Abstract
The normal cellular prion protein (PrP) is a glycosylphosphatidylinositol (GPI)-anchored cell surface glycoprotein. However, in pancreatic ductal adenocarcinoma cell lines, such as BxPC-3, PrP exists as a pro-PrP retaining its glycosylphosphatidylinositol (GPI) peptide signaling sequence. Here, we report the identification of another pancreatic ductal adenocarcinoma cell line, AsPC-1, which expresses a mature GPI-anchored PrP. Comparison of the 24 genes involved in the GPI anchor modification pathway between AsPC-1 and BxPC-3 revealed 15 of the 24 genes, including PGAP1 and PIG-F, were down-regulated in the latter cells. We also identified six missense mutations in DPM2, PIG-C, PIG-N, and PIG-P alongside eight silent mutations. When BxPC-3 cells were fused with Chinese hamster ovary (CHO) cells, which lack endogenous PrP, pro-PrP was successfully converted into mature GPI-anchored PrP. Expression of the individual gene, such as PGAP1, PIG-F, or PIG-C, into BxPC-3 cells does not result in phosphoinositide-specific phospholipase C sensitivity of PrP. However, when PIG-F but not PIG-P is expressed in PGAP1-expressing BxPC-3 cells, PrP on the surface of the cells becomes phosphoinositide-specific phospholipase C-sensitive. Thus, low expression of PIG-F and PGAP1 is the major factor contributing to the accumulation of pro-PrP. More importantly, BxPC-3 cells expressing GPI-anchored PrP migrate much slower than BxPC-3 cells bearing pro-PrP. In addition, GPI-anchored PrP-bearing AsPC-1 cells also migrate slower than pro-PrP bearing BxPC-3 cells, although both cells express filamin A. “Knocking out” PRNP in BxPC-3 cell drastically reduces its migration. Collectively, these results show that multiple gene irregularity in BxPC-3 cells is responsible for the formation of pro-PrP, and binding of pro-PrP to filamin A contributes to enhanced tumor cell motility.
Keywords: cell migration, cell surface, glycosylphosphatidylinositol (GPI anchor), pancreatic cancer, prion, GPI anchor synthesis, cancer cell motility, pancreatic ductal adenocarcinoma cell, prion protein
Introduction
PrP2 is a highly conserved and widely expressed glycoprotein tethered on the cell surface by a GPI anchor (1). Although PrP has been implicated in a plethora of biological functions (2–4), the physiologic function of PrP remains elusive, as Prnp knock-out mice and cattle show no obvious phenotype and PrP null sheep due to a stop codon mutation also occurs naturally (1, 5–7). The only well established function of PrP is that this protein is required for the pathogenesis of a group of fatal neurodegenerative diseases commonly referred to as prion diseases (8).
The expression of PrP is up-regulated in some cancer cells, which normally either lack PrP or have low levels of PrP (9–14). The up-regulation of PrP has been reported to contribute to tumor cell migration, proliferation, and multiple drug resistance (9, 15–17). More importantly, increased PrP expression is a biomarker for poor prognostics for patients with pancreatic cancer, breast cancer, or gastric cancer (11, 13, 18). Previously, in our studies of six PDAC cell lines and a melanoma cell line, we found that the PrP existed as a pro-PrP, as defined by retaining its normally cleaved GPI-PSS (11, 12). Sequencing of the open reading frame (ORF) of PRNP in these cell lines did not identify any mutations. Therefore, the retention of the PrP GPI-PSS is not due to mutation in the PRNP. Interestingly, the GPI-PSS of PrP contains an FLNa-binding motif and thus pro-PrP binds to FLNa (12). FLNa is a cytolinker protein that is important in linking cell surface receptors to the cytoskeleton (19, 20). Hence, binding of pro-PrP to FLNa alters the normal physiology of FLNa rendering the tumor cell to be more aggressive (11, 12).
The underlying mechanism for retaining the PrP GPI-PSS in PDAC is not clear. More than 20 genes are known to be important in the synthesis, assembly of the GPI anchor components, cleavage of the GPI-PSS, and eventual en bloc attachment of an assembled GPI anchor to its substrate (21). Mutations in GPI anchor synthesis enzymes are associated with many human diseases; most of these diseases affect neuronal development (22–35). Furthermore, a lack of GPI anchored protein in cancer cells has also been reported to be due to transcriptional silencing of the genes involved in biosynthesis of the GPI anchor (36). Interestingly, the efficiency of the GPI anchor modification is critical, depending on the sequence of the GPI-PSS. It is known that the GPI-PSS of PrP has the least efficiency among the 10 tested GPI-anchored proteins in an in vitro GPI anchor modification assay (37).
In this study, we reported the identification a PDAC cell line, AsPC-1, which expresses a GPI-anchored PrP. This cell line enables us to compare the expression of the 24 genes responsible for GPI anchor synthesis between GPI-anchored PrP bearing AsPC-1 cells and pro-PrP bearing BxPC-3 cells. We found that the expression levels of 15 of these genes were up-regulated in AsPC-1 cells compared with BxPC-3 cells. We also identified six missense mutations in DPM2, PIG-C, PIG-N, and PIG-P. To explore the functional contribution of these affected genes, we fused the Chinese hamster ovary (CHO) cell, which lacks endogenous PrP, with the BxPC-3 cell. We found that fusion of these two cells successfully converts the pro-PrP to become GPI-anchored PrP. We then tested whether expressing any of the genes with low expression levels in BxPC-3 can rescue the PrP phenotype. We found that expressing PGAP1, PIG-F, PIG-C, etc. alone did not generate GPI-anchored PrP. However, when PIG-F was expressed in PGAP1-expressing BxPC-3 cells, the cell surface PrP became GPI-anchored. The same phenomenon was not observed when PIG-P, PIG-N, etc. was expressed in PGAP1-expressing BxPC-3 cells, thus showing that the low expression levels of PGAP1 and PIG-F were the major factors contributing to the generation of pro-PrP in BxPC-3 cells. Furthermore, when compared with AsPC-1, whose PrP was GPI-anchored, BxPC-3 migrated faster, which supports the importance of interactions between FLNa and pro-PrP for cell motility. Finally, we showed that by knocking out PRNP in BxPC-3, the motility of the cells was greatly reduced. Together, these results provide strong evidence that defects in the GPI anchor synthesis machinery cause the accumulation of pro-PrP, which then contributes to the aggressive behavior of PDAC by disrupting the normal functions of FLNa.
Experimental Procedures
Cell Lines, Abs, and Reagents
AsPC-1, BxPC-3, and CHO-K1 cells were purchased from American Type Culture Collection (ATCC). AsPC-1 and BxPC-3 cells were cultured in RPMI 1640 medium (Life Technologies, Inc., catalog no. 31800-022) supplemented with 1.5 g/liter sodium bicarbonate, 10% fetal bovine serum (FBS) (Biological Industries, Kibbutz Beit Haemek, Israel), 1% sodium pyruvate, 1 mm HEPES, 4.5 g/liter glucose, 100 units/ml of penicillin, and 100 μg/ml streptomycin. CHO-K1 cells were cultured in α-minimal essential medium (Gibco, catalog no. 11900-024) supplemented with 1.67 g/liter sodium bicarbonate, 10% FBS, 12.6 mm HEPES, 1 g/liter glucose, 100 units/ml penicillin, and 100 μg/ml streptomycin. CHO-NC and CHO-hPrP cells were generated with lentivirus systems and were cultured in the same growth media as CHO-K1 cells. BxPC-3-CHO-NC was generated by fusing BxPC-3 and CHO-NC and was cultured in the same growth media as BxPC-3, except with 20% FBS.
Anti-PrP monoclonal antibodies (mAbs) (4H2, 8B4, and 5B2) were generated as described (38). Filamin A (FLNa) antibody was purchased from CHEMICON® International, Inc. (catalog no. mAb1678). Horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG-specific antibody was purchased from AntGene Biotech (Wuhan, China). Mouse anti-actin mAb was purchased from Tianjin Sungene Biotech, Tianjin, China (catalog no. KM9001). Mouse IgG1 isotype control, HRP-streptavidin, and APC conjugated goat anti-mouse IgG antibody were purchased from Biolegend (San Diego). 4′,6-Diamidine-2′-phenylindole dihydrochloride was purchased from Roche Diagnostics (Mannheim, Germany). Alexa Fluor® 488 and 555 dye-linked goat anti-mouse secondary antibody was purchased from Invitrogen. PGAP-1-expressing plasmid was a kind gift from Professor Taroh Kinoshita, Osaka University. All reagents purchased from commercial sources were used according to the suppliers' recommendations.
Quantitative Real Time PCR Analysis and Sequencing of the 24 Genes
Total RNA was isolated using an RNApure tissue kit (catalog no. CW0584, CWBiotech, Beijing, China). 1 μg of total RNA was used to synthesize first-strand complementary DNA (cDNA) using the PrimeScriptTM RT reagent kit with gDNA Eraser (catalog no. DRR047A, Takara Bio, Shiga, Japan). Quantitative PCR amplification was carried out with well characterized primer sets (supplemental Table S1) for the 24 genes involved in the biosynthesis of GPI anchor by using iQTM SYBR Green Supermix (product no. 170 l8882AP, Bio-Rad) on a Bio-Rad ConnectTM real time PCR instrument (CFX ConnectTM Optics Module). Each reaction was run in triplicate and contained 1 μl of cDNA template in a final reaction volume of 20 μl. Melting curves were performed to ensure that only a single product was amplified. The relative expression of 24 genes was normalized to the expression level of β-actin. Data shown represent the mean ± S.E. of the mean from three experiments. Extracted mRNAs were reverse-transcribed and submitted for sequencing with primers covering the full coding sequence of the 24 genes (supplemental Table S2).
