Abstract
p53 inactivation is a hallmark in non-small-cell lung cancer (NSCLC). It is therefore highly desirable to develop tumor-specific treatment for NSCLC therapy by restoring p53 function. Herein, a novel naphthalimide compound, NA-17, was identified as a promising drug candidate in view of both its anticancer activity and mechanism of action. NA-17 exhibited strong anticancer activity on a broad range of cancer cell lines but showed low toxicity to normal cell lines, such as HL-7702 and WI-38. Moreover, NA-17 showed p53-dependent inhibition selectivity in different NSCLC cell lines due to the activation state of endogenous p53 in the background level. Further studies revealed that NA-17 caused cell cycle arrest at the G1 phase, changed cell size, and induced apoptosis and cell death by increasing the proportion of sub-G1 cells. Molecular mechanism studies suggested that targeted accumulation of phospho-p53 in mitochondria and nuclei induced by NA-17 resulted in activation of Bak and direct binding of phospho-p53 to the target DNA sequences, thereby evoking cell apoptosis and cell cycle arrest and eventually leading to irreversible cancer cell inhibition. This work provided new insights into the molecular interactions and anticancer mechanisms of phospho-p53-dependent naphthalimide compounds.
Keywords: anticancer drug, apoptosis, cell cycle, lung cancer, p53
Introduction
Non-small cell lung cancer (NSCLC)3 continues to be the leading cause of cancer-related death worldwide with an overall 5-year survival rate no more than 15% (1, 2). The remarkable progress in targeted therapies (3, 4) has shown that sustained inactivation of key tumor suppressors and accumulation of mutations in oncogene pathways are the crucial factors in NSCLC malignant transformation (5). In the context of dysfunctional accumulation of tumor suppressors, constitutive inactivation of p53 is a common feature of NSCLC. Therapeutic strategies based on p53 restoration failed to achieve tumor regression due to the fact that the activation of p53 is only limited in relatively aggressive cancer cells within the tumors (6). Therefore, recruiting p53 function by persistent p53 activation should be an effective and tumor-specific treatment for NSCLC therapies.
Activation of p53 is mainly achieved via phosphorylation of its N-terminal domain by protein kinases belonging to the MAPK family (7), such as TP53-regulating kinase (8), checkpoint kinase (9), etc. Activated p53 translocates into the nucleus, binds to p53-responsive elements in the target DNA, and switches on the transcriptional program as a specific response to signal stimulation (10). Moreover, activated p53 can also accumulate in mitochondria to trigger apoptosis through a transcription-independent apoptotic mechanism (11–14). Transcriptional programs triggered by p53 activation in the nucleus ultimately lead to three major functional responses, i.e. cell cycle arrest, apoptosis, and senescence, resulting in proliferation inhibition and survival crisis due to altered gene expression (15–17). In contrast, targeted accumulation of activated p53 in mitochondria usually contributes to apoptosis by direct interaction with proapoptotic Bcl-2 family members and antiapoptotic Bcl-2 family members (18, 19). Bcl-xl, Bcl-2, and Mcl belong to the antiapoptotic Bcl-2 family, and members in this protein family can antagonize proapoptotic Bcl-2 family members, such as Bak and Bax, in normal cells for survival. Binding of phosphorylated p53 to Bak and Bax can induce a series of conformational rearrangements to expose the Bcl-2 homology 3 domains of Bak and Bax and alleviate antagonism of antiapoptotic proteins (18). Furthermore, phosphorylated p53 in the nuclei can also activate proapoptotic proteins, including Bad and Bim, to directly activate death effectors (20). Therefore, it will be beneficial to develop novel anticancer agents which persistently activate p53 for NSCLC therapies.
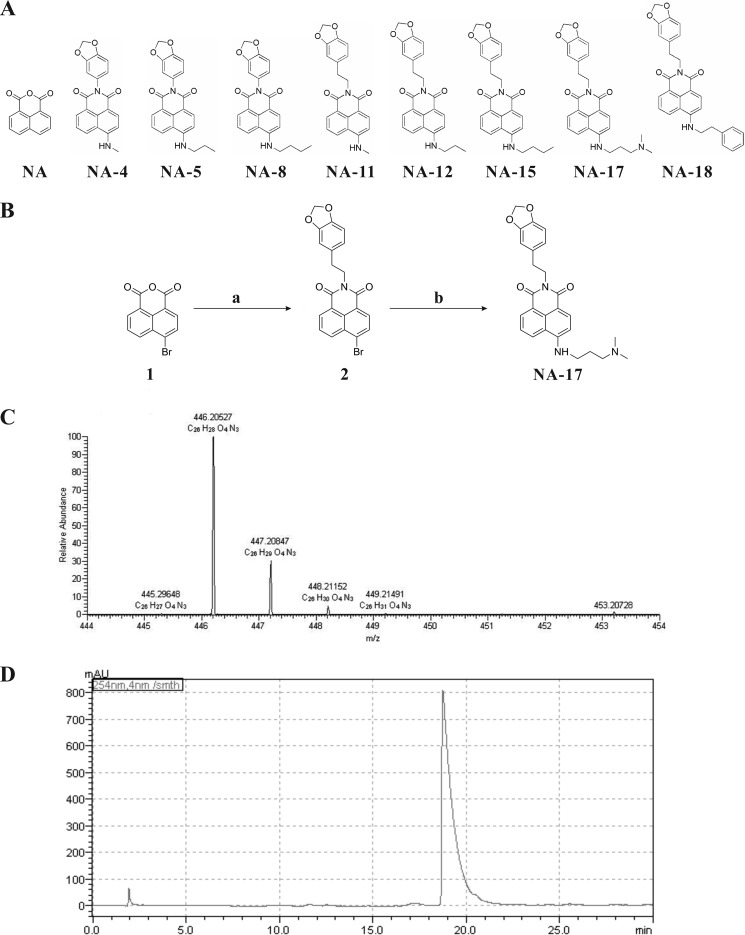
With the aim to develop tumor-specific anticancer agents, we screened eight naphthalimide derivatives synthesized in our laboratory (Fig. 1A). Naphthalimides exhibit anticancer activities preclinically and clinically (21–23), and the mechanism of action is possibly related to p53 activation (24). Notably, naphthalimides are fluorescent, allowing direct detection of their distribution in naphthalimide-treated cells (25). Among these compounds, a compound, NA-17, was identified as the most promising candidate for anticancer therapies based on its anticancer activity for NSCLC and mechanism of action. Our results showed that NA-17 induced accumulation of activated p53, which triggered complicated regulatory processes in cell apoptosis and proliferation inhibition. Furthermore, we found that NA-17 exhibited selective toxicity for cancer cells over normal cells. Our work suggested that NA-17 is a promising tumor-specific anticancer agent for NSCLC therapies and an efficient adjuvant in p53 restoration-mediated anticancer therapies.
FIGURE 1.
Synthesis of NA-17. A, structures of the eight naphthalimides compounds. B, synthetic route to NA-17. Reagents and conditions are as follows: a, 3,4-methylenedioxyphenethylamin, EtOH, 70 °C, 10 h; b, 3-dimethylaminopropylamine, DMSO, 135 °C. C, mass spectrum of NA-17. D, the purity of NA-17 determined by HPLC. The column used was a reversed-phase C18 column (YMC American Inc. HPLC column, 150 mm × 4.6-mm inner diameter). The column temperature was 35 °C. The mobile phase was methanol/H2O (from 10:90 to 100:0) with 0.01% TFA with a flow rate of 1.0 ml/min. The injection volume was 20 μl. mAU, milliabsorbance units.
Experimental Procedures
Reagents and Antibodies
All chemical reagents were commercially available and used without further purification unless noted specifically. RNase A, propidium iodide, and 3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were purchased from Sigma-Aldrich. Small interfering RNA (siRNA), scrambled RNA (scRNA), transfection reagents, and primers were obtained from Life Technologies. p53 siRNA* was purchased from Qiagen (catalogue number 1024849; Hilden, Germany). The Prime Script RT reagent kit and SYBR Premix Ex Taq were purchased from TaKaRa (Dalian, China). Anti-phospho-p53 (Ser-392), anti-p53, anti-p21, anti-Bim, anti-Bcl-2, anti-Bcl-xl, anti-apoptosis-inducing factor (AIF), anti-lamin B, anti-cytochrome c oxidase IV, anti-actin, and anti-Bax antibodies were purchased from Abcam (Cambridge, MA). Anti-Bak was purchased from Calbiochem. Anti-rabbit and anti-mouse secondary antibodies were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX). All chemicals for NA-17 synthesis were purchased from Alfa. Synthesized NA-17 was stored at −20 °C at a concentration of 10 mm in dimethyl sulfoxide (DMSO).
Synthesis of NA-17
Compounds 2 and NA-17 were synthesized as shown in Fig. 1B. The synthetic steps and spectral data of these compounds are described below. The purity of the synthesized compounds was determined by HPLC. NMR spectra were recorded using a Bruker Advance (500 MHz) with tetramethylsilane as an internal standard. High resolution mass spectra were recorded in a Thermo Exactive Orbitrap mass spectrometer.
4-Bromo-N-(3,4-methylenedioxyphenethylamin)-1,8-naphthalimide (2)
To a stirred ethanol solution of 1 (2.77 g; 10 mmol), 3,4-methylenedioxyphenethylamine (1.65 g; 10 mmol) was added. The reaction mixture was stirred at 70 °C for 10 h. Afterward, the solution was cooled to room temperature and filtered. The filter cake was recrystallized from CH2Cl2 to give 2 (2.97 g; yield, 70%). 1H NMR (500 MHz, DMSO-d6) δ: 8.53 (ddd, J = 9.5, 7.9, 1.0 Hz, 2H), 8.31 (d, J = 7.9 Hz, 1H), 8.20 (d, J = 7.9 Hz, 1H), 7.99–7.96 (m, 1H), 6.85 (d, J = 1.6 Hz, 1H), 6.80 (s, 1H), 6.69 (dd, J = 7.9, 1.7 Hz, 1H), 5.97 (s, 2H), 4.20–4.15 (m, 2H), 2.84 (t, J = 7.5 Hz, 2H). 13C NMR (126 MHz, DMSO-d6) δ: 162.86, 162.82, 147.36, 145.78, 132.78, 132.54, 131.72, 131.50, 131.11, 129.90, 129.30, 128.94, 128.36, 122.79, 122.01, 121.61, 109.09, 108.33, 100.82, 41.39, 33.19.
