Abstract
Selective exo-enzymatic labeling (or SEEL) uses recombinant glycosyltransferases and nucleotide-sugar analogues to allow efficient labeling of cell surface glycans. SEEL can circumvent many of the possible issues associated with metabolic labeling, including low incorporation of sugar precursors, and allows for sugars to be added selectively to different types of glycans by virtue of the inherent specificity of the glycosyltransferases. Here we compare the labeling of sialoglycoproteins in undifferentiated and differentiated human erythroleukemia cells (HEL) using SEEL using the sialyltransferases ST6Gal1 and ST3Gal1, which label N- and O-glycans, respectively. Our results show that the profile of glycoproteins detected varies between undifferentiated HEL cells and those differentiated to megakaryocytes, with a shift to more N-linked sialoglycoproteins in the differentiated cells. The efficiency of SEEL for both sialyltransferases in HEL cells was greatly increased with prior neuraminidase treatment highlighting the necessity for the presence of available acceptors with this labeling method. Following metabolic labeling or SEEL, tagged glycoproteins were enriched by immunoprecipitation and identified using mass spectrometry. The proteomic findings demonstrated that the detection of many glycoproteins is markedly improved by SEEL labeling, and that unique glycoproteins can be identified using either ST6Gal1 or ST3Gal1. Furthermore, this analysis enabled the identification of increased surface expression of several sialylated cell adhesion molecules, including the known megakaryocytic markers integrinβ3 and CD44, upon differentiation of HEL cells to adherent megakaryocytes.
Keywords: β-galactoside α-2,6-sialyltransferase 1 (ST6GAL1); cell differentiation; click chemistry; glycosyltransferase; proteomics; sialic acid
Introduction
The ability to label glycans using chemical glycobiology methods has ushered in new opportunities to investigate the functions of these molecules in living systems (1–3). These studies are beginning to yield insight into how glycan profiles changes during development and how the content, localization, and trafficking of glycans is altered in the context of many human diseases (4–9). Most methodologies utilize metabolic labeling as a way to incorporate functionalized sugars into glycans during biosynthesis. The extent of labeling in some cell types may be limited by the presence of endogenous non-labeled sugar precursors that can dilute out the azide-sugars available for incorporation. Variable distribution of azide-sugars into different glycan classes (e.g. glycoproteins versus glycolipids) following metabolic labeling can also skew or limit the types of glycans that can be analyzed. As an alternative approach, labeling methods have been developed that instead rely on the use of recombinant or purified glycosyltransferases to install functionalized sugars directly onto existing glycans (10–12). This method, recently termed SEEL (selective exo-enzymatic labeling),4 leverages the inherent selectivity of the recombinant glycosyltransferases to label certain types (e.g. N-linked or O-linked) of glycans. Prior work using the sialyltransferase ST6Gal1 demonstrated that only N-glycans were modified, confirming the established selectivity of this enzyme (10).
Because of the more selective nature of SEEL-based labeling, we propose that this method would allow glycoproteins bearing specific types of glycans to be enriched and identified by proteomic analysis. Such an approach would provide an advantageous and complementary way to identify glycoproteins whose glycosylation is sensitive in the context of human diseases or upon differentiation to various cell lineages. By virtue of the fact that neither the glycosyltransferases nor the modified nucleotide-sugars are capable of crossing the plasma membrane, only cell surface glycoproteins will be labeled by SEEL thus reducing the background typically associated with traditional methods of surface labeling. In the present study, we used SEEL with two different sialyltransferases in undifferentiated and differentiated human erythroleukemia (HEL) cells. We demonstrate that labeling with different sialyltransferases produces distinct profiles of sialoglycoproteins in HEL cells, and that SEEL facilitates the detection of glycoproteins that are not readily visualized by metabolic labeling. Moreover, we demonstrated that neuraminidase-coupled SEEL with the two different sialyltransferases improves the overall coverage and detection of sialoglycoproteins by mass spectrometry. An increase in cell surface expression of several adhesion proteins known to be markers of megakaryocytic differentiation was faithfully identified within the differentiated HEL cells using this approach. The potential of SEEL and neuraminidase-coupled SEEL to analyze the sialylation status and cell surface expression of sialoglycoproteins, respectively, is discussed.
Materials and Methods
Reagents
Recombinant rat α-(2,6)-sialyltransferase (ST6Gal1)or α-(2,3)-sialyltransferase (ST3Gal1) was prepared as previously reported (18). CMP-Neu5Ac9N3, S-DIBO-Biotin, Ac4ManNAc, and Ac4ManNAz were synthesized as previously described (16, 30). Vibrio cholerae neuraminidase (type II) was purchased from Sigma Aldrich (N6514). Alkaline phosphatase (FastAP) was purchased from Thermo Scientific (EF0651). PNGaseF and glycoprotein denaturatuion buffer was from Biolabs (P0704S). Mouse monoclonal anti-Biotin antibodies (200-032-211, HRP-conjugated from or 200-002-211, non-conjugated form) were purchased from Jackson ImmunoResearch Laboratories. HRP-conjugated β-actin antibody was from Abcam. Protease inhibitor mixture tablet (88666) and mass spectrometry compatible silver staining kit (24600) were from Thermo Scientific. Protein G beads were from Sigma Aldrich (Protein G-Sepharose, Fast Flow, P3296).
Cell Lines and Culture
Human erythroleukemia (HEL) cells were cultured in RPMI1640 media with l-glutamine (2.0 mm). Chinese hamster ovary (CHO) cells (Clone K1, ATCC) and mutant CHO cells (Lec2) were cultured in Minimum Essential Medium Alpha 1X (Cellgro) with Earle's salts, without ribonucleosides, deoxyribonucleosides, and l-glutamine. All medium was supplemented with 10% fetal bovine serum (FBS, BenchMark) and penicillin (100 IU/ml)/streptomycin (100 μg/ml, MediaTech). Cells were cultured in a 5% CO2 atmosphere, 37 °C humid incubator.
