Abstract
The TET enzymes are members of the 2-oxoglutarate-dependent dioxygenase family and comprise three isoenzymes in humans: TETs 1–3. These TETs convert 5-methylcytosine to 5-hydroxymethylcytosine (5-hmC) in DNA, and high 5-hmC levels are associated with active transcription. The importance of the balance in these modified cytosines is emphasized by the fact that TET2 is mutated in several human cancers, including myeloid malignancies such as acute myeloid leukemia (AML). We characterize here the kinetic and inhibitory properties of Tets and show that the Km value of Tets 1 and 2 for O2 is 30 μm, indicating that they retain high activity even under hypoxic conditions. The AML-associated mutations in the Fe2+ and 2-oxoglutarate-binding residues increased the Km values for these factors 30–80-fold and reduced the Vmax values. Fumarate and succinate, which can accumulate to millimolar levels in succinate dehydrogenase and fumarate hydratase-mutant tumors, were identified as potent Tet inhibitors in vitro, with IC50 values ∼400–500 μm. Fumarate and succinate also down-regulated global 5-hmC levels in neuroblastoma cells and the expression levels of some hypoxia-inducible factor (HIF) target genes via TET inhibition, despite simultaneous HIFα stabilization. The combination of fumarate or succinate treatment with TET1 or TET3 silencing caused differential effects on the expression of specific HIF target genes. Altogether these data show that hypoxia-inducible genes are regulated in a multilayered manner that includes epigenetic regulation via TETs and 5-hmC levels in addition to HIF stabilization.
Keywords: 5-hydroxymethylcytosine (5-hmC), epigenetics, hypoxia, hypoxia-inducible factor (HIF), leukemia, AML, TET, fumarate, succinate
Introduction
The 2-oxoglutarate-dependent dioxygenases (2-OGDDs)2 comprise an enzyme family of about 70 members in humans (1, 2). These enzymes all share the same basic reaction mechanism, in which the substrate is hydroxylated by molecular oxygen in the presence of a divalent metal cofactor (most commonly Fe2+) and the 2-oxoglutarate cosubstrate is decarboxylated to succinate and CO2 (1). The substrates for 2-OGDDs vary from proteins to DNA, RNA, and fatty acids (1). Interestingly, a large number of 2-OGDDs act on the chromatin structure, most notably the ten-eleven-translocation 5-methylcytosine dioxygenases (TETs) and the Jumonji domain-containing histone demethylases (1–3). The stability of the α subunit of the key regulator of the hypoxia response, the hypoxia-inducible factor (HIF), is also regulated by 2-OGDDs, namely the HIF-prolyl 4-hydroxylases (HIF-P4Hs), also known as PHDs and EglNs (1, 2, 4).
The TET enzymes convert the 5-methylcytosine (5-mC) in DNA sequentially to 5-hydroxymethylcytosine (5-hmC), 5-formylcytocine, and 5-carboxylcytocine, leading to DNA demethylation (3, 5–7). 5-hmC is also likely to have its own epigenetic function beyond simply being a demethylating base (3). The highest levels of 5-hmC are found in stem cells of various origins and in neural tissues (6, 7). There are three human TET isoenzymes. TET1 is highly expressed in embryonic stem cells, whereas TETs 2 and 3 are required for normal hematopoiesis (8, 9). Mutations in TET2 are frequently found in acute myeloid leukemia (AML) and in several other hematological malignances (9–12). The TETs are considered important epigenetic regulators of gene expression. 5-mC represses transcription when it is concentrated in promoters and CpG islands, whereas high 5-hmC levels are associated with active transcription (3). It was shown recently in a neuroblastoma cell culture system that 5-hmC accumulates at or near the HIF binding sites, associated with increased expression of HIF target genes under hypoxic conditions (13).
Mutations in genes encoding the Krebs cycle enzymes succinate dehydrogenase (SDH), fumarate hydratase (FH) and isocitrate dehydrogenase (IDH), and in its cytosolic isoenzymes, are found in paraganglioma, pheochromocytoma, uterine and skin leiomyoma, papillary renal carcinoma, glioma, and AML (14–19). These mutations result in accumulation of the 2-oxoglutarate analogues succinate, fumarate, and R-2-hydroxyglutarate (R-2HG), respectively (20, 21). Fumarate and succinate have been shown to inhibit several 2-OGDDs competitively with respect to 2-oxoglutarate, whereas R-2HG is an inhibitor of all the other 2-OGDDs studied except for the HIF-P4Hs, the activity of which is supported by R-2HG (22–25).
We produced and purified Tets as recombinant proteins and measured their enzyme kinetics in vitro with respect to substrate, cosubstrates, and the iron cofactor. We also studied the ability of 2-oxoglutarate analogues to inhibit the catalytic activity of the TETs in vitro and that of fumarate and succinate in cellulo. We showed in cellulo that fumarate and succinate play a role in the regulation of certain HIF target genes via TET inhibition, suggesting that 5-hmC has a role in regulation of the hypoxia response.
Experimental Procedures
Expression and Purification of Recombinant Enzymes
The catalytic domains for murine Tets 1–3 in the pFasbac-HTb vector with a N-terminal FLAG tag were a gift from Dr. Y. Zhang (8). Corresponding baculoviruses coding for Tet1 1367–2039 (NCBI reference sequence NP_001240786.1), Tet2 916–1921 (NCBI reference sequence NP_001035490.2), and Tet3 697–1668 (NCBI reference sequence NP_898961.2) were generated using the Bac-to-Bac TOPO expression system (Invitrogen) and used for expression of the corresponding recombinant proteins in Sf9 insect cells in TNM-FH media supplemented with 10% fetal bovine serum. The cells were infected with the respective baculoviruses and harvested 72 h after infection, washed with PBS, and homogenized in a buffer containing 40 mm Tris, 300 mm NaCl, 0.2% Nonidet P-40, 0.2% Triton, 5 mm DTT, and protease inhibitor mixture without EDTA (Roche Applied Science). The soluble fractions were subjected to purification with an anti-FLAG M2 affinity gel (Sigma), and the fractions collected were analyzed using 12% SDS-PAGE under reducing conditions followed by Coomassie Blue staining.
Mutagenesis
To generate Tet2 mutants H1302Y, D1304A, H1802R, R1817S, and R1817M, the plasmid containing the wild-type Tet2 cDNA was used as a template in mutagenesis performed using the QuickChange® Lightning site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. The primers for individual mutations are shown in Table 1. The corresponding baculoviruses were generated using the Bac-to-Bac TOPO expression system (Invitrogen) and used for expression and purification of the recombinant proteins as described for Tets 1–3.
TABLE 1.