Phosphatidylinositol Phospholipase C (PI-PLC) Treatment of Cells and Flow Cytometry Analysis
Cells were seeded in 6-cm Petri dishes overnight and then rinsed three times with ice-cold Dulbecco's modified phosphate-buffered saline (DPBS) followed by 0.25% trypsin/EDTA treatment. After rinsing twice with DPBS, cells were incubated with PI-PLC (0.1 unit/ml) (Sigma, catalog no. P5542) at 37 °C for 1 h) and then rinsed twice with DPBS followed by staining with control antibody or 4H2 for 20 min at 4 °C. Bound antibody was detected with an APC-conjugated goat anti-mouse IgG antibody and analyzed in a FACSTM C6 flow cytometer (San Jose, CA). The flow cytometry analysis was carried out at least twice with comparable results. To calculate the PI-PLC treatment on PrP, we use the following formula: (arithmetic mean after PI-PLC treatment − arithmetic mean of background)/(arithmetic mean of before PI-PLC treatment − arithmetic mean of background); to calculate the PI-PLC treatment on total GPI anchored proteins, we use following formula due to lack of background: (arithmetic mean of before PI-PLC treatment − arithmetic mean of after PI-PLC treatment)/(arithmetic mean of before PI-PLC treatment for FLARE staining).
Immunofluorescence Staining for Confocal Microscopy
Cells were cultured in poly-d-lysine-coated glass-bottomed Petri dishes (product no. P35GC-1.0–14-C, MatTek Corp., Ashland, MA) overnight, then rinsed three times with DPBS, and fixed in 4% paraformaldehyde for 12 min at room temperature. PrP was detected with anti-PrP mAbs 8B4 or 4H2 (10 μg/ml). Bound antibodies were detected using Alexa Fluor® 488 or 555 dye-conjugated goat anti-mouse IgG-specific antibody (Invitrogen). Nuclei were counter-stained with DAPI and observed using an inverted confocal microscope (PerkinElmer Life Sciences). To detect cell surface PrP after PI-PLC treatment of BxPC-3-CHO-NC, cells were cultured with brefeldin A (0.35 μm) and PI-PLC (0.1 units/ml) in a 37 °C incubator for 1 h. The subsequent steps were carried out as described above. All confocal microscopy experiments were carried out at least twice with comparable results. To detect PIG-F, PIG-C, PIC-N, and PIG-P expression in PGAP1-expressing BxPC-3 cells, transfected cells were subject to Olympus inverted microscopy (Olympus corporation, model no. IX73, Tokyo, Japan) for mCherry expression.
Generation and Purification of Rabbit Polyclonal Antibodies against GPI-PSS of hPrP
The specific pathogen-free rabbit was immunized four times with complete or incomplete Freund's adjuvant (Sigma) reconstituted and DMSO-dissolved synthesized polypeptide (SMVLFSSPPVILLISFLIFLIVG) corresponding to the GPI-PSS region of hPrP every 2 weeks for 1 month. 50 ml of blood were collected and clotted at room temperature for 1 h and then stored at 4 °C overnight. To collect serum, the supernatant was centrifuged and aliquoted before storing at −80 °C.
To purify-PrP specific IgG, the serum was precipitated with saturated ammonium sulfate and then centrifuged at 10,000 × g (4 °C). The pellet was resuspended in 100 mm Tris-HCl (pH 8.0) and dialyzed against three changes of 1× PBS (pH 7.4) at 4 °C and then centrifuged to remove remaining debris. The IgG was incubated with GST-dopple conjugated AminoLink® Plus coupling resin (Thermo Scientific) at 4 °C overnight to absorb nonspecific antibody. PrP GPI-PSS-specific IgG was collected by incubation-absorbed IgG with the GST-hPrP231–253-conjugated AminoLink® Plus coupling resin at 4 °C overnight.
Immunoblotting
Cell lysates were prepared in lysis buffer containing 20 mm Tris (pH 7.5), 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, 2.5 mm sodium pyrophosphate, 1 mm β-glycerol phosphate, 1 mm Na3VO4. 1 mm phenylmethylsulfonyl fluoride (PMSF), and EDTA-free protease inhibitor mixture were added just before cell lysis. Cell lysate was directly subjected to peptide N-glycosidase F (New England Biolabs, Inc., Ipswich, MA) treatment according to the provider's protocols. After treatment, samples were separated on SDS-PAGE and immunoblotted with anti-PrP mAb 4H2, and bound antibodies were detected with HRP-labeled goat anti-mouse IgG antibody. To detect the GPI-PSS region of hPrP, freshly prepared cell lysate was separated and detected with affinity-purified rabbit anti-GPI-PSS polyclonal antibodies or the rabbit preimmune serum. To detect PIG-C, PIG-F, PIG-N, or PIG-P in PGAP1-expressing BxPC-3 cells, freshly prepared cell lysate was separated and detected with Living Colors® DsRed polyclonal antibody (Clontech, catalog no. 632496). Experiments were repeated at least twice with comparable results.
Immunoaffinity Purification of PrP, Carboxypeptidase Treatment, and Co-immunoprecipitation of FLNa
To purify PrP from various cell lines, fresh cell lysate was prepared as above and affinity-purified with 200 μg of mAb 5B2-conjugated AminoLink® Plus coupling resin (Thermo Scientific) and incubated overnight at 4 °C. After elution and neutralization, purified PrP was subjected to carboxypeptidase Y treatment (0.5 units/20 μl of eluted and neutralized PrP) at room temperature for different periods of time. The samples were then separated on SDS-PAGE and immunoblotted with 4H2. Co-immunoprecipitation of FLNa was performed according to Li et al. (11).
Proaerolysin Overlays
To detect the GPI anchor on PrP, total cell lysate or freshly purified PrP was separated on a 10% SDS-polyacrylamide gel. The gel was then incubated in 50 mm Tris-HCl (pH 7.4), and 20% glycerol for 15 min at room temperature. Proteins were then blotted onto a nitrocellulose membrane (Thermo Scientific, Inc., catalog no. 88018, Rockford) for 1 h 20 min at 300 mA in the cold transfer buffer (10 mm NaHCO3, 3 mm Na2CO3 (pH 9.9), 20% methanol) using a wet transfer chamber (Bio-Rad). Membranes were then subjected to biotinylated proaerolysin overlay. Proaerolysin (Pinewood Scientific Services Inc., catalog no. FL2S, Victoria, British Columbia, Canada) was biotin-labeled with EZ-LinkTM Sulfo-NHS-biotinylation kit (Thermo Scientific, Inc., catalog no. 21425, Rockford) according to the supplier's instruction. The nitrocellulose membranes were incubated in the binding buffer (50 mm NaH2PO4 (pH 7.5), 0.3% Tween 20) for 20 min followed by a 2-h incubation in the presence of 5 nm biotinylated proaerolysin diluted in the same buffer at 37 °C. Bound proaerolysin was detected with HRP-streptavidin and revealed with chemiluminescence substrate (Millipore Corp., catalog no. WBKLS0500, Billerica, MA).
Cell Fusion and Sorting
The same number of BxPC-3 and CHO-NC was harvested and washed twice with ice-cold fusion medium (RPMI 1640 medium supplemented with 100 units/ml penicillin and 100 μg/ml streptomycin). The cells were then resuspended with warm fusion media and combined followed by centrifugation at 175 × g for 7 min. The supernatant was aspirated off, and 1 ml of pre-warmed 50% PEG 1500 was added followed by 25 ml of warm fusion media. The cells were kept at room temperature for several minutes and spun down at 150 × g for 7 min. The supernatant was completely removed before the cells were resuspended gently with 10 ml of warm complete hybridoma culture medium (RPMI 1640 medium supplemented with 1.5 g/liter sodium bicarbonate, 20% FBS (Gibco), 1% sodium pyruvate, 1 mm HEPES, 4.5 g/liter glucose, 100 units/ml penicillin, and 100 μg/ml streptomycin).
To enrich for BxPC-3-CHO-NC-fused cells, three 10-cm Petri dishes of fused cells were trypsinized and stained with mAb 4H2 (10 ng/μl). Bound primary antibody was detected with APC-labeled goat anti-mouse IgG antibody and then analyzed and sorted according to endogenous GFP signal from CHO-NC and APC dye signal from BxPC-3 in a FACS Aria III flow cytometer at Analysis and Test Center, Wuhan Institute of Virology, Chinese Academy of Science. The experiments were repeated twice with comparable results.
Membrane Protein Extraction
To detect cell surface GPI-anchored proteins, four 10-cm Petri dishes of confluent cells were harvested by scraping the cells off the surface of the plate with a cell scraper. The cells were gently rinsed with ice-cold 1× PBS, and the cell membrane protein was extracted with Beyotime® membrane protein extraction kit (product no. P0033, Beyotime Biotechnology, Shanghai, China). Separated membrane proteins were subjected to proaerolysin overlay assay to detect GPI-anchored proteins.
