErbB3 Translocation Assay
To induce nuclear translocation, cells were starved in FCS-free cultivation medium overnight and stimulated with 50 ng/ml of respective growth factors for the indicated time. Growth factors used were HRG β1 EGF domain, TGF-α, EGF, and amphiregulin (all R&D systems). To inhibit nuclear export cells were incubated with 20 ng/ml leptomycin B (Tocris) for 1 h before ligand stimulation. Subsequent subcellular fractionation was done as published previously (12). Briefly, ∼1 × 107 cells were lysed in 800 μl of hypotonic buffer (3 mm MgCl2, 20 mm KCl, 2 mm EDTA, in 40 mm HEPES, pH 8.0) supplemented with a mixture of protease and phosphatase inhibitors (Sigma) and homogenized by drawing through a 26-gauge needle of a syringe. After centrifugation at 700 × g for 10 min at 4 °C, the supernatants were analyzed for cytosolic proteins. The pellets, containing the nuclear fraction, were washed 3× with isotonic sucrose buffer (250 mm sucrose, 0.5% Triton X-100, 6 mm MgCl2 in 10 mm Tris-HCl, pH 7.4) at 700 × g for 10 min at 4 °C. The pellets containing the nuclear fractions were re-suspended in 150 μl of high salt buffer (hypertonic buffer plus 500 mm KCl) supplemented with 50 units of benzonase (Novagen) and protease/phosphatase inhibitors on a rotating wheel for 30 min at 4 °C. After centrifugation at 13,000 × g for 10 min at 4 °C, supernatants were collected as nuclear fractions. 50 μg of protein of the fractions were analyzed by immunoblotting.
Immunofluorescence Detecting ErbB3
Approximately 20000cells were seeded onto poly l-lysine-coated 14-mm coverslips. After 3 days of cultivation, cells were treated the same way as described for the translocation assay. Cell fixation was done with 4% PFA for 20 min at room temperature. After 3× washing with PBS, residual formaldehyde was quenched with 50 mm NH4Cl in PBS for 10 min. Cell permeabilization was done with 0.3% Triton X-100 for 15 min at room temperature and 3× washed with PBS. After blocking with 5% BSA, 1% Tween in PBS, cells were incubated with ErbB3 antibody (Ab-12708; 1:100, Cell Signaling) diluted in staining buffer (1% BSA, 0.1% Tween in PBS) overnight at 4 °C followed by 3x washing with PBS. The secondary antibody, goat anti-rabbit Alexa Fluor 647 (1:500, Invitrogen) was incubated for 1 h in staining buffer at room temperature. Cells were washed 3× with PBS, and nuclei were stained with DAPI before mounting. Z-stacks covering the cell nuclei were taken with a FluoViewTM FV1000 (Olympus) confocal microscope using a 63 × 1.35 NA oil immersion objective. Presented images are maximum intensity projections. Images were performed with FluoView Software (Olympus).
Immunoblotting
Protein samples analyzed for ErbB3 were resolved with SDS-PAGE using self-casted 8% acrylamide gels. Other samples were resolved in NuPAGE Novex 3–8% Tris Acetate gels (Invitrogen), according to manufacturer's instructions. Separated proteins were transferred to PVDF membranes (Perkin Elmer) with a semi-dry blotting system (Trans-Blot S.D., Bio-Rad) using 5 mA/cm2 for 40 min. For immunodetection, the following antibodies were used at 1:1000 dilution unless otherwise indicated: ErbB3 (Ab-1328, 1:500; Signalway); phospho-ErbB3 (Y1289), EGFR, ErbB2, cyclin D1, and calnexin (Cell Signaling); NUP358 and RNA polymerase (Bethyl Laboratories); HIF1-α (Novus Biological); dynamin (Calbiochem); clathrin, crm-1, importin β1, lamin A/C (BD Biosciences); α-tubulin, STAT 3, and STAT 5 (Santa Cruz Biotechnology); histone H3, E2F-1, and phospho-EGFR (Y1173) (Abcam). Secondary horseradish peroxidase-coupled antibodies were from Cell Signaling.
Endocytosis of Fluorescently Labeled HRG (HRG488)
Recombinant HRG β1 extracellular domain (R&D Systems) was fluorescently labeled with Alexa Fluor 488 microscale protein labeling kit (Invitrogen) according to the manufacturer's instructions. 20000 T47D cells were seeded onto poly l-lysine-coated 4 well μ-slides (Ibidi) and prepared for the translocation assay. Cells were incubated with 4 μg/ml Hoechst 33258 (Invitrogen) for 20 min at 37 °C and subsequently labeled with 2 μg/ml HRG488 at 4 °C for 1 h. Trafficking was initiated by medium change to 37 °C containing 0.4 μg/ml Hoechst. For time-lapse movies, a custom modified inverted LSM MP7 (Zeiss) with a 63× NA 1.2 water immersion objective was used. During recordings, cells were maintained at 37 °C and 5% CO2 in an incubation chamber (Solent Scientific). The frame rate was 5 s. For excitation, a Chameleon Ultra II laser (Coherent) tuned to 740 nm was used.
ErbB3 Activation Assay
For analysis of ErbB3 activation, the overnight starvation medium of ErbB3 translocation assay was supplemented with 20 μg/ml pertuzumab (Genentech) or 1 μm lapatinib (GlaxoSmithKline). Subsequently, ErbB3 translocation assay was performed in the presence of the inhibitors; the cells were fractionated and analyzed by immunoblotting.
Biotin Cell Surface Labeling
To prevent trafficking, all biotin labeling steps were carried out at 4 °C. To ∼1 × 106 T47D cells prepared for translocation assay, 10 ml of ice-cold PBS containing 250 μg/ml sulfo-NHS-SS-Biotin (Thermo Scientific) was added and incubated gently agitating for 30 min. Labeling was stopped by adding 800 μl of quenching buffer (137 mm NaCl, 5 mm KCl, 2.3 mm CaCl2, 0.5 mm MgCl2, 1 mm Na2HPO4 in 25 mm Tris, pH 8.0) for 10 min. After washing with ice-cold medium, membrane protein trafficking was initiated by adding 37 °C cultivation medium containing 50 ng/ml HRG for the indicated time points. Subcellular fractions were affinity purified with streptavidin magnetic beads (Thermo Scientific). 50 μl of beads were added to 800 μg of protein and rotated. Next day, beads were separated with a magnet and washed 3× with TBS containing 0.1% Tween 20. Proteins were eluted by boiling in sample SDS buffer and analyzed by immunoblotting.
ErbB3 Internalization Assay
To examine the endocytic pathway, T47D cells were incubated with either 100 μm chlorpromazin (Sigma) for 30 min or 1 μg/ml filipin III (Sigma) for 1 h. Cells were biotin-labeled as described, but after HRG stimulation at indicated time points, residual surface biotin was removed from the cells by incubating twice with 180 mm MesNa (Sigma) for 15 min at 4 °C. Excessive MesNa was quenched with 5 mg/ml iodoacetamide (Sigma) for 10 min. Subcellular fractions were affinity purified with MagStrep streptavidin beads as mentioned and analyzed by immunoblotting.
siRNA Knockdowns
The following RNAi (Invitrogen) oligonucleotides against crm-1 (s14938), clathrin (HSS174637), NUP358 (HSS109043), importin β1 (HSS180197) were used. For control experiments, scrambled siRNA was used (Invitrogen). Approximately 1 Mio cells were transfected with 20 nm siRNA oligoribonucleotides using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. Cultures were maintained for 72 h before analysis. Subcellular fractions were prepared and analyzed by immunoblotting.
