Abstract
Esophageal squamous cell carcinoma (ESCC) is the predominant pathological type of esophageal carcinoma in Asia. MicroRNAs (miRNAs) are a class of 19-22-nucleotide non-coding RNAs acting on target mRNAs that function as either oncogenes or anti-oncogenes. It has been confirmed that miR-373 expression varies among different tumor types. However, its mechanism is still unclear in ESCC. In our current study, we found that miR-373 expression was upregulated in ESCC tissues compared with matched adjacent normal tissues, as well as in the plasma of ESCC patients compared with that of healthy volunteers. Overexpression of miR-373 in ECA109 cells enhanced proliferation, G1-phase cell proportion, migration, and invasion. On the other hand, suppression of miR-373 in KYSE410 cells decreased proliferation, G1-phase cell proportion, migration, and invasion and also improved cell apoptosis. Moreover, we found that TIMP3, which was reported to suppress invasion and metastasis of ESCC, was a direct target of miR-373. Overexpression of miR-373 in ECA109 caused a reduction of TIMP3 mRNA and protein, whereas suppression of miR-373 in KYSE410 led to an increase of TIMP3 mRNA and protein. Introducing TIMP3 in miR-373 over-expressed cells or knocking down TIMP3 in miR-373 suppressed cells could partially abrogate the effect of miR-373 on migration and invasion. Therefore, these results prove that as an oncogene, miRNA-373 would be an important and reliable biomarker for ESCC diagnosis and treatment by targeting TIMP3.
Keywords: Esophageal cancer, microRNA-373, TIMP3, oncogene
Introduction
Esophageal carcinoma (EC) is the eighth leading cancer worldwide and is characterized by its high mortality rate, with a less than 15% 5-year survival rate [1]. Esophageal squamous cell carcinoma (ESCC) is the predominant pathological type of esophageal cancer in East Asia [2], which has a high tendency for recurrence and metastasis. The patients of intermediate and advanced stage have no other choice than to receive radiotherapy and chemotherapy, which themselves are not very effective. However, due to a lack of apparent symptoms and early detecting methods, the patients are often diagnosed at a late stage. Therefore, it is of great importance to find an effective molecular diagnostic method to detect subclinical esophageal carcinoma.
MicroRNAs (miRNAs) affect multiple pathways by binding to specific sites at 3’-UTR (3’-untranslated region) of target mRNAs [3]. miRNAs play an important role in regulating cellular differentiation [4], proliferation [5], aggressiveness [6], angiogenesis [7], and apoptosis [8]. Abnormity of miRNAs are closely related to the occurrence of a variety of tumors, such as bladder cancer [9], pancreatic cancer [5] and lung cancer [10]. miRNAs function as either oncogenes or anti-oncogenes and show variable expression in different types of human cancer. It was found that miR-373 was upregulated (tumor ≥ 1.5-fold normal) in ESCC tumors by microarray analysis [11]. As an oncogene, miR-373 can regulate cell cycle by targeting PPP6C and promotes the growth activity of HCC cells in vitro [12]. miR-373 suppresses invasion and metastasis by targeting Rab22a gene in human ovarian cancer, which serves as a new potential therapeutic target in epithelial ovarian cancer [13]. However, the mechanism of miR-373 in ESCC is still unclear.
Recent studies have shown that there is a close relationship between tissue inhibitor of metalloproteinases-3 (TIMP3) and malignant tumors. As a member of the TIMP family, TIMP3 is a tumor suppressor that inhibits matrix metalloproteinases (MMP). Abnormal expression level of TIMP3 can disrupt the balance between MMP and TIMP. Low expression of TIMP3 can facilitate tumor growth [14], angiogenesis [15], invasion [16], and metastasis [17] and suppress apoptosis [18]. TIMP3 could be a promising biomarker of independent prognostic factor in bladder cancer [19] and esophageal squamous cell carcinoma [20] as well as early detection of head and neck cancer recurrence [21]. It plays important roles on different cancers, such as breast cancer [22], colorectal cancer [23], hepatocellular carcinoma [24], and glioblastoma [25].
In this study, we showed that miR-373 was upregulated in ESCC tissues and ESCC patients’ plasma. There was a significant inverse correlation between miR-373 and TIMP3 mRNA. We identified TIMP3 as a direct target of miR-373 and showed that miR-373 promotes migration and invasion in human esophageal squamous cell carcinoma by inhibiting TIMP3 expression.
Materials and methods
Patients, samples, cell lines, and cultures
This study was approved by the Institutional Ethics Committee of Taian Central Hospital. Written informed consent was obtained from each participant. The study included 63 patients who were first diagnosed with ESCC in Taian Central Hospital (Taian, Shandong, China) from October 2012 to September 2014. Peripheral blood (10 ml) was collected from each patient before surgery. ESCC tissues and adjacent normal tissues (5 cm away from tumor) were collected from patients after resection in the operating room. All the patients had no chemotherapy or radiotherapy before surgery. Plasma samples were also collected as a control from 39 healthy volunteers. The samples were snap-frozen in liquid nitrogen after resection and stored at -80°C before processing.
Four human ESCC cell lines (EC9706, KYSE410, ECA109, TE-1) were cultured in RPMI-1640 medium (Hyclone, Logan, Utah) supplemented with 10% fetal bovine serum (FBS, Hyclone) and 0.2% penicillin-streptomycin (Invitrogen, Carlsbad, CA). All the cells were maintained in a 37°C, 5% CO2 incubator. ECA109 and TE-1 were maintained in our laboratory (Shandong Cancer Hospital and Institute, Shandong, China). KYSE410 and EC9706 were purchased from Shanghai Cell Bank of Chinese Academy of Sciences (Shanghai, China).
Cell transfection
Approximately 5-6×105 cells (0.2 ml) in the logarithmic phase of growth per well were seeded and cultured in 6-well plates with 2.2 ml appropriate culture medium containing serum and antibiotics shortly before transfection. For each well, 0.6-1.8 μl mimics (20 μM) (GenePharma, Shanghai, China) which would enhance the level of miR-373 was diluted in culture medium without serum to achieve a final concentration of 5-15 nM. And inhibitor (20 μM) which would lessen the level of miR-373 was diluted to reach a final concentration of 50-150 nM. To the mixture, 12 μl HiPerFect Transfection Reagent (Qiagen, Hilden, Germany) was added. Finally, the complexes were added to the cells 5-10 min after mixing. Transfection efficiency was measured by flow cytometry 48 h later. The optimal transfection efficiency was about 94% when the final concentration of mimics was 15 nM and the inhibitor’s final concentration was 150 nM. The cells were harvested for the next analysis after transfection for 48 h. All experiments were performed in triplicate.
RNA extraction
All the experimental apparatus were RNase-free. Total RNA was isolated from ESCC tissues, adjacent normal tissues, and processed cells using miRNeasy Mini Kit (Qiagen). Total plasma RNA was isolated before surgery using miRNeasy Serum/Plasma Kit (Qiagen) following the manufacturer’s protocols. The purity of total RNA was assessed by measuring A260/280 ratio. RNA integrity was checked by denaturing formaldehyde agarose gel electrophoresis.