Biotinylation of Cell Surface Proteins and Enrichment of Biotinylated Cell Surface Glycoprotein after PI-PLC Treatment
To detect GPI-anchored proteins released from the cell surface post PI-PLC treatment, three 10-cm Petri dishes of confluent cells were harvested with trypsin treatment and were biotinylated with EZ-LinkTM Sulfo-NHS-SS-Biotin (Thermo Scientific, Inc., catalog no. 21331), according to the manufacturer's procedure. The cells were then washed three times with ice-cold 1× PBS and subjected to PI-PLC treatment at room temperature for 1 h. GPI-anchored proteins were pulled down with lectin-agarose (Sigma, product no. L1394), at 4 °C overnight. The lectin-agarose beads were washed six times and collected by centrifugation. Separated proteins were detected with HRP-streptavidin for total GPI-anchored proteins and 4H2 for PrP.
Knocking Out PRNP from BxPC-3 Cells
To knock out PRNP in the BxPC-3 cells, oligonucleotides for the generation of PrP sgRNA expression plasmids were annealed and cloned into the BsaI sites of pGL3-U6-sgRNA-PGK-Puro (Addgene 51133) (knock-out target sequence 1 forward primer 5′-CACCGGGCCTTGGCGGCTACATGC-3′ and reverse primer 5′-AAACGCATGTAGCCGCCAAGGCCC-3′; knock-out target sequence 3 forward primer 5′-CACCGGTGGTGGCTGGGGTCAAGG-3′ and reverse primer 5′-AAACCCTTGACCCCAGCCACCACC-3′). The recombinant short guide RNA expression plasmid and Cas9 expression construct pST1374-Cas9-N-NLS-FLAG-linker (Addgene 44758) were transfected into 80% confluent BxPC-3 cells with Lipofectamine® 2000 reagent (Invitrogen 11668) in a 6-well plate according to Zhang and co-workers (39). The cells were cultured for 1 day and transferred to a 6-cm cell culture Petri dish and treated with 2 μg/ml puromycin and 1 μg/ml blasticidin. The positive clones (PrP knock-out) were identified by immunoblots.
Expression of PGAP1, PIG-N, PIG-P, PIG-C, and PIG-F in BxPC-3 Cells
PGAP1-expressing BxPC-3 cells (BxPC-3-PGAP1) were established by lentivirus expression system. PIG-N, PIG-P, PIG-C, and PIG-F were cloned from AsPC-1 cells into pmCherry-N1 plasmid with primers listed in supplemental Table S3. The mCherry fusion protein of PIG-N, PIG-P, PIG-C, and PIG-F was then transfected into BxPC-3 cells or BxPC-3-PGAP1 cells. Expression of fused proteins was sorted by FACS Aria III flow cytometer.
Densitometry
Densitometry analysis of the protein bands and the wound area of wound closure assay was performed using the National Institutes of Health Image J software (1.47 version) according to Li et al. (12). The gray level of band and average wound area were calculated (means ± S.E. of three experiments as indicated in the paper) for different time points.
Statistical Analysis
Student's t test was applied for experiments repeated three times. p < 0.05 was consider statistically significant different (*, p < 0.05; **, p < 0.01; ***, p < 0.001, respectively).
Results
PrP Expression in PDAC Cell Lines
We have previously reported the up-regulation of PrP expression in six PDAC cell lines, including BxPC-3 (11). Interestingly, the PrP detected in these cell lines existed as pro-PrP. To study the underlying mechanisms causing the aberrant processing of PrP, we continue to search for PDAC cell lines that may express PrP with a GPI anchor rather than pro-PrP. One additional PDAC cell line we identified is AsPC-1.
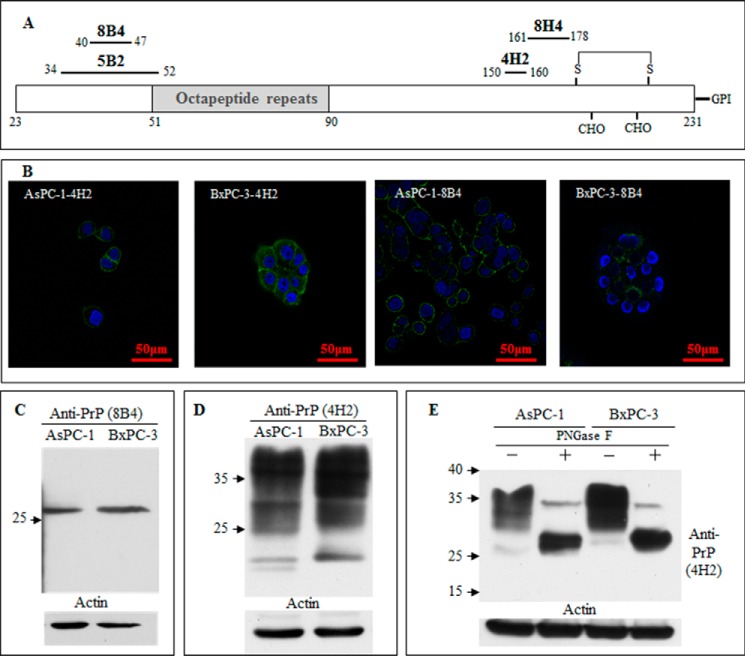
Because the processing of PrP is different in different cell models, we also used multiple anti-PrP monoclonal antibodies (mAbs) with distinct epitopes (Fig. 1A) to compare the expression of PrP in AsPC-1 cells with BxPC-3 cells. Although both BxPC-3 and AsPC-1 cells expressed PrP (Fig. 1B), the patterns of PrP distributions were differed between these two cell lines. In AsPC-1 cells, PrP was mainly localized on the cell surface. In contrast, in BxPC-3 cells, a significant amount of PrP was also detected in the cytosol (Fig. 1B). One interesting observation was that mAbs 4H2 and 8B4 reacted with different PrP species in these cell lines (Fig. 1C). It should be noted that although mAb 8B4 recognizes only full-length PrP, mAb 4H2 reacts with both full-length as well as some N-terminally truncated PrP (40). In immunoblots, mAb 8B4 reacted with one single band in both cell lines, corresponding to the unglycosylated full-length PrP. In contrast, mAb 4H2 reacted with multiple glycosylated PrP species (Fig. 1D). Upon treatment with peptide N-glycosidase F to remove the N-linked glycans, the majority of the PrP in both cell lines appeared as a single molecular species at around 27 kDa, which is the unglycosylated PrP. However, the unglycosylated PrP from BxPC-3 consistently migrated a bit slower than the unglycosylated PrP species from AsPC-1 cells, which is most likely due to the retention of the GPI-PSS (Fig. 1E).
FIGURE 1.
BxPC-3 and AsPC-1 pancreatic ductal adenocarcinoma cells express glycosylated prion protein. A, diagram of the mAbs used and their corresponding epitopes. B, immunofluorescence staining with anti-PrP-specific mAbs 4H2 and 8B4 showed PrP expression in two PDAC cell lines, AsPC-1 and BxPC-3. C, immunoblotting with mAb 8B4 identified PrP expression in AsPC-1 and BxPC-3. D, immunoblotting with mAb 4H2 identified PrP expression in AsPC-1 and BxPC-3. E, immunoblotting with monoclonal antibody 4H2 identified PrP was glycosylated after the peptide N-glycosidase F treatment of the cell lysate of AsPC-1 and BxPC-3.