Sequences of primers used for mutagenesis and qPCR
| Usage | Gene | Forward primer | Reverse primer |
|---|---|---|---|
| Mutagenesis | Tet2 H1302Y | GCTCATTCCTACAGAGACCAGCAGAACATGC | GCATGTTCTGCTGGTCTCTGTAGGAATGAGC |
| Mutagenesis | Tet2 D1304A | GCTCAATTCCCACAGAGCCCAGCAGAACATGC | GCATGTTCTGCTGGGCTCTGTGGGAATGAGC |
| Mutagenesis | Tet2 H1802R | GCAAAGTGTGAGGTTCGTGCCACAACC | GGTTGTGGCACGAACCTCACACTTTGC |
| Mutagenesis | Tet2 R1817M | CCCCACCATGATCTCACTTGTACTGTATAGG | CCTATACAGTACAAGTGAGATCATGGTGGGG |
| Mutagenesis | Tet2 R1817S | CCCCACCAGTATCTCACTTGTACTGTATAGG | CCTATACAGTACAAGTGAGATACTGGTGGGG |
| qPCR | TBP | GAATATAATCCCAAGCGGTTT | ACTTCACATCACAGCTCCCC |
| qPCR | TET1 | GAGAATAGGTATGGTCAAAA | CTTCATCACTGCTTCTTCTT |
| qPCR | TET2 | TGCCGTCTGGGTCTGAAG | CCTCAGGTTTTCCTCCAAAT |
| qPCR | TET3 | CGTCGAACAAATAGTGGAGA | CTTTCCCCTTCTCTCCATAC |
| qPCR | VEGFA | AGGAGGAGGGCAGAATCATCA | ATGTCCACCAGGGTCTCGATTG |
| qPCR | BNIP3 | CCACCTCGCTCGCAGACACCAC | GAGAGCAGCAGAGATGGAAGGAAAAC |
| qPCR | PGK1 | CTGTGGGGGTATTTGAATGG | CTTCCAGGAGCTCCAAACTG |
| qPCR | HK2 | ATTGTCCAGTGCATCGCGGA | AGGTCAAACTCCTCTCGCCG |
| qPCR | ENO1 | TGGTGTCTATCGAAGATCCCTT | CCTTGGCGATCCTCTTTGG |
| qPCR | IDH1 | TGTGGTAGAGATGCAAGGAGA | TTGGTGACTTGGTCGTTGGTG |
Kinetic Activity Assays
The catalytic activities of the Tet enzymes were assayed by a modified version of the previously reported method based on measurement of the hydroxylation-coupled stoichiometric release of 14CO2 from 2-oxo[1-14C]glutarate (26). Oligonucleotides containing a 5-mC (5′-CTATACCTCCTCAACTT[5-mC]GATCACCGTCTCCGGCG-3′ and 5′-Biotin-CGCCGGAGACGGTGAT[5-mC]GAAGTTGAGGAGGTATAG-3′) were annealed together to prepare a double-stranded synthetic DNA for use as a substrate. The activity assays were carried out in a final volume of 50 μl and the reaction mixture contained 2 μg/μl of bovine serum albumin, 50 mm Tris (pH 7.8), 0.1 mm DTT, 5 mm ascorbate, 0.05 mm FeSO4, 0.6 mm 2-oxo[1-14C]glutarate, and 1.8 μm DNA substrate. Km values for the substrate, cosubstrates, and cofactor were determined by varying the concentration of the component in question while keeping the concentrations of the others saturating and constant. The Km values for O2 were determined in an InVivo400 hypoxia work station (Ruskinn). The relative Vmax values for the Tet2 mutants were determined with respect to that obtained with wild-type Tet2. The IC50 values were studied by using 240 μm 2-oxoglutarate and saturating concentrations of the other substrates in the presence of increasing concentrations of the respective 2-oxoglutarate analogue in the reaction mixture. The reaction mixtures were incubated at 37 °C for 20 min.
Cell Culture Experiments with Diethyl Fumarate andDimethyl Succinate
SK-N-BE(2) neuroblastoma cells were maintained in RPMI 1640 media supplemented with 10% fetal bovine serum. The cells for the normoxic experiments were incubated under 21% O2 and 5% CO2, and the hypoxic experiments were conducted under 1% O2 and 5% CO2 in a Sci-tive hypoxia work station (Ruskinn). The cells for the diethyl fumarate (DEF) (Sigma) and dimethyl succinate (DMS) (Sigma) treatments were seeded 5–6 h prior to the initiation of the treatment at 35–45% confluence and treated with increasing concentrations of DEF or DMS (Sigma). The compounds were diluted in dimethyl sulfoxide (DMSO). The normoxic control cells and hypoxic cells were treated with an equivalent volume of DMSO. For the 48-h treatments, equal volumes and concentrations of the compounds were added every 24 h.
Determination of Intranuclear and Cytosolic Succinate Concentrations following DMS Treatment
Nuclear and cytosolic fractions were extracted from the cells using NEPER® Nuclear and Cytoplasmic Extraction Reagent Kit (Thermo Scientific). To extract organic acids, the supernatant containing the cytosolic fraction was subjected to an equal volume of ice-cold 8% perchloric acid and centrifuged. Similarly, the pellets containing the nuclear fractions were lysed in ice-cold 8% perchloric acid and centrifuged. The supernatants were neutralized by K2CO3 and analyzed using an UPLC-MS/MS method modified from a previous report (27). The chromatographic separation was performed at 30 °C using an ACQUITY HSS C18 column (100 × 2.1 mm, Waters Corp.) equipped with a ACQUITY UPLC pre-column filter. The eluents were A: 0.2% formic acid (Optima LC/MS, Fischer Scientific) in ultrapure H2O (v/v) and B: 0.2% formic acid in LC/MS-grade methanol (LiChrosolv GG). The chromatographic system used was Waters ACQUITYTM UPLC system (Waters Corp.). Detection of succinate was carried out using a Waters XevoTM TQ-S triple quadrupole tandem mass spectrometer (Waters Corp.) with a Z-spray electrospray ionization source. The MS/MS reaction used for quantitation of succinate was m/z 117 > 73 and an additional MS/MS-reaction m/z 117 > 99 using 10 eV collision energy for both multiple reaction monitoring reactions.
Immunoblotting
Total cell extracts were obtained by lysing the cells in 50 mm Tris (pH 7.5), 150 mm NaCl, and 0.5% Nonidet P-40. The extracts were analyzed in SDS-PAGE under reducing conditions followed by Western blotting with polyclonal rabbit antibodies against human HIF-1α (Novus, NB100-479), HIF-2α (Novus, NB100-122), and IDH1 (Cell Signaling, number 3997), followed by ECL immunodetection. Western blotting for β-actin (Novus, NB600-501) was used as a loading control.
Quantification of 5-mC and 5-hmC by High Performance Liquid Chromatography Coupled with Tandem Mass Spectrometry
Genomic DNA was extracted from the cells by lysing the cells with a buffer containing 0.1 m Tris (pH 8.5), 5 mm EDTA, 0.2% SDS, 0.2 m NaCl, and 0.1 mg/ml of Proteinase K and subjecting the lysates for phenol-chloroform extraction. Genomic DNA was hydrolyzed enzymatically to nucleosides and run on a Zorbax XDB-C18 2.1 × 50-mm column (1.8-mm particle size) attached to an Agilent 1200 Series liquid chromatography machine coupled to an Agilent 6410 Triple Quad Mass Spectrometer (13, 28, 29).
Quantitative Real Time RT-PCR (qPCR)
Total RNA was isolated from the cells and purified using an EZNA total RNA kit (OMEGA Bio-Tek). Reverse transcription was performed with an iScript cDNA synthesis kit (Bio-Rad) and qPCR was performed in a CFX96 Real-time System (Bio-Rad). TATA-binding protein (TBP) was used as a housekeeping gene. The sequences of the primers are presented in Table 1.
RNA Interference Experiments
SK-N-BE(2) neuroblastoma cells were transfected with TET1-targeting siRNA (Dharmacon ON-TARGETplus SMARTpool L-014635-03), TET3-targeting siRNA (Dharmacon ON-TARGETplus SMARTpool L-022722-02), or non-targeting control siRNA (Dharmacon ON-TARGETplus Non-targeting pool L001810) using Mirus TransIT-siQUEST transfection reagent (MIR 2114), according to the manufacturer's protocol 72 h before the initiation of treatment with DMSO, DEF, or DMS. The cells were passaged 48 h prior to the start of the treatment, and re-transfected with the siRNAs 16–18 h prior to the treatment.
Statistical Analyses
Student's two-tailed t test was used for comparisons between two groups. All data are shown as mean ± S.E. except for the enzyme kinetics and intranuclear and cytosolic succinate concentrations, which are shown as mean ± S.D. (*, p < 0.05; **, p < 0.01; ***, p < 0.005).