Differentiation of HEL Cells by Phorbol 12-Myristate 13-Acetate (PMA)
Typically, HEL cells (0.8∼0.9 × 106 cells/well) in a 6-well dish were treated with 16 nm PMA for 48 h. After removing the media together with floating cells, fresh medium containing 8 nm PMA was added, and cells were further cultured for 24 h. For metabolic labeling, the differentiated HEL cells were cultured with Ac4ManNAz (30 μm) in the presence of 8 nm PMA for additional 24 h. At the same time, cells for SEEL labeling were cultured in the presence of 8 nm PMA for additional 24 h without Ac4ManNAz.
Neuraminidase-coupled Selective Exo-enzymatic Labeling (SEEL) of HEL Cells
HEL cells (normal or differentiated) were collected in 1.5 ml Eppendorf tubes. Note that the differentiated HEL cells are adherent but they are easily lifted off by pipetting. After washing the cells with DPBS, cells were briefly centrifuged and then the resulting pellet was resuspended and incubated in serum-free RPMI 1640 media with or without Vibrio cholerae neuraminidase (50 mU/ml) for 2 h at 37 °C with rocking. Next, the resulting cells were washed with DPBS (1 ml × three times), and then the resulting pellet was re-suspended in SEEL reaction solution (typically 300 μl) containing 100 μg of sialyltransferase, CMP-Neu5Ac9N3, (100 μm unless otherwise noted), 2 μl of BSA (2 mg/ml), 2 μl of alkaline phosphatase, 46 μl of 3 m sucrose in serum-free RPMI 1640 medium, and then cells were incubated for 2 h at 37 °C with rocking. Based on earlier work, SEEL reaction can be performed across a range of concentration of CMP-Neu5Ac9N3 (50∼500 μm) without noticeable decreases in labeling. However, 100 μm CMP-Neu5Ac9N3 was found to be sufficient for maximal labeling with a defined concentration of sialyltransferase (100 μg in 300 μl reaction). Percent cell viability following the various steps in the SEEL procedure was determined as triplicate. Briefly, portions of HEL cells from SEEL reactions with ST6Gal1 in three Eppendorf tubes were collected over the course of SEEL reaction, 1) before sialidase treatment, 2) after sialidase treatment, 3) after ST6Gal1 and Neu5Ac9N3 reaction, 4) after S-DIBO-biotin treatment, and their viability was measured by Cellometer® Vision (Nexcelom Bioscience) using Trypan Blue exclusion method (Thermo Fisher Scientific).
Neuraminidase-coupled Selective Exo-enzymatic Labeling of CHO and Lec2 Cells
Neuraminidase treatment and SEEL of CHO or Lec2 cells were conducted in 12 well dishes. After the cells were confluent, cells were washed with DPBS (1 ml × two times) and then serum-free α-MEM (0.5 ml) was added to each well with or without V. cholerae neuraminidase (50 mU/ml). After incubating the dishes at 37 °C for 2 h, cells were washed with DPBS (1 ml × three times for each well), and then SEEL reaction solution (300 μl each) containing 100 μg of sialyltransferase, CMP-Neu5Ac9N3, (142 μm), BSA (2 mg/ml), 2 μl of alkaline phosphatase, 46 μl of 3 m sucrose in serum-free α-MEM, and then cells were incubated for 2 h at 37 °C without rocking.
S-DIBO-biotin Labeling
Cells metabolically labeled by Ac4ManNAz or SEEL were biotinylated with S-DIBO-biotin. Typically, 30 μm S-DIBO in DPBS containing 2% FBS was treated to cells either in Eppendorf tube (HEL cells) or 12-well dish (CHO or Lec2 cells) for 1 h at room temperature.
Western Blotting, Immunoprecipitation, and Silver Staining
Biotin-labeled cells were washed with DPBS and then lysed in RIPA buffer containing 50 mm Tris-HCl buffer, pH 8.0, 150 mm NaCl, 1.0% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycholate supplemented with protease inhibitor mixture (Thermo Scientific) on ice. After 30 min, lysates were spun down at 20,000 × g for 10 min, and then the supernatant was saved. Protein concentration was determined using the BCA Protein Assay Kit (Thermo Science Pierce) by following the manufacturer's protocol. Lysates were analyzed by SDS-PAGE and immunoblot using anti-biotin antibody conjugated with HRP.
In the case of treating lysate with PNGase F, cell lysate was denatured by boiling for 10 min with the supplementation of glycoproteins denaturation buffer. Then the denatured lysate was incubated with 10 units of PNGase F at 37 °C for 4 h. Next, the PNGase F-treated lysate was analyzed by SDS-PAGE and immunoblot using anti-biotin antibody conjugated with HRP.
For immunoprecipitation, anti-biotin-coated protein G beads (Sigma) were prepared by incubating anti-biotin antibody with protein G beads in the RIPA buffer followed by washing with the same buffer. 3∼4 mg of the lysates were precleared by incubating with protein G beads for 2 h at 4 °C and then spun down. The precleared lysate as supernatant was collected and then incubated with the antibody-coated protein G beads overnight at 4 °C. Next, the beads were washed five times with the RIPA buffer and then eluted with 2× sample loading buffer containing 10 mm DTT by boiling for 7 min. Eluted sample was separated in SDS-PAGE, and the gel was silver stained. Silver staining of the resulting gel was done with a mass spectrometry compatible silver staining kit from Thermo Scientific following the manufacturer's protocol.
In-gel Digestion of Silver-stained Proteins
Each lane of the silver-stained SDS-PAGE gel was cut into 4 parts above 50 kDa and subsequently processed for in-gel digestion. Briefly, destained gel bands were denatured by incubating with 10 mm dithiothreitol at 56 °C for 1 h and alkylated by 55 mm iodoacetamide for 45 min in dark prior to digestion with trypsin overnight. The resulting peptides were extracted, dried, and reconstituted in 0.1% formic acid.