6-(1,3-Propanediamine-N,N-dimethyl-N′-)-N″-(3,4-methylenedioxyphenethylamin)-1,8-naphthalimide (NA-17)
To a stirred solution of 2 (4.23 g; 10 mmol) in 50 ml of DMSO, 3-dimethylaminopropylamine (1.02 g, 10 mmol) was added. The reaction mixture was stirred at 135 °C for 8 h. Afterward, the solution was poured into ice-water, and a yellow precipitation was observed. The crude product was collected and purified using a silica gel column with 100:5 CH2Cl2:CH3OH as the eluent. The final product of NA-17 was a yellow solid, and the overall yield was 50%. 1H NMR (500 MHz, DMSO-d6) δ: 8.60 (d, J = 8.3 Hz, 1H), 8.41 (d, J = 7.2 Hz, 1H), 8.25 (d, J = 8.5 Hz, 1H), 7.96 (t, J = 5.0 Hz, 1H), 7.67 (t, J = 10.0 Hz, 1H), 6.84 (d, J = 1.5 Hz, 1H), 6.82 (d, J = 7.9 Hz, 1H), 6.75 (d, J = 8.7 Hz, 1H), 6.69 (dd, J = 7.9, 1.5 Hz, 1H), 5.98 (s, 2H), 4.20–4.13 (m, 2H), 3.40 (dd, J = 12.3, 6.6 Hz, 2H), 2.81 (t, J = 10 Hz, 2H), 2.38 (t, J = 6.7 Hz, 2H), 2.20 (s, 6H), 1.88–1.81 (m, 2H). 13C NMR (126 MHz, DMSO-d6) δ: 163.54, 162.66, 150.67, 147.17, 145.55, 134.21, 132.67, 130.53, 129.33, 128.33, 124.20, 121.73, 121.36, 120.02, 108.87, 108.11, 107.36, 103.61, 100.62, 56.90, 45.08, 41.38, 40.67, 33.33, 25.56. High resolution mass spectra m/z: 446.20527 ([M + H]+).
Cell Culture and Cancer Cell Lines
The MGC-803, NCI-1975, NCI-H1299, NCI-H460, NCI-H358, HCC-827, HCT-116, MCF-7, Hep-G2, BEL-7404, and WI-38 cell lines were purchased from ATCC (Manassas, VA), and the hepatic cell line HL-7702 was obtained from our laboratory stock. All cell lines but HL-7702 and WI-38 were cultured in RPMI 1640 medium (Gibco) supplemented with 10% fetal bovine serum (FBS). The HL-7702 and WI-38 cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 10% FBS.
Transfection of Cells with Plasmids
NCI-H460 cells were seeded into 6-well plates (3 × 105 cells/well) or 24-well plates (8 × 104 cells/well) at 37 °C in a moisturized environment supplied with 5% CO2. All transfections were performed using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. After transfection, the cells were grown in RPMI 1640 medium with 2.5% FBS to minimize cell death.
Reporter Gene Assay
The reporter gene assay was performed as described elsewhere (26). Briefly, NCI-H460 cells were seeded into 24-well tissue culture plates without antibiotics. After 24 h, cells reached 80–90% confluence and were washed twice with PBS. Then the cells were co-transfected with the firefly luciferase reporter (pp53-TA-Luc; Beyotime Biotechnology, China) and Renilla luciferase reporter (Promega) using LipofectamineTM 2000 in Opti-MEM I (Life Technologies) following the manufacturer's instructions. The luciferase activity was measured according to the manufacturer's protocol.
DNA Relaxation Assay
The supercoiled pBR322 DNA was treated with a range of concentrations of NA-17 (20–100 μm) in a buffer solution containing 5 mm Tris-HCl and 50 mm NaCl buffer, pH 7.2, and the sample solutions were incubated for 1 h. The samples were electrophoresed in a 1% agarose gel and stained with 0.5 μg ml−1 ethidium bromide for detection.
Cell Viability Assay
Cell viability was monitored using the MTT assay. MTT (5 mg ml−1) was added to the wells, and the plates were incubated for 4 h at 37 °C. The MTT reaction was stopped by adding DMSO (150 μl/well) followed by stirring for 10 min. The optical absorbance at 490 nm of each well was measured on a multiwell plate reader. Cell viability was calculated usingthe following formula: Cell viability (%) = (A490e/A490c) × 100%. A490e and A490c represent the absorbance values from the experimental and control groups, respectively.
Analyses of Cell Cycle and Cell Size
Cell cycle and cell size analyses were performed using flow cytometry. NCI-H460 cells were harvested from adherent cultures by trypsinization followed by centrifugation at 1000 rpm for 5 min. The harvested cells were washed with PBS and then fixed with 70% ethanol in PBS. The fixed cells were collected by centrifugation at 2500 rpm for 3 min and further washed with PBS. The resulting cell pellets were resuspended in PBS with 50 μg/ml propidium iodide for nucleic acid staining and treated with 100 μg/ml RNase A. Cell cycle distribution was analyzed in a BD Biosciences FACSAriaII flow cytometer using ModFit LT software. The mean FSC-H population was used to determine the relative cell size.
Fluorescence Confocal Microscopy
NCI-H460 cells were cultured on glass slides treated with NA-17. 48 h after the NA-17 treatment, the cells were washed with PBS, fixed in 4% paraformaldehyde, and permeabilized with 1% Triton X-100. The cells were washed for four times with PBS. A final concentration of 0.5 μg/ml Hoechst 33258 (Sigma) was included in the final wash to stain cell nuclei. The cell images were collected using a Carl Zeiss LSM 710 fluorescence confocal microscope with a Plan-Neofluar 20× or 40× objective. For immunostaining 48 h after the NA-17 treatment, the cells were washed with PBS, fixed with 4% paraformaldehyde, and incubated with appropriate primary antibodies followed by the addition of Cy3-conjugated secondary antibodies.
Western Blotting Analysis
NCI-H460 cells (3 × 105) were cultured in 6-well plates to 80–90% confluence. The cell culture medium was then replaced with 0.1% FBS, RPMI 1640 medium, and the cells were cultured for another 12 h. The cells were exposed to NA-17 (0–20 μm) for 24 h, then washed once with ice-cold PBS, and extracted with the sample buffer. The cell extracts were separated on polyacrylamide-SDS gels, transferred to nitrocellulose membrane, and probed with primary antibodies. The membrane was incubated with anti-rabbit IgG (alkaline phosphatase-linked) and developed by using an ECL Western blotting system (Eastman Kodak Co.).
siRNA Transfection
Lipofectamine 2000 was used to transfect siRNA. NCI-H460 cells were seeded in 6-well tissue culture plates without antibiotics. After 24 h, the cells reached 60–80% confluence. The cells were washed twice with PBS and then transfected with siRNA or scRNA (100 pmol/well) using Lipofectamine 2000 (5 μl/well) in Opti-MEM I following the manufacturer's instructions. After transfection, the cells were incubated with siRNA or scRNA for 6 h and then placed in fresh culture medium. 24 h after transfection, the cells were used for analysis. The siRNA target sequences are as follows (27): p53, 5′-AAGGAAATTTGCGTGTG GAGT-3′; p21, 5′-GTCACTGTCTTGTACCCTT-3′. The scRNA sequence is as follows: 5′-UUCUCCGAACGUGUCACG UT-3′.
RNA Extraction
Total RNA in NCI-H460 cells was extracted using the RNAsimple Total RNA kit (Tiangen). Briefly, cells grown on 10-cm dishes were treated with NA-17 (5 μm) for 24 h and then lysed. RNA was extracted from the NA-17-treated cells according to the manufacturer's protocol.
Quantitative Real Time PCR
Gene expression levels were examined using the quantitative real time PCR following the manufacturer's instructions (Applied Biosystems, Invitrogen). All real time reactions were performed in a 7500 Fast Real Time PCR System (Applied Biosystems). The amplification reaction solution (20 μl) consisted of 2× SuperReal PreMix (10 μl), the special primer set (1.2 μl), double distilled H2O (7.8 μl), and cDNA (1 μl), and the PCR was performed in the following condition: 40 cycles at 95 °C for 15 s and 60 °C for 60 s. Comparative real time PCR was performed in triplicate, including a no-template control. Relative expression levels were calculated using 2−ΔΔCt. Gene expression levels were analyzed using the 7500 Fast Real Time Sequence Detection System Software (Applied Biosystems). Primer sets used for these analyses are listed in Table 1.
TABLE 1.
Primer sequences
| Primer | Sequences (5′ to 3′) |
|---|---|
| β-Actin | Forward, GTGGGGCGCCCCAGGCACCA |
| Reverse, CTCCTTAATGTCACGCACGATTC | |
| p21 | Forward, CGATGGAACTTCGACTTTGTCA |
| Reverse, GCACAAGGGTACAAGACAGTG | |
| Bcl-xl | Forward, GAGCTGGTGGTTGACTTTCTC |
| Reverse, TCCATCTCCGATTCAGTCCCT | |
| Bax | Forward, CCCGAGAGGTCTTTTTCCGAG |
| Reverse, CCAGCCCATGATGGTTCTGAT | |
| Bak | Forward, GTTTTCCGCAGCTACGTTTTT |
| Reverse, GCAGAGGTAAGGTGACCATCTC | |
| Bim | Forward, TAAGTTCTGAGTGTGACCGAGA |
| Reverse, GCTCTGTCTGTAGGGAGGTAGG | |
| P1 | Forward, AGCCTTAAAAGAATGAATGAA |
| Reverse, GCTTGCTGTGTGACCTTGGGC | |
| P2 | Forward, CTATAGCCATTTACAAGTTAAAAG |
| Reverse, TCAGGAGGAAGAGGCAGTTTCTG | |
| P3 | Forward, AATAGAGCAACCATTAACAC |
| Reverse, TAATCTCTTGTGGACGGCC | |
| P4 | Forward, TGACATTAGGCATTAATCC |
| Reverse, ACTGCACTCCAGCCTGGGAG |
In Vitro Assay of Caspase-3 Activation
The assay was performed based on the ability of the active enzyme to cleave the chromophore of the enzyme substrate Red-DEVD-fmk. The inhibitor-treated cells and control cells were harvested at a density of 1 × 106 cells/ml in RPMI 1640 medium supplemented with 10% FBS. 300 μl of the inhibitor-treated culture or the control culture was incubated with 1 μl of Red-DEVD-fmk for 1 h in a 37 °C incubator with 5% CO2. Flow cytometric analysis was performed using a FACSAriaII flow cytometer equipped with a 543 nm argon laser. The results are presented as percent changes of the activity compared with the control sample.