Reverse transcriptase reactions and quantitative real time-PCR
The reverse transcription was carried out with miScript II RT Kit (Qiagen) according to the manufacturer’s protocol. The reaction was incubated for 60 min at 37°C, 5 min at 95°C to inactivate miScript Reverse Transcriptase Mix, and placed on ice. Then the cDNA was synthesized.
The miScript SYBR Green PCR Kit (Qiagen) was used for quantitative real time-PCR (qRT-PCR) with miScript miRNA PCR Array. 2×QuantiTect SYBR Green PCR Master Mix 10 μl, 10×miScript Precursor Assay 2 μl, cDNA template 2 μl, and RNase-free water variable were used to a final volume of 20 μl. Real-time PCR was run on LightCycler480 Real-time PCR system (Roche Molecular Systems, Indianapolis, USA) with miR-373-specific primers miR-373-3p (5’GAAGUGCUUCGAUUUUGGGGUGU). The reaction mixtures were incubated at 95°C for 15 min, followed by 40 cycles of 94°C for 15 s, 55°C for 30 s, and 70°C for 30 s. U6 (RUN6B) was used as an endogenous control in cell and tissue samples. U6 and miR-39 (5’UCACCGGGUGUAAAUCAGCUUG) were used as endogenous and external controls in plasma samples.
For analysis of the TIMP3 mRNA expression, quantitative real time-PCR was performed using 2×SYBR Green PCR Mix (Aidlab, Beijing, China) according to the manufacturer’s instructions. The primer sequence synthesized by Sangon (Shanghai, China) was as follows: TIMP3 5’TGCTCTCTGTCTCTTTTTTCAGCTT (F), 5’CTACAGTGTGTTGTCTGCTGCTTTT (R). GAPDH was assessed to standardize the expression level of TIMP3 mRNA. All the real time-PCRs were performed in triplicate. Data were presented based on calculating 2-ΔΔCt.
Cell proliferation, cell cycle, and cell apoptosis assay
In 96-well plates, 3-4×103 cells per well were seeded. After 24, 48, 72 h of transfection, 10 μl cell counting kit-8 (CCK-8, beyotime, China) was added to each well. The cells were kept at 37°C, 5% CO2 incubator for an additional 2 h by measuring absorbance at 450 nm using a microplate reader.
Cell cycle assay was analyzed by flow cytometry at 48 h after transfection. Cells were washed twice with PBS, and 70% ethanol was added after 48 h at 4°C. The cells were then stained with propidium iodide (PI; BD, Franklin, NJ) for more than 30 min. The results were obtained using a flow cytometer.
Cell apoptosis was assessed using FITC Annexin V Apoptosis Detection Kit I (BD, Franklin, NJ) at 48 h after transfection by flow cytometry. The cells seeded in 6-well plates were washed twice with cold PBS and resuspended in 1× Binding Buffer sufficiently, stained with 5 μl FITC Annexin V, and 5 μl PI incubating for 15 minutes in the dark, and the data were analyzed within 1 h using a flow cytometer.
Cell migration and invasion assay
Cell migration assay was performed using transwell chambers using 24-well plates with 8 µm pores (Corning, Corning, NY). The cells were starved in the serum-free 1640 medium for 12 h after being transfected for 24 h. 1×105 cells were plated to the upper chamber after being resuspended in the serum-free medium. The lower chamber was filled with 1640 medium plus 10% FBS. The chambers were stored in an incubator for 10 h. Then cells on the lower surface were fixed with glutaraldehyde solution and stained with Crystal violet. Ten random fields for each membrane were counted. A similar method was used in cell invasion assay and membranes were coated with Matrigel (BD, Franklin, NJ).
Western blotting
The cells were managed 72 h after transfection. Protein extracts were separated by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked with 5% fat-free milk for 1 h and incubated with anti-TIMP3 antibody (1:1000, Abcam Cambridge, England) at 4°C overnight. Anti-β-actin antibody was used as the internal reference. After washing thoroughly, the membranes were incubated with secondary antibodies for 30 min at room temperature. The results were detected by enhanced chemiluminescence technique (Amersham, Piscataway, NJ). The expression levels of these proteins were quantified using Image J software (National Institutes of Health, Bethesda, USA).
Luciferase reporter assay
Human HEK293T cells (1-2×104) were plated in a 96-well plate. The dual-luciferase reporter plasmids named TIMP3-3’UTR-WT (wild type) and TIMP3-3’UTR-MUT (mutant type) (Ribobio, Guangzhou, China) were co-transfected with miRNA mimics (50 nM) or NC (50 nM) by Attractene Transfection Reagent (Qiagen). The cells were harvested 48 h after transfection and luciferase activity was detected by the dual luciferase reporter system (Promega, Madison, WI). All transfection experiments were performed in triplicate.
Statistical analysis
All the statistical analyses were performed using SPSS 17.0 statistical software package. Results were presented as mean ± SEM. Student’s t-test was used to determine statistical difference between groups (ESCC tissues and adjacent normal tissues, ESCC plasma and healthy volunteer plasma). ANOVA was used to evaluate the differences between miRNA-373 and clinicopathological characteristics. Spearman analyses were used to evaluate the correlation between miR-373 expressions and clinicopathological characteristics. Other results were explored by independent samples T-test. Statistical significance was considered if P < 0.05.
Results
miR-373 expressions was upregulated in ESCC tissues and ESCC patients’ plasma
Table 1 shows the clinicopathological characteristics. Quantitative real time-PCR showed that miR-373 expression level was considerably increased in 63 ESCC tissues compared with matched adjacent normal tissues (6.373±0.318 vs. 1.616±0.109; P < 0.01) (Figure 1A). It was found that miR-373 expression level in tumor tissues was closely correlated with differentiation, tumor status, and lymph node metastasis (P < 0.05), but not with gender, age, TNM stage, smoking, and alcohol drinking (P > 0.05) (Table 2). We also assessed miR-373 level in the plasma from 63 ESCC patients and 39 healthy volunteers, which was much higher in ESCC patients than in healthy volunteers (0.454±0.017 vs. 0.174±0.020; P < 0.01) (Figure 1B). There was also a good correlation between miR-373 level in plasma and tumor status, lymph node metastasis (P < 0.05). Meanwhile, no significant association was found between miR-373 expression level in plasma and gender, age, differentiation, TNM stage, smoking, alcohol drinking (P > 0.05) (Table 2). In addition, there was a correlation between miR-373 level in ESCC tissues and that in the same individuals’ plasma using Spearman correlation test (r = 0.553; P < 0.01) (Figure 1C).
Table 1.
Clinicopathologic characteristics of ESCC patients
| Characteristics | Number of Case | |
|---|---|---|
| Gender | Male | 41 |
| Female | 22 | |
| Age | ≤ 60 | 38 |
| > 60 | 25 | |
| Differentiation | High | 17 |
| Moderate | 28 | |
| Low | 18 | |
| Tumor status | T1 | 15 |
| T2 | 22 | |
| T3 | 21 | |
| T4 | 5 | |
| Lymph node status | N0 | 29 |
| N1 | 34 | |
| TNM stage | I | 7 |
| II | 37 | |
| III | 17 | |
| IV | 2 | |
| Smoking | Yes | 35 |
| No | 28 | |
| Alcohol drinking | Yes | 38 |
| No | 25 |
Figure 1.
