Abstract
Fibroblast growth factor (FGF) signaling regulates a multitude of cellular processes, including cell proliferation, survival, migration, and differentiation. In the vertebrate lens, FGF signaling regulates fiber cell differentiation characterized by high expression of crystallin proteins. However, a direct link between FGF signaling and crystallin gene transcriptional machinery remains to be established. Previously, we have shown that the bZIP proto-oncogene c-Maf regulates expression of αA-crystallin (Cryaa) through binding to its promoter and distal enhancer, DCR1, both activated by FGF2 in cell culture. Herein, we identified and characterized a novel FGF2-responsive region in the c-Maf promoter (−272/−70, FRE). Both c-Maf and Cryaa regulatory regions contain arrays of AP-1 and Ets-binding sites. Chromatin immunoprecipitation (ChIP) assays established binding of c-Jun (an AP-1 factor) and Etv5/ERM (an Ets factor) to these regions in lens chromatin. Analysis of temporal and spatial expression of c-Jun, phospho-c-Jun, and Etv5/ERM in wild type and ERK1/2 deficient lenses supports their roles as nuclear effectors of FGF signaling in mouse embryonic lens. Collectively, these studies show that FGF signaling up-regulates expression of αA-crystallin both directly and indirectly via up-regulation of c-Maf. These molecular mechanisms are applicable for other crystallins and genes highly expressed in terminally differentiated lens fibers.
Keywords: crystallin, differentiation, fibroblast growth factor (FGF), lens, signaling, αA-crystallin, FGF signaling, c-Maf
Introduction
During embryonic development, the fibroblast growth factor (FGF)3 signal transduction pathway regulates a range of cellular processes including cell proliferation, survival, migration, and differentiation (1). The mammalian FGF signaling is mediated by the interaction of specific secreted FGFs (i.e. FGF1 to FGF10) that work in conjunction with a specialized class of transmembrane receptor tyrosine kinases, the FGF receptors (FGFR1 to FGFR4). Formation of a complex between the dimeric FGFR and its FGF ligand dimer triggers a cascade of intracellular processes relayed by mitogen-activated kinases (MAPKs) such as Erk1 (official gene name: Mapk3) and Erk2 (Mapk1), PI-3/Akt kinase system, and other kinases. Upon entering the nucleus, Erk1/2 kinases elicit transcription of specific DNA-binding transcription factors and/or their post-translational modifications. While the majority of FGF signaling output includes activation of cell proliferation, survival, and motility, FGF signaling also regulates lens, myoblast, and osteogenic terminal differentiation (1, 2).
The ocular lens has served as an advantageous model for studies of FGF signaling over many years (2). Primary rodent lens cell culture experiments showed that addition of a “high” concentration of bFGF/FGF2 (40 ng/ml) alone induced lens fiber cell terminal differentiation while “low” (0.15 ng/ml) and moderate (3 ng/ml) concentrations control cell survival and migration, respectively (3–5). FGF signaling is also modulated by the lens capsule, an extracellular matrix serving as an interface between the lens, aqueous and vitreous humor (6, 7). Subsequent genetic studies of FGF receptors (8, 9), components of the Frs2α/Ras/MAPK signaling arm (10–13), and the cooperating heparan sulfate biosynthesis pathway (14, 15) demonstrated in vivo roles of FGF signaling in mouse lens fiber cell survival and differentiation, and identified a set of lens regulatory genes, including c-Maf, Prox1, Etv1 (ER81), and Etv5 (ERM), whose expression was attenuated following genetic disruption of the FGF signaling pathway (9, 14, 15).
Among these factors, Etv1 and Etv5 are well-established nuclear components of FGF signaling during neural development (16). The bZIP nuclear oncogene c-Maf encodes an important DNA-binding transcription factor that controls lens fiber cell differentiation through crystallin target genes (17). In addition to the lens, c-Maf regulates T-cell (18) and chondrocyte differentiation (19). Up-regulation of MAF was found in multiple myeloma cells and is a potential therapeutic target to treat this cancer (20). Therefore, a thorough understanding of c-Maf transcriptional control relates not only to the basic question of embryonic development but also for dysregulated gene expression during oncogenesis.
Transcriptional control of c-Maf in lens and T cells is just beginning to be understood (21, 22). Expression of c-Maf in the lens is regulated by a 1.3 kb promoter in combination with a 5′-located distal enhancer through autoregulation by c-Maf and direct regulation by Pax6 (17). As this expression system recapitulates endogenous expression of c-Maf in differentiating lens fibers, we hypothesized that the c-Maf promoter/enhancer is regulated through FGF-regulated transcription factors. Up-regulation of c-Maf in the elongating cells of the lens vesicle is followed by expression of αA-crystallin (23). To understand the link between FGF signaling, crystallin gene expression, and lens fiber cell differentiation, we identified a 220-bp long FGF-responsive distal enhancer (DCR1) in the mouse Cryaa locus and demonstrated that DCR1 is sufficient for expression of αA-crystallin in the invaginating lens pit and is essential for αA-crystallin up-regulation in differentiating primary lens fiber cells (23, 24). Thus, it is possible that FGF signaling likely regulates αA-crystallin gene expression by multiple mechanisms that include c-Maf (indirectly) and DCR1 enhancer (directly). Earlier studies in different developmental and cellular systems have identified members of AP-1 (e.g. c-Jun) and Ets (e.g. Etv1 and Etv5, see above) families of transcription factors as primary nuclear effectors of FGF signaling (1, 25). However, it is not known which target genes are directly regulated by these AP-1 and Ets factors during lens differentiation.
Herein, we first examined whether FGF2 could augment c-Maf promoter activity in cultured lens cells. We next identified a critical region (−272/−70) of the c-Maf promoter containing arrays of multiple AP-1 and Ets-binding sites. Similarly, the enhancer of αA-crystallin (DCR1) also harbors these sites. Temporal and spatial analysis of c-Jun, Etv5/ERM, c-Maf, and αA-crystallin expression in mouse embryonic lenses coupled with ChIPs and co-transfection studies support the model of joint regulation of c-Maf by c-Jun and Etv5. Down-regulation of c-Maf and c-Jun proteins, as well as Etv5 mRNAs, was also found in Erk1/2 double conditional lens mutants. The results are summarized in a model comprised of the FGF/FGFR complex, MAPK signaling cascade, nuclear factors c-Jun and Etv5, and c-Maf and Cryaa target genes.
Materials and Methods
Reporter Gene Constructions
A parental 1.3-kb c-Maf promoter-EGFP reporter construct (−494/+866) is described elsewhere (17). A series of three c-Maf promoter deletions generating promoter fragments (Fig. 1) were synthesized by GenScript (Piscataway, NJ) in a pUC57 vector followed by their subcloning into a pEGFP-1 vector (Clontech). The −494/+210 c-Maf promoter fragment and its internal deletion (−272/−70) were also subcloned into a pGL3-luc vector (Promega, Madison, WI). Three copies of FRE (−272/−70) (3xFRE) fused to a minimal E4TATA promoter (26) were also synthesized by GenScript to generate a plasmid 3xFRE/luc. The mouse αA-crystallin promoter and promoter/DCR1-luciferase constructs are described elsewhere (24, 27).
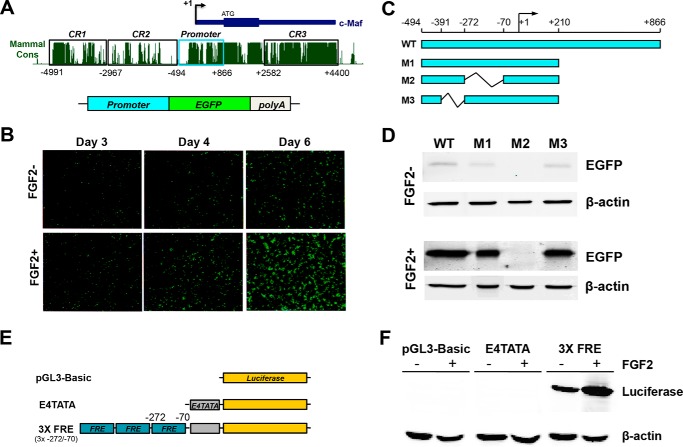
FIGURE 1.
