SUMMARY
Changes in bone size and shape are defining features of many vertebrates. Here we use genetic crosses and comparative genomics to identify specific regulatory DNA alterations controlling skeletal evolution. Armor bone size differences in sticklebacks maps to a major effect locus overlapping BMP family member GDF6. Freshwater fish express more GDF6 due in part to a transposon insertion, and transgenic overexpression of GDF6 phenocopies evolutionary changes in armor plate size. The human GDF6 locus also has undergone distinctive regulatory evolution, including complete loss of an enhancer that is otherwise highly conserved between chimps and other mammals. Functional tests show that the ancestral enhancer drives expression in hindlimbs but not forelimbs, in locations that have been specifically modified during the human transition to bipedalism. Both gain and loss of regulatory elements can localize BMP changes to specific anatomical locations, providing a flexible regulatory basis for evolving species-specific changes in skeletal form.
INTRODUCTION
Striking changes in vertebrate skeletal structures underlie interesting differences in locomotion, foraging, prey capture, competition, and body defense (Flower, 1870). BMPs are key signaling molecules that are known to be necessary, sufficient, and expressed at the right time and place to control local formation of bones and joints during normal development (Kingsley, 1994). Gain and loss of specific enhancers in BMP genes have long been proposed as plausible mechanisms for evolving new skeletal structures while preserving other essential functions (Kingsley, 1994; Abzhanov et al., 2004; Albertson et al., 2005; Guenther et al., 2008). However, the molecular changes that underlie different BMP expression are still unknown, and could map either to the BMP genes themselves, or to other trans-acting factors that alter BMP expression indirectly.
Genetic mapping of skeletal traits provides a powerful method for identifying the number and location of loci controlling interesting skeletal differences. Stickleback fish are particularly well suited for such studies, because many newly established freshwater populations have repeatedly evolved dramatic differences in skeletal structures following widespread melting of glaciers 10,000-20,000 years ago (Bell and Foster, 1994). Naturally occurring species with major changes in armor and trophic structures can still be crossed using artificial fertilization in the laboratory. Linkage analysis has mapped major quantitative trait loci (QTL) controlling a variety of skeletal traits (Kingsley and Peichel, 2007; Miller et al., 2014). Two of these QTL, which control two thirds or more of the variance in armor plate number and pelvic hindfin size, have now been traced to changes in the Ectodysplasin (EDA) and Pituitary homeodomain protein 1 (PITX1) genes (Colosimo et al., 2005; Chan et al., 2010).
Comparative sequencing and genome scanning provide another useful method for identifying genomic intervals with striking differences between species. For example, genome-wide sequence analysis has identified loci that show evidence of positive selection among modern humans that differentiate us from Neandertals, or that show unique sequence changes in humans compared to chimpanzees or more distantly related mammals (reviewed by Fu and Akey, 2013). Complete genome sequence analysis of replicate marine-freshwater pairs has identified a large number of loci with consistent changes seen during recent stickleback evolution. The overall set of repeatedly selected regions suggests that non-coding regulatory changes play a predominant role in stickleback evolution (Jones et al., 2012).
Despite the power of comparative sequence analysis to implicate particular genomic regions in evolutionary change, the particular traits controlled by most regions found by genome scanning are still unknown (Fu and Akey, 2013). Here we combine phenotypic information from QTL mapping and genome scan information from comparative sequencing to identify evolutionary mechanisms controlling armor plate size in sticklebacks. Our studies show that repeated evolution of armor plate size in freshwater sticklebacks has occurred in part by a cis-acting increase in expression of a particular BMP family member, Growth/differentiation factor 6 (GDF6) (Storm et al., 1994). We also show that the primate GDF6 locus had undergone cis-acting regulatory changes in humans, including the deletion of an enhancer sequence that normally controls expression in hindlimb structures that have undergone significant changes in the human transition to bipedalism. Regulatory changes in GDF6 expression may thus be a recurrent mechanism for evolving species-specific changes in skeletal structures, through both gain and loss of regulatory enhancers that control expression in particular anatomical domains of the body.
RESULTS
Fine Mapping of Stickleback Armor Plate Size
Marine sticklebacks are covered by large armor plates made of dermal bone. Freshwater fish have typically evolved both fewer and smaller plates, which may contribute to neutral buoyant density in freshwater, reduced metabolic demand for calcium and phosphate, or increased body flexibility and higher burst swimming speed (Giles, 1983; Bergstrom, 2002; Myhre and Klepaker, 2009). Previous mapping studies show that distinct loci control the number and the size of armor plates in a cross between heavily armored marine and armor-reduced freshwater sticklebacks (Colosimo et al., 2004). To further study the QTL with the largest effect on plate size, we separately measured both armor plate height and width in additional F2 progeny from the same marine x benthic cross, and typed fish with a dense set of microsatellite markers (Table S1) designed in the region surrounding previous peak marker on chromosome XX. Interestingly, armor plate height and width map to two distinct, but closely linked loci (Figure 1A), with 95% confidence intervals based on a 2- likelihood-of-odds (LOD) score criterion spanning approximately 460 kb and 320 kb (chrXX:3,280,000-3,740,000 and chrXX:3,700,000-4,020,000; respectively).
Figure 1. Genetic, Sequence, and Morphological Evidence for Dual Loci Controlling Armor Plate Dimensions in Sticklebacks.
(A) Two distinct, but closely-linked loci regulate armor plate height and width. Plate height (green) and width (red) were mapped in a large marine x benthic F2 cross using MapQTL. Likelihood-of-odds (LOD) scores and the percent variance explained (PVE) for peak markers for each trait are shown. The 2-LOD intervals for each QTL are shaded (green and red, overlap in yellow), and high significance cut offs (1000 permutations with MapQTL; p<0.001) are shown with dotted lines. Representative fish from the grandparental marine and benthic populations stained with Alizarin red are shown.
(B) There are two major peaks of repeated sequence divergence between freshwater and marine fish in the plate size interval. Sequence divergence between pairs (n=11) of geographically proximal marine and freshwater sticklebacks are plotted (colored lines). Populations are listed in Table S2. The 2-LOD score intervals for plate height and width (black rectangles), the GDF6 locus, and the region cloned for testing for enhancer activity in Figure 2 are shown.
(C and D) Geographic patterns in armor plate morphology match patterns of sequence divergence. Armor plate height (C) and width (D) were measured from adults (Table S2) from each of the twenty-one sequenced populations and were normalized for standard fish depth and length, respectively. Pacific basin freshwater (FW) populations are shown in black, Atlantic basin freshwater populations in gray, and Pacific and Atlantic basin marine populations in white. Two-sample Wilcoxon tests were used to examine the significances of plate size averages distribution differences between groups of fish populations (bracketed), and p-values are shown. Error bars represent SEM.
See also Figure S1 and Tables S1, S2 and S3.
To compare these high-resolution mapping results with patterns of genomic sequence changes in natural populations, we also analyzed patterns of DNA sequence divergence between 11 different pairs of marine and freshwater fish (Table S2) for which whole genome sequence is available (Jones et al., 2012). For each pair, we counted the number of single nucleotide polymorphisms (SNPs) between the marine and the freshwater sticklebacks in a sliding window with size 2500 bp and step 500 bp, and expressed the results as fraction of divergent nucleotides per window (Figure 1B). This analysis reveals two major signals of repeated sequence divergence between freshwater and marine sticklebacks, one in the plate height QTL, and the other in the plate width QTL (Figure 1B).