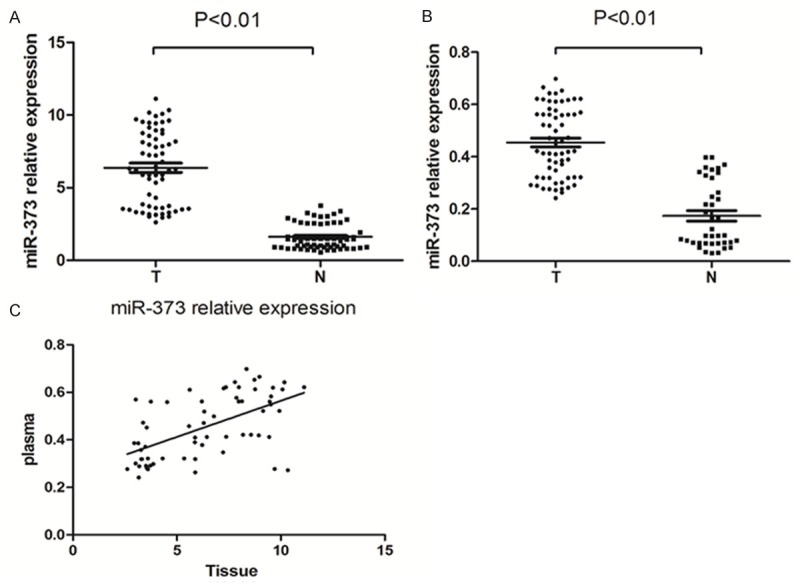
miR-373 expression in human ESCC tissue and plasma is analyzed. A. miR-373 levels were analyzed by quantitative real time-PCR. miR-373 was remarkably increased in human ESCC tissue. The levels of miR-373 in 63 ESCC tissues were compared with 63 matched adjacent normal tissues (*P < 0.01). (T = ESCC tissues, N = adjacent normal tissues). B. miR-373 was significantly increased in ESCC patients’ plasma. The levels of miR-373 in 63 ESCC patients’ plasma were compared with that in 39 healthy volunteers plasma (*P < 0.01). (T = ESCC plasma, N = healthy volunteers plasma). C. miR-373 levels in 63 ESCC tissues were consistent with that in the patients’ plasma.
Table 2.
The levels of miR-373 in tissue and plasma
| miR-373 in tissue | P value | miR-373 in plasma | P value | |
|---|---|---|---|---|
| Gender | ||||
| Male | 6.077±0.376 | 0.207 | 0.446±0.020 | 0.519 |
| Female | 6.924±0.578 | 0.469±0.029 | ||
| Age | ||||
| ≤ 60 | 6.394±0.403 | 0.937 | 0.435±0.020 | 0.151 |
| > 60 | 6.341±0.528 | 0.484±0.028 | ||
| Differentiation | ||||
| High | 5.234±0.611 | 0.023* | 0.405±0.029 | 0.200 |
| Moderate | 6.317±0.442 | 0.468±0.026 | ||
| Low | 7.537±0.571 | 0.479±0.300 | ||
| Tumor status | ||||
| T1 | 6.279±0.686 | 0.018* | 0.478±0.039 | 0.007** |
| T2 | 6.109±0.483 | 0.419±0.022 | ||
| T3 | 5.923±0.541 | 0.432±0.129 | ||
| T4 | 9.708±0.483 | 0.630±0.067 | ||
| Lymph node status | ||||
| N0 | 5.677±0.455 | 0.043* | 0.417±0.022 | 0.038* |
| N1 | 6.966±0.424 | 0.486±0.024 | ||
| TNM stage | ||||
| I | 5.264±1.052 | 0.460 | 0.403±0.057 | 0.304 |
| II | 6.304±0.382 | 0.451±0.020 | ||
| III | 6.793±0.712 | 0.465±0.037 | ||
| IV | 7.958±0.170 | 0.603±0.041 | ||
| Smoking | ||||
| Yes | 6.297±0.400 | 0.793 | 0.467±0.227 | 0.403 |
| No | 6.468±0.520 | 0.438±0.250 | ||
| Alcohol drinking | ||||
| Yes | 6.327±0.428 | 0.860 | 0.456±0.023 | 0.919 |
| No | 6.443±0.480 | 0.452±0.024 |
Indicates P < 0.05.
Indicates P < 0.01.
miR-373 promotes proliferation, G1 phase cell cycle arrest, and affects apoptosis
miR-373 expression levels in four different esophageal squamous carcinoma cell lines (KYSE410, EC9706, ECA109, TE-1) were evaluated by quantitative real time-PCR. As shown in Figure 2A, the highest expression level of miR-373 was observed in KYSE410 cells, and the lowest in ECA109 cells. As a result, KYSE410 and ECA109 were selected for further experiments. The transfection efficiency was shown in Figure 2B. We selected a final concentration of 15 nM in mimics and 150 nM in inhibitor for the further investigation. As shown in Figure 2C, 2D, the expression of miR-373 level was dramatically increased in the miR-373 mimics groups compared with the negative control groups (P < 0.01). However, miR-373 level was significantly decreased in miR-373 inhibitor groups (P < 0.01).
Figure 2.
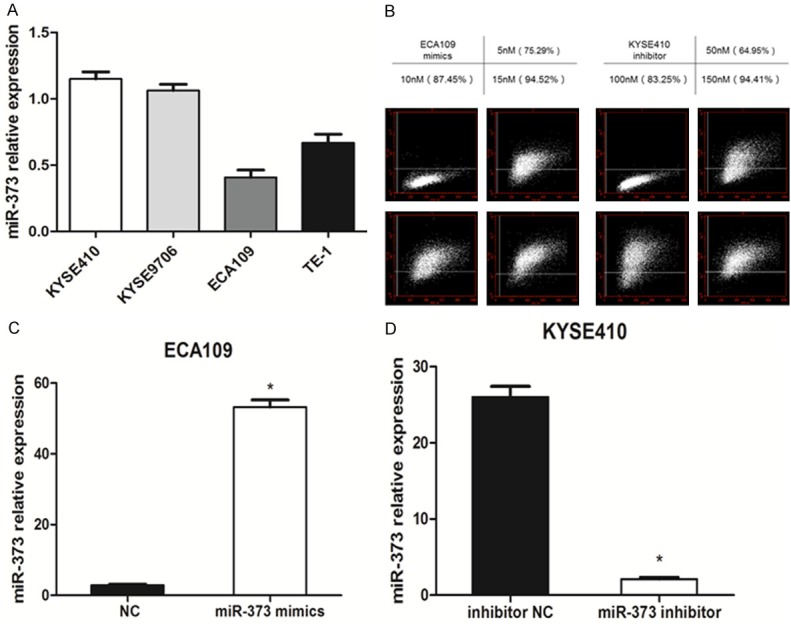
The levels of miR-373 in different groups were calculated. A. The levels of miR-373 expression in four ESCC cell lines were measured by quantitative real time-PCR. U6 was used as an internal control. B. The transfection efficiency was marked by FAM in different concentrations (50, 100, 150 nM) in ECA109 and KYSE410 by flow cytometer. C. miR-373 expression levels in miR-373 mimics ECA109 cells significantly increased compared with the negative control cells (*P < 0.01). D. miR-373 expression level in miR-373 inhibitor group significantly decreased compared with that in the inhibitor negative control group in KYSE410 cells (*P < 0.01).