Identification of an FGF-responsive element in the 1.3 kb mouse c-Maf promoter. A, schematic diagram of the mouse c-Maf locus, including the distal enhancer CR1 active in the lens, 1.3-kb promoter, and evolutionarily conserved blocks, and the EGFP reporter construct containing the 1.3 kb c-Maf promoter. B, expression of the 1.3-kb c-Maf promoter-EGFP reporter construct in transiently transfected primary cultures of chicken lens cells in the absence or presence of FGF2. C, diagram of WT and three deletion mutants (M1-M3) of the c-Maf promoter-EGFP reporter constructs. D, semi-quantitative EGFP reporter expression analysis (Western blot, β-actin used as loading control) after transient expression of constructs in primary cultures of chicken lens cells treated with (+FGF) or without (−FGF) FGF2 for 6 days. Note that the M2-reporter was tested at 2-fold DNA concentration. E, diagram of 3xFRE (−272/−70)/luc plasmid. F, analysis of 3XFRE-luc in the presence (+FGF) and absence (−FGF) FGF2 in DCDMLs.
Primary Lens Cell Culture, Transfections, and Western Blot
Primary cultures of embryonic chick lens epithelial cells (DCDMLs) were prepared from E10 chick lenses and plated at 1.2 × 105 cells/well onto laminin-coated 96-well tissue culture plates as previously described (28). Cells were cultured in the absence of serum in M199 medium plus BOTS (2.5 mg/ml bovine serum albumin, 25 mg/ml ovotransferrin, 30 nm selenium), penicillin G, and streptomycin (M199/BOTS). One day after plating, DCDML cultures were transfected in M199 medium using Lipofectamine 2000 (GibcoBRL) following the manufacturer's suggested protocol. Five hours after transfection, cells were cultured in the presence or absence of 10 ng/ml FGF-2 (R&D Systems; Minneapolis, MN). Six days later, the cells were solubilized directly in SDS-PAGE sample buffer and boiled. Equal amounts (10 μg) of total protein were transferred to polyvinylidene fluoride membranes, and the blots were probed with the JL-8 anti-GFP antibody from Clontech (MountainView, CA). Immunoreactive proteins were detected using secondary antibodies conjugated to Alexa Fluor 680 (Molecular Probes, Eugene, OR) and the LI-COR Biosciences Odyssey infrared imaging system (Lincoln, NE). The mutants of Ets binding sites in Cryaa DCR1 enhancer were generated by PCR mutagenesis. For these mutant constructs, primary lens explants were obtained from 3-day-old rat lenses, 2 μg of reporter, and 50 ng of CMV Renilla reference plasmids were transfected with Effecten system (Qiagen) as described elsewhere (29).
Cell Transfections and Reporter Assays
Transient co-transfections were conducted in a breast adenocarcinoma MCF-7 line previously used to study FGF signaling (30) and αTN4-1 mouse lens epithelial cells (27). c-Jun and Etv5 cDNAs in a pCMV6 vector were obtained from OriGene Technologies (Rockville, MD). Briefly, 0.5 μg of the reporter gene, 800 ng of cDNA plasmids (800 ng pCMV6, 400 ng of c-Jun, and 400 ng of pCMV6, 400 ng of ERM and 400 ng pCMV6, 400 ng of c-Jun and 400 ng of ERM), and 20 ng (MCF-7) or 0.25 ng (αTN4) of Renilla-TK were cotransfected into the cells using Lipofectamine 2000 (Invitrogen) in 24-well microplates. Transfection studies of the wild type and mutated DCR1 Cryaa enhancer in 3-day-old rat explants were conducted as described elsewhere (24). The dual luciferase reporter assay system (Promega) was used to measure promoter activity. The cells were harvested 36 h after the transfection, and the relative firefly luciferase reporter activities were measured by the dual luciferase reporter assay system (Promega). The firefly luciferase activities were normalized by Renilla luciferase as an internal control.
Transgenic Mouse Production and Analysis of EGFP Expression
The wild type (WT) 1.3-kb c-Maf promoter/EGFP reporter transgenic mouse was generated earlier (17). The EGFP/transgenic reporter construct with deletion of the −272/−70 FGF responsive element (ΔFRE) was generated by subcloning the synthesized fragment described above into a peGFP-1 vector (Clontech). The transgenic mice were generated by pronuclear injection of fertilized eggs at the Albert Einstein College of Medicine (AECOM) Transgenic Core Facility. EGFP expression was detected by immunofluorescence using a Leica SP5 confocal microscope as described elsewhere (17).
Analysis of Lens-specific Erk1/2 Conditional Mutants
Double conditional knock out (DCKO) mice with deletion of Erk1/2 were generated by crossing Le-CRE mice with Erk1−/−; Erk2F/F mice. The detailed procedures to obtain the embryos were described for the Erk2/Mapk1 model earlier (13). Lens fiber cell differentiation was analyzed in both E12.5 and E13.5 embryos. The Erk1−/−; Erk2F/F littermates with no detectable lens defects served as controls for the double conditional KO (DCKO) mice.
Immunofluorescence and in Situ Hybridizations
For staining of tissues on frozen sections, paraformaldehyde (4%) fixed embryos were cryoprotected with a PBS-buffered 30% sucrose and embedded in Optimal Cutting Temperature tissue freezing medium (Triangle Biomedical Sciences, Durham, NC) for cryosectioning. 8-μm transverse sections were collected, washed with PBS, and incubated for 30 min with Image iT™ FX signal enhancer (Invitrogen). For staining of DCKO Erk1/2 mice tissue, dissected embryos were fixed in 10% formalin overnight. Formalin-fixed embryos were dehydrated through an ethanol gradient. Tissues were processed, embedded in paraffin, and sectioned at 5 μm. Paraffin sections were incubated for 1 h at 60 °C, deparaffinized in xylene three times for 5 min, washed in 100% ethanol twice for 3 min, followed by incubation in 95, 80, and 70% ethanol for 3 min in each step. To retrieve the antigens, the slides were boiled in a 10 mm sodium citrate buffer (pH 6.9) for 20 min in a vegetable steamer. Slides were cooled for 20 min and washed twice with PBS for 10 min. From this stage, both paraffin and frozen sections were processed using the same procedure. Slides were then incubated overnight at 4 °C with the primary antibodies: rabbit anti-GFP (1:1000, Invitrogen, A-11122), rabbit anti-c-Maf (1:2000, Bethyl, A300–613A, or 1:1000, Santa Cruz, sc-7866), c-Jun (1:1000, Abcam, ab31419), phospho-c-Jun (1:50, Cell Signaling, cat. 9621), αB-crystallin (1:500, Enzo Life Science, ADI-SPA-223), β- and γ-crystallin antibodies (1:100, 1:50, Santa Cruz 22745 and 22746, respectively), ERM (1:1000, Santa Cruz, sc-22807) diluted in PBS containing 1% BSA and 0.05% Triton-X100. Antibodies against αA-crystallin (1:500) were described elsewhere (13). After washing with PBS, the slides were incubated for 45 min with goat anti-rabbit IgG secondary antibodies conjugated with Alexa Fluor 488 or 568 (1:500, Invitrogen), and with DAPI (1:50000, Invitrogen). Slides were then washed with PBS and mounted with Vectashield (Vector Laboratories, Burlingame, CA). Immunofluorescence was visualized by using a Zeiss fluorescence microscope and a Leica SP5 confocal microscope in the AECOM core facility. The intensities of the immunofluorescence signals of c-Maf, c-Jun, and phospho-c-Jun in the E12.5 and E13.5 lens were calculated by percentage of the staining positive cells in whole lens tissue (the number of staining positive cells/total number of lens cells stained by DAPI). The cells were counted three times. Student t-tests were performed by the R-project tool to establish the significance of changes of the protein expression in wild type and DCKO mouse lens. Cryosections (12 μm) of embryos were fixed by 4% PFA at 4 °C for overnight were used for in situ hybridization using standard protocols as described elsewhere (31). Briefly, Digoxin-labeled antisense RNA probes were generated via in vitro transcription using the linearized template plasmids Etv5 (Open Biosystems). Antigen retrieval was performed in 10 mm sodium citrate buffer for 40 min prior the use of crystallin-specific antibodies.
Quantification of Immunofluorescence
The c-Maf, c-Jun, ERM, and phospho-c-Jun immunofluorescence signals of control and Erk1/2 DCKO lenses from three sequential slides were measured by Image J software from NIH with a fixed threshold. The tissue areas were selected for measurements were based on the expression pattern in the control lens, and corresponding areas in WT and DCKO lens were analyzed. The software program quantifies the average fluorescence intensity of the selected area in each tissue. Student's t-tests were performed by R-project tool to establish the significance of differences in the intensity of staining between control and DCKO mouse lens as described elsewhere (32).