Results
Kinetic Analyses of Tets 1 and 2 Indicate High Activity under Hypoxia
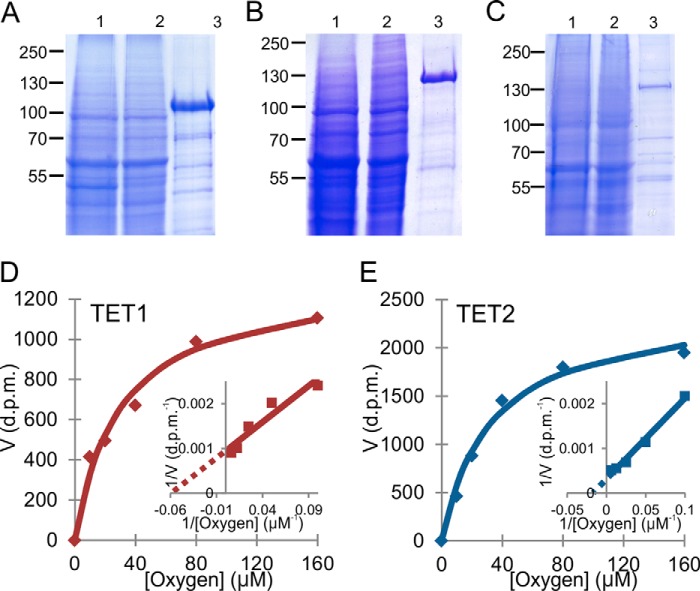
The catalytic domains of the murine Tets 1–3 were expressed in insect cells as FLAG-tagged proteins and affinity-purified using anti-FLAG affinity chromatography (Fig. 1, A–C). The elution fractions were run on SDS-PAGE followed by Coomassie Blue staining. Tets 1 and 2 gave a higher yield on purification than did Tet3 (Fig. 1, A–C).
FIGURE 1.
Expression, affinity purification, and kinetic analyses of Tets. A–C, SDS-PAGE and Coomassie Blue analysis of the expression and affinity purification of recombinant Tet1 (A), Tet2 (B), and Tet3 (C). Cell lysate (lane 1), unbound proteins (lane 2), and FLAG-affinity purified proteins (lane 3) are shown. D and E, Michaelis-Menten curves and Lineweaver-Burk plots (inset) of Tet1 and Tet2 for oxygen.
The catalytic activities of Tets 1–3 were assessed by a method based on measurement of the hydroxylation-coupled stoichiometric release of 14CO2 from 2-oxo-[1-14C]glutarate using a double-stranded DNA fragment containing 5-mC as a substrate. Tets 1 and 2 showed significant activity under these conditions, whereas the yield and activity of Tet3 was low (Fig. 1, A–C). We therefore concentrated the kinetic analyses on Tets 1 and 2.
The Km values of Tets 1 and 2 for the DNA substrate were 75 and 125 nm, respectively (Table 2). The Km values for iron for Tets 1 and 2 were 4.8 and 3.6 μm, respectively, being markedly higher than those for HIF-P4H-2, but in the range of those for the collagen prolyl 4-hydroxylase I (Table 2). The Km values of Tets 1–2 for 2-oxoglutarate were 55–60 μm, i.e. very similar to those for HIF-P4H-2 and slightly higher than those for the collagen prolyl 4-hydroxylase I (Table 2). Interestingly, the Km values of Tets 1 and 2 for oxygen were 30 μm (Fig. 1, D and E), in the same range of ∼40 μm as for the collagen prolyl 4-hydroxylase I but markedly lower than for HIF-P4H-2 (Table 2), indicating that Tets 1 and 2 also display significant activity under hypoxic conditions.
TABLE 2.
Km values of Tets 1 and 2 by comparison with HIF-P4H-2 and collagen prolyl 4-hydroxylase I (C-P4H-I) for the substrate, cosubstrates and cofactor
The values are mean ± S.D. of 3 to 7 independent assays.
Leukemic TET2 Mutations Impair Iron and 2-Oxoglutarate Binding
Many TET2 mutations are associated with myeloid malignances, including AML. We introduced point mutations into the three critical iron binding residues (His-1302, Asp-1304, and His-1802) and the 2-oxoglutarate coordinating residue (Arg-1817), which in three cases, H1802R, R1817S, and R1817M, served as models for reported AML-associated mutations in human TET2, namely H1881R, R1896S, and R1896M, respectively (9–12). We produced and purified the mutant proteins as recombinant proteins in insect cells (Fig. 2A) and studied the Km and Vmax values of the Tet2 mutants H1302Y, D1304A, and H1802R for iron and those of R1817M and R1817S for 2-oxoglutarate. The Km values for iron were increased by 30–56-fold for the Tet2 H1302Y, D1304A, and H1802R mutants compared with wild-type Tet2 and their Vmax values were reduced by about 50% (Table 3, Fig. 2B). The Tet2 R1817M mutant did not reach saturation at a 2-oxoglutarate concentration of 8 mm (Fig. 2C), making it impossible to determine the exact Km value for 2-OG or Vmax for this mutant or for its serine counterpart, although these values must have been at least 80-fold higher than that for wild-type Tet2 (Table 3).
FIGURE 2.
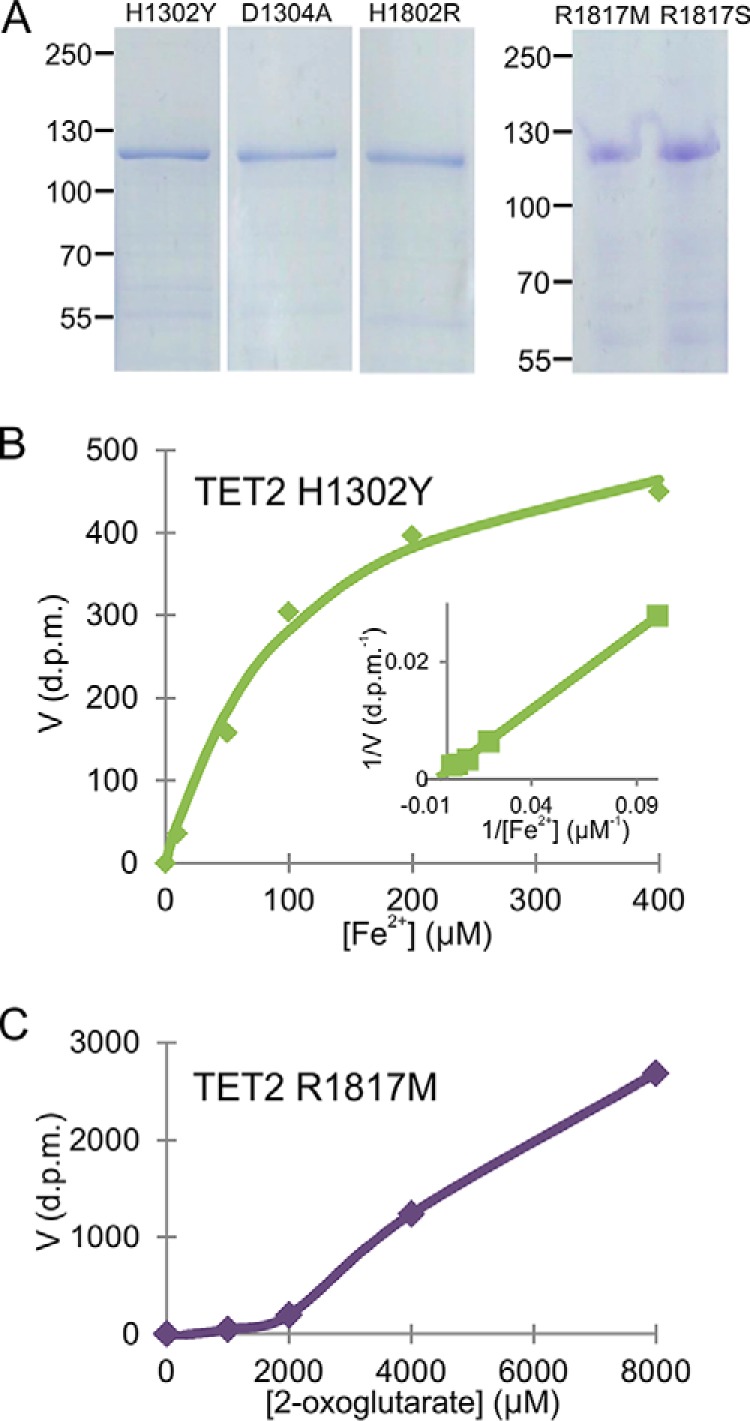
Kinetic analysis of AML-associated Tet2 mutants. A, SDS-PAGE analysis of purified recombinant Tet2 mutants H1302Y, D1304A, H1802R, R1817M, and R1817S. B and C, Michaelis-Menten curves and a Lineweaver-Burk plot (inset) of Tet2 mutants H1302Y and R1817M for Fe2+ and 2-oxoglutarate, respectively.