Interestingly, the prominent sequence divergence peak in the plate height region is observed in many Atlantic and Pacific population pairs, while the sequence divergence peak in the plate width region is only seen in sticklebacks from the Pacific Ocean basin (Figure 1B). To test for possible geographic differences in armor plate morphology, we also measured the average height and width of plates in multiple populations from both ocean basins. Sticklebacks from the Pacific have significant reductions in both plate height and width, while sticklebacks from the Atlantic have a significant reduction only in plate height (Figures 1C and 1D), a pattern that matches the pattern of sequence differentiation seen in genomic comparisons. Thus, high resolution genetic mapping, patterns of DNA sequence divergence, and patterns of armor plate morphological change all suggest that the height and width of armor plates can be controlled independently.
Coding and Regulatory Changes in Candidate Armor Plate Genes
Separate but closely linked morphological effects could be due either to distinct genes in the plate size region, or to distinct regulatory elements controlling a single key gene. Using the stickleback reference genome (Jones et al., 2012), and gene predictions based on ENSEMBL, GENSCAN, blastx, and stickleback expressed sequence tags (ESTs), we identify 17 genes in the plate height interval, and 11 genes in the plate width interval (Figure S1 and Table S3). A single gene encoding a bone morphogenetic protein, GDF6, is found in the small overlap between the two intervals (Figures 1B and S1, and Table S3). As expected, the QTL regions also include a large number of conserved non-coding elements (CNEs) (Figure S1A and Table S3), which often act as enhancers.
To look for possible coding region differences in these genes, we completely sequenced marine bacterial artificial chromosome clones (BACs) covering the plate size candidate regions, and compared the marine sequence to the reference freshwater genome and SNP information from previously sequenced populations (Jones et al., 2012). This analysis identified 82 SNPs in protein coding sequences that are found in many different stickleback populations (Table S4), most of which were synonymous mutations or missense mutations predicted to have little functional effects on proteins (Table S5).
The GDF6 gene showed no consistent marine freshwater amino acid changes. However it is a particularly interesting candidate gene for armor plate phenotypes since it is the only gene located in the overlap between the two armor plate size QTL intervals, and is known to be required for normal skeletal development in mice, humans, and fish (Settle et al., 2003; Tassabehji et al., 2007; Asai-Coakwell et al., 2009). To test for possible cis-acting regulatory changes in GDF6, we generated F1 hybrid fish from a cross between a large-plated marine fish and small-plated freshwater fish, extracted RNA from the developing armor plate regions, and compared the relative expression levels of freshwater and marine GDF6 alleles. The freshwater GDF6 allele was expressed at significantly higher levels than the marine allele in heterozygous sticklebacks (Figure 2A). In contrast, freshwater and marine alleles of the nearby Branched Chain Alpha-Keto Acid Dehydrogenase (BCKDHB) gene showed balanced expression in developing armor plates (Figure 2A). Since all assays were done in the same trans-acting environment of F1 hybrid individuals, these results show that significant cis-regulatory changes have evolved in the stickleback GDF6 gene.
Figure 2. Identification of cis-Regulatory Changes in the Plate Size Interval.
(A) The freshwater allele of GDF6 is expressed at higher level than the marine allele in F1 hybrids. RNA was extracted from the indicated region of F1 hybrid fish from a cross between a small-plated freshwater (FW) and large-plated marine fish, and the relative abundance of the freshwater and marine alleles was quantified by pyrosequencing. Significance of observed deviation from the fraction of freshwater allele of 0.5 (dashed line) was tested using one sample t-test, and “*” represents p<0.05. Error bars represent SEM.
(B) Freshwater but not marine enhancer drives expression along the flanks of developing stickleback embryos. Marine and freshwater (FW) sequences were cloned from the regulatory region shown in Figure 1B into a GFP reporter vector with a minimal hsp70 promoter, and were then injected in fertilized one-cell stage embryos from marine fish. Pictures of the flanks of developing fry (dashed white lines) were taken at 4 dpf.
(C) Map of the enhancer region and sequences tested in transgenic fish. Repeating vertical lines represent regions of alignment between marine (red) and freshwater (blue) sequence, with gaps shown in the region of a complex repeat and a L2 LINE transposable element (TE) present in the freshwater but not marine sequence. Tall vertical lines denote the edges of the region added or removed from the modified constructs also tested for enhancer activity.
Identification of an Armor Plate Enhancer
The recurrent sequence divergence peak that we observe in the plate height QTL region (Figure 1B) is located over non-coding sequences. To test whether this region may correspond to a regulatory enhancer, we separately cloned the region from both marine and freshwater fish and assayed for the ability to drive consistent GFP reporter patterns in transgenic sticklebacks. The cloned region is 4.2 kb in the freshwater reference genome (chrXX:3,610,110-3,614,309). The corresponding marine region is shorter, primarily due to the absence of a 1.3 kb transposable element belonging to the L2 LINE family and part of an adjacent high-complexity repeat, which together account for 89.9% of the differences between the marine and freshwater sequences (Table S6). The flanking regions share 94.1% sequence identity, differing by 18 small indels and 89 SNPs (Table S6), 58 of which show a greater than twofold difference in ecotypic allele frequency across many populations (Figure S2).
Constructs containing the marine or freshwater region were separately injected into fertilized one-cell stage marine embryos, and GFP expression was analyzed in developing fish. Many transgenic embryos injected with the freshwater allele expressed GFP along the developing flanks (6 out of 31 transgenic embryos) (Figure 2B). In contrast, flank expression was absent or weak when we injected the marine construct, with only 1 of 46 transgenic embryos showing a weak GFP signal anywhere along the flank (Fisher's Exact Test, p = 0.015). The plate size interval thus contains a regulatory enhancer for flank expression, and the multiple sequence changes seen between marine and freshwater fish lead to stronger expression from the freshwater enhancer.
To test the potential contribution of the transposon to increased expression from the freshwater enhancer, we modified the above transgenic constructs to either remove the transposable element (TE) from the freshwater enhancer or insert it into the corresponding location in the marine enhancer, and then compared the expression patterns of both modified and unmodified constructs in additional transgenic fish (Figure 2C). Flank GFP expression was again observed robustly in transgenic embryos injected with the pure freshwater construct (16/71), but not in embryos injected with either the pure marine construct (0/137), the “marine + TE” construct (0/372), or the “freshwater – TE” construct (0/291). Thus, the freshwater insertion is required, but not sufficient, to produce increased flank expression. These data suggest that the increased activity of the freshwater enhancer depends upon both the transposon as well as additional sequence differences flanking the insertion, consistent with the multi-step mutational changes also thought to contribute to other examples of evolved regulatory differences in both natural and laboratory populations (Rebeiz et al., 2009; Frankel et al., 2011; Blount et al., 2012).