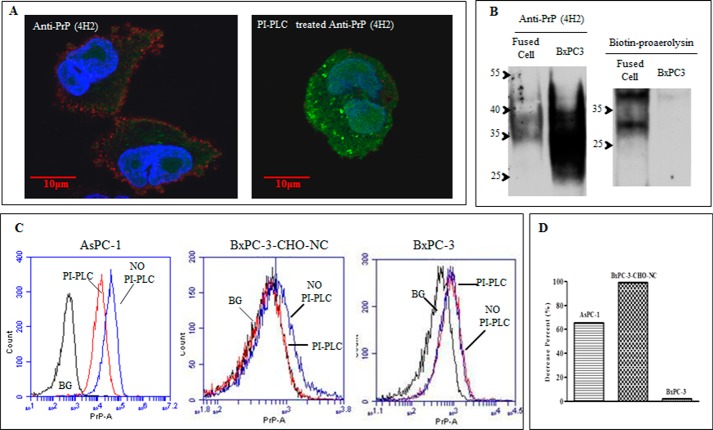
PrP from AsPC-1 but Not BxPC-3 Is Sensitive to PI-PLC Treatment and Binds Proaerolysin
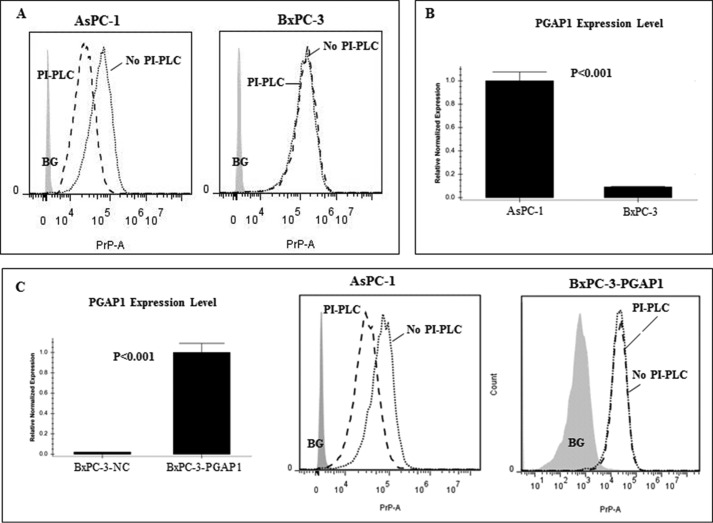
We next determined whether the PrP present in AsPC-1 is GPI-anchored or pro-PrP as in BxPC-3. We treated AsPC-1 and BxPC-3 cells with PI-PLC. PI-PLC treatment is a well established assay to determine whether a protein is GPI-anchored or not (11, 41). Using mAb 4H2, we found that although the PrP on the cell surface of BxPC-3 was highly resistant to PI-PLC, the PrP on the surface of AsPC-1 was much more susceptible to identical treatment (Fig. 2A). This implied that in contrast to BxPC-3 cells, the PrP expressed on the cell surface of AsPC-1 was GPI-anchored. However, this approach suffers if the GPI-anchored protein is inadequately deacylated due to a low PGAP1 expression level (42). To rule out the possibility that PI-PLC-resistant proteins from BxPC-3 cells were insufficiently deacylated due to low expression of PGAP1 (Fig. 2B), we transfected a human PGAP1 expression vector into BxPC-3 cells resulting in a 48-fold up-regulation of PGAP-1 based on Q-PCR data. However, the PrP is still resistant to PI-PLC treatment (Fig. 2C). These results provide additional evidence that the reason the PrP in BxPC-3 cells is PI-PLC-resistant is not simply due to lower expression of PGAP1.
FIGURE 2.
Reduced PGAP1 expression in BxPC-3 did not account for PI-PLC resistance of PrP. A, PrP on the AsPC-1 cell surface but not the BxPC-3 cell surface was sensitive to PI-PLC treatment. Cell surface PrP was detected with 4H2 with PI-PLC or without PI-PLC treatment. B, Q-PCR proved that PGAP1 expression level in BxPC-3 was significantly lower as compared with that in AsPC-1 cells. C, Q-PCR showed that the PGAP1 level was up-regulated 48-fold in BxPC-3 cells after forced expression. However, overexpression of PGAP1 in BxPC-3 cells did not result in PI-PLC susceptibility of PrP.
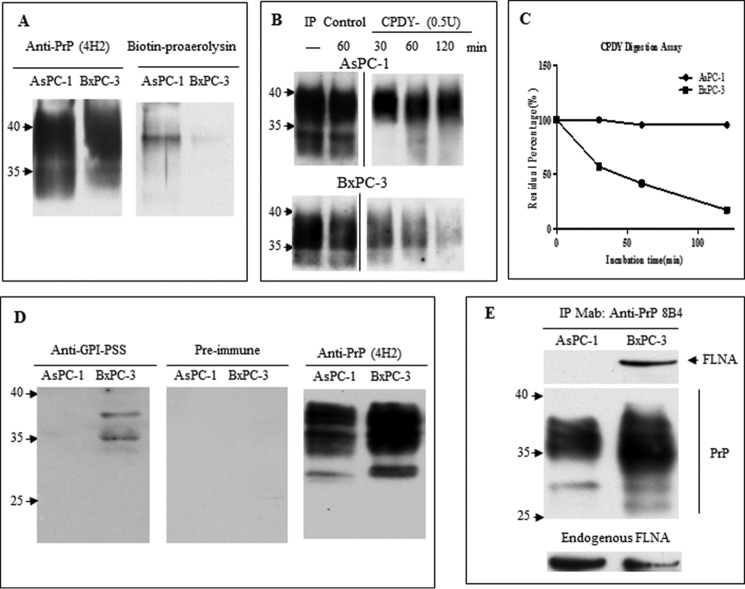
Proaerolysin reacts specifically with the GPI anchor component (43). If the PrP in AsPC-1 cell is GPI-anchored, it should react with proaerolysin. For this purpose, we first affinity-purified the PrP with mAb 5B2 and then immunoblotted the affinity-purified PrP with either mAb 4H2 or proaerolysin. (Fig. 3A). The authenticity of the affinity-purified PrP was confirmed by its reactivity with mAb 4H2 (Fig. 3A). More importantly, we found that proaerolysin only detected PrP from the AsPC-1 cell lysate but not from BxPC-3 cell lysate (Fig. 3A).
FIGURE 3.
PrP in AsPC-1 is GPI-anchored whereas PrP in BxPC-3 retains its GPI-PSS. A, PrP from AsPC-1 cell lysate but not BxPC-3 cell lysate purified by 5B2 reacted with proaerolysin (right panel). The blot was then re-probed with 4H2 after stripping to show that a similar amount of PrP was loaded for AsPC-1 and BxPC-3 (left panel). B, PrP from AsPC-1 but not BxPC-3 is more resistant to carboxypeptidase Y treatment. 5B2-purified PrP from AsPC-1 or BxPC-3 was treated with carboxypeptidase for different times and probed with 4H2. C, quantification of the total PrP after treatment showed PrP from AsPC-1 was more resistant to carboxypeptidase Y treatment. This is representative of three experiments with comparable results. D, prion protein from BxPC-3 but not from AsPC-1 reacted with rabbit polyclonal antibody against GPI-PSS (left panel), whereas the preimmune serum was negative for PrP from AsPC-1 and BxPC-3 cells (middle panel). The rabbit polyclonal antibody was stripped and reprobed with 4H2 to show similar loading of PrP (right panel). E, FLNa from BxPC-3 but not AsPC-1 could be co-purified with PrP. Loading of PrP was indicated. Both AsPC-1 and BxPC-3 expressed FLNa.
PrP from AsPC-1 Is Protected from Carboxypeptidase Digestion
To further validate that PrP in AsPC-1 cells is indeed GPI-anchored, we employed a complementary approach, using carboxypeptidase digestion. Because carboxypeptidase digests amino acids from the C terminus of a protein, it will not be able to cleave GPI-anchored PrP (11). We treated the immunopurified PrP with carboxypeptidase Y and found the enzyme degraded PrP from BxPC-3 much more readily than PrP from AsPC-1 (Fig. 3B). After treatment with the enzyme for 120 min, PrP immunoreactivity from BxPC-3 cells was reduced by more than 90% (Fig. 3C).
In addition, we generated a polyclonal antiserum that was specific for the GPI anchor peptide signal sequence on PrP. Rabbit IgG was purified by protein G beads, absorbed with recombinant human PrP (23–231), and then purified using the GPI-PSS peptide affinity chromatography. The purified antibody reacted with PrP from BxPC-3 but not the PrP from AsPC-1 cells, thus confirming that PrP from BxPC-3 was pro-PrP (Fig. 3D). Taken together, these results clearly showed that PrP from AsPC-1 but not BxPC-3 was modified with a GPI anchor at the C terminus.
Pro-PrP from BxPC-3 Binds FLNa
Because the GPI-PSS of pro-PrP binds to FLNa (11), we would expect to detect this protein from PrP pulldown from BxPC-3 but not from PrP in AsPC-1 cells. Prior to doing this experiment, we first confirmed that indeed both cell lines expressed FLNa (Fig. 3E). Co-immunoprecipitation experiments indeed showed that only PrP from BxPC3 cells co-immunoprecipitated with FLNa (Fig. 3E).
Differences in GPI Anchor Biosynthesis, Attachment, and Remodeling Pathway Genes between AsPC-1 and BxPC-3
More than 24 genes are involved in the GPI anchor biosynthesis, attachment, and remodeling pathway (44). The efficiency of the modification process also depends on the nature of the GPI-PSS; the efficiency is low for PrP GPI-PSS (37). To elucidate the mechanism(s) for the formation of pro-PrP in BxPC-3, we reasoned either the expression level of one or more of the 24 genes is/are low or there are mutations in some of these genes. Our sequencing of the mRNA for these 24 genes revealed six missense mutations in four of these genes compared with reported human genome sequences as follows: DPM2 (T76S), PIG-C (S266F), PIG-N (H229D; M416T; L460P), and PIG-P (P3S) (Table 1). However, whether any of these mutations results in loss of function is not known.
TABLE 1.