Identification of Biotinylated Glycoproteins by MS/MS
The peptides were separated on a 75 μm (I.D.) × 15 cm C18 capillary column (packed inhouse, YMC GEL ODS-AQ120ÅS-5, Waters) and eluted into the nano-electrospray ion source of an Orbitrap Fusion™ Tribrid™ mass spectrometer (Thermo Fisher Scientific) with a 180-min linear gradient consisting of 0.5–100% solvent B over 150 min at a flow rate of 200 nL/min. The spray voltage was set to 2.2 kV, and the temperature of the heated capillary was set to 280 °C. Full MS scans were acquired from m/z 300 to 2000 at 120k resolution, and MS2 scans following collision-induced fragmentation were collected in the ion trap for the most intense ions in the Top-Speed mode within a 3-s cycle using Fusion instrument software (v1.1, Thermo Fisher Scientific). The raw spectra were searched against the human protein database (UniProt, Oct. 2014) using SEQUEST (Proteome Discoverer 1.4, Thermo Fisher Scientific) with full MS peptide tolerance of 20 ppm and MS2 peptide fragment tolerance of 0.5 Da, and filtered using ProteoIQ (v2.7, Premier Biosoft) at the protein level to generate a 1% false discovery rate for protein assignments. UniProt was used to define cellular localization. Quantification was performed using spectral counts generated in ProteoIQ (v2.7, Premier Biosoft).
Results
Human erythroleukemia (HEL) cells grown in suspension can be differentiated to adherent megakaryocytes by phorbol 12-myristate 13-acetate (PMA) treatment, providing an opportunity to analyze how the profile of sialoglycoproteins changes upon differentiation with various labeling methods (13–15). Metabolic labeling of HEL cells with Ac4ManNAz showed predominantly O-sialoglycoproteins being labeled as demonstrated by the insensitivity of labeled glycoproteins to PNGase F treatment (Fig. 1). In contrast, PMA-derived differentiated HEL cells showed significantly increased N-sialoglycoprotein labeling as evidenced by an increase in specific bands (e.g. 80 kDa) and the subsequent disappearance of these bands following enzymatic removal of the N-glycans. A corresponding decrease in the major labeled O-sialoglycoprotein was also observed upon differentiation. Incubation of the cells with ManNAc resulted in no glycoprotein labeling as expected.
FIGURE 1.
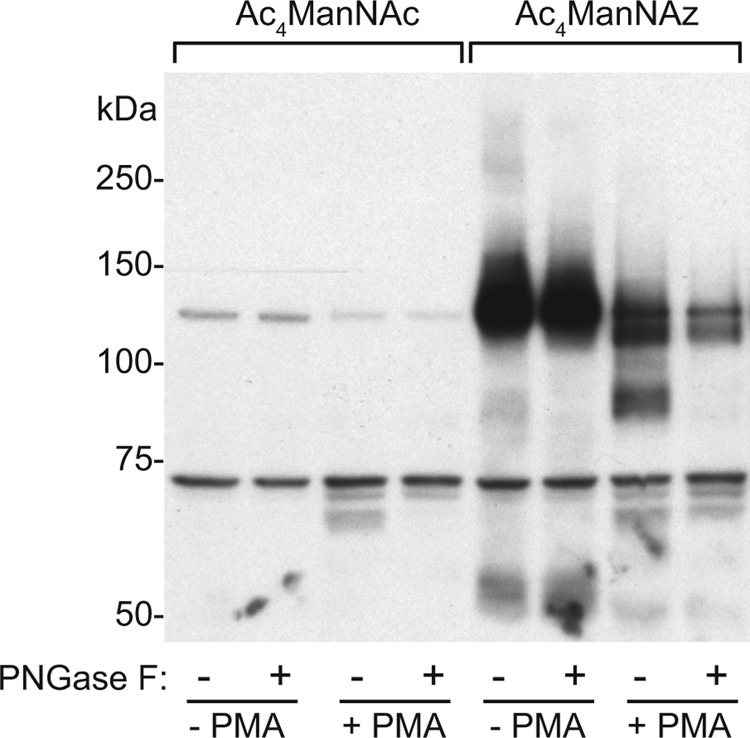
Differentiation of HEL cells showed increased metabolic labeling of N-sialoglycan. Non-treated or PMA-treated HEL cells were metabolically labeled with 30 μm Ac4ManNAc or Ac4ManNAz. The resulting cells were incubated with 100 μm S-DIBO-Biotin and the denatured cell lysates further treated with or without PNGase F. Samples were subjected to SDS-PAGE and WB analysis using an HRP-conjugated anti-biotin antibody. A representative image from four independent experiments is shown.
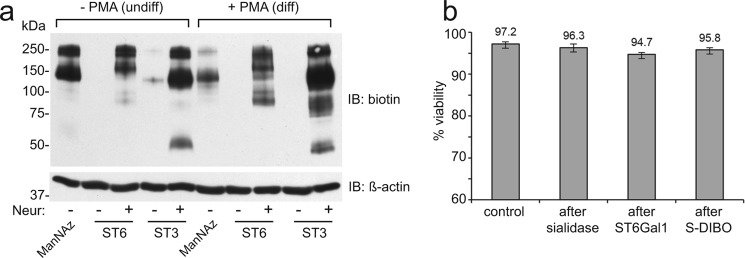
Next, HEL cells were either incubated with Ac4ManNAz for 24 h, or SEEL labeled with rat ST6Gal1 or human ST3Gal1 in the presence of CMP-Neu5Ac9N3 followed by treatment with S-DIBO for biotinylation (Fig. 2). S-DIBO was chosen since it does not cross the plasma membrane and therefore avoids conjugation with intracellular glycoproteins (16). In contrast to metabolically labeled cells, Western blot analysis with anti-biotin antibody showed only weak labeling following SEEL with either sialyltransferase. Increasing enzyme or nucleotide-sugar concentrations did not increase labeling, suggesting that HEL cells are heavily sialylated and lack available acceptors. To enhance SEEL, HEL cells were incubated with a bacterial neuraminidase (from Vibrio cholera) prior to SEEL reaction with ST6Gal1 or ST3Gal1. Immunoblotting of the resulting cell lysate showed dramatically enhanced SEEL sialylation of cell surface glycoproteins (Fig. 2a). These data demonstrate the importance of available acceptors as a prerequisite for SEEL labeling. All treatments resulted in only minimal effects on cell viability under the optimized conditions used (Fig. 2b). We also show that untreated WT CHO cells are poorly labeled by SEEL but that robust labeling can be achieved with both ST6Gal1 and ST3Gal1 following neuraminidase pre-treatment (Fig. 3). Lec2 cells, which bear a defect in the CMP-sialic acid transporter, showed strong SEEL labeling regardless of pre-treatment of neuraminidase due to the loss of any sialic acid incorporation in these cells.