Coimmunoprecipitation Assay
The coimmunoprecipitation assay was performed as described previously (28). Briefly, cells grown on 10-cm dishes were lysed in 300 μl of immunoprecipitation lysis buffer (20 mm Tris-HCl, pH 7.4, 135 mm NaCl, 1.5 mm MgCl2, 1 mm EDTA, 10% (v/v) glycerol, 1% (w/v) CHAPS with a protein inhibitor mixture (Roche Applied Science)). Immunoprecipitations were carried out using the Catch and Release v2.0 Reversible Immunoprecipitation System kit (Millipore). Columns were washed according to the protocol. The reaction mixture for immunoprecipitation contained 2 μg of immunoprecipitation antibody, 2000 μg of protein, and the antibody capture affinity ligand. Samples were incubated at 4 °C for 2 h and washed five times with 400 μl of 1× Catch and Release Washing Buffer. Antibody-captured proteins were eluted with 45 μl of Catch and Release Denaturing Buffer containing 200 μl of β-mercaptoethanol followed by centrifugation at 5000 rpm for 20 s.
In Situ NA-17 and p53 Interaction Assay
The direct interaction between NA-17 and p53 was detected by mass spectrometry (coimmunoprecipitation-MS). Briefly, cells growing on 10-cm dishes were lysed in 300 μl of immunoprecipitation lysis buffer, and then the whole proteins were incubated with NA-17 (10 μm) at 37 °C for 1 h. Immunoprecipitations were carried out using the Catch and Release v2.0 Reversible Immunoprecipitation System kit with the same protocol as described in the previous section. Finally, the precipitated sample was sonicated for 1 min and analyzed by electrospray ionization mass spectrometry.
Chromatin Immunoprecipitation (ChIP) Assay
The ChIP assay was performed as described previously (27). The histones of the cells grown on 10-cm dishes were cross-linked to DNA by adding formaldehyde directly to the culture medium to a final concentration of 1% followed by incubation for 10 min at 37 °C. The chromatin DNA was sheared into fragments in the range of 200–1000 base pairs by sonication. Immunoprecipitations were carried out using a ChIP assay kit (Millipore) with the anti-phospho-p53 antibody. The immunoprecipitated DNA was quantified by quantitative real time PCR. The primer sets used for these analyses are listed in Table 1.
In Vivo Xenograft Model Assay
Pathogen-free male BALB/c nude mice aged 6 weeks (Changzhou Cavens Experimental Animal Co., Ltd., Changzhou, China) were used to establish the NSCLC xenograft model. The mice were maintained under controlled environmental conditions of constant photoperiod (12-h light-dark cycle at 24 °C and 65–85% humidity). Solid tumors were constructed by subcutaneous injection of 5 × 106 NCI-H460 cells into the flank region of the nude mice (n = 6). The tumor-implanted mice were treated intraperitoneally with vehicle (5% DMSO in saline, v/v) or with 10 mg kg−1 NA-17 per 2 days. 10-Hydroxycamptothecin (6 mg kg−1; per 2 days) was used as a positive control. The body weight and tumor size of the mice were measured three times a week. The tumor size was determined by measuring the length (l) and width (w) and calculating the volume (V = lw2/2).
Statistical Analysis
All data are shown as mean ± standard deviation (S.D.) using two-tailed Student's t test and one-way analysis of variance with Bonferroni multiple comparison post-test. p < 0.05 was considered as a significant difference.
Results
Reporter Gene System Screen Identified NA-17 as a Novel p53 Activator
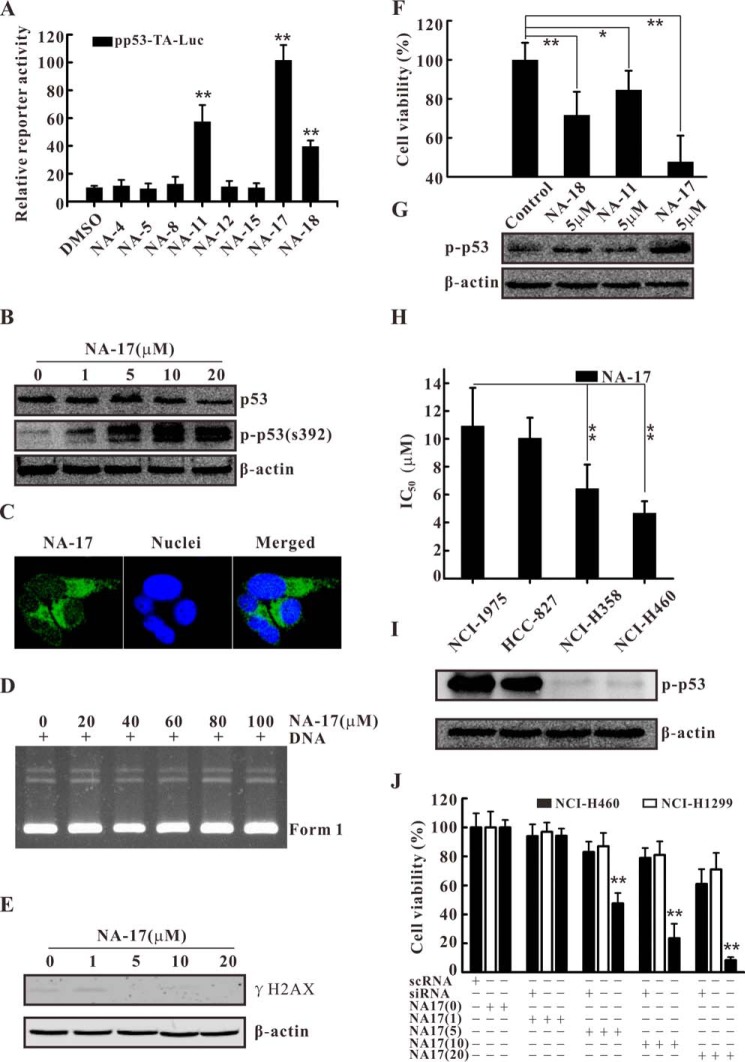
To obtain potential p53 activators, we performed reporter gene system screening. NCI-H460 cells were transiently transfected with pp53-TA-Luc reporter (a p53-responsive reporter) (26). Then the cells were treated with the candidate compounds (5 μm) for 24 h and analyzed by luciferase assay. As shown in Fig. 2A, the screening results showed that NA-17 caused the most significant increase in the p53-induced reporter gene activity. To further determine the mechanism of the p53-dependent reporter gene response, we checked the expression of p53 and the phosphorylation of p53 at serine 392 before and after NA-17 treatment. Phosphorylation of p53 at serine 392 (equivalent to mouse serine 389) is thought to result in conformational changes, thereby enhancing transcriptional activity and DNA-specific binding (29–31). As shown in Fig. 2B, p53 activation induced by NA-17 was mainly achieved via p53 phosphorylation in a dose-dependent manner with a minor contribution from the enhancement in p53 expression.
FIGURE 2.
Identification the novel p53 activator NA-17. A, reporter gene assay for p53 transactivation in NCI-H460 cells. NCI-H460 cells were transfected with the pp53-TA-Luc vector and further treated with a 5 μm concentration of each compound for 24 h. B, NA-17 significantly activated p53 with little influence on its expression. NCI-H460 cells were incubated with the indicated concentrations of NA-17 for 24 h. Whole-cell extracts were prepared and analyzed by Western blotting using antibodies against p53 and phospho-p53 (p-p53) (Ser-392). The same blots were stripped and reprobed with the β-actin antibody to show equal protein loading. C, NA-17 distributed in the cytoplasm of NCI-H460 cells. NCI-H460 cells were treated with 10 μm NA-17 for 1 h and stained with Hoechst 33258. Images were acquired using an LSM 710 microscope (magnification, 400×). D, gel electrophoresis of pBR322 DNA after NA-17 treatment at concentrations of 20–100 μm for 1 h. E, immunoblotting analysis of γH2AX related to NA-17-induced DNA damage. Whole-cell extracts were prepared and analyzed by Western blotting using antibody against γH2AX. F, cell proliferative inhibition effect of NA-11, NA-17, and NA-18 in NCI-H460 cells. G, immunoblotting analysis of phospho-p53 after treatment with NA-11, NA-17, and NA-18. H, IC50 values of NA-17 in four NSCLC cancer cell lines (NCI-1975, HCC-827, NCI-H358, and NCI-H460). I, p53 phosphorylation levels in four NSCLC cancer cell lines (NCI-1975, HCC-827, NCI-H358, and NCI-H460). Whole-cell extracts were prepared and analyzed by Western blotting using antibody against phospho-p53 (Ser-392). The same blots were stripped and reprobed with the β-actin antibody to show equal protein loading. J, selective inhibitory effect of NA-17 on cell proliferation was mainly dependent on p53 profiles. NCI-H460 cells were pre-exposed to EGF receptor siRNA or scRNA for 24 h and then treated with the indicated concentrations of NA-17 for 48 h. Error bars represent S.D. *, p < 0.05 and **, p < 0.01 versus control.
p53 activation is a common response to cellular stresses (32), especially in small molecule-induced p53 activation, with DNA damage as a main cause for p53 activation (33). To explore whether the p53 activation response triggered by NA-17 in NCI-H460 cells was due to DNA damage, we investigated the cellular distribution of NA-17. As depicted in Fig. 2C, NA-17 was mainly distributed in the cytoplasm, suggesting that DNA damage was not the major cause of p53 activation. To further rule out DNA damage-induced p53 activation, a DNA relaxation assay was performed to characterize the interaction between NA-17 and DNA directly. After NA-17 treatment at high concentrations (20–100 μm), there was nearly no change in the conformation or mobility rate of DNA (Fig. 2D). Moreover, a biomarker, γH2AX, was used to detect DNA damage. As depicted in Fig. 2E, no DNA damage was detected after the NA-17 treatment even at high concentrations. Collectively, these results suggested that NA-17 activated p53 without directly triggering DNA damage.