Several mutations occur in some of 24 genes from BxPC-3
Boldface is used for sense mutation to distinguish it from synonymous mutation.
| Gene | Nucleic acid conversion | Mutation in amino acid |
|---|---|---|
| DPM2 | 213U→C | Y71Y |
| 227C→G | T76S | |
| PIG-F | 327A→G | A109A |
| PIG-C | 267U→C | G89G |
| 797C→U | S266F | |
| PIG-N | 685C→G | H229D |
| 1247U→C | M416T | |
| 1379U→C | L460P | |
| 741C→U | H247H | |
| 939U→C | N313N | |
| 1752G→A | L584L | |
| 1962G→A | L654L | |
| 2010U→C | T670T | |
| PIG-P | 7C→U | P3S |
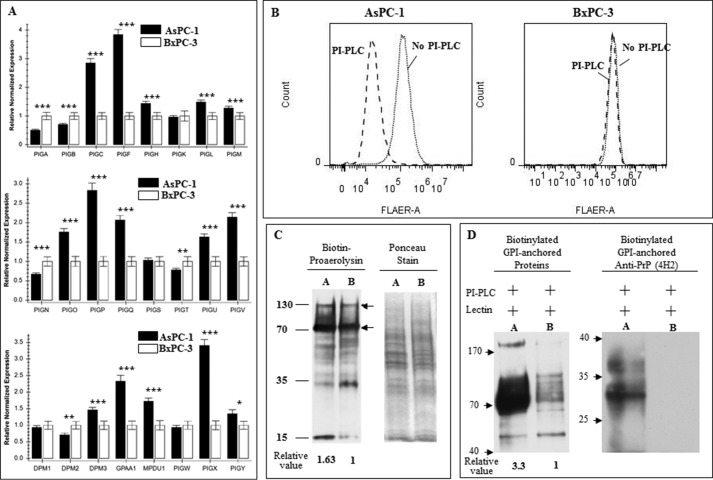
Next, we compared the expression levels of the 24 genes besides PGAP1 between AsPC-1 and BxPC-3. Q-PCR results revealed that there were seven genes whose mRNAs levels were at least 2.5-fold higher in AsPC-1 cells compared with BxPC-3 cells (Fig. 4A). Of these seven genes, two genes, PIG-F and PIG-X, have the largest differences (>3.5-fold) between these two cell lines. Differences in the expression of these genes were also observed in two other PDAC cell lines, CFPAC-1 and Panc10.05 cells, when compared with AsPC-1 cells (supplemental Fig. 1).
FIGURE 4.
Defects in GPI anchor synthesis did not completely disable the capability of GPI anchor synthesis in BxPC-3 cells. A, relative expression of the 24 genes for GPI anchor synthesis was quantified based on Q-PCR. 15 of the 24 genes in BxPC-3 expressed at a significantly lower level than those from AsPC-1 cells, among which seven showed at least 2-fold differences (results were from three experiments, *, p < 0.05; **, p < 0.01; ***, p < 0.001). B, FLARE staining of cells after PI-PLC treatment identified there were some GPI-anchored proteins on the cell surface of BxPC-3, and a few of them were PI-PLC-sensitive (right panel), although many more proteins on the cell surface of AsPC-1 were PI-PLC-sensitive (left panel). C, AsPC-1 (lane A) and BxPC-3 (lane B) cell membrane fractions were blotted with proaerolysin (left panel) to identify GPI-anchored proteins. Nonspecific reactions for streptavidin were indicated by two arrows on the right side. The cell membrane fractions were first stained with Ponceau S to show equal loading (right panel) before blotting with proaerolysin. The experiments were repeated with comparable results. D, AsPC-1 (lane A) expressed much more PI-PLC-sensitive GPI-anchored proteins on the cell surface than BxPC-3 (lane B). Surface biotinylated proteins were subjected to PI-PLC treatment and pulled down with lectin-agarose beads. Separated proteins were first reacted with streptavidin-HRP (left panel) and then blotted with 4H2 to show only PrP from AsPC-1 was sensitive to PI-PLC treatment (right panel).
We reported earlier that in BxPC-3 cells another normally GPI-anchored protein CD55 was still GPI-anchored; thus, the defects in GPI anchor modification in BxPC-3 cells is not absolute. In lights of our new results, one would expect that a reduction in the expression levels of 7 of the 24 genes responsible for GPI anchor modification pathway would have a more general effect on the GPI-anchored protein in BxPC-3 cell. To test this, we stained the cell surface of BxPC-3 cells and AsPC-1 cells with FLARE, which potentially should react with all cell surface proteins with GPI anchors. FLAER reacted stronger with AsPC-1 cells compared with BxPC-3 cells. Based on mean fluorescent intensity BxPC-3 cells appear to have about 30% less reactivity compared with AsPC-1 cells. This finding is consistent with our finding that a significantly larger portion of the FLAER immunoreactivity on AsPC-1 cells is sensitive to treatment with PI-PLC. However, identical treatment of BxPC-3 cells only resulted in a slight reduction in FLAER immunoreactivity (Fig. 4B).
To establish whether there are indeed more GPI-anchored proteins on the AsPC-1 cell membrane than on BxPC-3 cell membrane, we performed proaerolysin overlay of the membrane fractions from AsPC-1 and BxPC-3. We found that indeed there were about 60% more GPI-anchored proteins in AsPC-1 cells than in BxPC-3 cells (Fig. 4C).
Finally, we first biotinylated the cell surface of BxPC-3 and AsPC-1 cells, and after that, both cells were treated with PI-PLC to release the GPI-anchored proteins. Glycoproteins in the supernatant were then immunoprecipitated with lectin-conjugated beads to bring down the released glycoproteins. SDS-PAGE-separated proteins were then immunoblotted with avidin-conjugated enzyme to detect the total biotinylated, released proteins, or anti-PrP mAb to detect released PrP. It is clear that two times more GPI-anchored proteins were released from AsPC-1 cells compared with BxPC-3 cells. Most importantly, PrP is only detected in the supernatant of AsPC-1 cells but not in BxPC-3 cells (Fig. 4D). Collectively, these results suggest that the irregularities of the genes do not completely disable the synthesis of all GPI-anchored proteins in BxPC-3 cells.
Functional Rescue of Pro-PrP in BxPC-3 Cells
Multiple genes were expressed at lower levels in BxPC-3 as compared with AsPC-1, and several missense mutations also occurred in those genes. CHO-K1 cell lacks endogenous hamster PrP (45). We reasoned that if we fuse BxPC-3 cells with CHO cells, the GPI anchor synthesis machine from CHO-K1 cells might be able to process pro-PrP, resulting in the generation of a matured GPI-anchored PrP. For this propose, we first transfected a GFP expression construct into CHO-K1 cells as a marker. We then fused GFP-bearing CHO cells with BxPC-3 cells with PEG-1500. Next, we used flow cytometry to enrich for fused cells based on positive staining of cell surface PrP and GFP. Only about 0.1–0.3% of the fused cells was positive for GFP and cell surface PrP. We then performed confocal immunofluorescence staining of the sorted cells before and after PI-PLC treatment. It is apparent that the fused cells became PI-PLC-sensitive (Fig. 5A).
FIGURE 5.
PrP from BxPC-3-CHO-NC-fused cells were GPI-anchored. A, confocal immunofluorescence staining of PrP showed PrP from fused BxPC-3-CHO-NC cells were sensitive to PI-PLC treatment. Note the alteration of PrP distribution on the fused cells. B, proaerolysin overlay of PrP purified from BxPC-3-CHO-NC further proved that those PrP were GPI-anchored (right panel). The amount of PrP was revealed with 4H2 after the blot was treated with stripping buffer (left panel). C, flow cytometer staining proved that PrP from fused BxPC-3-CHO-NC cells was sensitive to PI-PLC treatment, thus proving that those PrPs were GPI-anchored. AsPC-1 cells were detected with 4H2 with or without PI-PLC treatment, and a significant shift was observed (left panel). BxPC-3-CHO-NC cells were stained with 4H2 with or without PI-PLC treatment. After treatment, no PrP could be detected on the surface of fused cells, thus proving the PrP was GPI-anchored (middle panel). BxPC-3 cells were detected with 4H2 with or without PI-PLC treatment, and no shift was observed (right panel). D, quantification of cell surface signal after PI-PLC treatment of those cells showed nearly 100% PrP from the fused cells was GPI-anchored. Relative value of PrP signal was based on mean value of staining (see under “Experimental Procedures” for details). The experiments were repeated with comparable results.
To determine whether the PrP expressed in the fused cells is GPI-anchored, we purified PrP from the fused cells with 5B2 and then immunoblotted with either mAb 4H2 or performed proaerolysin overlay. As expected, mAb 4H2 detected PrP from fused cells as well as BxPC-3, but only PrP from fused cells reacted with proaerolysin (Fig. 5B).
To further confirm that PrP on the surface of fused cells was GPI-anchored, we performed cell surface staining of PrP before and after PI-PLC treatment by flow cytometry. It is obvious that the cell surface PrP was sensitive to PI-PLC treatment. After PI-PLC treatment, no obvious PrP signal could be identified as compared with the background (Fig. 5C). Quantification of the flow cytometry results further revealed that although all the PrP on the surface of BxPC-3 cells was resistant to PI-PLC treatment, the PrP expressed on the surface of BxPC-3-CHO cells was PI-PLC-sensitive (Fig. 5D). Thus, fusion between CHO-K1 and BxPC-3 cells successfully rescued pro-PrP and converted it into GPI-anchored PrP in the fused cells.
