Abstract
Misfolding of Cu,Zn-superoxide dismutase (SOD1) is a pathological change in the familial form of amyotrophic lateral sclerosis caused by mutations in the SOD1 gene. SOD1 is an enzyme that matures through the binding of copper and zinc ions and the formation of an intramolecular disulfide bond. Pathogenic mutations are proposed to retard the post-translational maturation, decrease the structural stability, and hence trigger the misfolding of SOD1 proteins. Despite this, a misfolded and potentially pathogenic conformation of immature SOD1 remains obscure. Here, we show significant and distinct conformational changes of apoSOD1 that occur only upon reduction of the intramolecular disulfide bond in solution. In particular, loop regions in SOD1 lose their restraint and become significantly disordered upon dissociation of metal ions and reduction of the disulfide bond. Such drastic changes in the solution structure of SOD1 may trigger misfolding and fibrillar aggregation observed as pathological changes in the familial form of amyotrophic lateral sclerosis.
Keywords: amyotrophic lateral sclerosis (ALS) (Lou Gehrig disease), protein aggregation, protein folding, protein misfolding, superoxide dismutase (SOD)
Introduction
Mutations in Cu,Zn-superoxide dismutase (SOD1)2 are linked to familial forms of amyotrophic lateral sclerosis (fALS) (1). A major pathological change observed in SOD1-related fALS is the abnormal accumulation of misfolded mutant SOD1 proteins in affected motor neurons (2). Actually, many in vivo as well as in vitro studies have supported that pathogenic mutations facilitate the misfolding of SOD1 proteins (3); however, the molecular mechanism triggering the misfolding of SOD1 remains controversial.
SOD1 is known as one of the most stable proteins to the extent that its melting temperature (Tm) is >90 °C (4); therefore, a misfolding event appears quite unlikely for SOD1. Nonetheless, SOD1 was found to have acquired such high stability through several post-translational processes including copper and zinc binding and disulfide bond formation (Fig. 1A). Actually, disulfide-reduced apoSOD1 exhibits significantly decreased stability (Tm ∼ 42 °C) and is susceptible to unfolding/misfolding at physiological temperatures (5, 6). Intracellular deregulation of metal binding and/or disulfide formation will, hence, be a key event triggering the misfolding of SOD1.
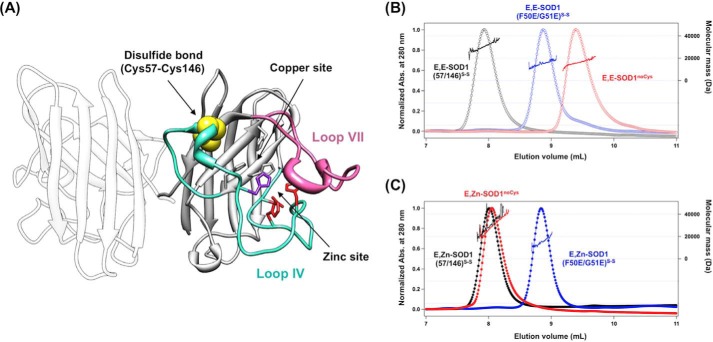
FIGURE 1.
The most immature state of SOD1 is monomeric, but its conformation is significantly different from that of an SOD1 protomer. A, a crystal structure of SOD1 homodimer (PDB ID 1HL5). Loops IV (cyan) and VII (pink) of SOD1 are indicated in the crystal structure, and the disulfide bond between Cys-57 and Cys-146 is shown in yellow. Ligands for binding copper and zinc ions are also shown: gray, His-46, His-48, and His-120 for copper binding; red, His-71, His-80, Asp-83 for zinc binding; purple, His63 for copper and zinc binding. B and C, gel filtration chromatograms of E,E-SOD1noCys (red open circles), E,E-SOD1(57/146)S-S (black open circles), and E,E-SOD1(F50E/G51E)S-S (blue open circles) (B) and E,Zn-SOD1noCys (red-filled circles), E,Zn-SOD1(57/146)S-S (black-filled circles), and E,Zn-SOD1(F50E/G51E)S-S (blue-filled circles) (C) were shown. A sample containing 60 μm protein in 100 mm Na-Pi, 100 mm NaCl at pH 7.0 was loaded on the gel filtration column, and the elution profile was monitored with absorbance (Abs.) change at 280 nm (left axis). Molecular mass obtained by MALS analysis is also shown in each chromatogram (right axis).
Notably, many pathogenic mutations are found to disturb the post-translational control of SOD1 maturation (7, 8) and thereby increase intracellular fractions of the apo- (9) and/or disulfide-reduced state (10). Only when both metal ions and disulfide bond are absent, SOD1 forms fibrillar aggregates (11). Given that SOD1 fibrillation is a pathological hallmark in SOD1-related fALS patients (12) as well as model mice (13), the most immature form of SOD1 will provide a clue to understand the molecular pathomechanism of this devastating disease. In a number of previous studies, the roles of metal binding and disulfide formation in the misfolding of SOD1 have been suggested by a variety of experimental methods (6, 8, 11, 14–19); however, conformational information on SOD1 in solution lacking both metal ions and the disulfide bond, which is the only state accessible to fibrillar aggregates (11), is still limited. Also, a crystal structure of apoSOD1 lacking the disulfide bond has implied little impacts of metal binding and disulfide formation on its overall folding pattern albeit with increased disorder at loop regions (19). This is probably because the crystallization process sorts the folded conformation out of many other conformations adopted by immature SOD1 in solution. Indeed, significant changes in chemical shifts were concentrated on loop regions (loops IV and VII; Fig. 1A) between the solid state and solution NMR spectra of apoSOD1 with a disulfide bond in crystalline and solution state, respectively (20). Nonetheless, the degree of disorder in the loop regions of immature SOD1 is still ambiguous. To understand the molecular mechanism of SOD1 misfolding in SOD1-related fALS, conformational features of apoSOD1 lacking a disulfide bond, which is prone to fibrillation, therefore need to be clarified in more detail.
Here, we have investigated conformational features of the most immature SOD1 lacking both metal ions and the disulfide bond by using spectroscopic and scattering methods. Although metallated SOD1 with a disulfide bond exists as a homodimer, apoSOD1 without the disulfide bond has been shown to favor a monomeric state and is considered to adopt a protomer conformation (21). In this study we nonetheless found that a conformation of disulfide-null apoSOD1 in solution was significantly distinct from that of the SOD1 protomer; in particular, loop regions usually connected via bound metal ions and a disulfide bond (loops IV and VII; Fig. 1A) lost their restraint and were significantly disordered upon both dissociation of metal ions and reduction of the disulfide bond. Such distinct conformational disorder of SOD1 realized only in its most immature state will be discussed in relation to the pathological changes observed in SOD1-related fALS cases.
Experimental Procedures
Preparation of SOD1 Proteins
Escherichia coli SHuffleTM (New England BioLabs) was transformed with a pET15b plasmid containing cDNA of human SOD1 with an N-terminal His6 tag, and the expression of His-tagged SOD1 proteins was induced by culturing the transformed cells in the presence of 0.1 mm isopropyl β-d-1-thiogalactopyranoside at 20 °C for 16 h. His-tagged SOD1 proteins in the lysates were purified with a HisTrap HP (1 ml, GE Healthcare) and dialyzed against 50 mm NaOAc, 100 mm NaCl, 50 mm EDTA, pH 4.0, at 4 °C for 16 h so as to remove the metal ions bound to SOD1. The resultant apoSOD1 was further dialyzed against 100 mm Na-Pi, 100 mm NaCl/5 mm EDTA, pH 7.0 (NNE buffer), and then incubated with thrombin at 37 °C for 3 h. Thrombin was removed from the sample solution using HiTrap benzamidine (1 ml, GE Healthcare), and the purified apoSOD1 was obtained using gel filtration column chromatography (Cosmosil 5Diol-300-II, Nacalai Tesque) with NNE buffer as the running buffer. For preparation of the Zn2+-bound form of SOD1, the NNE buffer of apoSOD1 samples was first exchanged to a Chelex-treated buffer without EDTA, and then an equimolar amount of ZnSO4 was added to apoSOD1.