Recapitulation of Armor Plate Phenotypes in GDF6 Transgenic Fish
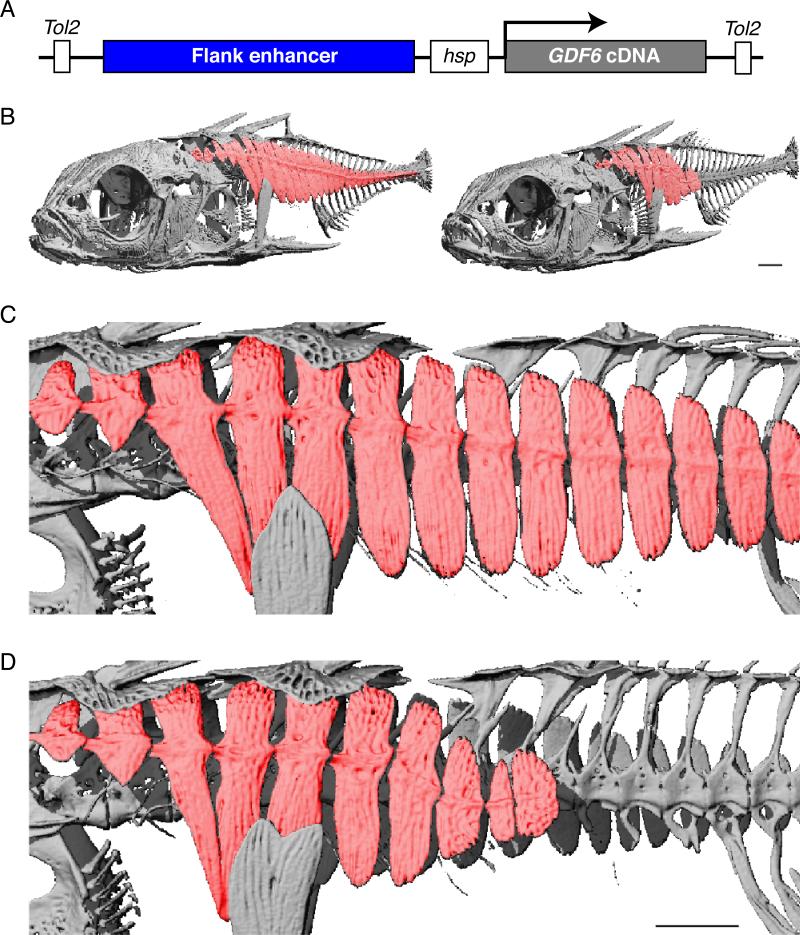
To directly test the functional effects of GDF6 expression on stickleback armor plate development, we cloned a stickleback GDF6 cDNA either under the control of the flank enhancer described above, or under the control of constitutive promoters from the pTK and pCMV genes. Constructs were injected into fertilized eggs from marine fish, and mosaic founder animals were aged to 6 months and scored for armor plate morphology. Multiple GDF6 transgenic sticklebacks showed either missing or smaller, shorter, plates (Figures 3 and S3), mimicking the reduction of armor plate seen in freshwater fish. In contrast, similar armor plate changes were never observed in control siblings from the same crosses (7/163 transgenic fish with plate phenotypes, versus 0/203 controls; Fisher's Exact Test p=0.003).
Figure 3. GDF6 Transgenic Fish have Armor Plate Phenotypes.
(A) Schematic of the construct used for generating GDF6 transgenic fish. The flank enhancer in the height QTL (Figures 1B and 2B) was cloned upstream of a minimal hsp70 promoter and GDF6 cDNA flanked by Tol2 transposase recognition sites. The construct was then co-injected with mRNA encoding Tol2 transposase into one-cell marine stickleback embryos.
(B) μCT-derived volumetric reconstructions of representative control (left) and GDF6 transgenic (right) fish at 219 dpf. The armored plates are colored in red. Note the absence of caudal plates from the left flank of the GDF6 transgenic fish.
(C and D) Close up views of control (C) and GDF6 (D) transgenic fish pictured in (B). The last three plates remaining in the GDF6 transgenic fish are noticeably smaller than the corresponding plates in the control fish. Scale bars in (B) and (D), 1 mm.
See also Figure S3.
A Human-Specific Deletion of a Hindlimb Enhancer in the GDF6 Locus
Our combined mapping, expression, and transgenic experiments together provide strong evidence that cis-regulatory evolution of the GDF6 gene contributes to a classic skeletal difference in sticklebacks. Previous studies have shown that the same loci that underlie repeated evolution of stickleback traits may also be reused when related traits evolve in other species, including humans (Shapiro et al., 2006; Miller et al., 2007; Guenther et al., 2014). Because GDF6 shares the structural and developmental properties of other loci that appear to be favorable substrates for repeated evolution (Knecht et al., 2007; Stern and Orgogozo, 2009), we examined whether lineage-specific regulatory changes may have also occurred in GDF6 during evolution of novel skeletal structures in primates.
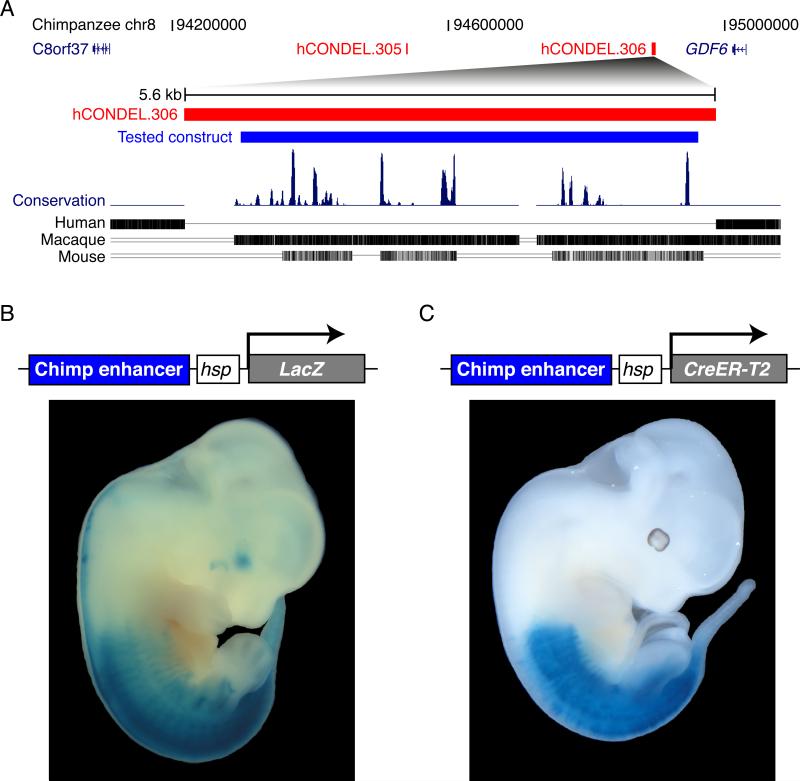
Previous genomic surveys have identified over 500 locations where humans have lost non-coding DNA sequences that are otherwise highly conserved between chimpanzees and other mammals (McLean et al., 2011). Two of the 510 confirmed human conserved sequence deletions (hCONDELs) are located in the noncoding regions adjacent to the GDF6 gene (Figure 4A and Table S7). To test the possible regulatory activity of the ancestral noncoding sequences normally found at these locations, we separately cloned the chimpanzee sequences surrounding hCONDEL.305 and hCONDEL.306 into expression vectors containing a minimal promoter and lacZ reporter gene. Transgenic mouse embryos carrying chimp sequences surrounding hCONDEL.305 did not drive any consistent patterns of lacZ expression at the embryonic day 14.5 and 16.5 stages tested. In contrast, transgenic mouse embryos carrying the chimp sequences found at the location of hCONDEL.306 (Figure 4A) showed consistent expression that was strikingly localized in the posterior region of embryos, including strong expression in hindlimbs but not in forelimbs (Figure 4B).
Figure 4. A Chimpanzee GDF6 Enhancer Missing in Humans Drives Expression in the Posterior of Mouse Embryos.
(A) There are two human-specific deletions of highly conserved chimpanzee sequences in the GDF6 locus. A 1.2 Mb region of the chimpanzee chromosome 8 is shown. Red bars show the positions of a 492 bp and a 5775 bp deletions in humans, hCONDEL.305 and hCONDEL.306, respectively (McLean et al, 2011). Below: multiple species comparison of the hCONDEL.306 region (red), showing sequences aligned between chimpanzee and other mammals. Blue bar represents the chimpanzee sequence tested for enhancer activity in transgenic mice.