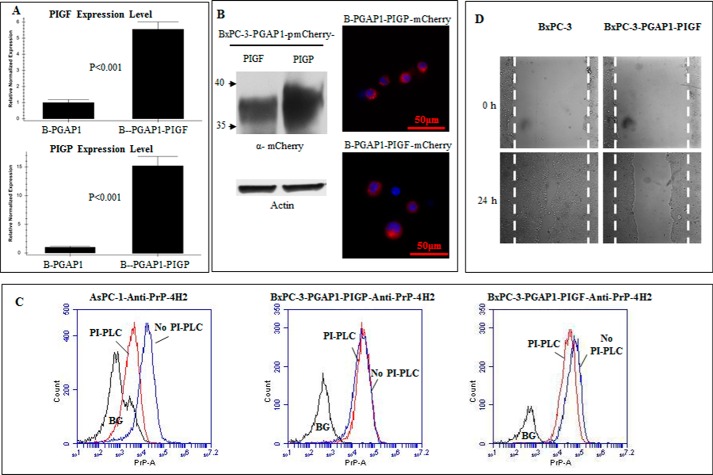
PIG-F Overexpression in BxPC-3-PGAP1 Cells Converts Pro-PrP to GPI-anchored PrP and Reduces Cancer Cell Motility
Successful rescuing of pro-PrP implicated that certain gene irregularities might be responsible for the production of pro-PrP in BxPC-3 cells. When either PIG-P or PIG-F was transfected into BxPC-3 cells, PrP on the BxPC-3 cell surface remained resistant to PI-PLC treatment (results not shown). We then transfected PIG-P or PIG-F into PGAP1-expressing BxPC-3 cells, and Q-PCR showed that PIG-P and PIG-F were expressed at 15- and 5.6-fold as compared with the BxPC-3-PGAP1 cells (Fig. 6A). Because all the proteins were mCherry-tagged, immunoblotting and fluorescence of mCherry proved the expression of these proteins (Fig. 6B). However, only PrP from PIG-F-expressing BxPC-3-PGAP1 cells was susceptible to PI-PLC treatment (Fig. 6C), thus proving that low expression of PIG-F and PGAP1 was the major defect in BxPC-3 cells responsible for the accumulation of pro-PrP. Expressing of either PIG-N or PIG-C into BxPC-3-PGAP1 cells also failed to convert pro-PrP to become GPI-anchored PrP (results not shown). More importantly, those BxPC-3 cells showed a much reduced cell motility in a wound healing assays than the BxPC-3 cells bearing pro-PrP (Fig. 6D).
FIGURE 6.
PrP from PIG-F-expressing BxPC-3-PGAP1 cells was susceptible to PI-PLC treatment and contributes to reduced cell motility. A, Q-PCR showed that PIG-P (labeled as B-PGAP1-PIGP) and PIG-F (labeled as B-PGAP1-PIGF) expression was up-regulated 15- and 5.6-fold in BxPC-3-PGAP1 cells compared with that from non-transfected cells (labeled as B-PGAP1). B, immunoblotting with Living Colors® DsRed polyclonal antibody (left panel) and immunofluorescence observation of mCherry (right panel) showed the expression of PIG-F and PIG-P. C, only PrP from PIG-F expression BxPC-3-PGAP1 cells was moderately susceptible to PI-PLC treatment. D, PIG-F expressing BxPC-3-PGAP1 cells had decreased motility compared with BxPC-3 cells expressing pro-PrP.
Expression of Pro-PrP in BxPC-3 Cells Is Responsible for Their Enhanced Cellular Migration
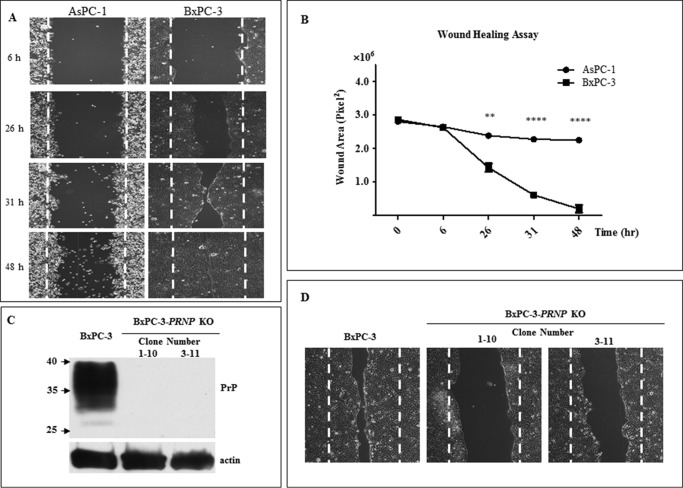
We had previously found that “knocking down” of PrP by shRNAi in BxPC-3 reduced their in vitro proliferation and invasion. This enhancement is most likely due to the binding of pro-PrP to FLNA (11). Because AsPC-1 cells expressed a GPI-anchored PrP and is unable to interact with FLNa, we next compared the migration between BxPC-3 and AsPC-1 cells using a wound-healing assay. It was obvious that BxPC-3 cells migrated significantly faster than AsPC-1 cells (Fig. 7A). Analysis of three different experiments showed that there was significant difference between the migration of BxPC-3 and AsPC-1 cells (Fig. 7B).
FIGURE 7.
Pro-PrP contributing to motility of BxPC-3 cells that migrated faster than AsPC-1 cells. A, wound healing of AsPC-1 and BxPC-3 cells after different time points showed that BxPC-3 cells migrated faster than AsPC-1 cells. B, quantification of wound healing of three different experiments showed that significant difference could be observed 26 h post-wound healing. C, knock-out of PrP from BxPC-3 cells with two different targets confirmed by 4H2 immunoblotting. 1–10 and 3–11 represent two different clones (clone 1–10 was targeted for sequence 1 and clone 3–11 was targeted for sequence 3). D, knocking out PrP significantly reduced motility of BxPC-3 cells. Pictures were taken 26 h post-wound healing. The experiments were repeated three times with comparable results.
To confirm that pro-PrP contributes to the enhanced migration, we knocked out PRNP in BxPC-3 cells by CRISPR/Cas9 approach. We were able to eliminate the PRNP genes in BxPC-3 cells and obtained multiple clones. The successful elimination of the PRNP was confirmed by sequencing (results not shown), and subsequently by immunoblotting with anti-PrP mAb, immunoblotting of two of the PRNP “knock-out” clones was shown (Fig. 7C). Furthermore, it was obvious that eliminating the PRNP gene also significantly alleviated the capability of BxPC-3 cells to migrate (Fig. 7D), thus proving interaction between pro-PrP and FLNa was important for cell migration.
Discussion
The PrP in the six PDAC cell lines we studied earlier existed as a pro-PrP, without a GPI anchor (11). In this study, we reported the detection of GPI-anchored PrP in an additional PDAC cell line, AsPC-1. The PrP in AsPC-1 cells is sensitive to PI-PLC treatment, binds proaerolysin, is resistant to carboxypeptidase treatment, and does not react with a rabbit polyclonal antiserum that is specific for GPI-PSS. On the contrary, the pro-PrP from BxPC-3 is resistant to PI-PLC treatment, does not bind proaerolysin, is sensitive to carboxypeptidase treatment, and reacts to the GPI-PSS specific polyclonal antiserum. Gene sequencing identifies six missense mutations in 4 of the 24 GPI anchor modification pathway genes in BxPC-3 cells. Furthermore, gene profiling of these two cell lines revealed that 15 of the GPI synthesis genes are down-regulated in BxPC-3 cells.
The most significant reduction (>3-fold) in this group of 15 genes are the PIG-F, PIG-X, and PGAP1 genes. PIG-F is required for transferring “bridging” phosphatidylethanolamine to the third mannose in glycosylphosphatidylinositol biosynthesis; PIG-X is a regulator for GPI-mannosyltransferase I complex, and PGAP1 is essential for inositol deacylation (46).
We found that the mature GPI-anchored PrP was detected if PIG-F was expressed in BxPC-3-PGAP1 cells. However, the PI-PLC susceptibility is only moderate (about 50% compared with that from AsPC-1 cells). Therefore, defects in other genes identified in BxPC-3 cells could contribute to the lower efficiency of GPI-anchored protein synthesis. The reason that mCherry-tagged PIG-F appeared with lower molecular weight than predicted is unknown. In fact, when PIG-F was expressed in 293T cells, we also observed the same phenomenon (results not shown). But the PI-PLC treatment experiment clearly demonstrated that the expression of PIG-F could partially rescue the processing of pro-PrP, resulting in the generation of GPI-anchored PrP, indicating that the expressed PIG-F is functional.
A few GPI anchor synthesis genes have been reported to be up-regulated in bladder, breast, and head and neck cancers (47–49). In contrast, the level of DPM2 is down-regulated in the saliva of PDAC patients (50). However, the pathological contribution of the affected genes to cancer cell physiology is unknown. One possibility is that by affecting GPI synthesis, some of the normally GPI-anchored proteins might lose their normal function(s) and/or gain abnormal function(s), thus contributing to tumorigenesis.
Previously, we showed that interactions between pro-PrP and FLNa were important in the proliferation and invasion of BxPC cells in vitro and their growth in vivo. In this study, we provided additional evidence that this interaction is also important in BxPC-3 cell migration in a wound-healing assay in vitro. Although both BxPC-3 and AsPC-1 express FLNa, the ability of BxPC-3 cells to cure a wound is much speedier than AsPC-1 cells. The underlying mechanism responsible for this difference is the expression of pro-PrP in BxPC-3 cells and the expression of a GPI-anchored PrP in AsPC-1 cells. More importantly, CRISPR/Cas9 elimination of the PRNP in BxPC3 cells greatly impaired their in vitro migration capability in the same assay. Taken together, these results indicate that dysfunction in the GPI synthesis machinery contributes to the aggressive behavior of PDAC.