TABLE 3.
Km and Vmax values of AML-associated Tet2 mutants for iron and 2-oxoglutarate
The values are mean ± S.D. of 3 to 7 independent assays.
| Variable measured | Unit | WT Tet2 | H1302Y | D1304A | H1802R | R1817M | R1817S |
|---|---|---|---|---|---|---|---|
| Km of Fe2+ | μm | 3.6 ± 3 | 110 ± 4 | 165 ± 90 | 200 ± 185 | NDa | ND |
| Km of 2-oxoglutarate | μm | 60 ± 15 | ND | ND | ND | >5000 | >5000 |
| Vmax | % of WT Tet2 | 100 | 45 | 55 | 50 | ND | ND |
a ND, not determined.
Succinate and Fumarate Are Efficient Inhibitors of Tets
We next studied the ability of the cancer-associated Krebs cycle 2-oxoglutarate analogues succinate and fumarate and an important metabolic regulator, citrate, to inhibit Tets 1–2 in vitro and compared the resulting IC50 values of these for R-2HG and its enantiomer S-2-hydroxyglutarate (S-2HG). The IC50 values of Tets 1 and 2 for fumarate were about 400 μm and for succinate about 550 μm (Table 4 and Fig. 3A and 3B), whereas this value for citrate was >5 mm (Table 4). These findings were similar to those for HIF-P4H-2 apart from the case of fumarate, which was a more efficient inhibitor of HIF-P4H-2 than Tets 1 and 2 (Table 4). Among the cancer-associated 2-oxoglutarate analogues, fumarate was the most efficient Tet1 and Tet2 inhibitor, with succinate the second most efficient (Table 4).
TABLE 4.
IC50 values of succinate, fumarate, citrate, and (R)- and (S)-2-hydroxyglutarate for Tets 1 and 2
The values are mean ± S.D. of 3 to 4 independent assays.
| Enzyme | Unit | Citrate | Succinate | Fumarate | R-2HGa | S-2HGa |
|---|---|---|---|---|---|---|
| Tet1 | μm | >5000 | 540 ± 100 | 390 ± 160 | 4000a | 1000a |
| Tet2 | μm | >5000 | 570 ± 190 | 400 ± 70 | 5000a | 1600a |
a Ref. 25.
FIGURE 3.
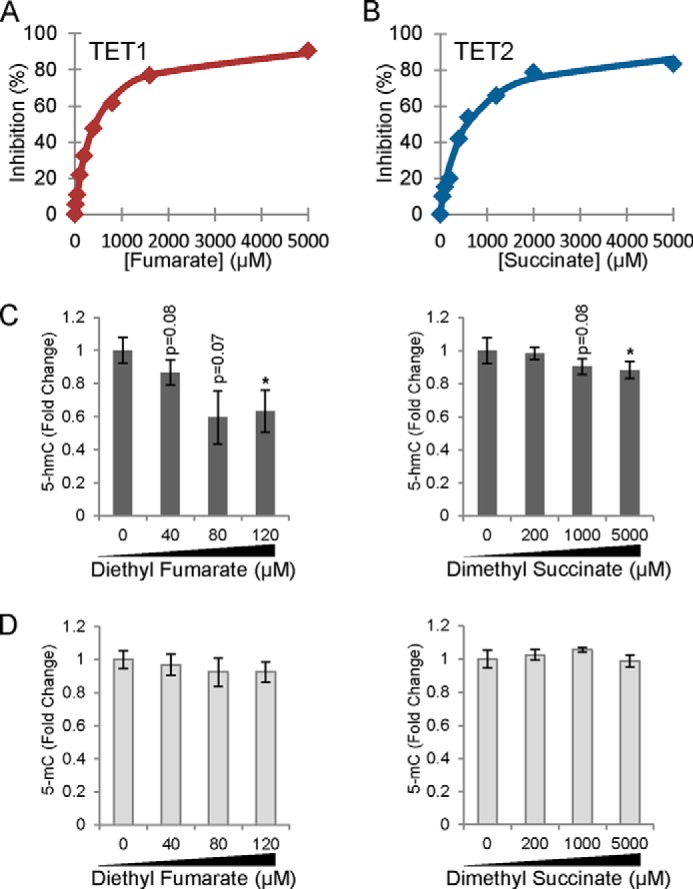
Tets are susceptible to competitive inhibition by fumarate and succinate, resulting in lower global 5-hmC levels in cells treated with cell-permeable forms of these compounds. A and B, IC50 curves of Tet1 and Tet2 for fumarate and succinate, respectively. C and D, HPLC-MS/MS quantitation of global 5-hmC (C) and 5-mC (D) levels in SK-N-BE(2) cells incubated with increasing concentrations of diethyl fumarate or dimethyl succinate for 48 h (n ≥ 3). 5-hmC and 5-mC quantitation graphs represent mean ± S.E. *, p < 0.05.
Treatment of SK-N-BE(2) Neuroblastoma Cells with Fumarate and Succinate Alters the Genomic 5-hmC Content and Expression of HIF Target Genes
We have shown earlier that treatment of cells with the cell-permeable form of fumarate (DEF) increased the intracellular fumarate concentration about 2-fold (24) (Table 5). We now analyzed the intracellular succinate levels following treatment with DMS and found significant increases to nearly 3-fold in the cytosol and nucleus (Table 5). To test whether fumarate and succinate alter the balance of 5-mC/5-hmC, we treated SK-N-BE(2) neuroblastoma cells that have been shown to gain 5-hmC density at or near HIF binding sites and across HIF target genes in response to hypoxia (13) with cell-permeable fumarate or succinate. The cells were exposed to increasing concentrations of fumarate or succinate for 48 h, and changes in global 5-hmC levels were determined by HPLC-MS/MS. We found that fumarate resulted in ∼40% lower 5-hmC levels, whereas succinate treatment resulted in ∼10% reduction, results that were statistically significant at the highest concentrations of the compounds (Fig. 3C). No significant changes were observed in the global 5-mC content (Fig. 3D).
TABLE 5.
Cellular concentrations of succinate in SK-N-BE(2) cells incubated with DMSO or DMS for 48 h in comparison to fumarate levels in cells incubated with DEF
The values are means ± S.D. of 3 independent assays.
| Cells treated | Unit | Succinate |
Fumarate cellular | |
|---|---|---|---|---|
| Nuclear | Cytosolic | |||
| DMSO | % of DMSO treated | 100 ± 10 | 100 ± 5 | 100a |
| 5 mm DMS | % of DMSO treated | 285 ± 65** | 255 ± 15*** | NDb |
| 40 μm DEF | % of DMSO treated | ND | ND | 180a |
a In HEK293 cells incubated for 20 h (24).
b ND, not determined.