FIGURE 2.
Prior neuraminidase treatment is required for efficient SEEL in HEL cells. a, HEL cells or differentiated HEL cells were labeled by Ac4ManNAz or SEEL. For SEEL, HEL cells were pre-treated with or without V. cholerae neuraminidase (50 mU/ml) and then the resulting cells were incubated with ST6Gal1 (ST6) or ST3Gal1 (ST3) in the presence of CMP-Neu5Ac9N3 as described in “Materials and Methods.” Cell lysates were subjected to SDS-PAGE and WB analysis using an HRP-conjugated anti-biotin antibody. Subsequent blotting with an HRP-conjugated anti-β-actin antibody was performed as a loading control. Images shown are representative of three independent labeling experiments. b, cell viability effects following various treatments in undifferentiated HEL cells (n = 3). Error bars represent S.D.
FIGURE 3.
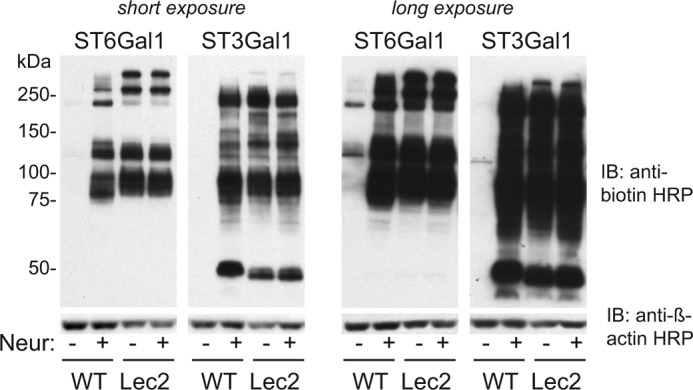
Neu-SEEL labeling of WT CHO and Lec2 cells. Western blot analysis of WT and Lec2 CHO cells following ST6Gal1 and ST3Gal1 SEEL (with or without prior neuraminidase treatment) is shown. The image depicted is representative of two independent experiments.
The specificity of the respective sialyltransferases was confirmed by treating SEEL-labeled lysates with PNGaseF prior to analysis. Glycoproteins labeled by ST6Gal1 completely disappeared upon treatment of PNGase F while glycoproteins labeled by ST3Gal1 were largely insensitive (Fig. 4). Among the glycoproteins labeled by ST3Gal1, some shifted to lower molecular weight range, presumably because they also bear N-glycans, but the intensity of labeling did not decrease. This finding is in good agreement with the known specificities of these two glycosyltransferases (17, 18), and establishes that these two sialyltransferases maintain their selectivity for different glycan classes when surface labeling of fully folded glycoproteins is performed.
FIGURE 4.
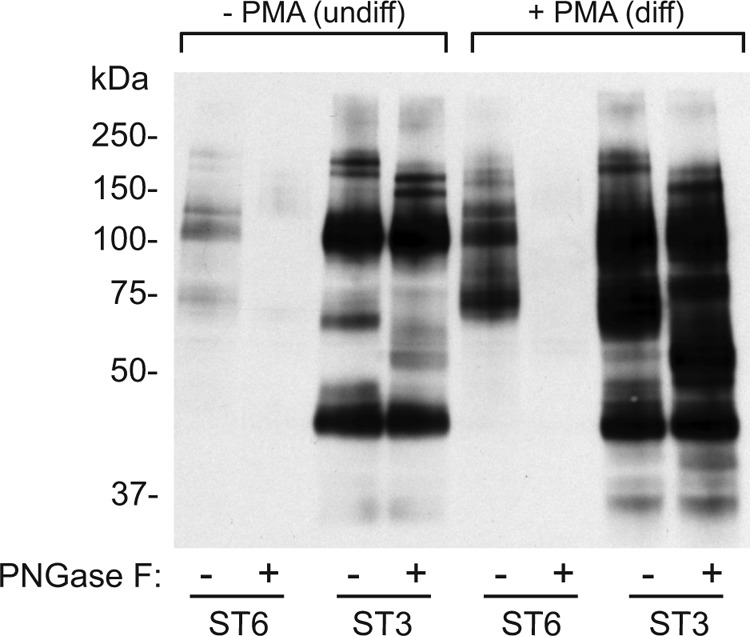
Selective glycan labeling using SEEL with ST6Gal1 and ST3Gal1. HEL cells or differentiated HEL cells were pre-treated with or without Vibrio cholerae neuraminidase (50 mU/ml) followed by SEEL with ST6Gal1 (ST6) or ST3Gal1 (ST3) and S-DIBO incubation. Cell lysates were incubated with or without PNGase F and analyzed by SDS-PAGE and WB analysis using an HRP-conjugated anti-biotin antibody. The image depicted here is representative of two independent experiments.