Immunostainings with HRG488
Approximately 20000 cells were seeded onto poly l-lysine coated 14 mm coverslips. After 2 days of cultivation, cells were treated the same way as described for the translocation assay. For HRG488 labeling, 2 μg/ml HRG488 was added at 4 °C for 1 h. Trafficking was initiated by medium change to 37 °C. After the indicated time points, cells were fixed with 4% PFA for 15 min at room temperature and washed 3× with PBS. Staining was done in PBS containing 1% BSA, rhodamin-phalloidin (1:1000, Invitrogen) and DAPI (1:20000; Invitrogen) for 20 min at room temperature. Cells were washed 3× with PBS and mounted with Fluor Preserve (Calbiochem). For endocytic inhibition, 1 μg/ml filipin III was used. Images were taken with a FluoViewTM FV1000 (Olympus) confocal microscope using a 63 × 1.35 NA oil immersion objective.
Image Processing for Vesicle Detection
Confocal images for all time-points were converted to 16-bit TIFF hyperstacks comprising 3-channels, which correspond to DAPI (nuclear stain, blue), anti-ErbB3-Alexa 488 (ErbB3-positive vesicle marker, green) and phalloidin (cell boundary marker, red). Each channel was subjected to isodata thresholding and binary hole filling to generate contiguous masks of nuclei, ErbB3-positive vesicles and cells. Watershed segmentation was applied to separate vesicles by limiting maximum region diameter to 400 nm. Similarly, cells were segmented by limiting the maximum region diameter to 30 μm. Object detection was performed on each channel to determine the centroids and extent of nuclei, vesicles and the extent of cells. Nuclei and vesicles were assigned to belong to specific cells if the respective centroids were located within the convex hull of the cell. Vesicles were further assigned to be within the nucleus if the centroids were located within the convex hull of the nucleus. The distances between all vesicle centroids and the parent nucleus centroids were calculated. The proportion of the nuclear diameter relative to the cell diameter was then determined. Histograms of vesicle frequency against the location over relative nuclear/cellular diameter were generated. The histograms represent the number of vesicles and their location inside the cell relative to the cell and nuclear boundaries.
ErbB3 Co-immunoprecipitations
Approximately 10 Mio cells were lysed in ice-cold 800 μl RIPA buffer (150 mm NaCl, 1 mm EDTA, 1% sodium deoxycholate, 1% Nonidet P-40, 0.1% SDS in 50 mm Tris, pH 7.4 containing protease/phosphatase inhibitors). Immunoprecipitations were done using 800 μg of protein sample incubated with the appropriate antibody coupled to either protein A or protein G Dynabeads (both Invitrogen) according to manufacturer's instructions rotating at 4 °C. On the next day, immunocomplexes were separated with a magnet and washed 3× with 200 μl of TBS containing 10% glycerol. Proteins were eluted by boiling in 25 μl 2× loading buffer (Bio-Rad). Proteins were separated with a magnet, and supernatants subsequently analyzed by immunoblotting. The following antibodies were used for immunoprecipitations: ErbB3 (Abcam), importin β1, clathrin (both BD Biosciences), NUP358, RNA polymerase (both Bethyl Laboratories), and dynamin (Calbiochem), in addition to control isotype antibodies (Santa Cruz Biotechnology).
Chromatin Precipitation
Chromatin precipitation was done as described previously (13). Harvested cells were incubated with ice-cold cell lysis buffer (0.34 m sucrose, 0.1% Triton X-100, 10 mm KCl, 1.5 mm MgCl2, 10% glycerol 1 mm DTT in 10 mm HEPES, pH 8.0 supplemented with protease/phosphatase inhibitors). After centrifugation at 1300 × g, nuclear pellets were washed with cell lysis buffer lacking Triton X-100 and resuspended in nuclei lysis buffer (3 mm EDTA, 0.2 mm EGTA, 1 mm DTT supplemented with protease/phosphatase inhibitors). Chromatin was obtained by centrifugation at 1300 × g. For immunoblot analysis, chromatin fractions were sonicated in nuclei lysis buffer.
Affinity Precipitations for Transcription Factors
The cytosolic part of human ErbB3 (gift from A. Ullrich laboratory) was cloned into the N-SF-TAP tag vector using the restriction sites, NheI and XbaI (gift from M. Ueffing laboratory (14)). HEK 293T cells were transfected with the resulting plasmid using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. After 48 h of expression, affinity precipitations were prepared analogous to the co-immunoprecipitation experiments, but with MagStrep beads (IBA-Lifesciences). Precipitates were analyzed by immunoblotting.
Normal Tissues and Carcinomas
For the liver regeneration model, 1.6 g/kg body weight CCl4 in olive oil was administered via intraperitoneal injection into C57BL/6N mice (Charles River). After 1 day, liver was excised and fixed in 4% PFA for 48 h at 4 °C. Other tissues were collected and prepared from C57BL/6N and pregnant CD1 control mice. For ErbB3 immunostaining in carcinomas, formalin-fixed paraffin embedded tissue was used. Breast carcinomas were available from a cohort of node-negative patients that was previously described (15, 16). 200 tissues were evaluated after ErbB3 immunostaining according to the following criteria: (i) no nuclear ErbB3 staining in the entire tumor tissue; (ii) nuclear ErbB3 staining in at least 90% of all tumor cells; (iii) mixed pattern with less than 90% of the tumor cells expressing ErbB3 in the nucleus. Statistical analysis was performed with SPSS software version 22. Ovarian carcinomas were from a cohort previously described by Ref. 17.
Immunohistochemistry
5 μm thick paraffin slices of human tumor and mouse tissues were cleared with Rotihistol (Carl Roth) for 2 × 5 min and rehydrated through graded ethanol to PBS. Antigen was retrieved by boiling in 10 mm citrate buffer, pH 6.0 for 3 × 7 min and subsequent peroxidase activity was quenched by treatment with hydrogen peroxide (3%) in methanol for 15 min at room temperature. After rinsing the slices 3× with PBS, blocking solution (5% swine serum (Vector Laboratories), 3% BSA, 0.1% Tween in PBS) was added overnight at 4 °C. Endogenous avidin and biotin were blocked with the avidin-biotin-blocking kit (Vector Laboratories) according to the manufacturer's instructions. Incubation with ErbB3 antibody (Ab-21510–2; 1:50, Signalway) in staining buffer (1% swine serum, 1% BSA, 0.1% Tween in PBS) was done overnight at 4 °C. After washing 3× with PBS-T (PBS containing 0.5% Tween), the horseradish peroxidase-labeled secondary antibody (1:25; Dako) was incubated in staining buffer for 2 h at room temperature. After washing 3× with PBS-T, samples were developed with diaminobenzidine (DAB) substrate (Vector Labs) for 1–5 min. Slides were washed with PBS and counter-stained with hematoxylin (Merck), dehydrated through graded ethanol steps, and embedded with entellan (Merck). Images were taken with a BX41 microscope (Olympus).
Tumor Microenvironment
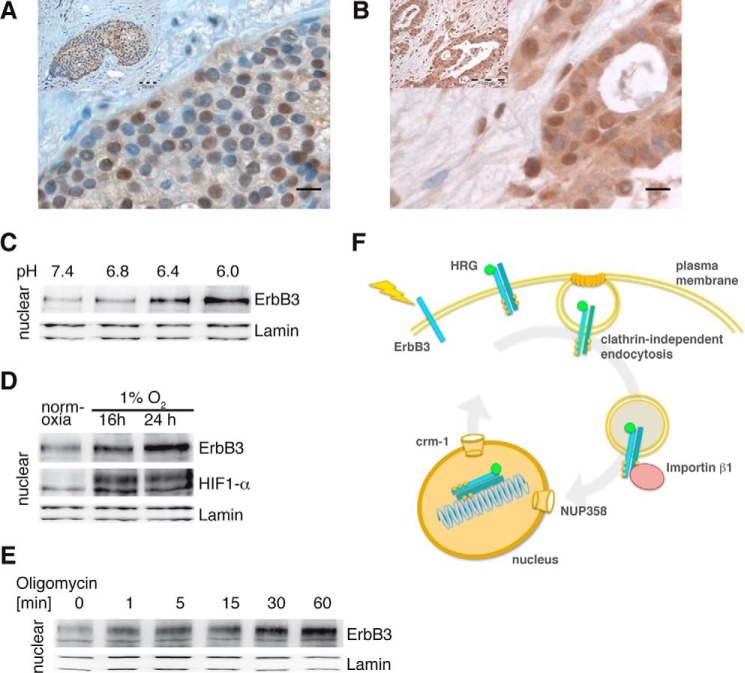
To produce acidified medium, bicarbonate-free DMEM/F12 medium was supplemented with 20 mm MES and adjusted to the indicated pH. Cells were incubated for 1 h in the acidified medium. The hypoxia experiment was done in a cell incubator set to 1% O2 using DMEM/F12 medium that was deoxygenized immediately before use. Cells were kept under these conditions for 16 and 24 h. In the ATP depletion experiment, a medium change with glucose-free DMEM/F12 containing 2.5 μm oligomycin B (Applichem) and 6 mm 2-deoxyglucose (Sigma) was done and maintained for the indicated time. For all conditions, cells were fractionated and analyzed by immunoblotting.