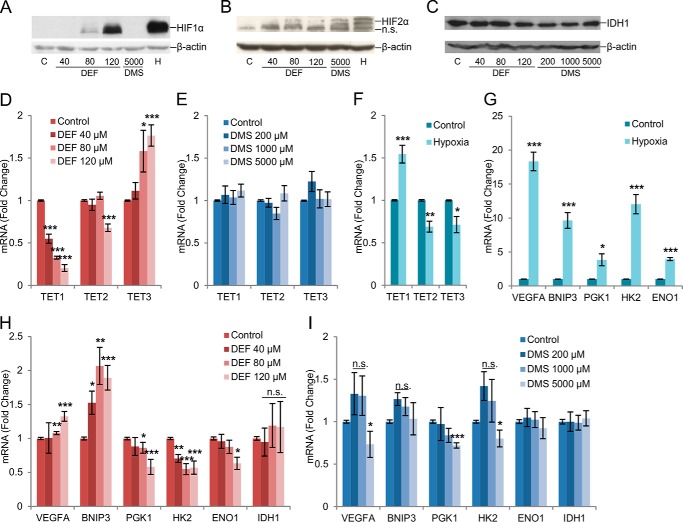
Fumarate, but not succinate, stabilized HIF-1α in the SK-N-BE(2) cells under normoxia when analyzed by Western blotting, whereas both fumarate and succinate modestly induced HIF-2α (Fig. 4, A and B). Fumarate and succinate did not alter the protein levels of IDH1 (Fig. 4C). Interestingly, the mRNA levels of TET1 and TET2 were down-regulated, whereas TET3 mRNA was up-regulated by fumarate (Fig. 4D). Succinate treatment did not alter mRNA levels of TET1/2/3 (Fig. 4E). Treatment of cells with 1% O2 significantly increased the expression of TET1 mRNA by about 150%, as reported earlier (13), but reduced the expression levels of TET2/3 by about 30% (Fig. 4F).
FIGURE 4.
Fumarate and succinate stabilize HIFαs and alter TET and HIF target gene expression. A and B, HIF-1α (A) and HIF-2α (B) protein levels determined by immunoblotting in SK-N-BE(2) cells exposed to hypoxia (1% O2) or incubated with increasing concentrations of DEF or 5 mm DMS for 48 h. β-Actin was used as a loading control. C, IDH1 protein levels were determined by immunoblotting in SK-N-BE(2) cells incubated with increasing concentrations of DEF or DMS for 48 h. β-Actin was used as a loading control. D–F, qPCR analysis of TET1–3 mRNA expression levels in cells treated with increasing concentrations of DEF (D), DMS (E), or exposed to hypoxia (1% O2) (F) for 48 h (n ≥ 3). G-I, qPCR analysis of selected HIF target genes in cells exposed to hypoxia (1% O2) (G) or treated with increasing concentrations of DEF (H) or DMS (I) for 48 h (n ≥ 3). All graphs represent mean ± S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.005.
We next analyzed the expression levels of selected HIF target genes in neuroblastoma cells following hypoxia and treatment with fumarate or succinate. All the genes analyzed were significantly increased by hypoxia, as expected (Fig. 4G). We observed that the expression level of vascular endothelial growth factor A (VEGFA) mRNA increased to 130% by fumarate (Fig. 4H). In contrast, succinate treatment decreased VEGFA mRNA levels by 30% at the highest compound concentration (Fig. 4I). The level of BNIP3 mRNA was also increased 200% by fumarate, whereas succinate did not affect its level (Fig. 4, H and I). The level of phosphoglycerate kinase 1 (PGK1) mRNA was decreased by 40% with fumarate and 30% with succinate (Fig. 4, H and I). Fumarate treatment also reduced the hexokinase 2 (HK2) and enolase 1 (ENO1) mRNA levels by ∼40% (Fig. 4H). Succinate treatment caused slightly less of a decrease in HK2 mRNA levels (Fig. 4I). Neither fumarate nor succinate altered the mRNA levels for IDH1 (Fig. 4, H and I). These data suggest that HIFα stabilization alone is not sufficient to induce all the HIF target genes, but that the TETs and 5-hmC are likely to provide another level of regulation for HIF target genes.
Silencing of TETs 1 and 3 Alters HIF Target Gene Expression
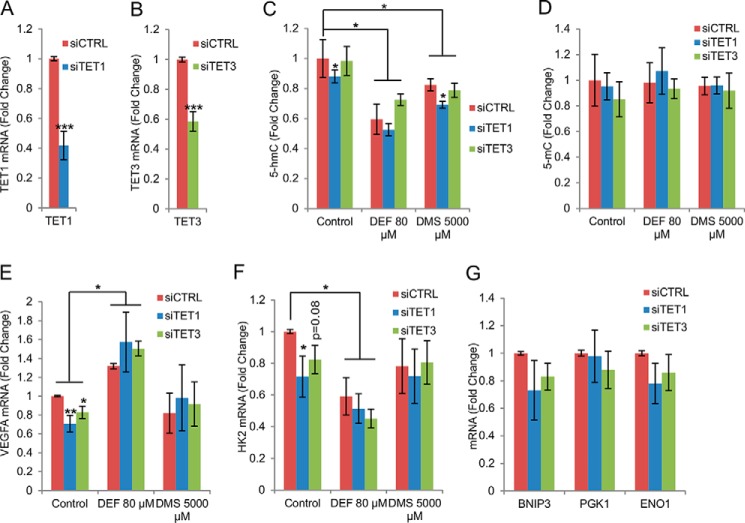
To understand the roles of the TETs in the regulation of the selected HIF target genes via 5-hmC we silenced TETs 1 or 3 with siRNA pools containing four siRNAs against each mRNA in the SK-N-BE(2) cells (Fig. 5, A and B). We also treated the cells in which TET1 or TET3 was silenced with selected concentrations of fumarate and succinate, shown in previous experiments to inhibit TET catalytic activity, and determined the levels of global genomic 5-hmC and 5-mC. As in the case of the non-transfected cells (Fig. 3C), a reduction in 5-hmC levels was observed in cells treated with a control siRNA together with either fumarate or succinate (Fig. 5C). No significant changes were observed in the global 5-mC content (Fig. 5D). Silencing of TET1 alone reduced the level of 5-hmC by about 15% (Fig. 5C), and the addition of fumarate or succinate to the TET1 siRNA further reduced the 5-hmC levels (Fig. 5C), whereas no significant differences were observed in the 5-mC levels (Fig. 5D). Silencing of TET3 alone did not significantly alter the 5-hmC or 5-mC levels (Fig. 5, C and D).
FIGURE 5.
TETs regulate HIF target gene expression. A and B, qPCR analysis of TET1 (A) and TET3 (B) mRNA expression levels in SK-N-BE(2) cells following siRNA knockdown of TET1 (A) or TET3 (B) (n = 3). C and D, HLPC-MS/MS quantitation of global 5-hmC (C) and 5-mC (D) levels in SK-N-BE(2) cells following siRNA knockdown of TET1 or 3 and DEF or DMS treatment (n = 3). E and F, qPCR analysis of VEGFA and HK2 mRNAs following knockdown of TET1 or TET3 and DEF or DMS treatment (n = 3). G, qPCR analysis of BNIP3, PGK1, and ENO1 mRNA expression in SK-N-BE(2) cells following siRNA knockdown of TET1 or TET3 (n = 3). All graphs represent mean ± S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.005.
We also studied the expression levels of the selected HIF target genes following TET1 and TET3 silencing. Our data show that silencing of TET1 and TET3 alone reduced the expression level of VEGFA mRNA significantly by about 30 and 20%, respectively (Fig. 5E). Similar reductions of ∼30 and ∼20% were observed in the expression of HK2 mRNA following the silencing of TET1 and TET3, respectively (Fig. 5F). Despite a trend for lower levels, no significant reductions were found in the expression levels of the other mRNAs studied, those for BNIP3, PGK1, and ENO1, following TET1 or TET3 silencing (Fig. 5G). When the cells in which TET1 or TET3 was silenced were treated with fumarate, VEGFA mRNA was induced (Fig. 5E), opposite to our observations when TET1 or TET3 only were silenced. Succinate treatment prevented the down-regulation of VEGFA mRNA observed with TET1 and TET3 siRNAs but did not itself induce VEGFA mRNA above baseline (Fig. 5E). Interestingly, fumarate treatment in combination with TET1 or TET3 silencing further reduced HK2 mRNA levels, whereas a combination treatment with succinate did not have any effect (Fig. 5F).
Discussion
Hypoxia has been shown to increase global 5-hmC levels via HIF-1-dependent induction of TET1 (13). Because TETs require O2 for their reaction, the increase in their product under hypoxia raised the question of their requirement for this cosubstrate. We therefore determined the Km values of Tets 1 and 2 for molecular oxygen and show that their activity is not highly dependent on oxygen, consistent with these enzymes remaining catalytically active when induced by hypoxia (13). The low Km value for O2 is also in line with the physiological role for the TETs in hypoxic environments such as bone marrow and during development, two settings where they are known to be highly expressed (6, 8).
