Abstract
The aim of this study was to characterize the interaction of KYSE-410, an esophageal squamous cell carcinoma cell line, and fibroblasts with respect to the extracellular matrix component hyaluronan (HA) and chemokine expression. KYSE-410 cells induced the mRNA expression of HA synthase 2 (Has2) in normal skin fibroblasts (SF) only in direct co-cultures. Parallel to Has2 mRNA, Has2 antisense RNA (Has2os2) was up-regulated in co-cultures. Knockdown of LEF1, a downstream target of Wnt signaling, abrogated Has2 and Has2os2 induction. After knockdown of Has2 in SF, significantly less α-smooth muscle actin expression was detected in co-cultures. Moreover, it was investigated whether the phenotype of KYSE-410 was affected in co-culture with SF and whether Has2 knockdown in SF had an impact on KYSE-410 cells in co-culture. However, no effects on epithelial-mesenchymal transition markers, proliferation, and migration were detected. In addition to Has2 mRNA, the chemokine CCL5 was up-regulated and CCL11 was down-regulated in SF in co-culture. Furthermore, co-cultures of KYSE-410 cells and cancer-associated fibroblasts (CAF) were investigated. Similar to SF, Has2 and Ccl5 were up-regulated and Ccl11 was down-regulated in CAF in co-culture. Importantly and in contrast to SF, inhibiting HA synthesis by 4-methylumbelliferone abrogated the effect of co-culture on Ccl5 in CAF. Moreover, HA was found to promote adhesion of CD4+ but not CD8+ cells to xenogaft tumor tissues. In conclusion, direct co-culture of esophageal squamous cell carcinoma and fibroblasts induced stromal HA synthesis via Wnt/LEF1 and altered the chemokine profile of stromal fibroblasts, which in turn may affect the tumor immune response.
Keywords: cancer, cell-cell interaction, chemokine, fibroblast, hyaluronan, CCL11, CCL5
Introduction
Tumors are complex tissues that are composed of neoplastic tumor cells, a variety of stromal cell types, and extracellular matrix (ECM).2 The various cell types form a microenvironment that actively contributes to tumorigenesis. Major cell types of the tumor-associated stroma are cancer-associated fibroblasts (CAF). In normal tissues, fibroblasts fulfill a variety of homeostatic functions, such as the maintenance of the structural integrity of the connective tissues and of the ECM. In addition, they interact with other cell types, such as endothelial cells, epithelial cells, and immune cells. CAF additionally promote cancer progression (1). They are known to secrete growth factors that support tumor cell proliferation, metastases, and angiogenesis (1–3). In addition, CAF influence cancer cell phenotypes, such as stemness, and they can induce epithelial-mesenchymal transition (EMT) (4). Moreover, they modulate the inflammatory response in tumor tissues. CAF can express interleukin (IL)-6, IL-1β, and TNFα and chemokines, such as CXCL1, -2, -12, and -14 and CCL2, -5, and -7 (1, 4, 5). They are involved in the recruitment of immune cells, such as macrophages, myeloid-derived suppressor cells, and T lymphocyte subsets, which may in turn show tumor-promoting properties (1). Furthermore, CAF are responsible for production and remodeling of the ECM in the tumor (2).
Hyaluronan (HA) is a component of the ECM, and stromal HA is associated with decreased disease-free survival in breast cancer and non-small cell lung adenocarcinoma (6, 7). Knock-out of HA synthase (HAS) isoforms in some cancer cells decreases the tumor volume of murine xenograft tumors and is associated with decreased proliferation or induced apoptosis (8–11). Inhibition of HA synthesis by 4-methylumbelliferone (4-MU) effectively attenuates the growth of subcutaneous xenograft tumors of esophageal squamous cell carcinoma (ESCC) cells in nude mice (8). In contrast, the huge amount of high molecular weight HA was discussed as contributing to the cancer resistance of the naked mole rat, and subcutaneous tumor growth of transformed rat 3Y1 fibroblasts was reduced upon HAS2 overexpression (12, 13). Additionally, HA synthesis may modulate cell migration, metastases, angiogenesis, and EMT (9, 10, 14–18).
Several in vitro experiments of cancer cells and fibroblasts showed that cancer cells are able to induce a fibroblast phenotype that can be characterized by the differential expression of growth factors, chemokines, cytokines, and components of the ECM (19–24). This study focuses on changes of chemokine expression and HA synthesis in fibroblasts in the interaction with an ESCC cell line. Primary normal skin fibroblasts (SF) displayed increased Has2 and Ccl5 as well as decreased Ccl11 mRNA expression upon direct co-culture with KYSE-410 cells. Parallel to Has2, the Has2 antisense RNA Has2os2 was up-regulated in co-culture. The observed increase in Has2 mRNA and Has2os2 expression was dependent on lymphoid enhancer binding factor 1 (LEF1) expression.
Interestingly, Has2 and chemokines were regulated in CAF in a similar way as in SF. In co-cultures of CAF and KYSE-410 cells, chemokine expression was in part dependent on HA synthesis. Moreover, HA was important for binding of CD4+ T-helper cells to xenograft tumor sections.
Experimental Procedures
Cell Culture
Human KYSE-410 cells were purchased from the Leibniz Institute DSMZ German collection of Microorganisms and Cell Cultures (Braunschweig, Germany) and maintained in RPMI 1640 GlutaMAXTM I medium (Gibco Life Technologies, Paisley, UK) supplemented with 10% fetal bovine serum (FBS; Gibco Life Technologies) and 1% penicillin and streptomycin (Gibco Life Technologies). HAS2flox/flox mice as described previously (25) were crossed with an ubiquitous Cre deleter mouse line (ROSA26CreERT2) (26) to establish a conditional tamoxifen-inducible Has2 deletion. Primary murine SF were isolated from the skin of NMRI nude mice (Taconic Biosciences, Inc.) or ROSA26CreERT2+/−/HAS2flox/flox mice. After digestion with 5 units/ml dispase II (Roche Diagnostics GmbH, Mannheim, Germany), the dermal tissue was scraped off of the epidermis and subsequently digested using 1000 units/ml collagenase from Clostridium histolyticum (Sigma-Aldrich). After the addition of ice-cold culture medium (DMEM, high glucose, GlutaMAXTM supplemented with 20% FBS, 1% minimum essential medium non-essential amino acid solution, and 1% penicillin and streptomycin), the cells were separated from remaining tissue fragments by the EASY-strainerTM cell strainer with mesh sizes of 70 or 100 μm (Greiner Bio-One, Frickenhausen, Germany). Has2 knockdown was induced in SF from ROSA26CreERT2+/−/HAS2flox/flox mice in vitro by treatment with 4-hydroxytamoxifen (500 nmol/liter) for 24 h minimum. Knockdown was confirmed by quantitative real-time PCR (qPCR).
CAF were prepared from subcutaneous xenograft ESCC tumors of NMRI nude mice. The procedure for ESCC cell injection was in accordance with the national guidelines for animal care and was approved by the local research board for animal experimentation (LANUV, Landesamt für Natur, Umwelt, und Verbraucherschutz NRW). Tumors were minced and subsequently digested twice for 20 min with 300 collagen-degrading units/ml collagenase from C. histolyticum and 0.96 units/ml dispase II. In between and at the end, it was processed in the gentleMACSTM dissociator (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany). Cells were then filtered in an EASY-strainerTM cell strainer with a mesh size of 40 μm. Afterward, murine fibroblasts were magnetically separated by Feeder Removal MicroBeads (Miltenyi Biotec GmbH) according to the manufacturer's protocol. SF and CAF were maintained in DMEM, high glucose, GlutaMAXTM supplemented with 20% FBS, 1% minimum essential medium non-essential amino acid solution, and 1% penicillin and streptomycin (all from Gibco Life Technologies). Cells were cultured in a humidified atmosphere at 37 °C and 5% CO2.
For standard direct co-culture experiments, cells were seeded in RPMI 1640 GlutaMAXTM I medium supplemented with 10% FBS and 1% penicillin and streptomycin at a total of 50,000 cells/well in a 6-well cell culture plate. For indirect co-culture, cell culture inserts of stainless steel were fixed in a 6-well plate with 0.8% agarose solution. Subsequently, cells were seeded in separate wells in 400 μl of medium. After adhesion, medium was changed to 2 ml. In addition, KYSE-410 or SF cells were seeded in ThinCertsTM cell culture inserts (Greiner Bio-One) with 0.4- and 1.0-μm pore sizes that were placed in a 6-well plate containing 50,000 SF for indirect co-culture.
In some experiments, cells were treated with 4-methylumbelliferone sodium salt (Sigma-Aldrich) at a final concentration of 300 μmol/liter. Actinomycin D (5 μg/ml; AppliChem GmbH, Darmstadt, Germany) was added to the cells for RNA stabilization experiments.
siRNA Transfection and Treatment with MEK1 Inhibitor
Cells were transfected with siRNA using Lipofectamine® RNAiMAX (Life Technologies, Inc.) according to the manufacturer's instructions for “reverse transfection.” Briefly, 24 pmol of Mm_Bsg_3 (catalog no. SI00187376, Qiagen, Hilden, Germany), Hs_BSG_1 (catalog no. SI00313733, Qiagen) or Has2os2 (target sequence 5′-GCCAAGGCCTGATGTTCAA-3′; Sigma) or 72 pmol of Mm_Lef1_5 (catalog no. SI02713025, Qiagen) or the respective amount of control (AllStars Negative Control siRNA, catalog no. SI03650318, Qiagen) were mixed with 4 μl of transfection reagent and 400 μl of medium free of serum or antibiotics in a 6-well plate. Cells were then added in medium containing 10% FBS. After 24 h, medium was changed, and KYSE-410 or SF cells were seeded together with the transfected cells for co-culture experiments. Knockdown was confirmed by qPCR. Cells were treated with 50 μmol/liter PD98059 (Calbiochem, Merck KGaA, Darmstadt, Germany) or vehicle (DMSO) 24 h after seeding for an additional 24 h.
Flow Cytometry
A total of 25,000 SF, CAF, or KYSE-410 cells were cultured for 3 days in monoculture or co-culture. After trypsinization, KYSE-410 cells were labeled with CD326 (EpCAM)-PE antibody (Miltenyi Biotech GmbH) at 4 °C for 30 min. KYSE-410 cells were quantified by the GalliosTM flow cytometer (Beckman Coulter, Inc.) after the addition of Flow CountTM Fluorospheres (Beckman Coulter Inc., Brea, CA). To determine the proliferation of fibroblasts, cells were incubated with 2.5 μmol/liter Cell TraceTM CFSE cell proliferation kit (Molecular Probes, Life Technologies, Inc.) before seeding. Cells were trypsinized, and mean fluorescence intensity was determined 1–4 days after seeding.
For analysis of macrophages in KYSE-410 xenograft tumors, tissue homogenization was performed as described above for CAF isolation. The following antibodies were used for staining: anti-F4/80-AlexaFluor®488 (clone BM8), CD45-PE (30-F11), CD206-AlexaFluor®647 (C068C2), and CD11b-PacificBlueTM (M1/70) (all from BioLegend®).
Time Lapse Microscopy
Time lapse microscopy was performed with the Zeiss Axio Observer Z.1 microscope (Carl Zeiss MicroImaging GmbH, Göttingen, Germany) using the definite focus assistance device. The mean speed of randomly migrating cells was determined by tracking of ≥5 cells/condition in three independent experiments. For this purpose, the Fiji manual tracking plugin (27) was used.
Fluorescence Microscopy
Before staining of frozen sections (10 μm) of subcutaneous ESCC xenograft tumors, nonspecific binding was blocked by the avidin/biotin blocking system (Thermo Fisher Scientific) and PBS supplemented with 10% FBS and 2% bovine serum albumin. Sections were stained for α-smooth muscle actin (αSMA, catalog no. ab5694, Abcam, Cambridge, UK; 1:100) or Mac2 (catalog no. CL8942AP, Cedarlane, Burlington, Canada; 1:100), cytokeratin 18 (CK-18, catalog no. GP-CK18, Progen Biotechnik GmbH, Heidelberg, Germany; 1:100), and HA (biotinylated HA-binding protein, Calbiochem; 1:50). The following fluorochrome-conjugated probes were used: Alexa Fluor® 568 goat anti-rabbit (αSMA, catalog no. A-11036, Life Technologies; 1:200), Rhodamine Red X anti-rat (Mac2, catalog no. 112-295-167, Dianova; 1:100), FITC anti-guinea pig (CK-18, catalog no. 106-096-003, Dianova; 1:200), and ZyMAXTMCyTM5 streptavidin (HA, Invitrogen; 1:50). Roti®-Mount Fluor Care DAPI (Carl Roth GmbH, Karlsruhe, Germany) was used for nuclei staining and mounting of sections.