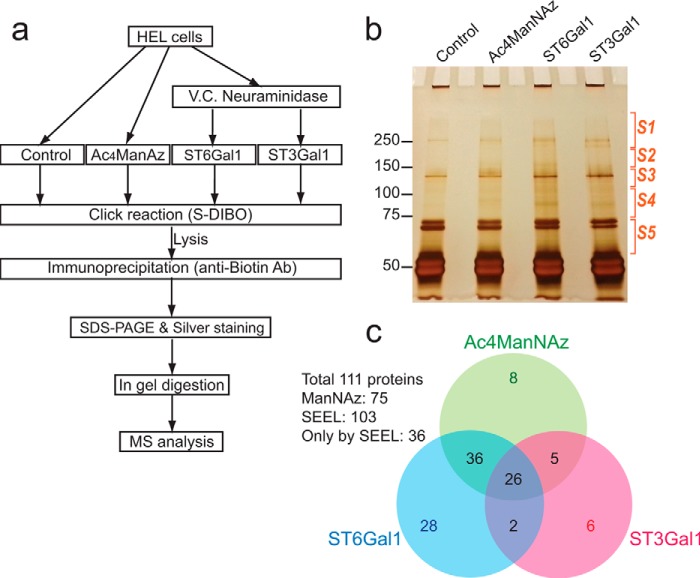
We next optimized an enrichment protocol for biotin-tagged glycoproteins using immunoprecipitation with an anti-biotin antibody and linked this enrichment to silver staining and proteomic analysis (Fig. 5a). For optimal pull-down of biotinylated proteins, we chose to use immunoprecipitation with anti-biotin antibody, which exhibits a minimal level of nonspecific interactions compared with streptavidin- or avidin-coated beads. Following labeling of sialoglycoproteins metabolically with ManNAz, or by neuraminidase-coupled SEEL with either ST6Gal1 or ST3Gal1, cell lysates were immunoprecipitated using the anti-biotin antibody and bound sialoglycoproteins were separated by SDS-PAGE and visualized by silver staining (Fig. 5b). Parallel analysis of unlabeled HEL cells was performed as a control for nonspecific binding to the agarose beads. In-gel digestion was performed and the digested peptides corresponding to each section were analyzed by LC-MS-MS. The results were quantified and a comparison of labeled glycoproteins presented (Fig. 5c, supplemental File S1). Overall, labeling with SEEL was more efficient than metabolic labeling for the identification of sialoglycoproteins. Out of 111 identified glycoproteins, 103 proteins were identified via SEEL while 75 proteins were identified via metabolic labeling. To eliminate nonspecific hits in MS analysis, common proteins that were pulled down in the control, proteins with spectral counts under 10 and nucleocytoplasmic proteins were not included. We observed selectivity when comparing the two SEEL enzymes. Out of total 103 protein assignments from the SEEL samples, 64 proteins were identified only from the ST6Gal1 SEEL sample, while 11 proteins were only assigned from the ST3Gal1 SEEL sample. The other 66 proteins were found in both SEEL samples, suggesting they have both N- and O-sialylation.
FIGURE 5.
Identification of sialogycome labeled by SEEL versus metabolic labeling. a, workflow diagram for analysis of sialoglycoproteins in undifferentiated HEL cells. b, silver stained gel of the immunoprecipitates. Each lane was cut into 5 sections, and each section was subjected to in gel digestion and MS analysis. c, Venn diagram of 111 proteins that were identified using the various methods. Proteins that were commonly found in control sample, those with spectral count under 10, and any cytosolic or nuclear proteins were omitted.
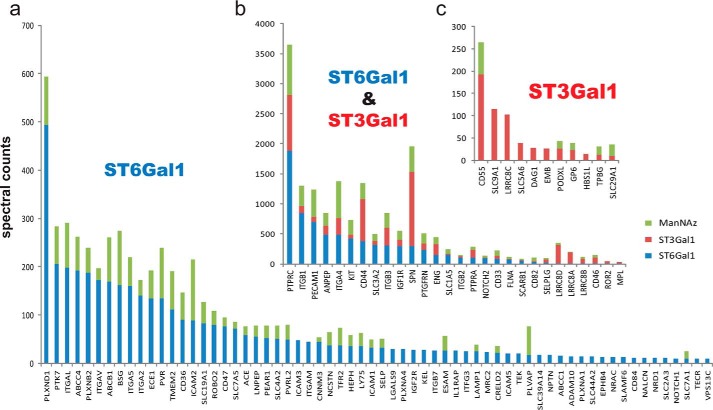
To compare the relative abundance of each protein hit, spectral counts were compared for the 103 proteins (Fig. 6). In nearly all cases, spectral counts were significantly higher in the SEEL sample compared with those from metabolic labeling. Major sialoglycoprotein hits identified were PTPRC (CD45), integrin β1, PECAM1, aminopeptidase N, plexin-D1, sialophorin (SPN, CD43), integrin β3, CD44, etc. Among them, proteins such as ITGB1, PECAM1 showed relatively higher spectral counts from the ST6Gal1 SEEL than spectral counts from ST3Gal1 SEEL, suggesting they are more highly N-sialylated than they are O-sialylated. SPN showed the highest spectral counts in the ST3Gal1 SEEL sample but relatively less spectral counts from ST6Gal1 SEEL sample, suggesting this protein has more O-sialoglycans than N-sialoglycans. This might be expected in light of the abundance of sialylated O-glycans on this protein (19, 20). Overall, the ability of SEEL to identify a distinct subset of glycoproteins is consistent with what was observed in the Western blot experiments above where specific glycoproteins were detected much more readily following SEEL.
FIGURE 6.
Spectral count comparison of the identified 103 proteins. a, 64 glycoproteins that were labeled by ST6Gal1 SEEL but not by ST3Gal1 SEEL. b, 28 glycoproteins that were labeled both by ST6Gal1 and ST3Gal1 SEEL. c, 11 glycoproteins that were labeled by ST3Gal1 but not by ST6Gal1. The spectral counts for specific glycoproteins tagged by SEEL generally exceeded those obtained by metabolic labeling. In addition, the identity of those glycoproteins only detected by SEEL is shown. Two independent biological experiments for SEEL and metabolic labeling in the undifferentiated cells were performed, with the same subset of major glycoproteins identified in both runs. A representative comparison is shown.