Mutations in TET2 iron and 2-oxoglutarate-binding residues have been reported in patients with hematological malignances including AML (9–12). We introduced some of these cancer-associated mutations into murine Tet2 and studied their effect on its catalytic activity and Fe2+ and 2-oxoglutarate requirements. The cancer-associated mutants had Km values for iron and 2-oxoglutarate that were substantially greater than those of wild-type Tet2, whereas their Vmax values were reduced by at least 50%, suggesting that these mutations resulted in a profound catalytic deficiency in TET2 activity. None of the mutants studied had completely lost its catalytic activity, however, raising the possibility that increasing the local concentration of iron or 2-oxoglutarate in the bone marrow could at least partially restore their activity and even reverse their oncogenic properties.
Mutations in SDH and FH are found in cases of paraganglioma, pheochromocytoma, uterine and skin leiomyoma, and papillary renal carcinoma (14–17). These mutations cause the accumulation of succinate and fumarate, respectively, and have been shown previously to signal at least partially via the HIF-P4Hs, resulting in HIF stabilization (20, 22–24). Our data show that fumarate and succinate are potent inhibitors of Tets 1 and 2, with IC50 values around 400–500 μm. We also show that the treatment of cells with fumarate and succinate reduced global 5-hmC levels, which is in agreement with our in vitro enzyme kinetic data. Despite the similar IC50 values, fumarate reduced the global 5-hmC levels of neuroblastoma cells more succinate. It is possible that this difference accounts for differences in intracellular succinate and fumarate metabolism. As fumarate reduced the mRNA levels for TET1 and TET2 and increased that for TET3 we cannot exclude the possibility that in addition to catalytic inhibition alteration in the proteins levels of TETs might have contributed to the changes in global 5-hmC levels. Knock-down of FH, SDHA, and SDHB that accumulate fumarate and succinate, respectively, have also shown to down-regulate 5hmC levels in HEK293T cells (30). Furthermore, it has been reported that fumarate and succinate can also inhibit the human histone H3K36 demethylase KDM4A, with IC50 values of 1.5 and 0.8 mm, respectively, and induce genome-wide histone methylation in cellulo, likely adding to the epigenetic regulation that occurs via these metabolites (30). This is also suggested by the differential outcome with respect to hypoxia-inducible gene expression following fumarate or succinate treatment and hypoxia, which both stabilize HIFα. Because fumarate and succinate accumulate to high millimolar levels in human SDH and FH mutant cancers (20), it is likely that TETs are inhibited in these tumors and that their inhibition, resulting in an alteration in 5-hmC levels, affects the biological behavior of these cancers. This reported hypermethylator phenotype in SDH mutant paragangliomas is consistent with this view (31). In contrast, reduced global 5-hmC levels have been reported in other cancers such as melanoma, glioma, prostate, colon, breast cancers, and in esophageal squamous cell carcinoma although the mechanism(s) underlying the loss of 5-hmC in these cancers is not known (32–35).
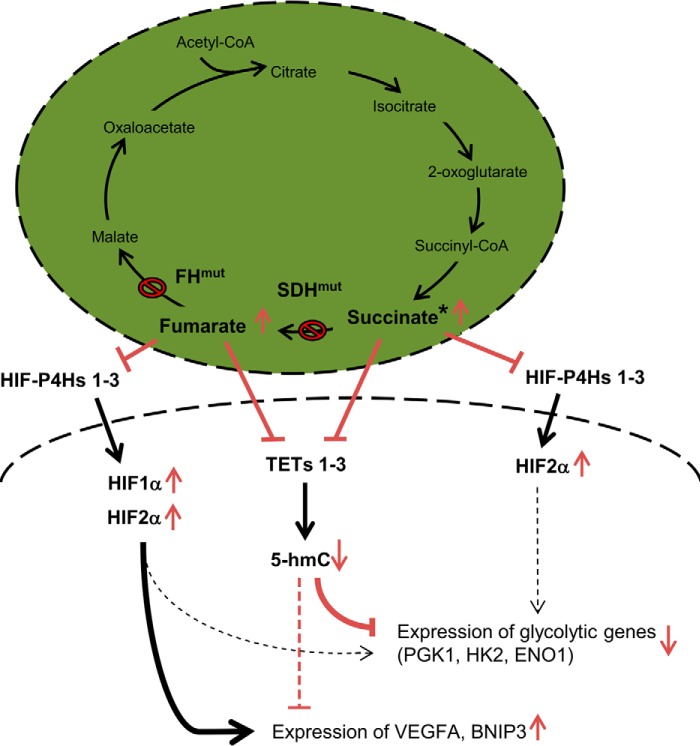
It has been shown recently that full induction of the hypoxia-responsive transcriptional program in aggressive neuroblastoma cells requires not only HIF stabilization but also TET1 induction and the resultant accumulation of 5-hmC in the hypoxia response elements (13). Consistent with these data, we show here that when the catalytic activity of the TETs is inhibited by fumarate the expression levels of the glycolytic HIF-1α target genes PGK1, HK2, and ENO1 (36) decline despite simultaneous HIF-1α and HIF-2α stabilization. Treatment of the same cells with succinate, which only stabilized HIF-2α and not HIF-1α, had a similar but smaller effect on the expression levels of PGK1 and HK2, but no statistically significant effect on ENO1. Altogether the data suggest that the local 5-hmC content plays a crucial role in the induction of these glycolytic HIF target genes (Fig. 6). However, not all hypoxia-responsive genes were down-regulated when the cells were treated with fumarate or succinate, as fumarate treatment increased the expression of VEGFA and BNIP3 mRNAs, whereas succinate did not affect the BNIP3 mRNA levels at all and reduced the VEGFA mRNA. These data suggest that lower 5-hmC levels were not sufficient to down-regulate the expression of BNIP3 and VEGFA mRNAs, but that this was principally driven by the fumarate-mediated stabilization of HIF-1α (Fig. 6).
FIGURE 6.
Fumarate and succinate regulate HIF target gene expression via TETs. FH and SDH mutations with impaired enzyme activity are found in various cancers, where they cause the accumulation of fumarate or succinate, respectively. Fumarate inhibits the TETs and HIF-P4Hs, leading to a reduction in global 5-hmC levels and HIF-1α/HIF-2α stabilization, respectively. Expression of the glycolysis-associated HIF1 target genes PGK1, HK2, and ENO1 was reduced in cells treated with fumarate, suggesting that the TETs and 5-hmC have a crucial role in the regulation of these genes. The expression of VEGFA and BNIP3 mRNAs was nevertheless, increased in cells treated with fumarate, suggesting that these genes are more driven by HIF-1α/HIF-2α stabilization than by the reduction in 5-hmC levels. *, cells treated with succinate showed reductions in VEGFA, PGK1, and HK2 mRNA expression, suggesting that the TETs and 5-hmC can regulate their expression when HIF-1α is not stabilized. HIF target genes are thus regulated in a multilayered manner in which 5-hmC acts as an additional layer of regulation. Not all HIF target genes are regulated equally by 5-hmC, suggesting that there could be promoter-specific gains and reductions in 5-hmC via TETs.
To verify the involvement of TETs in the regulation of these HIF target genes by fumarate and succinate, we silenced TET1 and TET3 and studied their expression. Loss of TET1 and TET3, similar to treatment with succinate, down-regulated VEGFA mRNA. In contrast, fumarate induced VEGFA mRNA, presumably because the robust induction of HIF by fumarate offsets its inhibitory effects on the TET enzymes. On the other hand, silencing of TET1 and TET3 alone significantly or almost significantly reduced, respectively, HK2 mRNA levels and the addition of fumarate potentiated the effects of TET1 or TET3 silencing. This suggests that the relative contributions of local 5-hmC levels and HIF protein levels differ for different HIF target genes. As TET knock-out cells were not available, and the levels of TET1 and TET3 knockdowns were moderate, we cannot exclude the possibility that other factors, in addition to TET inhibition, contributed to the observed changes in gene expression in vivo following fumarate or succinate treatment.