Cells were grown on glass coverslides in 24-well cell culture plates and fixed with 70% ethanol, 5% acetic acid, and 3.7% formaldehyde in PBS for HA staining. The staining of CK-18 and HA was performed using the probes mentioned above. For CK-18 staining, primary and secondary antibodies were used in a 1:500 dilution. Biotinylated HA-binding protein and ZyMAXTMCyTM5 streptavidin were diluted 1:250. Slides were mounted in ProLong® Gold Antifade Mountant with DAPI (Life Technologies). For staining of CK-18 and β-catenin, cells were fixed with 50% methanol, 50% acetone at −20 °C and permeabilized with 0.1% Triton X-100 in PBS for 5 min. After blocking, cells were incubated overnight at 4 °C with primary anti-β-catenin antibody (catalog no. 9582, Cell Signaling Technology, Danvers, MA; 1:500). Alexa Fluor® 568 goat anti-rabbit secondary antibody was used in a 1:1000 dilution. CK-18 staining was performed as described above. Fluorescence images were captured with the Zeiss Axio Observer Z.1 microscope and the ApoTome.2 module (Carl Zeiss MicroImaging).
Isolation of Human CD3+, CD4+, CD8+, and CD14+ Cells and Adhesion on Xenograft Tumor Sections
Human peripheral blood mononuclear cells were separated from buffy coat of healthy donors by density gradient centrifugation using Ficoll-PaqueTM PLUS (GE Healthcare Bio-Sciences AB, Uppsala, Sweden). Subsequently, cells were isolated with magnetic CD3, CD4, CD8, and CD14 MicroBeads according to manufacturer's protocol (Miltenyi Biotech GmbH). Before CD4+ and CD8+ cell isolation, CD14+ as well as CD56+ cells were removed with magnetic beads (Miltenyi Biotech GmbH). CD3+, CD4+, and CD8+ cells were activated using particles loaded with antibodies against human CD2, CD3, and CD28 (T cell activation kit, Miltenyi Biotech GmbH) for 3 days.
The cells were labeled with a final concentration of 10 μg/ml Calcein-AM (EMD Chemicals, Inc., San Diego, CA). A total of 6.5 × 105 cells were incubated on fixed frozen xenograft tumor sections for 30 min, shaking, at 70 rpm at room temperature. A consecutive section was digested with hyaluronidase from Streptomyces hyalurolyticus (Sigma) at a final concentration of 4 units/ml for 1 h at 37 °C before cells were added. Non-adherent cells were removed by washing with TBS, and subsequently, slides were fixed with acetone and mounted in Roti®-Mount Fluor Care DAPI. Mosaic images of whole tissue sections were acquired with the Zeiss Axio Observer Z.1 microscope. Bound cells were counted, and the area was determined by Fiji software (27).
Quantification of HA, CCL5, and CCL11
To quantify the amount of HA in cell culture supernatants, cells were grown as described above. Supernatants were harvested after 24 h, and HA content was determined with the hyaluronic acid test kit (Corgenix Medical Corp., Broomfield, CO) and normalized to 25,000 fibroblasts or KYSE-410 cells, respectively. CCL5 was quantified in supernatants conditioned for 48 h by 25,000 fibroblasts or 25,000 KYSE-410 cells in mono- and co-cultures with the RANTES (regulated on activation normal T cell expressed and secreted) (CCL5) mouse ELISA (Abcam). To obtain detectable amounts of CCL11, 25,000 cells of each cell type were cultured alone or in co-culture in serum-free DMEM for 60 h. For analysis with the eotaxin single analyte ELISArray kit (Qiagen), 100 μl of medium and 100 μl of antigen standard dilutions ranging from 15.6 to 500 pg/ml were used.
Western Blotting
After 48 h of co-culture, cells were lysed with a buffer containing 125 mmol/liter Tris, 4% SDS, 100 mmol/liter dithiothreitol, 20% glycerol, and protease inhibitors. Proteins were separated by standard 10% SDS-PAGE and blotted onto nitrocellulose membranes. They were probed with anti-αSMA antibody (catalog no. ab5694, Abcam; 1:2000) and anti-β-tubulin I antibody (catalog no. T7816, Sigma-Aldrich; 1:10,000). Anti-rabbit IRDye®800CW (αSMA) and anti-mouse IRDye®680LT (β-tubulin, both from LI-COR Biosciences, Lincoln, NE) were used as secondary antibodies in a 1:5000 dilution for visualization and quantitation with the Odyssey infrared imaging system (LI-COR Biosciences).
HA-Agarose Gel
Cell culture supernatants were collected after 48 h and digested with 250 μg/ml proteinase K (Invitrogen) for 45 min at 50 °C. Gycosaminoglycans were precipitated by the addition of two volumes of ethanol and incubation at −20 °C for 2 h. Pellets were dissolved in Tris/EDTA buffer as well as Healon 5 (Abbot Medical Optics Inc., Santa Ana, CA) solution finally containing 25 μg of HA as a positive control. Half of each sample was digested with hyaluronidase from S. hyalurolyticus (3 units/ml; Sigma) for 1 h at 37 °C. Next, a 0.5% TAE-agarose gel was loaded with 30 μl of the sample solution mixed with 5 μl of 4 mol/liter sucrose and 2 μl of bromphenol blue solved in 20% glycerol. A total of 5 μl of Select-HATM HiLadder and 5 μl of Select-HATM MegaLadder (Hyalose, LLC, Oklahoma City, OK) were used as HA molecular size markers. The gel was run at 80 V for 5 h, fixed in 50% ethanol, and stained with 0.05 mg/ml Stains All (Sigma) in 50% ethanol overnight. It was destained for 6 h in water followed by a 1-h exposure to ambient light.
qPCR
Total RNA was extracted with PeqGOLD TriFastTM (PEQLAB Biotechnologie GmbH, Erlangen, Germany) according to the manufacturer's instructions. The quantity and purity of RNA was determined by spectrophotometry (NanoDrop 1000, Thermo Scientific). A total of 1000 ng of RNA was used for reverse transcription with the QuantiTect reverse transcription kit (Qiagen), and a final concentration of 1.25 ng/μl cDNA (experiments with Has2 knockdown SF and respective controls) or 2.1 ng/μl cDNA (all other experiments) was used for quantitative real-time PCR. Forward and reverse primers were used at final concentrations of 0.625 μmol/liter. They were designed with the Primer3Plus software (28) and Primer-BLAST (29), and sequences are listed in Table 1. To distinguish mRNA expression of murine SF and human KYSE-410 cells, the primers were designed to be species-specific. Platinum®SYBR® Green qPCR SuperMix-UDG (Life Technologies) reagent was used, and qPCR was performed on the Applied Biosystems 7300 and on the Life Technologies StepOnePlus real-time PCR systems. Samples were measured in duplicates. Cq values were determined by the 7300 real-time PCR system RQ Study Software version 1.4 (Applied Biosystems) or by the StepOne Software version 2.3 (Life Technologies) and normalized to GAPDH as endogenous control (ΔCq). Relative quantity was calculated by 2−ΔCq.
TABLE 1.
Primers used for qPCR
| Gene | Forward (5′–3′) | Reverse (5′–3′) |
|---|---|---|
| Murine | ||
| Bsg | GTGGGCAGAAGCGAGATCAA | AGGGTAGGATGCATCGGACT |
| Ccl2 | TTCCTTCTTGGGGTCAGCAC | GGCTGGAGAGCTACAAGAGG |
| Ccl5 | AGCAGCAAGTGCTCCAATCT | CCCACTTCTTCTCTGGGTTG |
| Ccl7 | CGCTGCTTTCAGCATCCAAG | CTTCCCAGGGACACCGACTA |
| Ccl11 | CACGGTCACTTCCTTCACCT | GCTTTCAGGGTGCATCTGTT |
| Cxcl1 | GCACCCAAACCGAAGTCATA | AGGTGCCATCAGAGCAGTCT |
| Cxcl12 | TTCTTCGAGAGCCACATCGC | CTGTTGTTGTTCTTCAGCCGT |
| Gapdh | GGTGCTGAGTATGTCGTGGA | GTGGTTCACACCCATCACAA |
| Has1 | TATGCTACCAAGTATACCTCG | TCTCGGAAGTAAGATTTGGAC |
| Has2 | CGGAGGACGAGTCTATGAGC | TGTGATTCCGAGGAGGAGAG |
| Has3 | TCCCCAAGTAGGAGGTGTTG | CTCACACTGCTCAGGAAGGA |
| Has2os1 | TGGGAGTATTGATGCAGGCAAG | TCTTCCCGAGCGTCAAAGG |
| Has2os2 | AGCCAAGGCCTGATGTTCAA | GACACAGGAGCTTGTGACGA |
| Has2os3 | GAGGGTCGTGGAAGGAAGTT | GACACAGGAGCTTGTGACGA |
| Lef1 | GCACGGAAAGAGAGACAGCTA | TCTGGGACCTGTACCTGAAGT |
| Wnt2 | ATCAAGTTTGCCCGTGCCTT | GCCACTCACACCATGACACTT |
| Human | ||
| BSG | TCTGCAAGTCAGAGTCCGTG | TCACGAAGAACCTGCTCTCG |
| CCR1 | CACGGACAAAGTCCCTTGGAA | TCAAACTCTGTGGTCGTGTCA |
| CCR3 | ACCACTGGTCTTCTTGTGCTT | TGTGGTACCAAAGGTCTCAACT |
| CCR5 | GACATCCGTTCCCCTACAAGA | TGGCAGGGCTCCGATGTAT |
| CDH1 | GCCGAGAGCTACACGTTCA | ACACCATCTGTGCCCACTTT |
| FN1 | GCTGGGCGAGGGAGAATAAG | TGGTCTCCTCCAGGTGTCAC |
| GAPDH | CGAGATCCCTCCAAAATCAA | GGCAGAGATGATGACCCTTT |
| MKI67 | CTGCTCGACCCTACAGAGTG | GCGATGTGACATGTGCTTGT |
| SNAI1 | GCGAGCTGCAGGACTCTAAT | GGACAGAGTCCCAGATGAGC |
| VIM | GCAAAGCAGGAGTCCACTGA | GCAGCTTCAACGGCAAAGTT |
Transcription Factor Binding Sites
Sequences of murine Has2 mRNA and Has2 antisense (Has2os2), including the sequence of 1000 base pairs upstream and 500 base pairs downstream from the transcription start site, were retrieved from Ensembl database release 82 (30). The sequences were analyzed by Genomatix MatInsepctor version 8.0 (Genomatix Software GmbH) (31).
Statistical Analysis
Data are presented as mean ± S.E. Data were analyzed with GraphPad Prism version 6 software (GraphPad Software Inc., La Jolla, CA). Before statistical analysis, qPCR data were log-transformed. One outlier identified by Grubb's test was removed in Fig. 6C. However, statistical significance remained unaffected by the exclusion. An unpaired t test was used to compare two groups. For multiple comparisons, ordinary one-way analysis of variance and Sidak's multiple-comparison test were used. p values of <0.05 were considered as statistically significant.
FIGURE 6.