Inactivation of p53 commonly occurs in NSCLCs (6). Thus, we investigated whether NA-17 can inhibit cancer cell viability by restoring p53 function. We first checked the correlation between p53 activation and the inhibitory effect on NCI-H460 cells of the screened compounds NA-11, NA-17, and NA-18. As shown in Fig. 2, F and G, the inhibition in cell viability of these compounds was consistent with the levels of p53 phosphorylation, indicating that cell viability inhibition by these compounds was mainly dependent on p53 activation. Among the tested compounds, NA-17 was the most potent inhibitor and inhibited cell proliferation in a dose-dependent manner (Fig. 2J). Next, we sought to identify whether NA-17 can inhibit cell proliferation in other NSCLC cell lines. As shown in Fig. 2H, NA-17 also exhibited an anticancer effect in four other NSCLC cell lines. The inhibition effects in these NSCLC cell lines were distinctive to each other. This might be due to the fact that the activation states of endogenous p53 are different among NSCLC cell lines (34, 35). To investigate whether the activation state of endogenous p53 is essential for NA-17-induced inhibition in NSCLC cell lines, we examined the p53 phosphorylation level in different NSCLC cell lines. Our result showed that NSCLC cell lines insensitive to the NA-17 treatment showed high activation levels of endogenous p53 (Fig. 2I).
Moreover, the effect of NA-17 on the p53-null NSCLC cell line, NCI-H1299, and p53 siRNA-treated NCI-H460 cells was investigated. Both types of cells were exposed to different concentrations of NA-17 for 48 h. As shown in Fig. 2J, both the p53-null NCI-H1299 cells and the p53-ablated NCI-H460 cells were insensitive to the NA-17 treatment.
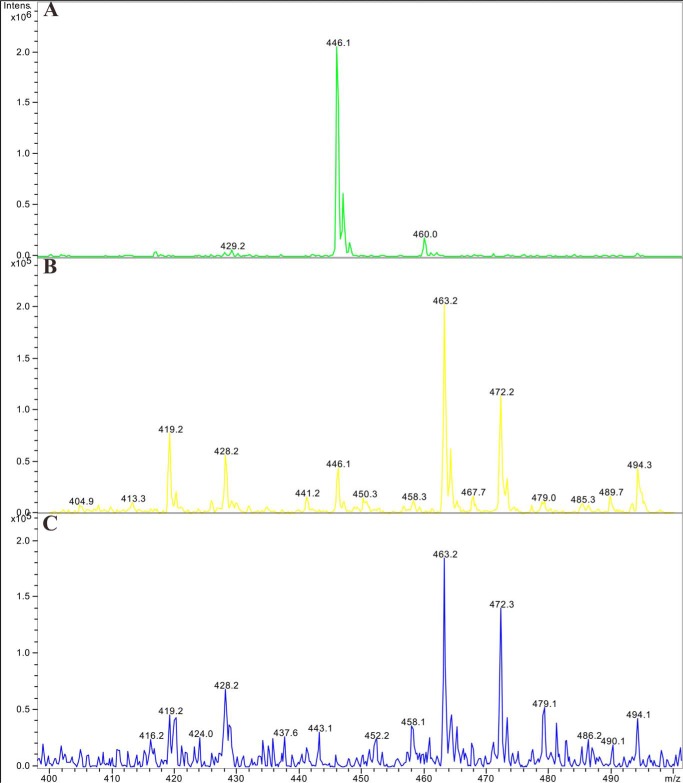
To examine whether there were direct interactions between NA-17 and p53, coimmunoprecipitation-MS experiments were performed with NCI-H460 cell extracts. As shown in Fig. 3, immunoprecipitation with antibodies against phospho-p53 (Fig. 3B) followed by detection of NA-17 using mass spectrometry (Fig. 3A) demonstrated that NA-17 directly interacted with p53 (Fig. 3B).
FIGURE 3.
Mass spectrometry detection of the interaction between NA-17 and p53. A, NA-17 was dissolved in DMSO and analyzed by electrospray ionization mass spectrometry (m/z 446.1; [M + H]+). B, NA-17 directly interacted with p53. The whole proteins extracted from NCI-H460 cells were incubated with NA-17 (10 μm) at 37 °C for 1 h. Immunoprecipitations were performed with anti-phospho-p53, and then the sample was sonicated for 1 min and finally analyzed by electrospray ionization mass spectrometry (m/z 446.1; [M + H]+). C, the negative control of the in situ NA-17 and p53 interaction assay. The whole proteins extracted from NCI-H460 cells were incubated with DMSO (1%) at 37 °C for 1 h. Immunoprecipitations were performed using the same method. Intens., intensity.
NA-17 Showed Antiproliferation Activity toward a Wide Range of Cancer Cell Lines
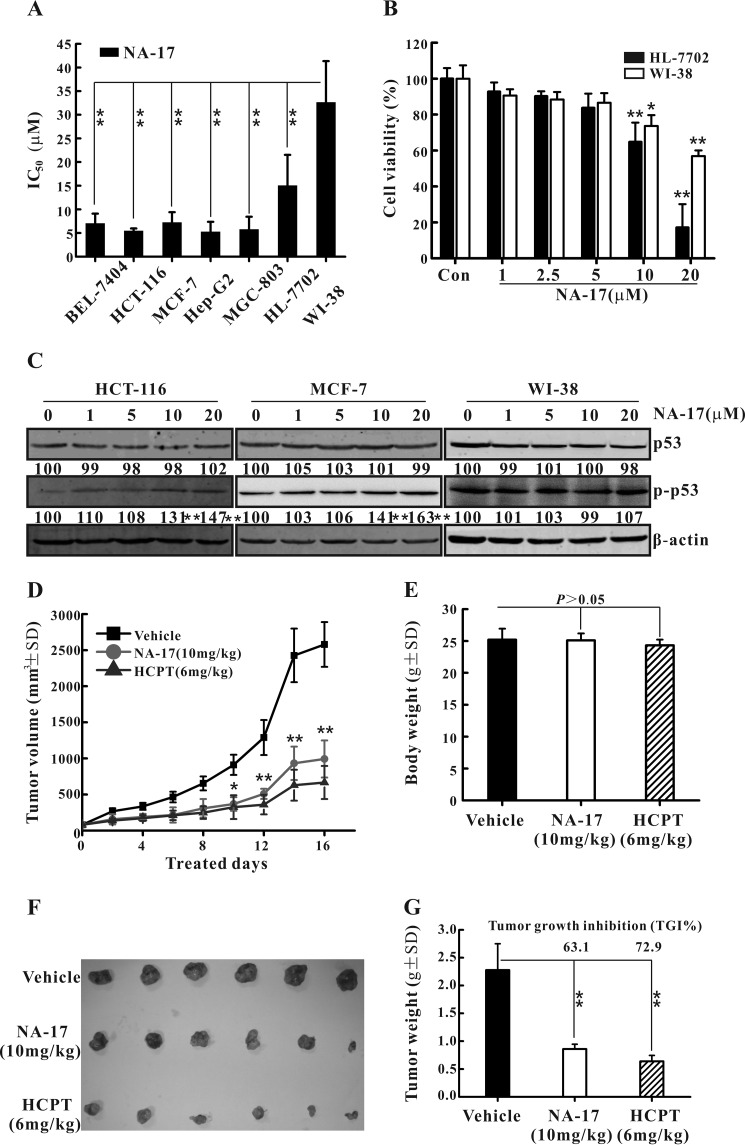
The anticancer effect of NA-17 against different cancer cell lines was characterized by the MTT assay. As depicted in Fig. 4A, NA-17 exhibited proliferation inhibition on a wide range of cancer cell lines with IC50 values in the micromolar range. To examine the effect of NA-17 on normal cells, we treated hepatic cells, HL-7702, and lung epithelial cells, WI-38, with NA-17. The cell viability of HL-7702 and WI-38 cells was reduced after NA-17 treatment for 48 h. However, inhibitory rates (no more than 50%) were much lesser than those of cancer cells (Fig. 4B). To demonstrate that the observed difference in the inhibition effects of NA-17 between cancer cells and normal cells was not caused by the difference in culturing conditions, the cytotoxicity of NA-17 on normal cell lines was also examined in RPMI 1640 medium. The IC50 values of NA-17 on normal cell lines in this culturing condition were almost identical to those in the DMEM culturing condition (data not shown; p > 0.05). These results suggested that NA-17 exhibited selective toxicity for cancerous over normal cells.
FIGURE 4.
NA-17 exhibited antiproliferative effects on a broad range of cancer cell lines but low toxicity to normal cells. A, IC50 values of NA-17 in different cell lines. B, cytotoxicity of NA-17 to normal cell lines HL-7702 and WI-38. C, phosphorylation levels of p53 after NA-17 treatment in HCT-116, MCF-7, and WI-38 cell lines. Cells were incubated with the indicated concentrations of NA-17 for 24 h. Whole-cell extracts were prepared and analyzed by Western blotting using antibodies against p53 and pospho-p53 (p-p53). The same blots were stripped and reprobed with the β-actin antibody to show equal protein loading. D, in vivo anticancer activity of NA-17 in NCI-H460 xenograft mice. Tumor-bearing mice were administered the vehicle (negative control), NA-17 (10 mg/kg/2 days) or 10-hydroxycamptothecin (HCPT) (6 mg/kg/2 days; positive control). The figure shows the average measured tumor volumes. E, body weights of the mice recorded at the end of the treatments. F, the images of the excised tumors from each group. G, the weight of the excised tumors from each group. Data are presented as the mean. Error bars represent S.D. n = 6. *, p < 0.05 and **, p < 0.01 versus control.
To investigate whether the anticancer effect of NA-17 against other cancer cell lines was due to p53 activation, we examined the phosphorylation level of p53 after NA-17 treatment in the breast cancer cell line MCF-7, the colon cancer cell line HCT-116, and the normal cell line WI-38. Our results showed that NA-17 increased the p53 phosphorylation level in the HCT-116 and MCF-7 cell lines in a dose-dependent manner. However, nearly no change in the p53 phosphorylation level was observed in WI-38 cells (Fig. 4C).