Concentrations of copper and zinc ions in the samples were determined by using Metallo Assay Copper LS (Metallogenics) and Metallo Assay Zinc LS (Metallogenics), respectively. Almost no contamination of copper and zinc ions (<1 mol % of total SOD1 concentrations) was confirmed in apoSOD1 samples examined here. The thiol-disulfide status of SOD1 was further confirmed by SDS-PAGE; after protection with a thiol-specific modifier, iodoacetamide, SOD1 samples were resolved by non-reducing SDS-PAGE. Electrophoretic mobility of SOD1 has been known to increase in the presence of the conserved intramolecular disulfide bond (22), and we thereby confirmed that our SOD1S-S samples had the intramolecular disulfide bond (data not shown).
Size-exclusion Chromatography with an Online Multi-angle Light Scattering (SEC-MALS)
60 μm (∼1 g/liter) SOD1 was loaded on a gel filtration column (TSKgel G2000SW, TOSOH) fitted to an HPLC system (Shimadzu), and the absorbance change at 280 nm of the elution was monitored. For the analysis of apoSOD1, 5 mm EDTA was included in the running buffer (100 mm Na-Pi, 100 mm NaCl, pH 7.0) to prevent the adventitious loading of metal ions to SOD1 during gel filtration. A molecular size of the protein eluted from the column was determined by multi-angle light scattering using miniDAWN TREOS (WYATT Technology) connected online to the HPLC system.
Aggregation Assay
The aggregation of SOD1 proteins was monitored by the increase of solution turbidity. 20 μm E,E-SOD1noCys and E,E-SOD1(57/146)S-S in NNE buffer were set in a 96-well plate and agitated with a POM ball (3/32 inch, SANPLATEC) at 1200 rpm at 37 °C. For the examination of Zn2+-bound SOD1, 5 mm EDTA in NNE buffer was replaced with 0.2 mm ZnSO4. Solution turbidity was monitored by absorbance change at 350 nm using a plate reader (Epoch, BioTek).
Spectroscopic Methods
For measurement of circular dichroism spectra, SOD1 samples (10 and 400 μm for far-UV and near-UV regions, respectively) were prepared in a Chelex-treated buffer (20 mm Na-Pi, 50 mm NaCl, pH 8.0), and a J-720WI spectropolarimeter (Jasco) was used. Fluorescence spectra of 5 μm SOD1 in NNE buffer were measured using fluorescence spectrophotometry (F-4500, Hitachi) with excitation at 282 nm. Fourier transform infrared (FTIR) spectra were measured by using an IRAffinity-1S spectrophotometer (Shimadzu) attached with an attenuated total reflection accessory (DuraSamplIR II, nine reflections). SOD1 proteins (∼1 mm) were demetallated by precipitation with trichloroacetic acid and redissolved in a deuterated buffer of 100 mm Tris, 100 mm NaCl at pD 7.0.
Nuclear Magnetic Resonance
15N-Labeled SOD1 proteins were prepared by culturing E. coli SHuffleTM harboring the plasmid for expression of His-tagged SOD1, and the protein expression was induced with 0.5 mm isopropyl β-d-1-thiogalactopyranoside in M9 minimal media containing 15NH4Cl, 3 μm ZnSO4, and 30 μm CuSO4 at 20 °C for 43 h. His tag-free demetallated SOD1 proteins were purified as described above. Using ZebaTM Spin Desalting Columns (ThermoFisher), the NNE buffer in purified SOD1 samples was exchanged to Chelex-treated buffer containing 100 mm Na-Pi and 100 mm NaCl at pH 7.4 (NN buffer).
NMR experiments were done at 283 K using Bruker AVANCE III 600 MHz spectrometer and Agilent UNITY Inova 600 MHz equipped with a cold probe. The spectra were processed using NMRPipe (23), and the data analysis was performed with Olivia 1.16.9.3 15N-Labeled E,E-SOD1(57/146)S-S and E,E-SOD1noCys in NN buffer (∼3 mm) were diluted 10× with NN buffer containing 10% D2O, and the 1H,15N heteronuclear single quantum correlation (HSQC) spectra were then measured as a reference. For hydrogen-deuterium exchange experiments, the 15N-labeled proteins (∼3 mm) were diluted 10× with a D2O buffer containing 100 mm Na-Pi and 100 mm NaCl at pD 7.4. The 1H,15N HSQC spectra were collected 5 min after dilution, and the spectral changes were monitored every 4 h. The NMR signals were analyzed based on a previously published assignment table (25–27). Using the resonance intensities before H/D exchange as a reference, relative intensities were calculated.
Small-angle X-ray Scattering (SAXS)
SAXS data were obtained using NANO-Viewer (Rigaku) equipped with a high brightness x-ray generator, RA-Micro7 (Rigaku), and a hybrid pixel array detector, PILATUS 200K (DECTRIS). SAXS measurements were performed at 10 °C using a series of samples serially diluted from 6.58 to 2.53 g/liter (SOD1noCys) or 8.64 to 3.11 g/liter (SOD1(57/146)) in 50 mm Tris, 100 mm NaCl, 5 mm EDTA at pH 7.0. To remove any oligomeric/aggregated species, the E,E-SOD1noCys samples were first loaded on the gel filtration column (G2000SW, TOSOH) equilibrated with 50 mm Tris, 100 mm NaCl, 5 mm EDTA at pH 7.0, and protein fractions of a symmetrical and sharp elution peak was collected and immediately used for SAXS measurements. A successive series of scattering images (3 min × 10 frames) was recorded, and then only the images free from radiation damages (10 frames in all experiments) were used to obtain scattering curves. The circular averaged data were normalized by exposure time (3 × 10 min) and protein concentration, thereby the scattering curves, I(Q), were obtained, where Q = 4πsin(θ)/λ, 2θ is the scattering angle, and λ is the wavelength of the x-ray (1.5418 Å). I(Q) at very low angle was fitted with the Guinier approximation using the equation,
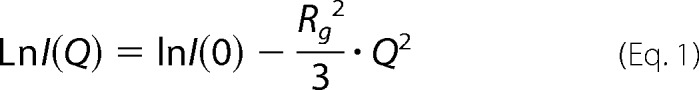 |
where I(0) and Rg are the forward scattering intensity (Q = 0) and the radius of gyration, respectively. Experimental SAXS data were manipulated using PRIMUS (28) and GNOM (29). The theoretical Rg value of the crystal structure of SOD1 (PDB ID 2C9V) was calculated using CRYSOL (30).
Shape Reconstructions
Low-resolution shapes of E,E-SOD1noCys were restored from SAXS data using GASBOR (31), scored with DAMAVER (32), and visualized with the SITUS package (33). The rigid-body and ensemble models of E,E-SOD1noCys were refined by using BUNCH (34) and EOM (35), respectively. During both refinements the β-barrel scaffold (residues 5–52 and 84–124) of E,E-SOD1noCys was treated as a rigid body harboring three flexible loops in its N terminus (residues 1–4), a region including loop IV (residues 53–83), and a region from loop VII to the C terminus (residues 125–157). Coordinates of the rigid body were taken from an x-ray crystal structure of SOD1 (PDB ID 2C9V). The flexible loops were first assumed as dummy residues, and the conformations were then refined against the experimental SAXS data while keeping the rigid body structure intact. In the ensemble modeling, 10,000 hypothetical rigid-body models were randomly generated, from which a plausible ensemble was suggested by refining populations and compositions of the hypothetical structures.
Results and Discussion
SOD1 has four Cys residues in total, among which Cys-57 and Cys-146 form an intramolecular disulfide bond (Fig. 1A). The other two Cys residues (Cys-6 and Cys-111) do not form disulfide bonds in native SOD1 but are susceptible to aberrant oxidation (36). To avoid oxidative modifications during experiments, Cys-6 and -111 of SOD1 in this study were mutated to Ser (SOD1(57/146)); the presence of a Cys-57–Cys-146 disulfide bond in SOD1(57/146) was confirmed electrophoretically (SOD1(57/146)S-S) (data not shown). Also, SOD1 in which all four Cys residues were mutated to Ser (SOD1noCys) was used as the model of SOD1 lacking the disulfide bond.