CCK-8 assay was used to evaluate the effect of miR-373 on ESCC cells’ proliferation ability. We measured cell proliferation at 24, 48, and 72 h after transfection with miR-373 mimics or miR-373 inhibitor. As shown in Figure 3A, 3B, miR-373 mimics increased proliferation ability in ECA109 cells compared with the negative control groups at 72 h (P < 0.05). The KYSE410 cells treated with miR-373 inhibitor showed a significant decrease in proliferation at 72 h (P < 0.05). These results indicated that miR-373 enhances proliferation in ESCC cell lines.
Figure 3.
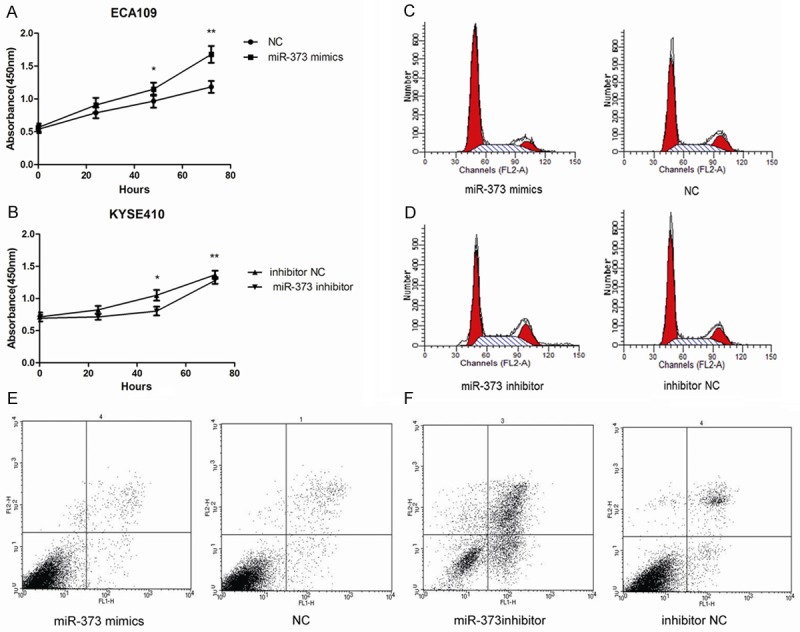
miR-373 promotes proliferation, G1 phase cell cycle arrest, and affects apoptosis. A, B. CCK-8 assay was used to evaluate the proliferation of ECA109 and KYSE410. miR-373 enhance proliferation ability in ESCC cell lines (*P < 0.05, **P < 0.01). C, D. Percentage of G1-phase cells in miR-373 mimics group was enhanced compared with negative control group in ECA109 (P < 0.01). The percentage of G1-phase in miR-373 inhibitor group was reduced compared with negative control group in KYSE410 (P < 0.01). E, F. Cell apoptosis was analyzed by Annexin V/PI assay. miR-373 can inhibit cell apoptosis (P < 0.01).
Flow cytometry was performed to measure the status of cell cycle. As shown in Figure 3C, 3D, G1-phase cell proportion in miR-373 mimics group (ECA109) was significantly increased compared with negative control groups (P < 0.01). Consistently, the proportion of G1-phase cells in miR-373 inhibitor group (KYSE410) decreased compared with the control group (P < 0.01). Therefore, miR-373 is able to induce G1-phase cell cycle arrest.
Next, cell apoptosis was measured after 48 h of transfection by flow cytometry. The percentage of apoptotic cells in miR-373 mimics treated ECA109 cells did not decrease significantly compared with the negative control groups (P > 0.05). However, the rate of apoptotic cells in miR-373 inhibitor treated KYSE410 cells was higher than that in the control cells (P < 0.01), which indicates suppression of miR-373 level may lead to apoptosis (Figure 3E, 3F).
miR-373 enhances migration and invasion in ESCC cell lines
To investigate whether miR-373 regulates migration and invasion of human ESCC cell lines, ECA109, which had the weakest migration and invasion ability, and KYSE410, which had the strongest, were selected for further research. After miR-373 transfection for 24 h, invasion and migration experiments were conducted using transwell chambers covered with or without Matrigel. As shown in Figure 4A, 4C, migration in ECA109 were enhanced significantly compared with the negative control group (128.400±4.675 vs. 44.200±3.484; P < 0.01). And the invasion ability was also dramatically increased (100.800±4.259 vs. 35.200±3.354; P < 0.01). As expected, migration and invasion in KYSE410 with miR-373 inhibitor declined significantly compared with the inhibitor negative control groups (Figure 4B, 4D). These observations demonstrate that miR-373 promotes migration and invasion in ESCC cell lines.
Figure 4.
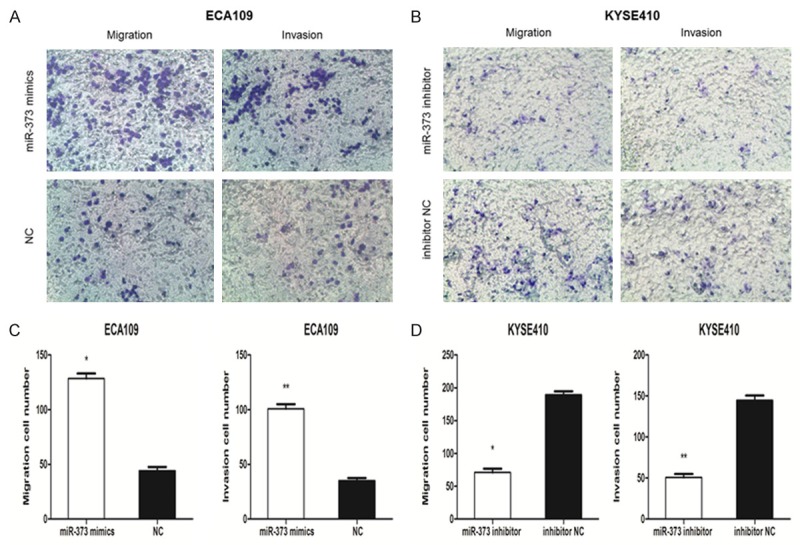
miR-373 promotes migration and invasion in ESCC cells. A, C. Cells that passed through the membrane were measured. The number of migrated cells and invasive cells was much higher in miR-373 mimics group than in negative control group (*P < 0.01, **P < 0.01). B, D. The number of migrated cells and invasive cells was lower in miR-373 inhibitor group than in inhibitor negative control group (*P < 0.01, **P < 0.01).
miR-373 directly regulates TIMP3 expression by targeting its 3’UTR
miRanda, miRDB, miRWalk and TargetScan were used to predict a potential target of miR-373. According to the database search results, TIMP3 stood out as a promising target (Figure 5A). ECA109, which had a low level of miR-373, expressed a high amount of TIMP3 mRNA (5.800±0.327) and protein. On the contrary, KYSE410 with high miR-373 level expressed a low level of TIMP3 mRNA (1.540±0.136) and protein (Figure 5B, 5C). Furthermore, as shown in Figure 6A-C, the mRNA (2.520±0.381 vs. 5.740±0.284; P < 0.01) and protein level of TIMP3 significantly decreased in ECA109 cells after miR-373 mimics transfection. And, miR-373 inhibitor increased TIMP3 expression in KYSE410 cells. For further verification, luciferase reporter assay was conducted by co-transfecting the reporter plasmids coalesced TIMP3-3’UTR-WT with miR-373 mimics in human HEK293T cells, which caused 37% decrease compared with the negative controls. However, there were no significant differences among TIMP3-WT/NC, TIMP3-Mut/NC, and TIMP3-Mut/miR-373. These results prove that TIMP3 is directly regulated by miR-373 (Figure 6D).