The in vivo anticancer activity of NA-17 was studied in NCI-H460 xenograft tumors in nude mice. The tumor-bearing mice treated with NA-17 (10 mg/kg/2 days) showed significant tumor growth inhibition over the treatment period compared with the negative control (Fig. 4, D, F, and G). The tumor growth inhibition values of NA-17 and the positive control, 10-hydroxycamptothecin, were 63.1 and 72.9%, respectively. In all experiments, except for a slight body weight decrease in 10-hydroxycamptothecin-treated animals, no differences in clinical signs of toxicity and food consumption were found between the control group and NA17-treated animals (Fig. 4E).
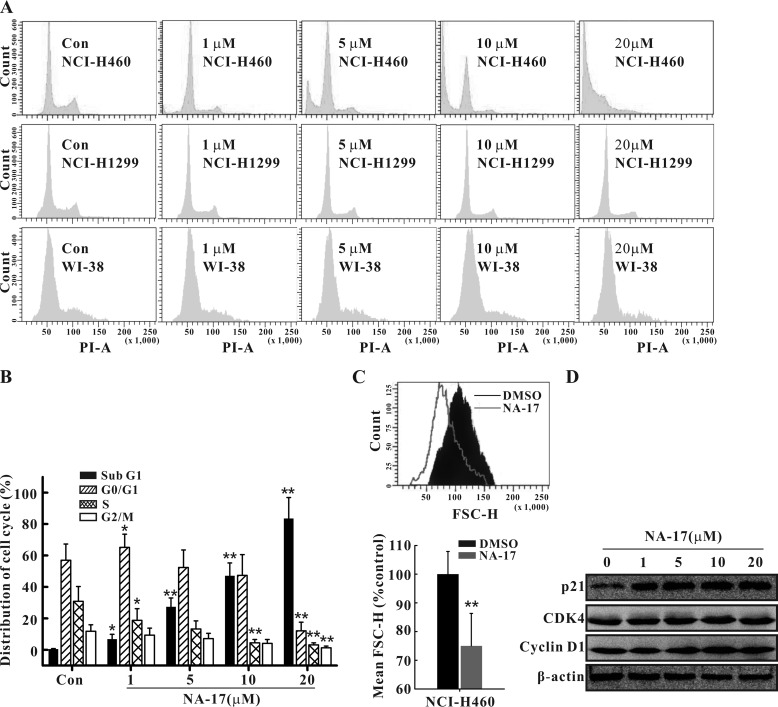
NA-17 Induced Cell Cycle Arrest and Cell Size Changes in NCI-H460 Cells
To study the potential mechanistic pathway responsible for cell proliferation inhibition of NA-17, we investigated NA-17-induced changes in the cell cycle. As shown in Fig. 5A, NA-17 treatment caused a pronounced increase in the cell population in the G0/G1 phase and the sub-G1 phase (a specific phase of cell death) as well as a marked decrease in the S and G2 phases (Fig. 5A and B). In contrast, there was no observable cell cycle arrest in the WI-38 cells or the p53-null NCI-H1299 cells (Fig. 5A).
FIGURE 5.
Effects of NA-17 on cell cycle distribution and cell size in NCI-H460 cells. A, the regulation of cell cycle arrest by NA-17 was examined by FACS analysis with propidium iodide (PI) staining in NCI-H460, WI-38, and NCI-H1299 cells. B, the distribution of cell cycles in NA-17-treated NCI-H460 cells was plotted. Data are presented as mean ± S.D. from three independent measurements. C, NA-17 treatment of NCI-H460 cells reduced the cell size. Cells were treated for 48 h with DMSO or 5 μm NA-17 and analyzed by FACS for relative cell size. Histograms of the forward scatter (FSC-H) for the treated cells and mean FSC-H values are shown. D, immunoblotting analysis of proteins related to NA-17-induced cell cycle arrest. Whole-cell extracts were prepared and analyzed by Western blotting using antibodies against the proteins indicated. Error bars represent S.D. *, p < 0.05 and **, p < 0.01 versus control (Con).
Moreover, NA-17 induced a reduction in the cell size (Fig. 5C, top panel). The relative cell size (mean FSC-H) of NA-17-treated NCI-H460 cells was 74.5% of that of the control cells (Fig. 5C, bottom panel). Collectively, these results showed that the antiproliferation activity of NA-17 was due to the G1 arrest-induced growth inhibition and cell size reduction.
To investigate the mechanism of NA-17-induced cell cycle arrest, we examined the expression level of p21, a protein inhibitor of the complexes of cyclin D and cyclin-dependent kinases (CDKs), which are involved in G1 to S phase progression (36). Our results showed that the NA-17 treatment led to a significant increase of p21 in a dose-dependent manner (Fig. 5D). However, almost no change was detected in the expression levels of cyclin D and CDK4. These results suggested that the observed G1 arrest induced by NA-17 resulted from inhibition of the CDK activity by p21 rather than reduced expression of cyclin D and CDK4.
NA-17 Induced Cell Apoptosis through the Intrinsic Apoptotic Pathway
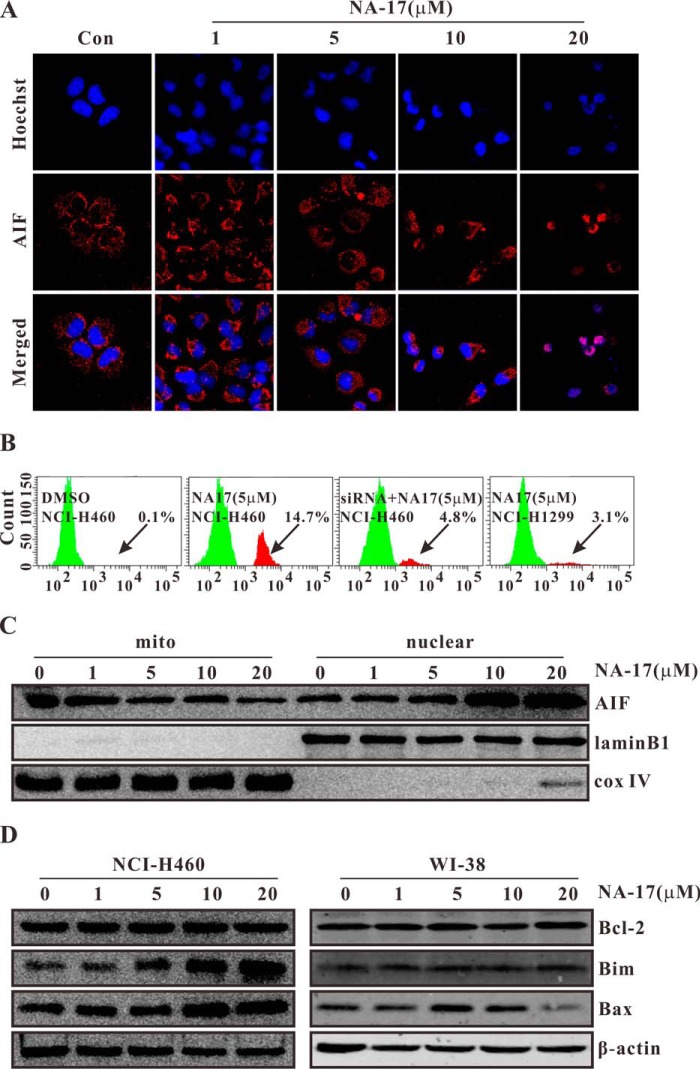
To obtain further insight into the mechanisms of the tumor suppression activity of NA-17, we investigated whether the sub-G1 phase cell cycle arrest induced by NA-17 was due to apoptosis. The NA-17-treated cells were studied via Hoechst 33258 staining and immunostaining of AIF for apoptosis identification. As shown in Fig. 6A, the cells treated with NA-17 displayed chromatin condensation and fragmentation. The condensed apoptotic nuclei were readily observed compared with the control cells without the NA-17 treatment. Furthermore, after treatment with NA-17, AIF was released from mitochondria and translocated to the nucleus to trigger apoptosis. To confirm the proapoptotic effect of NA-17 in NCI-H460 cells, cell apoptosis with activated caspase-3 was also studied. As shown in Fig. 6B, additional peaks for cells with activated caspase-3 were observed in the NA-17-treated cells compared with the control group. The proportion of caspase-3-activated cells after NA-17 treatment was 14.7% in the NCI-H460 cells. However, this value decreased to 4.8% in the p53-ablated NCI-H460 cells and to 3.1% in the p53-null NCI-H1299 cells.
FIGURE 6.
NA-17 induced cell apoptosis through the intrinsic apoptotic pathway. A, the induction of apoptosis by NA-17 was examined with Hoechst 33258 and AIF immunostaining in NCI-H460 cells. Images were acquired using an LSM 710 microscope (magnification, 400×). B, caspase-3 activation after NA-17 treatment of NCI-H460, p53-ablated NCI-H460, and p53-null NCI-H1299 cells. Cells were incubated with 5 μm NA-17 for 24 h. The arrows indicate the caspase-3-activited cells. C, subcellular localizations of AIF induced by NA-17. NCI-H460 cells treated with NA-17 were subjected to subcellular fractionation, and immunoblotting was performed with mitochondrial and nuclear fractions. Cytochrome c oxidase (cox) IV and lamin B1 were used as the mitochondrial and nuclear marker proteins, respectively. D, immunoblotting analysis of proteins related to the mitochondrially mediated intrinsic apoptotic pathway evoked by NA-17. Whole-cell extracts were prepared and analyzed by Western blotting using antibodies against the proteins indicated. The same blots were stripped and reprobed with the β-actin antibody to show equal protein loading. mito, mitochondrial.
Notably, during apoptotic events, AIF is usually released from mitochondria and translocated to cell nuclei to trigger apoptosis. To probe whether the NA-17-induced apoptosis was caused by the AIF-mediated pathway, we examined the protein level of AIF in cell nuclei and mitochondria before and after the NA-17 treatment. As depicted in Fig. 6C, as the concentration of NA-17 increased, the amount of AIF in the mitochondria was reduced after 24-h treatment due to the rapid translocation into cell nuclei.