(B) hCONDEL.306 chimpanzee sequence drives consistent expression of lacZ reporter in the posterior of E12.5 mouse embryos.
(C) Mice with the hCONDEL.306-hsp-CreER-T2 construct were bred to floxed-ROSA26 reporter mice. Tamoxifen was administered at E9.5 and embryos were lacZ stained at E12.5, showing consistent expression patterns in the posterior of the embryo.
For further studies of this expression pattern, we cloned the same chimp sequences upstream of a minimal-promoter tamoxifen-inducible Cre recombinase (CreER-T2) and generated stable transgenic mouse lines. The hCONDEL.306-hsp-CreER-T2 transgenic mice were then bred to floxed-ROSA26 reporter mice, and injected with tamoxifen at different developmental time points, in order to permanently mark by Cre-mediated recombination those cells expressing the GDF6 transgene at a particular developmental stage, as well as all of their descendants. Tamoxifen injections at E9.5, followed by lacZ staining at E12.5, revealed patterns very similar to those previously seen with the direct hCONDEL.306-hsp-lacZ transgene (compare Figures 4B and 4C; see also Figure S4). Tamoxifen injections at both earlier and later stages continued to give patterns of expression in caudal and hindlimb structures, without labeling of either cranial or forelimb structures (Figure S5). Within hindlimbs, stronger expression was always seen in posterior hindlimb digits compared to the first digit (Figures 5 and S5). Sectioning showed that lacZ expression domains included the cartilaginous rudiments of developing digits (Figure 5B), surrounding soft tissue, and a specific anterior stripe of expression that corresponds to the abductor hallicus muscle of the first toe (Figure S6).
Figure 5. The Chimpanzee GDF6 Enhancer Drives Expression in Posterior Digits of the Hindlimb.
(A) hCONDEL.306-hsp-CreER-T2 transgenic mice treated with tamoxifen at E8.5 show consistent lacZ localization in the hindlimbs but not forelimbs of E16.5 embryos, with stronger labeling in the posterior foot digits compared to the digit 1. Foot is shown in plantar view, with first digit on the left.
(B) The lacZ pattern in the developing foot is seen in transverse sections of the phalanges (outlined with white dashed line). Approximate section plane is indicated on the left (white line).
See also Figure S5.
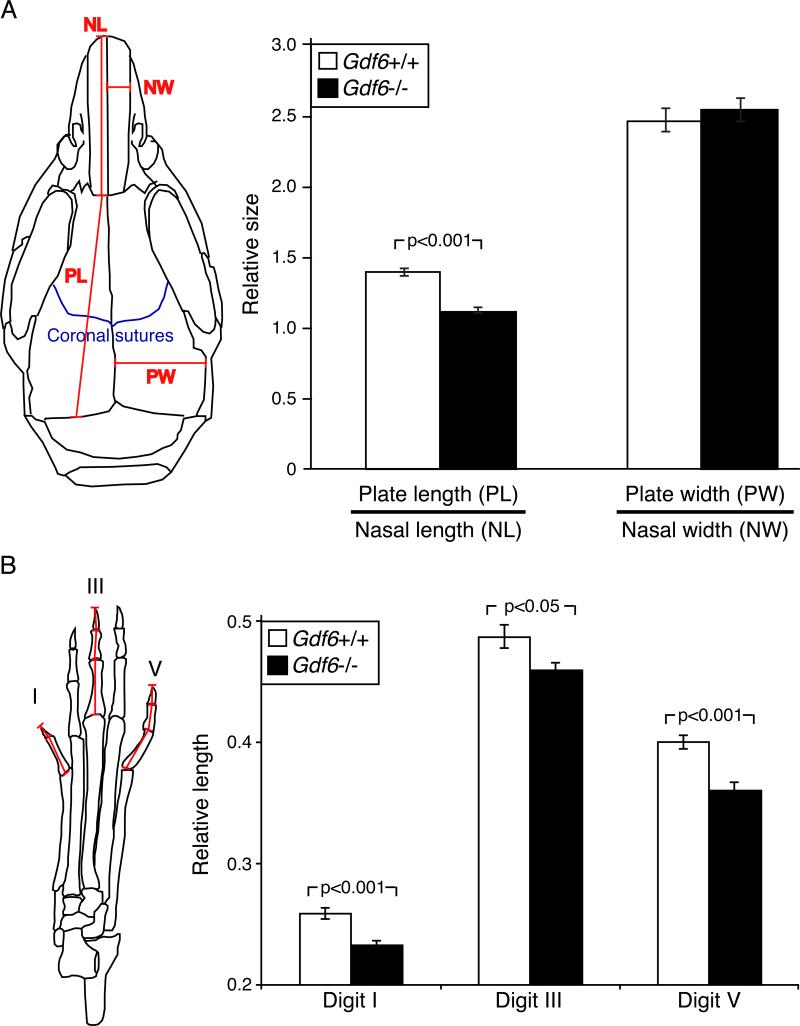
Skull and Digit Phenotypes in Gdf6 Knockout Mice
The mouse Gdf6 gene is known to be required for normal formation of particular joints in the wrist, ankles, middle ear, and vertebral column; and for formation of the coronal suture between the frontal and parietal bones in the skull (Settle et al., 2003). To test whether Gdf6 also controls the size of bones, we measured skull and digit dimensions in wild type and Gdf6 knockout mice. The length, but not width, of dermal flat bones in the skull were significantly shorter in Gdf6 null mutant than wild type mice (Figure 6A). In addition, digits of Gdf6 null mutant mice were significantly shorter than those of control littermates (Figure 6B), confirming that Gdf6 plays an important role in normal growth control of both skull and digit bones. Interestingly, the enhancer that has been deleted in the human lineage normally drives a posterior-specific expression that excludes both cranial regions and forelimbs. Loss of this enhancer would thus preserve normal GDF6 functions in the skull and forelimbs, while confining any digit changes to the posterior digits of the hindlimb (Figure 5). The spatial specificity of the enhancer shows a striking correlation with known changes in hindlimb digit length and musculature that have evolved during the transition to bipedal locomotion in the human lineage (Figure 7A).
Figure 6. Gdf6 Mutant Mice Have Altered Flat Bones and Shorter Toes.
(A) Average width and length of skull bones in wild type (n=12) and Gdf6−/− (n=16) mutant mice. Dimensions were measured for parietal bone width (PW), the combined length of the frontal and parietal bones (PL), nasal bone length (NL), nasal bone width (NW), and the relative length and width of the skulls.
(B) Gdf6 mutant mice have shorter hindlimb digits. The lengths of the sums of the phalanges of digit I, digit III, and digit V from wild type (n=8) and Gdf6−/− (n=9) mice were normalized to femur length and averaged for the two hindlimbs. We used two sample t-tests to examine the significance of skull plate (A) and digits size (B) differences between the wild type and Gdf6−/− mice, and p-values are shown. Error bars represent SEM.