Figure 5.
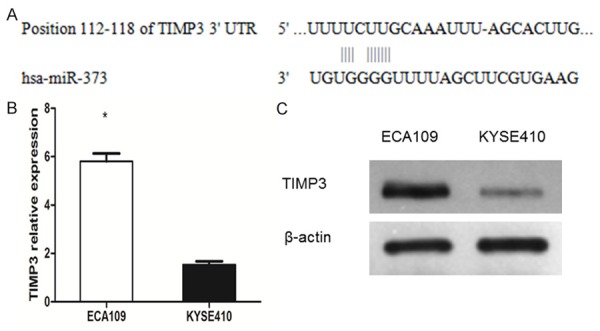
The relationship between miR-373 and TIMP3 were measured. A. Specific sequences and positions of the binding sites paired with miR-373 in 3’-UTR of TIMP3. B. TIMP3 mRNA in ECA109 was higher than that in KYSE410 (*P < 0.01). C. TIMP3 protein in ECA109 was higher than that in KYSE410 (*P < 0.01).
Figure 6.
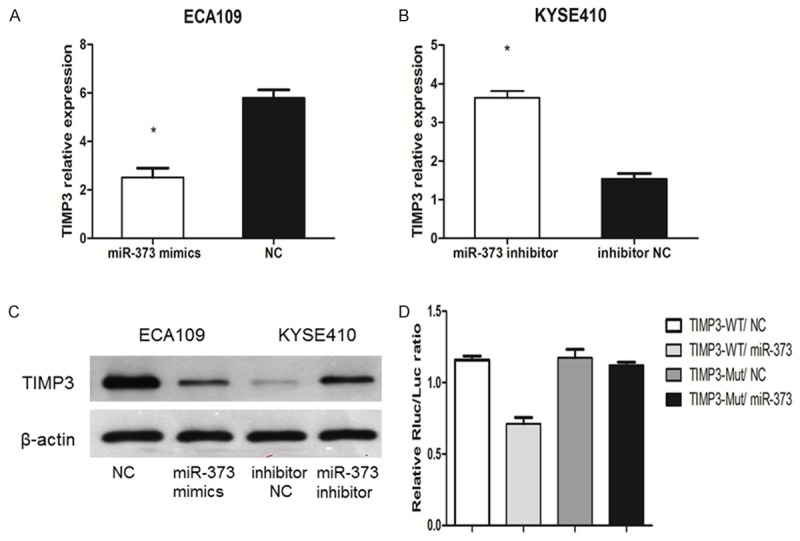
miR-373 inhibits TIMP3 expression by directly targeting its 3’-UTRs. A. TIMP3 mRNA decreased significantly in ECA109 cells transfected with miR-373 mimics (*P < 0.01). B. TIMP3 mRNA increased significantly in KYSE410 cells transfected with miR-373 inhibitor (*P < 0.01). C. TIMP3 protein was consistent with TIMP3 mRNA. D. TIMP3-3’UTR-WT co-transfected with miR-373 mimics caused 37% decrease compared with the negative control groups in relative luciferase activities.
TIMP3 expression level in 63 ESCC patients and 39 healthy volunteers were also assessed. TIMP3 level was considerably lower in ESCC tissues than in adjacent normal tissues (3.296±0.179 vs. 4.505±0.158; P < 0.05) (Figure 7A). Also, the TIMP3 was significantly less abundant in ESCC patients than in healthy volunteers (0.405±0.016 vs. 0.513±0.018; P < 0.01) (Figure 7B). According to Spearman correlation test (r = -0.832; P < 0.01) (Figure 7C), the level of TIMP3 was negatively correlated with that of miR-373 in ESCC tissues. A significant inverse correlation was observed between miR-373 and TIMP3 mRNA in the patients’ plasma (r = -0.479; P < 0.01) (Figure 7D).
Figure 7.
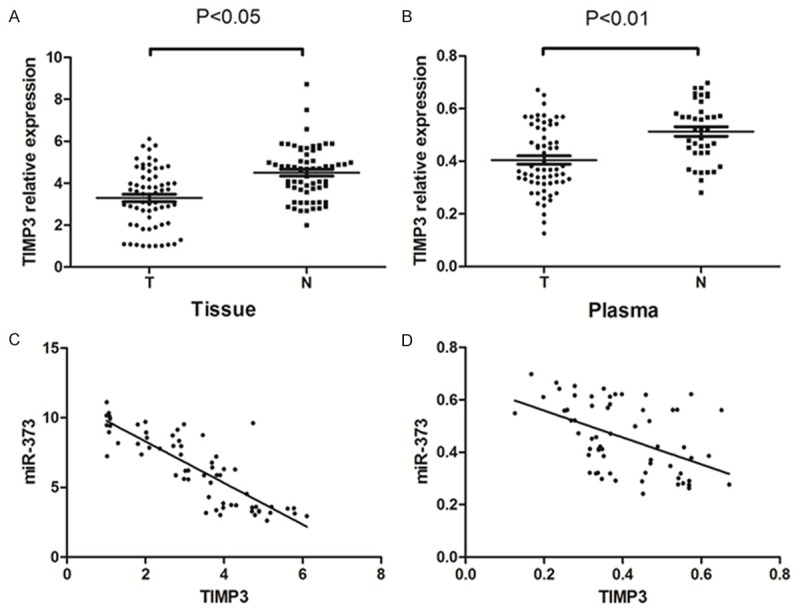
TIMP3 expression in human ESCC tissue and ESCC patients’ plasma was analyzed. A. TIMP3 significantly decreased in human ESCC tissue compared with matched adjacent normal tissues in 63 patients (P < 0.05). (T = ESCC tissues, N = adjacent normal tissues). B. TIMP3 significantly decreased in ESCC patients’ plasma compared with healthy volunteers’ plasma (P < 0.01). (T = ESCC plasma, N = healthy volunteers plasma). C. There was a negative correlation between miR-373 and TIMP3 in the same ESCC tissues. D. There was a negative correlation between miR-373 and TIMP3 in the same patient’s plasma.