Results
ErbB3 Localizes to the Nucleus in Response to Heregulin Stimulation
Nuclear localization of ErbB3 was investigated in the breast cancer cell lines MCF-7, T47D and BT474. Immunocytochemistry followed by confocal imaging showed ErbB3 localized in the nuclei of MCF-7 and T47D cells (Fig. 1A), which increased after heregulin (HRG) stimulation.
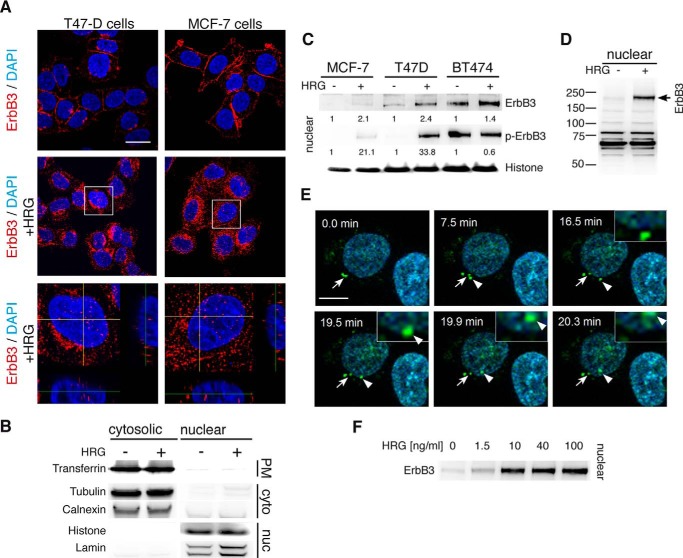
FIGURE 1.
Nuclear localization of ErbB3 in cancer cells. A, induction of nuclear ErbB3 in T47D and MCF-7 cells. Representative confocal images of starved cells (upper) and with HRG-stimulated cells (middle) immunostained for ErbB3 (red) and nuclei (blue), lower panel displays ortho-representations of confocal z-stacks of the nuclei of stimulated cells. B, validation of nuclear fractionation. Nuclear fractions and supernatants were prepared from unstimulated and HRG stimulated T47D cells. Fractions were immunoblotted and probed for marker proteins of the plasma membrane (PM; transferrin receptor), cytosol (cyto; tubulin and calnexin), and the nucleus (nuc; histone H3 and lamin A/C). C, induction of ErbB3 nuclear translocation by its ligand HRG. Starved MCF-7, T47D, and BT474 cells were stimulated for 10 min with 50 ng/ml heregulin (HRG). Nuclear fractions were immunoblotted for ErbB3, phosphorylated ErbB3, and histone. Shown are representative images from three independent experiments. D, ErbB3 translocates to the nucleus in its full-length form. Entire Western blot membrane immunoblotted for ErbB3 after HRG stimulation is shown. E, ErbB3 containing vesicles enter the nuclei. Starved T47D cells were stained with Hoechst (blue) and labeled with fluorescent HRG (HRG488; green) at 4 °C for 1 h. Trafficking was initiated by 37 °C medium change. Images are taken from Video1. F, identification of HRG concentrations stimulating ErbB3 nuclear transport. After stimulation of T47D cells using indicated HRG concentrations for 10 min, surface biotinylated proteins were purified from the nuclear fraction, immunoblotted and probed for ErbB3. Scale bars for (A) 20 μm and (E) 10 μm.
To confirm the immunohistochemistry results, we isolated nuclear fractions and performed Western blot analysis to localize ErbB3. Nuclear ErbB3 was detected along with validated nuclear marker, histone H3 thus confirming its nuclear localization in the investigated cell lines (Fig. 1, B and C). T47D and BT474 cells showed nuclear ErbB3 even in the absence of HRG stimulation; whereas, MCF-7 cells displayed almost no nuclear localization of ErbB3 prior to HRG stimulation. Despite the differences in basal nuclear ErbB3 expression, all three cell lines showed a clear increase in nuclear localized ErbB3 in response to HRG. The total nuclear ErbB3 level was associated with its phosphorylation state after stimulation with HRG (Fig. 1C). The BT474 cells are known to express high ErbB2 levels, which lead to ligand-independent activation of ErbB3 (18). The enhanced phosphorylation and concomitant increased nuclear translocation of ErbB3 observed in these cells are likely due to ErbB2-mediated trans-phosphorylation and are therefore not significantly affected by HRG stimulation. ErbB3 was found as a full-length molecule in the nuclear fractions, with a molecular mass of ∼185 kDa (Fig. 1D), ruling out that the nuclear fraction of ErbB3 represents truncated versions or alternative isoforms of the protein.
To directly observe ErbB3 nuclear trafficking in real time, fluorescently labeled HRG (HRG488) was used for live cell imaging in T47D cells. After the addition of HRG488, fluorescent vesicles accumulated within minutes around the perinuclear region, and a fraction of which entered the nucleus (Fig. 1E, video1.avi, video2.avi).
Activation of Membrane-bound ErbB3 Is a Precondition for Trafficking
Because of its impaired kinase activity, ErbB3 depends on heterodimerization for activation, mediated by its ligand HRG. In response to HRG stimulation, ErbB2 has been shown to be the preferred ErbB3 dimerization partner (19). To identify ligands able to trigger ErbB3 nuclear translocation and the contribution of dimerization partners, different growth factors of the EGFR family were tested. ErbB3 nuclear translocation in T47D cells was triggered by its ligand HRG in a concentration-dependent manner (Fig. 1F), but not by any of the other tested growth factors of the EGFR family, such as amphiregulin (AR), epidermal growth factor (EGF), and transforming growth factor (TGFα) (Fig. 2A). EGFR had been included as a control of the used growth factors.
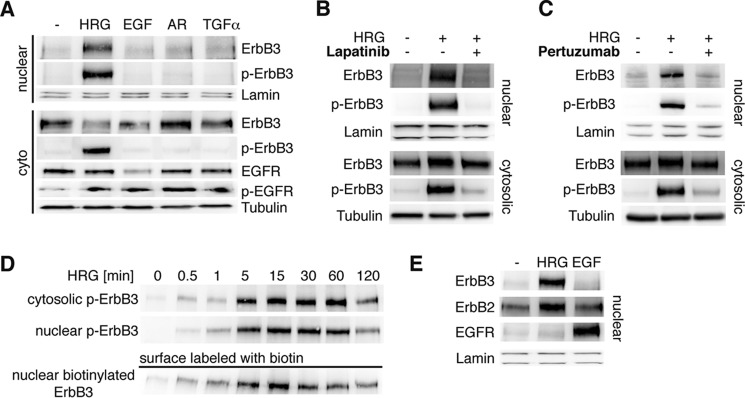
FIGURE 2.