Bioinformatics Tools
The AP-1 and Ets consensus binding sites and their logos were obtained from primary sources (33–35) and JASPAR database (36), respectively. The sequence alignment was conducted using the Institute Pasteur server.
Quantitative Chromatin Immunoprecipitation (qChIP)
Formaldehyde cross-linked chromatin was obtained from a pool of 400 mouse newborn lenses (CD1 mouse, Charles River Laboratories, Cambridge, MA). The sheared chromatin (average size 600 bp of DNA) was generated by sonication (24). Aliquots of chromatin representing 40 lenses were incubated with 5 μg anti-c-Jun or anti-Etv5/ERM antibodies bound to 20 μl of protein G-coated magnetic beads (Invitrogen). The immunoprecipitates were washed three times and resuspended in a buffer containing 10 mm Tris-HCl, pH 8.0, 100 mm NaCl, 25 mm EDTA supplemented with 0.1 mg/ml RNaseA and 0.2 mg/ml proteinase K. After a 2-h incubation at 55 °C, the crosslinks were reversed by overnight incubation at 65 °C. Genomic DNA was eluted into 250 μl of water using QIAquick Spin Gel Purification kit (Qiagen, Santa Clara, CA). The amounts of each specific DNA fragment (see supplemental Table S1) in immunoprecipitates were determined by quantitative PCR reactions using a standard curve generated for each primer set with 0.04, 0.2, and 1% input DNA samples. Using a standard curve, we transformed Ct values into DNA copy numbers. The copy number of a specific DNA fragment in each assay was compared with the copy number of that fragment before immunoprecipitation (“input DNA”). A control antibody (rabbit normal nonimmune IgG from Calbiochem) was included for each set of the qPCR experiments as described elsewhere (17). To determine a critical value to distinguish real specific binding signals from nonspecific background noise, statistical analysis was conducted using R Software (Version 2.13.1). Analysis of variance (ANOVA) was first performed for the signals obtained from all qChIP amplicon sites for each of the antibodies (ten at the c-Maf locus and twelve regions at the Cryaa locus), and indicated significant differences among the signals. Fisher's least significant difference (LSD) test was then performed to further analyze the data. According to the LSD values, signals obtained from all amplicon sites separated into several groups. The groups with lowest signals were defined as the background groups for each IP. We then performed a Student's t test for the background groups. By calculating the 99% confidence interval (CI), the background signal value is less than 0.124 (c-Jun), 0.132 (ERM), and 1.354 (H3K4me3), respectively. All regions having signals higher than these cutoffs (which were significantly higher than control IgG signals at the corresponding amplicon sites) were therefore considered to be specific binding regions.
Results
Transcriptional regulation of the mouse c-Maf gene in the lens in vivo is regulated by a combination of a 1.3-kb promoter (−494/+866) and a 5′-distal enhancer, CR1 (17). In contrast, in cell cultures the c-Maf promoter alone (i.e. in the absence of the CR1 enhancer) has a lens-preferred activity (37). To examine whether the c-Maf promoter (Fig. 1A) can be activated by FGF2, we used serum-free primary chick lens cell cultures (dissociated cell-derived monolayer cultures, DCDMLs) grown in the presence or absence of FGF2 as described elsewhere (38). The wild type c-Maf promoter activity was visualized via EGFP fluorescence and its “basal” activity was augmented in the presence of FGF2, evaluated at 3, 4, and 5 days following transfection (Fig. 1B). We next prepared a series of 5′- and 3′-truncated promoter fragments and tested these constructs as described above. We found that a 3′-truncated fragment −494/+210 (M1, Fig. 1C) had an activity similar to the WT −494/+866 fragment (data not shown). We next generated two internal deletions inside of the truncated M1 promoter (Fig. 1C) and evaluated their expression by immunofluorescence and Western immunoblotting as described in “Materials and Methods.” The c-Maf promoter activity was lost upon deletion of the −272/−70, but not the adjacent, −391/−272 region (Fig. 1D). We next generated a reporter plasmid driven by three copies of the −272/−70 region (3xFRE/luc) fused to the E4-TATA minimal promoter (Fig. 1E) and evaluated its expression in DCDMLs. We found that 3xFRE/luc reporter was active in lens cells in the absence of FGF2 and its “basal” activity was augmented in the presence of FGF2 (Fig. 1F). We conclude that the −272/−70 region of the mouse c-Maf promoter is important for its basal activity and mediates its inducibility by FGF2.
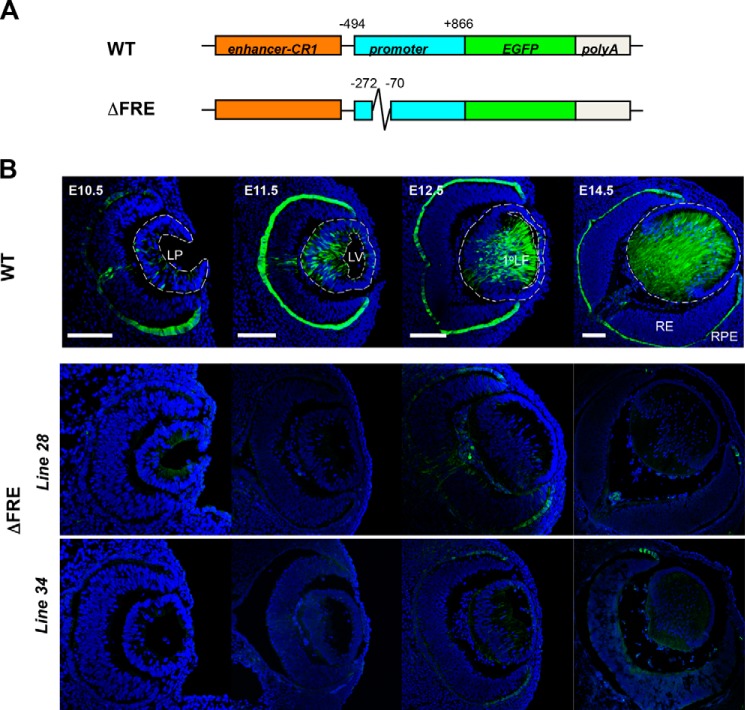
To analyze the function of this region in vivo, we compared activities of the wild type enhancer/promoter/EGFP reporter and its internal −272/−70 deletion (ΔFRE) (Fig. 2A) in transgenic mouse eyes. Expression of the wild type transgene in the lens, retinal pigmented epithelium (RPE), and optic nerve were evaluated in three independent lines as described elsewhere (17). Expression of this transgene recapitulates endogenous expression of c-Maf proteins in the lens (17). Here, five independent ΔFRE transgenic lines were established and analyzed for the EGFP expression. The detailed immunofluorescence data are shown for lines 28 and 34 (Fig. 2B). Deletion of the −272/−70 region resulted in the inactivation of lens expression from E10.5 to E14.5. In contrast, EGFP expression in RPE and optic nerve were disrupted but not abolished indicating that the transgenes were inserted into transcriptionally permissive genomic sites. The remaining three lines showed even lower EGFP expression levels in both lens and non-lens tissues. Taken together, the results demonstrate that the −272/−70 region of mouse c-Maf promoter is essential for its activity in vivo.
FIGURE 2.
The FGF-responsive element is essential for c-Maf transgenic promoter expression in mouse lens. A, schematic diagram of the wild type (WT) and −272/−70 deletion (ΔFRE) EGFP reporter constructs. The wild type (WT) construct containing a 5′-lens-preferred enhancer (CR1) and the 1.3-kb c-Maf promoter. The ΔFRE construct was generated by deleting the FRE region (−272/−70) within the c-Maf promoter. B, histological analysis of EGFP expression in transgenic mouse. Blue, DAPI staining of the nuclei. Green, anti-GFP staining. Bar, 100 μm.