We further explored the effect of TIMP3 on ESCC cells’ migration and invasion. It was found that TIMP3 overexpression could decrease cell migration and invasion in ECA109 (Figure 8A), whereas TIMP3 suppression could increase cell migration and invasion in KYSE410 (Figure 8B). To further investigate the functional causal relationship between miR-373 and TIMP3, we evaluated whether TIMP3 mediated the pro-migration and pro-invasion effect of miR-373 in ESCC. pcDNA3.1-TIMP3 without the 3’-UTR sequence and miR-373 mimics were co-transfected into ECA109 cells. It turned out that pcDNA3.1-TIMP3 could attenuate the migration and invasion ability induced by miR-373 mimics in ECA109 cells (Figure 8A, 8C). Then, shRNA-TIMP3 and miR-373 inhibitor were co-transfected into KYSE410 cells, which showed that shRNA-TIMP3 could partly reverted the migration and invasion ability inhibited by miR-373 inhibitor in KYSE410 (Figure 8B, 8D). These results demonstrate that, miR-373 promotes ESCC cell migration and invasion by targeting TIMP3.
Figure 8.
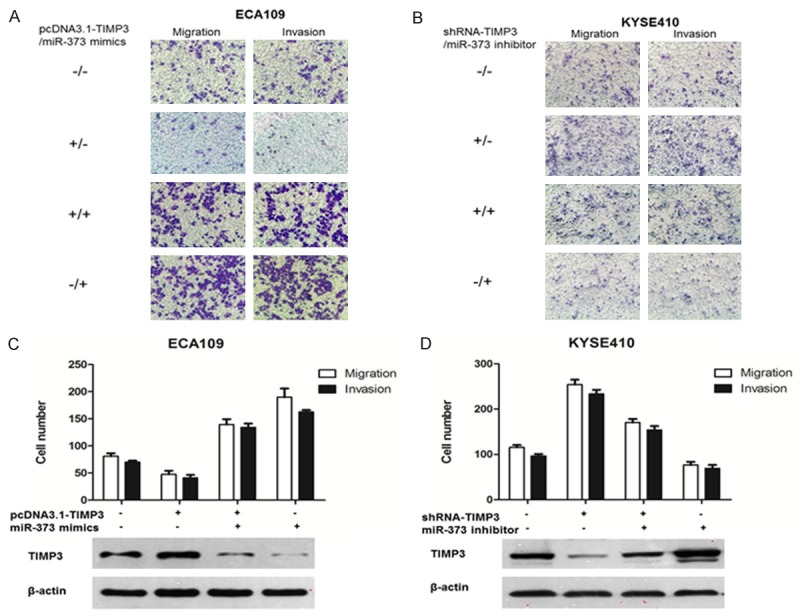
TIMP3 mediated the role of miR-373 in ESCC cell migration and invasion. A, C. pcDNA3.1-TIMP3 without the 3’-UTR sequence and miR-373 mimics were co-transfected into ECA109 cells. The average number of migration and invasion cells was higher evaluated by transwell assays. Western blot was used to evaluate TIMP3 protein levels. B, D. shRNA-TIMP3 and miR-373 inhibitor were co-transfected into KYSE410 cells. Transwell assays were used to assess the migration and invasion capacity of ESCC cells. TIMP3 protein level was estimated by Western blot.
Discussion
Recently, a large number of studies have shown that miRNAs can target a series of oncogenes or tumor suppressor genes. And there was a significant difference of miRNA level in tumor tissues and adjacent normal tissues [26], patients’ plasma and healthy volunteers’ plasma [27]. miRNA expression level was closely related to the biological and clinical characteristics of the tumor, such as pathological type, differentiation, tumor status, lymph node status, TNM stage, smoking and alcohol habits, prognosis, sensitivity of radiation, and chemotherapy [9,28,29]. It has been reported that a variety of miRNAs could participate in esophageal cancer occurrence and development process. Among them, miR-183 was upregulated in ESCC tissues compared with adjacent normal tissues, which promoted ESCC cell proliferation, migration, invasion and inhibited cell apoptosis by targeting the tumor suppressor gene programmed cell death 4 (PDCD4) [30]. High miR-142-3p expression was closely correlated to the poor-prognosis patients, including those patients with small tumor size, early stage, without lymph node metastasis, and early stage [31].
Studies have shown that the same miRNA has totally different expression level in different tumors. miR-373 was first discovered by Voorhoeve [32], who identified that miR-373 is a potential novel oncogene participating in the development of human testicular germ cell tumor by blocking the p53 pathway. miR-373 has been found to be upregulated in testicular germ cell tumors [33] and breast cancer [34], but down-regulated in colon cancer [35], non-small cell lung cancer [36]. Lee [11] found that miR-373 was an oncogene which was inversely correlated with LATS2 expression in ESCC tissues and esophageal cancer cell lines. There are few researches about miR-373 in esophageal cancer, which leaves its mechanism elusory. In this study, we investigated the biological function and identified the direct target of miR-373 in human ESCC.
In our current study, we investigated, for the first time, the high level of miR-373 in plasma and the relationship between miR-373 and biological and clinical characteristics of ESCC. miR-373 expression level in tissues was closely associated with differentiation, tumor status, and lymph node metastasis. There was also a good correlation between miR-373 level in plasma and tumor status, lymph node metastasis. Moreover, miR-373 expression in tissues was consistent with that in plasma. However, miR-373 is likely to exert diagonal effects on different tumors. For example, Chen [37] found that low expression of miR-373 in hilar cholangiocarcinom tissues was closely associated with poor cell differentiation and advanced clinical stages (stage III, IV vs I, II). While, Wang [38] investigated that miR-373 increased proliferation by directly targeting YOD1, a new potential therapeutic target in cervical cancer. Here, we show that miR-373, as an oncogene, might be a useful molecular marker in ESCC.
In addition, we further investigated the functional characteristics of two cell lines, The data suggested that the strong ability of proliferation, migration, and invasion in KYSE410 cell may attribute to high miR-373 level, on the contrary, ECA109 cells with a low miR-373 level, showed weak proliferation, migration, and invasion ability. It was found that miR-373 promoted proliferation, migration and invasion in human ESCC cells, which was consistent with that in breast tumor cells [39]. We also found that overexpression of miR-373 could induce G1-phase cell cycle arrest, but could not inhibit cell apoptosis. However, due to unknown reason, suppression of miR-373 could decrease G1-phase cell proportion and induce cell apoptosis.
According to Bioinformatics analysis, TIMP3 is a potential target of miR-373. Interestingly, we have found that TIMP3 is correlated with migration and invasion ability in ESCC [40]. It was confirmed that TIMP3 was indeed a bona fide target of miR-373 in our following study. TIMP3 was a potent tumor suppressor and an inhibitor of angiogenesis [41]. We showed that TIMP3 was negatively correlated to the level of miR-373 in plasma and tissues. The level of TIMP3 declined when we transfected miR-373 mimics into ECA109 and increased when we transfected miR-373 inhibitor in KYSE410. TIMP3-3’UTR-WT co-transfected with miR-373 mimics caused 37% decrease compared with the negative controls in the luciferase reporter assay. Ad-TIMP3 suppressed cells growth, migration and invasion in colorectal cancer cells. TIMP3 provided an alternative for comprehensive treatment strategy by suppressing cancer development [23]. It was also found that the breast cancer cells transfected with TIMP3 obviously inhibited the cells growth and proliferation [17]. These observations demonstrate that TIMP3 can inhibit migration and invasion, which counteract the role of miR-373. To further explore whether the pro-migration and pro-invasion effect of miR-373 is mediated by TIMP3, we performed remedial experiments by introducing TIMP3 or suppressing TIMP3 in miR-373 overexpressed or knocked down cell lines. It was found that manipulation of TIMP3 could abrogate the effect of miR-373 on esophageal cancer cell migration and invasion.