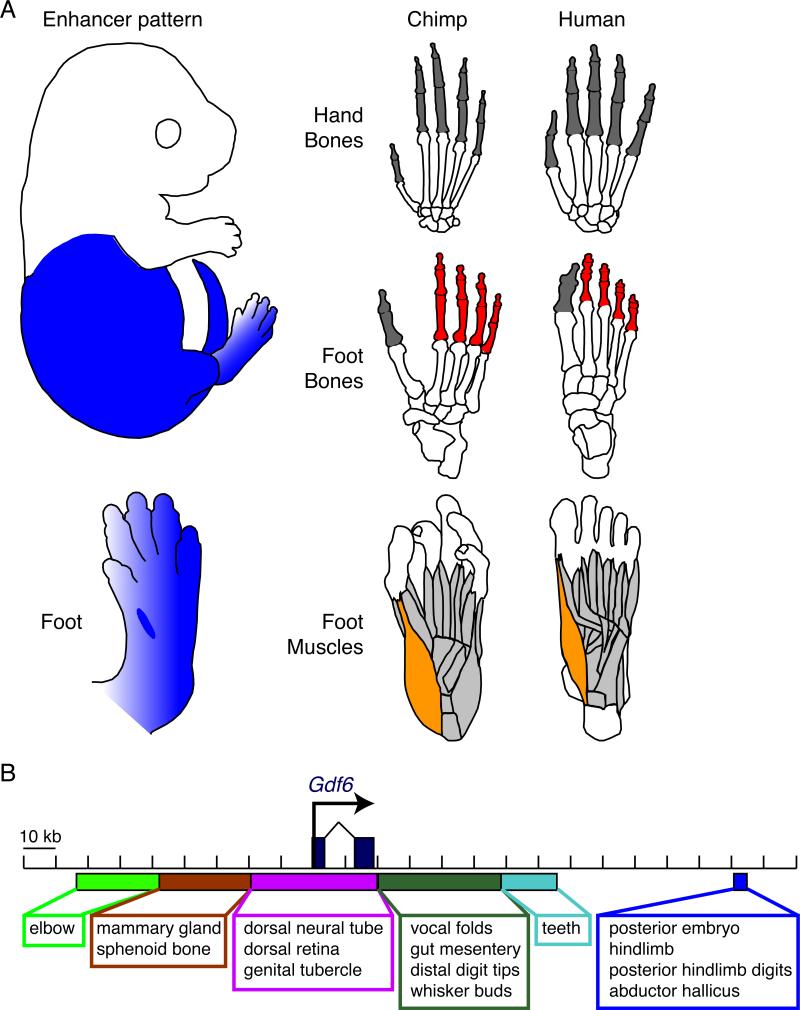
Figure 7. Region-Specific GDF6 Enhancer Compared to Limb-Specific Anatomical Modifications in Human Hindfeet and Forefeet.
(A) Human hindlimbs show multiple changes related to evolution of bipedalism, including shortening of posterior digits of the foot (red), presence of a large toe aligned with other digits, and reduction of the abductor hallicus muscle (orange), which is prominent in other primates with grasping feet and an opposable first toe. In contrast, the homologous digits of the human hand are still long and mobile (dark grey), illustrating the region-specific anatomical changes that have evolved in human hands and feet (modified from Swindler and Wood (1973) and skeletal reproductions of chimpanzee and human feet (BoneClones)). Enhancer sequences in the GDF6 gene also show striking regional specificity during development. The enhancer deleted in the human lineage shows prominent hindlimb but not forelimb expression, and stronger expression in posterior digits within the hindlimb. Loss of this enhancer would not disrupt GDF6 functions in cranial or forelimb regions, but could reduce GDF6 activity in many of the same hindlimb structures altered during the human transition to bipedalism.
(B) Our results add a distal regulatory region to a collection of tissue and region-specific enhancers surrounding Gdf6 (Mortlock et al., 2003). Gain and loss of modular enhancers in BMP genes provides a flexible genomic mechanism for altering skeletal morphology in particular regions of the body.
See also Figure S6.
DISCUSSION
Previous studies have shown that major morphological differences in sticklebacks can be mapped to QTL controlling anywhere from 2.5 to 100% of the variance in the corresponding trait (Kingsley and Peichel, 2007; Miller et al., 2014). To date, only a few of the QTL with largest effects have been traced to particular genes. The major QTL controlling armor plates, pelvis, pigment, and teeth all correspond to key developmental control genes (PITX1, EDA, KITLG, BMP6), each of which plays multiple roles during normal development (Shapiro et al., 2004; Colosimo et al., 2005; Miller et al., 2007; Cleves et al., 2014). In each of these examples, freshwater stickleback populations have made cis-acting regulatory alterations in the corresponding gene that reduce expression at some but not all body sites (Miller et al., 2007; Chan et al., 2010; Cleves et al., 2014; O'Brown et al., 2015). Our current studies show that armor plate size in sticklebacks is also controlled by regulatory changes in a major developmental signaling gene. However, in this case freshwater fish show increased rather than decreased expression, and transgenic experiments confirm that increased expression of GDF6 leads to reduction in armor plate size. These results show that QTL of intermediate effect size in sticklebacks are also associated with changes in essential developmental signaling molecules, and that either gain or loss of expression can contribute to evolutionary change in freshwater populations.
Adaptive Significance of Armor Plate Size
Armor plate reduction has evolved in parallel in many freshwater populations, strongly suggesting that smaller and fewer plates are adaptive in non-marine environments. Large protruding plates may facilitate stickleback capture by grappling insect predators (Reimchen, 1980). Fewer and smaller plates may facilitate faster burst swimming speed in freshwater sticklebacks (Bergstrom, 2002). Quantitative estimates suggest that the observed reduction in weight of mineralized bony armor structures in freshwater fish may help maintain neutral buoyancy after transition from marine to freshwater environments (Myhre and Klepaker, 2009). Smaller plates may also reduce metabolic demand for calcium and phosphate in ion-poor freshwater environments (Giles, 1983), an effect previously linked to higher growth rates for low-plated sticklebacks in freshwater (Barrett et al., 2008). Finally, it is possible that reductions in plate size are correlated with other traits that are actually under selection. For example, neuromast sensory structures form in close association with armor plates, and fish with reduced plates may show correlated changes in sensory structures or behavior (Wark et al., 2012; Greenwood et al., 2013).
Transition to Bidepalism in Humans
In contrast to the uncertain ecological factors that have led to selection of reduced plate size in sticklebacks, it is relatively easy to envision factors that have selected for regional control of digit length in the human lineage. The human lineage is characterized by a transition from partially arboreal to full bipedal locomotion. This transition involved multiple skeletal changes that differ between forelimbs and hindlimbs, including: reduction of posterior toe lengths in feet but not hands, maintenance or strengthening of the size of the first digit to generate the characteristic big toe, and loss of first digit lateral mobility in the lower limb as the hallux became permanently aligned with other toes to provide balance, weight-bearing, and propulsion during upright walking (Harcourt-Smith and Aiello, 2004; Lovejoy et al., 2009).
The regulatory change we have observed in the human GDF6 gene completely removes an interesting regional enhancer from a gene that is clearly required for normal skeletal development. The differences in anterior-posterior expression of the ancestral GDF6 enhancer would limit any limb effect of the regulatory deletion to hindlimbs but not forelimbs, and to particular digits and muscles within the limb. Previous studies have shown that the GDF6 gene plays essential roles in skull suture formation, eye development, and formation of joints (Settle et al., 2003; Tassabehji et al., 2008; Asai-Coakwell et al., 2009). Our studies show that Gdf6 is also required for normal digit growth, with loss of Gdf6 activity leading to shorter toes (Figure 6B). Deletion of a region-specific enhancer of the type identified here could limit the phenotypic consequences of a GDF6 regulatory mutation to posterior structures, avoiding negative pleiotropic consequences in the skull or forelimb, while modifying some of the same hindlimb bones and muscles that have undergone specific morphological changes during the evolution of bipedalism in the human lineage (Figure 7A).