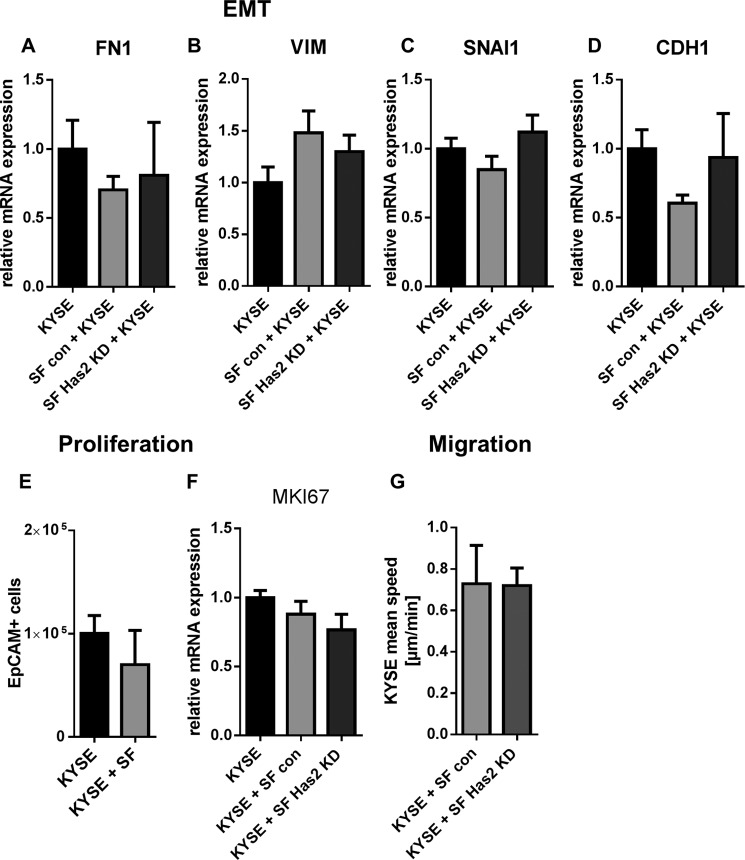
The KYSE-410 phenotype was not altered by increased HA synthesis in direct co-culture. A, B, C, and D, mRNA expression of mesenchymal markers fibronectin (FN1), vimentin (VIM), and SNAI1 and epithelial marker CDH1 in KYSE-410 cells after 96 h of monoculture or co-culture with either control SF or Has2 knockdown SF (n = 4). E, cell count of EpCAM+ KYSE-410 cells by flow cytometry after 3 days in monoculture and co-culture (n = 4). F, gene expression of the proliferation marker MKI67 in KYSE-410 cells in mono- and co-culture with either control SF or Has2 knockdown SF (n = 3). G, random migration of KYSE-410 cells in direct co-culture with control SF or Has2 knockdown SF (n = 3). Data are presented as mean ± S.E. (error bars).
Results
Has2 mRNA Expression Was Induced in SF by Direct Co-culture with KYSE-410 Cells
If KYSE-410 cells were used to induce xenograft tumors in nude mice, an HA-rich matrix was associated with αSMA-positive cells (Fig. 1A) and with the parenchyma, as identified by CK-18-positive cells. To investigate the regulation of HA synthesis and chemokine expression in the context of interacting cancer cells and fibroblasts, we established a direct co-culture of primary murine SF and human KYSE-410 cells.
FIGURE 1.
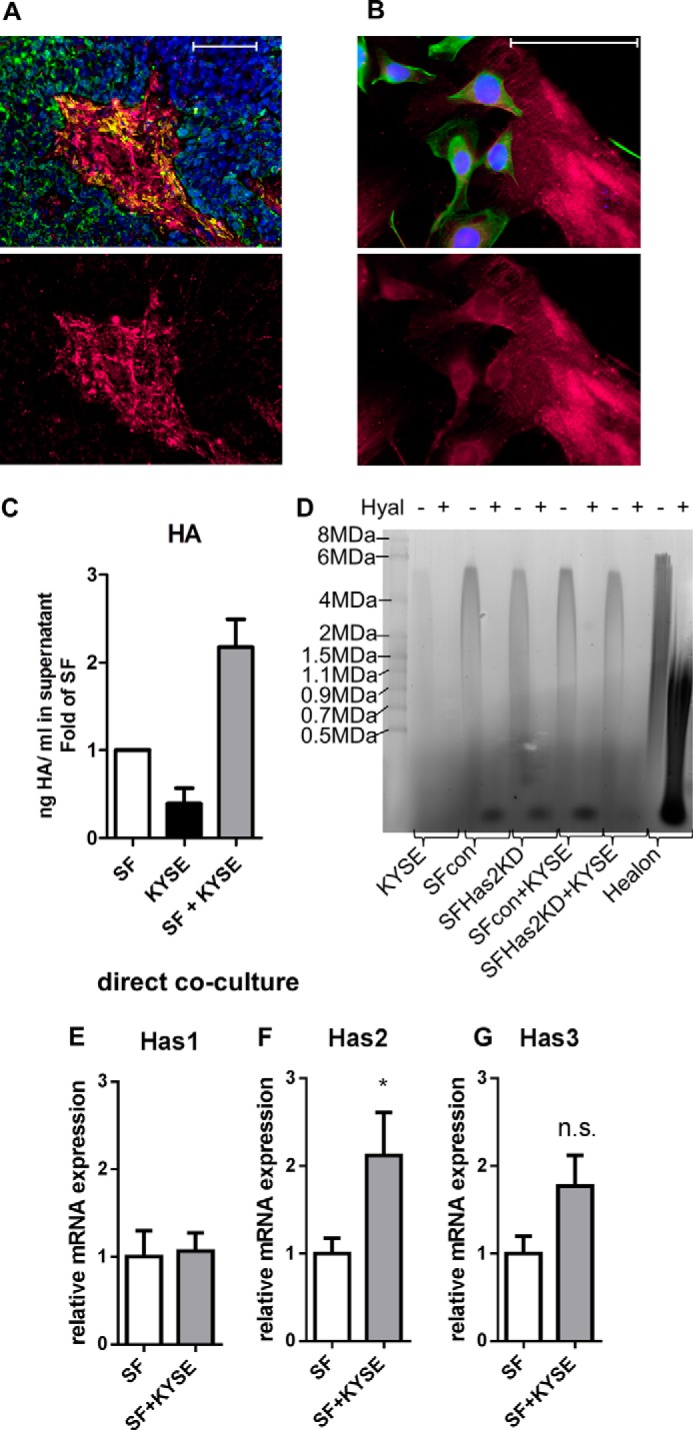
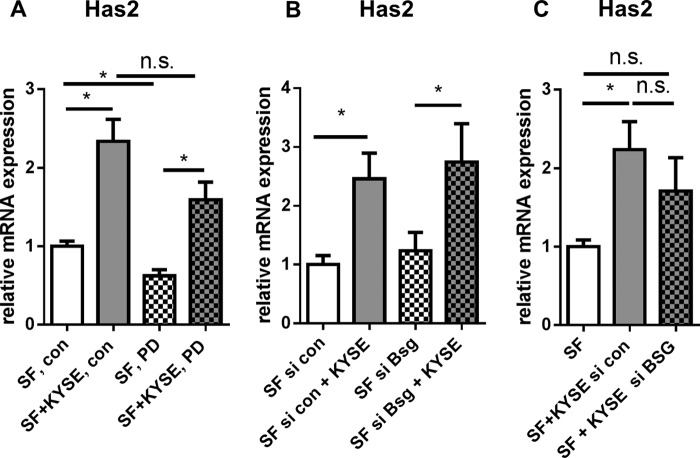
Direct co-culture of KYSE-410 cells and fibroblasts resulted in an HA-rich environment. A, fluorescent staining of HA (magenta), αSMA (yellow), and CK-18 (green) in KYSE-410 xenograft tumors. Scale bar, 100 μm. Bottom, HA staining alone. B, fluorescent staining of HA (magenta) in cell culture. KYSE-410 cells are labeled in green (CK-18). Scale bar, 100 μm. Bottom, HA staining alone. C, HA secreted into the supernatant of monocultures and co-cultures of SF and KYSE-410 cells after 24 h determined by an ELISA-like assay; n = 4. D, representative HA-agarose gel of supernatants of mono- and co-cultures with KYSE-410 cells and SF with and without Has2 knockdown conditioned for 48 h. E–G, Has1, -2, and -3 mRNA expression in SF after 24 h of monoculture or direct co-culture with KYSE-410 cells (n = 7). Error bars, S.E. *, p < 0.05; n.s., not significant.
Both KYSE-410 cells and SF were associated with HA in direct co-culture (Fig. 1B). Furthermore, more HA was secreted into the cell culture supernatant in co-cultures compared with the respective monocultures (Fig. 1C). HA produced by control SF in monoculture showed a strong signal of high molecular weight HA. The pattern of HA size in co-cultures resembled the SF monoculture (Fig. 1D). Because murine SF and a human cancer cell line were used for direct co-cultures, gene expression of each cell type could be analyzed using species-specific primers applying qPCR. In SF, Has2 mRNA expression was significantly induced by direct contact with KYSE-410 cells (Fig. 1F).
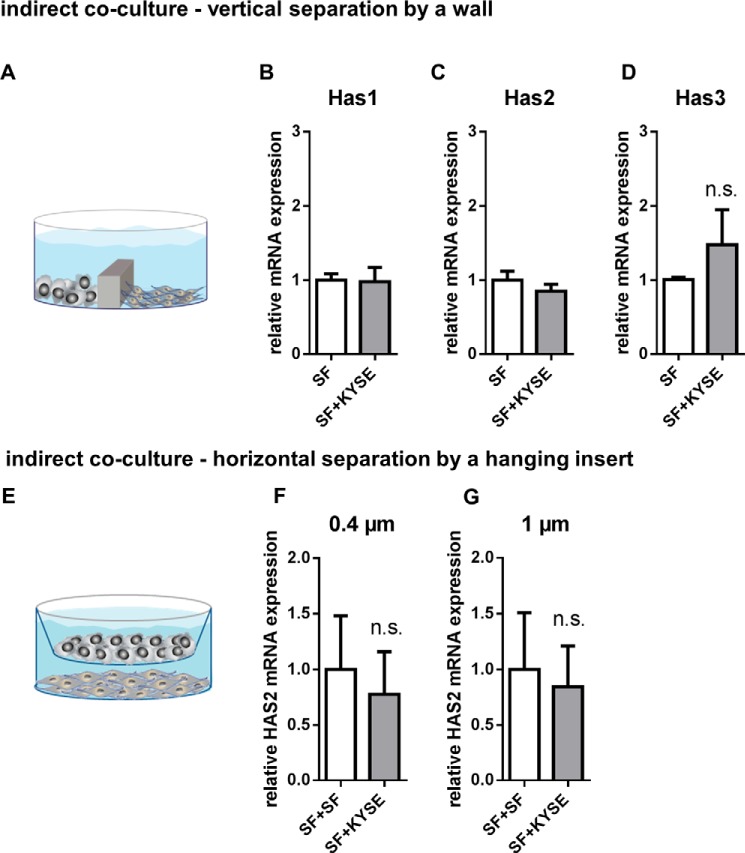
Importantly, if KYSE-410 cells and SF were seeded in a way that allowed exchange of media but prevented direct cell-cell contact, Has2 mRNA expression was no longer increased (Fig. 2). Therefore, in this set-up, direct cell-cell contact was required for Has2 induction in SF.
FIGURE 2.
Direct cell-cell contact was required for increased Has2 mRNA expression. A, scheme showing the set-up of indirect co-culture experiments where KYSE-410 cells and SF were separated by a wall but were able to exchange soluble factors by diffusion. B–D, Has1, -2, and -3 mRNA expression in SF after 24 h of indirect monoculture or co-culture (n = 3 for Has1, n = 4 for Has2 and -3). E, scheme showing a set-up of indirect co-culture using inserts. F, Has2 mRNA expression in SF separated from KYSE-410 cells by a membrane with 0.4-μm pore size, n = 4. G, Has2 mRNA expression in SF separated from KYSE-410 cells by a membrane with 1-μm pore size (n = 4). Data are presented as mean ± S.E. (error bars); n.s., not significant.