Overall, the present data show that hypoxia-inducible genes are regulated in a multilayered manner that includes HIF stabilization as well as epigenetic regulation via TETs and 5-hmC levels (Fig. 6). Not all HIF target genes are regulated equally via TETs. Fumarate and succinate can regulate global 5-hmC levels and the induction of HIF target genes via TET inhibition (Fig. 6). In the SDH and FH mutant tumors, and perhaps even in the IDH1 mutants that also associate with the DNA hypermethylator phenotype (37), inhibition of TETs by the accumulating 2-oxoglutarate analogues may well contribute to gene regulation and oncogenesis.
Author Contributions
T. L. designed, performed, and analyzed the experiments and participated in writing of the paper. C. J. M. and J. Z. C. performed and analyzed the experiments shown in Fig. 3, C and D. T. I. performed and analyzed the experiments shown in Figs. 1 and 2. J. H. performed and analyzed the experiments shown in Table 5. W. G. K. and L. A. G. took part in conception and design of the study and L. A. G. additionally contributed to analysis and interpretation of data. P. K. designed and coordinated the study and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank T. Aatsinki and E. Lehtimäki for excellent technical assistance.
This work was supported by Academy of Finland Grants 120156, 140765, 218129, and 266719 (to P. K.), and grants from the S. Jusélius Foundation (to P. K.), the Emil Aaltonen Foundation (to P. K.), the Jane and Aatos Erkko Foundation (to P. K.), and the Finnish Cancer Organizations (to P. K.). The authors declare that they have no conflicts of interest with the contents of this article.
- 2-OGDD
- 2-oxoglutarate-dependent dioxygenases
- TET
- ten-eleven-translocation 5-methylcytosine dioxygenases
- HIF
- hypoxia-inducible factor
- P4H
- prolyl 4-hydroxylases
- 5-mC
- 5-methylcytosine
- 5-hmC
- 5-hydroxy-methylcytosine
- AML
- acute myeloid leukemia
- SDH
- succinate dehydrogenase
- FH
- fumarate hydratase
- IDH
- isocitrate dehydrogenase
- DEF
- diethyl fumarate
- DMS
- dimethyl succinate
- DMSO
- dimethyl sulfoxide
- qPCR
- quantitative real time RT-PCR
- S-2HG
- S-2-hydroxyglutarate.
References
- 1. McDonough M. A., Loenarz C., Chowdhury R., Clifton I. J., and Schofield C. J. (2010) Structural studies on human 2-oxoglutarate dependent oxygenases. Curr. Opin. Struct. Biol. 20, 659–672 [DOI] [PubMed] [Google Scholar]
- 2. Losman J. A., and Kaelin W. G. Jr. (2013) What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 27, 836–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vasanthakumar A., and Godley L. A. (2015) 5-Hydroxymethylcytosine in cancer: significance in diagnosis and therapy. Cancer. Genet. 208, 167–177 [DOI] [PubMed] [Google Scholar]
- 4. Myllyharju J., and Koivunen P. (2013) Hypoxia-inducible factor prolyl 4-hydroxylases: common and specific roles. Biol. Chem. 394, 435–448 [DOI] [PubMed] [Google Scholar]
- 5. Pastor W. A., Aravind L., and Rao A. (2013) TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 14, 341–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tahiliani M., Koh K. P., Shen Y., Pastor W. A., Bandukwala H., Brudno Y., Agarwal S., Iyer L. M., Liu D. R., Aravind L., and Rao A. (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kriaucionis S., and Heintz N. (2009) The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ito S., D'Alessio A. C., Taranova O. V., Hong K., Sowers L. C., and Zhang Y. (2010) Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Abdel-Wahab O., Mullally A., Hedvat C., Garcia-Manero G., Patel J., Wadleigh M., Malinge S., Yao J., Kilpivaara O., Bhat R., Huberman K., Thomas S., Dolgalev I., Heguy A., Paietta E., Le Beau M. M., Beran M., Tallman M. S., Ebert B. L., Kantarjian H. M., Stone R. M., Gilliland D. G., Crispino J. D., and Levine R. L. (2009) Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood 114, 144–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Delhommeau F., Dupont S., Della Valle V., James C., Trannoy S., Massé A., Kosmider O., Le Couedic J. P., Robert F., Alberdi A., Lécluse Y., Plo I., Dreyfus F. J., Marzac C., Casadevall N., Lacombe C., Romana S. P., Dessen P., Soulier J., Viguié F., Fontenay M., Vainchenker W., and Bernard O. A. (2009) Mutation in TET2 in myeloid cancers. N. Engl. J. Med. 360, 2289–2301 [DOI] [PubMed] [Google Scholar]
- 11. Langemeijer S. M., Kuiper R. P., Berends M., Knops R., Aslanyan M. G., Massop M., Stevens-Linders E., van Hoogen P., van Kessel A. G., Raymakers R. A., Kamping E. J., Verhoef G. E., Verburgh E., Hagemeijer A., Vandenberghe P., de Witte T., van der Reijden B. A., and Jansen J. H. (2009) Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat. Genet. 41, 838–842 [DOI] [PubMed] [Google Scholar]
- 12. Tefferi A., Pardanani A., Lim K. H., Abdel-Wahab O., Lasho T. L., Patel J., Gangat N., Finke C. M., Schwager S., Mullally A., Li C. Y., Hanson C. A., Mesa R., Bernard O., Delhommeau F., Vainchenker W., Gilliland D. G., and Levine R. L. (2009) TET2 mutations and their clinical correlates in polycythemia vera, essential thrombocythemia and myelofibrosis. Leukemia 23, 905–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mariani C. J., Vasanthakumar A., Madzo J., Yesilkanal A., Bhagat T., Yu Y., Bhattacharyya S., Wenger R. H., Cohn S. L., Nanduri J., Verma A., Prabhakar N. R., and Godley L. A. (2014) TET1-mediated hydroxymethylation facilitates hypoxic gene induction in neuroblastoma. Cell. Rep. 7, 1343–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Astuti D., Latif F., Dallol A., Dahia P. L., Douglas F., George E., Sköldberg F., Husebye E. S., Eng C., and Maher E. R. (2001) Gene mutations in the succinate dehydrogenase subunit SDHB cause susceptibility to familial pheochromocytoma and to familial paraganglioma. Am. J. Hum. Genet. 69, 49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hao H. X., Khalimonchuk O., Schraders M., Dephoure N., Bayley J. P., Kunst H., Devilee P., Cremers C. W., Schiffman J. D., Bentz B. G., Gygi S. P., Winge D. R., Kremer H., and Rutter J. (2009) SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science. 325, 1139–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bayley J. P., Kunst H. P., Cascon A., Sampietro M. L., Gaal J., Korpershoek E., Hinojar-Gutierrez A., Timmers H. J., Hoefsloot L. H., Hermsen M. A., Suárez C., Hussain A. K., Vriends A. H., Hes F. J., Jansen J. C., Tops C. M., Corssmit E. P., de Knijff P., Lenders J. W., Cremers C. W., Devilee P., Dinjens W. N., de Krijger R. R., and Robledo M. (2010) SDHAF2 mutations in familial and sporadic paraganglioma and phaeochromocytoma. Lancet Oncol. 11, 366–372 [DOI] [PubMed] [Google Scholar]
- 17. Tomlinson I. P., Alam N. A., Rowan A. J., Barclay E., Jaeger E. E., Kelsell D., Leigh I., Gorman P., Lamlum H., Rahman S., Roylance R. R., Olpin S., Bevan S., Barker K., Hearle N., Houlston R. S., Kiuru M., Lehtonen R., Karhu A., Vilkki S., Laiho P., Eklund C., Vierimaa O., Aittomäki K., Hietala M., Sistonen P., Paetau A., Salovaara R., Herva R., Launonen V., Aaltonen L. A., and Multiple Leiomyoma Consortium (2002) Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat. Genet. 30, 406–410 [DOI] [PubMed] [Google Scholar]