Regulatory Control of Morphological Evolution
Powerful developmental control genes surrounded by large sets of modular regulatory regions have now been found underlying multiple examples of morphological evolution in natural populations. For example, evolution of specific pigmentation patterns in the wings of Drosophila and the coats of mice have been traced to regulatory changes surrounding the yellow and agouti genes (Gompel et al., 2005; Linnen et al., 2013). Similarly, the detailed patterns of trichomes in Drosophila are influenced by genetic changes in multiple enhancers of the ovo-shaven baby gene, each controlling trichome development in specific anatomical regions (McGregor et al., 2007). Mimicry patterns in butterflies have long been known to map to a few chromosome regions with large effects. Recent studies suggest that different evolved patterns of wing shape and color are due to multiple regulatory changes surrounding key developmental control genes in the mimicry regions (Reed et al., 2011; Martin et al., 2012).
Our results in both sticklebacks and mammals illustrate how detailed sizes and shapes of different bones in the vertebrate skeleton can also be controlled by specific regulatory changes in a BMP gene. Fine mapping of armor plate sizes reveals two distinct but closely linked genomic regions controlling different dimensions of plate size. These two regions map on either side of the GDF6 gene, and may correspond to distinct regulatory control regions affecting gene expression in different skeletal domains. The mammalian GDF6 gene is also surrounded by a large array of distinct regulatory sequences, many of which show striking specificity for particular tissues or regions of the body (Figure 7B). The regulatory region missing in humans would only alter GDF6 function in specific domains of the body, providing a simple genomic mechanism that would limit any effects on digit size for example, to posterior toes of the hindlimb.
Interestingly, both increases and decreases of GDF expression can lead to reductions in bone size in particular regions. Previous studies show that GDF6 can promote chondrogenesis (Erlacher et al., 1998; Gooch et al., 2002; Nochi et al., 2004), but can also act as inhibitor of osteogenesis (Shen et al., 2009; Clendenning et al., 2012). Digit bones form from cartilaginous precursors, and smaller toes in Gdf6 mutants likely reflects reduced chondrogenic activity in developing footplates, similar to the shortening of endochondral bones seen with mutations in other closely related GDF signaling molecules (Storm et al., 1994). In contrast, armor plates in sticklebacks, and skull bones in mammals, form directly as intramembranous bone without a cartilage template. Increased GDF6 expression in freshwater or transgenic fish may reduce the size of armor through its inhibitory effects on osteogenesis (Shen et al., 2009) rather than its stimulatory effects on chondrogenesis. Consistent with this model, loss of GDF6 activity in mutant mice actually expands ossification domains in the developing skull, leading to a failure to maintain undifferentiated suture mesenchyme between adjacent bones, as expected for decreased expression of an osteogenesis inhibitor (Clendenning et al., 2012). Expanded osteogensis then leads to early fusion of normally adjacent skull bones, with reduced postnatal skull expansion likely arising as a secondary consequence of premature bone fusion.
Over twenty years ago, the cloning and genetic studies of BMPs led to suggestions that gain and loss of specific regulatory elements within these genes may underlie the evolution of interesting skeletal features seen in different vertebrate species (Kingsley, 1994). While long distance regulatory elements were for many years difficult to identify in vertebrate genomes, subsequent studies have confirmed that many BMP genes, including GDF6, are surrounded by huge regulatory regions containing large numbers of modular enhancers (Mortlock et al., 2003; Pregizer and Mortlock, 2009). Some of these enhancers appear to function as “anatomy elements” that drive expression not in all bones or all cartilage structures, but rather in particular regions or the body, particular subsets of bones, or particular growth domains surrounding individual bones (Guenther et al., 2008).
Recent studies have also identified changes in BMP expression that are directly or indirectly linked to classic examples of skeletal evolution in vertebrates, including changes in the size and depth of beaks in Darwin's finches (Abzhanov et al., 2004), and altered mandibles and tooth patterns in cichlid and stickleback fish (Albertson et al., 2005; Cleves et al., 2014). However the actual molecular changes that lead to these changes in expression have remained unknown. The current studies identify particular enhancer sequences in BMP genes whose changes can be linked to corresponding alterations in the skeletal structures of natural species. Additional studies are still required to identify the complete set of base pair alterations within the fish enhancer that are responsible for the GDF6 expression changes and armor plate size alterations seen in sticklebacks. Future recreation of the human-specific deletion in mice will also make it possible to study the detailed phenotypes associated with complete loss of this interesting posterior-specific enhancer of the mammalian GDF6 gene. The combined examples from fish and humans suggest that both gain and loss of regional GDF6 expression can play an important role in modifying species-specific skeletal structures during vertebrate evolution.
EXPERIMENTAL PROCEDURES
QTL Mapping
Armor plate height and width were measured from Alizarin red stained F2 progeny from a marine by Paxton benthic cross (Colosimo et al., 2004). Residuals were calculated following correction for fish standard depth and length, respectively, and for sex using Minitab Release 13.31. Thirty new microsatellites (Table S1) were added to the LG20 meiotic map (Colosimo et al., 2004) using JoinMap 3.0. QTL mapping was performed using the Interval Mapping method in MapQTL 4.0. See Extended Experimental Procedures for further details.
Global Patterns in Sequence Divergence and Armor Plate Morphology
SNPs previously identified in 21 sequenced global stickleback populations (Table S2; (Jones et al., 2012)) were used to calculate the fraction of nucleotides that are divergent in pairs of freshwater and marine fish in a sliding window with size 2500 bp and step 500 bp. Armor plate height and width were measured for adult fish from the same 21 populations and were corrected for fish standard depth and length, respectively. See Extended Experimental Procedures for further details.
Gene Predictions and Coding Changes
The total number of genes in the plate size interval was predicted using ENSEMBL, GENSCAN, and blastx tools, and mapped ESTs. To identify coding changes in the plate size interval, BACs with genomic inserts from a marine fish population were completely sequenced and aligned to the coding regions of the reference freshwater stickleback genome. The observed SNPs were then intersected with the SNPs identified in the 21 stickleback genomes, and the effect of the non-synonymous sequence changes was evaluated using PolyPhen-2. See Extended Experimental Procedures for further details.
Allele-Specific Expression Studies
A Little Campbell River marine and a Matadero Creek freshwater fish were crossed using in vitro fertilization. Juvenile F1 hybrid fish were sacrificed, and developing armor plates and their surrounding skin tissues were collected. RNA was isolated using Tri reagent (Invitrogen) and was reverse transcribed using SuperScript III kit (Invitrogen) following manufacturer's recommendations. SNPs in the coding regions of GDF6 and BCKDHB were identified in the parental populations and were used for allele-specific assay design and pyrosequencing (EpigenDx). See Extended Experimental Procedures for further details.
Fish Transgenics
The region with highest level of sequence divergence in the plate size QTL (chrXX:3,610,110-3,614,309) was PCR amplified from a Little Campbell River marine and a Paxton Lake benthic fish. The alleles were then cloned into the pT2HE vector with minimal hsp70 promoter driving either EGFP alone or fused with GDF6 cDNA and flanked by Tol2 transposase recognition sites. The constructs were also modified to contain either the freshwater enhancer with the L2 LINE transposon removed, or the marine enhancer with the same transposon sequence inserted in the appropriate location. The constructs were then co-injected with mRNA encoding the Tol2 transposase into fertilized one-cell stage embryos. The embryos were allowed to develop and were monitored daily for GFP expression patterns. Adult transgenic fish injected with GDF6 cDNA were sacrificed, tested for the presence of the GDF6 transgene using PCR, and stained with Alizarin red to examine skeletal structures. Skeletal phenotypes were further evaluated by micro-computed tomography (μCT) using a Scanco μCT40 operated at 70kVp, 114 μA, at medium resolution and with 2x-averaging. See Extended Experimental Procedures for further details.