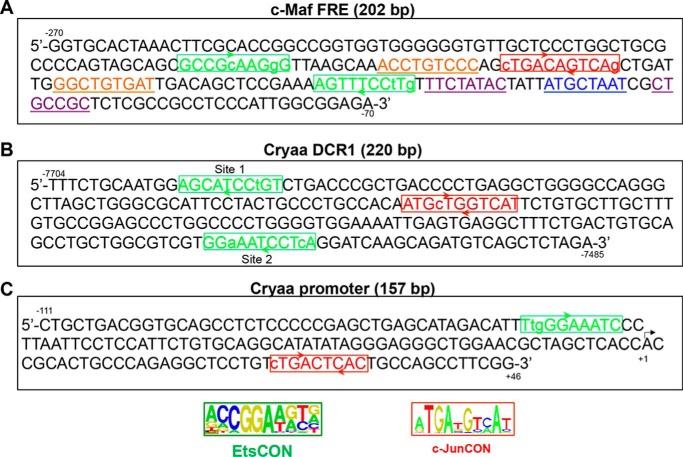
Studies of several FGF-responsive genes, including interstitial collagenase/MMP1 (39), bone sialoprotein (40), and proteoglycan syndecan (41), have shown that mutagenesis of the AP-1/Ets sites abrogated regulation of these genes by FGF signaling (1, 25). Based on AP-1 and Ets consensus binding sites (33, 34), we found an array of two Ets sites, along with an AP-1 site (Fig. 3A), within the −272/-70 region of the c-Maf promoter fragment. In addition, this region contains two candidate Smad-binding sites as well as Pou2f1 (Oct-1)-site (Fig. 3A, see “Discussion”). Earlier, we found that the mouse αA-crystallin gene, a downstream target of c-Maf, contains an FGF2-activated distal enhancer, DCR1(24). Multiple candidate AP-1 and Ets binding sites were also found in the DCR1 enhancer of the αA-crystallin gene (Fig. 3B). In addition, the αA-crystallin promoter also possesses both AP-1- and Ets-binding sites (Fig. 3C) and is stimulated by FGF2 albeit at lower fold change compared with the promoter/DCR1 system (24). These findings prompted us to examine the expression of AP-1/Ets factors in the lens in relationship to their presumptive target gene, c-Maf.
FIGURE 3.
Multiple AP-1- and Ets-binding sites are present in regulatory regions of mouse c-Maf and αA-crystallin genes. A, predicted Ets and AP-1 binding sites within the FRE (−272/−70) of the c-Maf promoter. B, Ets and AP-1 binding sites within the 220 bp DCR1 enhancer of the αA-crystallin locus. C, Ets and AP-1 binding sites within the αA-crystallin promoter (−111/+46). Note that the Ets-binding site matches to a cis-acting region of the mouse Cryaa promoter established earlier (61) and binding of c-Jun to the AP-1 site was reported elsewhere (62). The Ets binding sites were searched by allowing up to two mismatches in the Ets consensus sequence CCGGA(A/T)(A/G)(C/T) (33). AP-1 (c-Jun) binding sites were found by allowing up to two mismatches in the c-Jun palindromic consensus sequence ATGA(T/C)GTCAT (34) or TGA(G/C)T(A/C)A (35). Candidate Smad- (underlined/purple)-sites in the c-Maf FRE were predicted by using Smad consensus motifs 5′-GTCTAGAC-3′(58) and 5′-CWGSMGCY-3′ (57). The candidate Pou2f1 (underlined/blue)-site was predicted by using a consensus motif from the JASPAR database (motif ID MA0785.1) (63, 64).
Expression of the endogenous c-Maf proteins commences in the posterior part of the lens pit with subsequent abundance in differentiating primary and secondary lens fibers (17, 42, 43). To examine the temporal and spatial patterns of expression of the individual AP-1 and Ets factors, we focused on c-Jun and Etv5 since expression of phospho-c-Jun and Etv5 is perturbed in JNK1/2 double null (44) and Ndst1 (14) mutant lenses, respectively, consistent with disrupted lens fiber cell differentiation in both systems. In the E11.5 mouse eye, expression of c-Jun is confined to the posterior part of the lens vesicle from which the primary lens fibers differentiate (Fig. 4). Its expression is retained in the E12.5 and E14.5 differentiating primary lens fibers, and in the E14.5 newly forming secondary lens fiber cells (Fig. 4). Although expression studies of dozens of DNA-binding transcription factors are available in the embryonic lens (45), c-Jun expression is highly specific as it localizes to the posterior lens cells of the E11.5 lens vesicle undergoing early stages of differentiation and elongation (Fig. 4). Expression of Etv5 was examined by in situ hybridizations in the eye demonstrating its expression in differentiating lens fibers (Fig. 4). Taken together these expression studies raised the possibility that c-Jun and Etv5/ERM could serve as regulators of c-Maf gene expression in the embryonic lens.
FIGURE 4.
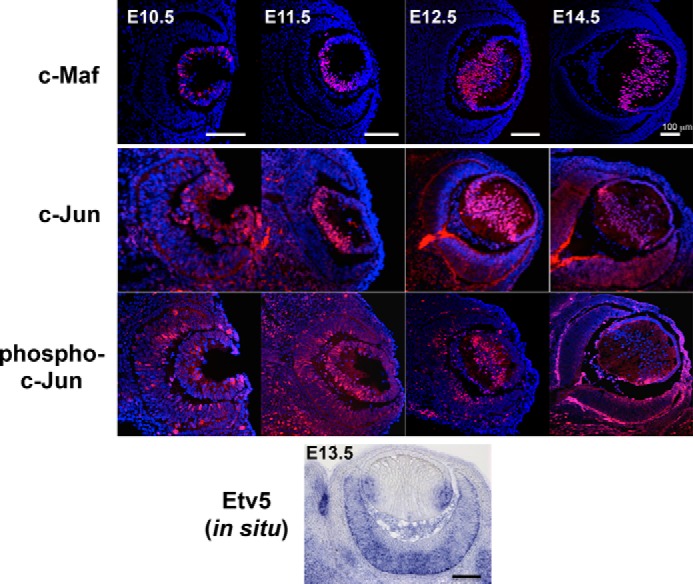
Expression of c-Maf, c-Jun, phospho-c-Jun and Etv5/ERM in the wild type embryonic mouse eye. Immunofluorescence staining of c-Maf, c-Jun, and phospho-c-Jun in developing lens from E10.5 to E14.5. In situ hybridization analysis of Etv5 in E13.5 mouse eye. Blue: DAPI staining of nuclei. Red, antibody staining. Bar, 100 μm.
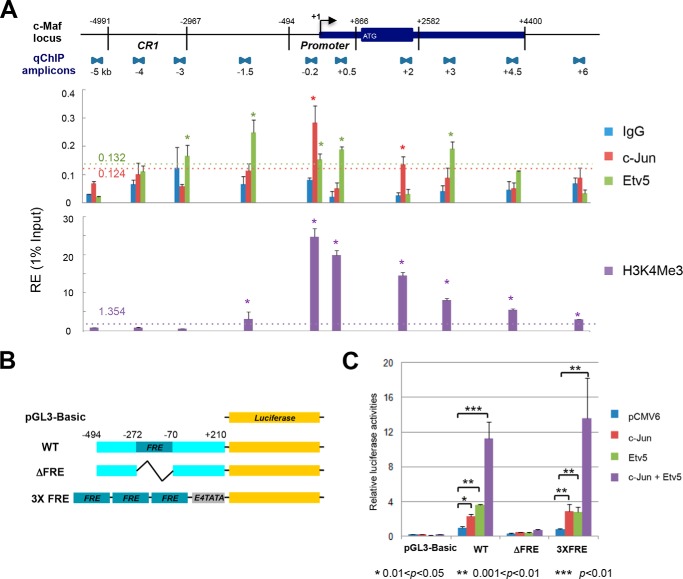
To test this model, we employed quantitative ChIP assays at the c-Maf locus using lens chromatin (17). Statistically significant binding of c-Jun was found in the c-Maf promoter region as well as in the +2 kb regulatory region (Fig. 5A), both of which are also bound by c-Maf as part of its autoregulation mechanism (17). Binding of Etv5 was detected in the promoter region and other locations across the ∼11 kb analyzed. As expected for a transcriptionally active gene, H3K4me3 post-translational modifications were detected in the promoter and body of the c-Maf gene (Fig. 5A).
FIGURE 5.
Transcriptional regulation of c-Maf by c-Jun and Etv5/ERM. A, distribution of c-Jun and Etv5/ERM factors and H3 K4me3 promoter marker along the mouse c-Maf locus in lens chromatin. The locations of the qChIP amplicons covering 11 kb (−5 kb/+6 kb) c-Maf locus is shown at top. c-Jun, Etv5/ERM, and H3K4me3 are presented in the middle and lower panel, respectively. Statistically significant enrichment of binding signals is indicated above the horizontal dotted line. The relative enrichments are shown as 1% of the input. B, diagram of firefly luciferase reporter constructs without promoter (pGL3-Basic), with the intact −494/+210 c-Maf promoter (WT), with the c-Maf promoter lacking the −272/-70 FRE region (ΔFRE), and with three copies of the FRE followed by a minimal E4TATA promoter (3XFRE). C, results of transient co-transfection/reporter assays. The firefly luciferase activities were normalized using Renilla luciferase as an internal control. The results shown are from two independent experiments with duplicates. The relative luciferase activities were calculated using the “WT reporter with pCMV6 cDNA vector” value set as 1.