Finally, we compared undifferentiated and differentiated HEL cells with neuraminidase-coupled SEEL followed by proteomic analysis to see if SEEL can be used to identify changes in cell surface sialoglycoprotein expression in these two different cell types (supplemental File S1). Interestingly, we found that the most abundant sialoglycoproteins such as PTPRC and SPN did not show any significant change upon differentiation when SEEL was performed with ST6Gal1 or ST3Gal1. In contrast, clear increases in several other proteins were specifically detected in the differentiated cells using both ST6Gal1 and ST3Gal1 (Fig. 7). Many of these glycoproteins such as integrinβ3 and CD44 are known cell adhesion proteins and megakaryocytic markers that facilitate attachment of the adherent megakaryocytes (21–23). Notably, the spectral counts of these glycoproteins far exceeded the level observed in the control cells whereas other proteins (HSP5a) could not be distinguished between control and SEEL-tagged lysates. This demonstrates the superior enrichment of glycoproteins that can be achieved using SEEL. While the integrins were primarily enriched by SEEL with ST6Gal1, other glycoproteins such as CD44 and SPN were primarily enriched using ST3Gal1, again highlighting the selectivity of the SEEL approach and its ability to enrich unique subsets of cell surface glycoproteins.
FIGURE 7.
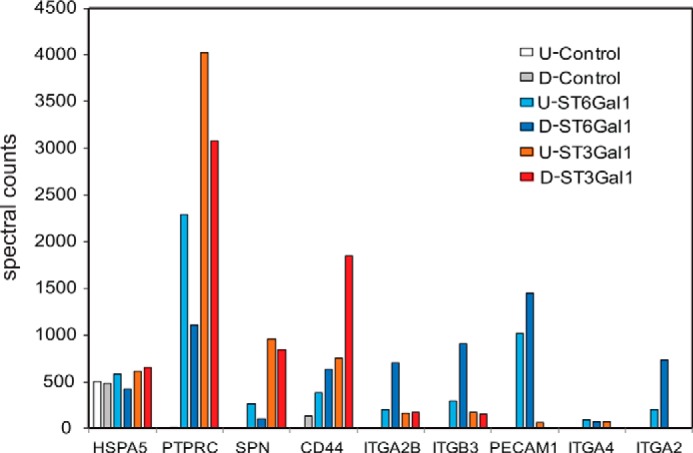
Changes of N- and O-sialoglycoprotein level of representative sialoglycoproteins upon differentiation of HEL cells. Spectral count analysis for selected glycoproteins enriched and detected by SEEL with either ST6Gal1 or ST3Gal1. Control: HEL cells that were only treated by S-DIBO; ST6Gal1: HEL cells only labeled by SEEL with ST6Gal1; ST3Gal1: HEL only labeled by SEEL with ST3Gal1. U refers to undifferentiated HEL cells, and D refers to HEL cells differentiated to megakaryocytes following PMA stimulation. The results from a representative analysis are shown. All SEEL labeling were done with prior neuraminidase treatment followed by S-DIBO-biotin. The data shown are derived from a single proteomic analyses of undifferentiated and differentiated HEL cells labeled at the same time under identical conditions.
Discussion
The present work extends the utility of the previously reported SEEL methodology by demonstrating the selectivity of a different SEEL enzyme (ST3Gal1) and showing that SEEL-tagged glycoproteins can be effectively enriched and identified by proteomic analysis. This work further provides a comparison of SEEL and metabolic labeling in a single cell system, revealing many consistent features between the two methods. For example, an increase in the presence of certain N-sialoglycoproteins could be detected following differentiation using both labeling methods (Figs. 1 and 4). However, several noteworthy differences between these two approaches were also identified and are considered below.
Neuraminidase-coupled SEEL with ST6Gal1 in undifferentiated HEL cells revealed a high level of N-glycan labeling, while metabolic labeling incorporated almost exclusively into O-glycans (Fig. 1, lane 5 versus Fig. 2, lane 3). Likewise, SEEL with ST3Gal1 also showed greater labeling of O-glycan while metabolic labeling showed significantly decreased O-glycan upon differentiation (Fig. 1, lane 7 versus Fig. 2, lane 10). We interpret the relative decrease in O-glycan tagging in the metabolically labeled differentiated HEL cells to reflect competition for common sialic acid precursors when the expression of N-sialoglycoproteins is increased. Metabolic labeling is influenced by the rate of glycoprotein biosynthesis and the rate of incorporation into these glycoproteins, which is often limited by competition with endogenous sugar precursor pools. Also, percentage of substitution of sialic acid into azide-tagged sialic acid varies among different cell lines (24, 25). Since SEEL does not rely on the biosynthetic machinery of the cell nor does it require new synthesis of glycoproteins, competition for substrates and differences in metabolic flux do not contribute to the incorporation of the modified NeuAc derivative. Thus, this approach provides an opportunity to more accurately survey the profile of cell surface glycoproteins. This is supported by the fact that SEEL with ST3Gal1 showed an increase (not the decrease noted with ManNAz labeling) in the labeling of O-sialoglycoproteins upon differentiation (Figs. 2 and 4). Furthermore, SEEL is capable of labeling glycoproteins with slow turnover. This advantage may be most relevant when analyzing cell adhesion glycoproteins or extracellular matrix proteins that are subject to slow internalization and turnover since they maintain stable interactions with other proteins outside the cell.
A few ER proteins, including oligosaccharyltransferase subunits and charperones, were also identified, albeit at low levels, following SEEL in the undifferentiated HEL cells. Based on the low level of spectral counts for these proteins and their appearance only in the non-SEEL labeled samples in the differentiated cells, these hits are likely nonspecific (supplemental File S1). Nonetheless, it is possible that a fraction of ER proteins escapes in these cells and traffics to the plasma membrane. Their presentation at the cell surface may also arise due to contacts made between the ER membrane and the plasma membrane (26, 27). Alternatively, ER-resident proteins may become exposed to the cell surface under specific circumstances including the onset of apoptosis (28, 29).