LEF1 Was Required for Has2 Up-regulation in SF
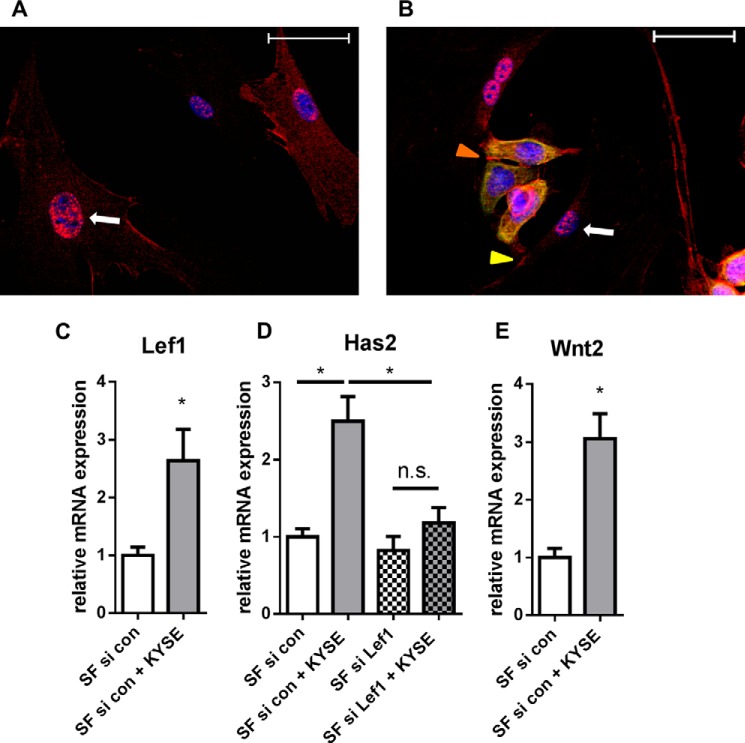
To further analyze the direct cell-cell contact between KYSE-410 cells and SF, the cells were stained for β-catenin. As shown in Fig. 3, β-catenin was found at cell-cell contacts of KYSE-410 and at some heterotypic cell-cell contacts between KYSE-410 and SF. Furthermore, it could be clearly detected in the nuclear region of SF in monoculture and co-culture. β-Catenin can bind to members of the T-cell factor/LEF family and subsequently promote the transcription of target genes, such as Lef1 (32). Lef1 was found to be up-regulated in SF in co-culture (Fig. 3C). Consequently, we analyzed whether LEF1 was involved in Has2 mRNA induction. Indeed, knockdown of Lef1 by siRNA (mRNA expression of Lef1: control, 1.0 ± 0.14; siRNA-transfected, 0.34 ± 0.09; mean ± S.E., n = 4) prevented the up-regulation of Has2 by co-culture with KYSE-410 cells (Fig. 3D). Nuclear translocation of β-catenin may result from active Wnt signaling (32). In this setup, direct co-culture induced Wnt2 mRNA expression in SF that in turn may autostimulate Lef1 and Has2 mRNA expression.
FIGURE 3.
β-catenin/LEF1 signaling was involved in Has2 mRNA up-regulation. A, immunocytochemistry of β-catenin (red) in SF monoculture. Nuclear DAPI staining is presented in blue. Scale bar, 50 μm. B, immunocytochemistry of β-catenin (red) and CK-18 (green) in co-culture. White arrow, positive β-catenin staining in the nuclear region (blue) in SF. Orange arrowhead, cell-cell contacts of KYSE-410 cells; yellow arrowhead, cell-cell contacts of KYSE-410 cells and SF. Scale bar, 50 μm. C, Lef1 mRNA expression in SF after 24 h of monoculture or direct co-culture (n = 4). D, Has2 mRNA expression in monocultures or direct co-cultures after transfection with siRNA directed against Lef1 and control siRNA (n = 4). E, Wnt2 mRNA expression in SF in monocultures or direct co-cultures (n = 4). Data are presented as mean ± S.E. (error bars). *, p < 0.05; n.s., not significant.
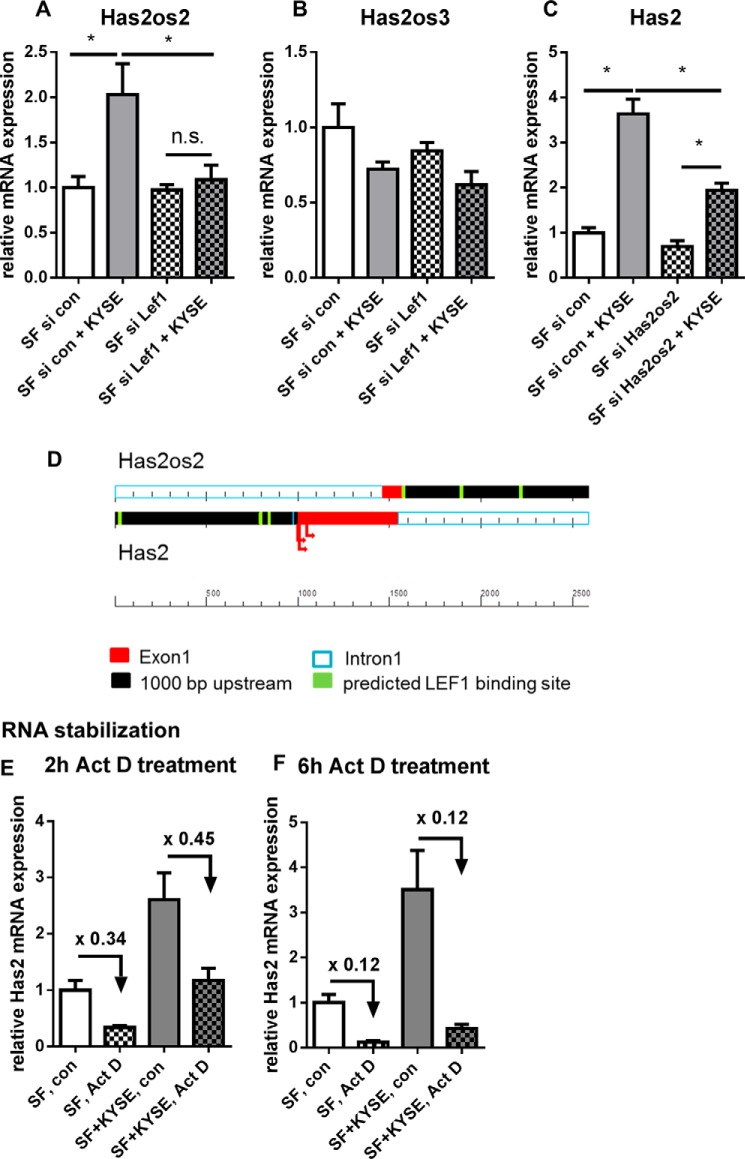
Recently, HAS2 antisense RNA was reported to increase HAS2 transcription in human aortic smooth muscle cells (33). The online database Ensembl (30) describes three Has2 opposite strand transcripts (Has2os1, -2, and -3) for murine Has2. Interestingly, Has2os2 RNA shows coordinated up-regulation with Has2 mRNA in co-cultures. Likewise, up-regulation of Has2os2 was prevented by siRNA directed against Lef1 (Fig. 4A). Has2os2 expression was effectively down-regulated by siRNA (control, 1.0 ± 0.24; siRNA-transfected, 0.07 ± 0.02; mean ± S.E., n = 4). In line with a stimulatory role of Has2os2, knockdown of Has2os2 reduced Has2 mRNA expression in co-culture (Fig. 4C). Hence, among others, Has2os2 potentially contributed to Has2 mRNA induction in co-culture.
FIGURE 4.
Has2os2 RNA was regulated parallel to Has2 mRNA in co-cultures. Has2 antisense RNA Has2os1, Has2os2, and Has2os3 were measured by qPCR. Has2os1 was below the limit of detection in both monocultures and co-cultures (n = 2). A and B, Has2os2 and Has2os3 RNA expression in monocultures or direct co-cultures after transfection with siRNA directed against Lef1 or control siRNA (n = 4). C, Has2 mRNA expression in SF after knockdown of Has2os2 or control treatment (n = 4). D, scheme shows predicted LEF1 binding sites (green) in Has2 (lower strand) and Has2os2 (upper strand) promotor regions (black). Exons are depicted in red, and arrows show transcription start sites of Has2 according to the Ensembl, Uniprot, and NCBI gene databases. E and F, Has2 mRNA expression after treatment with transcriptional inhibitor actinomycin D (Act D) or vehicle control for 2 and 6 h (n = 3). Data are presented as mean ± S.E. (error bars). *, p < 0.05; n.s., not significant.
LEF1 binding sites were predicted in both Has2 and Has2os2 promotor regions. This may be an explanation for the parallel regulation of both genes (Fig. 4D). Because, as was discussed, HAS2 antisense RNA may stabilize HAS2 mRNA (34), cells were treated with actinomycin D to inhibit transcription, and subsequently mRNA expression was measured by qPCR. However, the co-culture setting did not result in Has2 mRNA stabilization (Fig. 4, E and F).
Because it has been reported that Has2 mRNA expression in fibroblasts depends on ERK signaling (24, 35, 36), we additionally addressed the question of whether PD98059, a MEK1 inhibitor, was able to prevent Has2 induction in co-culture. However, Has2 mRNA expression was still increased in the co-culture setting. Still, Has2 was significantly reduced when comparing PD98059-treated and vehicle-treated monocultures (Fig. 5A). Furthermore, basigin was described previously as influencing HAS expression and fibroblast phenotypes (37, 38). Knockdown of basigin in either SF or KYSE-410 cells did not suggest a major role of basigin in Has2 mRNA induction in co-culture (Fig. 5C). The knockdown efficiencies of siRNA targeting human and murine basigin were determined by qPCR (murine Bsg control, 1.0 ± 0.26; siRNA-transfected SF, 0.12 ± 0.07 (n = 5); human BSG control, 1.0 ± 0.19; siRNA-transfected KYSE-410 cells, 0.01 ± 0.002 (n = 4); mean ± S.E.).
FIGURE 5.
Has2 mRNA was up-regulated despite MEK1 inhibition or basigin knockdown in SF. A, Has2 mRNA expression in SF in monoculture or direct co-culture after 24 h of treatment with the MEK1 inhibitor PD98059 (n = 6). B, Has2 mRNA expression in SF after transfection with control siRNA and siRNA targeting murine basigin (Bsg) (n = 5). C, Has2 mRNA expression in SF in co-culture with si BSG- or si control-transfected KYSE-410 cells (n = 4). Data are presented as mean ± S.E. (error bars). *, p < 0.05; n.s., not significant.
Has2 Induction in SF Did Not Induce Phenotypic Changes in KYSE-410 Cells
Both the co-culture setting with fibroblasts and HA actions were reported to have an impact on cancer cell phenotypes. Therefore, the mRNA expression of EMT markers fibronectin, vimentin, snai1 (SNAI1), and the epithelial marker e-cadherin (CDH1) were determined in KYSE-410 cells in co-culture and compared with the monoculture. To analyze the impact of Has2 up-regulation in fibroblasts, KYSE-410 cells were seeded in monocultures and in co-cultures with either control SF or SF in which Has2 had been knocked down (mRNA expression of Has2: control, 1.0 ± 0.15; 4-hydroxytamoxifen treatment, 0.15 ± 0.02; mean ± S.E., n = 4). Neither after 48 h nor after 96 h of co-culture was a significant change of EMT markers observed (Fig. 6; data not shown for 48 h). There was a trend toward reduced CDH1 mRNA expression in co-cultures of KYSE-410 cells and control SF.
We further investigated whether the co-culture setting enhanced the proliferation of KYSE-410 cells. After 3 days in monoculture or co-culture, respectively, KYSE-410 cells were labeled with EpCAM antibody and counted by flow cytometry. Additionally, the proliferation marker MKI67 was measured after 48 h by qPCR using species-specific primers. As shown in Fig. 6, E and F, the proliferation of KYSE-410 cells was not significantly changed in co-culture compared with cells in monoculture. Furthermore, the random migration of KYSE-410 cells did not differ in co-cultures with Has2 knockdown SF compared with co-cultures with control SF (Fig. 6G).