To find whether c-Jun and Etv5 could activate the c-Maf promoter in transfected cells, we first examined the intact M1 promoter −494/+210, followed by the ΔFRE mutant and 3xFRE/luc reporters (Fig. 5B). Both c-Jun and Etv5 alone activated the wild type M1 promoter and the 3xFRE/luc reporters, but not the ΔFRE mutant (Fig. 5C). In addition, strong synergistic effects of co-expressed c-Jun and Etv5 proteins were found in these experiments when the −272/−70 c-Maf FRE region was present. Taken together, localization of c-Jun and Etv5 in c-Maf promoter in lens chromatin, their ability to activate reporter gene expression driven by wild type c-Maf but not its ΔFRE mutant, and activation of 3xFRE by FGF2 in primary lens cultures support the model in which c-Jun and Etv5 serve as transcription factors that regulate c-Maf expression in the lens.
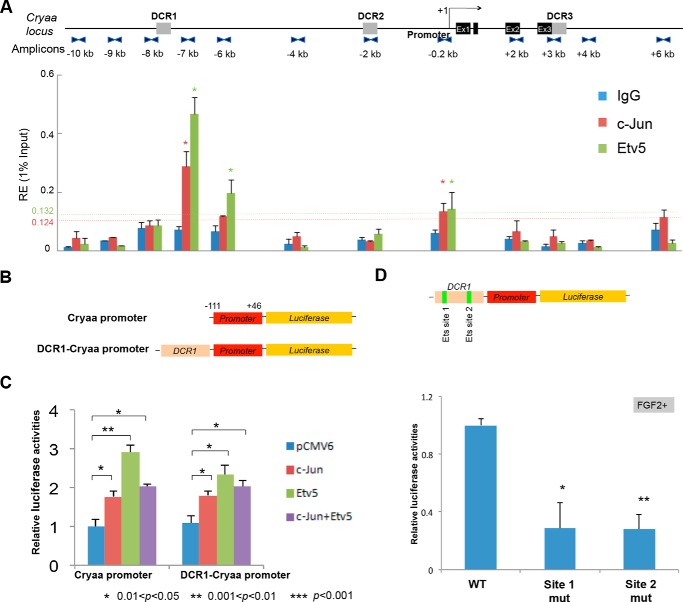
We next analyzed binding of c-Jun, Etv5, and c-Maf at the ∼16 kb Cryaa locus (24). Both c-Jun and Etv5 occupied the 5′-distal enhancer DCR1 and the promoter (Fig. 6A). Binding of c-Maf was mostly found in the promoter region as shown earlier (24). Co-transfection of the mouse αA-crystallin promoter (Fig. 6B) with c-Jun and Etv5 resulted in moderate activation of the promoter in cultured lens cells (Fig. 6C). Similarly, the DCR1/αA-crystallin promoter was also stimulated by these individual factors. Importantly, site-directed mutagenesis of two predicted Ets sites (Fig. 3B) reduced expression of the reporter gene in lens cells treated by FGF2 (Fig. 6D). From data shown in Figs. 5 and 6 we conclude that both mouse c-Maf and Cryaa loci are occupied in vivo by c-Jun and Etv5/ERM. Their most prominent binding regions include the c-Maf promoter, DCR1, and promoter regions of αA-crystallin in agreement with multiple predicted binding sites within these regions (Fig. 3).
FIGURE 6.
Transcriptional regulation of αA-crystallin by c-Jun and Etv5/ERM. A, distribution of c-Jun and Etv5/ERM in the mouse Cryaa locus in lens chromatin. The locations of the qChIP amplicons covering 16 kb (−10 kb/+6 kb) Cryaa locus are shown in the upper panel. c-Jun and ERM binding and IgG background binding are presented in the lower panel. The significant enrichments of binding signals were calculated as described in the legend to Fig. 5A. B, diagram of αA-crystallin promoter and DCR1 enhancer firefly luciferase reporter constructs. C, results of transient co-transfection reporter assays. The results shown are means ± S.D. (n = 3). The relative luciferase activities were calculated using the “Cryaa promoter with pCMV6 cDNA vector” value set as 1. D, results of transient transfections using wild type (WT) and mutated DCR1 (Ets sites 1 and 2, see Fig. 3B) in rat explants grown in the presence of FGF2. Significance was determined using paired Student's t tests between reactions using Cryaa promoter with the empty cDNA control. ** indicates p values ≤ 0.05 and * indicates p values ≤ 0.5.
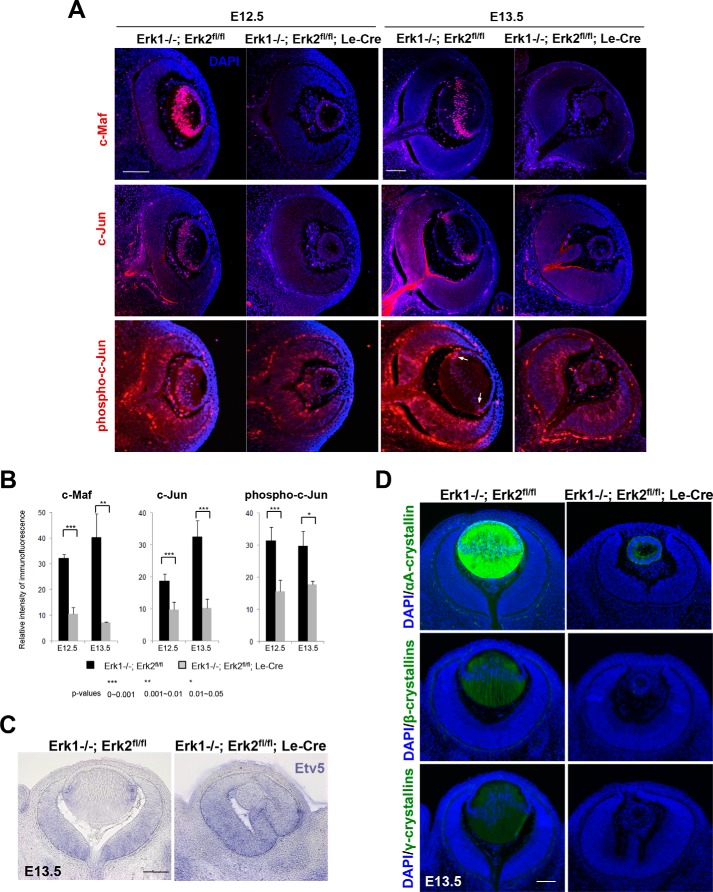
Mitogen-activated protein kinases Erk1/2 are FGF-regulated nuclear kinases expressed in the embryonic lens (10, 13, 46, 47). Inhibiting Erk1/2 function with small molecule inhibitor U0126 impairs lens differentiation (47, 48). Disrupting their function is expected to abrogate FGF signaling, by potentially dysregulating expression and posttranslational modifications of their target DNA-binding transcription factors. To address this possibility, we analyzed mouse embryonic eyes following lens-specific conditional inactivation of both Erk1/Mapk3 and Erk2/Mapk1 (referred here as DCKO Erk1/2) mediated by Le-Cre (49). Inactivation of Erk1/2 in DCKO Erk1/2 animals resulted in arrested lens growth and differentiation that was accompanied by reduced expression of c-Jun in the lens and of its direct target gene, c-Maf (Fig. 7). The reduction in c-Jun in DCKO lenses was also detectable when assessed by anti-phospho-c-Jun immunostaining. In addition, Etv5 expression, evaluated by in situ hybridization, was also reduced at the mRNA level in DCKO lenses but not in the retina (Fig. 7C). Finally, we analyzed expression of α-, β-, and γ-crystallins. The mutated lenses failed to upregulate αA-crystallin expression in the lens rudiment (Fig. 7D). In wild type lens, expression of αA-crystallin is detectable at this stage of lens morphogenesis (23). In contrast, expression of αB-crystallin was not reduced, even at more advanced ages. Previous studies showed that expression of αA- but not αB-crystallin is reduced in c-Maf null lenses (43). Expression of β- and γ-crystallins is also reduced in these mutated lenses (Fig. 7D). Taken together, these data provide genetic evidence that expression of c-Jun, Etv5, and c-Maf is downstream of the Erk1/2 effector nuclear kinases.