Because AIF is a mitochondrial protein (37), we next examined the death effectors in mitochondria that participated in the death signaling. The proteins from the Bcl-2 family were chosen as the candidates. As shown in Figs. 6D (left) and 7B, upon NA-17 treatment, we observed a significant increase in the protein levels of Bak, Bim, and Bax and a pronounced decrease in the protein level of Bcl-xl. There was no significant change in the protein level of Bcl-2. In comparison, there was nearly no change in the protein levels of the Bcl-2 family members after NA-17 treatment in the normal WI-38 cells (Fig. 6D, right). These results indicated that the NA-17-induced cell apoptosis was achieved via the mitochondrially mediated intrinsic apoptotic pathway.
FIGURE 7.
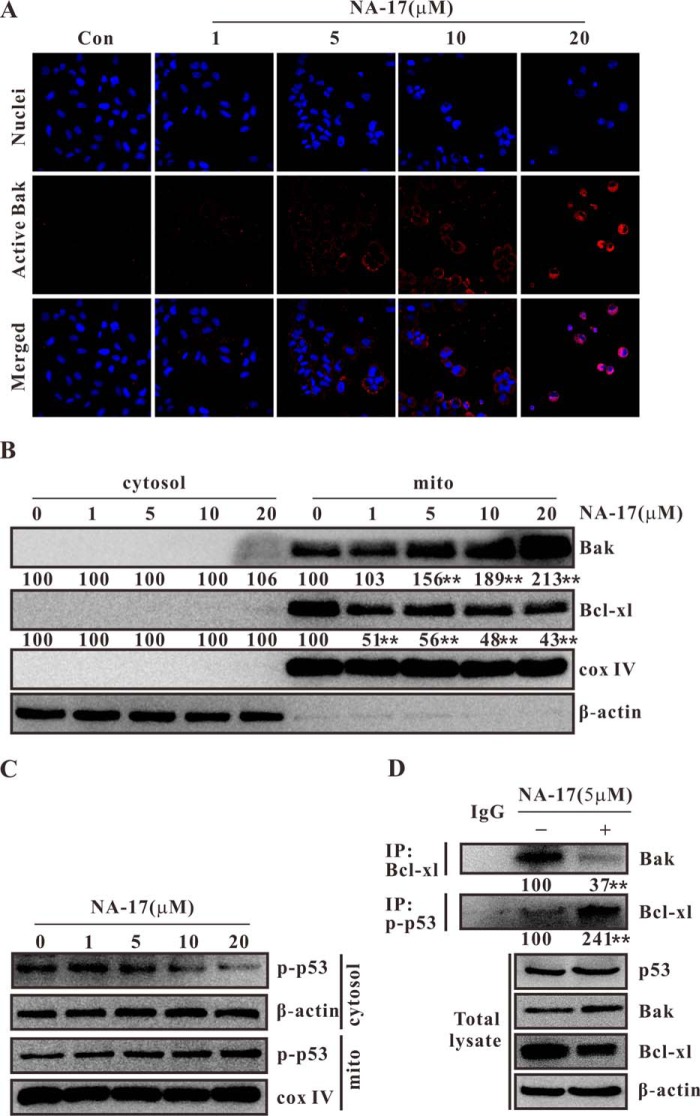
NA-17-induced mitochondrial accumulation of phospho-p53 conformationally activated Bak by reorganizing the Bak·Bcl-xl complex. A, NA-17 induces conformational Bak activation. Cells were immunostained with antibody for active Bak. Images were acquired using an LSM 710 microscope (magnification, 400×). B, subcellular localization and variation of Bak and Bcl-xl after NA-17 treatment. NCI-H460 cells treated with NA-17 were subjected to subcellular fractionation, and immunoblotting was performed with cytosolic (cytosol) and mitochondrial (mito) fractions. β-Actin and cytochrome c oxidase (cox) IV were used as the cytosolic and mitochondrial marker proteins, respectively. C, phospho-p53 (p-p53) accumulation in mitochondria after NA-17 treatment. NCI-H460 cells treated with NA-17 were subjected to subcellular fractionation, and immunoblotting was performed with cytosolic and mitochondrial fractions. β-Actin and cytochrome c oxidase IV were used as the cytosolic and mitochondrial marker proteins, respectively. D, status of the Bak/Bcl-xl interaction and phospho-p53/Bcl-xl interaction in NCI-H460 cells treated with 5 μm NA-17 for 24 h. **, p < 0.01 versus control (Con). IP, immunoprecipitation.
NA-17 Induced Conformational Activation of Bak for Cell Apoptosis by Reorganizing the Bak·Bcl-xl Complex
Conformational activation of Bak via N-terminal exposure is a distinct marker of the sensitized mitochondrial apoptotic pathway (28). NA-17 treatment induced apoptosis in the NCI-H460 cells and caused an increase in the protein level of Bak. An immunohistochemistry method was used to examine whether NA-17 activated Bak through conformational changes. As shown in Fig. 7A, there was nearly no activated Bak in the control group. Notably, inactivated Bak cannot function as a death effector. After NA-17 treatment, conformationally altered Bak was generated and further captured at the N terminus by the Bak antibody. Thus, NA-17 was able to conformationally activate Bak for cell apoptosis.
We further examined the cellular distribution of Bak. Confocal microscopy examination showed that Bak was mainly located in cytoplasm, not in cell nuclei (Fig. 7A). A subcellular fractionation assay suggested that Bak was present only in mitochondria and that NA-17 was able to up-regulate the Bak expression in mitochondria in a dose-dependent manner (Fig. 7B).
Previous studies have shown that an antiapoptotic Bcl-2 family member, Bcl-xl, controls the conformational status of Bak via forming the Bak·Bcl-xl complex (28). Conformational activation of Bak triggers dissociation of such complexes, thereby generating Bak homodimers (18). We performed coimmunoprecipitation analysis to probe whether the NA-17-induced Bak activation was caused by disruption of the Bak·Bcl-xl complex. Our result showed that NA-17 disrupted the Bak/Bcl-xl interaction (Fig. 7D) and down-regulated the Bcl-xl expression in mitochondria (Fig. 7B).
NA-17 was demonstrated to be a p53 activator in this study. It significantly promoted p53 phosphorylation without altering the expression of p53 (Fig. 2B). It is noteworthy that phospho-p53 was mainly accumulated in mitochondria (Fig. 7C). Previous studies have shown that activated p53 is usually translocated into mitochondria to trigger apoptosis through the transcription-independent p53 pathways (18, 19). These results guided us to study the potential role of p53 in NA-17-induced Bak activation. To analyze the role of p53 in perturbing the Bak/Bcl-xl interaction after NA-17 treatment, we coimmunoprecipitated phospho-p53 with Bcl-xl and Bcl-xl with Bak in NCI-H460 cells. As shown in Fig. 7D, direct interaction between phospho-p53 and Bcl-xl was observed after the NA-17 treatment, which was consistent with a previous report (38). It is highly possible that the binding of phosopho-p53 to the Bcl-xl molecules in the Bak·Bcl-xl complexes induced the dissociation of the Bak·Bcl-xl complexes, thereby leading to the activation of Bak.
NA-17 Regulated Cell Cycle and Cell Apoptosis-related Targets via Transcriptional Regulation
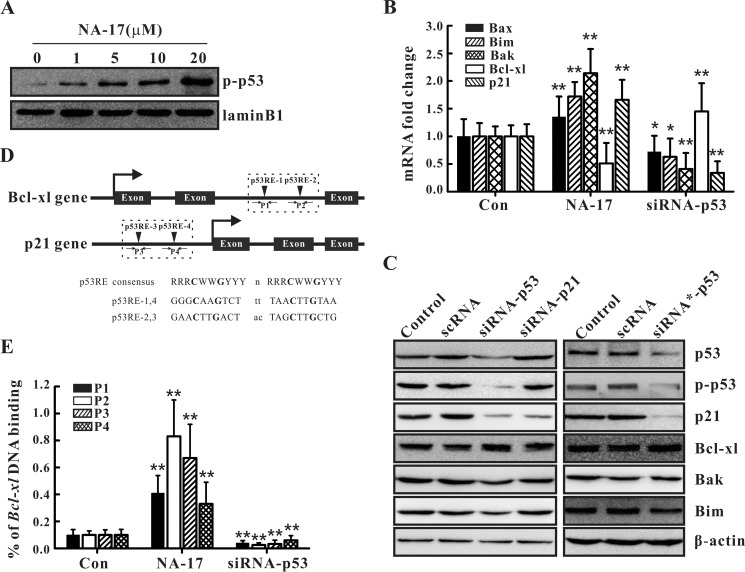
As evident from the experimental results presented in this work, the NA-17 treatment caused alterations in the expression levels of several proteins (Figs. 5D, 6D, and 7B). Thus, NA-17-induced p53 activation might play an important role in transcriptionally regulating related genes as p53 usually activates the transcriptional cascade in response to signal stimulation (10). To test this hypothesis, we examined whether phospho-p53 was translocated into cell nuclei from the cytoplasm after NA-17 treatment. As shown in Fig. 8A, with increasing concentrations of NA-17, the amount of phospho-p53 in cell nuclei was significantly increased along with a pronounced reduction in the amount in the cytoplasm (Fig. 7C).
FIGURE 8.
NA-17-induced nuclear accumulation of phospho-p53 transcriptionally regulated the related targets by directly binding to DNA. A, phospho-p53 (p-p53) accumulation in cell nuclei after NA-17 treatment. NCI-H460 cells treated with NA-17 were subjected to subcellular fractionation, and immunoblotting was performed with nuclear fractions. Lamin B1 was used as the nuclear marker protein. B, quantitative real time PCR assays were performed in non-treated (control (Con)), NA-17-treated (5 μm), or p53 siRNA-treated NCI-H460 cells. C, immunoblotting analysis of p21 and the Bcl-2 family proteins after p53 siRNA or p21 siRNA treatment. D, putative p53 response elements were located in the intron of the Bcl-xl gene and in the promoter of the p21 gene. D, ChIP assays based on an anti-phospho-p53 antibody and primer pairs P1, P2, P3, or P4 were performed in non-treated, NA-17-treated (5 μm), or p53 siRNA-treated NCI-H460 cells. Error bars represent S.D.