Knockdown of Has2 Decreased the Myofibroblast Marker αSMA in Fibroblasts in Co-culture
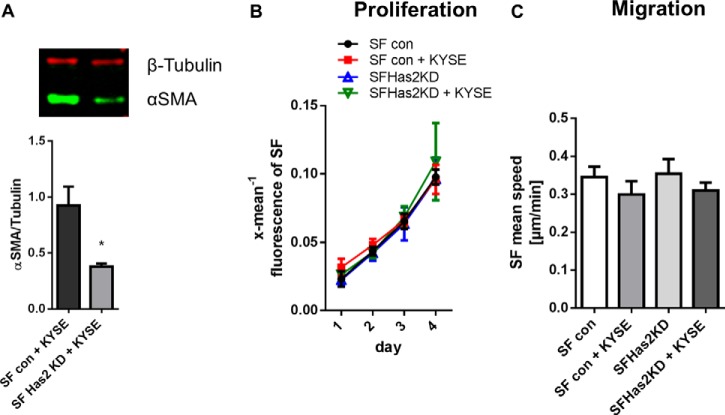
The impact of Has2 on the fibroblast phenotype was also determined. In co-cultures of SF with Has2 knockdown and KYSE-410 cells, reduced αSMA expression was found compared with control co-cultures (Fig. 7A). Therefore, HAS2 potentially promoted the development of a myofibroblast phenotype. For analysis of proliferation, SF were labeled with CFSE. No difference in fibroblast proliferation was observed comparing mono- and co-cultures and comparing Has2 knockdown SF with controls (Fig. 7B). Also, random migration of SF was affected neither by the co-culture with KYSE-410 cells nor by knockdown of Has2 (Fig. 7C).
FIGURE 7.
Has2 up-regulation in co-culture may modulate the myofibroblast phenotype. A, Western blotting analysis of αSMA expression in direct co-cultures of KYSE-410 cells and control SF or Has2 knockdown SF 48 h after seeding (n = 4). B, SF proliferation as measured by decreasing fluorescence of CFSE-labeled SF (n = 3). C, random migration speed of SF determined by time lapse microscopy in SF monocultures and direct co-cultures of KYSE-410 cells and control SF or Has2 knockdown SF (n = 3). Data are presented as mean ± S.E. (error bars). *, p < 0.05.
Secretion of Chemokines CCL5 and CCL11 by Fibroblasts Was Modulated in Co-culture
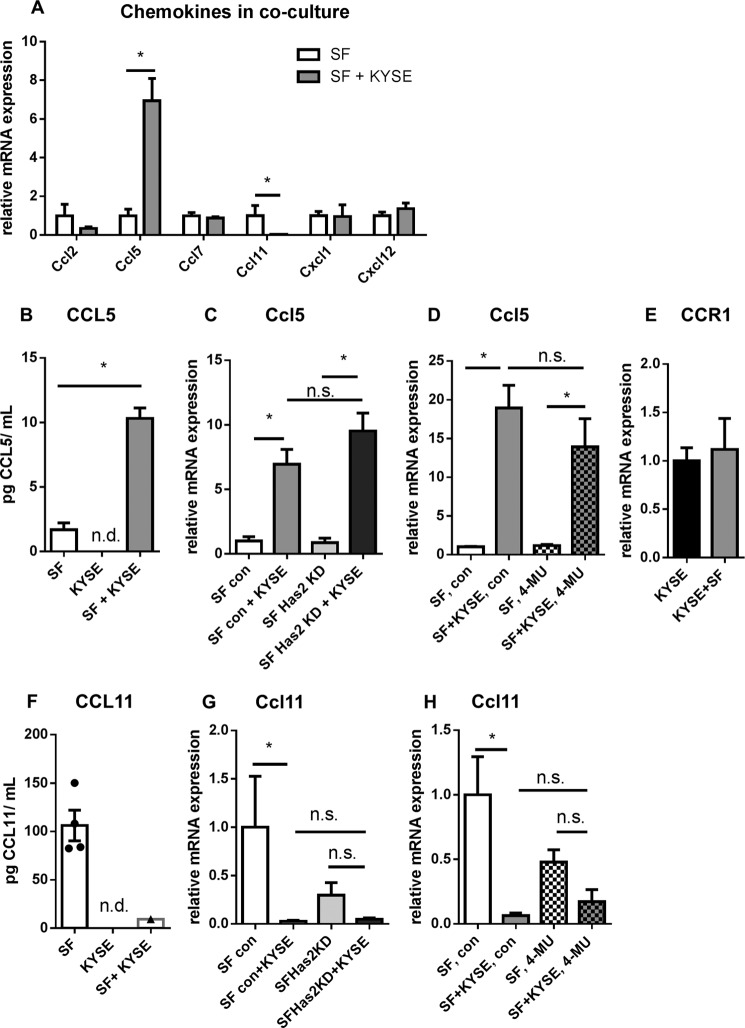
Stromal HAS2 is known to play a role in the recruitment of tumor-associated macrophages (39), and low molecular weight HA increases the number of CD3+ lymphocytes in a murine colorectal carcinoma model (40). Therefore, we analyzed whether other chemoattractants are modulated by the interaction of KYSE-410 cells and SF. As shown in Fig. 8, the chemokine CCL5 was up-regulated, whereas CCL11 was down-regulated in SF in co-culture. Ccl2 and -7 and Cxcl1 and -12 mRNA expression in SF were not significantly modulated by co-culture (Fig. 8A).
FIGURE 8.
Direct co-culture with KYSE-410 cells induced CCL5 and reduced CCL11 chemokine expression by fibroblasts. A, mRNA expression of murine chemokines Ccl2, Ccl5, Ccl7, Ccl11, Cxcl1, and Cxcl12 in SF after 48 h in monoculture or co-culture (n = 4). B, murine CCL5 protein concentration in cell culture supernatants of SF and KYSE-410 monoculture and direct co-cultures 48 h after seeding, determined by ELISA (n = 4). C, Ccl5 mRNA expression in control SF and Has2 knockdown SF after 48 h of co-culture with KYSE-410 cells (n = 4). D, Ccl5 mRNA expression in SF after treatment with 300 μm 4-MU or vehicle for 48 h (n = 4). E, expression of chemokine receptor CCR1 mRNA in KYSE-410 cells after 48 h in mono- and co-culture (n = 3). F, murine CCL11 protein concentration in supernatants of SF and KYSE-410 monocultures and direct co-cultures conditioned for 60 h (n = 4). G, Ccl11 mRNA expression in control SF and Has2 knockdown SF after 48 h in monoculture or co-culture (n = 4). H, Ccl11 mRNA expression in SF after treatment with 300 μm 4-MU or vehicle for 48 h (n = 4). Data are depicted as mean ± S.E. (error bars). *, p < 0.05; n.s., not significant; n.d., below the limit of detection.
Murine CCL5 protein was detected in cell culture supernatants of SF monocultures and of co-cultures. It was significantly elevated in the co-culture compared with the monoculture setting (Fig. 8B). Likewise, Ccl5 mRNA expression was increased in SF co-cultured with KYSE-410 cells (Fig. 8C). To investigate whether this up-regulation was dependent on the observed induction of Has2 mRNA in SF, we conducted co-culture experiments with Has2 knockdown SF or treated cells with the HA synthesis inhibitor 4-MU. Impaired HA synthesis did not affect Ccl5 mRNA induction in co-culture (Fig. 8, C and D). CCL5 can affect cancer cell behavior through its receptor CCR-1, -3, or -5 (41). However, in KYSE-410 cells, only CCR1 mRNA was consistently expressed in monocultures and co-cultures as measured by qPCR (Fig. 8E). CCR5 mRNA expression was below the limit of detection, and CCR3 could only be detected in one of the six lysates of monocultures and co-cultures.
For quantification of murine CCL11 by ELISA, cells had to be cultured in serum-free medium for 60 h. Under these conditions, CCL11 was detected in all supernatants of SF monocultures but only in one of four supernatants of co-cultures (Fig. 8F). This finding corresponds to the decreased Ccl11 mRNA expression in co-culture. Again, a potential effect of impaired HA synthesis on Ccl11 regulation was evaluated using Has2 knockdown SF or treatment with 4-MU. Ccl11 mRNA showed a trend toward decreased expression in co-cultures with Has2 knockdown SF and in cells treated with 4-MU (Fig. 8, G and H).
Has2 and CCL5 Were Increased and CCL11 Was Decreased in CAF in Co-culture
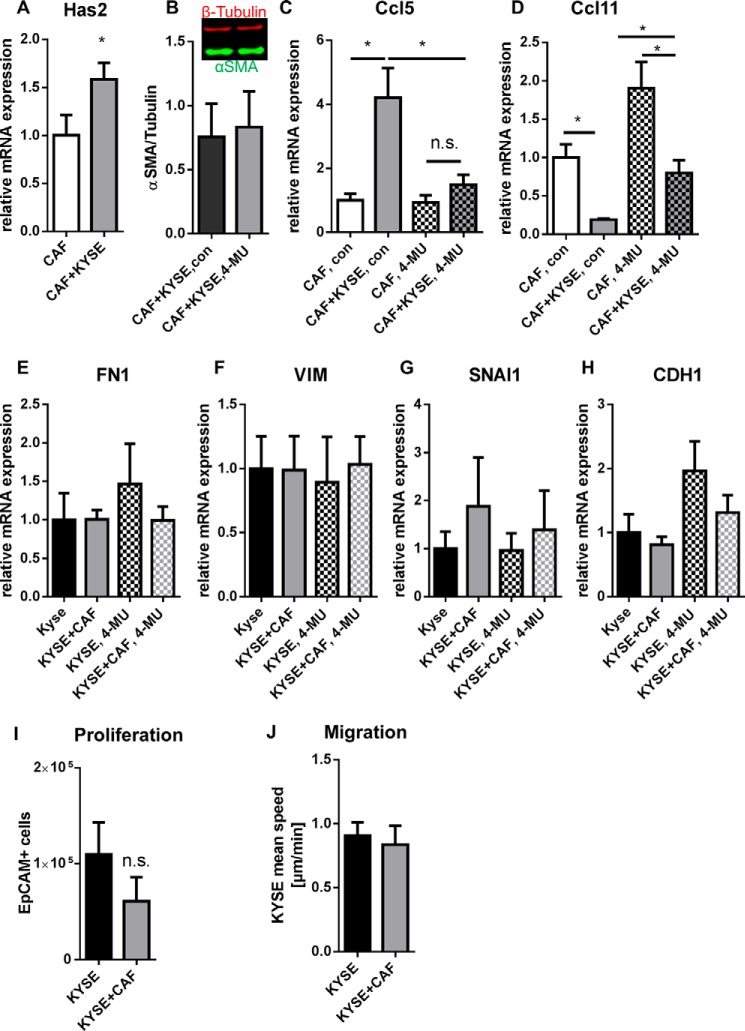
The findings presented above sought to model the reprogramming of normal tissue fibroblasts upon initial contact with cancer cells. It was of interest to determine whether Has2, Ccl5, and Ccl11 mRNA expression were also modulated in differentiated CAF. Therefore, CAF were isolated from subcutaneous ESCC xenograft tumors and seeded in direct co-cultures with KYSE-410 cells. Subsequent qPCR analysis revealed that Has2 (Fig. 9A) and Ccl5 mRNA expression were also induced in CAF, whereas Ccl11 mRNA expression was reduced (Fig. 9, C and D). However, impaired HA synthesis by 4-MU treatment did not result in decreased αSMA in Western blotting analysis (Fig. 9B). Furthermore, interfering with HA synthesis by 4-MU treatment reduced the regulation of chemokines in CAF. Specifically, in 4-MU-treated co-cultures, mRNA expression of Ccl5 was significantly lower and mRNA expression of Ccl11 was significantly higher as compared with vehicle-treated co-cultures (Fig. 9, C and D). Additionally, treatment with 4-MU successfully inhibited a significant up-regulation of Ccl5 mRNA in co-culture.
FIGURE 9.