FIGURE 7.
Expression of c-Maf, c-Jun, phospho-c-Jun, and Etv5/ERM in Erk1/2 mutant lens. A, immunofluorescence staining in E12.5 and E13.5 DCKO mutant and control lens for c-Maf, c-Jun, and phospho c-Jun. B, quantitative analysis of these immunofluorescence signals showed a significant reduction of c-Maf, c-Jun, and phospho-c-Jun in Erk1/2 DCKO mutant lens compared with control lens. The arrow indicates the restricted areas (transition zones) where high levels of c-Jun phosphorylation were observed in E13.5 lens of the control mice. C, analysis of Etv5 expression (blue) in Erk1/2 DCKO mouse embryos. D, analysis of αA-, β-, and γ-crystallins expression (green) in Erk1/2 DCKO mouse embryos. Blue, DAPI staining of nuclei. Red, antibody staining of the transcription factors. Bar, 100 μm.
Discussion
The present data suggest that FGF signaling directly regulates expression of c-Maf and αA-crystallin genes via c-Jun and Etv5. These DNA-binding transcription factors recognize two critical regulatory regions: the −272/−70 promoter region of c-Maf identified here, and the DCR1 enhancer of the αA-crystallin (24). The present data show that the −272/−70 c-Maf promoter contains an FGF2-responsive region, is occupied by c-Jun and Etv5 proteins in lens chromatin, and is synergistically activated by c-Jun and Etv5/ERM in co-transfection experiments. These findings provide a direct link between the previously identified “core” GRN, comprised of Pax6, c-Maf, and αA-crystallin, that controls crystallin gene expression (17)and FGF signaling, thereby filling the gap between the known roles of FGF signaling in lens fiber cell differentiation and its hallmark process, crystallin gene expression. Given the multiple roles of FGF signaling during hematopoiesis and bone differentiation, it is possible that this c-Maf promoter region also functions in non-lens cells to regulate c-Maf expression.
Expression of αA-crystallin first appears in the invaginating lens pit of the E10.5-E11.5 mouse embryo (23). This “initial” low expression is regulated by a feed-forward loop between Pax6, c-Maf, and Cryaa (17). Following up-regulation of c-Maf, increase of αA-crystallin gene expression is detected in the posterior part of the lens vesicle from which the primary lens fiber cells are formed (E12.5-E14.5) and requires the presence of the 5′-distal enhancer DCR1 (24). We show here that at least two specific DNA-binding factors, c-Jun and Etv5, bind to the FGF-responsive regions in c-Maf and its direct target gene, Cryaa. Increased expression of c-Maf in differentiating lens fibers correlates with its increased abundance at the αA-crystallin gene promoter in lens chromatin compared with the chromatin obtained from non-differentiating lens epithelial cells (24). Use of the DCR1 enhancer, occupied by the FGF-regulated proteins c-Jun and Etv5, provides a mechanism to both initiate and augment the expression of αA-crystallin. αA-crystallin ranks among the most highly expressed genes in mammalian tissues (50, 65).
Both the AP-1 (35) and Ets families have members other than the c-Jun and Etv5 (1), which were also examined in lens. Among the Ets factors, expression of Etv1/ER81, Ets2, Etv4/Pea3 (51, 52), and Elf1 (53) was established in the embryonic lens. In contrast to the spatially constrained expression of c-Jun in differentiating primary lens fibers, five of these Ets factors are expressed both in the anterior and posterior parts of the lens vesicle as well as in the lens epithelium and fibers. Although mice with conditional deletion of Frs2α, α lipid-anchored docking protein (12) show reduced expression of Etv1 in E10.5 lens pit, preliminary ChIP experiments did not find this transcription factor at either c-Maf or Cryaa loci. The expression pattern of c-Jun in the lens vesicle, the presence of c-Jun in the key regulatory regions of c-Maf and Cryaa loci, the ability of c-Jun and Etv5 to activate Cryaa promoter, and the disrupted expression of c-Jun in Erk1/2-mutated lens all support the idea that c-Jun is an FGF signaling-regulated transcription factor which controls expression of both the c-Maf and αA-crystallin genes. Among all known DNA-binding factors expressed in the lens (45), only c-Jun (Fig. 4) and Gata3 (54) exhibit an expression pattern confined to the post-mitotic differentiating cells. Additional experiments will be needed to determine if Etv1, Etv4, and Ets1 factors interact with c-Maf and crystallin loci in lens chromatin.
Genetic studies of selected AP-1 and Ets factors have been conducted in mice. Germline knock-out of c-Jun is lethal between E11.5-E15.5; the eye phenotype of these animals was not reported (56). JNK1/2 double null embryos show reduced expression of phospho-c-Jun, accompanied by disrupted lens growth and lens fiber cell differentiation and reduced expression of α- and β/γ-crystallins (44). The AP-1/Ets factors might have additional roles in lens development. Recent studies have shown that both CBP and p300 histone acetyltransferases are required for lens induction (32). Reduced expression of c-Jun and Etv5 were found in the lens prospective ectoderm of animals defective in expression of CBP and p300 histone acetyltransferases. In addition, gene dosage effect studies revealed that reducing CBP or p300 histone expression to a single functional allele resulted in abnormal lens fiber cell differentiation (32), and both AP-1 and Ets factors are known to recruit CBP/p300 proteins (56).
Although the −272/−70 c-Maf promoter region has been identified here as an FGF2-response element (Fig. 1F), it is possible that additional signal-dependent regulatory mechanisms (e.g. BMP signaling) operate using this regulatory region. In preliminary experiments we found that both noggin (inhibitor of BMP signaling) and PD173074 (a drug that inhibits FGF and VEGF receptors) independently inhibited FGF2-mediated up-regulation of the 3xFRE/luc reporter system in DCDMLs when cells were grown on laminin but not on fibronectin. Two potential Smad-binding sites are found downstream of the Ets site (Fig. 3A), 5′-TTCTATAC-3′ and 5′-CTGCCGC-3′ predicted by Smad-consensus sites (57, 58). Alternatively, it is possible that AP-1 proteins form complexes with Smad3/4 to augment their roles as activators (59). It has been recently shown that αA-crystallin reporter gene expression driven by the DCR1 enhancer is also inhibited by noggin (60) raising the possibility that c-Maf FRE and Cryaa DCR1 are under joint control of FGF and BMP signaling.
In conclusion, in a sequence of temporally and spatially coordinated events, expression of a small group of DNA-binding transcription factors including c-Jun, c-Maf, Gata3, Prox1, Sox1, and Hsf4 is highly increased in the posterior compartment of the embryonic lens vesicle. Among this group, expression of c-Jun and Gata3 is confined to the postmitotic posterior cells of the lens vesicle. Expression of c-Jun, c-Maf, and Prox1 is abrogated in mutants that attenuate FGF signaling, including deletion of Erk1/2, FGFRs, Frs2α, and Ndst1. A combinatorial binding of c-Jun and Etv5/ERM to the regulatory regions of c-Maf promoter and its αA-crystallin target gene, found in lens chromatin, reveals a general molecular mechanism that links FGF signaling to crystallin gene expression.
Author Contributions
A. C. conceived and coordinated the study and wrote the paper. A. C. and Q. X. designed, performed, and analyzed the experiments shown in all figures. L. S. M. performed the transfection experiments shown in Fig. 1. R. M. performed the transfection in Fig. 6C. C. Y. G. performed the experiments in Fig. 5D. R. H. and W. L. performed experiments for in situ and crystallin staining. L. W. R. provided the ERK1/2 double knockout mice.
This work was supported by National Institutes of Health Grants R01 EY014237 (to A. C.), EY016422 and EY022113 (to L. S. M.), EY013146 (to L. W. R.), EY022645 (to W. L.), and an unrestricted grant from Research to Prevent Blindness to the Department of Ophthalmology and Visual Sciences. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Table S1.
- FGF
- fibroblast growth factor
- Cryaa
- αA-crystallin
- DCKO
- double conditional knock out.