In summary, it has been shown that miR-373 is frequently up-regulated in ESCC tissues and ESCC patients’ plasma, which is likely to be an oncogene in ESCC. Proliferation, migration, and invasion abilities of ESCC cells were positively correlated to miR-373 expression level. We further confirmed that TIMP3 is a target of miR-373 and miR-373 promotes esophageal cancer migration and invasion by targeting TIMP3, which might be a promising target for esophageal cancer diagnosis and treatment.
Acknowledgements
This work was supported by grants from Natural Science Foundation of China (No. 81272501). This work was supported in part by the National Natural Science Foundation of China under Grants 81272501 and Taishan Scholars Program of Shandong Province, China (Grant No. ts20120505).
Disclosure of conflict of interest
None.
References
- 1.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 2.Matsushima K, Isomoto H, Kohno S, Nakao K. MicroRNAs and esophageal squamous cell carcinoma. Digestion. 2010;82:138–144. doi: 10.1159/000310918. [DOI] [PubMed] [Google Scholar]
- 3.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38(Suppl):S8–13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 4.Kumar P, Luo Y, Tudela C, Alexander JM, Mendelson CR. The c-Myc-regulated microRNA-17~92 (miR-17~92) and miR-106a~363 clusters target hCYP19A1 and hGCM1 to inhibit human trophoblast differentiation. Mol Cell Biol. 2013;33:1782–1796. doi: 10.1128/MCB.01228-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sarkar S, Dubaybo H, Ali S, Goncalves P, Kollepara SL, Sethi S, Philip PA, Li Y. Downregulation of miR-221 inhibits proliferation of pancreatic cancer cells through up-regulation of PTEN, p27(kip1), p57(kip2), and PUMA. Am J Cancer Res. 2013;3:465–477. [PMC free article] [PubMed] [Google Scholar]
- 6.Ben Gacem R, Ben Abdelkrim O, Ziadi S, Ben Dhiab M, Trimeche M. Methylation of miR-124a-1, miR-124a-2, and miR-124a-3 genes correlates with aggressive and advanced breast cancer disease. Tumour Biol. 2014;35:4047–4056. doi: 10.1007/s13277-013-1530-4. [DOI] [PubMed] [Google Scholar]
- 7.He J, Jing Y, Li W, Qian X, Xu Q, Li FS, Liu LZ, Jiang BH, Jiang Y. Roles and mechanism of miR-199a and miR-125b in tumor angiogenesis. PLoS One. 2013;8:e56647. doi: 10.1371/journal.pone.0056647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He Y, Wang J, Wang J, Yung VY, Hsu E, Li A, Kang Q, Ma J, Han Q, Jin P, Xing R, Lu Y, Sheng J. MicroRNA-135b regulates apoptosis and chemoresistance in colorectal cancer by targeting large tumor suppressor kinase 2. Am J Cancer Res. 2015;5:1382–1395. [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Yu G, Shi R, Lang B, Chen X, Xia D, Xiao H, Guo X, Guan W, Ye Z, Xiao W, Xu H. Cisplatin-induced epigenetic activation of miR-34a sensitizes bladder cancer cells to chemotherapy. Mol Cancer. 2014;13:8. doi: 10.1186/1476-4598-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du L, Zhao Z, Ma X, Hsiao TH, Chen Y, Young E, Suraokar M, Wistuba I, Minna JD, Pertsemlidis A. miR-93-directed downregulation of DAB2 defines a novel oncogenic pathway in lung cancer. Oncogene. 2014;33:4307–4315. doi: 10.1038/onc.2013.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee KH, Goan YG, Hsiao M, Lee CH, Jian SH, Lin JT, Chen YL, Lu PJ. MicroRNA-373 (miR-373) post-transcriptionally regulates large tumor suppressor, homolog 2 (LATS2) and stimulates proliferation in human esophageal cancer. Exp Cell Res. 2009;315:2529–2538. doi: 10.1016/j.yexcr.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 12.Wu N, Liu X, Xu X, Fan X, Liu M, Li X, Zhong Q, Tang H. MicroRNA-373, a new regulator of protein phosphatase 6, functions as an oncogene in hepatocellular carcinoma. FEBS J. 2011;278:2044–2054. doi: 10.1111/j.1742-4658.2011.08120.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Zhao FJ, Chen LL, Wang LQ, Nephew KP, Wu YL, Zhang S. MiR-373 targeting of the Rab22a oncogene suppresses tumor invasion and metastasis in ovarian cancer. Oncotarget. 2014;5:12291–12303. doi: 10.18632/oncotarget.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen YY, Brown NJ, Jones R, Lewis CE, Mujamammi AH, Muthana M, Seed MP, Barker MD. A peptide derived from TIMP-3 inhibits multiple angiogenic growth factor receptors and tumour growth and inflammatory arthritis in mice. Angiogenesis. 2014;17:207–219. doi: 10.1007/s10456-013-9389-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das AM, Seynhaeve AL, Rens JA, Vermeulen CE, Koning GA, Eggermont AM, Ten Hagen TL. Differential TIMP3 expression affects tumor progression and angiogenesis in melanomas through regulation of directionally persistent endothelial cell migration. Angiogenesis. 2014;17:163–177. doi: 10.1007/s10456-013-9385-2. [DOI] [PubMed] [Google Scholar]
- 16.Wu DW, Tsai LH, Chen PM, Lee MC, Wang L, Chen CY, Cheng YW, Lee H. Loss of TIMP-3 promotes tumor invasion via elevated IL-6 production and predicts poor survival and relapse in HPV-infected non-small cell lung cancer. Am J Pathol. 2012;181:1796–1806. doi: 10.1016/j.ajpath.2012.07.032. [DOI] [PubMed] [Google Scholar]
- 17.Han X, Zhang H, Jia M, Han G, Jiang W. Expression of TIMP-3 gene by construction of a eukaryotic cell expression vector and its role in reduction of metastasis in a human breast cancer cell line. Cell Mol Immunol. 2004;1:308–310. [PubMed] [Google Scholar]
- 18.Kallio JP, Hopkins-Donaldson S, Baker AH, Kahari VM. TIMP-3 promotes apoptosis in nonadherent small cell lung carcinoma cells lacking functional death receptor pathway. Int J Cancer. 2011;128:991–996. doi: 10.1002/ijc.25404. [DOI] [PubMed] [Google Scholar]
- 19.Hoque MO, Begum S, Brait M, Jeronimo C, Zahurak M, Ostrow KL, Rosenbaum E, Trock B, Westra WH, Schoenberg M, Goodman SN, Sidransky D. Tissue inhibitor of metalloproteinases-3 promoter methylation is an independent prognostic factor for bladder cancer. J Urol. 2008;179:743–747. doi: 10.1016/j.juro.2007.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ninomiya I, Kawakami K, Fushida S, Fujimura T, Funaki H, Takamura H, Kitagawa H, Nakagawara H, Tajima H, Kayahara M, Ohta T. Quantitative detection of TIMP-3 promoter hypermethylation and its prognostic significance in esophageal squamous cell carcinoma. Oncol Rep. 2008;20:1489–1495. [PubMed] [Google Scholar]