Has2 and Ccl5 mRNA expression were also induced, and Ccl11 mRNA expression was reduced in CAF in direct co-culture with KYSE-410 cells. A, Has2 mRNA expression in CAF after 24 h of direct co-culture with KYSE-410 cells (n = 10). B, Western blotting analysis of αSMA expression in direct co-cultures of KYSE-410 cells and CAF treated with 300 μm 4-MU or vehicle for 48 h (n = 3). C, Ccl5 mRNA expression in CAF after 48 h of treatment with 300 μm 4-MU or vehicle (n = 4). D, Ccl11 mRNA expression in CAF after 48 h of treatment with 300 μm 4-MU or vehicle (n = 4). E–H, mRNA expression of EMT markers in KYSE-410 cells co-cultured with CAF for 96 h and treated with 4-MU or vehicle for the last 72 h (n = 4). I, cell count of KYSE-410 as determined by EpCAM+ cells in flow cytometry after 3 days in culture (n = 5). J, average migration speed of KYSE-410 cells in mono- and co-culture with CAF (n = 3). Data depicted as mean ± S.E. (error bars). *, p < 0.05; n.s., not significant.
There is evidence that CAF can affect cancer cells more potently than normal fibroblasts (42–44). Consequently, mRNA expression of EMT markers as well as proliferation and migration were also measured in co-cultures with KYSE-410 cells and CAF. However, no significant differences were detected (Fig. 9, E–J). There was a trend toward induced CDH1 mRNA expression in 4-MU-treated KYSE-410 cells compared with vehicle-treated cells.
HA Increased CD3+ and CD4+ Cell Adhesion
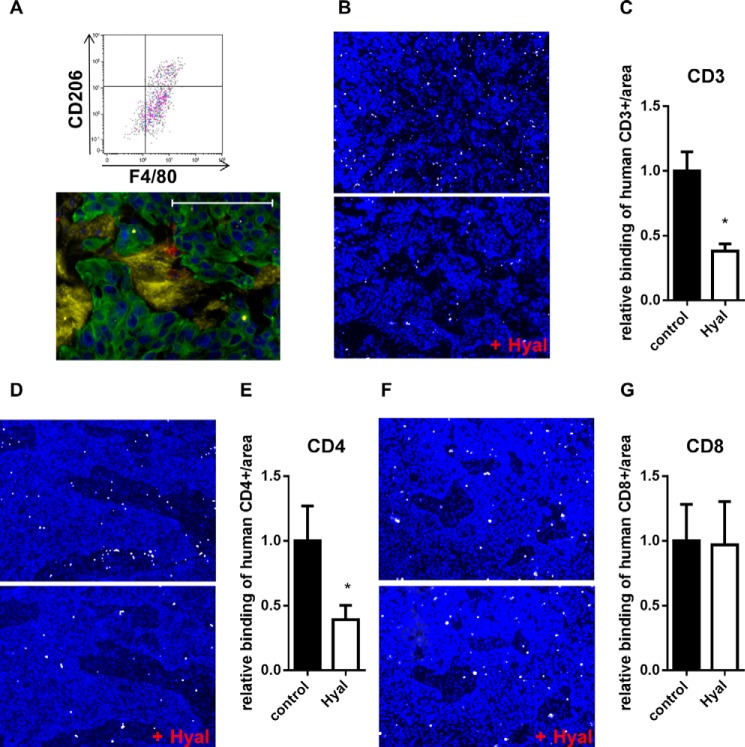
In xenograft tumors, some macrophages were detected by flow cytometry (CD11b+/F4/80+/CD206+ cells) and immunohistochemical staining of Mac2 (Fig. 10A). Because direct contact to cancer cells led to increased HA synthesis and chemokine secretion in fibroblasts, we analyzed whether this could be a synergistic mechanism with respect to the tumor immune response. Therefore, we investigated whether HA promotes the adhesion of immune cells that may be recruited to the tumor. CD14+, CD3+, CD4+, and CD8+ cells were isolated from human buffy coat, and their binding to xenograft tumors was quantified after hyaluronidase digestion or control treatment. The adhesion of CD14+ cells to the tissue did not depend on HA (data not shown). Interestingly, CD3+ lymphocyte subsets showed distinct adhesion patterns. Whereas HA increased CD4+ binding, CD8+ binding was independent of HA (Fig. 10, B and G).
FIGURE 10.
HA enhanced the adhesion of CD4+ cells to tumor tissues. A, detection of F4/80+/CD206+ cells in dissociated KYSE-410 xenograft tumors. Gating was performed on living CD45+/CD11b+ cells. Bottom, fluorescent staining of HA (yellow), Mac2 (red), and CK-18 (green) in KYSE-410 xenograft tumors. Scale bar, 100 μm. B, adhesion of activated CD3+ cells (white) to xenograft tumor sections with or without prior hyaluronidase (Hyal) digestion. Nuclei were stained with DAPI (blue). C, number of CD3+ cells bound to xenograft tumor sections normalized to the area and depicted as -fold of control (n = 6). D, binding of activated CD4+ cells (white) to xenograft tumor sections with or without prior Hyal digestion. Nuclei were stained with DAPI (blue). E, number of CD4+ cells bound to xenograft tumor sections normalized to the area and depicted as -fold of control (n = 6). F, adhesion of activated CD8+ cells (white) to control xenograft tumor sections or to sections that were digested with Hyal. DAPI staining is shown in blue. G, number of bound CD8+ cells to xenograft tumor sections normalized to the area and depicted as -fold of control (n = 6). Data are presented as mean ± S.E. (error bars). *, p < 0.05.
Discussion
This study shows that direct co-culture of KYSE-410 cells and fibroblasts induced Has2 mRNA expression in fibroblasts. In SF, this induction was mediated by direct cell-cell contact and dependent on LEF1, and it was associated with a myofibroblast phenotype. Additionally, CCL5 expression was up-regulated and CCL11 expression was down-regulated in fibroblasts. Similar effects of co-culture of CAF and KYSE-410 cells on chemokine expression were detected that were, however, modulated by 4-MU treatment.
It was shown previously (23, 24) that melanoma cell conditioned medium induces Has2 mRNA expression in fibroblasts. This is mediated by platelet-derived growth factor receptor-PI3K-AKT and MAPK pathways (23, 24). In this study, we show for the first time that LEF1 was required for the induction of Has2 mRNA expression in fibroblasts. LEF1 is both a target gene and downstream transcription factor of, for example, Wnt signaling. The fact that Wnt2 and Lef1 were up-regulated and the detection of nuclear β-catenin in SF could be signs of activated Wnt signaling in direct co-cultures. In line with this, it was reported previously (45) that highly active mutant β-catenin induces Has2 mRNA expression in prostate cancer cells. Moreover, in Madin-Darby canine kidney cells, β-catenin overexpression leads to the formation of a pericellular HA coat (18). In addition, coordinated induction of Has2os2 that was also dependent on LEF1 probably contributed to increased Has2 transcription, as described for human aortic smooth muscle cells (33).
In the experimental set-up presented in this paper, the induction of Has2 mRNA expression was dependent on direct cell-cell contact. Mechanistically, KYSE-410 cells may induce fibroblasts to produce autostimulatory Wnt proteins, such as Wnt2, upon direct cell-cell contact. Indeed, CAF isolated from ESCC tissue show increased Wnt2 and Wnt5a expression compared with normal fibroblasts (44). The reduced amount of αSMA in co-cultures with Has2 knockdown SF underlines the role of HA in myofibroblast differentiation. HAS2 overexpression alone does not induce the differentiation of fibroblasts (46). However, HA is required for a TGFβ-dependent αSMA stimulation in fibroblasts (47). It was shown that ESCC cell lines are able to induce a myofibroblast phenotype in fetal esophageal fibroblasts by TGFβ (48). Interestingly, modulation of αSMA by HA was only detected in SF. In co-cultures of KYSE-410 cells and CAF, 4-MU treatment did not affect the amount of αSMA. This may be due to an already differentiated phenotype of CAF.
Besides an up-regulation of Has2, induction of CCL5 was observed in co-cultures. These results are in agreement with the finding that CAF isolated from ESCC tissue have an increased CCL5 gene expression compared with normal fibroblasts (44). It was shown for co-cultures with MDA-MB-231 breast cancer cells or ovarian cancer cells that these cells can induce Ccl5 mRNA expression in mesenchymal stromal cells and normal fibroblasts, respectively (21, 49).
Importantly, the inhibition of HA synthesis by 4-MU decreased Ccl5 mRNA expression in co-cultures with CAF. Thus, in CAF, the induction of Has2/HA synthesis may modulate the chemokine expression. It was shown that low molecular weight HA induces CCL5 in macrophages (50). HA can signal via several receptors, such as CD44, RHAMM, and TLR-2 and -4. In keratinocytes, TLR-2 and -4 modulate CCL5 expression, because a knock-out of these receptors leads to decreased CCL5 in mouse tissue (51). Additionally, CD44 may be involved in CCL5 expression. IL-6 increases Has1, Has2, and Ccl5 mRNA expression in cardiac fibroblasts. By knockdown of CD44 in these cells, CCL5 was no longer significantly induced (52).
It was reported that CCL5 can directly affect cancer cells by promoting invasion and proliferation (41, 53). We assume that direct CCL5 effects on KYSE-410 cells are of minor relevance because the mRNA of the chemokine receptors CCR3 and -5 was not consistently expressed in the mono- and co-culture of KYSE-410 cells, and CCR1 mRNA expression was low and not affected by co-culture. This finding is supported by the fact that migration and proliferation of KYSE-410 cells were not increased in co-culture.
Potentially, the parallel regulation of Has2 and CCL5 acts synergistically on immune cells. CCL5 recruits mainly T lymphocytes and monocytes into the tumor (41). Also, HA is important for macrophage recruitment (39, 54), and low molecular weight HA leads to increased numbers of CD3+ lymphocytes (40). Low molecular weight HA can induce chemokine and cytokine expression of macrophages and act as an adjuvant to enhance the T cell response (50, 55, 56). Furthermore, cells can produce leukocyte-adhesive HA in cable-like structures upon various inflammatory stimuli (57). In xenograft tumor sections, HA did enhance the binding of CD3+ cells, particularly of CD4+ subsets. It is known that HA produced by poly(I:C)-stimulated lung fibroblasts plays a role in CD4+ T lymphocyte retention in vitro (58). In contrast, the binding of CD8+ cells was not affected by HA digestion. In many tumors, CD8+ cells are associated with a better prognosis because they mediate cytotoxic anti-tumor effects. CD4+ cells comprise heterogeneous subsets displaying differential influence on tumor progression. For example, T helper 1 (Th1) cells support cytotoxic anti-tumor effects, whereas regulatory T-cells (Treg cells) are described as being tumor-promoting by inhibiting cytotoxic immune responses, although in a meta-analysis of human studies, Treg cell infiltration was not associated with a poorer prognosis (59). For Treg cells, it was shown that high molecular weight HA enhances their function (60, 61), thereby promoting immune tolerance. This is another aspect of how HA may modulate the immune responses in tumors.
CCL5 is often described to exhibit tumor-promoting capacity. In a murine breast cancer model, CCL5 deletion results in attenuated tumor and metastasis burden and impairs Th2 polarization (62). However, in a murine breast cancer model with CCL5 knockdown in tumor cells only, no differences in tumor growth or metastasis occurred (63). However, in squamous cell carcinoma, knockdown of CCL5 leads to tumor regression and reduced numbers of recruited Treg cells (64). Further, knockdown of CCL5 in murine colorectal carcinoma cells delays tumor growth and decreases both Treg cell infiltration and CD8+ T-cell apoptosis (65). In contrast, in human ESCC tissue samples, CCL5 is correlated with increased CD8+ lymphocyte markers and a prolonged overall survival of patients (66). In this study, 4-MU decreased the Ccl5 mRNA expression in CAF co-cultures. This could be a novel mechanism of how 4-MU can exhibit anti-cancer activity.
The chemokine CCL11 was decreased in fibroblasts by co-culture. CCL11, also known as eotaxin-1, is involved in the recruitment of eosinophils. Additionally, it can recruit basophils and T helper 2 lymphocytes (67, 68). There are sparse data on the impact of CCL11 on cancer progression. On one hand, CCL11 was shown to promote the proliferation and invasion of ovarian cancer cells and to recruit endothelial cells that support angiogenesis (69). On the other hand, CCL11 overexpression results in decreased tumor growth in a murine model of hepatocellular carcinoma (70). In BALB/c CCL11 knock-out mice, growth of fibrosarcoma was induced compared with control (71). In CCL11 knock-out mice, IL-5 knock-out T helper 2 lymphocytes were unable to reduce melanoma lung metastasis (72).