Enhancer Assays
The chimpanzee versions of sequences missing in the human GDF6 locus were PCR amplified and cloned into hsp-lacZ and hsp-CreER-T2 minimal promoter expression vectors and injected into the pronuclei of fertilized FVB embryos (Xenogen and Stanford Transgenic Facility). The hCONDEL.306-hsp-CreER-T2 transgenic mice were then bred to floxed-ROSA26 reporter mice, and injected with tamoxifen at different developmental time points. For both types of constructs, functional enhancer assays were then carried out by staining for lacZ expression activity at multiple developmental stages. For more detailed analysis, lacZ stained hindlimbs were embedded in gelatin, sectioned in transverse orientation using a Leica CM3050 S, and counterstained with Nuclear Fast Red (Vector Laboratories, CA). See Extended Experimental Procedures for further details.
Supplementary Material
ACKNOWLEDGEMENTS
We thank D. Schluter for the stickleback cross used in the Colosimo et al., 2004 and subsequent QTL mapping studies, G. Bejerano and C. McLean for useful discussions about the human deletions in the GDF6 locus, C. Miller for advice on QTL mapping, C. Lowe for advice on statistics, T. Howes for advice on stickleback transgenics, and M. Bell, A. Pollen, K. Xie and all members of the Kingsley lab for helpful comments and suggestions. This work was supported in part by US National Institutes of Health grant R01-AR42236 (D.M.K.) and a Center of Excellence in Genomic Science award 5P50HG2568 (D.M.K. and R.M.M.). D.M.K. is a Howard Hughes Medical Institute investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
V.B.I. and D.M.K. conceived and oversaw the project. G.A.K. analyzed L2 effects on the armor plate enhancer. F.C.J. identified repeated sequence divergence peaks in the armor QTL intervals. C.A.G. performed the μCT analysis of the GDF6 transgenic sticklebacks. J.G., J.S., and R.M.M. sequenced and assembled the marine BACs in the plate size interval. V.B.I. carried out high resolution QTL mapping and sequence and expression analysis of genes in the armor plate size interval, generated transgenic sticklebacks for both GFP and phenotypic rescue experiments, tested the functions of the two hCONDELs in the GDF6 locus using transient and staging series of stable mouse lines, and analyzed Gdf6 null mouse phenotypes. V.B.I. and D.M.K. wrote the manuscript with input from all authors.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, six Supplemental Figures and seven Supplemental Tables and can be found with this article online.
REFERENCES
- Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Bmp4 and morphological variation of beaks in Darwin's finches. Science. 2004;305:1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- Albertson RC, Streelman JT, Kocher TD, Yelick PC. Integration and evolution of the cichlid mandible: the molecular basis of alternate feeding strategies. Proc. Natl. Acad. Sci. USA. 2005;102:16287–16292. doi: 10.1073/pnas.0506649102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai-Coakwell M, French CR, Ye M, Garcha K, Bigot K, Perera AG, Staehling-Hampton K, Mema SC, Chanda B, Mushegian A, et al. Incomplete penetrance and phenotypic variability characterize Gdf6-attributable oculo-skeletal phenotypes. Hum. Mol. Genet. 2009;18:1110–1121. doi: 10.1093/hmg/ddp008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RD, Rogers SM, Schluter D. Natural selection on a major armor gene in threespine stickleback. Science. 2008;322:255–257. doi: 10.1126/science.1159978. [DOI] [PubMed] [Google Scholar]
- Bell MA, Foster SA. The evolutionary biology of the threespine stickleback. Oxford University Press; New York: 1994. [Google Scholar]
- Bergstrom CA. Fast-start swimming performance and reduction in lateral plate number in threespine stickleback. Can. J. Zool. 2002;80:207–213. [Google Scholar]
- Blount ZD, Barrick JE, Davidson CJ, Lenski RE. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature. 2012;489:513–518. doi: 10.1038/nature11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YF, Marks ME, Jones FC, Villarreal G, Jr., Shapiro MD, Brady SD, Southwick AM, Absher DM, Grimwood J, Schmutz J, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clendenning DE, Mortlock DP. The BMP ligand Gdf6 prevents differentiation of coronal suture mesenchyme in early cranial development. PLoS One. 2012;7:e36789. doi: 10.1371/journal.pone.0036789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleves PA, Ellis NA, Jimenez MT, Nunez SM, Schluter D, Kingsley DM, Miller CT. Evolved tooth gain in sticklebacks is associated with a cis- regulatory allele of Bmp6. Proc. Natl. Acad. Sci. USA. 2014;111:13912–13917. doi: 10.1073/pnas.1407567111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo PF, Hosemann KE, Balabhadra S, Villarreal G, Jr., Dickson M, Grimwood J, Schmutz J, Myers RM, Schluter D, Kingsley DM. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science. 2005;307:1928–1933. doi: 10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- Colosimo PF, Peichel CL, Nereng K, Blackman BK, Shapiro MD, Schluter D, Kingsley DM. The genetic architecture of parallel armor plate reduction in threespine sticklebacks. PLoS Biol. 2004;2:E109. doi: 10.1371/journal.pbio.0020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlacher L, McCartney J, Piek E, ten Dijke P, Yanagishita M, Oppermann H, Luyten FP. Cartilage-derived morphogenetic proteins and osteogenic protein-1 differentially regulate osteogenesis. J. Bone. Miner. Res. 1998;13:383–392. doi: 10.1359/jbmr.1998.13.3.383. [DOI] [PubMed] [Google Scholar]
- Flower WH. An introduction to the osteology of the mammalia. MacMillan and Co.; London: 1870. [Google Scholar]
- Frankel N, Erezyilmaz DF, McGregor AP, Wang S, Payre F, Stern DL. Morphological evolution caused by many subtle-effect substitutions in regulatory DNA. Nature. 2011;474:598–603. doi: 10.1038/nature10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Akey JM. Selection and adaptation in the human genome. Ann. Rev. Genom. Hum. Genet. 2013;14:467–489. doi: 10.1146/annurev-genom-091212-153509. [DOI] [PubMed] [Google Scholar]
- Giles N. The possible role of environmental calcium levels during the evolution of phenotypic diversity in Outer Hebridean populations of the Three-spined stickleback, Gasterosteus aculeatus. J. Zool. Lond. 1983;199:535–544. [Google Scholar]
- Gompel N, Prud'homme B, Wittkopp PJ, Kassner VA, Carroll SB. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433:481–487. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- Gooch KJ, Blunk T, Courter DL, Sieminski AL, Vunjak-Novakovic G, Freed LE. Bone morphogenetic proteins-2, -12, and -13 modulate in vitro development of engineered cartilage. Tissue Eng. 2002;8:591–601. doi: 10.1089/107632702760240517. [DOI] [PubMed] [Google Scholar]