Activation by HRG triggers ErbB3's nuclear translocation. A, stimulation with ligands of the EGF receptor family. Starved T47D cells were stimulated with HRG, amphiregulin (AR), EGF, or TGF β. 50 μg of cytosolic and nuclear extracts were immunoblotted for ErbB3, phosphorylated ErbB3, phosphorylated EGFR and lamin. B and C, blocking of ErbB3 dimerization or phosphorylation by pertuzumab or lapatinib antagonizes nuclear translocation. Starved T47D cells were incubated with (B) lapatinib or (C) pertuzumab and stimulated with HRG. Cytosolic and nuclear extracts were immunoblotted for ErbB3, phosphorylated ErbB3, lamin and tubulin. D, nuclear ErbB3 originates from the plasma membrane. Starved T47D cells were surface-labeled with biotin. After the stimulation with HRG for the indicated time periods, nuclear fractions and supernatants were immunoblotted for ErbB3 and phosphorylated ErbB3. In addition, nuclear fractions were affinity purified using streptavidin-conjugated beads. Precipitates were analyzed by immunoblotting using an antibody against ErbB3. E, ErbB2 is the favorite trafficking partner of ErbB3 in T47D cells. Starved T47D cells were stimulated with HRG or EGF. Nuclear extracts were immunoblotted for ErbB3, ErbB2, EGFR, and lamin. A representative immunoblot from three independent experiments is shown.
To study the relevance of the activation state of ErbB3 for nuclear translocation, T47D cells were incubated with lapatinib, which inhibits the kinase activity of EGFR and ErbB2, thus preventing cross-phosphorylation of ErbB3. Both ErbB3 phosphorylation and HRG-induced nuclear translocation were blocked by lapatinib (Fig. 2B), establishing a clear relationship between nuclear translocation and the phosphorylation state of ErbB3. Similarly, lapatinib treatment also blocked nuclear translocation of ErbB3 in BT474 cells (data not shown), underscoring its dependence on receptor phosphorylation.
To confirm that nuclear translocation of ErbB3 is indeed mediated by trans-phosphorylation by other RTKs, we inhibited the dimerization of ErbB3 with its preferred partner ErbB2 using pertuzumab, a therapeutic antibody against ErbB2 (20). As with lapatinib, pertuzumab inhibited ErbB3 phosphorylation, and completely blocked its nuclear translocation after HRG stimulation (Fig. 2C).
Nuclear ErbB3 Originates from the Plasma Membrane-bound ErbB3 Fraction
To track ErbB3 phosphorylated at the plasma membrane, we utilized surface protein labeling with biotin in T47D cells. The location of surface-labeled ErbB3 was investigated over time using Western blot analysis. Biotin-labeled ErbB3 was detected in nuclear fractions as early as 1 min after stimulation with HRG, which correlated with the phosphorylation of the receptor in the cytosol (Fig. 2D). Nuclear ErbB3 levels peaked at 15 min, and gradually decreased over the next 90 min.
Analysis of the nuclear fraction for EGFR or ErbB2 showed that ErbB2 levels but not EGFR levels, increased in the nucleus along with ErbB3 (Fig. 2E). Alternatively, stimulation with EGF instead of HRG, led to an increase in the nuclear translocation of EGFR and ErbB2, but not ErbB3. Together, these results show that ErbB3 co-migrates to the nucleus with ErbB2 following ligand-specific stimulation and phosphorylation.
Nuclear Transport of ErbB3 Is Mediated by Clathrin-independent Endocytosis
To identify the pathway by which the ErbB3-loaded vesicles are derived, we investigated clathrin-dependent and clathrin-independent endocytosis as common mechanisms for transmembrane receptor internalization (21). Pharmaceutical inhibition with chlorpromazine: an inhibitor of clathrin-dependent endocytosis (22), and filipin: a general inhibitor of clathrin-independent endocytosis (23), revealed that both pathways translocate ErbB3 to the nucleus with distinct kinetics. Following surface-biotinylation and HRG-stimulation, biotinylated ErbB3 was tracked in the nuclear and cytosolic fractions of T47D cells. Inhibition of clathrin-mediated endocytosis resulted in a significant delay in ErbB3's translocation to the cytosol as well as the nucleus 1 min following HRG-stimulation (Fig. 3, A and B). Immunoprecipitation in total cell extracts followed by Western blotting also showed that ErbB3 co-precipitated with clathrin and dynamin, indicating the involvement of clathrin-mediated endocytosis in ErbB3 translocation (Fig. 3C). Nevertheless, knocking down clathrin using siRNA did not influence the nuclear transport of ErbB3 (Fig. 3D).
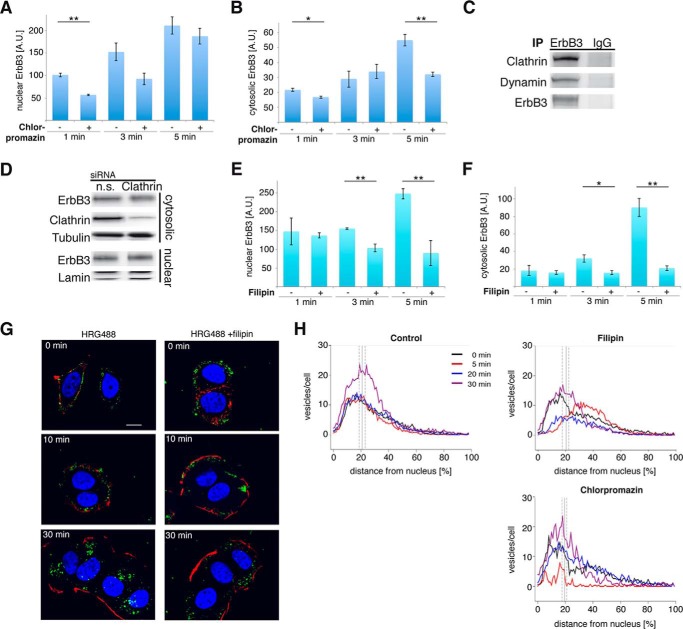
FIGURE 3.
Clathrin-independent trafficking translocates ErbB3 to the nucleus. A and B, blocking clathrin-dependent pathways reduced ErbB3 nuclear translocation at early time points. Starved T47D cells were incubated with chlorpromazine. Surface proteins were labeled with biotin and stimulated with HRG for the indicated time periods. Remaining surface biotin was removed and nuclear (A) and cytosolic (B) fractions were analyzed for ErbB3 in immunoblotting and quantified. C, ErbB3 binds to clathrin and dynamin. Total cell lysates from T47D cells were immunoprecipitated (IP) with an antibody against ErbB3 and isotype IgG for control. Precipitates were analyzed by immunoblotting for clathrin, dynamin, and ErbB3. D, knock-down of clathrin does not influence nuclear levels of ErbB3. T47D cells were transfected with siRNA against clathrin and nonspecific siRNA (n.s.). To determine the influence of clathrin down-regulation, the cytosolic fraction was analyzed by immunoblotting for ErbB3, clathrin, and tubulin. The T47D cells were stimulated with HRG and the nuclear fraction immunoblotted and analyzed for ErbB3 and lamin. E and F, inhibition of clathrin-independent endocytosis prevented nuclear ErbB3. Same procedure as for A and B but with filipin. Nuclear (E) and cytosolic (F) fractions were analyzed for ErbB3 in immunoblotting and quantified. G and H, Filipin induced delayed nuclear entry of ErbB3 vesicles. Starved T47D cells were labeled with fluorescent HRG (HRG488; green) at 4 °C for 1 h. Trafficking was initiated by 37 °C medium change without (left) and in the presence of filipin (right). H, starved T47D cells without inhibitor (upper left), incubated with filipin (upper right), or chlorpromazin (lower) were stimulated with HRG for the indicated time periods. Cells were stained with an antibody specific for ErbB3, phalloidin, and DAPI and confocal images were taken. Plots are representing the quantified distances of ErbB3 containing vesicles between the point of the nearest plasma membrane and the centroid of the corresponding nucleus in percent. Nuclear boundaries were quantified and indicated with the gray solid vertical line; vesicles within the boundary were considered to be within the nucleus. Scale bar 10 μm. The numbers represent the mean ± S.E. of quantified ErbB3 immunoblot signals (n = 3; arbitrary units A.U.). A representative immunoblot and images from three independent experiments are shown. Results are shown in mean ± STD; *, p < 0.05; **, p < 0.01.