Next, we investigated whether NA-17 regulated the related gene targets at the transcriptional level by performing quantitative real time PCR assays in NCI-H460 cells. Compared with the control group, the NA-17 treatment increased the transcription levels of p21, Bim, Bax, and Bak but decreased the transcriptional level of Bcl-xl, consistent with the immunoblotting results (Figs. 5D, 6D, and 7B). In contrast, ablation of p53 in NCI-H460 cells by p53 siRNA significantly decreased the transcription levels of p21, Bim, Bax, and Bak but increased the transcriptional level of Bcl-xl compared with the control group (Fig. 8, B and C). To rule out off-target effects, p21 siRNA and p53 siRNA* (a commercially purchased siRNA) were used in parallel experiments. As depicted in the left panel of Fig. 8C, ablation of p21 in NCI-H460 cells significantly decreased p21 expression. However, there was nearly no influence on the expression levels of Bim, Bax, Bcl-xl, and Bak. Meanwhile, ablation of p53 in NCI-H460 cells by p53 siRNA* produced a similar effect on gene expression as that generated by p53 siRNA (Fig. 8C, right panel). Collectively, these results suggested that NA-17-induced nuclear accumulation of phospho-p53 is a critical cause for cell cycle- and cell apoptosis-related changes.
To determine whether phospho-p53 directly bound to the target genes, ChIP assays were performed. Previous studies have documented that p53 can directly bind to the genes of Bax, p21, and Bim (10, 39, 40). Herein, we investigated its interactions with Bak and Bcl-xl. A survey of the p53FamTag database showed that there was no putative p53-responsive element (p53RE) in the intron and promoter region of Bak, but two putative p53REs (p53RE-1 and p53RE-2) were identified within the Bcl-xl intron (Fig. 8D). To eliminate false positive results in the ChIP assays, the canonical p53 target, p21, which possesses two p53REs (p53RE-3 and p53RE-4) in its promoter (Fig. 8D), was adopted as a positive control. The ChIP assays demonstrated that NA-17 treatment increased p53 binding to the intron of Bcl-xl at p53RE-1 and p53RE-2 and to the promoter of p21 at p53RE-3 and p53RE-4. Ablation of p53 in NCI-H460 cells, however, caused a pronounced reduction in p53 binding to the intron of Bcl-xl and to the promoter of p21 (Fig. 8E). These results indicated that NA-17-induced p53 activation enhanced p53 binding to the intron of Bcl-xl directly, resulting in suppression of the transcription of Bcl-xl.
Discussion
In this study, we discovered a novel naphthalimide compound, NA-17, that can directly active p53 in NSCLC cells without causing DNA damage. Our results showed that NA-17 effectively suppressed cell proliferation of lung cancer cell lines in which p53 is inactivated. Moreover, we demonstrated that the antitumor effect of NA-17 is likely due to the induction of apoptosis and cell cycle arrest by phospho-p53.
Conventional chemotherapeutic agents usually fail clinical tests due to the incapability to distinguish between healthy and cancer cells. Our studies on the cellular performance of NA-17 revealed that it strongly inhibited the cell viability of a broad range of cancer cell lines but exhibited low toxicity to normal cell lines. Notably, NA-17 displayed distinctive inhibitory effects toward different NSCLC cell lines. This phenomenon might be due to the difference in the activation state of endogenous p53 between different NSCLC cell lines (33, 34) as NA-17 was potent to the NSCLC cell lines with inactivated p53.
Cell cycle arrest is a common response to p53 activation (15–17). NA-17 induced cell cycle arrest in the G1 phase. It transcriptionally up-regulated the expression of p21 with no influence on the expression levels of cyclin D and CDK4. Further studies indicated that NA-17 induced accumulation of phospho-p53 in cell nuclei, which triggered cell cycle arrest through transcriptional regulation.
We further examined the cell apoptosis induced by NA-17 in NCI-H460 cells. We found that NA-17-induced cell apoptosis was accompanied by chromatin condensation and fragmentation, caspase-3 activation, and AIF release from mitochondria. The mitochondrially mediated intrinsic apoptotic pathway always involves small molecule-induced cell apoptosis (41). We identified the death effectors in mitochondria that participated in the NA-17-induced apoptosis, including Bak and Bim. The ChIP assays showed that the NA-17-induced phospho-p53 molecules accumulated in cell nuclei directly bound to the Bcl-xl gene and inhibited its transcription.
Conformational activation of Bak is a characteristic marker of the sensitized mitochondrial apoptotic pathway (28). Here we found that NA-17-induced mitochondrial accumulation of phospho-p53 conformationally activated Bak by reorganizing the Bak·Bcl-xl complex. Coimmunoprecipitation analysis suggested that NA-17-induced phospho-p53 directly bound to the Bcl-xl molecules in the Bak·Bcl-xl complexes, causing dissociation of Bak from the complexes. This process activated Bak for cell apoptosis. It has been shown previously that Bcl-xl is one of the direct targets of p53, and it can bind cytosolic p53 at its DNA-binding domain (38, 42). The regulation of Bcl-xl by p53 is not a cell-specific phenomenon as endogenous p53·Bcl-xl complexes have been detected in various cell lines, such as MCF-10A, MCF-7, and HeLa cells (28).
The activation of p53 is usually achieved via disrupting the interaction between p53 and the MDM2 protein due to DNA damage (7–9) or ADP-ribosylation factor function (43, 44), thereby preventing the degradation of the p53 protein. Alternatively, phosphorylation of p53 at Ser-392 can cause conformational changes that enhance DNA binding and increase transcriptional activity (29–31). In this study, NA-17 might activate p53 through a third mechanism as Ser-392 can be phosphorylated only by UV radiation according to previous reports (30, 31). NA-17 might directly interact with p53 (Fig. 3) and trigger phosphorylation of p53.
In conclusion, we synthesized NA-17, characterized its anticancer properties, and investigated the mechanism of action. Our results demonstrated that NA-17 is a promising tumor-specific anticancer agent for NSCLC therapies and an efficient adjuvant in p53 restoration-mediated anticancer therapies. This work also highlighted the importance of establishing molecular regulatory networks for mechanistic pharmacological studies.
Author Contributions
G. Z. and Y. A. planned and performed all experiments, carried out data analyses, and drafted the article. G. Z., Y. P., and Z.-F. C. designed the research. X. L., H. Z., and Y. Z. synthesized the compounds used in this study. Y. W., F. M., and J. Y. performed and analyzed the experiments shown in Figs. 1 and 2. Y. L. and Z. Z. contributed valuable discussions during the study.
This work was supported by Natural Science Foundation of China Grant 81401912; The Ministry of Education Innovation Team Grant IRT1225; Natural Science Foundation of Guangxi Province Grants 2013GXNSFBA019159, 2014GXNSFAA118165, and 2015GXNSFDA139010; Guangxi Scientific Research and Technology Development Project Grant 13349002; Guangxi Pharmaceutical Industry Talent Small Highland Project Grants 1309 and 1312; the Foundation of State Key Laboratory Cultivation Base for the Chemistry and Molecular Engineering of Medicinal Resources; Ministry of Science and Technology of China Grants CMEMR2015-A09 and CMEMR2013-A07; and the Bagui scholar program (to Z. Z.). The authors declare that they have no conflicts of interest with the contents of this article.
- NSCLC
- non-small cell lung cancer
- Bcl-2
- B-cell lymphoma/leukemia-2
- Bak
- Bcl-2 antagonist/killer
- Bax
- Bcl-2-associated X protein
- Bim
- Bcl-2-like 1
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- AIF
- apoptosis-inducing factor
- p53RE
- p53-responsive element
- scRNA
- scrambled RNA
- DMSO
- dimethyl sulfoxide
- FSC-H
- forward scatter height
- fmk
- fluoromethyl ketone
- CDK
- cyclin-dependent kinase.