SOD1 Becomes a Monomer with Decreased Contents of Secondary Structures upon Losing Both Metal Ions and the Disulfide Bond
To understand conformational features of the most immature SOD1, a quaternary structure of apoSOD1noCys (E,E-SOD1noCys) was examined by SEC-MALS. Native SOD1 is a homodimeric protein (Fig. 1A), but the monomer-dimer equilibrium has been known to be regulated by the binding of metal ions and the formation of the disulfide bond (21, 37). Indeed, we have successfully reproduced our previous findings (21); E,E-SOD1noCys was monomeric with a molecular mass of 16.0 kDa and that either the introduction of a disulfide bond (E,E-SOD1(57/146)S-S) or the binding of a Zn2+ ion (E,Zn-SOD1noCys) was sufficient to render SOD1 dimeric with a 32.0-kDa molecular mass (Fig. 1, B and C). Previous studies have shown that a protomer of the SOD1 dimer can be prepared with F50E/G51E double mutations (38); indeed, our SEC-MALS analysis also showed that both apo- and Zn2+-bound forms of SOD1(F50E/G51E) with a disulfide bond were monomeric with a 16.0-kDa molecular mass (Fig. 1, B and C). Quite notably, however, the elution volume of SOD1(F50E/G51E)S-S (8.87 ml) was significantly smaller than that of E,E-SOD1noCys (9.39 ml) even though both SOD1 species were monomeric (Fig. 1B). It is thus expected that monomeric SOD1 in the most immature state adopts a conformation that is significantly altered from that of a protomer of native SOD1 dimer.
To further confirm the roles of metal binding and disulfide formation in the conformation of SOD1, we have performed secondary structural analysis on the distinct metallation/thiol-disulfide status of SOD1 using circular dichroism (CD) spectroscopy. Matured SOD1 (i.e. a copper- and zinc-bound state with a disulfide bond, Cu,Zn-SOD1(57/146)S-S) exhibits a negative CD signal at 208 nm with a distinct shoulder around 230 nm, consistent with the β-barrel structure of SOD1 (data not shown and Ref. 39). Removing copper ion from Cu,Zn-SOD1(57/146)S-S did not change the CD spectrum (E,Zn-SOD1(57/146)S-S), suggesting limited effects of copper ion to the secondary structure of SOD1 (data not shown and Fig. 2A). In contrast, the binding of a zinc ion and the formation of a disulfide bond were found to play significant roles in attaining the secondary structures (Fig. 2A); upon dissociation of a zinc ion, the peak around 230 nm became vague, and a negative CD signal around 200 nm increased its intensity. In E,E-SOD1noCys (red open circles, Fig. 2A) in particular, a negative CD signal at 208 nm that was evident in other SOD1 species examined in this study was lost, and an additional signal was observed around 200 nm, implying the presence of random coils (40).
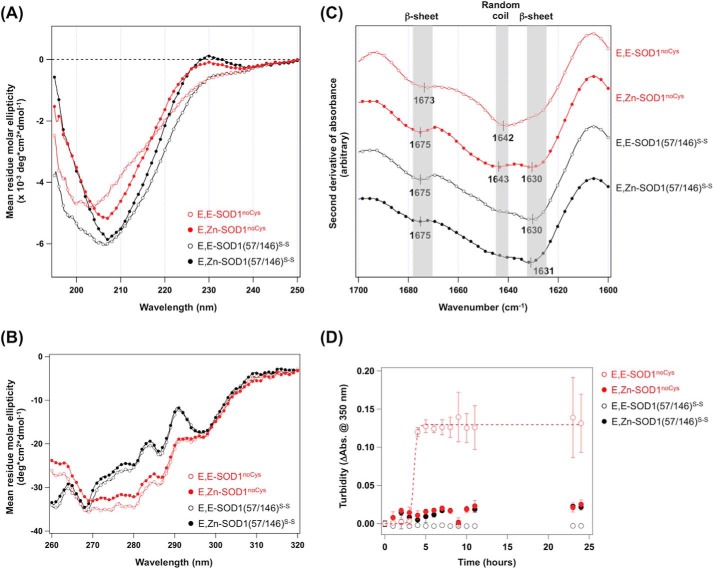
FIGURE 2.
Contents of secondary structures were decreased in the most immature and aggregation-prone state of SOD1. Far-UV CD spectra (A), near-UV CD spectra (B), FTIR spectra (in second derivatives) (C), and aggregation kinetics of E,E-SOD1noCys (red open circles), E,Zn-SOD1noCys (red-filled circles), E,E-SOD1(57/146)S-S (black open circles), and E,Zn-SOD1(57/146)S-S (black-filled circles) (D) are shown. The samples for CD measurements contained 10 μm (far UV) and 400 μm (near UV) proteins in 20 mm Na-Pi, 50 mm NaCl at pH 8.0, whereas FTIR spectra were measured by using 1 mm protein in 100 mm Tris, 100 mm NaCl, pD 7.0. Aggregation kinetics was examined by using 20 μm SOD1 and monitored by the increase of solution turbidity. Abs., absorbance.
We also measured the CD spectra in the near-UV region and thereby attempted to characterize the effects of disulfide formation and Zn2+ binding on the tertiary structure of SOD1. A near-UV CD spectrum reflects the environments around aromatic side chains (Trp, Phe, Tyr) and disulfide bonds (41). As shown in Fig. 2B, significant differences in spectral shape were observed between SOD1(57/146)S-S and SOD1noCys, which probably reflects the contribution of the disulfide bond to CD signals in this region (240–290 nm) (41). On the other hand, the effects of Zn2+ ion on the spectrum were minimal in SOD1(57/146)S-S and SOD1noCys. Although interpretation of near-UV CD spectra has not been well established (41), the results would support no drastic changes of SOD1 in the regions near Trp and Phe residues (no Tyr residue in SOD1) upon losing a Zn2+ ion.
The presence of random coils in E,E-SOD1noCys was further confirmed with FTIR spectroscopy. As shown in Fig. 2C (red open circles), the second derivative of an IR spectrum of E,E-SOD1noCys showed an absorption peak at 1642 cm−1, which is characteristic to random coils (42). Upon the addition of an equimolar Zn2+ ion to E,E-SOD1noCys, an absorption peak at 1630 cm−1 that corresponds to β-sheet structures (42) emerged, whereas random coil structures indicated by an absorption peak at 1643 cm−1 still remained (Fig. 2C, red-filled circles). In contrast, both apo- and Zn2+-bound SOD1 with its disulfide bond exhibited an absorption peak at 1630 cm−1 but not 1642 cm−1, suggesting the formation of β-sheet structures (Fig. 2C, black-filled and open circles). Although SOD1 has already been shown to become fibrillated only in the apo- and disulfide-reduced state (11), we have confirmed again in this study that E,E-SOD1noCys but not E,E-SOD1(57/146)S-S is fibrillogenic and also that the addition of excess Zn2+ ions suppresses the fibrillation of SOD1 proteins (Fig. 2D). Accordingly, a distinct conformational disorder of apoSOD1 that is realized only in the absence of the disulfide bond may have the potential to trigger the fibrillation of SOD1.
The β-Barrel-like Scaffold of SOD1 Is Not Significantly Affected in the Most Immature State
Whereas increased fractions of random coils were evident in apoSOD1 upon losing the disulfide bond, it is notable that the absorption peak at 1673 cm−1, corresponding to β-sheet structures, were still observed in E,E-SOD1noCys (Fig. 2C, red open circles). The structure of SOD1 consists of a β-barrel-like fold with two major loop regions (loop IV and loop VII; also see Fig. 1A). As suggested by previous studies (14, 19, 25–27), upon dissociation of metal ions and/or reduction of the disulfide bond, the two major loops appear to increasingly fluctuate with a mostly retained structure of the β-barrel-like core region.
To probe the effects of the disulfide reduction on the protein folding of our apoSOD1 samples, we took advantage of the fact that SOD1 has a single Trp residue in the β3 strand of the β-barrel-like scaffold and examined its fluorescence properties in E,E-SOD1(57/146)S-S and E,E-SOD1noCys. In particular, the wavelength of maximum fluorescence emission of Trp has been known to be an indicator of changes in the environment surrounding Trp residues (43). Actually, the fluorescence peak was red-shifted (∼6 nm) when E,E-SOD1noCys and E,E-SOD1(57/146)S-S were unfolded in the presence of guanidine hydrochloride (data not shown); nonetheless, the spectra were almost completely overlapped between E,E-SOD1(57/146)S-S and E,E-SOD1noCys (data not shown). These results hence further support that the β-barrel structure in apoSOD1 is not largely affected upon reduction of the disulfide bond.