In contrast to the chlorpromazin experiment, inhibition of clathrin-independent pathways showed significant retardation of ErbB3 internalization as well as nuclear translocation, albeit at later time points after stimulation. Filipin-treatment prevented internalization of ErbB3, reflected by reduced (>60%) cytosolic and nuclear biotinylated ErbB3 at 3–5 min following HRG stimulation (Fig. 3, E and F). This inhibition occurs much later compared with the clathrin-mediated pathway, which appears to make a greater contribution toward ErbB3 internalization at 1 min following HRG stimulation. Control experiments using fluorescently labeled transferrin showed that the applied filipin concentration was not affecting clathrin-dependent uptake (data not shown). These data show that both clathrin-dependent and independent pathways contribute to ErbB3 internalization, but the fast clathrin-dependent endocytosis is responsible for ErbB3's cytoplasmic trafficking, while the high capacity clathrin-independent pathway is responsible for ErbB3's nuclear translocation.
In accordance with the result of the biotin-based internalization assay, immunocytochemistry along with Alexa-488 labeled HRG confirmed retardation of ErbB3 uptake in T47D cells incubated with filipin (Fig. 3G). HRG488-bound ErbB3 was localized to the perinuclear region and nucleus in the control cells 30 min following HRG488 stimulation. However, in the filipin-treated cells, the HRG488-bound ErbB3 was localized in vesicles close to the plasma membrane, distant from nuclei.
In addition to the pulse-chase experiments, ErbB3 endocytosis in response to HRG was further addressed by immunocytochemistry using ErbB3-specific antibodies in order to visualize all ErbB3-containing vesicles. Confocal images were segmented and the spatial distribution of each ErbB3-positive vesicle was quantified. Thereby, the ratio of the distance of each ErbB3 containing vesicle between the centroids of the nucleus and the closest plasma membrane was determined. In absence of HRG, ErbB3 staining represents the steady-state localization of ErbB3 vesicles, which represents the balance between nuclear entry as well as nuclear exit (Fig. 3H). The increase in the fraction of ErbB3-containing vesicles inside the nucleus due to HRG stimulation was confirmed in the control experiment without inhibitors. Analogous to the biotin-based internalization assay, blocking of the clathrin-dependent endocytosis with chlorpromazin inhibited very efficiently the net nuclear transport of ErbB3 positive vesicles at the early time points after HRG stimulation. At later time points, the fractions of ErbB3 containing vesicles increased comparable to the control experiment. In contrast to chlorpromazin, filipin treatment hindered the nuclear entry of ErbB3 containing vesicles in comparison to control samples without pharmacological inhibitors. Blocking clathrin-independent pathway decreased ErbB3 nuclear translocation after HRG stimulation, despite ErbB3 vesicles accumulated at the perinuclear area. These data show that the clathrin-dependent pathway contributes initially to the ErbB3 internalization. But at later time points, clathrin-independent endocytosis becomes the main pathway responsible for the major fraction of ErbB3 transported to the nucleus.
ErbB3 Shuttles Across the Nuclear Membrane through the Nuclear Pore Complex
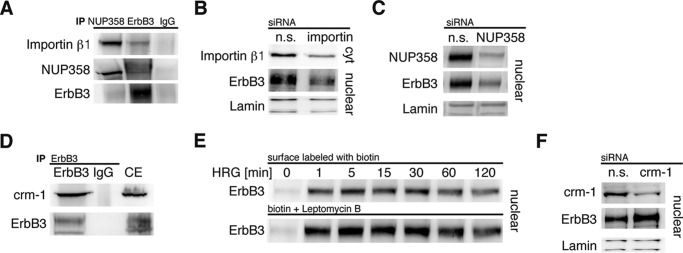
The shuttling mechanism of ErbB3 through the nuclear membrane was investigated by examining the roles of two major proteins involved in nuclear import; importin β and nuclear pore component NUP358 (24). Immunoprecipitations using antibodies against ErbB3 and NUP358 revealed the interaction between ErbB3, nuclear pore complex, and importin β (Fig. 4A). Furthermore, siRNA-mediated knock-down of either importin β or NUP358 led to reduced nuclear transport of ErbB3 (Fig. 4B+C). Western blots of nuclear fractions showed that ErbB3 is gradually depleted from the nucleus over time (Fig. 2D), suggesting nuclear export via the nuclear export receptor, crm-1. Indeed, ErbB3 was shown to interact with crm-1 by immunoprecipitation (Fig. 4D). Pharmacological inhibition of crm-1 by leptomycin B, a specific crm-1 inhibitor, induced a strong enrichment of nuclear ErbB3 compared with controls (Fig. 4E). Finally, these data were corroborated by siRNA-mediated knock down of crm-1, which also showed increased nuclear ErbB3 levels (Fig. 4F).
FIGURE 4.
Nuclear import and export of ErbB3. A–C, importin β1 and NUP358 are required for nuclear import of ErbB3. A, total cell lysates from T47D cells were immunoprecipitated (IP) with antibodies against NUP358, ErbB3, and isotype IgG for control. Precipitates were analyzed by immunoblotting reciprocally for NUP358, ErbB3, and importin β1. B, same cells were transfected with siRNA against importin β1 and nonspecific (n.s.) siRNA. To confirm knock-down, cytosolic fractions were analyzed by immunoblotting for importin β1. The nuclear fractions were immunoblotted and analyzed for ErbB3 and lamin. C, cells were transfected with siRNA against NUP358 and unspecific control siRNA. After HRG stimulation nuclear fractions were analyzed for the knock-down efficiency of NUP358 and the effect on ErbB3 nuclear translocation. D–F, ErbB3 nuclear export by crm-1. D, total cell extracts (CE) from the same cells were immunoprecipitated with an antibody against ErbB3 or serotype IgG for control. Precipitates were analyzed for crm-1 and ErbB3. E, surface proteins of starved cells incubated with leptomycin B were biotin labeled and stimulated for the indicated periods with HRG. Nuclear proteins were affinity-purified with streptavidin beads and analyzed for ErbB3 by immunoblotting. F, crm-1 was knocked-down by siRNA in T47D cells. Nuclear fractions were analyzed by immunoblotting for crm-1 and ErbB3. A representative immunoblot from three independent experiments is shown.
Nuclear ErbB3 Is Associated with Cell Proliferation
To determine the function of ErbB3 localized to the nucleus, we investigated its interaction with DNA using isolated chromatin fractions from T47D cells. Western blot analysis following HRG-stimulation with an ErbB3 antibody confirmed ErbB3's ability to bind DNA, which was not the case in lapatinib-treated cells (Fig. 5A). Furthermore, immunoprecipitation revealed binding of ErbB3 to RNA polymerase II and histone H3 (Fig. 5B). In addition, ErbB3 co-precipitated with the transcription factors E2F-1, STAT 3, STAT 5, but not cyclin D1 (Fig. 5C), suggesting its role as a transcriptional co-regulator. The transcription factors STAT3, STAT5 are involved in cell proliferation, indicative of a role for ErbB3 in this process.
FIGURE 5.