In conclusion, KYSE-410 cells stimulated both normal fibroblasts and established CAF to produce an HA-rich and immunomodulatory microenvironment. This interaction may therefore result in tumor-promoting inflammation, and 4-MU treatment in CAF potentially disrupts these processes. In addition, interfering with Wnt signaling may result in an inhibition of Has2 induction.
Author Contributions
I. K. and J. W. F. designed the experiments and wrote the manuscript. Experiments were performed by I. K., T. F., S. T., and M. G., and data were analyzed by I. K. In addition, M. G. designed and analyzed the data shown in Fig. 10A. Y. Y. provided mice for the HAS2 knockdown model. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgment
We thank Petra Rompel for excellent technical assistance.
This study was supported by Deutsche Forschungsgemeinschaft Grants FI682/4-1 and GRK 1739. The authors declare that they have no conflicts of interest with the contents of this article.
- ECM
- extracellular matrix
- αSMA
- α smooth muscle actin
- CAF
- cancer-associated fibroblast(s)
- CDH1
- E-cadherin
- CK-18
- cytokeratin 18
- EMT
- epithelial-mesenchymal transition
- ESCC
- esophageal squamous cell carcinoma
- HA
- hyaluronan
- HAS
- HA synthase
- HAS2os
- HAS2 opposite strand
- LEF1
- lymphoid enhancer binding factor 1
- 4-MU
- 4-methylumbelliferone
- qPCR
- quantitative real-time PCR
- SF
- skin fibroblasts.
References
- 1. Servais C., and Erez N. (2013) From sentinel cells to inflammatory culprits: cancer-associated fibroblasts in tumour-related inflammation. J. Pathol 229, 198–207 [DOI] [PubMed] [Google Scholar]
- 2. Öhlund D., Elyada E., and Tuveson D. (2014) Fibroblast heterogeneity in the cancer wound. J. Exp. Med. 211, 1503–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cirri P., and Chiarugi P. (2011) Cancer associated fibroblasts: the dark side of the coin. Am. J. Cancer Res. 1, 482–497 [PMC free article] [PubMed] [Google Scholar]
- 4. Augsten M. (2014) Cancer-associated fibroblasts as another polarized cell type of the tumor microenvironment. Front. Oncol. 4, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Harper J., and Sainson R. C. (2014) Regulation of the anti-tumour immune response by cancer-associated fibroblasts. Semin. Cancer Biol. 25, 69–77 [DOI] [PubMed] [Google Scholar]
- 6. Auvinen P., Tammi R., Parkkinen J., Tammi M., Agren U., Johansson R., Hirvikoski P., Eskelinen M., and Kosma V. M. (2000) Hyaluronan in peritumoral stroma and malignant cells associates with breast cancer spreading and predicts survival. Am. J. Pathol 156, 529–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pirinen R., Tammi R., Tammi M., Hirvikoski P., Parkkinen J. J., Johansson R., Böhm J., Hollmén S., and Kosma V. M. (2001) Prognostic value of hyaluronan expression in non-small-cell lung cancer: Increased stromal expression indicates unfavorable outcome in patients with adenocarcinoma. Int. J. Cancer 95, 12–17 [DOI] [PubMed] [Google Scholar]
- 8. Twarock S., Freudenberger T., Poscher E., Dai G., Jannasch K., Dullin C., Alves F., Prenzel K., Knoefel W. T., Stoecklein N. H., Savani R. C., Homey B., and Fischer J. W. (2011) Inhibition of oesophageal squamous cell carcinoma progression by in vivo targeting of hyaluronan synthesis. Mol. Cancer 10, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Golshani R., Lopez L., Estrella V., Kramer M., Iida N., and Lokeshwar V. B. (2008) Hyaluronic acid synthase-1 expression regulates bladder cancer growth, invasion, and angiogenesis through CD44. Cancer Res. 68, 483–491 [DOI] [PubMed] [Google Scholar]
- 10. Udabage L., Brownlee G. R., Waltham M., Blick T., Walker E. C., Heldin P., Nilsson S. K., Thompson E. W., and Brown T. J. (2005) Antisense-mediated suppression of hyaluronan synthase 2 inhibits the tumorigenesis and progression of breast cancer. Cancer Res. 65, 6139–6150 [DOI] [PubMed] [Google Scholar]
- 11. Teng B. P., Heffler M. D., Lai E. C., Zhao Y. L., LeVea C. M., Golubovskaya V. M., and Bullarddunn K. M. (2011) Inhibition of hyaluronan synthase-3 decreases subcutaneous colon cancer growth by increasing apoptosis. Anticancer Agents Med. Chem. 11, 620–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tian X., Azpurua J., Hine C., Vaidya A., Myakishev-Rempel M., Ablaeva J., Mao Z., Nevo E., Gorbunova V., and Seluanov A. (2013) High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature 499, 346–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Itano N., Sawai T., Atsumi F., Miyaishi O., Taniguchi S., Kannagi R., Hamaguchi M., and Kimata K. (2004) Selective expression and functional characteristics of three mammalian hyaluronan synthases in oncogenic malignant transformation. J. Biol. Chem. 279, 18679–18687 [DOI] [PubMed] [Google Scholar]
- 14. Li Y., Li L., Brown T. J., and Heldin P. (2007) Silencing of hyaluronan synthase 2 suppresses the malignant phenotype of invasive breast cancer cells. Int. J. Cancer 120, 2557–2567 [DOI] [PubMed] [Google Scholar]
- 15. Liu N., Gao F., Han Z., Xu X., Underhill C. B., and Zhang L. (2001) Hyaluronan synthase 3 overexpression promotes the growth of TSU prostate cancer cells. Cancer Res. 61, 5207–5214 [PubMed] [Google Scholar]
- 16. Koyama H., Hibi T., Isogai Z., Yoneda M., Fujimori M., Amano J., Kawakubo M., Kannagi R., Kimata K., Taniguchi S., and Itano N. (2007) Hyperproduction of hyaluronan in neu-induced mammary tumor accelerates angiogenesis through stromal cell recruitment: possible involvement of versican/PG-M. Am. J. Pathol 170, 1086–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Simpson M. A., Wilson C. M., and McCarthy J. B. (2002) Inhibition of prostate tumor cell hyaluronan synthesis impairs subcutaneous growth and vascularization in immunocompromised mice. Am. J. Pathol 161, 849–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zoltan-Jones A., Huang L., Ghatak S., and Toole B. P. (2003) Elevated hyaluronan production induces mesenchymal and transformed properties in epithelial cells. J. Biol. Chem. 278, 45801–45810 [DOI] [PubMed] [Google Scholar]
- 19. Vicent S., Sayles L. C., Vaka D., Khatri P., Gevaert O., Chen R., Zheng Y., Gillespie A. K., Clarke N., Xu Y., Shrager J., Hoang C. D., Plevritis S., Butte A. J., and Sweet-Cordero E. A. (2012) Cross-species functional analysis of cancer-associated fibroblasts identifies a critical role for CLCF1 and IL-6 in non-small cell lung cancer in vivo. Cancer Res. 72, 5744–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rajaram M., Li J., Egeblad M., and Powers R. S. (2013) System-wide analysis reveals a complex network of tumor-fibroblast interactions involved in tumorigenicity. PLoS Genet. 9, e1003789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mitra A. K., Zillhardt M., Hua Y., Tiwari P., Murmann A. E., Peter M. E., and Lengyel E. (2012) MicroRNAs reprogram normal fibroblasts into cancer-associated fibroblasts in ovarian cancer. Cancer Discov. 2, 1100–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sakko A. J., Ricciardelli C., Mayne K., Tilley W. D., Lebaron R. G., and Horsfall D. J. (2001) Versican accumulation in human prostatic fibroblast cultures is enhanced by prostate cancer cell-derived transforming growth factor β1. Cancer Res. 61, 926–930 [PubMed] [Google Scholar]
- 23. Pasonen-Seppänen S., Takabe P., Edward M., Rauhala L., Rilla K., Tammi M., and Tammi R. (2012) Melanoma cell-derived factors stimulate hyaluronan synthesis in dermal fibroblasts by upregulating HAS2 through PDGFR-PI3K-AKT and p38 signaling. Histochem. Cell Biol. 138, 895–911 [DOI] [PubMed] [Google Scholar]
- 24. Willenberg A., Saalbach A., Simon J. C., and Anderegg U. (2012) Melanoma cells control HA synthesis in peritumoral fibroblasts via PDGF-AA and PDGF-CC: impact on melanoma cell proliferation. J. Invest. Dermatol. 132, 385–393 [DOI] [PubMed] [Google Scholar]
- 25. Matsumoto K., Li Y., Jakuba C., Sugiyama Y., Sayo T., Okuno M., Dealy C. N., Toole B. P., Takeda J., Yamaguchi Y., and Kosher R. A. (2009) Conditional inactivation of Has2 reveals a crucial role for hyaluronan in skeletal growth, patterning, chondrocyte maturation and joint formation in the developing limb. Development 136, 2825–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Seibler J., Zevnik B., Küter-Luks B., Andreas S., Kern H., Hennek T., Rode A., Heimann C., Faust N., Kauselmann G., Schoor M., Jaenisch R., Rajewsky K., Kühn R., and Schwenk F. (2003) Rapid generation of inducible mouse mutants. Nucleic Acids Res. 31, e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., and Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B. C., Remm M., and Rozen S. G. (2012) Primer3: new capabilities and interfaces. Nucleic Acids Res. 40, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., and Madden T. L. (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Flicek P., Amode M. R., Barrell D., Beal K., Billis K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fitzgerald S., Gil L., Girón C. G., Gordon L., Hourlier T., Hunt S., Johnson N., Juettemann T., Kähäri A. K., Keenan S., Kulesha E., Martin F. J., Maurel T., McLaren W. M., Murphy D. N., Nag R., Overduin B., Pignatelli M., Pritchard B., Pritchard E., Riat H. S., Ruffier M., Sheppard D., Taylor K., Thormann A., Trevanion S. J., Vullo A., Wilder S. P., Wilson M., Zadissa A., Aken B. L., Birney E., Cunningham F., Harrow J., Herrero J., Hubbard T. J., Kinsella R., Muffato M., Parker A., Spudich G., Yates A., Zerbino D. R., and Searle S. M. (2014) Ensembl 2014. Nucleic Acids Res. 42, D749–D755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., and Werner T. (2005) MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21, 2933–2942 [DOI] [PubMed] [Google Scholar]
- 32. Jamieson C., Sharma M., and Henderson B. R. (2012) Wnt signaling from membrane to nucleus: β-catenin caught in a loop. Int. J. Biochem. Cell Biol. 44, 847–850 [DOI] [PubMed] [Google Scholar]
- 33. Vigetti D., Deleonibus S., Moretto P., Bowen T., Fischer J. W., Grandoch M., Oberhuber A., Love D. C., Hanover J. A., Cinquetti R., Karousou E., Viola M., D'Angelo M. L., Hascall V. C., De Luca G., and Passi A. (2014) Natural antisense transcript for hyaluronan synthase 2 (HAS2-AS1) induces transcription of HAS2 via protein O-GlcNAcylation. J. Biol. Chem. 289, 28816–28826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Michael D. R., Phillips A. O., Krupa A., Martin J., Redman J. E., Altaher A., Neville R. D., Webber J., Kim M. Y., and Bowen T. (2011) The human hyaluronan synthase 2 (HAS2) gene and its natural antisense RNA exhibit coordinated expression in the renal proximal tubular epithelial cell. J. Biol. Chem. 286, 19523–19532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maeda-Sano K., Gotoh M., Morohoshi T., Someya T., Murofushi H., and Murakami-Murofushi K. (2014) Cyclic phosphatidic acid and lysophosphatidic acid induce hyaluronic acid synthesis via CREB transcription factor regulation in human skin fibroblasts. Biochim. Biophys. Acta 1841, 1256–1263 [DOI] [PubMed] [Google Scholar]