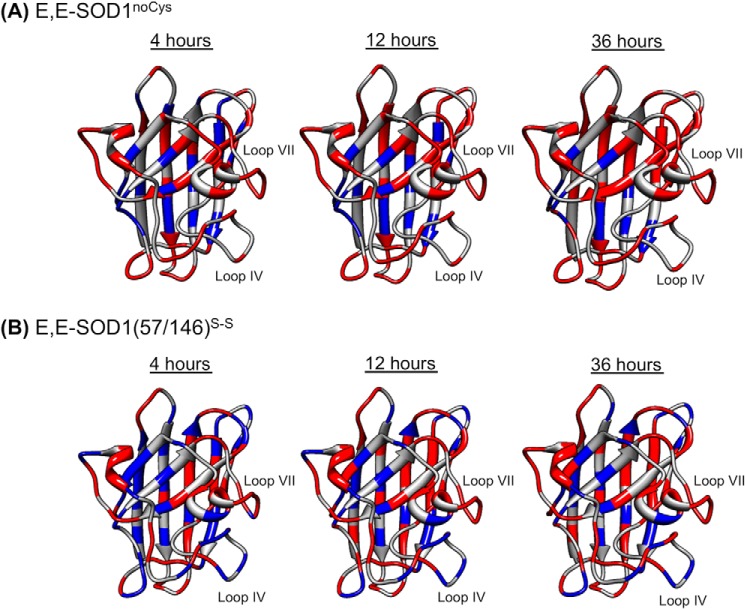
Actually, previous NMR studies on several immature states of SOD1 have shown that the β-barrel structure is well maintained and that the flexible loop regions (loop IV and VII) experience extensive mobility upon removal of metal ions and/or reduction of the disulfide bond (14, 25–27). To confirm the distinct behaviors of the β-barrel structure and the loop regions, we performed a hydrogen-deuterium (H/D) exchange analysis on the 1H,15N HSQC spectra of our E,E-SOD1noCys and E,E-SOD1(57/146)S-S proteins at 283 K. After dilution of the protein sample with deuterated buffer, intensities of the resonances from amide protons decreased due to the H/D exchange. Fig. 3 represents time-dependent and heterogeneous changes of the resonance intensities in E,E-SOD1noCys and E,E-SOD1(57/146)S-S over the entire proteins. In this study, 20% of resonance intensities relative to those before H/D exchange were set as a threshold; namely, the resonances of the residues colored red almost disappeared with <20% of the original intensities before dilution, whereas significant intensities (>20%) of resonances were maintained in the residues colored blue. In both E,E-SOD1noCys and E,E-SOD1(57/146)S-S, most of the residues with relatively slow H/D exchange (colored blue) are found in β-strands, consistent with a robust β-barrel scaffold of SOD1 (Fig. 3). Moreover, all of the observed resonances from the residues in loop IV and VII already disappeared in E,E-SOD1noCys at 4 h after H/D exchange (Fig. 3A), but notably, resonances from some of the residues in loop IV and VII of E,E-SOD1(57/146)S-S (Gly-61, Arg-69, His-71, Asp-76, Glu-78 in loop IV; Thr-135 and Ser-142 in loop VII) retained significant intensities even after 36 h of exchange (Fig. 3B). These data are thus consistent with the flexible nature of those loop regions and also imply that the loop regions in E,E-SOD1 would become more disordered upon losing the disulfide bond. To get more insight into the effects of disulfide reduction on the conformation of demetallated SOD1 species, we further evaluated their molecular size and shape by utilizing SAXS.
FIGURE 3.
A robust β-barrel structure of SOD1 with flexible loop regions. After dilution of the protein samples E,E-SOD1noCys (A) and E,E-SOD1(57/146)S-S (B) with a deuterated buffer, 1H,15N HSQC spectra were taken every 4 h, and the changes in the intensities of resonances from amide protons were analyzed. Relative intensities of resonances at 4, 12, and 36 h after dilution were calculated using those before H/D exchange as a reference. Amino acid residues in the structure of SOD1 protomer are colored based upon the relative intensities (red, <0.2; blue, >0.2).
The Most Immature SOD1 Has a Significantly Distinct Conformation from That of Dimeric SOD1 in Solution
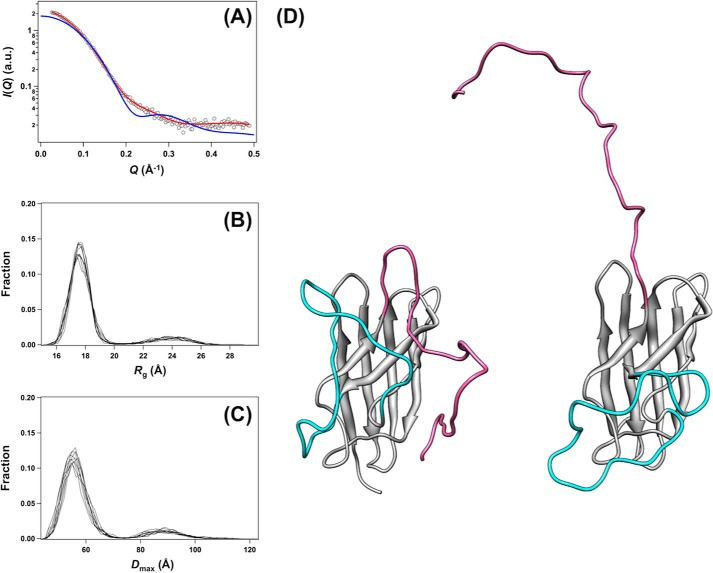
Fig. 4A shows scattering curves of E,E-SOD1(57/146)S-S and E,E-SOD1noCys at infinite dilution. From the Guinier plot of the scattering curve, forward scattering intensity, I(0), was first determined from the intercept of the linear fit (Fig. 4B) and then used for estimation of a relative molecular mass (Table 1). Based upon the estimates, E,E-SOD1(57/146)S-S and E,E-SOD1noCys were found to be a dimer and a monomer in solution, respectively. These estimated masses are also consistent with the ones empirically calculated from the Porod volume (Vp, Table 1), further strengthening the fact that apoSOD1 became a monomer upon reduction of the disulfide bond (Fig. 1, B and C).
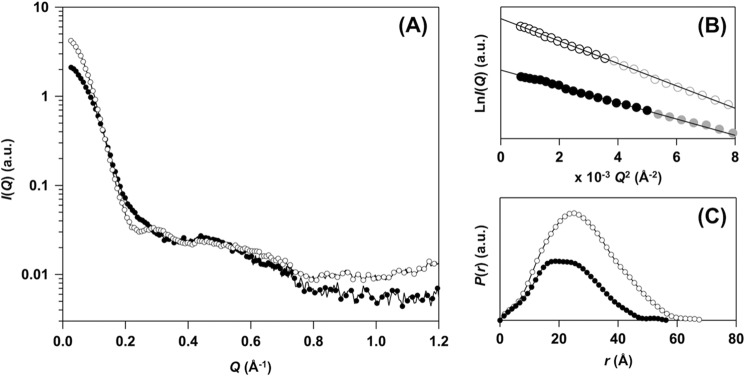
FIGURE 4.
SAXS experiments support distinct conformations between E,E-SOD1noCys and E,E-SOD1(57/146)S-S. A, SAXS curves of E,E-SOD1noCys (filled circles) and E,E-SOD1(57/146)S-S (open circles). Scattering intensity, I(Q), is plotted against scattering angular momentum, Q, in a logarithmic scale. The indicated curves were obtained by merging the two curves taken with different camera lengths, one at infinite dilution (0.02592 Å−1 < Q < 0.70975 Å−1) and the other at finite concentration (0.06994 Å−1 < Q < 1.48845 Å−1). B, Guinier plots of the scattering curves obtained in E,E-SOD1noCys (filled circles) and E,E-SOD1(57/146)S-S (open circles) at infinite dilution. Each line shows the linear fit to ln I(Q) using the Q range from 0.02588 to Qmax < 1.3/Rg (black data points), which gave the estimated values of Rg and I(0) (Table 1). C, the pair-distance distribution function, P(r), from the scattering profile is shown: E,E-SOD1noCys (filled circles) and E,E-SOD1(57/146)S-S (open circles). Dmax values obtained from P(r) are summarized in Table 1.
TABLE 1.