Author Contributions
C. L., M. S. Y., and L. Y. conceived and coordinated the study and designed the experiments. C. L. and L. Y. wrote the manuscript. L. Y., L. H., Z. G., and G. W. carried out experiments. C. L., M. S. Y., B. S. W., Y. Z., X. Y., L. Z., and W. X. revised and polished the manuscript. All authors reviewed and refined the manuscript for important intellectual content.
Acknowledgments
We thank Professor Taroh Kinoshita, Osaka University, for PGAP1 expression plasmid. We also thank The Core Facility and Technical Support, Wuhan Institute of Virology, for technique support in Confocal Microscopy (Dr. Gao) and Flow Cytometry (J. Min).
This work was supported by Chinese National Science Foundation Grants 81172376 and 31270209, Nature Science Foundation of Hubei Province Grant 2015CFA087, Chinese Academy of Sciences 100-Talents Program, State Key Laboratory of Virology of China Grant 2015IOV005, and WIV “One-Three-Five” Strategic Programs. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table S1–S3 and Fig. S1.
- PrP
- prion protein
- PDAC
- pancreatic ductal adenocarcinoma cell
- GPI
- glycosylphosphatidylinositol
- GPI-PSS
- GPI peptide signaling sequence
- PGAP1
- post-GPI attachment to proteins 1
- PI-PLC
- phosphoinositide-specific phospholipase C
- FLNa
- filamin A
- DPBS
- Dulbecco's modified phosphate-buffered saline
- Q-PCR
- quantitative PCR
- APC
- allophycocyanin.
References
- 1. Prusiner S. B., Groth D., Serban A., Koehler R., Foster D., Torchia M., Burton D., Yang S. L., and DeArmond S. J. (1993) Ablation of the prion protein (PrP) gene in mice prevents scrapie and facilitates production of anti-PrP antibodies. Proc. Natl. Acad. Sci. U.S.A. 90, 10608–10612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brown D. R., Qin K., Herms J. W., Madlung A., Manson J., Strome R., Fraser P. E., Kruck T., von Bohlen A., Schulz-Schaeffer W., Giese A., Westaway D., and Kretzschmar H. (1997) The cellular prion protein binds copper in vivo. Nature 390, 684–687 [DOI] [PubMed] [Google Scholar]
- 3. Pauly P. C., and Harris D. A. (1998) Copper stimulates endocytosis of the prion protein. J. Biol. Chem. 273, 33107–33110 [DOI] [PubMed] [Google Scholar]
- 4. Mouillet-Richard S., Ermonval M., Chebassier C., Laplanche J. L., Lehmann S., Launay J. M., and Kellermann O. (2000) Signal transduction through prion protein. Science 289, 1925–1928 [DOI] [PubMed] [Google Scholar]
- 5. Sailer A., Büeler H., Fischer M., Aguzzi A., and Weissmann C. (1994) No propagation of prions in mice devoid of PrP. Cell 77, 967–968 [DOI] [PubMed] [Google Scholar]
- 6. Richt J. A., Kasinathan P., Hamir A. N., Castilla J., Sathiyaseelan T., Vargas F., Sathiyaseelan J., Wu H., Matsushita H., Koster J., Kato S., Ishida I., Soto C., Robl J. M., and Kuroiwa Y. (2007) Production of cattle lacking prion protein. Nat. Biotechnol. 25, 132–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Benestad S. L., Austbø L., Tranulis M. A., Espenes A., and Olsaker I. (2012) Healthy goats naturally devoid of prion protein. Vet. Res. 43, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Miller M. W., Swanson H. M., Wolfe L. L., Quartarone F. G., Huwer S. L., Southwick C. H., and Lukacs P. M. (2008) Lions and prions and deer demise. PLoS One 3, e4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pan Y., Zhao L., Liang J., Liu J., Shi Y., Liu N., Zhang G., Jin H., Gao J., Xie H., Wang J., Liu Z., and Fan D. (2006) Cellular prion protein promotes invasion and metastasis of gastric cancer. FASEB J. 20, 1886–1888 [DOI] [PubMed] [Google Scholar]
- 10. Antonacopoulou A. G., Grivas P. D., Skarlas L., Kalofonos M., Scopa C. D., and Kalofonos H. P. (2008) POLR2F, ATP6V0A1 and PRNP expression in colorectal cancer: new molecules with prognostic significance? Anticancer Res. 28, 1221–1227 [PubMed] [Google Scholar]
- 11. Li C., Yu S., Nakamura F., Yin S., Xu J., Petrolla A. A., Singh N., Tartakoff A., Abbott D. W., Xin W., and Sy M. S. (2009) Binding of pro-prion to filamin A disrupts cytoskeleton and correlates with poor prognosis in pancreatic cancer. J. Clin. Invest. 119, 2725–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li C., Yu S., Nakamura F., Pentikäinen O. T., Singh N., Yin S., Xin W., and Sy M. S. (2010) Pro-prion binds filamin A, facilitating its interaction with integrin β1, and contributes to melanomagenesis. J. Biol. Chem. 285, 30328–30339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Déry M. A., Jodoin J., Ursini-Siegel J., Aleynikova O., Ferrario C., Hassan S., Basik M., and LeBlanc A. C. (2013) Endoplasmic reticulum stress induces PRNP prion protein gene expression in breast cancer. Breast Cancer Res. 15, R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang X., Zhang Y., Zhang L., He T., Zhang J., and Li C. (2014) Prion protein and cancers. Acta Biochim. Biophys. Sin. 46, 431–440 [DOI] [PubMed] [Google Scholar]
- 15. Meslin F., Conforti R., Mazouni C., Morel N., Tomasic G., Drusch F., Yacoub M., Sabourin J. C., Grassi J., Delaloge S., Mathieu M. C., Chouaib S., Andre F., and Mehrpour M. (2007) Efficacy of adjuvant chemotherapy according to Prion protein expression in patients with estrogen receptor-negative breast cancer. Ann. Oncol. 18, 1793–1798 [DOI] [PubMed] [Google Scholar]
- 16. Meslin F., Hamaï A., Gao P., Jalil A., Cahuzac N., Chouaib S., and Mehrpour M. (2007) Silencing of prion protein sensitizes breast adriamycin-resistant carcinoma cells to TRAIL-mediated cell death. Cancer Res. 67, 10910–10919 [DOI] [PubMed] [Google Scholar]
- 17. Liang J., Ge F., Guo C., Luo G., Wang X., Han G., Zhang D., Wang J., Li K., Pan Y., Yao L., Yin Z., Guo X., Wu K., Ding J., and Fan D. (2009) Inhibition of PI3K/Akt partially leads to the inhibition of PrP(C)-induced drug resistance in gastric cancer cells. FEBS J. 276, 685–694 [DOI] [PubMed] [Google Scholar]
- 18. Zhou L., Shang Y., Liu C., Li J., Hu H., Liang C., Han Y., Zhang W., Liang J., and Wu K. (2014) Overexpression of PrPc, combined with MGr1-Ag/37LRP, is predictive of poor prognosis in gastric cancer. Int. J. Cancer 135, 2329–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stossel T. P., Condeelis J., Cooley L., Hartwig J. H., Noegel A., Schleicher M., and Shapiro S. S. (2001) Filamins as integrators of cell mechanics and signalling. Nat. Rev. Mol. Cell Biol. 2, 138–145 [DOI] [PubMed] [Google Scholar]
- 20. Stossel T. P., and Hartwig J. H. (2003) Filling gaps in signaling to actin cytoskeletal remodeling. Dev. Cell 4, 444–445 [DOI] [PubMed] [Google Scholar]
- 21. Maeda Y., Ashida H., and Kinoshita T. (2006) CHO glycosylation mutants: GPI anchor. Methods Enzymol. 416, 182–205 [DOI] [PubMed] [Google Scholar]
- 22. Takeda J., Miyata T., Kawagoe K., Iida Y., Endo Y., Fujita T., Takahashi M., Kitani T., and Kinoshita T. (1993) Deficiency of the GPI anchor caused by a somatic mutation of the PIG-A gene in paroxysmal nocturnal hemoglobinuria. Cell 73, 703–711 [DOI] [PubMed] [Google Scholar]
- 23. Krawitz P. M., Schweiger M. R., Rödelsperger C., Marcelis C., Kölsch U., Meisel C., Stephani F., Kinoshita T., Murakami Y., Bauer S., Isau M., Fischer A., Dahl A., Kerick M., Hecht J., et al. (2010) Identity-by-descent filtering of exome sequence data identifies PIGV mutations in hyperphosphatasia mental retardation syndrome. Nat. Genet. 42, 827–829 [DOI] [PubMed] [Google Scholar]
- 24. Maydan G., Noyman I., Har-Zahav A., Neriah Z. B., Pasmanik-Chor M., Yeheskel A., Albin-Kaplanski A., Maya I., Magal N., Birk E., Simon A. J., Halevy A., Rechavi G., Shohat M., Straussberg R., and Basel-Vanagaite L. (2011) Multiple congenital anomalies-hypotonia-seizures syndrome is caused by a mutation in PIGN. J. Med. Genet. 48, 383–389 [DOI] [PubMed] [Google Scholar]