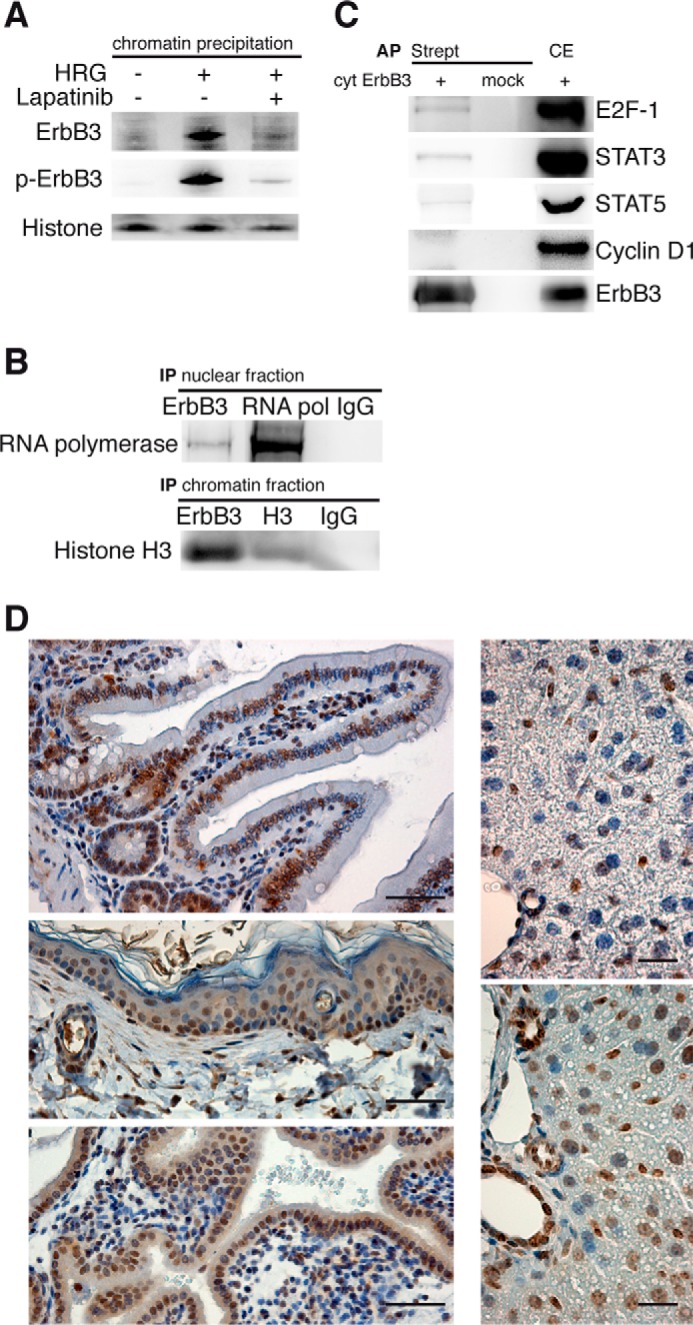
ErbB3 interacts with the transcription machinery and its nuclear expression in proliferating tissues. A and B, ErbB3 binds to chromatin and RNA polymerase II. A, starved T47D cells were incubated with lapatinib and HRG. Chromatin precipitates were analyzed using immunoblots for ErbB3 and histone H3. B, nuclear or chromatin fractions from T47D cells were immunoprecipitated (IP) with antibodies against ErbB3, RNA polymerase II, histone H3, and control isotype IgGs. Precipitates were analyzed using immunoblots for RNA polymerase and histone H3. C, ErbB3 binds to transcription factors. HEK 293T cells were transfected with the cytosolic part of ErbB3 fused to N-SF-TAP tag (cyt ErbB3). Affinity precipitations (AP) with streptavidin beads were performed using total cell extracts and analyzed for ErbB3 binding by immunoblotting for the indicated transcription factors. D, proliferating tissues exhibit nuclear ErbB3. Mouse tissues were immunostained for ErbB3 and counterstained with hematoxylin in the following tissues: intestine (upper left panel), skin (center left panel), uterus of pregnant mouse (bottom left panel), normal liver (upper right panel) and regenerating liver (lower right panel: Regenerating liver was obtained 1 day after intraperitoneal injection of the liver toxicant, CCl4). Scale bars for left panel 50 μm, right panel 20 μm. A representative immunoblot and images from three independent experiments are shown.
To investigate the role of nuclear ErbB3 in cell proliferation, we tested for its nuclear presence in murine tissue-types with high basal proliferation - intestinal, epidermal, and uterine epithelia. Nuclear ErbB3 was found in the crypts of the small intestine, in cells of the germinal layer of the skin, and the uterine epithelial layer of a pregnant mouse (day 16.5 of pregnancy) (Fig. 5D). Furthermore, regenerating mouse liver, collected on day 1 after treatment with the liver toxicant, carbon tetrachloride (25), showed nuclear ErbB3 in more than 95% of the hepatocytes compared with less than 5% in untreated livers. Together, these data strongly correlate the presence of nuclear ErbB3 in proliferating cells, possibly functioning as a transcriptional co-regulator.
Extracellular pH, pO2, and Intracellular ATP Affect ErbB3 Nuclear Localization
The role of nuclear ErbB3 in proliferating cells led us to investigate its implications in oncogenic cell proliferation. We studied ErbB3 localization in a previously described cohort (15) of 200 node-negative breast carcinomas by immunohistochemistry. ErbB3 was predominantly localized to the nucleus of the cancer cells in 19% of the carcinomas, and further 19.5% of the carcinomas exhibited a mixed ErbB3 localization with both positive and negative nuclei (Fig. 6A). Nuclear ErbB3 was also observed in epithelial ovarian carcinoma tissues (Fig. 6B) from an independent cohort (17).
FIGURE 6.
Tumor microenvironment causes nuclear ErbB3. A and B, nuclear expression of ErbB3 in tumor tissue. A, representative examples of a node-negative estrogen receptor positive breast carcinoma, and (B) a serous, epithelial ovarian carcinoma. Both tissue specimens were immunostained for ErbB3 and nuclei were counterstained with hematoxylin. Tumor environment triggered nuclear translocation of ErbB3. C, acidic environment. T47D cells were maintained in medium with the indicated pH for 1 h. Nuclear fractions were immunoblotted for ErbB3 and lamin. D, hypoxic conditions. T47D cells were maintained at 21% O2 (normoxia) for 24 h and at 1% O2 for the indicated time periods. Isolated nuclei were subjected to Western blotting and immunostained for ErbB3, HIF1-α, and lamin. E, ATP deprivation. T47D cells were incubated with oligomycin B for the indicated time periods. Nuclear proteins were extracted, subjected to immunoblotting, and labeled for ErbB3 and lamin. A representative immunoblot from three independent experiments is shown. F, proposed model for ErbB3 nuclear translocation. ErbB3 is activated at the plasma membrane by its ligand HRG, dimerizes with ErbB2 and becomes cross-phosphorylated. ErbB3 is endocytosed via a clathrin-independent mechanism and is guided by importin β1 to the nucleus. At the nuclear membrane, ErbB3 binds to NUP358 which facilitates its transport through the nuclear pores. Once inside the nucleus, ErbB3 binds to chromatin and serves as a transcription regulator together with transcription factors. Crm-1 exports ErbB3 out of the nucleus to the cytoplasm.
We have shown earlier that HRG stimulation induces nuclear translocation of ErbB3. A characteristic feature of tumors is high cell densities, which results in decreased nutrient and oxygen availability as well as low pH. To investigate if tumor micro-environments trigger ErbB3 nuclear translocation, T47D cells were maintained either in acidified medium, under hypoxic conditions, or upon depletion of ATP. ErbB3 levels in the nuclei of T47D cells increased in proportion to acidity of the medium over 1 h (Fig. 6C). Similarly, in T47D cells maintained under hypoxic conditions (1% O2), ErbB3 levels in the nucleus increased with time over 24 h correlating with the induction of the hypoxia marker, HIF1-α (Fig. 6D). Finally, ATP depletion using oligomycin B led to a rapid (∼1 min) increase in nuclear ErbB3 lasting up to 60 min (Fig. 6E). Together, these data suggest that nuclear ErbB3 plays important roles in oncogenicity that are related to transcriptional modulation that are distinct from its well established roles in canonical MAPK/PI3K signaling.