SAXS structural parameters
| Proteins | Rga | Rgb | I(0)a | I(0)b | Dmaxc | Vpd | Mr from I(0)e | Mr from Vpf | Mrg |
|---|---|---|---|---|---|---|---|---|---|
| Å | Å | a.u. | a.u. | Å | Å3 | kDa | kDa | kDa | |
| E,E-SOD1(57/146)S-Sh | 21.5 ± 0.2 | 21.2 ± 0.1 | 4.67 ± 0.03 | 4.63 ± 0.01 | 68 | 5.85 × 104 | 38.7 | 35.4 | 16.2 |
| E,E-SOD1noCysh | 18.4 ± 0.2 | 17.7 ± 0.1 | 2.31 ± 0.02 | 2.26 ± 0.01 | 58 | 3.00 × 104 | 19.1 | 18.2 | 16.2 |
| BSAi | 27.6 ± 0.3 | ND. j) | 8.01 ± 0.05 | NDj | NDj | NDj | NDj | NDj | 66.4 |
a Guinier analysis using the Q range from 0.02588 to Qmax < 1.3/Rg.
b Estimates in real space upon P(r) determination.
c Maximum dimension estimated by using GNOM package.
d Porod volume.
e Mr, molecular mass calculated by using the I(0) value for BSA as the standard.
f Mr calculated according to an empirical relationship (Mr = Vp/1.65) (53).
g Theoretical Mr calculated according to amino acid sequences.
h Extrapolated to infinite dilution.
i At finite concentration of 3.06 mg/ml (54).
j Not determined.
Furthermore, in the Guinier plot (Fig. 4B) the slope of the linear fit contained information on the radius of gyration, Rg, of a molecule. As summarized in Table 1, the Rg value of E,E-SOD1(57/146)S-S was estimated from the plot as 21.5 Å, which is comparable with the one calculated based upon the crystal structure of matured human SOD1 (PDB ID 2C9V, 20.9 Å) (30). In contrast, E,E-SOD1noCys showed an Rg of 18.4 Å, which was significantly larger than the one calculated from the protomer in the crystal structure of SOD1 dimer (15.5 Å) (30). A less compact structure in E,E-SOD1noCys was further supported by the pair-distance distribution function, P(r); E,E-SOD1noCys and E,E-SOD1(57/146)S-S had a maximum dimension, Dmax, of 58 and 68 Å, respectively (Fig. 4C, Table 1). The observed Dmax of E,E-SOD1(57/146)S-S was comparable with the one calculated from the crystal structure of matured SOD1 dimer (70.7 Å), but again, E,E-SOD1noCys in solution had a Dmax that was larger than of a protomer of SOD1 in its crystal structure (47.7 Å). Collectively, these results clearly indicate that monomeric E,E-SOD1noCys adopts a relatively extended conformation compared with a monomer unit of the dimer.
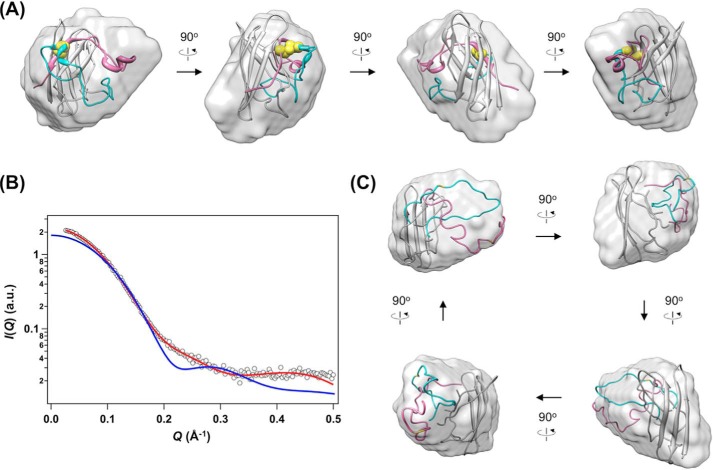
Actually, a low resolution model of E,E-SOD1noCys, which was restored from the scattering curve using GASBOR (31), could not be superimposed well to a monomer unit of the native SOD1 dimer and was more ellipsoidal (Fig. 5A, Table 2). More precisely speaking, our experimental SAXS curve of E,E-SOD1noCys was significantly deviated from the theoretical one calculated using a protomer of the SOD1 crystal structure (χ = 7.29; Fig. 5B). These observations may be reconciled with our results to point to the conformational disorder of loops IV and VII upon losing the disulfide bond in apoSOD1. To confirm this, we refined the conformations of loops IV and VII in the SOD1 crystal structure against the experimental SAXS curve of E,E-SOD1noCys while keeping the β-barrel scaffold intact. As shown in Fig. 5, B and C, a rigid body model of disulfide-null apoSOD1, in which loops IV and VII were “peeled” off from the β-barrel scaffold, gave a scattering curve quite similar to the experimentally observed one (χ = 1.98).
FIGURE 5.
Significant disorder of loops IV and VII describes a distinct conformation of the most immature SOD1. A, an overall shape of E,E-SOD1noCys (surface model) that was restored from the scattering curve is visualized using the SITUS package, onto which a monomer unit of the SOD1 crystal structure (ribbon model) is superimposed (normalized spatial discrepancy = 0.9203). Cys-57 and -146, which are involved in the formation of an intramolecular disulfide bond, are shown in yellow. Regions allowed for conformational changes during refinements are colored in cyan (loop IV) and pink (from loop VII to the C terminus). B, an experimentally observed scattering curve of E,E-SOD1noCys (open circles) is compared with the one calculated from a monomer unit of the SOD1 crystal structure (blue, χ = 7.29). A theoretical curve of the most representative model of E,E-SOD1noCys with peeled loops IV and VII is shown in red (χ = 1.98), which is superimposed in panel C onto the low resolution envelope of E,E-SOD1noCys (normalized spatial discrepancy = 0.8867). The details and statistics of the modeling are summarized in Table 2.
TABLE 2.
SAXS shape reconstruction statistics
| Shape reconstruction | E,E-SOD1noCys |
|---|---|
| GASBOR2.2i | |
| Q range (Å−1) | 0.02618-1.110 |
| Symmetry | P1 |
| Search space | Sphere |
| Number of Shannon channels | 20.49 |
| Total number of dummy residues | 157 |
| Square root of χ2 (mean ± S.D.) | 0.958–1.584 (1.198 ± 0.113) |
| Number of models averaged | 96 |
| DAMAVER NSD (mean ± S.D.) | 0.980–1.061 (1.015 ± 0.023) |
| BUNCH08 | |
| Q range (Å−1) | 0.02618–0.49974 |
| Symmetry | P1 |
| Total number of residues | 157 |
| Total number of dummy residues | 68 |
| Dummy residues | 1–4, 53–83, 125–157 |
| Total number of known residues | 89 |
| Known residues treated as rigid bodies | 5–52, 84–124 |
| Square root of χ2 (mean ± S.D.) | 1.978–3.321 (2.378 ± 0.383) |
| Number of models reconstructed | 10 |
| DAMAVER NSD (mean ± S.D.) | 1.049–1.166 (1.103 ± 0.033) |
| EOM2.0 | |
| Q range (Å−1) | 0.02618–0.49974 |
| Symmetry | P1 |
| Total number of residues | 157 |
| Total number of dummy residues | 68 |
| Dummy residues | 1–4, 53–83, 125–157 |
| Total number of known residues | 89 |
| Known residues treated as rigid bodies | 5–52, 84–124 |
| Number of theoretical curves (pool structures) | 10,000 |
| Square root of χ2 (mean ± S.D.) | 1.138–1.147 (1.143 ± 0.002) |
| Number of ensembles reconstructed | 10 |
Nevertheless, loops IV and VII in the rigid body model are highly extended (Fig. 5C) and thus seem to be inappropriate for the description of a single distinct conformer. Rather, taking into account that previous studies showed significant fluctuations at loops IV and VII in immature forms of SOD1 (14, 25–27), E,E-SOD1noCys is considered to adopt multiple conformations with highly mobile loops IV and VII. Indeed, compared with the rigid body model, the experimental SAXS curve of E,E-SOD1noCys was a better fit to the theoretical scattering curve from an ensemble of conformations carrying the flexed loops IV and VII (χ = 1.14) (Fig. 6A, Table 2). Most of the simulated conformations exhibited Rg of 16–19 Å and Dmax of 40–70 Å, whereas minor conformations also populated at Rg 21–27 Å and Dmax 70–100 Å (Fig. 6, B and C). It is, therefore, possible that the most immature apoSOD1 without the disulfide bond is monomeric with significant fluctuations in loops IV and VII (Fig. 6D), resulting in a conformation distinct from that of a protomer of the native SOD1 dimer.
FIGURE 6.