While the affinity-based analysis of sialoglycoproteins is powerful method, lectins or antibodies have not been in broad use with proteomic profiling of cellular sialoglycan or in search of disease biomarkers, because of their limited affinity, selectivity and availability for pan-specific sialoglycans. In this respect, SEEL provides an efficient strategy to assess the level of sialylation of glycoproteins for many reasons. First, SEEL can be applied to normally cultured cells without perturbation of the cellular metabolism. Second, since the SEEL signal reflects the level of available vacant sites for additional sialylation, the difference of SEEL signals between control cells and neuraminidase treated cells is proportional to the pre-existing sialic acid on cell surface, as was demonstrated using SEEL with CHO and Lec2 cells (Fig. 3). Third, the sialyltransferases such as ST6Gal1 or ST3Gal1 can add Neu5Ac9N3 to N-glycan or O-glycan selectively, yielding an opportunity to limit the type of glycoproteins analyzed.
SEEL with concomitant use of two sialyltransferases provided the relative level of N-sialoglycoproteins with 2,6 linkage and that of O-sialoglycoproteins with 2,3 linkage. It is important to note that ST6Gal1 covers only 2,6-linked N-sialoglycoproteins. For covering the 2,3-linked N-sialoglycan, other sialyltransferase such as ST3Gal4 or ST3Gal6 could be employed. As another advantage, SEEL allows the degree of sialylation of cells to be analyzed at the level of individual protein. Although we only showed the relative level of sialylation of proteins by comparing spectral count of each protein, if desired, more precise comparison of the sialylation level is possible via the added step of stable isotope labeling of peptides. It should be noted that the signal in the anti-biotin Western blot is proportional to the total number of sialic acid residues, however, the spectral counts in the MS analysis is not. Rather the spectral count is proportional to the number of protein molecules that are pulled-down. Overall, 37% more spectral counts for sialoglycoproteins were identified by SEEL compared with metabolic labeling in MS analysis.
In light of its ability to increase overall labeling above what can typically be achieved with metabolic labeling, and its ability to label unique glycoproteins, SEEL should prove advantageous for the analysis of glycoprotein levels at the cell surface as well as assessing the sialylation status of these glycoproteins. We envision that SEEL using sialyltransferases without prior neuraminidase treatment will represent a convenient method to survey the sialylation status of cells via its ability to modify available acceptors. In this case, an increase in SEEL labeling would reflect a greater availability of acceptors, an indirect measure of sialic acid occupancy. Neuraminidase-coupled SEEL, on the other hand, is ideal for the detection of cell surface sialoglycoproteins since the extent of tagging will depend on the amount of the glycoprotein at the cell surface instead of its original sialylation status.
Author Contributions
S. Y., P. Z., L. W., and R. S. designed the study. S. Y. and P. Z. carried out the biochemical and proteomic studies. T. S., G. J. B., Z. G., and K. M. provided the substrates and enzymes for SEEL labeling. S. Y. and R. S. wrote the paper. All authors analyzed the results and approved the final version of the manuscript.
This work was supported by National Institutes of Health P01 Grant GM-107012 from NIGMS. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental File S1.
- SEEL
- selective exo-enzymatic labeling
- HEL
- human erythroleukemia cell
- ST6GAL1
- β-galactoside α-2,6-sialyltransferase 1
- PMA
- phorbol 12-myristate 13-acetate
- HRP
- horseradish peroxidase.
References
- 1. Cook B. N., and Bertozzi C. R. (2002) Chemical approaches to the investigation of cellular systems. Bioorg. Med. Chem. 10, 829–840 [DOI] [PubMed] [Google Scholar]
- 2. Prescher J. A., and Bertozzi C. R. (2006) Chemical technologies for probing glycans. Cell 126, 851–854 [DOI] [PubMed] [Google Scholar]
- 3. Boons G. J. (2010) Bioorthogonal chemical reporter methodology for visualization, isolation and analysis of glycoconjugates. Carbohydr. Chem. 36, 152–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mbua N. E., Flanagan-Steet H., Johnson S., Wolfert M. A., Boons G. J., and Steet R. (2013) Abnormal accumulation and recycling of glycoproteins visualized in Niemann-Pick type C cells using the chemical reporter strategy. Proc. Natl. Acad. Sci. U.S.A. 110, 10207–10212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baskin J. M., Dehnert K. W., Laughlin S. T., Amacher S. L., and Bertozzi C. R. (2010) Visualizing enveloping layer glycans during zebrafish early embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 10360–10365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dehnert K. W., Baskin J. M., Laughlin S. T., Beahm B. J., Naidu N. N., Amacher S. L., and Bertozzi C. R. (2012) Imaging the sialome during zebrafish development with copper-free click chemistry. Chembiochem 13, 353–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jiang H., Feng L., Soriano del Amo D., Seidel Iii R. D., Marlow F., and Wu P. (2011) Imaging glycans in zebrafish embryos by metabolic labeling and bioorthogonal click chemistry. J. Vis. Exp. 10.3791/2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tian Y., Almaraz R. T., Choi C. H., Li Q. K., Saeui C., Li D., Shah P., Bhattacharya R., Yarema K. J., and Zhang H. (2015) Identification of sialylated glycoproteins from metabolically oligosaccharide engineered pancreatic cells. Clin. Proteomics 12, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Almaraz R. T., Tian Y., Bhattarcharya R., Tan E., Chen S. H., Dallas M. R., Chen L., Zhang Z., Zhang H., Konstantopoulos K., and Yarema K. J. (2012) Metabolic flux increases glycoprotein sialylation: implications for cell adhesion and cancer metastasis. Mol. Cell Proteomics 11, M112 017558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mbua N. E., Li X., Flanagan-Steet H. R., Meng L., Aoki K., Moremen K. W., Wolfert M. A., Steet R., and Boons G. J. (2013) Selective exo-enzymatic labeling of N-glycans on the surface of living cells by recombinant ST6Gal I. Angewandte Chemie 52, 13012–13015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Whiteheart S. W., and Hart G. W. (1987) Sialyltransferases as specific cell surface probes of terminal and penultimate saccharide structures on living cells. Anal. Biochem. 163, 123–135 [DOI] [PubMed] [Google Scholar]