Discussion
Nuclear translocation of RTKs represents an alternative to canonical signaling, which interacts with the transcriptional machinery of cells (26). However, the mechanism by which membrane bound RTK translocate to the nucleus remains poorly understood. In the present study, we identified a trafficking mechanism of ErbB3 from the cell surface to the nucleus in breast cancer cell lines. The results support a model whereby ligand-activated phosphorylated ErbB3 is endocytosed via a clathrin-independent pathway; followed by its transfer across the nuclear membrane through interaction with importin β1, which then initiates ErbB3 passage through the nuclear pore complex with the aid of NUP358 (Fig. 6F). Nuclear ErbB3 binds to DNA and interacts with proteins of the transcription machinery, such as RNA polymerase II and transcription factors involved in cell survival and proliferation, for example STAT 3, STAT 5, and E2F-1. Using several orthogonal approaches, including subcellular fractionation, surface protein labeling, and direct live cell imaging techniques, a mechanism of ErbB3 nuclear translocation was derived. Trafficking of ErbB3 to the nucleus was induced by its ligand HRG and by conditions typical for the tumor microenvironment; hypoxia, ATP depletion, and low pH. Whether stress conditions also induce nuclear transport via autocrine HRG stimulation or by a different mechanism remains elusive.
Upon ligand binding, dimerization of ErbB3 takes place and tyrosine residues are cross-phosphorylated, which our data shows to be an important prerequisite for subsequent nuclear translocation. It may be noted that ErbB3 was found in its full-length form in the nucleus, where it retains its phosphorylation state and accumulates along with its dimerization partner ErbB2, possibly serving as a docking site for interacting proteins in the nucleus. Extracellular biotin-labeling of ErbB3 showed that nuclear ErbB3 originated at the cell surface and accumulated over time. This contradicts a previous report that ErbB3 is exported out of the nucleus in response to HRG (27). Our results show that ErbB3 is shuttled out of the nucleus by the nuclear export protein crm-1, suggesting a transient nuclear localization. The exact mechanism of ErbB3 internalization remains unclear. Initially ErbB3 has been suggested to be endocytosis-impaired (28, 29); not only because it showed slower rate of internalization in comparison to EGFR but also because it is unable to bind AP2, an important player in clathrin-dependent endocytosis. Recently it was reported that a constitutive internalization of ErbB3 from the plasma membrane occurred in the absence of the added ligand in a clathrin-dependent manner (30). Our results further demonstrate that a clathrin-independent pathway is important for the endocytic internalization and subsequent nuclear translocation of the ligand-activated receptor. A strong decrease in ErbB3 nuclear translocation was observed over time upon inhibiting non-clathrin pathways with filipin. Inhibiting the clathrin-dependent pathway with chlorpromazine did not show any influence on ErbB3 nuclear translocation at later time points where filipin showed clear inhibitory effects. The fast kinetics, but saturable nature of the clathrin-dependent pathway (31) explain its contribution to ErbB3 nuclear translocation at early time points and its limited ability to accommodate HRG-induced ErbB3 internalization, where it is taken over by the slow high capacity non-clathrin pathway. This observation might explain why an interaction between ErbB3 and clathrin was observed, but clathrin knock-down had no influence on nuclear levels of ErbB3.
Previous work showed that RTKs can bind to the promoter regions of various genes and may control transcription. In this context it may be of interest that RON, another RTK, translocates to the nucleus and activates the c-Jun promoter in response to hypoxia (32). However, the molecular mechanism underlying RON internalization and its nuclear translocation remains unknown.
Previous reports showed that the C-terminal domain of ErbB3, distal to the tyrosine kinase domain contained a potent transactivation activity (33) and that an 80 kDa nuclear variant of ErbB3 was able to activate Cyclin D1 promoter (34). In this regard our study shows that ErbB3 binds the transcription factors STAT 3 and 5, both of which are important for proliferation. Moreover our observation that nuclear ErbB3 is found in several proliferating tissues supports the concept that nuclear ErbB3 promotes cell proliferation via co-regulation of gene transcription, and is consistent with the nuclear expression of ErbB3 in a number of proliferating tissues, including tumors, and during the process of tissue regeneration which had been also proposed by previous studies (15, 17, 35, 36). Taken together, ErbB3 responds to extracellular stimuli by nuclear trafficking, which is driven by a clathrin-independent mechanism and leads to the binding of transcription factors involved in cell proliferation.
Overall, the present study demonstrates the mechanism of ErbB3's translocation from the cell surface to the nucleus, including a characterization of the stimuli that result in its cytoplasmic internalization, and nuclear entry. Furthermore, ErbB3 binding to factors of the transcriptional machinery and its expression in the nucleus of proliferating tissues highlight that ErbB3 nuclear translocation occurs more frequently than previously expected, not only in tumors, but also in healthy proliferating tissues.
Author Contributions
R. R., A. A., and J. G. H. designed the study; R. R., A. A., J. S., G. G., A. G., and W. S. performed experiments; R. R., A. A., J. S., G. G., A. G., M. S., W. S., N. V., and J. G. H. analyzed and interpreted data; R. R., A. A., and J. G. H. prepared the manuscript. All authors approved the final version of the manuscript.
Acknowledgments
We thank Frauke Melchior from ZMBH, University of Heidelberg, Germany for the NUP358 antibody, Axel Ullrich from Max Planck Institute for Biochemistry, Munich, Germany for the ErbB3 construct, Marius Ueffing from Helmholtz Zentrum München, Neuherberg, Germany for the SF-TAP vector, and Rosemarie Marchan for critically reading the manuscript.
This work was supported in part by the BMBF project Oncoprofile (Grant Number 01GR0816). The authors declare that they have no conflicts of interest with the contents of this article.
- EGFR
- epidermal growth factor receptor
- RTK
- receptor tyrosine kinase family
- HRG
- heregulin.