- Greenwood AK, Wark AR, Yoshida K, Peichel CL. Genetic and neural modularity underlie the evolution of schooling behavior in threespine sticklebacks. Curr. Biol. 2013;23:1884–1888. doi: 10.1016/j.cub.2013.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther C, Pantalena-Filho L, Kingsley DM. Shaping skeletal growth by modular regulatory elements in the Bmp5 gene. PLoS Genet. 2008;4:e1000308. doi: 10.1371/journal.pgen.1000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther CA, Tasic B, Luo L, Bedell MA, Kingsley DM. A molecular basis for classic blond hair color in Europeans. Nat. Genet. 2014;46:748–752. doi: 10.1038/ng.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt-Smith WE, Aiello LC. Fossils, feet and the evolution of human bipedal locomotion. J. Anat. 2004;204:403–416. doi: 10.1111/j.0021-8782.2004.00296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, Swofford R, Pirun M, Zody MC, White S, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley DM. What do BMPs do in mammals? Clues from the mouse short-ear mutation. Trends Genet. 1994;10:16–21. doi: 10.1016/0168-9525(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Kingsley DM, Peichel CL. The molecular genetics of evolutionary change in sticklebacks. In Biology of the three-spined stickleback. In: Ostlund-Nilsson S, Mayer I, Huntingford FA, editors. Taylor & Francis Group; Boca Raton: 2007. pp. 41–81. [Google Scholar]
- Knecht AK, Hosemann KE, Kingsley DM. Constraints on utilization of the EDA signaling pathway in threespine stickleback evolution. Evolution and Devel. 2007;9:141–154. doi: 10.1111/j.1525-142X.2007.00145.x. [DOI] [PubMed] [Google Scholar]
- Linnen CR, Poh YP, Peterson BK, Barrett RD, Larson JG, Jensen JD, Hoekstra HE. Adaptive evolution of multiple traits through multiple mutations at a single gene. Science. 2013;339:1312–1316. doi: 10.1126/science.1233213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy CO, Latimer B, Suwa G, Asfaw B, White TD. Combining prehension and propulsion: the foot of Ardipithecus ramidus. Science. 2009;326:72e71–78. [PubMed] [Google Scholar]
- Martin A, Papa R, Nadeau NJ, Hill RI, Counterman BA, Halder G, Jiggins CD, Kronforst MR, Long AD, McMillan WO, et al. Diversification of complex butterfly wing patterns by repeated regulatory evolution of a Wnt ligand. Proc. Natl. Acad. Sci. USA. 2012;109:12632–12637. doi: 10.1073/pnas.1204800109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor AP, Orgogozo V, Delon I, Zanet J, Srinivasan DG, Payre F, Stern DL. Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature. 2007;448:587–590. doi: 10.1038/nature05988. [DOI] [PubMed] [Google Scholar]
- McLean CY, Reno PL, Pollen AA, Bassan AI, Capellini TD, Guenther C, Indjeian VB, Lim X, Menke DB, Schaar BT, et al. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature. 2011;471:216–219. doi: 10.1038/nature09774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CT, Beleza S, Pollen AA, Schluter D, Kittles RA, Shriver MD, Kingsley DM. cis-Regulatory changes in Kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell. 2007;131:1179–1189. doi: 10.1016/j.cell.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CT, Glazer AM, Summers BR, Blackman BK, Norman AR, Shapiro MD, Cole BL, Peichel CL, Schluter D, Kingsley DM. Modular skeletal evolution in sticklebacks is controlled by additive and clustered quantitative trait Loci. Genetics. 2014;197:405–420. doi: 10.1534/genetics.114.162420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortlock DP, Guenther C, Kingsley DM. A general approach for identifying distant regulatory elements applied to the Gdf6 gene. Genome Res. 2003;13:2069–2081. doi: 10.1101/gr.1306003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myhre F, Klepaker T. Body armour and lateral-plate reduction in freshwater three-spined stickleback Gasterosteus aculeatus: adaptations to a different buoyancy regime? J. Fish Biol. 2009;75:2062–2074. doi: 10.1111/j.1095-8649.2009.02404.x. [DOI] [PubMed] [Google Scholar]
- Nochi H, Sung JH, Lou J, Adkisson HD, Maloney WJ, Hruska KA. Adenovirus mediated BMP-13 gene transfer induces chondrogenic differentiation of murine mesenchymal progenitor cells. J. Bone. Miner. Res. 2004;19:111–122. doi: 10.1359/jbmr.2004.19.1.111. [DOI] [PubMed] [Google Scholar]
- O'Brown NM, Summers BR, Jones FC, Brady SD, Kingsley DM. A recurrent regulatory change underlying altered expression and Wnt response of the stickleback armor plates gene EDA. Elife. 2015;4:e05290. doi: 10.7554/eLife.05290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pregizer S, Mortlock DP. Control of BMP gene expression by long-range regulatory elements. Cytokine Growth Factor Rev. 2009;20:509–515. doi: 10.1016/j.cytogfr.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M, Pool JE, Kassner VA, Aquadro, C.F., Carroll SB. Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science. 2009;326:1663–1667. doi: 10.1126/science.1178357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed RD, Papa R, Martin A, Hines HM, Counterman BA, Pardo-Diaz C, Jiggins CD, Chamberlain NL, Kronforst MR, Chen R, et al. optix drives the repeated convergent evolution of butterfly wing pattern mimicry. Science. 2011;333:1137–1141. doi: 10.1126/science.1208227. [DOI] [PubMed] [Google Scholar]
- Reimchen TE. Spine deficiency and polymorphism in a population of Gasterosteus aculeatus: an adaptation to predators? Can. J. Zool. 1980;58:1232–1244. [Google Scholar]
- Settle SH, Jr., Rountree RB, Sinha A, Thacker A, Higgins K, Kingsley DM. Multiple joint and skeletal patterning defects caused by single and double mutations in the mouse Gdf6 and Gdf5 genes. Dev. Biol. 2003;254:116–130. doi: 10.1016/s0012-1606(02)00022-2. [DOI] [PubMed] [Google Scholar]
- Shapiro MD, Marks ME, Peichel CL, Blackman BK, Nereng KS, Jonsson B, Schluter D, Kingsley DM. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature. 2004;428:717–723. doi: 10.1038/nature02415. [DOI] [PubMed] [Google Scholar]
- Shapiro MD, Bell MA, Kingsley DM. Parallel genetic origins of pelvic reduction in vertebrates. Proc. Natl. Acad. Sci. USA. 2006;103:13753–13758. doi: 10.1073/pnas.0604706103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Bhargav D, Wei A, Williams LA, Tao H, et al. BMP-13 emerges as a potential inhibitor of bone formation. Int. J. Biol. Sci. 2009;5:192–200. doi: 10.7150/ijbs.5.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V. Is genetic evolution predictable? Science. 2009;323:746–751. doi: 10.1126/science.1158997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm EE, Huynh TV, Copeland NG, Jenkins NA, Kingsley DM, Lee S-L. “Limb alterations in brachypodism mice due to mutations in a new member of the TGF-beta superfamily.”. Nature. 1994;368:639–643. doi: 10.1038/368639a0. [DOI] [PubMed] [Google Scholar]
- Swindler DR, Wood CD. An atlas of primate gross anatomy: Baboon, Chimpanzee, and Man. University of Washington Press; Seattle: 1973. [Google Scholar]
- Tassabehji M, Fang ZM, Hilton EN, McGaughran J, Zhao Z, de Bock CE, Howard E, Malass M, Donnai D, Diwan A, et al. Mutations in GDF6 are associated with vertebral segmentation defects in Klippel-Feil syndrome. Hum. Mutat. 2008;29:1017–1027. doi: 10.1002/humu.20741. [DOI] [PubMed] [Google Scholar]
- Wark AR, Mills MG, Dang LH, Chan YF, Jones FC, Brady SD, Absher DM, Grimwood J, Schmutz J, Myers RM, et al. Genetic architecture of variation in the lateral line sensory system of threespine sticklebacks. G3. 2012;2:1047–1056. doi: 10.1534/g3.112.003079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.