References
- 1. DeSantis C. E., Lin C. C., Mariotto A. B., Siegel R. L., Stein K. D., Kramer J. L., Alteri R., Robbins A. S., and Jemal A. (2014) Cancer treatment and survivorship statistics. CA Cancer J. Clin. 64, 252–271 [DOI] [PubMed] [Google Scholar]
- 2. Siegel R., Ma J., Zou Z., and Jemal A. (2014) Cancer statistics. CA Cancer J. Clin. 64, 9–29 [DOI] [PubMed] [Google Scholar]
- 3. Mok T. S., Wu Y. L., Thongprasert S., Yang C. H., Chu D. T., Saijo N., Sunpaweravong P., Han B., Margono B., Ichinose Y., Nishiwaki Y., Ohe Y., Yang J. J., Chewaskulyong B., Jiang H., Duffield E. L., Watkins C. L., Armour A. A., and Fukuoka M. (2009) Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 361, 947–957 [DOI] [PubMed] [Google Scholar]
- 4. Mitsudomi T., Morita S., Yatabe Y., Negoro S., Okamoto I., Tsurutani J., Seto T., Satouchi M., Tada H., Hirashima T., Asami K., Katakami N., Takada M., Yoshioka H., Shibata K., Kudoh S., Shimizu E., Saito H., Toyooka S., Nakagawa K., Fukuoka M., and West Japan Oncology Group (2010) Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 11, 121–128 [DOI] [PubMed] [Google Scholar]
- 5. Feldser D. M., Kostova K. K., Winslow M. M., Taylor S. E., Cashman C., Whittaker C. A., Sanchez-Rivera F. J., Resnick R., Bronson R., Hemann M. T., and Jacks T. (2010) Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature 468, 572–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Junttila M. R., Karnezis A. N., Garcia D., Madriles F., Kortlever R. M., Rostker F., Brown Swigart L., Pham D. M., Seo Y., Evan G. I., and Martins C. P. (2010) Selective activation of p53-mediated tumour suppression in high-grade tumours. Nature 468, 567–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eliaš J., Dimitrio L., Clairambault J., and Natalini R. (2014) The p53 protein and its molecular network: modelling a missing link between DNA damage and cell fate. Biochim. Biophys. Acta 1844, 232–247 [DOI] [PubMed] [Google Scholar]
- 8. Abe Y., Matsumoto S., Wei S., Nezu K., Miyoshi A., Kito K., Ueda N., Shigemoto K., Hitsumoto Y., Nikawa J., and Enomoto Y. (2001) Cloning and characterization of a p53-related protein kinase expressed in interleukin-2-activated cytotoxic T-cells, epithelial tumor cell lines, and the testes. J. Biol. Chem. 276, 44003–44011 [DOI] [PubMed] [Google Scholar]
- 9. Zhang Y., and Hunter T. (2014) Roles of Chk1 in cell biology and cancer therapy. Int. J. Cancer 134, 1013–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Riley T., Sontag E., Chen P., and Levine A. (2008) Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9, 402–412 [DOI] [PubMed] [Google Scholar]
- 11. Chipuk J. E., Kuwana T., Bouchier-Hayes L., Droin N. M., Newmeyer D. D., Schuler M., and Green D. R. (2004) Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 303, 1010–1014 [DOI] [PubMed] [Google Scholar]
- 12. Brady C. A., Jiang D., Mello S. S., Johnson T. M., Jarvis L. A., Kozak M. M., Kenzelmann Broz D., Basak S., Park E. J., McLaughlin M. E., Karnezis A. N., and Attardi L. D. (2011) Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell 145, 571–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Green D. R., and Kroemer G. (2009) Cytoplasmic functions of the tumour suppressor p53. Nature 458, 1127–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vaseva A. V., Marchenko N. D., Ji K., Tsirka S. E., Holzmann S., and Moll U. M. (2012) p53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell 149, 1536–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mollereau B., and Ma D. (2014) The p53 control of apoptosis and proliferation: lessons from Drosophila. Apoptosis 19, 1421–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wawryk-Gawda E., Chylińska-Wrzos P., Lis-Sochocka M., Chłapek K., Bulak K., Jędrych M., and Jodłowska-Jędrych B. (2014) P53 protein in proliferation, repair and apoptosis of cells. Protoplasma 251, 525–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yao G., Qi M., Ji X., Fan S., Xu L., Hayashi T., Tashiro S., Onodera S., and Ikejima T. (2014) ATM-p53 pathway causes G2/M arrest, but represses apoptosis in pseudolaric acid B-treated HeLa cells. Arch. Biochem. Biophys. 558, 51–60 [DOI] [PubMed] [Google Scholar]
- 18. Westphal D., Dewson G., Czabotar P. E., and Kluck R. M. (2011) Molecular biology of Bax and Bak activation and action. Biochim. Biophys. Acta 1813, 521–531 [DOI] [PubMed] [Google Scholar]
- 19. Llambi F., Moldoveanu T., Tait S. W., Bouchier-Hayes L., Temirov J., McCormick L. L., Dillon C. P., and Green D. R. (2011) A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol. Cell 44, 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen L., Willis S. N., Wei A., Smith B. J., Fletcher J. I., Hinds M. G., Colman P. M., Day C. L., Adams J. M., and Huang D. C. (2005) Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol. Cell 17, 393–403 [DOI] [PubMed] [Google Scholar]
- 21. Ingrassia L., Lefranc F., Kiss R., and Mijatovic T. (2009) Naphthalimides and azonafides as promising anti-cancer agents. Curr. Med. Chem. 16, 1192–1213 [DOI] [PubMed] [Google Scholar]
- 22. Van Quaquebeke E., Mahieu T., Dumont P., Dewelle J., Ribaucour F., Simon G., Sauvage S., Gaussin J. F., Tuti J., El Yazidi M., Van Vynckt F., Mijatovic T., Lefranc F., Darro F., and Kiss R. (2007) 2,2,2-Trichloro-N-({2-[2-(dimethylamino)ethyl]-1,3-dioxo-2,3-dihydro-1H-benzo[de]isoquinolin-5-yl}carbamoyl)acetamide (UNBS3157). J. Med. Chem. 50, 4122–4134 [DOI] [PubMed] [Google Scholar]
- 23. Freeman C. L., Swords R., and Giles F. J. (2012) Amonafide: a future in treatment of resistant and secondary acute myeloid leukemia? Expert Rev. Hematol. 5, 17–26 [DOI] [PubMed] [Google Scholar]
- 24. Zhu H., Miao Z. H., Huang M., Feng J. M., Zhang Z. X., Lu J. J., Cai Y. J., Tong L. J., Xu Y. F., Qian X. H., and Ding J. (2009) Naphthalimides induce G2 arrest through the ATM-activated Chk2-executed pathway in HCT116 cells. Neoplasia 11, 1226–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Walls T. H., Grindrod S. C., Beraud D., Zhang L., Baheti A. R., Dakshanamurthy S., Patel M. K., Brown M. L., and MacArthur L. H. (2012) Synthesis and biological evaluation of a fluorescent analog of phenytoin as a potential inhibitor of neuropathic pain and imaging agent. Bioorg. Med. Chem. 20, 5269–5276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li H., Sun L., Tang Z., Fu L., Xu Y., Li Z., Luo W., Qiu X., and Wang E. (2012) Overexpression of TRIM24 correlates with tumor progression in non-small cell lung cancer. PLoS One 7, e37657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Marcel V., Ghayad S. E., Belin S., Therizols G., Morel A. P., Solano-Gonzàlez E., Vendrell J. A., Hacot S., Mertani H. C., Albaret M. A., Bourdon J. C., Jordan L., Thompson A., Tafer Y., Cong R., Bouvet P., Saurin J. C., Catez F., Prats A. C., Puisieux A., and Diaz J. J. (2013) p53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell 24, 318–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nieminen A. I., Eskelinen V. M., Haikala H. M., Tervonen T. A., Yan Y., Partanen J. I., and Klefström J. (2013) Myc-induced AMPK-phospho p53 pathway activates Bak to sensitize mitochondrial apoptosis. Proc. Natl. Acad. Sci. U.S.A. 110, E1839–E1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hao M., Lowy A. M., Kapoor M., Deffie A., Liu G., and Lozano G. (1996) Mutation of phosphoserine 389 affects p53 function in vivo. J. Biol. Chem. 271, 29380–29385 [DOI] [PubMed] [Google Scholar]
- 30. Hoogervorst E. M., Bruins W., Zwart E., van Oostrom C. T., van den Aardweg G. J., Beems R. B., van den Berg J., Jacks T., van Steeg H., and de Vries A. (2005) Lack of p53 Ser389 phosphorylation predisposes mice to develop 2-acetylaminofluorene-induced bladder tumors but not ionizing radiation-induced lymphomas. Cancer Res. 65, 3610–3616 [DOI] [PubMed] [Google Scholar]
- 31. Bruins W., Bruning O., Jonker M. J., Zwart E., van der Hoeven T. V., Pennings J. L., Rauwerda H., de Vries A., and Breit T. M. (2008) The absence of Ser389 phosphorylation in p53 affects the basal gene expression level of many p53-dependent genes and alters the biphasic response to UV exposure in mouse embryonic fibroblasts. Mol. Cell. Biol. 28, 1974–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kruse J. P., and Gu W. (2009) Modes of p53 regulation. Cell 137, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vousden K. H., and Prives C. (2009) Blinded by the light: the growing complexity of p53. Cell 137, 413–431 [DOI] [PubMed] [Google Scholar]
- 34. Serrano M., Lin A. W., McCurrach M. E., Beach D., and Lowe S. W. (1997) Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602 [DOI] [PubMed] [Google Scholar]
- 35. Bourouba M., Benyelles-Boufennara A., Terki N., Baraka-Kerboua E., Bouzid K., and Touil-Boukoffa C. (2011) Epidermal growth factor receptor (EGFR) abundance correlates with p53 and Bcl-2 accumulation and patient age in a small cohort of North African nasopharyngeal carcinoma patients. Eur. Cytokine Netw. 22, 38–44 [DOI] [PubMed] [Google Scholar]
- 36. Gartel A. L., and Radhakrishnan S. K. (2005) Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 65, 3980–3985 [DOI] [PubMed] [Google Scholar]
- 37. Otera H., Ohsakaya S., Nagaura Z., Ishihara N., and Mihara K. (2005) Export of mitochondrial AIF in response to proapoptotic stimuli depends on processing at the intermembrane space. EMBO J. 24, 1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sot B., Freund S. M., and Fersht A. R. (2007) Comparative biophysical characterization of p53 with the pro-apoptotic BAK and the anti-apoptotic BCL-xL. J. Biol. Chem. 282, 29193–29200 [DOI] [PubMed] [Google Scholar]
- 39. Miyashita T., and Reed J. C. (1995) Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 80, 293–299 [DOI] [PubMed] [Google Scholar]
- 40. Wei C. L., Wu Q., Vega V. B., Chiu K. P., Ng P., Zhang T., Shahab A., Yong H. C., Fu Y., Weng Z., Liu J., Zhao X. D., Chew J. L., Lee Y. L., Kuznetsov V. A., Sung W. K., Miller L. D., Lim B., Liu E. T., Yu Q., Ng H. H., and Ruan Y. (2006) A global map of p53 transcription-factor binding sites in the human genome. Cell 124, 207–219 [DOI] [PubMed] [Google Scholar]
- 41. Zhang G. H., Cai L. J., Wang Y. F., Zhou Y. H., An Y. F., Liu Y. C., Peng Y., Chen Z. F., and Liang H. (2013) Novel compound PS-101 exhibits selective inhibition in non-small-cell lung cancer cell by blocking the EGFR-driven antiapoptotic pathway. Biochem. Pharmacol. 86, 1721–1730 [DOI] [PubMed] [Google Scholar]
- 42. Follis A. V., Llambi F., Ou L., Baran K., Green D. R., and Kriwacki R. W. (2014) The DNA-binding domain mediates both nuclear and cytosolic functions of p53. Nat. Struct. Mol. Biol. 21, 535–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Busch S. E., Moser R. D., Gurley K. E., Kelly-Spratt K. S., Liggitt H. D., and Kemp C. J. (2014) ARF inhibits the growth and malignant progression of non-small-cell lung carcinoma. Oncogene 33, 2665–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bates S., Phillips A. C., Clark P. A., Stott F., Peters G., Ludwig R. L., and Vousden K. H. (1998) p14ARF links the tumour suppressors RB and p53. Nature 395, 124–125 [DOI] [PubMed] [Google Scholar]