References
- 1. Hynes N. E., and MacDonald G. (2009) ErbB receptors and signaling pathways in cancer. Curr. Opin. Cell Biol. 21, 177–184 [DOI] [PubMed] [Google Scholar]
- 2. Sergina N. V., and Moasser M. M. (2007) The HER family and cancer: emerging molecular mechanisms and therapeutic targets. Trends Mol. Med. 13, 527–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Reschke M., Mihic-Probst D., van der Horst E. H., Knyazev P., Wild P. J., Hutterer M., Meyer S., Dummer R., Moch H., and Ullrich A. (2008) HER3 is a determinant for poor prognosis in melanoma. Clin. Cancer Res. 14, 5188–5197 [DOI] [PubMed] [Google Scholar]
- 4. Sithanandam G., and Anderson L. M. (2008) The ERBB3 receptor in cancer and cancer gene therapy. Cancer Gene Therapy 15, 413–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Engelman J. A., Zejnullahu K., Mitsudomi T., Song Y., Hyland C., Park J. O., Lindeman N., Gale C. M., Zhao X., Christensen J., Kosaka T., Holmes A. J., Rogers A. M., Cappuzzo F., Mok T., Lee C., Johnson B. E., Cantley L. C., and Jänne P. A. (2007) MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316, 1039–1043 [DOI] [PubMed] [Google Scholar]
- 6. Hamburger A. W. (2008) The role of ErbB3 and its binding partners in breast cancer progression and resistance to hormone and tyrosine kinase directed therapies. J. Mammary Gland Biol. Neoplasia 13, 225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jain A., Penuel E., Mink S., Schmidt J., Hodge A., Favero K., Tindell C., and Agus D. B. (2010) HER kinase axis receptor dimer partner switching occurs in response to EGFR tyrosine kinase inhibition despite failure to block cellular proliferation. Cancer Res. 70, 1989–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sergina N. V., Rausch M., Wang D., Blair J., Hann B., Shokat K. M., and Moasser M. M. (2007) Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature 445, 437–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marti U., Burwen S. J., Wells A., Barker M. E., Huling S., Feren A. M., and Jones A. L. (1991) Localization of epidermal growth factor receptor in hepatocyte nuclei. Hepatology 13, 15–20 [PubMed] [Google Scholar]
- 10. Giri D. K., Ali-Seyed M., Li L. Y., Lee D. F., Ling P., Bartholomeusz G., Wang S. C., and Hung M. C. (2005) Endosomal transport of ErbB-2: mechanism for nuclear entry of the cell surface receptor. Mol. Cell Biol. 25, 11005–11018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ni C. Y., Murphy M. P., Golde T. E., and Carpenter G. (2001) gamma -Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science 294, 2179–2181 [DOI] [PubMed] [Google Scholar]
- 12. Ledoux S., Yang R., Friedlander G., and Laouari D. (2003) Glucose depletion enhances P-glycoprotein expression in hepatoma cells: role of endoplasmic reticulum stress response. Cancer Res. 63, 7284–7290 [PubMed] [Google Scholar]
- 13. Narita M., Narita M., Krizhanovsky V., Nuñez S., Chicas A., Hearn S. A., Myers M. P., and Lowe S. W. (2006) A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell 126, 503–514 [DOI] [PubMed] [Google Scholar]
- 14. Gloeckner C. J., Boldt K., Schumacher A., Roepman R., and Ueffing M. (2007) A novel tandem affinity purification strategy for the efficient isolation and characterisation of native protein complexes. Proteomics 7, 4228–4234 [DOI] [PubMed] [Google Scholar]
- 15. Schmidt M., Böhm D., von Törne C., Steiner E., Puhl A., Pilch H., Lehr H. A., Hengstler J. G., Kölbl H., and Gehrmann M. (2008) The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 68, 5405–5413 [DOI] [PubMed] [Google Scholar]
- 16. Schmidt M., Micke P., Gehrmann M., and Hengstler J. G. (2012) Immunoglobulin kappa chain as an immunologic biomarker of prognosis and chemotherapy response in solid tumors. Oncoimmunology 1, 1156–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tanner B., Hasenclever D., Stern K., Schormann W., Bezler M., Hermes M., Brulport M., Bauer A., Schiffer I. B., Gebhard S., Schmidt M., Steiner E., Sehouli J., Edelmann J., Läuter J., Lessig R., Krishnamurthi K., Ullrich A., and Hengstler J. G. (2006) ErbB-3 predicts survival in ovarian cancer. J. Clin. Oncol. 24, 4317–4323 [DOI] [PubMed] [Google Scholar]
- 18. Citri A., Skaria K. B., and Yarden Y. (2003) The deaf and the dumb: the biology of ErbB-2 and ErbB-3. Exp. Cell Res. 284, 54–65 [DOI] [PubMed] [Google Scholar]
- 19. Holbro T., Beerli R. R., Maurer F., Koziczak M., Barbas C. F. 3rd, and Hynes N. E. (2003) The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc. Natl. Acad. Sci. U.S.A. 100, 8933–8938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Franklin M. C., Carey K. D., Vajdos F. F., Leahy D. J., de Vos A. M., and Sliwkowski M. X. (2004) Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer Cell 5, 317–328 [DOI] [PubMed] [Google Scholar]
- 21. Kumari S., Mg S., and Mayor S. (2010) Endocytosis unplugged: multiple ways to enter the cell. Cell Res. 20, 256–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang L. H., Rothberg K. G., and Anderson R. G. (1993) Mis-assembly of clathrin lattices on endosomes reveals a regulatory switch for coated pit formation. J. Cell Biol. 123, 1107–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kitajima Y., Sekiya T., and Nozawa Y. (1976) Freeze-fracture ultrastructural alterations induced by filipin, pimaricin, nystatin and amphotericin B in the plasmia membranes of Epidermophyton, Saccharomyces and red complex-induced membrane lesions. Biochim. Biophys. Acta 455, 452–465 [DOI] [PubMed] [Google Scholar]
- 24. Tran E. J., and Wente S. R. (2006) Dynamic nuclear pore complexes: life on the edge. Cell 125, 1041–1053 [DOI] [PubMed] [Google Scholar]
- 25. Slater T. F., Cheeseman K. H., and Ingold K. U. (1985) Carbon tetrachloride toxicity as a model for studying free-radical mediated liver injury. Phil. Trans. R. Soc. Lond. B, Biol. Sci. 311, 633–645 [DOI] [PubMed] [Google Scholar]
- 26. Wang Y. N., and Hung M. C. (2012) Nuclear functions and subcellular trafficking mechanisms of the epidermal growth factor receptor family. Cell Biosci. 2, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Offterdinger M., Schöfer C., Weipoltshammer K., and Grunt T. W. (2002) c-erbB-3: a nuclear protein in mammary epithelial cells. J. Cell Biol. 157, 929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baulida J., and Carpenter G. (1997) Heregulin degradation in the absence of rapid receptor-mediated internalization. Exp. Cell Res. 232, 167–172 [DOI] [PubMed] [Google Scholar]
- 29. Baulida J., Kraus M. H., Alimandi M., Di Fiore P. P., and Carpenter G. (1996) All ErbB receptors other than the epidermal growth factor receptor are endocytosis impaired. J. Biol. Chem. 271, 5251–5257 [DOI] [PubMed] [Google Scholar]
- 30. Sak M. M., Breen K., Rønning S. B., Pedersen N. M., Bertelsen V., Stang E., and Madshus I. H. (2012) The oncoprotein ErbB3 is endocytosed in the absence of added ligand in a clathrin-dependent manner. Carcinogenesis 33, 1031–1039 [DOI] [PubMed] [Google Scholar]
- 31. Lund K. A., Opresko L. K., Starbuck C., Walsh B. J., and Wiley H. S. (1990) Quantitative analysis of the endocytic system involved in hormone-induced receptor internalization. J. Biol. Chem. 265, 15713–15723 [PubMed] [Google Scholar]
- 32. Chang H. Y., Liu H. S., Lai M. D., Tsai Y. S., Tzai T. S., Cheng H. L., and Chow N. H. (2014) Hypoxia promotes nuclear translocation and transcriptional function in the oncogenic tyrosine kinase RON. Cancer Res. 74, 4549–4562 [DOI] [PubMed] [Google Scholar]
- 33. Brand T. M., Iida M., Luthar N., Wleklinski M. J., Starr M. M., and Wheeler D. L. (2013) Mapping C-terminal transactivation domains of the nuclear HER family receptor tyrosine kinase HER3. PLoS ONE 8, e71518. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34. Andrique L., Fauvin D., El Maassarani M., Colasson H., Vannier B., and Séité P. (2012) ErbB3(80 kDa), a nuclear variant of the ErbB3 receptor, binds to the Cyclin D1 promoter to activate cell proliferation but is negatively controlled by p14ARF. Cell Signal. 24, 1074–1085 [DOI] [PubMed] [Google Scholar]
- 35. Koumakpayi I. H., Diallo J. S., Le Page C., Lessard L., Gleave M., Bégin L. R., Mes-Masson A. M., and Saad F. (2006) Expression and nuclear localization of ErbB3 in prostate cancer. Clin. Cancer Res. 12, 2730–2737 [DOI] [PubMed] [Google Scholar]
- 36. Raabe T. D., Deadwyler G., Varga J. W., and Devries G. H. (2004) Localization of neuregulin isoforms and erbB receptors in myelinating glial cells. Glia. 45, 197–207 [DOI] [PubMed] [Google Scholar]