- 21.Righini CA, de Fraipont F, Timsit JF, Faure C, Brambilla E, Reyt E, Favrot MC. Tumor-specific methylation in saliva: a promising biomarker for early detection of head and neck cancer recurrence. Clin Cancer Res. 2007;13:1179–1185. doi: 10.1158/1078-0432.CCR-06-2027. [DOI] [PubMed] [Google Scholar]
- 22.Lui EL, Loo WT, Zhu L, Cheung MN, Chow LW. DNA hypermethylation of TIMP3 gene in invasive breast ductal carcinoma. Biomed Pharmacother. 2005;59(Suppl 2):S363–365. doi: 10.1016/s0753-3322(05)80079-4. [DOI] [PubMed] [Google Scholar]
- 23.Lin H, Zhang Y, Wang H, Xu D, Meng X, Shao Y, Lin C, Ye Y, Qian H, Wang S. Tissue inhibitor of metalloproteinases-3 transfer suppresses malignant behaviors of colorectal cancer cells. Cancer Gene Ther. 2012;19:845–851. doi: 10.1038/cgt.2012.70. [DOI] [PubMed] [Google Scholar]
- 24.Defamie V, Sanchez O, Murthy A, Khokha R. TIMP3 controls cell fate to confer hepatocellular carcinoma resistance. Oncogene. 2015;34:4098–4108. doi: 10.1038/onc.2014.339. [DOI] [PubMed] [Google Scholar]
- 25.Saraiva-Esperon U, Ruibal A, Herranz M. The contrasting epigenetic role of RUNX3 when compared with that of MGMT and TIMP3 in glioblastoma multiforme clinical outcomes. J Neurol Sci. 2014;347:325–331. doi: 10.1016/j.jns.2014.10.043. [DOI] [PubMed] [Google Scholar]
- 26.Hedback N, Jensen DH, Specht L, Fiehn AM, Therkildsen MH, Friis-Hansen L, Dabelsteen E, von Buchwald C. MiR-21 expression in the tumor stroma of oral squamous cell carcinoma: an independent biomarker of disease free survival. PLoS One. 2014;9:e95193. doi: 10.1371/journal.pone.0095193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng EK, Li R, Shin VY, Jin HC, Leung CP, Ma ES, Pang R, Chua D, Chu KM, Law WL, Law SY, Poon RT, Kwong A. Circulating microRNAs as specific biomarkers for breast cancer detection. PLoS One. 2013;8:e53141. doi: 10.1371/journal.pone.0053141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ni Y, Meng L, Wang L, Dong W, Shen H, Wang G, Liu Q, Du J. MicroRNA-143 functions as a tumor suppressor in human esophageal squamous cell carcinoma. Gene. 2013;517:197–204. doi: 10.1016/j.gene.2012.12.031. [DOI] [PubMed] [Google Scholar]
- 29.Xu X, Chen Z, Zhao X, Wang J, Ding D, Wang Z, Tan F, Tan X, Zhou F, Sun J, Sun N, Gao Y, Shao K, Li N, Qiu B, He J. MicroRNA-25 promotes cell migration and invasion in esophageal squamous cell carcinoma. Biochem Biophys Res Commun. 2012;421:640–645. doi: 10.1016/j.bbrc.2012.03.048. [DOI] [PubMed] [Google Scholar]
- 30.Ren LH, Chen WX, Li S, He XY, Zhang ZM, Li M, Cao RS, Hao B, Zhang HJ, Qiu HQ, Shi RH. MicroRNA-183 promotes proliferation and invasion in oesophageal squamous cell carcinoma by targeting programmed cell death 4. Br J Cancer. 2014;111:2003–2013. doi: 10.1038/bjc.2014.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin RJ, Xiao DW, Liao LD, Chen T, Xie ZF, Huang WZ, Wang WS, Jiang TF, Wu BL, Li EM, Xu LY. MiR-142-3p as a potential prognostic biomarker for esophageal squamous cell carcinoma. J Surg Oncol. 2012;105:175–182. doi: 10.1002/jso.22066. [DOI] [PubMed] [Google Scholar]
- 32.Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A, Zlotorynski E, Yabuta N, De Vita G, Nojima H, Looijenga LH, Agami R. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 33.Syring I, Bartels J, Holdenrieder S, Kristiansen G, Muller SC, Ellinger J. Circulating serum miRNA (miR-367-3p, miR-371a-3p, miR-372-3p and miR-373-3p) as biomarkers in patients with testicular germ cell cancer. J Urol. 2015;193:331–337. doi: 10.1016/j.juro.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 34.Eichelser C, Flesch-Janys D, Chang-Claude J, Pantel K, Schwarzenbach H. Deregulated serum concentrations of circulating cell-free microRNAs miR-17, miR-34a, miR-155, and miR-373 in human breast cancer development and progression. Clin Chem. 2013;59:1489–1496. doi: 10.1373/clinchem.2013.205161. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka T, Arai M, Wu S, Kanda T, Miyauchi H, Imazeki F, Matsubara H, Yokosuka O. Epigenetic silencing of microRNA-373 plays an important role in regulating cell proliferation in colon cancer. Oncol Rep. 2011;26:1329–1335. doi: 10.3892/or.2011.1401. [DOI] [PubMed] [Google Scholar]
- 36.Seol HS, Akiyama Y, Shimada S, Lee HJ, Kim TI, Chun SM, Singh SR, Jang SJ. Epigenetic silencing of microRNA-373 to epithelial-mesenchymal transition in non-small cell lung cancer through IRAK2 and LAMP1 axes. Cancer Lett. 2014;353:232–241. doi: 10.1016/j.canlet.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen YJ, Luo J, Yang GY, Yang K, Wen SQ, Zou SQ. Mutual regulation between microRNA-373 and methyl-CpG-binding domain protein 2 in hilar cholangiocarcinoma. World J Gastroenterol. 2012;18:3849–3861. doi: 10.3748/wjg.v18.i29.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang LQ, Zhang Y, Yan H, Liu KJ, Zhang S. MicroRNA-373 functions as an oncogene and targets YOD1 gene in cervical cancer. Biochem Biophys Res Commun. 2015;459:515–520. doi: 10.1016/j.bbrc.2015.02.138. [DOI] [PubMed] [Google Scholar]
- 39.Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, Gimotty PA, Katsaros D, Coukos G, Zhang L, Pure E, Agami R. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 40.Miyazaki T, Kato H, Nakajima M, Faried A, Takita J, Sohda M, Fukai Y, Yamaguchi S, Masuda N, Manda R, Fukuchi M, Ojima H, Tsukada K, Kuwano H. An immunohistochemical study of TIMP-3 expression in oesophageal squamous cell carcinoma. Br J Cancer. 2004;91:1556–1560. doi: 10.1038/sj.bjc.6602185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qi JH, Anand-Apte B. Tissue inhibitor of metalloproteinase-3 (TIMP3) promotes endothelial apoptosis via a caspase-independent mechanism. Apoptosis. 2015;20:523–534. doi: 10.1007/s10495-014-1076-y. [DOI] [PMC free article] [PubMed] [Google Scholar]