- 12. Zeng Y., Ramya T. N., Dirksen A., Dawson P. E., and Paulson J. C. (2009) High-efficiency labeling of sialylated glycoproteins on living cells. Nature Methods 6, 207–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barnes J., Warejcka D., Simpliciano J., Twining S., and Steet R. (2012) Latency-associated peptide of transforming growth factor-β1 is not subject to physiological mannose phosphorylation. J. Biol. Chem. 287, 7526–7534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zauli G., Bassini A., Catani L., Gibellini D., Celeghini C., Borgatti P., Caramelli E., Guidotti L., and Capitani S. (1996) PMA-induced megakaryocytic differentiation of HEL cells is accompanied by striking modifications of protein kinase C catalytic activity and isoform composition at the nuclear level. Br. J. Haematol. 92, 530–536 [DOI] [PubMed] [Google Scholar]
- 15. Long M. W., Heffner C. H., Williams J. L., Peters C., and Prochownik E. V. (1990) Regulation of megakaryocyte phenotype in human erythroleukemia cells. J. Clin. Invest. 85, 1072–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Friscourt F., Ledin P. A., Mbua N. E., Flanagan-Steet H. R., Wolfert M. A., Steet R., and Boons G. J. (2012) Polar dibenzocyclooctynes for selective labeling of extracellular glycoconjugates of living cells. J. Am. Chem. Soc. 134, 5381–5389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kiwamoto T., Brummet M. E., Wu F., Motari M. G., Smith D. F., Schnaar R. L., Zhu Z., and Bochner B. S. (2014) Mice deficient in the St3gal3 gene product alpha2,3 sialyltransferase (ST3Gal-III) exhibit enhanced allergic eosinophilic airway inflammation. J. Allergy Clin. Immunol. 133, 240–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Meng L., Forouhar F., Thieker D., Gao Z., Ramiah A., Moniz H., Xiang Y., Seetharaman J., Milaninia S., Su M., Bridger R., Veillon L., Azadi P., Kornhaber G., Wells L., Montelione G. T., Woods R. J., Tong L., and Moremen K. W. (2013) Enzymatic basis for N-glycan sialylation: structure of rat α2,6-sialyltransferase (ST6GAL1) reveals conserved and unique features for glycan sialylation. J. Biol. Chem. 288, 34680–34698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fukuda M., and Carlsson S. R. (1986) Leukosialin, a major sialoglycoprotein on human leukocytes as differentiation antigens. Med. Biol. 64, 335–343 [PubMed] [Google Scholar]
- 20. Maemura K., and Fukuda M. (1992) Poly-N-acetyllactosaminyl O-glycans attached to leukosialin. The presence of sialyl Le(x) structures in O-glycans. J. Biol. Chem. 267, 24379–24386 [PubMed] [Google Scholar]
- 21. Hagiwara T., Nagasawa T., Nagahisa H., Takizawa M., Osada M., and Abe T. (1996) Expression of adhesion molecules on cytoplasmic processes of human megakaryocytes. Exp. Hematol. 24, 690–695 [PubMed] [Google Scholar]
- 22. Koshiishi I., Shizari M., and Underhill C. B. (1994) CD44 can mediate the adhesion of platelets to hyaluronan. Blood 84, 390–396 [PubMed] [Google Scholar]
- 23. Molla A., Berthier R., Chapel A., Schweitzer A., and Andrieux A. (1995) β1 integrins mediate adherent phenotype of human erythroblastic cell lines after phorbol 12-myristate 13-acetate induction. Biochem. J. 309, 491–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chang P. V., Chen X., Smyrniotis C., Xenakis A., Hu T., Bertozzi C. R., and Wu P. (2009) Metabolic labeling of sialic acids in living animals with alkynyl sugars. Angewandte Chemie 48, 4030–4033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luchansky S. J., Argade S., Hayes B. K., and Bertozzi C. R. (2004) Metabolic functionalization of recombinant glycoproteins. Biochemistry 43, 12358–12366 [DOI] [PubMed] [Google Scholar]
- 26. Henne W. M., Liou J., and Emr S. D. (2015) Molecular mechanisms of inter-organelle ER-PM contact sites. Curr. Opin. Cell Biol. 35, 123–130 [DOI] [PubMed] [Google Scholar]
- 27. Phillips M. J., and Voeltz G. K. (2015) Structure and function of ER membrane contact sites with other organelles. Nature Reviews. Molecular Cell Biology, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bilyy R. O., Shkandina T., Tomin A., Muñoz L. E., Franz S., Antonyuk V., Kit Y. Y., Zirngibl M., Fürnrohr B. G., Janko C., Lauber K., Schiller M., Schett G., Stoika R. S., and Herrmann M. (2012) Macrophages discriminate glycosylation patterns of apoptotic cell-derived microparticles. J. Biol. Chem. 287, 496–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Franz S., Herrmann K., Führnrohr B. G., Sheriff A., Frey B., Gaipl U. S., Voll R. E., Kalden J. R., Jäck H. M., and Herrmann M. (2007) After shrinkage apoptotic cells expose internal membrane-derived epitopes on their plasma membranes. Cell Death Differ. 14, 733–742 [DOI] [PubMed] [Google Scholar]
- 30. Mbua N. E., Guo J., Wolfert M. A., Steet R., and Boons G. J. (2011) Strain-promoted alkyne-azide cycloadditions (SPAAC) reveal new features of glycoconjugate biosynthesis. Chembiochemistry 12, 1912–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]