- 18. Yan H., Parsons D. W., Jin G., McLendon R., Rasheed B. A., Yuan W., Kos I., Batinic-Haberle I., Jones S., Riggins G. J., Friedman H., Friedman A., Reardon D., Herndon J., Kinzler K. W., Velculescu V. E., Vogelstein B., and Bigner D. D. (2009) IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 360, 765–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ward P. S., Patel J., Wise D. R., Abdel-Wahab O., Bennett B. D., Coller H. A., Cross J. R., Fantin V. R., Hedvat C. V., Perl A. E., Rabinowitz J. D., Carroll M., Su S. M., Sharp K. A., Levine R. L., and Thompson C. B. (2010) The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17, 225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pollard P. J., Brière J. J., Alam N. A., Barwell J., Barclay E., Wortham N. C., Hunt T., Mitchell M., Olpin S., Moat S. J., Hargreaves I. P., Heales S. J., Chung Y. L., Griffiths J. R., Dalgleish A., McGrath J. A., Gleeson M. J., Hodgson S. V., Poulsom R., Rustin P., and Tomlinson I. P. (2005) Accumulation of Krebs cycle intermediates and over-expression of HIF1α in tumours which result from germline FH and SDH mutations. Hum. Mol. Genet. 14, 2231–2239 [DOI] [PubMed] [Google Scholar]
- 21. Dang L., White D. W., Gross S., Bennett B. D., Bittinger M. A., Driggers E. M., Fantin V. R., Jang H. G., Jin S., Keenan M. C., Marks K. M., Prins R. M., Ward P. S., Yen K. E., Liau L. M., Rabinowitz J. D., Cantley L. C., Thompson C. B., Vander Heiden M. G., and Su S. M. (2009) Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Selak M. A., Armour S. M., MacKenzie E. D., Boulahbel H., Watson D. G., Mansfield K. D., Pan Y., Simon M. C., Thompson C. B., and Gottlieb E. (2005) Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-α prolyl hydroxylase. Cancer Cell 7, 77–85 [DOI] [PubMed] [Google Scholar]
- 23. Isaacs J. S., Jung Y. J., Mole D. R., Lee S., Torres-Cabala C., Chung Y. L., Merino M., Trepel J., Zbar B., Toro J., Ratcliffe P. J., Linehan W. M., and Neckers L. (2005) HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell 8, 143–153 [DOI] [PubMed] [Google Scholar]
- 24. Koivunen P., Hirsilä M., Remes A. M., Hassinen I. E., Kivirikko K. I., and Myllyharju J. (2007) Inhibition of hypoxia-inducible factor (HIF) hydroxylases by citric acid cycle intermediates: possible links between cell metabolism and stabilization of HIF. J. Biol. Chem. 282, 4524–4532 [DOI] [PubMed] [Google Scholar]
- 25. Koivunen P., Lee S., Duncan C. G., Lopez G., Lu G., Ramkissoon S., Losman J. A., Joensuu P., Bergmann U., Gross S., Travins J., Weiss S., Looper R., Ligon K. L., Verhaak R. G., Yan H., and Kaelin W. G. Jr. (2012) Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature 483, 484–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hirsilä M., Koivunen P., Günzler V., Kivirikko K. I., and Myllyharju J. (2003) Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J. Biol. Chem. 278, 30772–30780 [DOI] [PubMed] [Google Scholar]
- 27. Birkler R. I., Støttrup N. B., Hermannson S., Nielsen T. T., Gregersen N., Bøtker H. E., Andreasen M. F., and Johannsen M. (2010) A UPLC-MS/MS application for profiling of intermediary energy metabolites in microdialysis samples: a method for high-throughput. J. Pharm. Biomed. Anal. 53, 983–990 [DOI] [PubMed] [Google Scholar]
- 28. Madzo J., Liu H., Rodriguez A., Vasanthakumar A., Sundaravel S., Caces D. B., Looney T. J., Zhang L., Lepore J. B., Macrae T., Duszynski R., Shih A. H., Song C. X., Yu M., Yu Y., Grossman R., Raumann B., Verma A., He C., Levine R. L., Lavelle D., Lahn B. T., Wickrema A., and Godley L. A. (2014) Hydroxymethylation at gene regulatory regions directs stem/early progenitor cell commitment during erythropoiesis. Cell. Rep. 6, 231–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vasanthakumar A., Lepore J. B., Zegarek M. H., Kocherginsky M., Singh M., Davis E. M., Link P. A., Anastasi J., Le Beau M. M., Karpf A. R., and Godley L. A. (2013) Dnmt3b is a haploinsufficient tumor suppressor gene in Myc-induced lymphomagenesis. Blood 121, 2059–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xiao M., Yang H., Xu W., Ma S., Lin H., Zhu H., Liu L., Liu Y., Yang C., Xu Y., Zhao S., Ye D., Xiong Y., and Guan K. L. (2012) Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 26, 1326–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Letouzé E., Martinelli C., Loriot C., Burnichon N., Abermil N., Ottolenghi C., Janin M., Menara M., Nguyen A. T., Benit P., Buffet A., Marcaillou C., Bertherat J., Amar L., Rustin P., De Reyniès A., Gimenez-Roqueplo A. P., and Favier J. (2013) SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell 23, 739–752 [DOI] [PubMed] [Google Scholar]
- 32. Lian C. G., Xu Y., Ceol C., Wu F., Larson A., Dresser K., Xu W., Tan L., Hu Y., Zhan Q., Lee C. W., Hu D., Lian B. Q., Kleffel S., Yang Y., Neiswender J., Khorasani A. J., Fang R., Lezcano C., Duncan L. M., Scolyer R. A., Thompson J. F., Kakavand H., Houvras Y., Zon L. I., Mihm M. C. Jr, Kaiser U. B., Schatton T., Woda B. A., Murphy G. F., and Shi Y. G. (2012) Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell 150, 1135–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Orr B. A., Haffner M. C., Nelson W. G., Yegnasubramanian S., and Eberhart C. G. (2012) Decreased 5-hydroxymethylcytosine is associated with neural progenitor phenotype in normal brain and shorter survival in malignant glioma. PLoS ONE 7, e41036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Haffner M. C., Chaux A., Meeker A. K., Esopi D. M., Gerber J., Pellakuru L. G., Toubaji A., Argani P., Iacobuzio-Donahue C., Nelson W. G., Netto G. J., De Marzo A. M., and Yegnasubramanian S. (2011) Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget 2, 627–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Murata A., Baba Y., Ishimoto T., Miyake K., Kosumi K., Harada K., Kurashige J., Iwagami S., Sakamoto Y., Miyamoto Y., Yoshida N., Yamamoto M., Oda S., Watanabe M., Nakao M., and Baba H. (2015) TET family proteins and 5-hydroxymethylcytosine in esophageal squamous cell carcinoma. Oncotarget 6, 23372–23382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Semenza G. L. (2009) Regulation of oxygen homeostasis by hypoxia-inducible factor 1. Physiology (Bethesda) 24, 97–106 [DOI] [PubMed] [Google Scholar]
- 37. Turcan S., Rohle D., Goenka A., Walsh L. A., Fang F., Yilmaz E., Campos C., Fabius A. W., Lu C., Ward P. S., Thompson C. B., Kaufman A., Guryanova O., Levine R., Heguy A., Viale A., Morris L. G., Huse J. T., Mellinghoff I. K., and Chan T. A. (2012) IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483, 479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hirsilä M., Koivunen P., Xu L., Seeley T., Kivirikko K. I., and Myllyharju J. (2005) Effect of desferrioxamine and metals on the hydroxylases in the oxygen sensing pathway. FASEB J. 19, 1308–1310 [DOI] [PubMed] [Google Scholar]
- 39. Myllyharju J., and Kivirikko K. I. (1997) Characterization of the iron- and 2-oxoglutarate-binding sites of human prolyl 4-hydroxylase. EMBO J. 16, 1173–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]