References
- 1. Turner N., and Grose R. (2010) Fibroblast growth factor signalling: from development to cancer. Nat. Rev. Cancer 10, 116–129 [DOI] [PubMed] [Google Scholar]
- 2. Lovicu F. J., McAvoy J. W., and de Iongh R. U. (2011) Understanding the role of growth factors in embryonic development: insights from the lens. Philos Trans. R. Soc. Lond B. Biol. Sci. 366, 1204–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McAvoy J. W., and Chamberlain C. G. (1989) Fibroblast growth factor (FGF) induces different responses in lens epithelial cells depending on its concentration. Development 107, 221–228 [DOI] [PubMed] [Google Scholar]
- 4. West-Mays J. A., Pino G., and Lovicu F. J. (2010) Development and use of the lens epithelial explant system to study lens differentiation and cataractogenesis. Prog Retin Eye. Res. 29, 135–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zelenka P. S., Gao C. Y., and Saravanamuthu S. S. (2009) Preparation and culture of rat lens epithelial explants for studying terminal differentiation. J. Vis. Exp. 10.3791/1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tholozan F. M., Gribbon C., Li Z., Goldberg M. W., Prescott A. R., McKie N., and Quinlan R. A. (2007) FGF-2 release from the lens capsule by MMP-2 maintains lens epithelial cell viability. Mol. Biol. Cell 18, 4222–4231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu W., Tholozan F. M., Goldberg M. W., Bowen L., Wu J., and Quinlan R. A. (2014) A gradient of matrix-bound FGF-2 and perlecan is available to lens epithelial cells. Exp. Eye. Res. 120, 10–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Garcia C. M., Yu K., Zhao H., Ashery-Padan R., Ornitz D. M., Robinson M. L., and Beebe D. C. (2005) Signaling through FGF receptor-2 is required for lens cell survival and for withdrawal from the cell cycle during lens fiber cell differentiation. Dev. Dyn 233, 516–527 [DOI] [PubMed] [Google Scholar]
- 9. Zhao H., Yang T., Madakashira B. P., Thiels C. A., Bechtle C. A., Garcia C. M., Zhang H., Yu K., Ornitz D. M., Beebe D. C., and Robinson M. L. (2008) Fibroblast growth factor receptor signaling is essential for lens fiber cell differentiation. Dev. Biol. 318, 276–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gotoh N., Ito M., Yamamoto S., Yoshino I., Song N., Wang Y., Lax I., Schlessinger J., Shibuya M., and Lang R. A. (2004) Tyrosine phosphorylation sites on FRS2alpha responsible for Shp2 recruitment are critical for induction of lens and retina. Proc. Natl. Acad. Sci. U.S.A. 101, 17144–17149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li H., Tao C., Cai Z., Hertzler-Schaefer K., Collins T. N., Wang F., Feng G. S., Gotoh N., and Zhang X. (2014) Frs2alpha and Shp2 signal independently of Gab to mediate FGF signaling in lens development. J. Cell Sci., 127(Pt 3), 571–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Madakashira B. P., Kobrinski D. A., Hancher A. D., Arneman E. C., Wagner B. D., Wang F., Shin H., Lovicu F. J., Reneker L. W., and Robinson M. L. (2012) Frs2alpha enhances fibroblast growth factor-mediated survival and differentiation in lens development. Development 139, 4601–4612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Upadhya D., Ogata M., and Reneker L. W. (2013) MAPK1 is required for establishing the pattern of cell proliferation and for cell survival during lens development. Development 140, 1573–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pan Y., Woodbury A., Esko J. D., Grobe K., and Zhang X. (2006) Heparan sulfate biosynthetic gene Ndst1 is required for FGF signaling in early lens development. Development 133, 4933–4944 [DOI] [PubMed] [Google Scholar]
- 15. Qu X., Hertzler K., Pan Y., Grobe K., Robinson M. L., and Zhang X. (2011) Genetic epistasis between heparan sulfate and FGF-Ras signaling controls lens development. Dev. Biol. 355, 12–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guillemot F., and Zimmer C. (2011) From cradle to grave: the multiple roles of fibroblast growth factors in neural development. Neuron 71, 574–588 [DOI] [PubMed] [Google Scholar]
- 17. Xie Q., and Cvekl A. (2011) The orchestration of mammalian tissue morphogenesis through a series of coherent feed-forward loops. J. Biol. Chem. 286, 43259–43271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ho I. C., and Glimcher L. H. (2002) Transcription: tantalizing times for T cells. Cell 109, S109–S120 [DOI] [PubMed] [Google Scholar]
- 19. MacLean H. E., Kim J. I., Glimcher M. J., Wang J., Kronenberg H. M., and Glimcher L. H. (2003) Absence of transcription factor c-maf causes abnormal terminal differentiation of hypertrophic chondrocytes during endochondral bone development. Dev. Biol. 262, 51–63 [DOI] [PubMed] [Google Scholar]
- 20. Chang H., Qi Q., Xu W., and Patterson B. (2007) c-Maf nuclear oncoprotein is frequently expressed in multiple myeloma. Leukemia 21, 1572–1574 [DOI] [PubMed] [Google Scholar]
- 21. Ise W., Kohyama M., Schraml B. U., Zhang T., Schwer B., Basu U., Alt F. W., Tang J., Oltz E. M., Murphy T. L., and Murphy K. M. (2011) The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nature Immunology 12, 536–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rani A., Afzali B., Kelly A., Tewolde-Berhan L., Hackett M., Kanhere A. S., Pedroza-Pacheco I., Bowen H., Jurcevic S., Jenner R. G., Cousins D. J., Ragheb J. A., Lavender P., and John S. (2011) IL-2 regulates expression of C-MAF in human CD4 T cells. J. Immunol. 187, 3721–3729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wolf L., Yang Y., Wawrousek E., and Cvekl A. (2008) Transcriptional regulation of mouse alpha A-crystallin gene in a 148kb Cryaa BAC and its derivates. BMC Dev. Biol. 8, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang Y., Stopka T., Golestaneh N., Wang Y., Wu K., Li A., Chauhan B. K., Gao C. Y., Cveklová K., Duncan M. K., Pestell R. G., Chepelinsky A. B., Skoultchi A. I., and Cvekl A. (2006) Regulation of αA-crystallin via Pax6, c-Maf, CREB and a broad domain of lens-specific chromatin. EMBO J. 25, 2107–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Oikawa T., and Yamada T. (2003) Molecular biology of the Ets family of transcription factors. Gene 303, 11–34 [DOI] [PubMed] [Google Scholar]
- 26. Chauhan B. K., Yang Y., Cveklová K., and Cvekl A. (2004) Functional interactions between alternatively spliced forms of Pax6 in crystallin gene regulation and in haploinsufficiency. Nucleic Acids Res. 32, 1696–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang Y., and Cvekl A. (2005) Tissue-specific regulation of the mouse αA-crystallin gene in lens via recruitment of Pax6 and c-Maf to its promoter. J. Mol. Biol. 351, 453–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Le A. C., and Musil L. S. (1998) Normal differentiation of cultured lens cells after inhibition of gap junction-mediated intercellular communication. Dev. Biol. 204, 80–96 [DOI] [PubMed] [Google Scholar]
- 29. Golestaneh N., Fan J., Fariss R. N., Lo W. K., Zelenka P. S., and Chepelinsky A. B. (2004) Lens major intrinsic protein (MIP)/aquaporin 0 expression in rat lens epithelia explants requires fibroblast growth factor-induced ERK and JNK signaling. J. Biol. Chem. 279, 31813–31822 [DOI] [PubMed] [Google Scholar]
- 30. Suyama K., Shapiro I., Guttman M., and Hazan R. B. (2002) A signaling pathway leading to metastasis is controlled by N-cadherin and the FGF receptor. Cancer Cell 2, 301–314 [DOI] [PubMed] [Google Scholar]
- 31. Liu W., Lagutin O., Swindell E., Jamrich M., and Oliver G. (2010) Neuroretina specification in mouse embryos requires Six3-mediated suppression of Wnt8b in the anterior neural plate. J. Clin. Invest. 120, 3568–3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wolf L., Harrison W., Huang J., Xie Q., Xiao N., Sun J., Kong L., Lachke S. A., Kuracha M. R., Govindarajan V., Brindle P. K., Ashery-Padan R., Beebe D. C., Overbeek P. A., and Cvekl A. (2013) Histone posttranslational modifications and cell fate determination: lens induction requires the lysine acetyltransferases CBP and p300. Nucleic Acids Res. 41, 10199–10214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wei G. H., Badis G., Berger M. F., Kivioja T., Palin K., Enge M., Bonke M., Jolma A., Varjosalo M., Gehrke A. R., Yan J., Talukder S., Turunen M., Taipale M., Stunnenberg H. G., Ukkonen E., Hughes T. R., Bulyk M. L., and Taipale J. (2010) Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 29, 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li M., Ge Q., Wang W., Wang J., and Lu Z. (2011) c-Jun binding site identification in K562 cells. J. Genet. Genomics 38, 235–242 [DOI] [PubMed] [Google Scholar]