An experimental SAXS curve of E,E-SOD1noCys was reasonably fit with an ensemble of conformations. From 10,000 hypothetical rigid-body models with the randomly flexed loops IV and VII, 10 independent ensembles of conformations were reconstructed to fit the experimental SAXS curve of E,E-SOD1noCys (Fig. 4A). A, a theoretical curve obtained from a representative ensemble of conformations with the flexed loops IV and VII is shown in red and well matched with the observed scattering curve (open squares, χ = 1.14). A scattering curve calculated from a monomer unit of the SOD1 crystal structure is again shown for comparison (blue, χ = 7.29). A distribution of Rg (B) and Dmax of conformations (C) in each of those 10 ensembles is shown. D, representative conformations in a population with (left) 17.9 Å and (right) 24.2 Å of Rg are shown. Regions allowed for conformational changes during refinements are colored cyan (loop IV) and pink (from loop VII to the C terminus).
Pathological Significance of the Most Immature SOD1
Misfolding/aggregation of SOD1 proteins is a major pathological change in SOD1-related fALS patients (44). It has been suggested that demetallation and/or disulfide reduction are involved in misfolding of SOD1 for the formation of insoluble aggregates (3). Actually, only the most immature state of SOD1 (modeled by E,E-SOD1noCys in this study) was accessible to the formation of fibrillar aggregates (Fig. 2D) (11). In other words, E,E-SOD1noCys can be regarded as a precursor for fibrillation, and we have found here that its structural conformation is unique and distinct from that of the dimeric SOD1 in the metallated and/or disulfide-bonded states. In particular, severe disorder at loops IV and VII is considered to be responsible for such a unique conformation of fibrillation-prone E,E-SOD1noCys (Figs. 5 and 6).
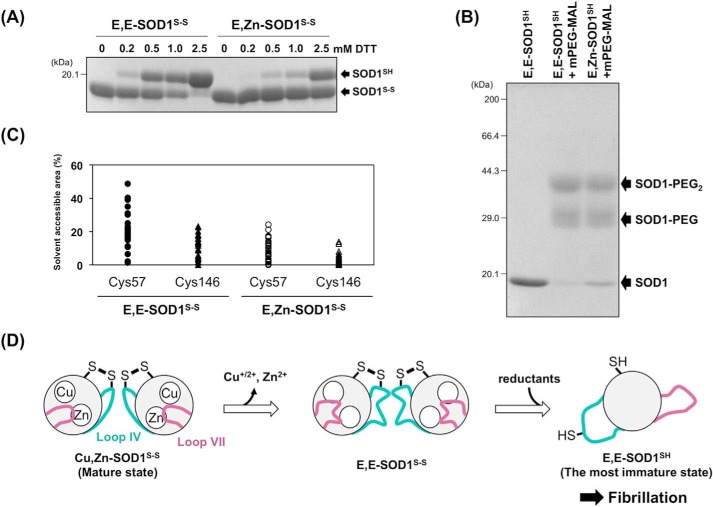
Given that loop IV contains all three zinc ligands (His-71, His-80, Asp-83) and the copper-zinc bridging ligand (His-63) (Fig. 1A), demetallation is expected to cause increased disorder around the loop. Indeed, it has been suggested that in the 1H,15N HSQC spectra of apoSOD1SH/S-S, the amino acid residues at loops IV and VII exhibit significant changes in their chemical shifts upon binding of a Zn2+ ion (25, 26). Many of the amino acid residues in loop IV have not been identified both in the crystal and solution structures of metal-deficient SOD1SH/S-S due to severe thermal fluctuations (14, 19, 45). Moreover, the disulfide bond tethers loop IV (Cys-57) to the β-strand after loop VII (Cys-146) (Fig. 1A); therefore, lost of the disulfide bond is also a significant contribution to the increased fluctuation of the loops. Actually, the absence of the disulfide bond facilitated the H/D exchange of amide protons at loops IV and VII of apoSOD1 (Fig. 3). Given that either demetallation or disulfide reduction alone is not sufficient for triggering fibrillation of SOD1 proteins (Fig. 2D) (11), the increased disorder of loops IV and VII, which are realized only in the most immature state of SOD1, could facilitate fibrillation.
As described above, loop IV includes both the disulfide bonding Cys residue (Cys-57) and the Zn2+ binding ligands (Fig. 1A); therefore, fluctuations in loop IV caused by the removal of metal ions would affect the conformation around the disulfide bond. Indeed, lesser amounts of dithiothreitol were required to reduce the disulfide bond in E,E-SOD1S-S compared with its Zn2+-bound form, E,Zn-SOD1S-S (Fig. 7A), suggesting that the disulfide bond becomes increasingly accessible upon losing the metal ions. Also, the chemical modification of Cys-57 and -146 in the disulfide-reduced state (SOD1SH) was found to proceed more efficiently in the apo form than in its Zn2+-bound form (Fig. 7B); certain amounts of the unmodified state remained in E,Zn-SOD1SH, but the modification was almost completed in E,E-SOD1SH. This is also supported by the NMR-derived solution structures of apo- and Zn2+-bound SOD1 with a disulfide bond (14, 46) in which Cys-57 and Cys-146 are found to become more exposed to the solvent upon dissociation of a Zn2+ ion (47) (Fig. 7C). Collectively, therefore, these data show that the disulfide bond as well as its constituents, Cys-57 and -146, are more exposed to the solvent upon losing metal ions. SOD1 is localized mostly in the cytoplasm, where the redox environment is highly reducing; therefore, exposure of the disulfide bond to the solvent would increase the chance of being attacked by endogenous reductants such as glutathione. Once the disulfide bond is reduced, apoSOD1 became a monomer with an ellipsoidal conformation in which loops IV and VII are possibly wide-open (Figs. 5 and 6).
FIGURE 7.
A proposed mechanism of SOD1 fibrillation triggered with conformational disorder of loops IV and VII. A, E,E-SOD1(57/146)S-S and E,Zn-SOD1(57/146)S-S (20 μm) were incubated with the indicated concentrations of dithiothreitol (DTT) at 20 °C for 1 h. To quench the reduction reaction, the proteins were precipitated with trichloroacetic acid and reacted with a thiol-specific modifier, iodoacetamide, in the presence of 2% SDS. SOD1 proteins with and without the disulfide bond were then separated by non-reducing SDS-PAGE. B, E,E-SOD1(57/146)SH and E,Zn-SOD1(57/146)SH (20 μm) were reacted with mPEG-MAL (0.1 mm, Nanocs Inc.) at 10 °C for 1 h. The modification reaction was quenched by adding 10 mm DTT, and the proteins were analyzed with reducing SDS-PAGE. C, solvent-accessible areas of Cys-57 and Cys-146 in apo- and Zn2+-bound SOD1 with a disulfide bond. Each model in the NMR structures of E,Zn-SOD1S-S (35 models: PDB ID, 1KMG) and E,E-SOD1S-S (30 models: PDB ID, 1RK7) was used for calculation of the accessible areas of Cys residues using GetArea. The areas are expressed as a percentage ratio of the surface area in proteins to that in the random coil state. D, a proposed scheme of SOD1 fibrillation. Dissociation of metal ions resulted in significant disorder of loops IV and VII, which allows intracellular reductants to attack the disulfide bond and thereby facilitate the formation of the most immature SOD1. The most immature SOD1 had a unique ellipsoidal conformation, which is accessible to further fibrillation.
Although both concentration and specific activities of SOD1 were decreased in erythrocytes from affected SOD1-related fALS patients (48), it is notable that enzymatic activity of SOD1 is almost fully retained in transgenic mice expressing human SOD1 with pathogenic mutations (49). In physiological conditions, therefore, mutant SOD1 could exist initially as a matured state: i.e. a copper, zinc-bound state with a disulfide bond. Given that affinity for Zn2+ ion has been shown to decrease in mutant SOD1 proteins in vitro (7), mutant SOD1 in the matured state would nevertheless gradually lose its bound metal ions in the cytoplasm with high chelating capacity for metal ions. In motor neurons with a meter-long axon in particular, more than a year will be necessary for SOD1 to be anterogradely transported to the nerve termini (50, 51); therefore, it may be difficult for mutant SOD1 to survive in a matured form during the axonal transport. Losing metal ions from the matured state of mutant SOD1 could gradually occur in a year-long scale and contribute to the accumulation of the most immature SOD1 proteins (Fig. 7D).