- 25. Horn D., Krawitz P., Mannhardt A., Korenke G. C., and Meinecke P. (2011) Hyperphosphatasia-mental retardation syndrome due to PIGV mutations: expanded clinical spectrum. Am. J. Med. Genet. A 155A, 1917–1922 [DOI] [PubMed] [Google Scholar]
- 26. Krawitz P. M., Murakami Y., Hecht J., Krüger U., Holder S. E., Mortier G. R., Delle Chiaie B., De Baere E., Thompson M. D., Roscioli T., Kielbasa S., Kinoshita T., Mundlos S., Robinson P. N., and Horn D. (2012) Mutations in PIGO, a member of the GPI-anchor-synthesis pathway, cause hyperphosphatasia with mental retardation. Am. J. Hum. Genet. 91, 146–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ng B. G., Hackmann K., Jones M. A., Eroshkin A. M., He P., Wiliams R., Bhide S., Cantagrel V., Gleeson J. G., Paller A. S., Schnur R. E., Tinschert S., Zunich J., Hegde M. R., and Freeze H. H. (2012) Mutations in the glycosylphosphatidylinositol gene PIGL cause CHIME syndrome. Am. J. Hum. Genet. 90, 685–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kvarnung M., Nilsson D., Lindstrand A., Korenke G. C., Chiang S. C., Blennow E., Bergmann M., Stödberg T., Mäkitie O., Anderlid B. M., Bryceson Y. T., Nordenskjöld M., and Nordgren A. (2013) A novel intellectual disability syndrome caused by GPI anchor deficiency due to homozygous mutations in PIGT. J. Med. Genet. 50, 521–528 [DOI] [PubMed] [Google Scholar]
- 29. Krawitz P. M., Murakami Y., Rieß A., Hietala M., Krüger U., Zhu N., Kinoshita T., Mundlos S., Hecht J., Robinson P. N., and Horn D. (2013) PGAP2 mutations, affecting the GPI-anchor-synthesis pathway, cause hyperphosphatasia with mental retardation syndrome. Am. J. Hum. Genet. 92, 584–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hansen L., Tawamie H., Murakami Y., Mang Y., ur Rehman S., Buchert R., Schaffer S., Muhammad S., Bak M., Nöthen M. M., Bennett E. P., Maeda Y., Aigner M., Reis A., Kinoshita T., et al. (2013) Hypomorphic mutations in PGAP2, encoding a GPI-anchor-remodeling protein, cause autosomal-recessive intellectual disability. Am. J. Hum. Genet. 92, 575–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Murakami Y., Tawamie H., Maeda Y., Büttner C., Buchert R., Radwan F., Schaffer S., Sticht H., Aigner M., Reis A., Kinoshita T., and Jamra R. A. (2014) Null mutation in PGAP1 impairing GPI-anchor maturation in patients with intellectual disability and encephalopathy. PLoS Genet. 10, e1004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kato M., Saitsu H., Murakami Y., Kikuchi K., Watanabe S., Iai M., Miya K., Matsuura R., Takayama R., Ohba C., Nakashima M., Tsurusaki Y., Miyake N., Hamano S., Osaka H., et al. (2014) PIGA mutations cause early-onset epileptic encephalopathies and distinctive features. Neurology 82, 1587–1596 [DOI] [PubMed] [Google Scholar]
- 33. Howard M. F., Murakami Y., Pagnamenta A. T., Daumer-Haas C., Fischer B., Hecht J., Keays D. A., Knight S. J., Kölsch U., Krüger U., Leiz S., Maeda Y., Mitchell D., Mundlos S., Phillips J. A., 3rd, et al. (2014) Mutations in PGAP3 impair GPI-anchor maturation, causing a subtype of hyperphosphatasia with mental retardation. Am. J. Hum. Genet. 94, 278–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Belet S., Fieremans N., Yuan X., Van Esch H., Verbeeck J., Ye Z., Cheng L., Brodsky B. R., Hu H., Kalscheuer V. M., Brodsky R. A., and Froyen G. (2014) Early frameshift mutation in PIGA identified in a large XLID family without neonatal lethality. Hum. Mutat. 35, 350–355 [DOI] [PubMed] [Google Scholar]
- 35. Swoboda K. J., Margraf R. L., Carey J. C., Zhou H., Newcomb T. M., Coonrod E., Durtschi J., Mallempati K., Kumanovics A., Katz B. E., Voelkerding K. V., and Opitz J. M. (2014) A novel germline PIGA mutation in Ferro-Cerebro-Cutaneous syndrome: a neurodegenerative X-linked epileptic encephalopathy with systemic iron-overload. Am. J. Med. Genet. A 164A, 17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hu R., Mukhina G. L., Lee S. H., Jones R. J., Englund P. T., Brown P., Sharkis S. J., Buckley J. T., and Brodsky R. A. (2009) Silencing of genes required for glycosylphosphatidylinositol anchor biosynthesis in Burkitt lymphoma. Exp. Hematol. 37, 423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen R., Knez J. J., Merrick W. C., and Medof M. E. (2001) Comparative efficiencies of C-terminal signals of native glycophosphatidylinositol (GPI)-anchored proproteins in conferring GPI-anchoring. J. Cell. Biochem. 84, 68–83 [DOI] [PubMed] [Google Scholar]
- 38. Zanusso G., Liu D., Ferrari S., Hegyi I., Yin X., Aguzzi A., Hornemann S., Liemann S., Glockshuber R., Manson J. C., Brown P., Petersen R. B., Gambetti P., and Sy M. S. (1998) Prion protein expression in different species: analysis with a panel of new mAbs. Proc. Natl. Acad. Sci. U.S.A. 95, 8812–8816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protocols 8, 2281–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang L., Zhang Y., Hu L., Zhu Y., Sy M. S., and Li C. (2014) A panel of monoclonal antibodies against the prion protein proves that there is no prion protein in human pancreatic ductal epithelial cells. Virol. Sin. 29, 228–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moss J., Balducci E., Cavanaugh E., Kim H. J., Konczalik P., Lesma E. A., Okazaki I. J., Park M., Shoemaker M., Stevens L. A., and Zolkiewska A. (1999) Characterization of NAD:arginine ADP-ribosyltransferases. Mol. Cell. Biochem. 193, 109–113 [PubMed] [Google Scholar]
- 42. Fujita M., and Kinoshita T. (2010) Structural remodeling of GPI anchors during biosynthesis and after attachment to proteins. FEBS Lett. 584, 1670–1677 [DOI] [PubMed] [Google Scholar]
- 43. Diep D. B., Nelson K. L., Raja S. M., Pleshak E. N., and Buckley J. T. (1998) Glycosylphosphatidylinositol anchors of membrane glycoproteins are binding determinants for the channel-forming toxin aerolysin. J. Biol. Chem. 273, 2355–2360 [DOI] [PubMed] [Google Scholar]
- 44. Fujita M., and Kinoshita T. (2012) GPI-anchor remodeling: potential functions of GPI-anchors in intracellular trafficking and membrane dynamics. Biochim. Biophys. Acta 1821, 1050–1058 [DOI] [PubMed] [Google Scholar]
- 45. Lehmann S., and Harris D. A. (1995) A mutant prion protein displays an aberrant membrane association when expressed in cultured cells. J. Biol. Chem. 270, 24589–24597 [DOI] [PubMed] [Google Scholar]
- 46. Hong Y., Maeda Y., Watanabe R., Inoue N., Ohishi K., and Kinoshita T. (2000) Requirement of PIG-F and PIG-O for transferring phosphoethanolamine to the third mannose in glycosylphosphatidylinositol. J. Biol. Chem. 275, 20911–20919 [DOI] [PubMed] [Google Scholar]
- 47. Ho J. C., Cheung S. T., Patil M., Chen X., and Fan S. T. (2006) Increased expression of glycosyl-phosphatidylinositol anchor attachment protein 1 (GPAA1) is associated with gene amplification in hepatocellular carcinoma. Int. J. Cancer 119, 1330–1337 [DOI] [PubMed] [Google Scholar]
- 48. Jiang W. W., Zahurak M., Zhou Z. T., Park H. L., Guo Z. M., Wu G. J., Sidransky D., Trink B., and Califano J. A. (2007) Alterations of GPI transamidase subunits in head and neck squamous carcinoma. Mol. Cancer 6, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nagpal J. K., Dasgupta S., Jadallah S., Chae Y. K., Ratovitski E. A., Toubaji A., Netto G. J., Eagle T., Nissan A., Sidransky D., and Trink B. (2008) Profiling the expression pattern of GPI transamidase complex subunits in human cancer. Mod. Pathol. 21, 979–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang L., Farrell J. J., Zhou H., Elashoff D., Akin D., Park N. H., Chia D., and Wong D. T. (2010) Salivary transcriptomic biomarkers for detection of resectable pancreatic cancer. Gastroenterology 138, 949–957 [DOI] [PMC free article] [PubMed] [Google Scholar]