- 35. Yang Y., and Cvekl A. (2007) Large Maf Transcription Factors: Cousins of AP-1 Proteins and Important Regulators of Cellular Differentiation. The Einstein Journal of Biology and Medicine 23, 2–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bryne J. C., Valen E., Tang M. H., Marstrand T., Winther O., da Piedade I., Krogh A., Lenhard B., and Sandelin A. (2008) JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 36, D102–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sakai M., Serria M. S., Ikeda H., Yoshida K., Imaki J., and Nishi S. (2001) Regulation of c-maf gene expression by Pax6 in cultured cells. Nucleic Acids Res. 29, 1228–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Musil L. S. (2012) Primary cultures of embryonic chick lens cells as a model system to study lens gap junctions and fiber cell differentiation. J. Membr Biol. 245, 357–368 [DOI] [PubMed] [Google Scholar]
- 39. Newberry E. P., Willis D., Latifi T., Boudreaux J. M., and Towler D. A. (1997) Fibroblast growth factor receptor signaling activates the human interstitial collagenase promoter via the bipartite Ets-AP1 element. Mol. Endocrinol 11, 1129–1144 [DOI] [PubMed] [Google Scholar]
- 40. Takai S., Hanai Y., Matsushima-Nishiwaki R., Minamitani C., Otsuka T., Tokuda H., and Kozawa O. (2008) P70 S6 kinase negatively regulates fibroblast growth factor 2-stimulated interleukin-6 synthesis in osteoblasts: function at a point downstream from protein kinase C. J Endocrinol 197, 131–137 [DOI] [PubMed] [Google Scholar]
- 41. Jaakkola P., and Jalkanen M. (1999) Transcriptional regulation of Syndecan-1 expression by growth factors. Prog. Nucleic Acids Res. Mol. Biol. 63, 109–138 [DOI] [PubMed] [Google Scholar]
- 42. Kawauchi S., Takahashi S., Nakajima O., Ogino H., Morita M., Nishizawa M., Yasuda K., and Yamamoto M. (1999) Regulation of lens fiber cell differentiation by transcription factor c-Maf. J. Biol. Chem. 274, 19254–19260 [DOI] [PubMed] [Google Scholar]
- 43. Ring B. Z., Cordes S. P., Overbeek P. A., and Barsh G. S. (2000) Regulation of mouse lens fiber cell development and differentiation by the Maf gene. Development 127, 307–317 [DOI] [PubMed] [Google Scholar]
- 44. Weston C. R., Wong A., Hall J. P., Goad M. E., Flavell R. A., and Davis R. J. (2003) JNK initiates a cytokine cascade that causes Pax2 expression and closure of the optic fissure. Genes Dev. 17, 1271–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cvekl A., and Ashery-Padan R. (2014) The cellular and molecular mechanisms of vertebrate lens development. Development 141, 4432–4447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Iyengar L., Patkunanathan B., Lynch O. T., McAvoy J. W., Rasko J. E., and Lovicu F. J. (2006) Aqueous humour- and growth factor-induced lens cell proliferation is dependent on MAPK/ERK1/2 and Akt/PI3-K signalling. Exp. Eye. Res. 83, 667–678 [DOI] [PubMed] [Google Scholar]
- 47. Le A. C., and Musil L. S. (2001) FGF signaling in chick lens development. Dev. Biol. 233, 394–411 [DOI] [PubMed] [Google Scholar]
- 48. Lovicu F. J., and McAvoy J. W. (2001) FGF-induced lens cell proliferation and differentiation is dependent on MAPK (ERK1/2) signalling. Development 128, 5075–5084 [DOI] [PubMed] [Google Scholar]
- 49. Ashery-Padan R., Marquardt T., Zhou X., and Gruss P. (2000) Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes Dev. 14, 2701–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sun J., Rockowitz S., Xie Q., Ashery-Padan R., Zheng D., and Cvekl A. (2015) Identification of in vivo DNA-binding mechanisms of Pax6 and reconstruction of Pax6-dependent gene regulatory networks during forebrain and lens development. Nucleic Acids Res. 43, 6827–6846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Maroulakou I. G., Papas T. S., and Green J. E. (1994) Differential expression of ets-1 and ets-2 proto-oncogenes during murine embryogenesis. Oncogene 9, 1551–1565 [PubMed] [Google Scholar]
- 52. Chotteau-Lelièvre A., Desbiens X., Pelczar H., Defossez P. A., and de Launoit Y. (1997) Differential expression patterns of the PEA3 group transcription factors through murine embryonic development. Oncogene 15, 937–952 [DOI] [PubMed] [Google Scholar]
- 53. Wolf L., Gao C. S., Gueta K., Xie Q., Chevallier T., Podduturi N. R., Sun J., Conte I., Zelenka P. S., Ashery-Padan R., Zavadil J., and Cvekl A. (2013) Identification and characterization of FGF2-dependent mRNA: microRNA networks during lens fiber cell differentiation. G3 3, 2239–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Maeda A., Moriguchi T., Hamada M., Kusakabe M., Fujioka Y., Nakano T., Yoh K., Lim K. C., Engel J. D., and Takahashi S. (2009) Transcription factor GATA-3 is essential for lens development. Dev. Dyn. 238, 2280–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hilberg F., Aguzzi A., Howells N., and Wagner E. F. (1993) c-jun is essential for normal mouse development and hepatogenesis. Nature 365, 179–181 [DOI] [PubMed] [Google Scholar]
- 56. Kasper L. H., Fukuyama T., Biesen M. A., Boussouar F., Tong C., de Pauw A., Murray P. J., van Deursen J. M., and Brindle P. K. (2006) Conditional knockout mice reveal distinct functions for the global transcriptional coactivators CBP and p300 in T-cell development. Mol. Cell. Biol. 26, 789–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Morikawa M., Koinuma D., Tsutsumi S., Vasilaki E., Kanki Y., Heldin C. H., Aburatani H., and Miyazono K. (2011) ChIP-seq reveals cell type-specific binding patterns of BMP-specific Smads and a novel binding motif. Nucleic Acids Res. 39, 8712–8727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shi Y., Wang Y. F., Jayaraman L., Yang H., Massagué J., and Pavletich N. P. (1998) Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-β signaling. Cell 94, 585–594 [DOI] [PubMed] [Google Scholar]
- 59. Liberati N. T., Datto M. B., Frederick J. P., Shen X., Wong C., Rougier-Chapman E. M., and Wang X. F. (1999) Smads bind directly to the Jun family of AP-1 transcription factors. Proc. Natl. Acad. Sci. U.S.A. 96, 4844–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Boswell B. A., and Musil L. S. (2015) Synergistic interaction between the fibroblast growth factor and bone morphogenetic protein signaling pathways in lens cells. Mol. Biol. Cell 26, 2561–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nakamura T., Donovan D. M., Hamada K., Sax C. M., Norman B., Flanagan J. R., Ozato K., Westphal H., and Piatigorsky J. (1990) Regulation of the mouse alpha A-crystallin gene: isolation of a cDNA encoding a protein that binds to a cis sequence motif shared with the major histocompatibility complex class I gene and other genes. Mol. Cell. Biol. 10, 3700–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ilagan J. G., Cvekl A., Kantorow M., Piatigorsky J., and Sax C. M. (1999) Regulation of αA-crystallin gene expression. Lens specificity achieved through the differential placement of similar transcriptional control elements in mouse and chicken. J. Biol. Chem. 274, 19973–19978 [DOI] [PubMed] [Google Scholar]
- 63. Mathelier A., Fornes O., Arenillas D. J., Chen C. Y., Denay G., Lee J., Shi W., Shyr C., Tan G., Worsley-Hunt R., Zhang A. W., Parcy F., Lenhard B., Sandelin A., and Wasserman W. W. (2015) JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Verrijzer C. P., Alkema M. J., van Weperen W. W., Van Leeuwen H. C., Strating M. J., and van der Vliet P. C. (1992) The DNA binding specificity of the bipartite POU domain and its subdomains. EMBO J. 11, 4993–5003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sun J., Rockowitz S., Chauss D., Wang P., Kantorow M., Zheng D., and Cvekl A. (2015) Chromatin features, RNA polymerase II and the comparative expression of lens genes encoding crystallins, transcription factors, and autophagy mediators. Mol. Vis. 21, 955–973 [PMC free article] [PubMed] [Google Scholar]