Actually, the amount of SOD1-positive inclusions formed in model mice appear to be negatively correlated with the affinity of mutant SOD1 for metal ions. For example, human SOD1 with mutation abrogating (H46R) or significantly reducing (G85R) metal binding have been found to accumulate significant amounts of inclusions, whereas formation of inclusions was quite limited in the model mice expressing mutant SOD1 retaining the metal binding ability (e.g. G37R, G93A) (24, 52). These pathological observations are thus consistent with our proposed mechanism where failure of metal acquisition in SOD1 increased the amounts of the disulfide-reduced state in the cytoplasm (Fig. 7D), and a unique conformation of the most immature SOD1 will be a promising target for controlling the pathogenicity of mutant SOD1 in ALS.
Author Contributions
Y. F. directed the project, performed the experiments, analyzed most of the data, and wrote the manuscript. S. A. performed the SAXS experiments and analyzed the data. M. I., F. J. C. C., T. S., and K. I. performed the NMR experiments and analyzed the data. K. N., I. A., and T. N. contributed to the preparation and measurements of protein samples for MALS, SAXS, fluorescence, and NMR. All authors review the manuscript.
This work was supported by Grants-in-aid 25291028 for Scientific Research (B) and 15H01566 for Scientific Research on Innovative Areas (to Y. F.) and a Nanotechnology Platform Program (Molecule and Material Synthesis) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and in part by Cooperative Research in Joint Studies at Institute for Molecular Science, Japan. The authors declare that they have no conflicts of interest with the contents of this article.
Olivia project. Yokochi, M., Sekiguchi S., and Inagaki, F., Hokkaido University, Sapporo, Japan.
- SOD1
- Cu,Zn-superoxide dismutase
- SOD1(57/146)
- SOD1 with C6S/C111S mutations
- SOD1noCys
- SOD1 with C6S/C57S/C111S/C146S mutations
- E,E
- an apo state
- E,Zn
- a Zn2+-bound state
- fALS
- familial form of amyotrophic lateral sclerosis
- SEC
- size-exclusion chromatography
- MALS
- multi-angle light scattering
- SAXS
- small-angle x-ray scattering
- a.u.
- arbitrary units
- HSQC
- heteronuclear single quantum correlation.
References
- 1. Rosen D. R., Siddique T., Patterson D., Figlewicz D. A., Sapp P., Hentati A., Donaldson D., Goto J., O'Regan J. P., and Deng H. X. (1993) Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362, 59–62 [DOI] [PubMed] [Google Scholar]
- 2. Bruijn L. I., Houseweart M. K., Kato S., Anderson K. L., Anderson S. D., Ohama E., Reaume A. G., Scott R. W., and Cleveland D. W. (1998) Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science 281, 1851–1854 [DOI] [PubMed] [Google Scholar]
- 3. Furukawa Y. (2012) Protein aggregates in pathological inclusions of amyotrophic lateral sclerosis. In Amyotrophic Lateral Sclerosis (Maurer M. H., ed.) pp. 335–356 [Google Scholar]
- 4. Forman H. J., and Fridovich I. (1973) On the stability of bovine superoxide dismutase: the effects of metals. J. Biol. Chem. 248, 2645–2649 [PubMed] [Google Scholar]
- 5. Furukawa Y., and O'Halloran T. V. (2005) Amyotrophic lateral sclerosis mutations have the greatest destabilizing effect on the apo, reduced form of SOD1, leading to unfolding and oxidative aggregation. J. Biol. Chem. 280, 17266–17274 [DOI] [PubMed] [Google Scholar]
- 6. Lindberg M. J., Normark J., Holmgren A., and Oliveberg M. (2004) Folding of human superoxide dismutase: disulfide reduction prevents dimerization and produces marginally stable monomers. Proc. Natl. Acad. Sci. U.S.A. 101, 15893–15898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hayward L. J., Rodriguez J. A., Kim J. W., Tiwari A., Goto J. J., Cabelli D. E., Valentine J. S., and Brown R. H. Jr. (2002) Decreased metallation and activity in subsets of mutant superoxide dismutases associated with familial amyotrophic lateral sclerosis. J. Biol. Chem. 277, 15923–15931 [DOI] [PubMed] [Google Scholar]
- 8. Tiwari A., and Hayward L. J. (2003) Familial amyotrophic lateral sclerosis mutants of copper/zinc superoxide dismutase are susceptible to disulfide reduction. J. Biol. Chem. 278, 5984–5992 [DOI] [PubMed] [Google Scholar]
- 9. Lelie H. L., Liba A., Bourassa M. W., Chattopadhyay M., Chan P. K., Gralla E. B., Miller L. M., Borchelt D. R., Valentine J. S., and Whitelegge J. P. (2011) Copper and zinc metallation status of copper-zinc superoxide dismutase from amyotrophic lateral sclerosis transgenic mice. J. Biol. Chem. 286, 2795–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jonsson P. A., Graffmo K. S., Andersen P. M., Brännström T., Lindberg M., Oliveberg M., and Marklund S. L. (2006) Disulphide-reduced superoxide dismutase-1 in CNS of transgenic amyotrophic lateral sclerosis models. Brain 129, 451–464 [DOI] [PubMed] [Google Scholar]
- 11. Furukawa Y., Kaneko K., Yamanaka K., O'Halloran T. V., and Nukina N. (2008) Complete loss of post-translational modifications triggers fibrillar aggregation of SOD1 in familial form of ALS. J. Biol. Chem. 283, 24167–24176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kato S., Takikawa M., Nakashima K., Hirano A., Cleveland D. W., Kusaka H., Shibata N., Kato M., Nakano I., and Ohama E. (2000) New consensus research on neuropathological aspects of familial amyotrophic lateral sclerosis with superoxide dismutase 1 (SOD1) gene mutations: inclusions containing SOD1 in neurons and astrocytes. Amyotroph. Lateral Scler. Other Motor Neuron Disord. 1, 163–184 [DOI] [PubMed] [Google Scholar]
- 13. Wang J., Xu G., Gonzales V., Coonfield M., Fromholt D., Copeland N. G., Jenkins N. A., and Borchelt D. R. (2002) Fibrillar inclusions and motor neuron degeneration in transgenic mice expressing superoxide dismutase 1 with a disrupted copper-binding site. Neurobiol. Dis. 10, 128–138 [DOI] [PubMed] [Google Scholar]
- 14. Banci L., Bertini I., Cramaro F., Del Conte R., and Viezzoli M. S. (2003) Solution structure of Apo Cu,Zn superoxide dismutase: role of metal ions in protein folding. Biochemistry 42, 9543–9553 [DOI] [PubMed] [Google Scholar]
- 15. Doucette P. A., Whitson L. J., Cao X., Schirf V., Demeler B., Valentine J. S., Hansen J. C., and Hart P. J. (2004) Dissociation of human copper-zinc superoxide dismutase dimers using chaotrope and reductant: insights into the molecular basis for dimer stability. J. Biol. Chem. 279, 54558–54566 [DOI] [PubMed] [Google Scholar]
- 16. Potter S. Z., Zhu H., Shaw B. F., Rodriguez J. A., Doucette P. A., Sohn S. H., Durazo A., Faull K. F., Gralla E. B., Nersissian A. M., and Valentine J. S. (2007) Binding of a single zinc ion to one subunit of copper-zinc superoxide dismutase apoprotein substantially influences the structure and stability of the entire homodimeric protein. J. Am. Chem. Soc. 129, 4575–4583 [DOI] [PubMed] [Google Scholar]
- 17. Rodriguez J. A., Valentine J. S., Eggers D. K., Roe J. A., Tiwari A., Brown R. H. Jr., and Hayward L. J. (2002) Familial amyotrophic lateral sclerosis-associated mutations decrease the thermal stability of distinctly metallated species of human copper/zinc superoxide dismutase. J. Biol. Chem. 277, 15932–15937 [DOI] [PubMed] [Google Scholar]
- 18. Shaw B. F., Durazo A., Nersissian A. M., Whitelegge J. P., Faull K. F., and Valentine J. S. (2006) Local unfolding in a destabilized, pathogenic variant of superoxide dismutase 1 observed with H/D exchange and mass spectrometry. J. Biol. Chem. 281, 18167–18176 [DOI] [PubMed] [Google Scholar]
- 19. Strange R. W., Antonyuk S., Hough M. A., Doucette P. A., Rodriguez J. A., Hart P. J., Hayward L. J., Valentine J. S., and Hasnain S. S. (2003) The structure of holo and metal-deficient wild-type human Cu,Zn superoxide dismutase and its relevance to familial amyotrophic lateral sclerosis. J. Mol. Biol. 328, 877–891 [DOI] [PubMed] [Google Scholar]