- 36. Röck K., Grandoch M., Majora M., Krutmann J., and Fischer J. W. (2011) Collagen fragments inhibit hyaluronan synthesis in skin fibroblasts in response to ultraviolet B (UVB): new insights into mechanisms of matrix remodeling. J. Biol. Chem. 286, 18268–18276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marieb E. A., Zoltan-Jones A., Li R., Misra S., Ghatak S., Cao J., Zucker S., and Toole B. P. (2004) Emmprin promotes anchorage-independent growth in human mammary carcinoma cells by stimulating hyaluronan production. Cancer Res. 64, 1229–1232 [DOI] [PubMed] [Google Scholar]
- 38. Xu J., Lu Y., Qiu S., Chen Z. N., and Fan Z. (2013) A novel role of EMMPRIN/CD147 in transformation of quiescent fibroblasts to cancer-associated fibroblasts by breast cancer cells. Cancer Lett. 335, 380–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kobayashi N., Miyoshi S., Mikami T., Koyama H., Kitazawa M., Takeoka M., Sano K., Amano J., Isogai Z., Niida S., Oguri K., Okayama M., McDonald J. A., Kimata K., Taniguchi S., and Itano N. (2010) Hyaluronan deficiency in tumor stroma impairs macrophage trafficking and tumor neovascularization. Cancer Res. 70, 7073–7083 [DOI] [PubMed] [Google Scholar]
- 40. Alaniz L., Rizzo M., Malvicini M., Jaunarena J., Avella D., Atorrasagasti C., Aquino J. B., Garcia M., Matar P., Silva M., and Mazzolini G. (2009) Low molecular weight hyaluronan inhibits colorectal carcinoma growth by decreasing tumor cell proliferation and stimulating immune response. Cancer Lett. 278, 9–16 [DOI] [PubMed] [Google Scholar]
- 41. Aldinucci D., and Colombatti A. (2014) The inflammatory chemokine CCL5 and cancer progression. Mediators Inflamm. 2014, 292376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Soon P. S., Kim E., Pon C. K., Gill A. J., Moore K., Spillane A. J., Benn D. E., and Baxter R. C. (2013) Breast cancer-associated fibroblasts induce epithelial-to-mesenchymal transition in breast cancer cells. Endocr. Relat. Cancer 20, 1–12 [DOI] [PubMed] [Google Scholar]
- 43. Kim S. H., Choe C., Shin Y. S., Jeon M. J., Choi S. J., Lee J., Bae G. Y., Cha H. J., and Kim J. (2013) Human lung cancer-associated fibroblasts enhance motility of non-small cell lung cancer cells in co-culture. Anticancer Res. 33, 2001–2009 [PubMed] [Google Scholar]
- 44. Zhang C., Fu L., Fu J., Hu L., Yang H., Rong T. H., Li Y., Liu H., Fu S. B., Zeng Y. X., and Guan X. Y. (2009) Fibroblast growth factor receptor 2-positive fibroblasts provide a suitable microenvironment for tumor development and progression in esophageal carcinoma. Clin. Cancer Res. 15, 4017–4027 [DOI] [PubMed] [Google Scholar]
- 45. Wan X., Liu J., Lu J. F., Tzelepi V., Yang J., Starbuck M. W., Diao L., Wang J., Efstathiou E., Vazquez E. S., Troncoso P., Maity S. N., and Navone N. M. (2012) Activation of β-catenin signaling in androgen receptor-negative prostate cancer cells. Clin. Cancer Res. 18, 726–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Simpson R. M., Wells A., Thomas D., Stephens P., Steadman R., and Phillips A. (2010) Aging fibroblasts resist phenotypic maturation because of impaired hyaluronan-dependent CD44/epidermal growth factor receptor signaling. Am. J. Pathol. 176, 1215–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Webber J., Jenkins R. H., Meran S., Phillips A., and Steadman R. (2009) Modulation of TGFβ1-dependent myofibroblast differentiation by hyaluronan. Am. J. Pathol. 175, 148–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Noma K., Smalley K. S., Lioni M., Naomoto Y., Tanaka N., El-Deiry W., King A. J., Nakagawa H., and Herlyn M. (2008) The essential role of fibroblasts in esophageal squamous cell carcinoma-induced angiogenesis. Gastroenterology 134, 1981–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mi Z., Bhattacharya S. D., Kim V. M., Guo H., Talbot L. J., and Kuo P. C. (2011) Osteopontin promotes CCL5-mesenchymal stromal cell-mediated breast cancer metastasis. Carcinogenesis 32, 477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hodge-Dufour J., Noble P. W., Horton M. R., Bao C., Wysoka M., Burdick M. D., Strieter R. M., Trinchieri G., and Puré E. (1997) Induction of IL-12 and chemokines by hyaluronan requires adhesion-dependent priming of resident but not elicited macrophages. J. Immunol. 159, 2492–2500 [PubMed] [Google Scholar]
- 51. Suga H., Sugaya M., Fujita H., Asano Y., Tada Y., Kadono T., and Sato S. (2014) TLR4, rather than TLR2, regulates wound healing through TGF-beta and CCL5 expression. J. Dermatol. Sci. 73, 117–124 [DOI] [PubMed] [Google Scholar]
- 52. Müller J., Gorressen S., Grandoch M., Feldmann K., Kretschmer I., Lehr S., Ding Z., Schmitt J. P., Schrader J., Garbers C., Heusch G., Kelm M., Scheller J., and Fischer J. W. (2014) Interleukin-6-dependent phenotypic modulation of cardiac fibroblasts after acute myocardial infarction. Basic Res. Cardiol. 109, 440. [DOI] [PubMed] [Google Scholar]
- 53. Karnoub A. E., Dash A. B., Vo A. P., Sullivan A., Brooks M. W., Bell G. W., Richardson A. L., Polyak K., Tubo R., and Weinberg R. A. (2007) Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature 449, 557–563 [DOI] [PubMed] [Google Scholar]
- 54. Jameson J. M., Cauvi G., Sharp L. L., Witherden D. A., and Havran W. L. (2005) Gammadelta T cell-induced hyaluronan production by epithelial cells regulates inflammation. J. Exp. Med. 201, 1269–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Scheibner K. A., Lutz M. A., Boodoo S., Fenton M. J., Powell J. D., and Horton M. R. (2006) Hyaluronan fragments act as an endogenous danger signal by engaging TLR2. J. Immunol. 177, 1272–1281 [DOI] [PubMed] [Google Scholar]
- 56. Black K. E., Collins S. L., Hagan R. S., Hamblin M. J., Chan-Li Y., Hallowell R. W., Powell J. D., and Horton M. R. (2013) Hyaluronan fragments induce IFNβ via a novel TLR4-TRIF-TBK1-IRF3-dependent pathway. J. Inflamm. 10, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tammi R. H., Passi A. G., Rilla K., Karousou E., Vigetti D., Makkonen K., and Tammi M. I. (2011) Transcriptional and post-translational regulation of hyaluronan synthesis. FEBS J. 278, 1419–1428 [DOI] [PubMed] [Google Scholar]
- 58. Evanko S. P., Potter-Perigo S., Bollyky P. L., Nepom G. T., and Wight T. N. (2012) Hyaluronan and versican in the control of human T-lymphocyte adhesion and migration. Matrix Biol. 31, 90–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gooden M. J., de Bock G. H., Leffers N., Daemen T., and Nijman H. W. (2011) The prognostic influence of tumour-infiltrating lymphocytes in cancer: a systematic review with meta-analysis. Br. J. Cancer 105, 93–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bollyky P. L., Falk B. A., Long S. A., Preisinger A., Braun K. R., Wu R. P., Evanko S. P., Buckner J. H., Wight T. N., and Nepom G. T. (2009) CD44 costimulation promotes FoxP3+ regulatory T cell persistence and function via production of IL-2, IL-10, and TGF-β. J. Immunol. 183, 2232–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bollyky P. L., Lord J. D., Masewicz S. A., Evanko S. P., Buckner J. H., Wight T. N., and Nepom G. T. (2007) Cutting edge: high molecular weight hyaluronan promotes the suppressive effects of CD4+CD25+ regulatory T cells. J. Immunol. 179, 744–747 [DOI] [PubMed] [Google Scholar]
- 62. Zhang Q., Qin J., Zhong L., Gong L., Zhang B., Zhang Y., and Gao W. Q. (2015) CCL5-mediated Th2 immune polarization promotes metastasis in luminal breast cancer. Cancer Res. 75, 4312–4321 [DOI] [PubMed] [Google Scholar]
- 63. Jayasinghe M. M., Golden J. M., Nair P., O'Donnell C. M., Werner M. T., and Kurt R. A. (2008) Tumor-derived CCL5 does not contribute to breast cancer progression. Breast Cancer Res. Treat. 111, 511–521 [DOI] [PubMed] [Google Scholar]
- 64. Serrels A., Lund T., Serrels B., Byron A., McPherson R. C., von Kriegsheim A., Gómez-Cuadrado L., Canel M., Muir M., Ring J. E., Maniati E., Sims A. H., Pachter J. A., Brunton V. G., Gilbert N., Anderton S. M., Nibbs R. J., and Frame M. C. (2015) Nuclear FAK controls chemokine transcription, Tregs, and evasion of anti-tumor immunity. Cell 163, 160–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chang L. Y., Lin Y. C., Mahalingam J., Huang C. T., Chen T. W., Kang C. W., Peng H. M., Chu Y. Y., Chiang J. M., Dutta A., Day Y. J., Chen T. C., Yeh C. T., and Lin C. Y. (2012) Tumor-derived chemokine CCL5 enhances TGF-β-mediated killing of CD8(+) T cells in colon cancer by T-regulatory cells. Cancer Res. 72, 1092–1102 [DOI] [PubMed] [Google Scholar]
- 66. Liu J., Li F., Ping Y., Wang L., Chen X., Wang D., Cao L., Zhao S., Li B., Kalinski P., Thorne S. H., Zhang B., and Zhang Y. (2015) Local production of the chemokines CCL5 and CXCL10 attracts CD8+ T lymphocytes into esophageal squamous cell carcinoma. Oncotarget 6, 24978–24989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Menzies-Gow A., Ying S., Sabroe I., Stubbs V. L., Soler D., Williams T. J., and Kay A. B. (2002) Eotaxin (CCL11) and eotaxin-2 (CCL24) induce recruitment of eosinophils, basophils, neutrophils, and macrophages as well as features of early- and late-phase allergic reactions following cutaneous injection in human atopic and nonatopic volunteers. J. Immunol. 169, 2712–2718 [DOI] [PubMed] [Google Scholar]
- 68. Sallusto F., Mackay C. R., and Lanzavecchia A. (1997) Selective expression of the eotaxin receptor CCR3 by human T helper 2 cells. Science 277, 2005–2007 [DOI] [PubMed] [Google Scholar]
- 69. Salcedo R., Young H. A., Ponce M. L., Ward J. M., Kleinman H. K., Murphy W. J., and Oppenheim J. J. (2001) Eotaxin (CCL11) induces in vivo angiogenic responses by human CCR3+ endothelial cells. J. Immunol. 166, 7571–7578 [DOI] [PubMed] [Google Scholar]
- 70. Kataoka S., Konishi Y., Nishio Y., Fujikawa-Adachi K., and Tominaga A. (2004) Antitumor activity of eosinophils activated by IL-5 and eotaxin against hepatocellular carcinoma. DNA Cell Biol. 23, 549–560 [DOI] [PubMed] [Google Scholar]
- 71. Simson L., Ellyard J. I., Dent L. A., Matthaei K. I., Rothenberg M. E., Foster P. S., Smyth M. J., and Parish C. R. (2007) Regulation of carcinogenesis by IL-5 and CCL11: a potential role for eosinophils in tumor immune surveillance. J. Immunol. 178, 4222–4229 [DOI] [PubMed] [Google Scholar]
- 72. Mattes J., Hulett M., Xie W., Hogan S., Rothenberg M. E., Foster P., and Parish C. (2003) Immunotherapy of cytotoxic T cell-resistant tumors by T helper 2 cells: an eotaxin and STAT6-dependent process. J. Exp. Med. 197, 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]