- 20. Banci L., Bertini I., Blaževitš O., Cantini F., Lelli M., Luchinat C., Mao J., and Vieru M. (2011) NMR characterization of a “fibril-ready” state of demetalated wild-type superoxide dismutase. J. Am. Chem. Soc. 133, 345–349 [DOI] [PubMed] [Google Scholar]
- 21. Arnesano F., Banci L., Bertini I., Martinelli M., Furukawa Y., and O'Halloran T. V. (2004) The unusually stable quaternary structure of human SOD1 is controlled by both metal occupancy and disulfide status. J. Biol. Chem. 279, 47998–48003 [DOI] [PubMed] [Google Scholar]
- 22. Toichi K., Yamanaka K., and Furukawa Y. (2013) Disulfide scrambling describes the oligomer formation of superoxide dismutase (SOD1) proteins in the familial form of amyotrophic lateral sclerosis. J. Biol. Chem. 288, 4970–4980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., and Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 24. Watanabe M., Dykes-Hoberg M., Culotta V. C., Price D. L., Wong P. C., and Rothstein J. D. (2001) Histological evidence of protein aggregation in mutant SOD1 transgenic mice and in amyotrophic lateral sclerosis neural tissues. Neurobiol. Dis. 8, 933–941 [DOI] [PubMed] [Google Scholar]
- 25. Banci L., Bertini I., Cantini F., D'Amelio N., and Gaggelli E. (2006) Human SOD1 before harboring the catalytic metal: solution structure of copper-depleted, disulfide-reduced form. J. Biol. Chem. 281, 2333–2337 [DOI] [PubMed] [Google Scholar]
- 26. Sekhar A., Rumfeldt J. A., Broom H. R., Doyle C. M., Bouvignies G., Meiering E. M., and Kay L. E. (2015) Thermal fluctuations of immature SOD1 lead to separate folding and misfolding pathways. eLife 4, e07296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Teilum K., Smith M. H., Schulz E., Christensen L. C., Solomentsev G., Oliveberg M., and Akke M. (2009) Transient structural distortion of metal-free Cu/Zn superoxide dismutase triggers aberrant oligomerization. Proc. Natl. Acad. Sci. U.S.A. 106, 18273–18278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., and Svergun D. I. (2003) PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 36, 1277–1282 [Google Scholar]
- 29. Svergun D. I. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 25, 495–503 [Google Scholar]
- 30. Svergun D., Barberato C., and Koch M. H. J. (1995) CRYSOL: a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 28, 768–773 [Google Scholar]
- 31. Svergun D. I., Petoukhov M. V., and Koch M. H. (2001) Determination of domain structure of proteins from x-ray solution scattering. Biophys. J. 80, 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Volkov V. V., and Svergun D. I. (2003) Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Cryst. 36, 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wriggers W., and Chacón P. (2001) Modeling tricks and fitting techniques for multiresolution structures. Structure 9, 779–788 [DOI] [PubMed] [Google Scholar]
- 34. Petoukhov M. V., and Svergun D. I. (2005) Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 89, 1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bernadó P., Mylonas E., Petoukhov M. V., Blackledge M., and Svergun D. I. (2007) Structural characterization of flexible proteins using small-angle x-ray scattering. J. Am. Chem. Soc. 129, 5656–5664 [DOI] [PubMed] [Google Scholar]
- 36. Fujiwara N., Nakano M., Kato S., Yoshihara D., Ookawara T., Eguchi H., Taniguchi N., and Suzuki K. (2007) Oxidative modification to cysteine sulfonic acid of Cys-111 in human copper-zinc superoxide dismutase. J. Biol. Chem. 282, 35933–35944 [DOI] [PubMed] [Google Scholar]
- 37. Furukawa Y., Torres A. S., and O'Halloran T. V. (2004) Oxygen-induced maturation of SOD1: a key role for disulfide formation by the copper chaperone CCS. EMBO J. 23, 2872–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bertini I., Piccioli M., Viezzoli M. S., Chiu C. Y., and Mullenbach G. T. (1994) A spectroscopic characterization of a monomeric analog of copper, zinc superoxide dismutase. Eur. Biophys. J. 23, 167–176 [DOI] [PubMed] [Google Scholar]
- 39. Kayatekin C., Zitzewitz J. A., and Matthews C. R. (2010) Disulfide-reduced ALS variants of Cu,Zn superoxide dismutase exhibit increased populations of unfolded species. J. Mol. Biol. 398, 320–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Greenfield N. J. (2006) Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 1, 2876–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kelly S. M., Jess T. J., and Price N. C. (2005) How to study proteins by circular dichroism. Biochim. Biophys. Acta 1751, 119–139 [DOI] [PubMed] [Google Scholar]
- 42. Byler D. M., and Susi H. (1986) Examination of the secondary structure of proteins by deconvolved FTIR spectra. Biopolymers 25, 469–487 [DOI] [PubMed] [Google Scholar]
- 43. Vivian J. T., and Callis P. R. (2001) Mechanisms of tryptophan fluorescence shifts in proteins. Biophys. J. 80, 2093–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bruijn L. I., Miller T. M., and Cleveland D. W. (2004) Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu. Rev. Neurosci. 27, 723–749 [DOI] [PubMed] [Google Scholar]
- 45. Elam J. S., Taylor A. B., Strange R., Antonyuk S., Doucette P. A., Rodriguez J. A., Hasnain S. S., Hayward L. J., Valentine J. S., Yeates T. O., and Hart P. J. (2003) Amyloid-like filaments and water-filled nanotubes formed by SOD1 mutant proteins linked to familial ALS. Nat. Struct. Biol. 10, 461–467 [DOI] [PubMed] [Google Scholar]
- 46. Banci L., Bertini I., Cantini F., D'Onofrio M., and Viezzoli M. S. (2002) Structure and dynamics of copper-free SOD: the protein before binding copper. Protein Sci. 11, 2479–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fraczkiewicz R., and Braun W. (1998) Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J. Comput. Chem. 19, 319–333 [Google Scholar]
- 48. Bowling A. C., Barkowski E. E., McKenna-Yasek D., Sapp P., Horvitz H. R., Beal M. F., and Brown R. H. Jr. (1995) Superoxide dismutase concentration and activity in familial amyotrophic lateral sclerosis. J. Neurochem. 64, 2366–2369 [DOI] [PubMed] [Google Scholar]
- 49. Jonsson P. A., Graffmo K. S., Brännström T., Nilsson P., Andersen P. M., and Marklund S. L. (2006) Motor neuron disease in mice expressing the wild type-like D90A mutant superoxide dismutase-1. J. Neuropathol. Exp. Neurol. 65, 1126–1136 [DOI] [PubMed] [Google Scholar]
- 50. Borchelt D. R., Wong P. C., Becher M. W., Pardo C. A., Lee M. K., Xu Z. S., Thinakaran G., Jenkins N. A., Copeland N. G., Sisodia S. S., Cleveland D. W., Price D. L., and Hoffman P. N. (1998) Axonal transport of mutant superoxide dismutase 1 and focal axonal abnormalities in the proximal axons of transgenic mice. Neurobiol. Dis. 5, 27–35 [DOI] [PubMed] [Google Scholar]
- 51. Rakhit R., Cunningham P., Furtos-Matei A., Dahan S., Qi X. F., Crow J. P., Cashman N. R., Kondejewski L. H., and Chakrabartty A. (2002) Oxidation-induced misfolding and aggregation of superoxide dismutase and its implications for amyotrophic lateral sclerosis. J. Biol. Chem. 277, 47551–47556 [DOI] [PubMed] [Google Scholar]
- 52. Nagai M., Aoki M., Miyoshi I., Kato M., Pasinelli P., Kasai N., Brown R. H. Jr., and Itoyama Y. (2001) Rats expressing human cytosolic copper-zinc superoxide dismutase transgenes with amyotrophic lateral sclerosis: associated mutations develop motor neuron disease. J. Neurosci. 21, 9246–9254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Petoukhov M. V., Franke D., Shkumatov A. V., Tria G., Kikhney A. G. Gajda M., Gorba C., Mertens H. D. T., Konarev P. V., and Svergun D. I. (2012) New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Cryst. 45, 342–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Akiyama S. (2010) Quality control of protein standards for molecular mass determinations by small-angle x-ray scattering. J. Appl. Cryst. 43, 237–243 [Google Scholar]